Abstract
The diagnosis of bloodstream infections presents numerous challenges, in part, due to the low concentration of pathogens present in the peripheral bloodstream. As an alternative to existing time-consuming, culture-based diagnostic methods for organism identification, microfluidic devices have emerged as rapid, high-throughput and integrated platforms for bacterial and fungal enrichment, detection, and characterization. This focused review serves to highlight and compare the emerging microfluidic platforms designed for the isolation of sepsis-causing pathogens from blood and suggest important areas for future research.
Keywords: microfluidics, sepsis, infectious disease diagnostics, hematology, lab-on-a-chip
1.0. Introduction
Sepsis is a life-threatening condition that occurs when infectious organisms are present in the peripheral bloodstream resulting in a host shock response. Despite the severity and prevalence of sepsis, current diagnostic methods are extremely time-consuming. This is, in part, due to the extremely low concentration of infectious organisms in the bloodstream 115, making pathogen detection technically challenging.
Microfluidic platforms have emerged as a popular alternative to traditional macro-scale diagnostic methods 30, 34, 55, 74, 90. These micro-scale systems have demonstrated their ability to isolate and detect rare cells from simple fluids (e.g. water, liquid media) by harnessing a variety of physical and chemical separation methods 12, 14, 17, 22, 32, 33, 47, 52, 58, 59, 70, 73, 85, 91, 92, 96, 103, 104, 108. More specifically, prior work has demonstrated the feasibility of employing microfluidic platforms for bacterial isolation and detection of waterborne 4, 8, 60–62, 101, 120 and foodborne 1, 9, 24,54, 57, 65, 75, 80, 87, 117, 121 pathogens. Additionally, a smaller subset of studies has evaluated bacterial capture and detection in urine 83, 116 and joint synovial fluid 16. That said, most studies in this space circumvent a pressing technical challenge: the use of micro-scale systems for the isolation, detection, and characterization of sepsis-causing organisms in complex biological mediums, such as blood. Blood contains a wide variety of biological material, including red blood cells, white blood cells, platelets, and other small molecules 38, making targeted isolation and characterization of sepsis-causing organisms a more challenging technical problem.
Our discussion begins by providing the necessary background on the epidemiology and etiology of bloodstream infections, as well as highlighting the existing diagnostic workflow. Next, we will summarize and review the primary literature that describes the use of microfluidic tools and approaches for the enrichment of bacteria and fungi from blood. In addition, some of these studies employ novel and integrated pathogen detection strategies, which will also be discussed. We will also briefly highlight some examples of emerging microfluidic platforms for pathogen identification and characterization, which could potentially be employed for downstream analysis. Finally, we will discuss key areas for future work that are needed to help address this important diagnostic need.
2.0. Epidemiology and Etiology of Bloodstream Infections
Sepsis ranks among the top seven causes of death in North America and Europe 35. In these regions, the average annual mortality rate averages 29 per 100,000 individuals, with a total case fatality rate ranging from 13% to 20% 35. Further, among all patients admitted to the intensive care unit (ICU), approximately 7.7% will develop a bloodstream infection (Figure 1) 110. Decreasing time-to-diagnosis is uniquely critical for bloodstream infections. Specifically, it is estimated that mortality from sepsis increases by 7.6% for every hour that treatment is delayed 31. Therefore, decreasing time-to-diagnosis is critical to improving overall patient outcomes 66.
Figure 1.
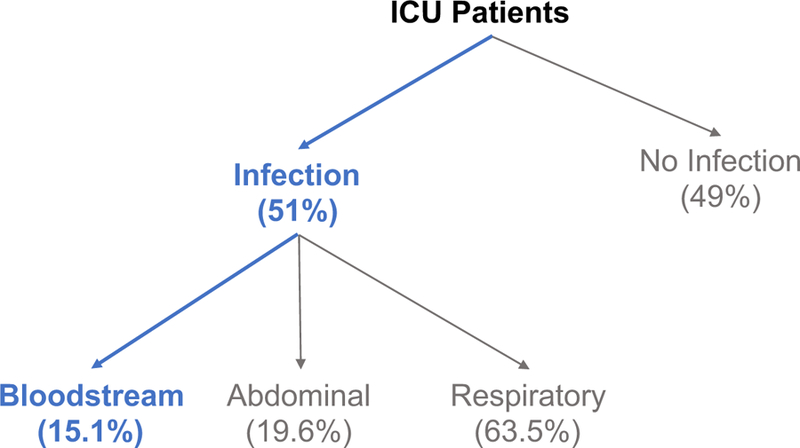
Classification of ICU patients as a function of infection and infection type110.
Sepsis can be caused by a wide-range of pathogens (Table 1) 79. As is listed in Table 1, gram-positive bacteria are identified in approximately 46.8% of infections, gram-negative bacteria are identified in approximately 62.2% of infections, and fungi are identified in approximately 19.4% of infections. Of note is the relatively high frequency of methicillin-resistant Staphylococcus aureus (MRSA) observed in bloodstream infections. Additionally, in approximately 17% of cases, there is more than one pathogenic species causing an infection, resulting in a co-infection 79. As will be discussed in detail below, both rapid species identification and antibiotic susceptibility profiling of the sepsis-causing organism(s) are critical variables in informing the appropriate antibiotic treatment 23, 67, 69, 81.
Table 1:
Observed frequency of sepsis-causing pathogens. Table adapted from Mayr et al. 79.
| Frequency % | Odds Ratio | |
|---|---|---|
| Gram-positive | 46.8 | - |
| Staphylococcus aureus | 20.5 | 0.8 |
| Methicillin-resistant Staphylococcus aureus | 10.2 | 1.3 |
| Enterococcus | 10.9 | 1.6 |
| Staphylococcus epidermis | 10.8 | 0.9 |
| Streptococcus pneumoniae | 4.1 | 0.8 |
| Other | 6.4 | 0.9 |
| Gram-negative | 62.2 | - |
| Pseudomonas spp. | 19.9 | 1.4 |
| Escherichia coli | 16.0 | 0.9 |
| Acinetobacter spp. | 8.8 | 1.0 |
| Enterobacter | 7.0 | 1.5 |
| Other | 17.0 | 1.2 |
| Anaerobes | 4.5 | 0.9 |
| Other bacteria | 1.5 | 1.1 |
| Fungi | 19.4 | - |
| Parasites | 0.7 | - |
| Other organisms | 3.9 | - |
3.0. Current Diagnostic Workflow for Bloodstream Infections
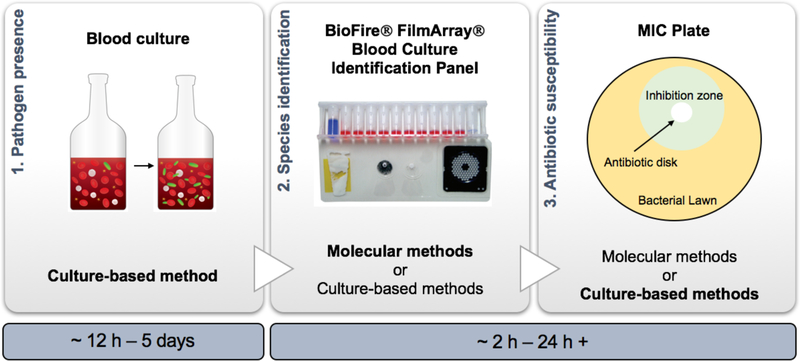
The diagnosis of sepsis involves three primary benchmarks: 1) pathogen presence, 2) species identification, and 3) antibiotic susceptibility profiling (Figure 2) 23, 67, 69, 81. First, a blood culture is conducted in order to confirm the presence of the pathogen. To do this, a primary patient blood specimen is cultured in a nutrient rich media. As the pathogen proliferates, carbon dioxide is emitted, and generally, detected via a carbon dioxide sensor that is integrated into the blood culture bottle. The time to blood culture positivity is usually greater than twelve hours, and can take upwards of five days, depending on the specific growth rate of the sepsis-causing organism 37, 86. After establishing the presence of a pathogen through a blood culture, additional tests are then needed to identify the specific pathogen causing the infection, as well as characterize the antibiotic susceptibility of the pathogen 5, 29, 53, 88. Traditional culture-based characterization methods require at least 24 hours 67. Newer, molecular methods can perform rapid species-level identification and characterize antibiotic resistance profiles for select organisms in as little as 2 – 7.5 hours following blood culture positivity 5, 76, 99.
Figure 2.
Overview schematic of current bloodstream infection diagnostic workflow. First, a 5 – 10 mL patient blood sample is cultured in 25 mL of nutrient rich media to determine if a pathogen is present13(1). Next, either culture-based or molecular methods are used to identify the pathogen(s) causing the infection (2). Above, we show a rapid (~2 h), molecular testing system for pathogen identification, the BioFire® FilmArray® Blood Culture Identification Panel (BCID). Reprinted under permission of the Creative Commons Attribution License93. Lastly, the organism’s susceptibility to antibiotics is determined (3). Here, we highlight the minimum inhibitory concentration (MIC) plate, which is a widely employed culture-based methodology.
Given the strong correlation between shortened time-to-diagnosis and deceased mortality rates, more rapid diagnostic methods are needed to identify and characterize the sepsis-causing organism(s)66. By pinpointing the etiology of the organism(s) at an earlier time-point, therapy can more rapidly transition from a broad-spectrum to a targeted antibiotic. In effect, this reduces the likelihood of super infection, lowers healthcare costs, and minimizes contributions to antibiotic resistance 23. Bacterial enrichment and detection using microfluidics offers the potential to eliminate the current universal need for blood culture. Further, use of an integrated microfluidic platform to 1) diagnose sepsis and 2) characterize the sepsis-causing organism(s) directly from whole blood could serve to optimize and refine the current diagnostic workflow, and contribute dramatically to improved patient outcomes.
4.0. Field Overview: Microfluidic Platforms as a Diagnostic Tool for Bloodstream Infections
In recent years, microfluidic devices have emerged as promising platforms for the diagnosis of bacteremia (sepsis). Microfluidic systems enable extremely precise fluid control and manipulation. This becomes especially relevant when processing samples in complex sample mediums, such as whole blood. Additionally, these high-throughput systems can be readily multiplexed, enabling the simultaneous isolation and detection of multiple bacterial targets. Table 2 outlines the major blood components and some of the physical characteristics that are relevant to consider in the design of microfluidic platforms. As shown, many blood components have similar physical characteristics to bacteria (E. coli). Further, the concentration of sepsis-causing organisms in blood is significantly less than the concentration of other cellular blood components. In combination, these two factors make microfluidic isolation of pathogenic organisms from whole blood a much more challenging process than microfluidic isolation of pathogens from simple fluids.
Table 2:
Physical characteristics of main blood components relative to example sepsis-causing pathogen (E. coli).
| Component | Size | Concentration (cells/mL) | Density (g/mL) | References |
|---|---|---|---|---|
| Red blood cells (RBCs) | 7.5 – 8.7 µm (diameter) | 3.7 – 5.8 × 109 | 1.086 – 1.122 | 25, 38, 90 |
| White blood cells (WBCs) | 7 – 20 µm (diameter) | 3.0 – 11.7 × 106 | 1.057 – 1.092 | 38, 90, 95 |
| Platelets | 1.5 – 3 µm (diameter) | 2.0 – 4.0 × 108 | 1.072 – 1.077 | 38, 90, 107 |
| Plasma | - | - | 1.024 | 90 |
| E. coli | 1 – 3 µm (length) | < 10 | 1.105 | 78, 98, 115 |
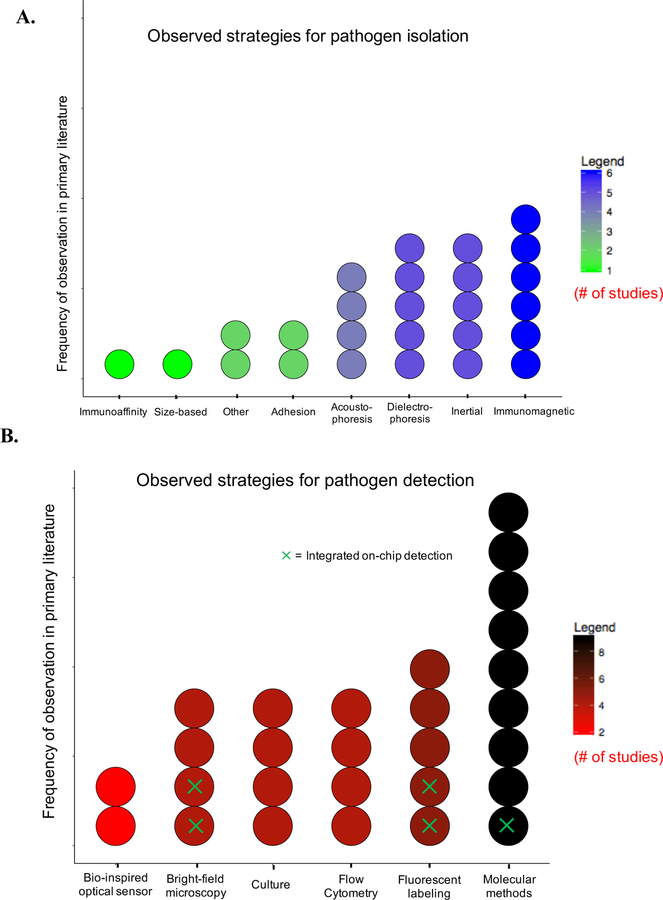
Table 3 summarizes studies that have employed microfluidic approaches to isolate sepsis-causing pathogens from whole blood. To the best of our knowledge, this matrix captures the current state of the field. As shown, a variety of microfluidic approaches have been employed for rapid pathogen isolation, and include acoustophoresis, dielectrophoresis, immunoaffinity-based methods, inertial fractionation, and adhesion-based separation 2, 6, 10, 15, 18, 20, 21, 27, 28, 40, 42–44, 49, 50, 56, 63, 72, 82, 97, 111–114, 118, 119. Figure 3A highlights the observed frequency in the primary literature of various physical and chemical strategies for pathogen isolation. Notably, only a limited number of studies evaluate bacterial isolation at clinically relevant bacterial concentrations (<10 CFU/mL), with most studies assessing separation performance at bacterial loads greater than 1000 CFU/mL. Additionally, the majority of existing studies limit their analysis to only a single pathogen: Escherichia coli, and only one study tackles the need for rapid antibiotic susceptibility profiling 40. Figure 3B summarizes the observed frequency of the various strategies for pathogen detection described in the primary literature. To date, most studies (>85%) fail to integrate on-chip detection. Alternatively, these studies rely on well-established off-chip detection and enumeration strategies, including flow cytometry, microscopy techniques, and molecular methods (Figure 3B) 2, 6, 10, 15, 18, 20, 21, 27, 28, 40, 42–44, 49, 50, 56, 63, 72, 82, 97, 111–114, 118, 119.
Table 3:
Comparison matrix for primary literature reporting pathogen isolation from whole blood 2, 6, 10, 15, 18, 20, 21, 27, 28, 40, 42–44, 49, 50, 56, 63, 72, 82, 97, 111–114, 118, 119.
| AUTHOR, PUBLICATION YEAR | SEPARATION MECHANISM | MINIMUM CELL LOAD (CELLS/ML)* | CAPTURE EFFICIENCY* | PATHOGENS EVALUATED | SAMPLE FLOW RATE | ON-CHIP DETECTION? | SPECIES ID? | AST? |
|---|---|---|---|---|---|---|---|---|
| AI ET AL., 20032 | Acoustophoresis | 3 × 106 | 95.65% (purity) |
E. coli | 0.5 µL/min | ● | ● | ● |
| ARAZ ET AL., 20036 | Acoustophoresis | Not specified | Not specified | E. coli | n/a | ● | ● | ● |
| BISCEGLIA ET AL., 201510 | Dielectrophoresis | 1 × 104 | 97% (E. coli) |
E. coli, S. epidermis, C. albicans | 10 – 20 µL/h | ● | ● | ● |
| CAI ET AL., 201415 | Dielectrophoresis | 1 × 10^3 | 70.9–94.8% | E. coli | 1µL/min | ● |
● (On-Chip PCR) |
● |
| CHO ET AL., 200718 | Immunomagnetic | 1 × 105 | 93.40% | E. coli | 12 min (Total analysis time) |
● |
● (Integrated in separation) |
● |
| COOPER ET AL., 201420 | Immunomagnetic | 1 × 100–1 | 98% | C. albicans | 10 mL/h | ● | ● | ● |
| D’AMICO ET AL., 201721 | Dielectrophoresis | 1 × 103 | 78–79% | E. coli, S. aureus | 10 µL/min | ● |
● (Off-Chip PCR) |
● |
| DOW ET AL., 201827 | Acoustophoresis | 1.2 × 102 | 68% (P. aeruginosa) |
E. coli, P. aeruginosa, S. aureus | 50 µL/min | ● |
● (Off-Chip Luminescence Assay) |
● |
| FARIDI ET AL., 201728 | Inertial | 1 × 106 | 76% | E. coli | 30 – 60 µL/h | ● | ● | ● |
| HOU ET AL., 201540 | Inertial | 1 × 101 | >10% | E. coli, P. aeruginosa, K. pneumoniae, S. aureus, E. faecalis | 50 µL/min | ● |
● (Off-Chip PCR) |
● |
| HUANG ET AL., 200342 | Dielectrophoresis | 4.6 × 103 | 89% | E. coli, B. cereus, L. monocytogenes | n/a | ● |
● (Off-Chip PCR) |
● |
| HWANG ET AL., 200843 | Adhesion | 1 × 107 | 40% | E. coli | 200 µL/min | ● |
● (Off-Chip PCR) |
● |
| HWANG ET AL., 201144 | Adhesion | 1 × 104 | 40% | E. coli | 200 µL/min | ● |
● (Off-Chip PCR) |
● |
| KANG ET AL., 201449 | Nanodroplets | 1 × 100 | 77% | E. coli | 0.5 – 3 µL/min | ● |
● (Nature of Target Complex) |
● |
| KANG ET AL., 201450 | Immunomagnetic | 1 × 104 | >90% | S. aureus, E. coli, C. albicans | 10 mL/h | ● | ● | ● |
| KUCXSENSKI ET AL., 201156 | Dielectrophoresis | 1 × 106 | 30% | E. coli | 35 µL/h | ● | ● | ● |
| LEE ET AL., 201363 | Immunomagnetic | 5 × 106 | >95% | E. coli | 60 mL/h | ● | ● | ● |
| MACH ET AL., 201072 | Inertial | 1 × 108 | >80% | E. coli | 200 µL/min | ● | ● | ● |
| OHLSSON ET AL., 201682 | Acoustophoresis | 1 × 103 | 10% | P. putida | 80 µL/min | ● |
● (Off-Chip PCR) |
● |
| RAUB ET AL., 201497 | Size-exclusion | 2.2 × 106 | 30% | E. coli | ~100 µL/min | ● | ● | ● |
| WANG ET AL., 2012111 | Immuno-affinity | 5 × 101 | 70.7% | E. coli | 2 µL/min | ● |
● (Integrated in separation) |
● |
| WEI-HOU ET AL., 2012112 | Inertial | 1 × 106 | 80–90% | E. coli, S. cerevisiae | 15 µL/min | ● | ● | ● |
| WU ET AL., 2009113 | Inertial | 1 × 107.2 | 62% | E. coli | 2 – 18 µL/min | ● | ● | ● |
| XIA ET AL., 2006114 | Immunomagnetic | 5 × 106 | 78% | E. coli | 25 µL/h | ● |
● (Integrated in separation) |
● |
| YUNG ET AL., 2009118 | Immunomagnetic | 1 × 106 | 80% | C. albicans | 20 mL/h | ● |
● (Integrated in separation) |
● |
| ZELENIN ET AL., 2014119 | Targeted cell lysis | 1 × 107 | 100% | E. coli, M. luteus | 48 µL/min | ● | ● | ● |
Data for lowest reported cell concentration evaluated with statistically different results from control. Pathogens evaluated include only those where separation from whole blood was reported. ID = identification; AST = antibiotic susceptibility testing; ● = no; ● = yes.
Figure 3.
Breakdown of strategies for pathogen isolation and detection, and their observed frequency in primary literature. A. Strategies for pathogen isolation from whole blood 2, 6, 10, 15, 18, 20, 21, 27, 28, 40, 42–44, 49, 50, 56, 63, 72, 82, 97, 111–114, 118, 119. Each dot represents one study in the primary literature. “Other” category includes 1) isolation using nanodroplets 49 and 2) selective lysis of red blood cells 119. B. Strategies for pathogen detection following isolation2, 6, 10, 15, 18, 20, 21, 27, 28, 40, 42–44, 49, 50, 56, 63, 72, 82, 97, 111–114, 118, 119. Primary detection method is indicated. Green ×’s indicates on-chip detection. In some cases, (n=2), two methods are specified. “Culture” is only listed as the detection method if no other method was employed. (In many cases, culture-base methods are used as a control and/or gold standard; these cases are not specified above if more rapid methods were employed).
Below, we will discuss the primary literature in more detail. First, we provide an overview of notable strategies for on-chip pathogen isolation and enrichment. Next, we describe reported integrated methods for on-chip pathogen isolation and detection. Lastly, we highlight our conclusions and suggest areas for future research and development.
5.0. Microfluidic Methods for Pathogen Isolation and Enrichment
5.1. Acoustophoresis
Acoustophoresis is a separation method that employs high-intensity sound waves to enable contact-free migration of target cells towards low pressure nodes. Contact-free manipulation of cells limits cell stress and preserves cell viability making acoustophoresis an attractive option for whole-cell pathogen separation 109. To date, a few studies have used this separation tool for the purpose of separating bacteria from whole blood. Ai et al. separated E. coli from peripheral blood mononuclear cells with a resulting sample purity of 95.65%. Following on-chip separation using two identical surface acoustic waves (Figure 4A), the separation efficiency was characterized off-chip using flow cytometry 2. Although this study provides an initial proof-of-concept for the use of acoustic waves for the separation of bacteria from whole blood, the bacterial concentration used for this analysis was on the order of 106 cell/mL, which is significantly higher than the concentration of bacteria found in a primary human blood sample 2.
Figure 4.
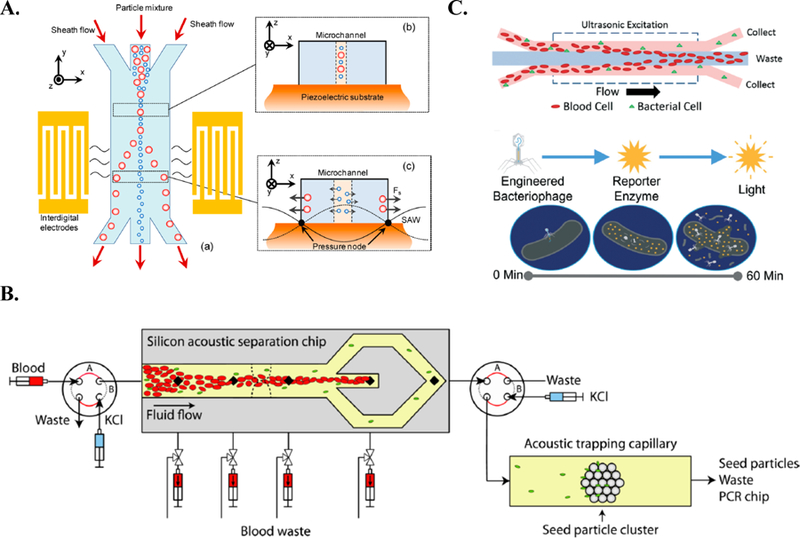
Acoustic separation platforms for the isolation of bacteria from blood. A. Overview of Ai et al. platform, which employed two parallel acoustic waves to focus bacteria in the center of the microchannel 2. Reprinted with permission from https://pubs.acs.org/doi/10.1021/ac4017715. Copyright 2013 American Chemical Society. Further permissions related to the material excerpted should be directed to the ACS. B. Overview of Ohlsson et al. workflow. First, RBCs are focused in the center of the channel. Next, bacteria are acoustically trapped on a seeded particle and profiled using PCR 82. Reprinted with permission from https://pubs.acs.org/doi/abs/10.1021%2Facs.analchem.6b00323. Copyright 2016 American Chemical Society. Further permissions related to the material excerpted should be directed to the ACS. C. Methodology proposed by Dow et al. First, the bacterial sample is enriched on a plastic microdevice. Enriched sample is exposed to bacteriophage luminescence assay 27. Republished with permission of Royal Society of Chemistry from, Acoustic separation in plastic microfluidics for rapid detection of bacteria in blood using engineered bacteriophage, Dow et al., 18, 2018; permission conveyed through Copyright Clearance Center, Inc.
Ohlsson et al. conducted a very comprehensive study using acostophoretic separation. Experiments from this study included the separation of bacteria from plasma, the separation of bacteria from whole blood, and the separation and identification of bacteria from ex vivo clinical blood cultures 82. Specifically, for their work involving the separation of bacteria from whole blood, Ohlsson et al. reported the successful detection of 1000 bacteria/mL using their microchip coupled with an external thermocycler 82. First, Ohlsson et al. acoustically focused red blood cells in the center of the microchannel to be removed. Following this, bacteria (Pseudomonas putida) were trapped on 12 μm polystyrene seeding particles in a sequential capillary channel (Figure 4B) 82. These acoustically trapped bacteria-seeding particle complexes were then washed and released into a secondary polymer microchip contain dry PCR reagents. The solution was then transferred to a thermocycler to continue the molecular analysis 82. Although the detection limit of their first-generation system is not yet capable of detecting low-grade sepsis, the results presented in this study are extremely promising, and suggest the feasibility of eliminating the need for the time-consuming blood culture.
Most recently, Dow et al. coupled acoustic separation in a plastic micro-device to a bacteriophage-based luminescence assay (Figure 4C) 27. At a clinically relevant bacterial input concentration of 120 cells/mL, their acoustic separation device achieved a 68% bacteria yield (P. aeruginosa). As a result of this enrichment step, they observed a 33-fold improvement on the limit of detection for their novel bacteriophage-based luminescence assay 27. Notably, the host-phage specificity required for the functionality of the luminescence assay allows for the potential incorporation of rapid pathogen species identification into the proposed workflow. Dow et al. also evaluated the separation efficiency of E. coli and S. aureus from whole blood to demonstrate the versatility of a non-specific separation platform in isolating both Gram-positive and Gram-negative organisms 27.
5.2. Dielectrophoresis
Dielectrophoresis is a technique that uses an electric field gradient to isolate target cells as a function of their dielectric characteristics 89. Kuczenski et al. presented the critical hydrodynamic and electrokinetic theories for effective sorting of spheroid cells. The team then applied this theory to the design of an electrophoretic microfluidic device 56. With an initial bacterial load (E. coli) of 106 cell/mL, their device had a capture efficiency of 30%. Further improving upon these results, Bisceglia et al. demonstrated a 97% capture rate of E. coli using dielectrophoresis with an initial bacterial load of 1 × 104 cells/mL 56. Bisceglia et al. also demonstrated the feasibility of this methodology in isolating Gram-positive (S. aureus) and fungal (C. albicans) pathogens 10. Additionally, this work assessed the relative effects of isotonic versus hypotonic buffer conditions on the conductivity and relative permeability of the sample components, in addition to the overall efficacy of the separation 10.
More recently, D’Amico et al. reported on a microfluidic system that used dielectrophoresis to isolate E. coli and S. aureus from whole blood 21. At bacterial concentrations as low as 1000 cells/mL, capture efficiencies were 79% and 78%, respectively 21. Specifically, they reported on an integrated microfluidic dialysis-dielectrophoresis isolation system (Figure 5A) 21. This system involves two membraneless microdialysis devices (MMDs) arranged in series, followed by the active dielectrophoresis microchip. The MMD devices are necessary to help reduce the electrical conductivity of the blood sample via rapid diffusion prior to the sample entering the dielectrophoresis microchip. In the dielectrophoresis microchip, bacteria are drawn towards the electrodes, while other blood components are repelled and washed away (Figure 5B) 21. Following separation, the dielectric field is turned off, the captured sample washed out of the microchip, and 16S PCR is conducted for species identification and characterization (Figure 5C) 21. This study by D’Amico et al. demonstrates the potential feasibility of dielectrophoresis as a method for rapid bloodstream infection diagnosis.
Figure 5.
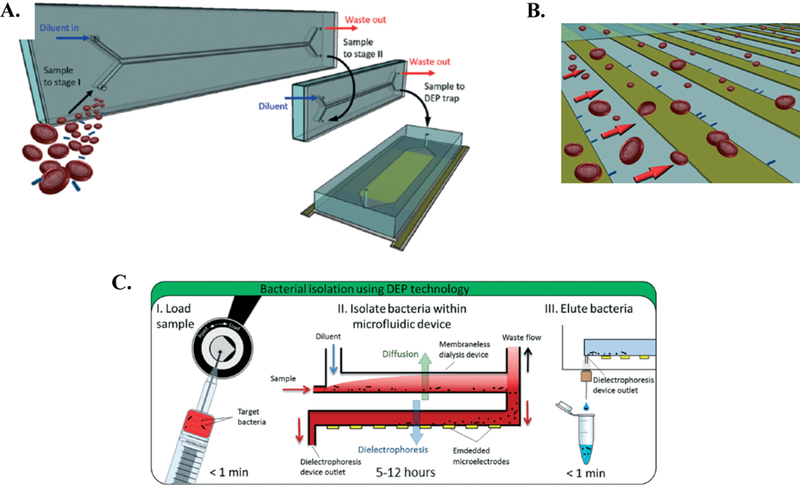
Dielectrophoretic platform for isolation of bacteria from blood proposed by D’Amico et al 21. A. Schematic of the integrated microfluidic dialysis-dielectrophoresis isolation system. Two MMDs are connected in series, followed by the active dielectrophoresis device. B. Sample flow through separation microchip. Bacteria (green) are attracted to electrodes, while other blood components are repelled and washed through the device. C. Overview of proposed workflow. Following separation, sample is eluted from device for molecular analysis. Republished with permission of Royal Society of Chemistry from, Isolation and concentration of bacteria from blood using microfluidic membraneless dialysis and dielectrophoresis, D’Amico et al., 17, 2017; permission conveyed through Copyright Clearance Center, Inc.
5.3. Immunoaffinity-Based Methods
Immunoaffinity-based methods use recognition antibodies to specifically bind a target antigen on a cell surface. This method is desirable due to its unparalleled specificity. However, major drawbacks include high cost and the relative instability of antibodies. One example is described in a study by Wang et al., which reports the selective capture of E. coli using immobilized capture antibodies on the surface of the microchip 111. By employing a biotinylated anti-lipopolysaccharide binding protein (anti-LBP) and NeutrAvidin-based surface chemistry, Wang et al. reports a limit of detection of 50 cells/mL, with a mean capture efficiency of 70.7% 111. Following capture and wash steps, detection was carried out on chip via fluorescent tagging and imaging of E. coli 111.
Immunomagnetic separation is one of the most commonly reported immunoaffinity-based separation methods used to isolate pathogens from whole blood. In this approach, magnetic particles are functionalized with antibodies to enable highly specific binding to a target antigen on the cell surface. The primary sample is incubated with these functionalized magnetic nanoparticles, and target cells (e.g. pathogens) are labeled and retained within the microfluidic chip (positive selection) by an external magnet. In one example, Cho et al. used immunomagnetic separation to isolate E. coli from whole blood, enabling on-chip DNA extraction using a centrifugal microfluidic device on a compact disc (CD) platform (Figure 6) 18. Specifically, primary biotinylated anti-E. coli antibodies were bound to 1 μm streptavidin-coated magnetic beads for E. coli isolation. Of note, incubation of the sample and functionalized magnetic particles was performed on-chip. At a starting pathogen concentration of 1 × 105 cells/mL, capture efficiency was measured to be 93.4% 18. Following capture, the magnetic particles served a second function as micro-scale heaters. The enriched sample was exposed to an 808 nm laser. The conductive nature of the magnetic particle enabled rapid on-chip cell lysis, allowing for downstream (off-chip) molecular analysis 18. This entire sample enrichment and preparation process was reported to take less than 12 min 18.
Figure 6.
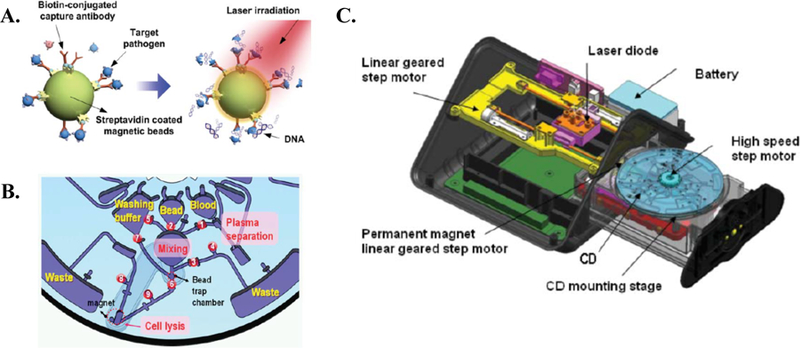
Immunomagnetic platforms for the isolation of bacteria from blood proposed by Cho et al 18. A. Image displays dual-functionality of bound magnetic particles (separation & micro-heaters). B. Schematic of on-chip functionality. C. Overview of CD centrifugal platform. Republished with permission of Royal Society of Chemistry from, One-step pathogen specific DNA extraction from whole blood on a centrifugal microfluidic device, Cho et al., 7, 2007; permission conveyed through Copyright Clearance Center, Inc.
A few studies have proposed the use of immunomagnetic separation of pathogens from blood as feasible strategy for sepsis therapy (i.e. blood cleansing). These studies are included because although the application is derivative from the current discussion, the same fundamental technical strategies of bacterial capture and isolation from a primary blood sample are explored. For example, Lee et al. designed a novel functional group that can non-specifically bind both Gram-negative and Gram-positive bacteria 63. By modifying magnetic nanoparticles with a non-specific functional group, they demonstrated >95% clearance of E. coli at a starting concentration of 5 × 106 cells/mL, and a flow rate of 60 mL/h 63. Additionally, Xia et al. attempted the immunomagnetic enrichment of E. coli. In this case, the chosen capture antibody was specific to E. coli, and bound to 125 nm magnetic particles 114. The group observed that increasing magnetic bead concentration relative to pathogen concentration improved the separation efficiency. At 25 μL/h and a starting cell concentration of 5 × 106 cells/mL, separation efficiency of E. coli from blood was approximately 78% 114. Yung et al. explored a similar approach, but with a different pathogenic target: C. albicans 118. Using 1 μm magnetic nanoparticles functionalized with antibodies specific to C. albicans, Yung et al. demonstrated 80% clearance of the pathogen from whole blood at a starting cell concentration of 106 cells/mL 118. Lastly, Kang et al. engineered a broad spectrum human opsonin (mannose-binding lectin) to enable the non-specific capture of a Gram-positive bacteria, Gram-negative bacteria, and fungi. The in vitro results demonstrate >90% clearance of multiple pathogens (e.g. E. coli, S. aureus, C. albicans) 50.
5.4. Inertial Focusing
Inertial focusing relies on three fundamental forces that exist in the microchannel environment: 1) channel wall interactions, 2) fluid drag, and 3) and lift 77. These forces, and how they relate to one another, can be tuned by a variety of experimental factors including microchannel design, flowrate, and the physical and chemical characteristics of the liquid medium. With appropriate tuning, bacterial cells can be separated from other blood components. For example, Faridi et al. reported the inertial separation of E. coli (1 × 106 cells/mL) from blood with 76% separation efficiency 28, and Mach et al. demonstrated >80% margination of E. coli with a starting bacterial load of 1 × 108 cells/mL 72. More specifically, Wu et al. explored an alternative approach that combines an asymmetrical sheath flow with a specifically designed channel geometry 113. This approach resulted in the generation of a soft inertial force proportional to the fluid Reynolds number, and a particle deflection distance proportion to the particle Reynolds number. In other words, larger particles were deflected away from the sample streamline, while smaller particles were retained near the original flow line. Using this design, Wu et al. reported a 62% separation recovery of E. coli 113.
Of note, is a study conducted by Hou et al. In this work, use of a spiral microchannel creates a lateral drag force due to Dean Flow fractionation, resulting in the separation of bacteria (outer channel wall) from red blood cells and white blood cells (inner channel wall) (Figure 7A)40. Hou et al. evaluated this platform on four different bacterial species (E. coli, S. aureus, P. aeruginosa, Enterococcus faecalis) in order to adequately represent the different shapes and surface characteristics of sepsis-causing organisms40. At bacterial concentrations as low as 10 cells/mL, bacteria were successfully recovered. Following bacterial isolation, the team reported on a detailed methodology for the downstream molecular profiling of the isolated pathogens, including antibiotic susceptibility testing (Figure 7B)40. This study is notable due to its success in isolating a variety of representative sepsis-causing pathogens at clinically relevant concentrations, as well as its holistic consideration of the entire diagnostic workflow.
Figure 7.
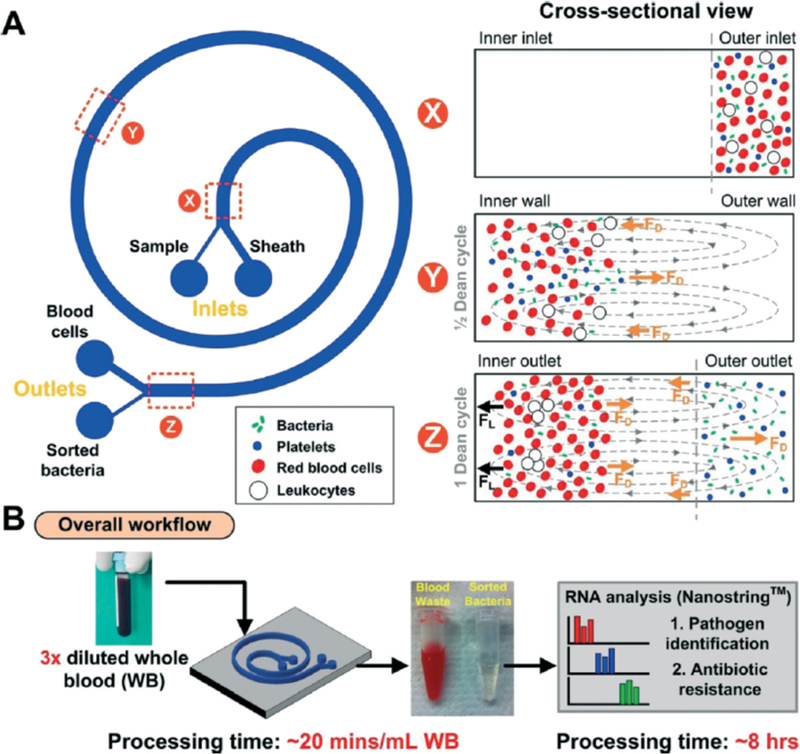
Inertial separation of spiral microchannel as described by Hou et al40. A. Spiral microchannel (left) and cross-sectional view of the sample at indicated points along the microchannel (right). B. Overview of proposed workflow for pathogen isolation, species identification, and antibiotic susceptibility profiling. Republished with permission of Royal Society of Chemistry from, Direct detection and drug-resistance profiling of bacteremias using inertial microfluidics, Hou et al., 15, 2015; permission conveyed through Copyright Clearance Center, Inc.
5.5. Adhesion-Based Methods
One of the more unique approaches to bacterial enrichment from blood has been pioneered by Hwang et al. 43, 44. Their approach involved constructing an array of surface-modified silicon micropillars with surface conditions adapted to optimize bacterial adhesion. Specifically, silane compounds were coated on silicon micropillars. The silicon micropillar structure was employed in effort to maximize surface to volume ratio. The micropillars were 25 μm2 with 12 μm pillar spacing, and the microchip had a total internal volume of a 5 μL 44. When coupled with an optimized media pH, the capture efficiency of E. coli (1 × 107 cells/mL) was about 40% in a 50% whole blood matrix 44. It was also observed that during the washing step, E. coli were retained in the microchip, while remaining RBCs were washed out. Additionally, Hwang et al. demonstrated in situ DNA extraction. Following the capture and wash steps, PCR reagents were injected into the microchip and the mixture was moved to an external thermocycler to continue the analysis 44. An overview of the experimental setup and an SEM image of the micropillar array is shown in Figure 8 44.
Figure 8.
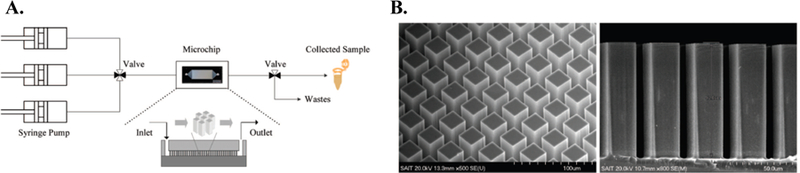
Adhesion-based bacterial isolation from whole blood as described in by Hwang et al. 44. A. Experimental setup. B. SEM images of functionalized silicon microarray. Reprinted with permission from Hwang K.-Y. et al. Bacterial DNA Sample Preparation from Whole Blood Using Surface-Modified Si Pillar Arrays. Analytical Chemistry 80: 7786–7791, 2008. Copyright 2008 American Chemical Society.
Overall, the method proposed and tested by Hwang et al. is effective in decreasing the blood volume in a highly concentrated E. coli sample. Sample preparation prior to molecular analysis took less than 1 hour, and detection sensitivity was improved by more than 100-fold. That said, given the high concentration of a bacteria in the initial sample, this platform has not yet demonstrated its utility in bacterial isolation and detection from a primary human blood sample. This technology could be a viable platform for rapid sample preparation for organism characterization following blood culture positivity, where concentrations of bacteria range from approximately 107 to 109 cells/mL 19.
6.0. Integrated On-Chip Pathogen Enrichment and Detection Platforms
To the best of our knowledge, there have been very few reports of integrated pathogen and enrichment and detection platforms. Many studies have come close, but still require some type of off-chip imaging and/or analysis. Below, we will highlight two studies that have successfully integrated on-chip pathogen enrichment and detection. (See Table 1 for additional integrated platforms). First, Cooper et al. designed a micro-device that employs an immunomagnetic isolation strategy, followed by an on-chip optical detection strategy 20. Because the number of magnetic beads required is over 1000-fold the pathogen concentration, it can be challenging to optically detect pathogens captured immunomagnetically, as dense piles of bead-pathogen complexes form in the microchannel. Cooper et al. optimized the magnetic capture field to promote the formation of a homogenous layer of captured cells within the microchannel, allowing for automated optical detection (Figure 9A) 20. Using a generic blood opsonin capture antibody, 98% percent of the fungal pathogens (C. albicans) were captured from initial starting sample concentrations ranging from 10 – 100 cells/mL. Further, by employing the optimized magnetic field concentrator, the optical detection rate increased from 43% to 67% 20.
Figure 9.
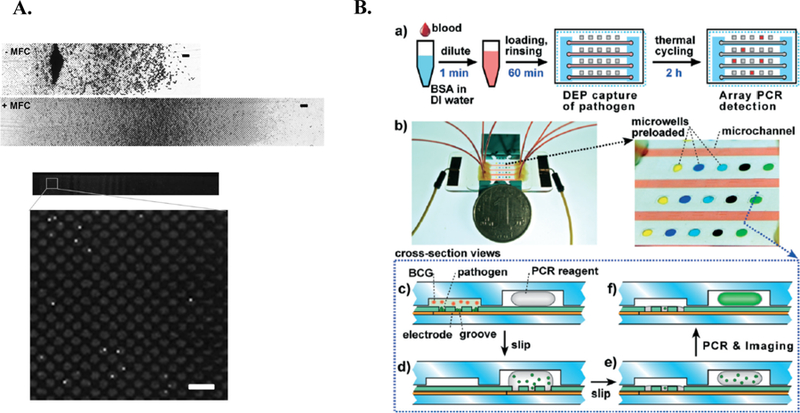
Integrated on-chip isolation and detection platform. A. Overview of Cooper et al. Image of microchannel without (top) and with (middle) magnetic field concentrator (MFC). First half of channel is shown (scale bar = 50μm). Fluorescent image displaying captured C. albicans following immunofluorescent staining (bottom) 20. Further, the magnetic field concentrator (MFC) minimizes magnetic bead clumping (middle), allowing for optical detection (bottom)20. Republished with permission of Royal Society of Chemistry from, A microdevice for rapid optical detection of magnetically captured rare blood pathogens, Cooper et al., 14, 2014; permission conveyed through Copyright Clearance Center, Inc. B. Overview of Cai et al. integrated methodology. a) Schematic of workflow. b) relative size of microdevice (left) with zoomed in image of microchannel and preloaded microwells. c-f) cross-sectional view of device operation. Pathogen is retained in grooves, while blood cells (BCG) are washed away. Device is “slipped” to expose liquid PCR reagents to pathogen. Device is returned to original position avoid contaminated surface. Device is thermocycled in situ and imaged 15. Republished with permission of Royal Society of Chemistry from, An integrated microfluidic device utilizing dielectrophoresis and multiplex array PCR for point-of-care detection of pathogens, Cai et al., 14, 2014; permission conveyed through Copyright Clearance Center, Inc.
Second, Cai et al. developed an integrated microfluidic device that coupled dielectrophoresis with on-chip multiplex array PCR 15. Using dielectrophoresis as the separation method, E. coli were subjected to a positive dielectrophoretic force and were retained in grooves along the base of the microchip (Figure 9B) 15. At a starting sample concentration of 1.6 × 107 cells/mL, E. coli capture efficiency ranged from 70.9% (10-fold diluted blood) to 91.5% (100-fold diluted blood) 15. Following capture, the remaining blood components were flushed out of the channel. The device was “slipped” to mix retained pathogens with pre-loaded PCR reagents. Following mixing, the device was returned to its original position to avoid unnecessary contamination. After in situ thermocycling, pathogen presence and identification was determined via a fluorescent readout 15. Although the isolation of other pathogens was not reported in the study, the on-chip multiplex array PCR system was also validated with Candida tropicalis, P. aeruginosa, S. aureus, and Streptococcus mutans 15. Although clinically relevant pathogen concentrations were not assessed in this work, Cai et al. creatively demonstrates the efficacy of integrated dielectrophoretic isolation and species-level pathogen detection.
7.0. Microfluidic platforms for downstream pathogen characterization
We would like to briefly highlight a few examples of microfluidic platforms that have demonstrated success in 1) pathogen identification, or 2) pathogen antibiotic susceptibility profiling. Following successful organism isolation from whole blood, these systems could have potential downstream utility in enabling comprehensive characterization of the disease-causing organism(s). As discussed, both species-level information and antibiotic susceptibility information are critical data needed to inform a targeted antibiotic therapy regimen, and to improve patient outcomes.
First, to obtain species-level information, molecular methods are widely employed for rapid classification. For example, Jiang et al. designed a rapid, continuous-flow PCR and DNA hybridization microfluidic platform for bacterial identification. The platform demonstrated successful identification of clinically relevant pathogens, such as E. coli, S. aureus, and P. aeruginosa46. In another example, Sun et al. proposed a bacterial classification platform that circumvents the use of bacterial DNA. The group developed a microfluidic platform to differentiate bacteria as a function of cell wall structural differences. By employing a tapered channel design, cell stiffness could be determined as function of distance traveled in the microchannel (Figure 10A).
Figure 10.
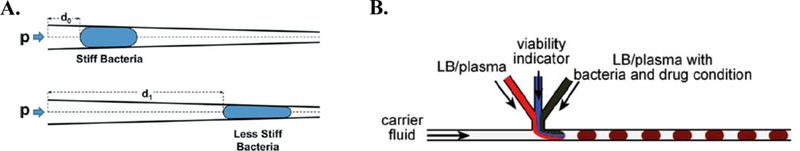
Strategies for downstream pathogen identification and antibiotic susceptibility profiling. A. Basic bacterial differentiation principle proposed by Sun et al. The distance (d) traveled in the tapered microchannel is inversely proportional to cell stiffness. Force is drive by fluid pressure, p105. Republished with permission of Royal Society of Chemistry from, A microfluidic platform for profiling biomechanical properties of bacteria, Sun et al., 14, 2014; permission conveyed through Copyright Clearance Center, Inc. B. Plug-based microfluidic platform proposed by Boedicker et al11. Schematic of channel shows formation of nanoliter droplets containing bacteria, drug condition, and viability indicator. Republished with permission of Royal Society of Chemistry from, Detecting bacteria and determining their susceptibility to antibiotics by stochastic confinement in nanoliter droplets using plug-based microfluidics, Boedicker et al., 8, 2008; permission conveyed through Copyright Clearance Center, Inc.
Second, the growing problem of antibiotic resistance has led to increased research efforts surrounding the development of rapid antibiotic susceptibility profiling platforms84. Boedicker et al. demonstrated on-chip antibiotic susceptibility testing by employing plug-based microfluidics to form nanoliter droplets containing a single cell, an antibiotic-of-interest, and a viability indicator11. This platform allowed for the rapid determination and quantification of antibiotic efficacy and minimum inhibitory concentration (MIC), respectively (Figure 10B)11. In another example, Kalashnikov et al. described a stress-based microfluidic platform for evaluating antibiotic susceptibility48. Further, it was observed that shear flow potentiated the efficacy of antibiotics. In this system, bacteria were covalently-bound to the floor of the microchannel and exposed to the antibiotic-of-interest and shear flow. Bacterial viability was observed via automated fluorescent microscopy. This study reports comprehensive antibiotic susceptibility profiling of clinically-relevant Gram-negative bacteria within 2 h48.
8.0. Conclusions and Future Work
In this review, we discussed emerging micro-scale platforms for the isolation of sepsis-causing pathogens from whole blood. The highlighted research represents a relatively new and emerging field, though the basic notion of employing microfluidic platforms for rare-cell capture from whole blood is not entirely new 94. A large body of work exists describing rare-cell capture from whole blood for cancer screening, diagnosis, and monitoring. More specifically, the field of liquid biopsy describes numerous strategies for the capture and detection of circulating tumor cells (CTCs) and exosomes 3, 7, 26, 36, 39, 41, 45, 51, 63, 64, 68, 71, 100, 102, 106. The existing and emerging technologies in this space should be considered for their potential translation to whole-cell capture of infectious organisms in whole blood samples.
Upon thorough consideration of the reported work in the area of sepsis-causing organisms’ isolation from whole blood, we would like to offer the following recommendations for future studies conducted in this space. First, clinically relevant bacterial concentrations must be evaluated. Given that the primary technical hurdle to sepsis diagnosis is the relatively low concentration of bacteria in the blood (1 – 10 CFU/mL), micro-scale systems must demonstrate their ability to detect bacteria at these low levels. As at minimum, experimental studies should report a limit of detection. Second, the field needs to move beyond non-specific organism isolation and detection, and towards a platform that can provide a comprehensive etiological characterization of the specific sepsis-causing organism(s). Both species-level information and antibiotic susceptibility information are critical data needed to inform a targeted antibiotic therapy regimen and improve patient outcomes. Creative methods that can provide more a more specific and multi-dimensional diagnostic output should also be explored. Finally, it is extremely important to design with downstream integration in mind. Although this paper focused on microfluidic capture strategies and platforms, some of the most attractive platforms successfully integrate organism capture and detection on a single chip. A holistic analysis of diagnostic workflow is critical for future clinical translation and commercial integration.
Acknowledgements
The authors acknowledge partial financial support from the National Institute of Health (NIH) Director’s Transformative Research Award (R01HL137157), and support from the Thayer School of Engineering Ph.D. Innovation Program and Holekamp Family Innovation Fellowship. We would like to acknowledge the Clinical Microbiology staff at Dartmouth Hitchcock Medical Center in Lebanon, NH for their support and access to their facility. We would also like to acknowledge the guidance of Dr. Pierre-Hugues Stefanuto and Dr. Christiaan A. Rees.
Footnotes
Conflicts of interest
There are no personal or financial conflicts of interest to declare.
References
- 1.Abdel-Hamid I, Ivnitski D, Atanasov P and Wilkins E. Flow-through immunofiltration assay system for rapid detection of E. coli O157:H7. Biosensors and Bioelectronics 14: 309–316, 1999. [DOI] [PubMed] [Google Scholar]
- 2.Ai Y, Sanders CK and Marrone BL. Separation of Escherichia coli bacteria from peripheral blood mononuclear cells using standing surface acoustic waves. Anal Chem 85: 9126–9134, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alix-Panabieres C and Pantel K. Challenges in circulating tumour cell research. Nat Rev 14: 623–631, 2014. [DOI] [PubMed] [Google Scholar]
- 4.Altintas Z, Akgun M, Kokturk G and Uludag Y. A fully automated microfluidic-based electrochemical sensor for real-time bacteria detection. Biosens Bioelectron 100: 541–548, 2018. [DOI] [PubMed] [Google Scholar]
- 5.Altun O, M. Almuhayawi M Fau - Ullberg, V. Ullberg M Fau - Ozenci and Ozenci V. Clinical evaluation of the FilmArray blood culture identification panel in identification of bacteria and yeasts from positive blood culture bottles. J Clin Microbiol 51: 4130–4136, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Araz MK, Chung-Hoon L and Lal A. Ultrasonic separation in microfluidic capillaries. In: IEEE Symposium on Ultrasonics, 20032003, p. 1066–1069 Vol.1061. [Google Scholar]
- 7.Au SH, Edd J, Stoddard AE, Wong KHK, Fachin F, Maheswaran S, Haber DA, Stott SL, Kapur R and Toner M. Microfluidic Isolation of Circulating Tumor Cell Clusters by Size and Asymmetry. Scientific Reports 7: 2433, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Balasubramanian AK, Soni KA, Beskok A and Pillai SD. A microfluidic device for continuous capture and concentration of microorganisms from potable water. Lab Chip 7: 1315–1321, 2007. [DOI] [PubMed] [Google Scholar]
- 9.Beyor N, Yi L, Seo TS and Mathies RA. Integrated Capture, Concentration, Polymerase Chain Reaction, and Capillary Electrophoretic Analysis of Pathogens on a Chip. Analytical Chemistry 81: 3523–3528, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bisceglia E, Cubizolles M, Trainito CI, Berthier J, Pudda C, Français O, Mallard F and Le Pioufle B. A generic and label free method based on dielectrophoresis for the continuous separation of microorganism from whole blood samples. Sensors and Actuators B: Chemical 212: 335–343, 2015. [Google Scholar]
- 11.Boedicker JQ, Li L, Kline TR and Ismagilov RF. Detecting bacteria and determining their susceptibility to antibiotics by stochastic confinement in nanoliter droplets using plug-based microfluidics. Lab Chip 8: 1265–1272, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boehm DA, Gottlieb PA and Hua SZ. On-chip microfluidic biosensor for bacterial detection and identification. Sensors and Actuators B: Chemical 126: 508–514, 2007. [Google Scholar]
- 13.Bouza E, Sousa D, Rodríguez-Créixems M, Lechuz JG and Muñoz P. Is the Volume of Blood Cultured Still a Significant Factor in the Diagnosis of Bloodstream Infections? Journal of clinical microbiology 45: 2765, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cabrera CR Continuous concentration of bacteria in a microfluidic flow cell using electrokinetic techniques. Electrophoresis 22: 355–362. [DOI] [PubMed] [Google Scholar]
- 15.Cai D, Xiao M, Xu P, Xu YC and Du W. An integrated microfluidic device utilizing dielectrophoresis and multiplex array PCR for point-of-care detection of pathogens. Lab Chip 14: 3917–3924, 2014. [DOI] [PubMed] [Google Scholar]
- 16.Chang WH, Wang CH, Lin CL, Wu JJ, Lee MS and Lee GB. Rapid detection and typing of live bacteria from human joint fluid samples by utilizing an integrated microfluidic system. Biosens Bioelectron 66: 148–154, 2015. [DOI] [PubMed] [Google Scholar]
- 17.Cho YK, Kim S, Lee K, Park C, Lee JG and Ko C. Bacteria concentration using a membrane type insulator-based dielectrophoresis in a plastic chip. Electrophoresis 30: 3153–3159, 2009. [DOI] [PubMed] [Google Scholar]
- 18.Cho YK, Lee JG, Park JM, Lee BS, Lee Y and Ko C. One-step pathogen specific DNA extraction from whole blood on a centrifugal microfluidic device. Lab Chip 7: 565–573, 2007. [DOI] [PubMed] [Google Scholar]
- 19.Christner M, Rohde H, Wolters M, Sobottka I, Wegscheider K and Aepfelbacher M. Rapid identification of bacteria from positive blood culture bottles by use of matrix-assisted laser desorption-ionization time of flight mass spectrometry fingerprinting. J Clin Microbiol 48: 1584–1591, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cooper RM, Leslie DC, Domansky K, Jain A, Yung C, Cho M, Workman S, Super M and Ingber DE. A microdevice for rapid optical detection of magnetically captured rare blood pathogens. Lab Chip 14: 182–188, 2014. [DOI] [PubMed] [Google Scholar]
- 21.D’Amico L, Ajami NJ, Adachi JA, Gascoyne PR and Petrosino JF. Isolation and concentration of bacteria from blood using microfluidic membraneless dialysis and dielectrophoresis. Lab Chip 17: 1340–1348, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Delehanty JB and Ligler FS. A Microarray Immunoassay for Simultaneous Detection of Proteins and Bacteria. Analytical Chemistry 74: 5681–5687, 2002. [DOI] [PubMed] [Google Scholar]
- 23.Dellinger RP, A. Levy Mm Fau - Rhodes, D. Rhodes A Fau - Annane, H. Annane D Fau - Gerlach, S. M. Gerlach H Fau - Opal, J. E. Opal Sm Fau - Sevransky, C. L. Sevransky Je Fau - Sprung, I. S. Sprung Cl Fau - Douglas, R. Douglas Is Fau - Jaeschke, T. M. Jaeschke R Fau - Osborn, M. E. Osborn Tm Fau - Nunnally, S. R. Nunnally Me Fau - Townsend, K. Townsend Sr Fau - Reinhart, R. M. Reinhart K Fau - Kleinpell, D. C. Kleinpell Rm Fau - Angus, C. S. Angus Dc Fau - Deutschman, F. R. Deutschman Cs Fau - Machado, G. D. Machado Fr Fau - Rubenfeld, S. A. Rubenfeld Gd Fau - Webb, R. J. Webb Sa Fau - Beale, J.-L. Beale Rj Fau - Vincent, R. Vincent Jl Fau – Moreno and Moreno R. Surviving sepsis campaign: international guidelines for management of severe sepsis and septic shock: 2012. Intensive Care Med 39: 165–228, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dharmasiri U, Witek MA, Adams AA, Osiri JK, Hupert ML, Bianchi TS, Roelke DL and Soper SA. Enrichment and Detection of Escherichia coli O157:H7 from Water Samples Using an Antibody Modified Microfluidic Chip. Analytical Chemistry 82: 2844–2849, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Diez-Silva M, Dao M, Han J, Lim C-T and Suresh S. Shape and Biomechanical Characteristics of Human Red Blood Cells in Health and Disease. MRS bulletin / Materials Research Society 35: 382–388, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dong Y, Skelley AM, Merdek KD, Sprott KM, Jiang C, Pierceall WE, Lin J, Stocum M, Carney WP and Smirnov DA. Microfluidics and Circulating Tumor Cells. The Journal of Molecular Diagnostics 15: 149–157, 2013. [DOI] [PubMed] [Google Scholar]
- 27.Dow P, Kotz K, Gruszka S, Holder J and Fiering J. Acoustic separation in plastic microfluidics for rapid detection of bacteria in blood using engineered bacteriophage. Lab Chip 18: 923–932, 2018. [DOI] [PubMed] [Google Scholar]
- 28.Faridi MA, Ramachandraiah H, Banerjee I, Ardabili S, Zelenin S and Russom A. Elasto-inertial microfluidics for bacteria separation from whole blood for sepsis diagnostics. J Nanobiotechnology 15: 3, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ferreira L, I. Sanchez-Juanes F Fau - Porras-Guerra, M. I. Porras-Guerra I Fau - Garcia-Garcia, J. E. Garcia-Garcia Mi Fau - Garcia-Sanchez, J. M. Garcia-Sanchez Je Fau - Gonzalez-Buitrago, J. L. Gonzalez-Buitrago Jm Fau - Munoz-Bellido and Munoz-Bellido JL. Microorganisms direct identification from blood culture by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Clin Microbiol Infect 17: 546–551, 2010. [DOI] [PubMed] [Google Scholar]
- 30.Foudeh AM, Fatanat Didar T, Veres T and Tabrizian M. Microfluidic designs and techniques using lab-on-a-chip devices for pathogen detection for point-of-care diagnostics. Lab Chip 12: 3249–3266, 2012. [DOI] [PubMed] [Google Scholar]
- 31.Frost R, Newsham H, Parmar S and Gonzalez-Ruiz A. Impact of delayed antimicrobial therapy in septic ITU patients. Critical Care 14: P20–P20, 2010. [Google Scholar]
- 32.Gagnon Z and Chang HC. Aligning fast alternating current electroosmotic flow fields and characteristic frequencies with dielectrophoretic traps to achieve rapid bacteria detection. Electrophoresis 26: 3725–3737, 2005. [DOI] [PubMed] [Google Scholar]
- 33.Gomez-Sjoberg R, Morisette DT and Bashir R. Impedance microbiology-on-a-chip: microfluidic bioprocessor for rapid detection of bacterial metabolism. Journal of Microelectromechanical Systems 14: 829–838, 2005. [Google Scholar]
- 34.Gossett DR, Weaver WM, Mach AJ, Hur SC, Tse HT, Lee W, Amini H and Di Carlo D. Label-free cell separation and sorting in microfluidic systems. Anal Bioanal Chem 397: 3249–3267, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goto M and Al-Hasan MN. Overall burden of bloodstream infection and nosocomial bloodstream infection in North America and Europe. Clinical Microbiology and Infection 19: 501–509, 2013. [DOI] [PubMed] [Google Scholar]
- 36.Hajba L and Guttman A. Circulating tumor-cell detection and capture using microfluidic devices. TrAC Trends in Analytical Chemistry 59: 9–16, 2014. [Google Scholar]
- 37.Hardy DJ, Hulbert BB and Migneault PC. Time to detection of positive BacT/Alert blood cultures and lack of need for routine subculture of 5- to 7-day negative cultures. Journal of clinical microbiology 30: 2743–2745, 1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hollowell J. G. V. A. O. W. G. E. e. a. Hematological and iron-related analytes--reference data for persons aged 1 year and over: United States, 1988–94. National Center for Health Statistics Vital Health Stat 11: 1–156, 2005. [PubMed] [Google Scholar]
- 39.Hoshino K, N. Huang Yy Fau - Lane, M. Lane N Fau - Huebschman, J. W. Huebschman M Fau - Uhr, E. P. Uhr Jw Fau - Frenkel, X. Frenkel Ep Fau – Zhang and Zhang X. Microchip-based immunomagnetic detection of circulating tumor cells. Lab Chip 11: 3449–3457, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hou HW, Bhattacharyya RP, Hung DT and Han J. Direct detection and drug-resistance profiling of bacteremias using inertial microfluidics. Lab Chip 15: 2297–2307, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hou HW, Warkiani ME, Khoo BL, Li ZR, Soo RA, Tan DS-W, Lim W-T, Han J, Bhagat AAS and Lim CT. Isolation and retrieval of circulating tumor cells using centrifugal forces. Scientific Reports 3: 1259, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang Y, Yang JM, Hopkins PJ et al. Separation of Simulants of Biological Warfare Agents from Blood by a Miniaturized Dielectrophoresis Device. Biomedical Microdevices 5: 217, 2003. [Google Scholar]
- 43.Hwang K-Y, Jeong S-Y, Kim Y-R, Namkoong K, Lim H-K, Chung W-S, Kim J-H and Huh N. Rapid detection of bacterial cell from whole blood: Integration of DNA sample preparation into single micro-PCR chip. Sensors and Actuators B: Chemical 154: 46–51, 2011. [Google Scholar]
- 44.Hwang K-Y, Lim H-K, Jung S-Y, Namkoong K, Kim J-H, Huh N, Ko C and Park J-C. Bacterial DNA Sample Preparation from Whole Blood Using Surface-Modified Si Pillar Arrays. Analytical Chemistry 80: 7786–7791, 2008. [DOI] [PubMed] [Google Scholar]
- 45.Hyun KA, Lee TY, Lee SH and Jung HI. Two-stage microfluidic chip for selective isolation of circulating tumor cells (CTCs). Biosens Bioelectron 67: 86–92, 2015. [DOI] [PubMed] [Google Scholar]
- 46.Jiang X, Shao N, Jing W, Tao S, Liu S and Sui G. Microfluidic chip integrating high throughput continuous-flow PCR and DNA hybridization for bacteria analysis. Talanta 122: 246–250, 2014. [DOI] [PubMed] [Google Scholar]
- 47.Jung JH, Kim GY and Seo TS. An integrated passive micromixer-magnetic separation-capillary electrophoresis microdevice for rapid and multiplex pathogen detection at the single-cell level. Lab Chip 11: 3465–3470, 2011. [DOI] [PubMed] [Google Scholar]
- 48.Kalashnikov M, Mueller M, McBeth C, Lee JC, Campbell J, Sharon A and Sauer-Budge AF. Rapid phenotypic stress-based microfluidic antibiotic susceptibility testing of Gram-negative clinical isolates. Scientific Reports 7: 8031, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kang DK, Ali MM, Zhang K, Huang SS, Peterson E, Digman MA, Gratton E and Zhao W. Rapid detection of single bacteria in unprocessed blood using Integrated Comprehensive Droplet Digital Detection. Nat Commun 5: 5427, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kang JH, Super M, Yung CW, Cooper RM, Domansky K, Graveline AR, Mammoto T, Berthet JB, Tobin H, Cartwright MJ, Watters AL, Rottman M, Waterhouse A, Mammoto A, Gamini N, Rodas MJ, Kole A, Jiang A, Valentin TM, Diaz A, Takahashi K and Ingber DE. An extracorporeal blood-cleansing device for sepsis therapy. Nat Med 20: 1211–1216, 2014. [DOI] [PubMed] [Google Scholar]
- 51.Karabacak NM, Spuhler PS, Fachin F, Lim EJ, Pai V, Ozkumur E, Martel JM, Kojic N, Smith K, Chen P.-i., Yang J, Hwang H, Morgan B, Trautwein J, Barber TA, Stott SL, Maheswaran S, Kapur R, Haber DA and Toner M. Microfluidic, marker-free isolation of circulating tumor cells from blood samples. Nature Protocols 9: 694, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim U and Soh HT. Simultaneous sorting of multiple bacterial targets using integrated dielectrophoretic-magnetic activated cell sorter. Lab Chip 9: 2313–2318, 2009. [DOI] [PubMed] [Google Scholar]
- 53.Kok J, Thomas LC, Olma T, Chen SCA and Iredell JR. Identification of Bacteria in Blood Culture Broths Using Matrix-Assisted Laser Desorption-Ionization Sepsityper™ and Time of Flight Mass Spectrometry. PLOS ONE 6: e23285, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Koo OK, Liu Y, Shuaib S, Bhattacharya S, Ladisch MR, Bashir R and Bhunia AK. Targeted Capture of Pathogenic Bacteria Using a Mammalian Cell Receptor Coupled with Dielectrophoresis on a Biochip. Analytical Chemistry 81: 3094–3101, 2009. [DOI] [PubMed] [Google Scholar]
- 55.Kothari A, Morgan M and Haake DA. Emerging technologies for rapid identification of bloodstream pathogens. Clin Infect Dis 59: 272–278, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kuczenski RS, Chang HC and Revzin A. Dielectrophoretic microfluidic device for the continuous sorting of Escherichia coli from blood cells. Biomicrofluidics 5: 32005–3200515, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Laczka O, Maesa JM, Godino N, del Campo J, Fougt-Hansen M, Kutter JP, Snakenborg D, Munoz-Pascual FX and Baldrich E. Improved bacteria detection by coupling magneto-immunocapture and amperometry at flow-channel microband electrodes. Biosens Bioelectron 26: 3633–3640, 2011. [DOI] [PubMed] [Google Scholar]
- 58.Lagally ET, Lee SH and Soh HT. Integrated microsystem for dielectrophoretic cell concentration and genetic detection. Lab Chip 5: 1053–1058, 2005. [DOI] [PubMed] [Google Scholar]
- 59.Lam B, Fang Z, Sargent EH and Kelley SO. Polymerase chain reaction-free, sample-to-answer bacterial detection in 30 minutes with integrated cell lysis. Anal Chem 84: 21–25, 2012. [DOI] [PubMed] [Google Scholar]
- 60.Lapizco-Encinas BH, Davalos RV, Simmons BA, Cummings EB and Fintschenko Y. An insulator-based (electrodeless) dielectrophoretic concentrator for microbes in water. J Microbiol Methods 62: 317–326, 2005. [DOI] [PubMed] [Google Scholar]
- 61.Lapizco-Encinas BH, Simmons BA, Cummings EB and Fintschenko Y. Insulator-based dielectrophoresis for the selective concentration and separation of live bacteria in water. Electrophoresis 25: 1695–1704, 2004. [DOI] [PubMed] [Google Scholar]
- 62.Lay C, Teo CY, Zhu L, Peh XL, Ji HM, Chew BR, Murthy R, Feng HH and Liu WT. Enhanced microfiltration devices configured with hydrodynamic trapping and a rain drop bypass filtering architecture for microbial cells detection. Lab Chip 8: 830–833, 2008. [DOI] [PubMed] [Google Scholar]
- 63.Lee JJ, Jeong KJ, Hashimoto M, Kwon AH, Rwei A, Shankarappa SA, Tsui JH and Kohane DS. Synthetic ligand-coated magnetic nanoparticles for microfluidic bacterial separation from blood. Nano Lett 14: 1–5, 2014. [DOI] [PubMed] [Google Scholar]
- 64.Lee SW, Hyun KA, Kim SI, Kang JY and Jung HI. Continuous enrichment of circulating tumor cells using a microfluidic lateral flow filtration chip. J Chromatogr A 1377: 100–105, 2015. [DOI] [PubMed] [Google Scholar]
- 65.Lee W, Kwon D, Choi W, Jung GY and Jeon S. 3D-printed microfluidic device for the detection of pathogenic bacteria using size-based separation in helical channel with trapezoid cross-section. Sci Rep 5: 7717, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Leibovici L, M. Shraga I Fau - Drucker, H. Drucker M Fau - Konigsberger, Z. Konigsberger H Fau - Samra, S. D. Samra Z Fau – Pitlik and Pitlik SD. The benefit of appropriate empirical antibiotic treatment in patients with bloodstream infection. J. Intern. Med 244: 379–386, 1998. [DOI] [PubMed] [Google Scholar]
- 67.Lever A and Mackenzie I. Sepsis: definition, epidemiology, and diagnosis 335: 879–883, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li P, Mao Z, Peng Z, Zhou L, Chen Y, Huang P-H, Truica CI, Drabick JJ, El-Deiry WS, Dao M, Suresh S and Huang TJ. Acoustic separation of circulating tumor cells. Proceedings of the National Academy of Sciences 112: 4970, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Liesenfeld O, Lehman L, Hunfeld KP and Kost G. Molecular diagnosis of sepsis: New aspects and recent developments. European journal of microbiology & immunology 4: 1–25, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Liu RH, Yang J, Lenigk R, Bonanno J and Grodzinski P. Self-Contained, Fully Integrated Biochip for Sample Preparation, Polymerase Chain Reaction Amplification, and DNA Microarray Detection. Analytical Chemistry 76: 1824–1831, 2004. [DOI] [PubMed] [Google Scholar]
- 71.Liu Z, Zhang W, Huang F, Feng H, Shu W, Xu X and Chen Y. High throughput capture of circulating tumor cells using an integrated microfluidic system. Biosensors and Bioelectronics 47: 113–119, 2013. [DOI] [PubMed] [Google Scholar]
- 72.Mach AJ and Di Carlo D. Continuous scalable blood filtration device using inertial microfluidics. Biotechnol Bioeng 107: 302–311, 2010. [DOI] [PubMed] [Google Scholar]
- 73.Mai J, Abhyankar VV, Piccini ME, Olano JP, Willson R and Hatch AV. Rapid detection of trace bacteria in biofluids using porous monoliths in microchannels. Biosens Bioelectron 54: 435–441, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mairhofer J, Roppert K and Ertl P. Microfluidic systems for pathogen sensing: a review. Sensors (Basel) 9: 4804–4823, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mannoor MS, Zhang S, Link AJ and McAlpine MC. Electrical detection of pathogenic bacteria via immobilized antimicrobial peptides. Proc Natl Acad Sci U S A 107: 19207–19212, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Marschal M, Bachmaier J, Autenrieth I, Oberhettinger P, Willmann M and Peter S. Evaluation of the Accelerate Pheno System for Fast Identification and Antimicrobial Susceptibility Testing from Positive Blood Cultures in Bloodstream Infections Caused by Gram-Negative Pathogens. J Clin Microbiol 55: 2116–2126, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Martel JM and Toner M. Inertial focusing in microfluidics. Annu Rev Biomed Eng 16: 371–396, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Martínez-Salas E, Martín JA and Vicente M. Relationship of Escherichia coli density to growth rate and cell age. Journal of Bacteriology 147: 97–100, 1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mayr FB, Yende S and Angus DC. Epidemiology of severe sepsis. Virulence 5: 4–11, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mujika M, Arana S, Castano E, Tijero M, Vilares R, Ruano-Lopez JM, Cruz A, Sainz L and Berganza J. Magnetoresistive immunosensor for the detection of Escherichia coli O157:H7 including a microfluidic network. Biosens Bioelectron 24: 1253–1258, 2009. [DOI] [PubMed] [Google Scholar]
- 81.Murray PR and Masur H. Current approaches to the diagnosis of bacterial and fungal bloodstream infections in the intensive care unit. Crit Care Med 40: 3277–3282, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ohlsson P, Evander M, Petersson K, Mellhammar L, Lehmusvuori A, Karhunen U, Soikkeli M, Seppa T, Tuunainen E, Spangar A, von Lode P, Rantakokko-Jalava K, Otto G, Scheding S, Soukka T, Wittfooth S and Laurell T. Integrated Acoustic Separation, Enrichment, and Microchip Polymerase Chain Reaction Detection of Bacteria from Blood for Rapid Sepsis Diagnostics. Anal Chem 88: 9403–9411, 2016. [DOI] [PubMed] [Google Scholar]
- 83.Olanrewaju AO, Ng A, DeCorwin-Martin P, Robillard A and Juncker D. Microfluidic Capillaric Circuit for Rapid and Facile Bacteria Detection. Anal Chem 89: 6846–6853, 2017. [DOI] [PubMed] [Google Scholar]
- 84.Özenci V and Rossolini GM. Rapid microbial identification and antimicrobial susceptibility testing to drive better patient care: an evolving scenario. Journal of Antimicrobial Chemotherapy 74: i2–i5, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Park S, Zhang Y, Wang TH and Yang S. Continuous dielectrophoretic bacterial separation and concentration from physiological media of high conductivity. Lab Chip 11: 2893–2900, 2011. [DOI] [PubMed] [Google Scholar]
- 86.Peralta G, Roiz MP, Sánchez MB, Garrido JC, Ceballos B, Rodríguez-Lera MJ, Mateos F and De Benito I. Time-to-positivity in patients with Escherichia coli bacteraemia. Clinical Microbiology and Infection 13: 1077–1082, 2007. [DOI] [PubMed] [Google Scholar]
- 87.Pereiro I, Bendali A, Tabnaoui S, Alexandre L, Srbova J, Bilkova Z, Deegan S, Joshi L, Viovy JL, Malaquin L, Dupuy B and Descroix S. A new microfluidic approach for the one-step capture, amplification and label-free quantification of bacteria from raw samples. Chem Sci 8: 1329–1336, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Peters RP, A. M. Savelkoul Ph Fau - Simoons-Smit, S. A. Simoons-Smit Am Fau - Danner, C. M. J. E. Danner Sa Fau - Vandenbroucke-Grauls, M. A. Vandenbroucke-Grauls Cm Fau - van Agtmael and van Agtmael MA. Faster identification of pathogens in positive blood cultures by fluorescence in situ hybridization in routine practice. J Clin Microbiol 44: 119–123, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Pethig R Review article-dielectrophoresis: status of the theory, technology, and applications. Biomicrofluidics 4: 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Pitt WG, Alizadeh M, Husseini GA, McClellan DS, Buchanan CM, Bledsoe CG, Robison RA, Blanco R, Roeder BL, Melville M and Hunter AK. Rapid separation of bacteria from blood-review and outlook. Biotechnol Prog 32: 823–839, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Pivetal J, Toru S, Frenea-Robin M, Haddour N, Cecillon S, Dempsey NM, Dumas-Bouchiat F and Simonet P. Selective isolation of bacterial cells within a microfluidic device using magnetic probe-based cell fishing. Sensors and Actuators B: Chemical 195: 581–589, 2014. [Google Scholar]
- 92.Podszun S, Vulto P, Heinz H, Hakenberg S, Hermann C, Hankemeier T and Urban GA. Enrichment of viable bacteria in a micro-volume by free-flow electrophoresis. Lab Chip 12: 451–457, 2012. [DOI] [PubMed] [Google Scholar]
- 93.Poritz MA, Blaschke AJ, Byington CL, Meyers L, Nilsson K, Jones DE, Thatcher SA, Robbins T, Lingenfelter B, Amiott E, Herbener A, Daly J, Dobrowolski SF, Teng DHF and Ririe KM. FilmArray, an Automated Nested Multiplex PCR System for Multi-Pathogen Detection: Development and Application to Respiratory Tract Infection. PLOS ONE 6: e26047, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Pratt ED, Huang C, Hawkins BG, Gleghorn JP and Kirby BJ. Rare Cell Capture in Microfluidic Devices. Chem Eng Sci 66: 1508–1522, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Prinyakupt J and Pluempitiwiriyawej C. Segmentation of white blood cells and comparison of cell morphology by linear and naive Bayes classifiers. Biomed Eng Online 14: 63, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Puchberger-Enengl D, Podszun S, Heinz H, Hermann C, Vulto P and Urban GA. Microfluidic concentration of bacteria by on-chip electrophoresis. Biomicrofluidics 5: 44111–4411110, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Raub CB, Lee C and Kartalov E. Sequestration of bacteria from whole blood by optimized microfluidic cross-flow filtration for Rapid Antimicrobial Susceptibility Testing. Sensors and Actuators B: Chemical 210: 120–123, 2015. [Google Scholar]
- 98.Reshes G, Vanounou S, Fishov I and Feingold M. Cell shape dynamics in Escherichia coli. Biophys J 94: 251–264, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Salimnia H, Fairfax MR, Lephart PR, Schreckenberger P, DesJarlais SM, Johnson JK, Robinson G, Carroll KC, Greer A, Morgan M, Chan R, Loeffelholz M, Valencia-Shelton F, Jenkins S, Schuetz AN, Daly JA, Barney T, Hemmert A and Kanack KJ. Evaluation of the FilmArray Blood Culture Identification Panel: Results of a Multicenter Controlled Trial. Journal of clinical microbiology 54: 687, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Sarioglu AF, Aceto N, Kojic N, Donaldson MC, Zeinali M, Hamza B, Engstrom A, Zhu H, Sundaresan TK, Miyamoto DT, Luo X, Bardia A, Wittner BS, Ramaswamy S, Shioda T, Ting DT, Stott SL, Kapur R, Maheswaran S, Haber DA and Toner M. A microfluidic device for label-free, physical capture of circulating tumor cell clusters. Nature methods 12: 685–691, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Schwartz O and Bercovici M. Microfluidic assay for continuous bacteria detection using antimicrobial peptides and isotachophoresis. Anal Chem 86: 10106–10113, 2014. [DOI] [PubMed] [Google Scholar]
- 102.Sequist LV, Nagrath S, Toner M, Haber DA and Lynch TJ. The CTC-chip: an exciting new tool to detect circulating tumor cells in lung cancer patients. Journal of thoracic oncology : official publication of the International Association for the Study of Lung Cancer 4: 281–283, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Suehiro J, Noutomi D, Shutou M and Hara M. Selective detection of specific bacteria using dielectrophoretic impedance measurement method combined with an antigen–antibody reaction. Journal of Electrostatics 58: 229–246, 2003. [Google Scholar]
- 104.Suehiro J, Ohtsubo A, Hatano T and Hara M. Selective detection of bacteria by a dielectrophoretic impedance measurement method using an antibody-immobilized electrode chip. Sensors and Actuators B: Chemical 119: 319–326, 2006. [Google Scholar]
- 105.Sun X, Weinlandt WD, Patel H, Wu M and Hernandez CJ. A microfluidic platform for profiling biomechanical properties of bacteria. Lab on a Chip 14: 2491–2498, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Tadimety A, Syed A, Nie Y, Long CR, Kready KM and Zhang JX. Liquid biopsy on chip: a paradigm shift towards the understanding of cancer metastasis. Integr Biol 9: 22–49, 2017. [DOI] [PubMed] [Google Scholar]
- 107.Thon JN, Macleod H, Begonja AJ, Zhu J, Lee K-C, Mogilner A, Hartwig JH and Italiano JE. Microtubule and cortical forces determine platelet size during vascular platelet production. Nature Communications 3: 2012. [DOI] [PubMed] [Google Scholar]
- 108.Tokel O, Yildiz UH, Inci F, Durmus NG, Ekiz OO, Turker B, Cetin C, Rao S, Sridhar K, Natarajan N, Shafiee H, Dana A and Demirci U. Portable microfluidic integrated plasmonic platform for pathogen detection. Sci Rep 5: 9152, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Urbansky A, Ohlsson P, Lenshof A, Garofalo F, Scheding S and Laurell T. Rapid and effective enrichment of mononuclear cells from blood using acoustophoresis. Scientific Reports 7: 17161, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Vincent JL, J. Rello J Fau - Marshall, E. Marshall J Fau - Silva, A. Silva E Fau - Anzueto, C. D. Anzueto A Fau - Martin, R. Martin Cd Fau - Moreno, J. Moreno R Fau - Lipman, C. Lipman J Fau - Gomersall, Y. Gomersall C Fau - Sakr, K. Sakr Y Fau – Reinhart and Reinhart K. International study of the prevalence and outcomes of infection in intensive care units. JAMA 302: 2323–2329, 2009. [DOI] [PubMed] [Google Scholar]
- 111.Wang S, Inci F, Chaunzwa TL, Ramanujam A, Vasudevan A, Subramanian S, Chi Fai Ip A, Sridharan B, Gurkan UA and Demirci U. Portable microfluidic chip for detection of Escherichia coli in produce and blood. Int J Nanomedicine 7: 2591–2600, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wei Hou H, Gan HY, Bhagat AA, Li LD, Lim CT and Han J. A microfluidics approach towards high-throughput pathogen removal from blood using margination. Biomicrofluidics 6: 24115–2411513, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wu Z, Willing B, Bjerketorp J, Jansson JK and Hjort K. Soft inertial microfluidics for high throughput separation of bacteria from human blood cells. Lab Chip 9: 1193–1199, 2009. [DOI] [PubMed] [Google Scholar]
- 114.Xia N, Hunt TP, Mayers BT, Alsberg E, Whitesides GM, Westervelt RM and Ingber DE. Combined microfluidic-micromagnetic separation of living cells in continuous flow. Biomed Microdevices 8: 299–308, 2006. [DOI] [PubMed] [Google Scholar]
- 115.Yagupsky P and Nolte FS. Quantitative aspects of septicemia. Clinical microbiology reviews 3: 269–279, 1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Yang Y, Kim S and Chae J. Separating and Detecting Escherichia Coli in a Microfluidic Channel for Urinary Tract Infection Applications. Journal of Microelectromechanical Systems 20: 819–827, 2011. [Google Scholar]
- 117.Yu JJ, Xiao LD and Yang M. A PDMS microfluidic chip with nanostructures for bacteria concentration and fast detection. In: 2008 IEEE 21st International Conference on Micro Electro Mechanical Systems 2008, p. 272–275.
- 118.Yung CW, Fiering J, Mueller AJ and Ingber DE. Micromagnetic-microfluidic blood cleansing device. Lab Chip 9: 1171–1177, 2009. [DOI] [PubMed] [Google Scholar]
- 119.Zelenin S, Hansson J, Ardabili S, Ramachandraiah H, Brismar H and Russom A. Microfluidic-based isolation of bacteria from whole blood for sepsis diagnostics. Biotechnol Lett 37: 825–830, 2015. [DOI] [PubMed] [Google Scholar]
- 120.Zhu L, Zhang Q, Feng H, Ang S, Chau FS and Liu WT. Filter-based microfluidic device as a platform for immunofluorescent assay of microbial cells. Lab Chip 4: 337–341, 2004. [DOI] [PubMed] [Google Scholar]
- 121.Zhu Z, Zhang W, Leng X, Zhang M, Guan Z, Lu J and Yang CJ. Highly sensitive and quantitative detection of rare pathogens through agarose droplet microfluidic emulsion PCR at the single-cell level. Lab Chip 12: 3907–3913, 2012. [DOI] [PubMed] [Google Scholar]