Abstract
Acute myeloid leukemias (AML) are characterized by mutations of tumor suppressor and oncogenes, involving distinct genes in adults and children. While certain mutations have been associated with the increased risk of AML relapse, the genomic landscape of primary chemotherapy resistant AML is not well defined. As part of the TARGET initiative, we performed whole-genome DNA and transcriptome RNA and miRNA sequencing analysis of pediatric AML with failure of induction chemotherapy. We identified at least three genetic groups of patients with induction failure, including those with NUP98 rearrangements, somatic mutations of WT1 in the absence of apparent NUP98 mutations, and additional recurrent variants including those in KMT2C and MLLT10. Comparison of specimens before and after chemotherapy revealed distinct and invariant gene expression programs. While exhibiting overt therapy resistance, these leukemias nonetheless showed diverse forms of clonal evolution upon chemotherapy exposure. This included selection for mutant alleles of FRMD8, DHX32, PIK3R1, SHANK3, MKLN1, as well as persistence of WT1 and TP53 mutant clones, and elimination of FLT3, PTPN11, and NRAS mutant clones. These findings delineate genetic mechanisms of primary chemotherapy resistance in pediatric AML, which should inform improved approaches for its diagnosis and therapy.
Introduction
Overall survival for children with acute myeloid leukemia (AML) remains low, due principally to the failure to achieve durable disease remission after initial induction therapy. Failure rate of primary induction remission therapy in pediatric AML is 10–15%, and only about a third of patients for whom primary induction therapy fails are ultimately cured (1). A recently completed Children’s Oncology Group study of pediatric AML treatment had remission induction rates of only 85%, with less than 65% of children alive at three years (12). Reasons for the lack of response to initial chemotherapy in pediatric AML remain unclear, and a molecular understanding of this process is needed.
Since the first AML genome was sequenced (2, 3), numerous genomic studies have revealed diverse disease subtypes and distinct modes of disease relapse (4). For example, whole-genome sequencing of AML specimens from adults with relapsed disease revealed broad patterns of clonal evolution, suggesting that founding clones gained mutations upon relapse, or that subclones persisted with acquisition of additional mutations upon therapy (5). Analysis of exome capture sequencing from matched diagnosis, remission, and relapse trio pediatric specimens showed that responses of specific genetically-defined clones were associated with disease relapse (6). Similarly, clonal persistence after induction chemotherapy was found to be associated with disease relapse in adult AML (7).
Recent study of primary chemotherapy resistance in a cohort of 107 children and adults with AML using targeted gene sequencing demonstrated that few patients exhibited specific individual mutations associated with primary chemotherapy resistance and failure of induction chemotherapy (8). In addition, at least for some pediatric and adult AML subtypes, chemotherapy resistance is caused by the epigenetic activation of the transcription factor MEF2C (9–10). This suggests that there are additional genetic or molecular mechanisms mediating primary chemotherapy resistance in pediatric and adult AML. Importantly, pediatric AML is characterized by distinct genetic mutations and genomic rearrangements, with relative paucity of the recurrent mutations frequently observed in adult AML (11). Thus, direct study of primary chemotherapy resistance is needed for pediatric AML.
Here, we assembled a cohort of pediatric patients with primary chemotherapy resistance and failure of induction chemotherapy, as part of the TARGET initiative. We analyzed whole-genome DNA, mRNA, and miRNA sequence data, obtained at diagnosis and upon chemotherapy administration. These studies revealed distinct classes of genetic mutations and their clonal evolution in chemotherapy resistant disease, which should inform future approaches for the diagnosis, risk stratification and therapeutic interventions for pediatric AML.
Methods
Complete methodological details are provided in the Supplementary Methods. Briefly, all specimens and clinical data were obtained from patients enrolled on biology studies and clinical trials through the Children’s Oncology Group (protocols AAML0531 and AAML03P1). Patient samples with adequate amounts of high-quality nucleic acids were sequentially identified, and selected for genomic profiling. All available specimens with induction failure were included in our analyses. Patient samples were collected as matched trios: bone marrow aspirates pre- and post-induction, and matched fibroblasts isolated from the bone marrow. Details of sample preparation protocols and clinical annotations and all primary data are available through the TARGET Data Matrix (https://ocg.cancer.gov/programs/target/data-matrix). The details of additional specimens from patients without induction failure were performed as previously published (11). Whole-genome paired-end sequencing libraries were prepared using the genomic 350–450bp insert Illumina library construction protocol with Biomek FX robot (Beckman-Coulter, USA), sequenced with the average coverage of 30-fold using Illumina HiSeq2500. Sequence reads were mapped to the GRCh37 (hg19) genome using bwa-mem, and analyzed to identify single nucleotide variants (SNVs), insertions/deletions (indels), gene fusions, and structural variants. For mRNA-sequencing, extracted RNAs was used to generate cDNAs using the SMART cDNA synthesis protocol with the SMARTScribe reverse transcriptase (Clontech) and resultant libraries were sequenced with 75 bp paired reads using Illumina HiSeq2500. RNA-seq reads were aligned with STAR (version 2.4.2a), and genes annotated in Gencode v18 were quantified with featureCounts (v1.4.3-p1). Fusion genes were detected using FusionCatcher and STAR-Fusion. Resultant variant call files (VCFs) were subsequently aggregated using an integrated script, available from https://github.com/kentsisresearchgroup/TargetInductionFailure. VCFs were parsed to assemble single nucleotide variants, indels, copy number variation, structural variants, and gene fusions in a master table, and filtered to identify high-confidence calls. Normalization and differential expression was done with the Bioconductor package DESeq2. Gene set enrichment analysis was performed using GSEA v2.2.1 with MSigDB v6.0. All raw sequencing data are available via dbGaP accession numbers phs000465, phs000178 and phs000218, with the processed mutational and expression data available from Zenodo (http://doi.org/10.5281/zenodo.1403737).
Results
Genomic landscape of pediatric induction failure AML.
A total of 28 patients with primary chemotherapy resistance and failure of induction chemotherapy were studied. The patients were uniformly treated as part of the COG AAML0531 study, having received cytarabine, daunorubicin and etoposide (ADE10+3) chemotherapy. Demographic features of the study cohort are listed in Supplemental Table 1, and are representative of the entire patient cohort enrolled in the AAML0531 study (12). Failure of induction chemotherapy was defined as morphologic persistence of at least 5% of AML blasts in the bone marrow at least 28 days after therapy initiation, but prior to the second course of induction chemotherapy (12). Given the persistence of circulating leukemia cells in patients with induction failure, we cultured fibroblasts from the bone marrow to isolate non-tumor germline DNA to identify somatically acquired mutations. Using supervised analysis based on genes currently known to have cancer predisposition potential, we did not identify any apparent pathogenic germline variants in this cohort (Supplemental Table 2). For whole-genome DNA sequencing, we obtained mean coverage of 39 (range 23–69). mRNA and miRNA sequencing data had on average 59% and 19% mapping coverage, respectively.
In agreement with prior studies (11), we found that this cohort of pediatric AML with induction failure had relatively few mutations commonly observed in adult AML, including DNMT3A, TET2, IDH1/2 and others (13). The most common mutations observed in our cohort involved NUP98, WT1, RUNX1, MLLT10, SPECC1, and KMT2C, predominantly as a result of genomic rearrangements and somatic structural variants (Figure 1A). All annotated identified somatic variants are provided in Supplemental Table 3, with specific mutations highlighted in Supplemental Figure 1, such as the truncating mutations in WT1. In particular, we identified NUP98-NSD1 fusions, as well as a number of additional genomic rearrangements, leading to the production of chimeric fusion genes, as evidenced by the combined genomic rearrangements in DNA, and the presence of supporting mRNA sequencing reads in RNA-seq data (Figure 1B). While mutations of FLT3 and KMT2A, and t(8;21), inv(16) and trisomy 8 alterations were the five most common events in the analysis of the entire cohort of pediatric AML patients (11), these abnormalities were relatively depleted in our induction failure cohort.
Figure 1: Recurrently mutated genes in pediatric induction failure AML identified by whole-genome and RNA sequencing analysis.
(a) Top 20 most commonly called mutated genes, with the mutation type indicated by color, observed at diagnosis, enumerated by the number of total calls, independent of patient assignment. (b) Circos plot of high-confidence gene fusions, identified from combined analysis of RNA and whole-genome sequencing data, observed at diagnosis. All variants are tabulated as fusions for calls from RNA-seq, structural variants when called from whole-genome sequencing, and both when both the fusion and supporting genomic structural variants match. For example, NUP98-NSD1 translocations are identified as fusions when found by RNA-seq, and structural variants when identified in whole-genome sequencing.
The relative enrichment for NUP98 chimeric gene rearrangements and WT1 mutations in this induction failure cohort is consistent with the reported poor prognosis of these mutations, with a reported 4-year event-free survival of less than 10% (14). By combining our results with 197 additional specimens from the TARGET cohort for whom whole genome or exome sequencing was available, we also confirmed the poor prognostic significance of these mutations (Supplemental Figure 2). In particular, 18% of specimens for whom remission could be achieved had WT1 mutations, as compared to 54% (15 of 28 patients) in the induction failure group. Similarly, NUP98 rearrangements were observed in 21% (6 of 28) of the induction failure cohort, in contrast to less than 4% of the remission induction pediatric AML patients. Consistent with the notion that chemotherapy resistance can be mediated by specific combinations of mutations, patients with distinct combinations of mutations had significantly varied overall survival (Supplemental Figure 2). While some of the recurrent mutations were shared between this induction failure cohort and the previously published TARGET remission patients (Supplemental Table 4), mutations unique to the induction failure cohort at diagnosis included KMT2C mutations, PTRPRC deletions and ELF1 mutations among others, suggesting potential new mechanisms of primary chemotherapy resistance.
Genetic subtypes of pediatric AML with primary chemotherapy resistance.
Although diverse mutations were observed in our cohort, unsupervised hierarchical clustering was unable to segregate the observed cohort into distinct classes. Therefore, we divided the patients into three groups based on the most common recurrent mutations (Figure 2A). Group 1 (6 patients) was defined by the presence of NUP98 rearrangements, and additional mutations including WT1, ELF1, and FRMD8. No specimens exhibited chromosomal monosomies or complex karyotypes. Although FLT3 mutations are often observed in NUP98-NSD1 leukemias, group 1 did not appear to have FLT3 mutations, consistent with the relative survival benefit of FLT3 mutations in this cohort (Supplemental Figure 2). The association of NUP98 rearrangements with WT1 mutations in AML induction failure may be due to the functional interaction between these two factors, since patients with both alterations are known to have a much worse prognosis than either alone (Supplemental Figure 2) (11, 14, 15). In addition, we observed an association of NUP98 rearrangements with deletions of the ETS transcription factor ELF1 and copy number gains of the gene encoding cell adhesion signaling factor FRMD8, both of which have also been observed to be mutated in myeloid malignancies (16, 17). This association may involve similar cooperating interactions that presumably cause intrinsic chemotherapy resistance. We also identified gains of MYC in two patients from group 1, in agreement with prior finding of activating MYC mutations in NUP98-NSD1 rearranged AML (18).
Figure 2: Three groups of pediatric induction failure AML identified by whole-genome and RNA sequencing analysis.
(a) Tile plot of recurrently mutated genes and gene expression profiles by patient, showing three disease groups, as labeled, with each row listing the mutant gene, and each column representing an individual patient specimen: Group 1, defined by NUP98 alterations (patients with NUP98-NSD1 fusions except patient 1 with NUP98 gain); Group 2, defined by WT1 mutations, and Group 3, defined by the apparent absence of NUP98 or WT1 mutations. (b) Gene set enrichment analysis (GSEA) of the three patient groups, listing significantly enriched (red) and downregulated (blue) gene sets, as a function of their normalized enrichment.
Group 2 (11 patients) was defined by the presence of WT1 mutations without apparent NUP98 rearrangements, and also involved additional mutations including tyrosine kinase domain (TKD, SNV) and internal tandem duplication (ITD, indel) of FLT3, and various other copy number changes and genomic rearrangements. We observed both missense and nonsense WT1 mutations (Supplemental Figure 1), consistent with previous reports in AML (19, 20, 21). In addition, group 2 included cases with copy number alterations involving the BCL11B, AKT1, and ARID1B loci, among others (Figure 2A), as well as one patient specimen PATISD with monosomy 7 (Supplemental Table 1). BCL11B is a known tumor suppressor gene mutated in refractory forms of T-cell acute lymphoblastic leukemias (T-ALL) (22), including a subtype that may share common origins with refractory AML (20). In addition, both BCL11B and ARID1B are components of the SWI/SNF/BAF chromatin remodeling complex that is disrupted in diverse human cancers (24).
Group 3 (11 patients) was defined by the apparent absence of NUP98 rearrangements and WT1 mutations, and instead included leukemias with mutations of KMT2C and MLLT10 as well as other genes (Figure 2A). KMT2C is the tumor suppressor gene that encodes the MLL3 chromatin remodeling factor, that is also inactivated in myeloid malignancies as a result of losses of chromosome 7q (23). Similarly, MLLT10 is frequently rearranged as part of KMT2A/MLL1 and other chromosomal translocations in acute leukemias, including refractory forms of T-ALL in particular (24) (25). Given the involvement of additional genes and loci recurrently mutated or rearranged in this cohort of patients, it is probable that additional subtypes of chemotherapy resistant disease exist.
Using mRNA sequencing, we analyzed gene expression programs associated with the primary chemotherapy resistant AML, as assessed using gene set enrichment analysis in diagnostic samples (Figure 2B). Unsupervised hierarchical clustering of gene expression profiles did not segregate with the genetically defined groups (Supplemental Figure 3), with the complete RNA expression data provided in Supplemental Table 5. This suggests that diverse genetic subtypes of induction failure AML may engage common gene expression programs. This notion is consistent with the recent study implicating epigenetic signaling by the transcription factor MEF2C in AML chemotherapy resistance (9).
Intriguingly, while only one patient in group 1 remained alive at 6 years after therapy, and three patients remained alive in group 2, five survivors were observed in group 3. However, the size of this cohort is not sufficiently powered to detect statistically significant differences in survival (log-rank p = 0.39, 0.70, and 0.55 for group 1 vs 2, 1 vs 3, and 2 vs 3, respectively). Comparison of gene expression profiles in alive versus deceased patients revealed differentially expressed genes both at diagnosis and after induction chemotherapy (Supplemental Figure 4, Supplemental Table 6). For example, deceased patients had relatively higher expression of PREX2, which has previously been reported to be overexpressed in EVI1-rearranged AML and encodes a regulator of PI3K signaling (47). Similarly, we observed overexpression of MPL among deceased patients, which encodes the thrombopoietin receptor and regulates cytokine signaling (48). In addition, we found several differentially expressed gene sets in alive versus deceased patients, such as those associated with cell cycle control (Supplemental Figure 5). These results suggest that the apparent diversity of genetic subtypes of induction failure may also be associated with survival outcomes.
Lastly, we surveyed microRNA expression in diagnostic samples from this cohort, with the most highly expressed miRNAs listed in Supplemental Table 7. We observed that miR-21 was highly expressed among all 3 subgroups of induction failure patients, consistent with its reported association with inferior clinical outcomes (26). Similarly, we observed high levels of expression of miR-10a, particularly in group 1. Upregulation of miR-10a has been reported in NPM1-mutant AML with associated MDM4 downregulation, potentially interfering with TP53 signaling (27). We also found high expression of miR-103 in group 1 patients, which has been reported to downregulate RAD51, leading to dysregulated DNA damage response (28). In addition, we found upregulation of miR-181a in groups 2 and 3, which has been reported to be overexpressed and mediate ATM downregulation in AML cell lines (29). In all, these findings are consistent with the proposed mechanisms of regulation of chemotherapy response by miRNAs in AML (30). Overall miRNA expression profiles for highly expressed miRNAs were not significantly different across the three groups (Supplemental Figure 6).
Diverse models of clonal evolution by induction chemotherapy.
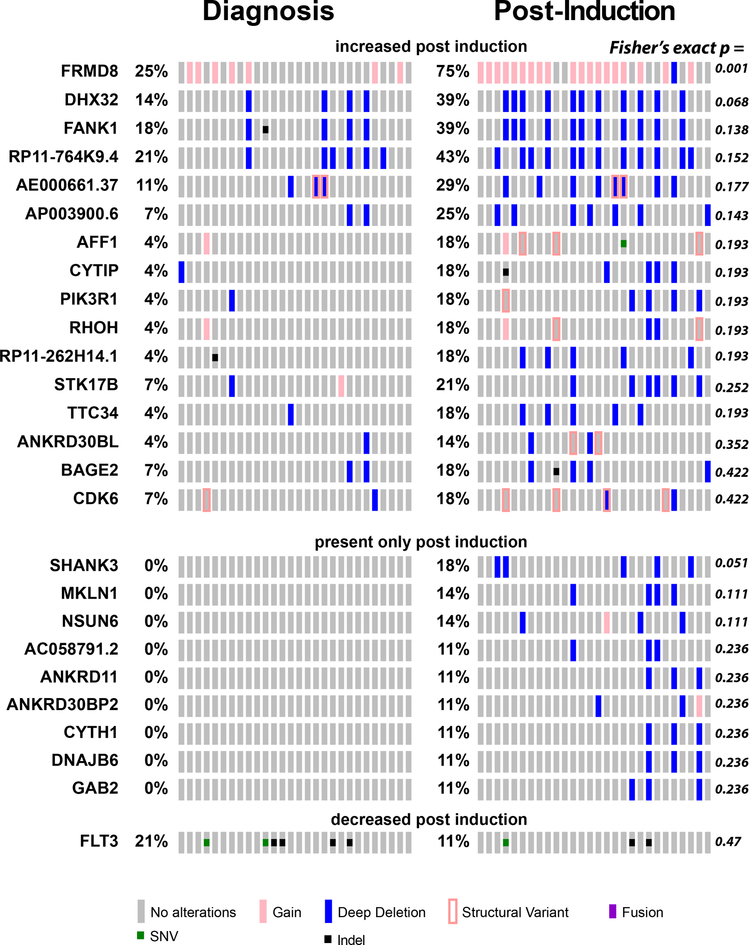
We reasoned that exposure to chemotherapy would lead to the selection of genetic clones with mutations conferring chemotherapy resistance, and contraction or elimination of clones that are susceptible to ADE chemotherapy. Thus, we compared the prevalence of mutations among different patients in specimens collected before and after induction chemotherapy (Figure 3). We found numerous genomic rearrangements and mutations that were increased in prevalence upon induction chemotherapy exposure. For example, we observed that gains of the FRMD8 locus were present in 7 of 28 (25%) patients at diagnosis, as compared to 20 of 28 (71%) patients post-chemotherapy (Fisher’s exact test p = 1.0e-3). This suggests that genomic rearrangement involving FRMD8 or linked genes may contribute to chemotherapy resistance. FRMD8 mutations were not identified in the patients without induction failure in the complete TARGET cohort (11) (Supplemental Table 4), consistent with a specific association with primary chemotherapy resistance. FRMD8 encodes a plasma membrane-associated FERM domain that can contribute to Wnt signaling and processing of transmembrane precursors of inflammatory cytokines (31, 32). In addition, increased FRMD8 gene expression was found to be a marker of poor prognosis in adult AML (33).
Figure 3: Clonal selection upon chemotherapy treatment.
Tile plot showing recurrently mutated genes with changes in apparent allele frequencies upon induction chemotherapy. For FLT3 mutations, TKD mutations are listed as SNVs, and ITDs as indel mutations.
Mutations and rearrangements of various additional genes with functions in cell adhesion and signaling, including FANK1, PIK3R1, SHANK3, and MKLN1, also appear to be selected upon chemotherapy exposure, suggesting that they may also contribute to therapy resistance. Consistent with a specific association of FANK1 mutations with primary chemotherapy resistance, we did not observe them in specimens from remission patients in the TARGET study (11)(Supplemental Table 4). In contrast, mutations of FLT3 exhibited depletion upon chemotherapy administration, although the difference in mutation prevalence post-induction did not reach statistical significance (Figure 3A). Aberrant activation of FLT3 kinase signaling is a known oncogenic event in AML pathogenesis, contributing to the enhanced proliferation and survival of AML cells, and is associated with inferior prognosis when present at sufficiently high allelic frequencies (34–38). Its relative depletion by chemotherapy in AML induction failure suggests that its subclonal evolution in and of itself does not cause chemotherapy resistance. Rather, its activation in combination with specific other pathogenic events as part of distinct clones, such as those with mutations of WT1 or NUP98 rearrangements or others (6, 38), may cause resistance to chemotherapy.
In addition to the marked changes in overall clonal architecture associated with induction chemotherapy, we also observed multiple modes of clonal evolution within individual leukemias. In general, induction chemotherapy induced a relative contraction of the overall AML cell population, as evidenced by the reduction of the apparent variant allele frequencies (VAF) of mutant genes (mean 0.40 versus 0.28 for pre- and post-chemotherapy, respectively, Bonferroni adjusted t-test p = 2.6e-4). While VAF contraction was common to most mutations, closer examination of individual patients revealed distinct potential modes of clonal evolution (Figure 4). For example, specimen PASFHK exhibited significant expansion of the WT1;PTCH1;ZNF785-mutant clone, and elimination of the FLT3;SERPIN2-mutant subclones, upon chemotherapy exposure (Figure 4A). This is consistent with the prior reports of elimination of FLT3-mutant subclones upon AML relapse (5), supporting the proposal that activated FLT3 contributes to chemotherapy resistance only when present with specific cooperating mutations, such as WT1. For specimen PATJMY, we observed evolution of a new loss-of-function nonsense mutation of CHMP6, which emerged either upon chemotherapy exposure or was selected as a pre-existing subclone, present at less than 2% fraction at diagnosis, given the 50-fold sequencing coverage for CHMP6 (Figure 4B). Reduced CHMP6 gene expression has been associated with inferior survival of elderly AML patients (39), and its function in endosomal cell surface receptor recycling may contribute to chemotherapy resistance (40). In agreement with prior reports (7), specimen PASTZK exhibited subclonal evolution of mutant TP53 at diagnosis, which led to its clonal expansion in combination with clonal PHF6 mutation upon chemotherapy administration, in contrast to mutation of NRAS which remained subclonal (Figure 4C). Finally, specimen PARXYR exhibited relative contraction of the WT1;PTPN11-mutant subclone, and relative expansion of the GPR137B-mutant subclone that additionally acquired a CD82 mutation (Figure 4D). Other leukemias showed similar subclonal composition pre- and post-induction, such as for specimen PARBTV, which demonstrated the likely pathogenic IDH2 R172K (VAF 0.58 pre and 0.45 post) and H3F3A K27M mutations (0.46 pre and 0.54 post). These findings demonstrate distinct modes of clonal selection upon chemotherapy exposure, which are expected to inform future targeting of specific molecular mechanisms to overcome or block chemotherapy resistance.
Figure 4: Mutant allele frequencies suggest diverse modes of clonal evolution upon chemotherapy exposure.
Variant allele frequencies (VAF) in four specimens with WT1 mutant clones are represented as the height of color-coded clones, with full height of allele frequency axis corresponding to a mutant allele frequency of 1. Simplest models of clonal architecture are shown, with additional models possible. Each hypothesized subclone is represented by a different color. (a) Specimen PASFHK in Group 2, largest VAF at diagnosis was 0.96 for WT1 R430P; largest VAF at induction failure was 0.38 for WT1 R430P. (b) Specimen PATJMY in Group 1, largest VAF at diagnosis was 0.53 for WT1 S381, largest VAF at induction failure was 0.47 for CHMP6 E108stop. (c) Specimen PASTZK in Group 2, largest VAF at diagnosis was 0.77 for PHF6 R370stop, largest VAF at induction failure was 0.37 for TP53 R273C. (d) Specimen PARXYR in Group 1, largest VAF at diagnosis was 0.57 for RBAK V708I, largest VAF at induction failure was 0.43 for GPR137B T182M.
Discussion
This study defines the genomic landscape of pediatric AML with primary chemotherapy resistance and failure of induction remission therapy. Importantly, primary chemotherapy resistant pediatric AML involves multiple distinct genetic mechanisms. Most notably, we found substantial prevalence of structural rearrangements, at least some of which are associated with the expression of chimeric fusion genes. In particular, we observed at least three distinct genetic groups of patients with induction failure, including those with NUP98 rearrangements, somatic mutations of WT1, ELF1, KMT2C, MLLT10, and additional recurrent gene mutations, fusions, and structural rearrangements, some of which have been observed in other malignancies. Given the known technical challenges with the detection of genomic rearrangements and gene fusions (41), it is possible that additional pathogenic structural variants or chimeric gene fusions may contribute to AML and primary chemotherapy resistance.
In our prior study of primary chemotherapy resistance, we identified individual mutations of ASXL1, SETBP1 and RELN to be significantly enriched in a subset of pediatric AML with primary induction failure (8). Insofar as ASXL1 and WT1 mutations are mutually exclusive in both pediatric and adult AML, the prevalence of WT1 mutations and absence of apparent ASXL1 mutations in our current cohort suggest that additional genetic mechanisms of primary chemotherapy resistance likely exist. Our results also suggest that varied genetic mechanisms of chemotherapy resistance may converge on coherent gene expression programs, at least insofar as they cannot be statistically decomposed by matrix factorization used as part of this gene set enrichment analysis.
Importantly, our study identified distinct combinations of mutations that appear to be associated with primary chemotherapy resistance. In particular, we observed an association between NUP98-NSD1 fusions and mutations of WT1, ELF1 and FRMD8, suggesting possible cooperativity in their pathogenic functions. Similarly, we observed an association between WT1 mutations and rearrangements of BCL11B and ARID1B loci, both of which encode components of the SWI/SNF/BAF chromatin remodeling complex. Notably, BCL11B is recurrently mutated in refractory forms of T-ALL, which may share common origins with subsets of AML (20). Evidently, these combinatorial mechanisms in pediatric AML are distinguished from other mechanisms of chemotherapy resistance, such as inactivation of TP53 in adult AML (7).
Our study identified additional mutations associated with pediatric primary chemotherapy resistance. This includes loss-of-function mutations of KMT2C, which encodes a component of the MLL3 chromatin remodeling complex, potentially similar to the deletions of chromosome 7q observed in high-risk AML that involve this locus and have been found to confer susceptibility to epigenetic therapies (23). We also observed deletions of MLLT10, which is recurrently rearranged as gene fusions in subsets of T-ALL. Insofar as MLLT10 is a cofactor of the DOT1L methyltransferase, this may be associated with the susceptibility to emerging DOT1L methyltransferase inhibitors such as pinometostat (EPZ-5686), which will need to be tested in future studies. Naturally, additional mutations associated with primary chemotherapy resistance may be found in larger studies. For instance, the presence of likely pathogenic IDH2 R172K and H3K27M K27M mutations in one specimen in our cohort suggests additional potential mechanisms of chemotherapy resistance (42–45), which may confer susceptibility to emerging therapies such as the IDH inhibitor enasidenib (AG-221) for example.
Our findings also suggest that the diversity of genetic chemotherapy resistance mechanisms may be associated with variable outcomes of intense combination chemotherapy in AML. Importantly, increase in the apparent prevalence and allelic frequency of genetic clones with mutations of FRMD8, FANK1, PIK3R1, WT1 and others indicate that these alleles, in cooperation with NUP98-NSD1 and other initiating mutations, may directly cause chemotherapy resistance. In contrast, subclonal mutations of FLT3, PTPN11 and NRAS were substantially reduced or eliminated by chemotherapy, suggesting that these secondary mutations in and of themselves do not cause chemoresistance. Indeed, subclonal mutations of FLT3 or NRAS were not significantly associated with primary chemotherapy resistance in our prior study (8). Diverse genetic mechanisms of chemotherapy resistance may be associated with clonal evolution (5, 6), as also evidenced by our findings (Figures 3 and 4). On the other hand, common gene expression programs may be associated with shared molecular dependencies, substantiating the development of targeted therapies, as recently evidenced by molecular therapy of MEF2C in chemotherapy resistant AML (46).
In all, our study demonstrates that primary chemotherapy resistance and failure of induction chemotherapy in pediatric AML are associated with multiple genetic mechanisms, and exhibit diverse clonal dynamics, dependent on distinct combinations of mutations. Future functional studies will be needed to assess the mechanisms of cooperativity among the observed chemotherapy-associated mutations and their specific pharmacologic targeting. Similarly, additional studies will be needed to define the prognostic significance of the observed chemotherapy-associated mutations. This is expected to delineate molecular mechanisms of primary chemotherapy resistance in pediatric AML, which should inform improved approaches for its diagnosis and therapy.
Supplementary Material
Acknowledgements.
This paper is dedicated to the memory of Dr. Robert Arceci. This work was supported by the NCI U10 CA98543 (TARGET), U24 CA114766 (COG), U10 CA180886, U10 CA098413, U10 CA180899, P30 CA008748, R01 CA204396, by the Damon Runyon-Richard Lumsden Foundation Clinical Investigator and St. Baldrick’s Foundation Arceci Innovation Awards (A.K.), and the Charles E. Trobman Scholarship (N.M.). We thank T. Davidsen and P. Gesuwan for their support of the TARGET Data Coordinating Center, and Alejandro Gutierrez, Gila Spitzer, and Maria Luisa Sulis for comments on the manuscript.
Footnotes
Conflicts of Interest. A.K. is a consultant for Novartis. All other authors declare no relevant conflicts of interest.
References
- 1.Rasche M, Zimmermann M, Borschel L, Bourquin JP, Dworzak M, Klingebiel T, et al. Successes and challenges in the treatment of pediatric acute myeloid leukemia: a retrospective analysis of the AML-BFM trials from 1987 to 2012. Leukemia. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ley TJ, Ding L, Walter MJ, McLellan MD, Lamprecht T, Larson DE, et al. DNMT3A mutations in acute myeloid leukemia. N Engl J Med. 2010;363(25):2424–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mardis ER, Ding L, Dooling DJ, Larson DE, McLellan MD, Chen K, et al. Recurring mutations found by sequencing an acute myeloid leukemia genome. N Engl J Med. 2009;361(11):1058–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Papaemmanuil E, Gerstung M, Bullinger L, Gaidzik VI, Paschka P, Roberts ND, et al. Genomic Classification and Prognosis in Acute Myeloid Leukemia. N Engl J Med. 2016;374(23):2209–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ding L, Ley TJ, Larson DE, Miller CA, Koboldt DC, Welch JS, et al. Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature. 2012;481(7382):506–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Farrar JE, Schuback HL, Ries RE, Wai D, Hampton OA, Trevino LR, et al. Genomic Profiling of Pediatric Acute Myeloid Leukemia Reveals a Changing Mutational Landscape from Disease Diagnosis to Relapse. Cancer Res. 2016;76(8):2197–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Klco JM, Miller CA, Griffith M, Petti A, Spencer DH, Ketkar-Kulkarni S, et al. Association Between Mutation Clearance After Induction Therapy and Outcomes in Acute Myeloid Leukemia. JAMA. 2015;314(8):811–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown FC, Cifani P, Drill E, He J, Still E, Zhong S, et al. Genomics of primary chemoresistance and remission induction failure in paediatric and adult acute myeloid leukaemia. Br J Haematol. 2017;176(1):86–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brown FC, Still E, Koche RP, Yim CY, Takao S, Cifani P, et al. MEF2C Phosphorylation Is Required for Chemotherapy Resistance in Acute Myeloid Leukemia. Cancer Discov. 2018;8(4):478–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Laszlo GS, Alonzo TA, Gudgeon CJ, Harrington KH, Kentsis A, Gerbing RB, et al. High expression of myocyte enhancer factor 2C (MEF2C) is associated with adverse-risk features and poor outcome in pediatric acute myeloid leukemia: a report from the Children’s Oncology Group. J Hematol Oncol. 2015;8:115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bolouri H, Farrar JE, Triche T Jr., Ries RE, Lim EL, Alonzo TA, et al. The molecular landscape of pediatric acute myeloid leukemia reveals recurrent structural alterations and age-specific mutational interactions. Nat Med. 2018;24(1):103–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gamis AS, Alonzo TA, Meschinchi S, Sung L, Berbing RB, Raimondi SC, et al. Gemtuzumab Ozogamicin in children and adolescents with de novo acute myeloid leukemia improves event-free survival by reducing relapse risk: results from the randomized phase III chidren’s oncology group trial AAML0531. Journal of Clinical Oncology. 2014;32(27):3021–3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cancer Genome Atlas Research N, Ley TJ, Miller C, Ding L, Raphael BJ, Mungall AJ, et al. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368(22):2059–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hollink IH, van den Heuvel-Eibrink MM, Arentsen-Peters ST, Pratcorona M, Abbas S, Kuipers JE, et al. NUP98/NSD1 characterizes a novel poor prognostic group in acute myeloid leukemia with a distinct HOX gene expression pattern. Blood. 2011;118(13):3645–56. [DOI] [PubMed] [Google Scholar]
- 15.Ostronoff F, Othus M, Gerbing RB, Loken MR, Raimondi SC, Hirsch BA, et al. NUP98/NSD1 and FLT3/ITD coexpression is more prevalent in younger AML patients and leads to induction failure: a COG and SWOG report. Blood. 2014;124(15):2400–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ando K, Tsushima H, Matsuo E, Horio K, Tominaga-Sato S, Imanishi D, et al. Mutations in the nucleolar phosphoprotein, nucleophosmin, promote the expression of the oncogenic transcription factor MEF/ELF4 in leukemia cells and potentiates transformation. J Biol Chem. 2013;288(13):9457–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.da Silva-Coelho P, Kroeze LI, Yoshida K, Koorenhof-Scheele TN, Knops R, van de Locht LT, et al. Clonal evolution in myelodysplastic syndromes. Nat Commun. 2017;8:15099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lavallee VP, Lemieux S, Boucher G, Gendron P, Boivin I, Girard S, et al. Identification of MYC mutations in acute myeloid leukemias with NUP98-NSD1 translocations. Leukemia. 2016;30(7):1621–4. [DOI] [PubMed] [Google Scholar]
- 19.Miyagi T, Ahuja H, Kubota T, et al. Expression of the Candidate Wilms-Tumor Gene, Wt1, in Human Leukemia-Cells. Leukemia. 1993;7(7):970–977. [PubMed] [Google Scholar]
- 20.Bergmann L, Miething C, Maurer U, et al. High levels of Wilms’ tumor gene (wt1) mRNA in acute myeloid leukemias are associated with a worse long-term outcome. Blood. 1997;90(3):1217–1225. [PubMed] [Google Scholar]
- 21.King-Underwood L, Renshaw J, PritchardJones K. Mutations in the Wilms’ tumor gene WT1 in leukemias. Blood. 1996;87(6):2171–2179. [PubMed] [Google Scholar]
- 22.Gutierrez A, Kentsis A, Sanda T, Holmfeldt L, Chen SC, Zhang J, et al. The BCL11B tumor suppressor is mutated across the major molecular subtypes of T-cell acute lymphoblastic leukemia. Blood. 2011;118(15):4169–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen C, Liu Y, Rappaport AR, Kitzing T, Schultz N, Zhao Z, et al. MLL3 is a haploinsufficient 7q tumor suppressor in acute myeloid leukemia. Cancer Cell. 2014;25(5):652–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gutierrez A, Kentsis A. Acute myeloid/T-lymphoblastic leukaemia (AMTL): a distinct category of acute leukaemias with common pathogenesis in need of improved therapy. Br J Haematol. 2018;180(6):919–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brandimarte L, Pierini V, Di Giacomo D, Borga C, Nozza F, Gorello P, et al. New MLLT10 gene recombinations in pediatric T-acute lymphoblastic leukemia. Blood. 2013;121(25):5064–7. [DOI] [PubMed] [Google Scholar]
- 26.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103(7):2257–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ovcharenko D, Stolzel F, Poitz D, Fierro F, Schaich M, Neubauer A, et al. miR-10a overexpression is associated with NPM1 mutations and MDM4 downregulation in intermediate-risk acute myeloid leukemia. Exp Hematol. 2011;39(10):1030–42 e7. [DOI] [PubMed] [Google Scholar]
- 28.Huang JW, Wang Y, Dhillon KK, Calses P, Villegas E, Mitchell PS, et al. Systematic screen identifies miRNAs that target RAD51 and RAD51D to enhance chemosensitivity. Mol Cancer Res. 2013;11(12):1564–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu X, Liao W, Peng H, Luo X, Luo Z, Jiang H, et al. miR-181a promotes G1/S transition and cell proliferation in pediatric acute myeloid leukemia by targeting ATM. J Cancer Res Clin Oncol. 2016;142(1):77–87. [DOI] [PubMed] [Google Scholar]
- 30.Gabra MM, Salmena L. microRNAs and Acute Myeloid Leukemia Chemoresistance: A Mechanistic Overview. Front Oncol. 2017;7:255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kunzel U, Grieve AG, Meng Y, Sieber B, Cowley SA, Freeman M. FRMD8 promotes inflammatory and growth factor signalling by stabilising the iRhom/ADAM17 sheddase complex. Elife. 2018;7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kategaya LS, Changkakoty B, Biechele T, Conrad WH, Kaykas A, Dasgupta R, et al. Bili inhibits Wnt/beta-catenin signaling by regulating the recruitment of axin to LRP6. PLoS One. 2009;4(7):e6129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bou Samra E, Klein B, Commes T, Moreaux J. Development of gene expression-based risk score in cytogenetically normal acute myeloid leukemia patients. Oncotarget. 2012;3(8):824–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Abu-Duhier FM, Goodeve AC, Wilson GA, Gari MA, Peake IR, Rees DC, et al. FLT3 internal tandem duplication mutations in adult acute myeloid leukaemia define a high-risk group. Br J Haematol. 2000;111(1):190–5. [DOI] [PubMed] [Google Scholar]
- 35.Meshinchi S, Woods WG, Stirewalt DL, Sweetser DA, Buckley JD, Tjoa TK, et al. Prevalence and prognostic significance of Flt3 internal tandem duplication in pediatric acute myeloid leukemia. Blood. 2001;97(1):89–94. [DOI] [PubMed] [Google Scholar]
- 36.Thiede C, Steudel C, Mohr B, Schaich M, Schakel U, Platzbecker U, et al. Analysis of FLT3-activating mutations in 979 patients with acute myelogenous leukemia: association with FAB subtypes and identification of subgroups with poor prognosis. Blood. 2002;99(12):4326–35. [DOI] [PubMed] [Google Scholar]
- 37.Schlenk RF, Kayser S, Bullinger L, Kobbe G, Casper J, Ringhoffer M, et al. Differential impact of allelic ratio and insertion site in FLT3-ITD-positive AML with respect to allogeneic transplantation. Blood. 2014;124(23):3441–9. [DOI] [PubMed] [Google Scholar]
- 38.Garg M, Nagata Y, Kanojia D, Mayakonda A, Yoshida K, Haridas Keloth S, et al. Profiling of somatic mutations in acute myeloid leukemia with FLT3-ITD at diagnosis and relapse. Blood. 2015;126(22):2491–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Visani G, Ferrara F, Di Raimondo F, Loscocco F, Fuligni F, Paolini S, et al. Low-dose lenalidomide plus cytarabine in very elderly, unfit acute myeloid leukemia patients: Final result of a phase II study. Leuk Res. 2017;62:77–83. [DOI] [PubMed] [Google Scholar]
- 40.Yorikawa C, Shibata H, Waguri S, Hatta K, Horii M, Katoh K, et al. Human CHMP6, a myristoylated ESCRT-III protein, interacts directly with an ESCRT-II component EAP20 and regulates endosomal cargo sorting. Biochem J. 2005;387(Pt 1):17–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kumar S, Vo AD, Qin F, Li H. Comparative assessment of methods for the fusion transcripts detection from RNA-Seq data. Sci Rep. 2016;6:21597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang F, Travins J, Delabarre B, Penard-Lacronique V, Schalm S, et al. Targeted inhibition of mutant IDH2 in leukemia cells induces cellular differentiation. Science. 2013;340(6132):622–6. [DOI] [PubMed] [Google Scholar]
- 43.Lewis PW, Muller M, Koletsky MS, Cordero F, Lin S, et al. Inhibition of PRC2 activity by gain-of-function H3 mutation found in pediatric glioblastoma. Science. 2013;340(6134):857–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lehnertz B, Zhang Y, Boivin I, Mayotte N, Tomellini E, et al. H3K27M mutations promote context-dependent transformation in acute myeloid leukemia with RUNX1 alterations. Blood. 2017;130(20):2204–14. [DOI] [PubMed] [Google Scholar]
- 45.Kernytsky A, Wang F, Hansen E, Schalm S, Straley K, et al. IDH2 mutation-induced histone and DNA hypermethylation is progressively reversed by small-molecule inhibition. Blood. 2015;125(2):296–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vakoc CR, Kentsis A. Disabling an oncogenic transcription factor by targeting of control kinases. Oncotarget. 2018;9(64):32276–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lavallee P, Gendron P, Lemieux S, D’Angelo G, Hebert J, and Sauvageau G. EVI1-rearranged acute myeloid leukemias are characterized by distinct molecular alterations. Blood. 2015;125(1): 140–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li H, Rao Q, Yu P, Chen S, Li Z, et al. Expression of MPL in Leukemia Stem Cells and Its Role in Stemness Maintenance. Blood. 2016;128(22), 172327330000 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.