Abstract
Background
Multiple genetic and environmental factors contribute to the individual‐level heterogeneity in stroke. This study aimed to assess how the genetic interactions confer risk of stroke.
Methods
In a Chinese case‐control study including 1,405 strokes and 1,263 controls who were followed up (range, 0.1–6.0 years), eight genes, including apolipoprotein(a) (APOA1), methylenetetrahydrofolate reductase (MTHFR), vitamin K epoxide reductase complex subunit 1 (VKORC1), arachidonate 5‐lipoxygenase‐activating protein (ALOX5AP), NOTCH3, chromosome 9p21.3(Chr.9p21.3), vascular endothelial growth factor (VEGFA), and kinase insert domain‐containing receptor (KDR), were analyzed for interactions by the generalized multifactor dimensionality reduction method and validated by the multivariate logistic regression models. The genetic associations with carotid artery intima‐media thickness (IMT) were examined.
Results
The interaction of VKORC1 and Chr.9p21.3 was identified for stroke and its worse prognosis, and subjects having the VKORC1 rs2359612C and Chr.9p21.3 rs10757274G alleles had higher risks for stroke (OR = 1.83, 95% CI = 1.32–2.52) as well as for stroke recurrence (HR = 1.84, 95% CI = 1.24–2.73), cardiovascular events (HR = 1.65, 95% CI = 1.15–2.38), and cardiovascular mortality (HR = 2.16, 95% CI = 1.24–3.79). Supporting, they were associated with higher IMT. Hypertension or physical inactivity increased the risk effect. The interaction of VEGFA rs833061C and KDR rs2305948T was identified for hemorrhagic stroke.
Conclusions
Our findings identified two novel genetic interactions of VKORC1 and Chr.9p21.3 and of VEGFA and KDR for risk of stroke and subtypes as well as future stroke prognosis.
Keywords: case‐control study, gene–gene interaction, genetics, risk factors, stroke
1. INTRODUCTION
Stroke is a major cause of death and disability worldwide and in China, and survivors are usually at high risk of recurrence and poststroke death (Wang, Hu, Sang, Luo, & Yu, 2017). Although traditional risk factors potentially explain most of stroke risk, it is generally accepted that multiple genetic variants contribute to the individual‐level heterogeneity in stroke and clinical phenotypes (Markus, 2011). Recent evidence has also shown that among individuals at high genetic risk of coronary diseases, adherence to a healthy lifestyle would markedly reduce the relative risk of cardiovascular events and the prevalent subclinical burden of atherosclerosis (Khera et al., 2016). Due to the complex nature of stroke, single‐locus method may not be appropriate to study common complex disorders.
Traditionally, the logistic regression model is most often used to explore the risk effects of multiple genetic variants in complex disease, but it has limited power to reveal the interactions between genes only with weak effects (Culverhouse, Suarez, Lin, & Reich, 2002; Hoh & Ott, 2003). Here, the generalized multifactor dimensionality reduction (GMDR) method (Lou et al., 2007) was used in this study, which can not only explore the complex interaction between genetic and environmental factors by collapsing high‐dimensional interactions of genetic data into a single dimension, but also avoid the biased results associated with disease risk by adjusting for the confounding covariates.
In this study, we included a total of 16 variants in eight susceptibility genes which have been assessed in our previous studies to be associated with stroke risk, including apolipoprotein(a) (APOA1, OMIM 107680; Sun et al., 2003), methylenetetrahydrofolate reductase (MTHFR, OMIM 607093; Li et al., 2003), vitamin K epoxide reductase complex subunit 1 (VKORC1, OMIM 608547; (Wang et al., 2006), arachidonate 5‐lipoxygenase‐activating protein (ALOX5AP, OMIM 603700; Zhang, Yang, Shi, Sun, & Hui, 2006), NOTCH3 (OMIM 600276), vascular endothelial growth factor (VEGFA, OMIM 192240), and kinase insert domain‐containing receptor (KDR, also called VEGF receptor‐2, OMIM 191306; Zhang et al., 2009), and antisense noncoding RNA in the INK4 locus (ANRIL, OMIM 613149) on chromosome 9p21.3 (Chr.9p21.3; Zhang et al., 2012). These risk genes play critical roles in inflammation, lipid metabolism, endothelial dysfunction, and abnormal vascular remodeling during the development of atherosclerosis. Therefore, in a large case‐control study including 2000 stroke patients and 2000 age‐, gender‐, and demographic‐matched community controls in China, we investigated the risk model of genetic interactions for stroke and subtypes by the GMDR method and then validated these models by the traditional logistic regression model. We further assessed the relationship between genetic interactions and stroke recurrence among patients who have been prospectively followed up for a median of 4.5 years.
In addition, much evidence has shown that carotid artery intima‐media thickness (IMT) is a useful measure of subclinical atherosclerosis and predicts the incidence of cardiovascular diseases (CVD) and stroke (Chambless et al., 2000; Polak, Pencina, O'Leary, & D'Agostino, 2011). In a meta‐analysis with data from 37,197 subjects, Lorenz et al showed that the difference of 0.1 mm in carotid IMT is associated with increased risk of stroke by 13%–18% (Lorenz, Markus, Bots, Rosvall, & Sitzer, 2007). Thus, we examined the genetic association with IMT in 1,123 subjects who were randomly selected from a community‐based population without a history of stroke (Zhang et al., 2009), so as to explore the possible underlying mechanisms.
Since stroke is a multifactorial disorder, a comprehensive prevention program has been advocated such as hypertension control and lifestyle modification, including not smoking, avoiding obesity, regular physical activity, and a healthy diet pattern (Lloyd‐Jones et al., 2010). However, the extent to which genetic risk can be affected by environmental factors remains undetermined. Therefore, in this study, we also analyzed whether environmental factors (including hypertension, smoking, drinking, overweight/obesity, and physical inactivity) could affect the risk of stroke among people at high genetic risk.
2. MATERIALS AND METHODS
2.1. Ethical compliance
This study was approved by the institutional ethics committees of Fu Wai Hospital and the collaborating hospitals, and all participants signed the informed consent.
2.2. Study population
The Multicenter Chinese Stroke Study is a large‐scale case‐control study that has been reported in details in our previous studies, including the definition of diagnosis, criteria of inclusion and exclusion, recruitment of case and control subjects, collection of clinical characteristics, and measurement of plasma biochemical variables (Li et al., 2003; Sun et al., 2003; Wang et al., 2006; Zhang et al., 2009; Zhang et al., 2012). Initially, a total of 2000 consecutive stroke cases (aged 35–74 years) were recruited from seven clinic centers in China from November 2000 to November 2001, and 2000 age‐, gender‐, and resident area‐matched healthy controls were recruited at the same time and at the same demographic area from the local community‐based inhabitants. All cases with first‐onset stroke were diagnosed by neurologists on the basis of clinical symptoms, and brain imaging examination such as computed tomography (CT) or magnetic resonance imaging ([MRI]; Adams et al., 1993). Three subtypes were recruited: cerebral thrombosis (atherothrombotic stroke), intracerebral hemorrhage, and lacunar infarction. The baseline characteristics of participants were collected by direct patient interviews using a structured questionnaire, including demographic factors, medical history, and lifestyle behaviors, and family history.
In this study, 595 cases and 737 control subjects were excluded because of no plasma data, insufficient DNA quantity, poor DNA quality, or incomplete genotyping data, and finally, 1,405 cases (607 diagnosed as atherothrombotic stroke, 415 as lacunar infarction, and 383 as hemorrhagic stroke) and 1,263 controls were included for analyzing the effects of gene–gene and gene–environment interactions on risk of stroke. No significant differences in clinical characteristics were observed between those excluded and included (Table S1).
2.3. Follow‐up and outcome assessment
Stroke cases recruited in this study were prospectively followed up for a median of 4.5 years (from January 2001 to May 2006) by physician investigators via face‐to‐face interview and/or telephone contact with a standard questionnaire as described in our previous studies (Zhang et al., 2009; Zhang et al., 2012). The end‐points included recurrent events of stroke, all‐cause mortality, and mortality due to CVD. The recurrent stroke events were independently confirmed by the Clinical Endpoint Committee based on the patient medical records and brain imaging. Information on the cause of death was obtained from death certificates and/or medical records when possible, or from family members or care givers.
2.4. Selection and genotyping for genetic risk variants
Genomic DNA was extracted from peripheral blood leukocytes of the participants using FlexGen Blood DNA Kit according to a standard procedure (Beijing ComWin Biotech Co., Ltd.). In this study, we examined 16 functional variants in eight susceptible genes which have been investigated to be associated with stroke risk in previous studies, and information on the features of variants and genotyping methods is shown in the Table S2. Briefly, we included a pentanucleotide TTTTA repeat (PNTR) variant in the APOA1 (NM_000039.1) gene that correlates inversely with plasma Lipoprotein (a) [Lp(a)] level (Jurgens et al., 1995; Sun et al., 2003). The PNTR variation was determined by the method of polymerase chain reaction (PCR)–nondenaturing polyacrylamide gels and DNA sequencing analysis. Single‐nucleotide polymorphisms (SNPs) were the 677C>T (rs1801133) in MTHFR (NM_005957.4) gene that affects the homocysteine metabolism (Li et al., 2003; Zhao et al., 2017), the 2255T>C (rs2359612) in VKORC1 (NM_024006.5) gene that affects the formation of thrombosis and vascular calcification (Wang et al., 2013, 2006), SG13S114T>A (rs10507391) and SG13S89G>A (rs4769874) in the ALOX5AP gene (NM_001629.3) that regulates the biosynthesis of proinflammatory leukotrienes (Helgadottir et al., 2004; Zhang et al., 2006), the 684G>A (rs1043994) in the NOTCH3 (NM_000435.2) gene that affects vascular development (Ross et al., 2013), six functional SNPs in VEGFA (NM_001025366.2) and its receptor KDR (NM_002253.3) gene that affects the transcriptional activity and expression (Eichmann & Simons, 2012; Zhang et al., 2009). These SNPs were genotyped by the method of PCR and restriction fragment length polymorphism (RFLP) analysis. In addition, four SNPs (rs2383206, rs2383207, rs10757278, and rs10757274) at the Chr.9p21.3 loci that correlate with the ANRIL (NM_078487.2) expression and atherosclerosis development (Johnson et al., 2013; Smith et al., 2009) were included and genotyped by a ligase detection reaction method (Zhang et al., 2012).
2.5. Measurements of carotid IMT
The association between genetic interactions and IMT was tested in 1,123 subjects aged 45–83 years (871 men and 252 women) who were randomly selected from a community‐based population without a history of stroke (Zhang et al., 2009). The carotid ultrasonography was performed and images were analyzed according to a standardized protocol by a trained physician. The ultrasonography scanning included the right and left common carotid artery (CCA) and internal carotid artery (ICA) at the following locations: proximal CCA (15–30 mm proximal to the carotid bulb), distal CCA (<15 mm proximal to the carotid bulb), and proximal ICA (carotid bulb, identified by loss of the parallel wall present in the CCA and the initial 10 mm of the vessel above the flow divider between external and internal carotid arteries) (Kiechl & Willeit, 1999; Zhang et al., 2009). In the analysis, the mean maximum IMT of the left and right arteries was used. The details of IMT measurement were shown in Supplementary materials.
2.6. Statistical analysis
The Hardy–Weinberg equilibrium (HWE) of variants was tested for the genotypes distribution in control subjects to assess possible selection bias and genotyping errors. Clinical features between cases and control subjects were compared by Student's t test for quantitative variables and Chi‐squared test for categorical variables. In the GMDR model, the genotypes of each studied variant were classified into wild‐type genotype (with 0 risk allele), heterozygous genotype (with 1 risk allele), and high‐risk genotype (with 2 risk alleles). Briefly, the GMDR calculates the maximum likelihood estimates and the cumulative score values for all individuals, and if the average score is equal to or higher than a preassigned threshold of 0, the identified genotype combination would be determined as high‐risk model; on the other hand, if the score is lower than 0, it would be determined as low‐risk model (Figure S1). The score‐based GMDR method uses the data‐reduction strategy to reduce the dimensionality from multidimensional to one‐dimensional, and then identify the specific combinations of genetic factors that show the strongest association with phenotype from all potential combinations. The test accuracy and cross‐validation consistency reach their maxima when the appropriate multi‐locus models are obtained (Hou et al., 2019; Lou et al., 2007). The algorithm of GMDR was shown in Supplemental methods.
Next, for genetic interaction models identified using the GMDR method, we further assessed their associations with the risk of stroke and its subtypes using the multivariate logistic regression models, so as to ensure not missing some other potentially interesting and important interactions, and odds ratio (OR) with 95% confidence intervals (CIs) were obtained after adjustment for age, gender, and vascular risk factors mentioned above. Multiple comparisons were corrected by false discovery rate at the 0.05 level which was achieved by the Benjamini–Hochberg procedure (Benjamini & Hochberg, 2000).
For the gene–environment analysis, persons at low genetic risk with favorable environmental factors served as the reference group, and ORs (95% CI) for the risk of stroke and its subtypes were calculated by the multivariate logistic regression models. The significances of multiplicative interaction between genetic risk model and hypertension as well as each unhealthy lifestyle factor (including smoking, drinking, overweight/obesity, and physical inactivity) were determined by the likelihood ratio test. Hypertension was defined as systolic BP ≥140 mm Hg and/or diastolic BP ≥90 mm Hg, and/or receiving antihypertensive medications, and/or history of hypertension (Liu, 2011). Smoking and drinking were classified as “never,” “former,” or “current”. Physical activity was evaluated from responses to questions and classified as “regular exercise (at least once weekly, more than 30 min)” or “inactive (exercise < once/week)”. BMI was classified as normal‐weight (18.5–24 kg/m2), overweight (24–28 kg/m2), or obesity (≥28 kg/m2; He et al., 2014).
The Cox proportional model was used to assess the hazards ratio (HR) for relative risks of recurrent stroke events and poststroke mortality associated with recommended genetic model from GMDR analysis. The association between genetic model and carotid IMT (dependent variable) was analyzed by multiple linear regression models. Differences in carotid IMT across the genotype groups were examined by one‐way analysis of variance. Analyses were conducted with spss software (version 20.0, SPSS Inc). All reported P values are two‐sided, and p ≤ .05 was considered statistically significant.
3. RESULTS
3.1. Clinical characteristics of participants
The present study included 1,405 stroke cases and 1,263 controls, and clinical features are shown in Table 1. As expected, stroke patients had a higher prevalence of comorbidities including hypertension, diabetes, coronary heart disease, and family history of stroke. These patients were further prospectively followed up for a median of 4.5 years (range, 0.1–6.0 years) to assess the prognosis of stroke. Among them, 72 (5.1%) were lost due to emigration, but no substantial differences were found in baseline characteristics between the follow‐up and lost‐to‐follow‐up subjects (Table S3). During the follow‐up, 280 recurrent stroke events, 325 cardiovascular disease events, and 261 deaths from all causes (146 from stroke or CHD and 115 from other causes) were recorded.
Table 1.
Clinical characteristics of stroke cases and control subjects in the present study
| Characteristics | Control subjects (n = 1,263) | Stroke patients | |||
|---|---|---|---|---|---|
| Total cases (n = 1,405) | Hemorrhagic stroke (n = 383) | Atherothrombotic stroke (n = 607) | Lacunar infarction (n = 415) | ||
| Age, years | 60.0 ± 8.0 | 60.5 ± 9.2 | 58.3 ± 9.6** | 61.4 ± 9.1** | 61.1 ± 8.5* |
| Men, n (%) | 770 (61.0%) | 902 (64.2%) | 250 (65.3%) | 389 (64.1%) | 263 (63.4%) |
| BMI (kg/m2) | 24.16 ± 3.28 | 24.25 ± 3.45 | 24.01 ± 3.56 | 24.21 ± 3.57 | 24.53 ± 3.13* |
| Systolic BP (mmHg) | 129 ± 17 | 147 ± 22** | 152 ± 23** | 147 ± 23** | 143 ± 20** |
| Diastolic BP (mmHg) | 79 ± 10 | 88 ± 13** | 92 ± 13** | 86 ± 13** | 86 ± 11** |
| Fasting glucose (mmol/L) | 5.88 ± 1.72 | 6.61 ± 2.62** | 6.57 ± 2.40** | 6.77 ± 2.80** | 6.42 ± 2.55** |
| Lipids (mmol/L) | |||||
| Total cholesterol | 5.00 ± 0.99 | 4.74 ± 1.01** | 4.52 ± 0.99** | 4.84 ± 1.00** | 4.80 ± 1.00** |
| Triglycerides | 1.49 (1.05–2.18) | 1.66 (1.19–2.41)** | 1.50 (1.12–2.02) | 1.71 (1.21–2.50)** | 1.75 (1.23–2.52)** |
| HDL‐cholesterol | 1.06 ± 0.31 | 0.90 ± 0.28** | 0.89 ± 0.33** | 0.90 ± 0.26** | 0.92 ± 0.26** |
| LDL‐cholesterol | 3.10 ± 0.91 | 2.96 ± 0.92** | 2.86 ± 0.90** | 3.03 ± 0.92 | 2.94 ± 0.93** |
| Current smoking, n (%) | 337 (26.7%) | 381 (27.1%) | 104 (27.2%) | 177 (29.2%) | 100 (24.1%) |
| Current alcohol intake, n (%) | 318 (25.2%) | 306 (21.8%)* | 100 (26.1%) | 126 (20.8%)* | 80 (19.3%)* |
| Exercise, n (%) | 872 (69.0%) | 646 (46.0%)** | 135 (35.2%)** | 267 (44.0%)** | 244 (58.8%)** |
| History of hypertension, n (%) | 342 (27.1%) | 868 (61.8%)** | 241 (62.9%)** | 376 (61.9%)** | 251 (60.5%)** |
| History of diabetes, n (%) | 136 (10.8%) | 334 (23.8%)** | 85 (22.2%)** | 158 (26.0%)** | 91 (21.9%)** |
| History of CHD, n (%) | 147 (11.6%) | 206 (14.7%)* | 32 (8.4%) | 121 (19.9%)** | 53 (12.8%) |
| Family history of stroke, n (%) | 298 (23.6%) | 451 (32.1%)** | 124 (32.4%)** | 196 (32.3%)** | 131 (31.6%)** |
Values are mean ± SD, number (percentage), or median (interquartile range), versus control subjects.
Abbreviations: BMI, Body mass index; BP, blood pressure; CHD, coronary heart disease; HDL, high‐density lipoprotein; LDL, low‐density lipoprotein.
p < 0.05.
p < 0.01.
3.2. GMDR analysis of the gene–gene interactions associated with stroke risk
The genotype frequencies of all 16 studied variants in cases and controls are shown in Table 2 and they did not deviate from the HWE (p > .05). The high‐order interactions were explored for thrombotic stroke, lacunar infarction, hemorrhagic stroke, and overall stroke by the GMDR method. After adjustment for age, gender, and vascular risk factors, the best gene–gene model for overall stroke risk was found to be the interaction between VKORC1 rs2359612T>C and Chr.9p21.3 rs10757274A>G, which scored 7/10 for cross‐validation consistency and 10 for sign test (p = .001; Table 3). In addition, one‐locus model of VKORC1 rs2359612T>C was also identified for stroke risk, with scores of 9/10 for cross‐validation consistency and 9 for sign test (p = .01; Table 3). It was reported in our previous study that the presence of the rs2359612C allele of VKORC1 conferred almost twice the risk of vascular disease (OR 1.95, 95% CI 1.58–2.41, p < .001 for stroke; OR 1.72, 95% CI 1.24–2.38, p < .01 for coronary heart disease; and OR 1.90, 95% CI 1.04–3.48, p < .05 for aortic dissection; Wang et al., 2006).
Table 2.
The genotypes frequency of all 16 studied variants in stroke cases and control subjects
| Gene and variants | Controls (n = 1,263) | Stroke cases | ||||
|---|---|---|---|---|---|---|
| Total cases (n = 1,405) | Hemorrhagic stroke (n = 383) | Atherothrombotic stroke (n = 607) | Lacunar infarction (n = 415) | |||
| APOA1 PNTR | ||||||
| LRN | 125 (9.9%) | 1709 (12.1%) | 48 (12.5%) | 73 (12.0%) | 49 (11.8%) | |
| HRN | 1,138 (90.1%) | 1,235 (87.9%) | 335 (87.5%) | 534 (88.0%) | 366 (88.2%) | |
| MTHFR rs1801133 | ||||||
| CC | 410 (32.5%) | 412 (29.3%) | 116 (30.3%) | 173 (28.5%) | 123 (29.6%) | |
| CT | 585 (46.3%) | 664 (47.3%) | 182 (47.5%) | 287 (47.3%) | 195 (47.0%) | |
| TT | 268 (21.2%) | 329 (23.4%) | 85 (22.2%) | 147 (24.2%) | 97 (23.4%) | |
| VKORC1 rs2359612 | ||||||
| TT | 1,099 (87.0%) | 1,129 (80.4%) | 314 (82.0%) | 496 (81.7%) | 319 (76.9%) | |
| TC | 156 (12.4%) | 259 (18.4%) | 62 (16.2%) | 107 (17.6%) | 90 (21.7%) | |
| CC | 8 (0.6%) | 17 (1.2%) | 7 (1.8%) | 4 (0.7%) | 6 (1.4%) | |
| ALOX5AP rs4769874 | ||||||
| GG | 1,162 (92.0%) | 1,277 (90.9%) | 350 (91.4%) | 551 (90.8%) | 376 (90.6%) | |
| GA | 96 (7.6%) | 122 (8.7%) | 32 (8.4%) | 52 (8.6%) | 38 (9.2%) | |
| AA | 5 (0.4%) | 6 (0.4%) | 1 (0.3%) | 4 (0.7%) | 1 (0.2%) | |
| ALOX5AP rs10507391 | ||||||
| TT | 635 (50.3%) | 703 (50.0%) | 205 (53.5%) | 283 (46.6%) | 215 (51.8%) | |
| TA | 505 (40.0%) | 568 (40.4%) | 145 (37.9%) | 257 (42.3%) | 166 (40.0%) | |
| AA | 123 (9.7%) | 134 (9.5%) | 33 (8.6%) | 67 (11.0%) | 34 (8.2%) | |
| NOTCH3 rs1043994 | ||||||
| GG | 1,027 (81.3%) | 1,169 (83.2%) | 317 (82.8%) | 502 (82.7%) | 350 (84.3%) | |
| GA | 218 (17.3%) | 211 (15.0%) | 62 (16.2%) | 94 (15.5%) | 55 (13.3%) | |
| AA | 18 (1.4%) | 25 (1.8%) | 4 (1.0%) | 11 (1.8%) | 10 (2.4%) | |
| VEGFA rs2010963 | ||||||
| GG | 386 (30.6%) | 436 (31.0%) | 135 (35.2%) | 190 (31.3%) | 111 (26.7%) | |
| GC | 619 (49.0%) | 684 (48.7%) | 179 (46.7%) | 288 (47.4%) | 217 (52.3%) | |
| CC | 258 (20.4%) | 285 (20.3%) | 69 (18.0%) | 129 (21.3%) | 87 (21.0%) | |
| VEGFA rs1570360 | ||||||
| GG | 357 (28.3%) | 401 (28.5%) | 123 (32.1%) | 166 (27.3%) | 112 (27.0%) | |
| GA | 628 (49.7%) | 709 (50.5%) | 184 (48.0%) | 300 (49.4%) | 225 (54.2%) | |
| AA | 278 (22.0%) | 295 (21.0%) | 76 (19.8%) | 141 (23.2%) | 78 (18.8%) | |
| VEGFA rs833061 | ||||||
| TT | 561 (44.4%) | 616 (43.8%) | 156 (40.7%) | 266 (43.8%) | 194 (46.7%) | |
| TC | 566 (44.8%) | 628 (44.7%) | 176 (46.0%) | 273 (45.0%) | 179 (43.1%) | |
| CC | 136 (10.8%) | 161 (11.5%) | 51 (13.3%) | 68 (11.2%) | 42 (10.1%) | |
| KDR rs2071559 | ||||||
| TT | 624 (49.4%) | 713 (50.7%) | 168 (43.9%) | 342 (56.3%) | 203 (48.9%) | |
| TC | 505 (40.0%) | 567 (40.4%) | 175 (45.7%) | 219 (36.1%) | 173 (41.7%) | |
| CC | 134 (10.6%) | 125 (8.9%) | 40 (10.4%) | 46 (7.6%) | 39 (9.4%) | |
| KDR rs1870377 | ||||||
| AA | 473 (37.5%) | 519 (36.9%) | 115 (30.0%) | 249 (41.0%) | 155 (37.3%) | |
| AT | 573 (45.4%) | 632 (45.0%) | 192 (50.1%) | 254 (41.8%) | 186 (44.8%) | |
| TT | 217 (17.2%) | 254 (18.1%) | 76 (19.8%) | 104 (17.1%) | 74 (17.8%) | |
| Chr.9p21.3 rs2383206 | ||||||
| AA | 390 (30.9%) | 417 (29.7%) | 106 (27.7%) | 170 (28.0%) | 141 (34.0%) | |
| AG | 631 (50.0%) | 674 (48.0%) | 187 (48.8%) | 289 (47.6%) | 198 (47.7%) | |
| GG | 242 (19.2%) | 314 (22.3%) | 90 (23.5%) | 148 (24.4%) | 76 (18.3%) | |
| Chr.9p21.3 rs2383207 | ||||||
| AA | 171 (13.5%) | 192 (13.7%) | 59 (15.4%) | 72 (11.9%) | 61 (14.7%) | |
| AG | 591 (46.8%) | 629 (44.8%) | 155 (40.5%) | 280 (46.1%) | 194 (46.7%) | |
| GG | 501 (39.7%) | 584 (41.6%) | 169 (44.1%) | 255 (42.0%) | 160 (38.6%) | |
| Chr.9p21.3 rs10757278 | ||||||
| AA | 303 (24.0%) | 312 (22.2%) | 75 (19.6%) | 132 (21.7%) | 105 (25.3%) | |
| AG | 633 (50.1%) | 658 (46.8%) | 184 (48.0%) | 274 (45.1%) | 200 (48.2%) | |
| GG | 327 (25.9%) | 435 (31.0%) | 124 (32.4%) | 201 (33.1%) | 110 (26.5%) | |
| Chr.9p21.3 rs10757274 | ||||||
| AA | 364 (28.8%) | 378 (26.8%) | 89 (23.2%) | 158 (26.0%) | 131 (31.6%) | |
| AG | 642 (50.8%) | 667 (47.5%) | 186 (48.6%) | 284 (46.8%) | 197 (47.5%) | |
| GG | 257 (20.3%) | 360 (25.6%) | 108 (28.2%) | 165 (27.2%) | 87 (21.0%) | |
Abbreviations: ALOX5AP, arachidonate 5‐lipoxygenase‐activating protein; APOA1, apolipoprotein (a); Chr.9p21.3, Chromosome 9p21.3; HRN, high repeat number (sum of both alleles ≥16); KDR, kinase insert domain‐containing receptor; LRN, low repeat number (sum of both alleles <16); MTHFR, methylenetetrahydrofolate reductase; PNTR, a pentanucleotide TTTTA repeat; VEGFA, vascular endothelial growth factor; VKORC1, vitamin K epoxide reductase complex subunit 1.
Table 3.
The multi‐loci interactions with stroke risk identified by GMDR analysis
| No. of loci | GMDR model | Balanced Accuracy | Sign test (p)* | Cross‐validation consistency | |
|---|---|---|---|---|---|
| Training set | Testing set | ||||
| 1 | 9 | 0.5364 | 0.5266 | 9 (.0107) | 9/10 |
| 2 | 9, 16 | 0.5476 | 0.5328 | 10 (.0010) | 7/10 |
| 3 | 3, 5, 16 | 0.5587 | 0.5021 | 5 (.6230) | 3/10 |
| 4 | 2, 8, 10, 16 | 0.5869 | 0.5112 | 7 (.1719) | 5/10 |
| 5 | 2, 3, 4, 10, 15 | 0.6311 | 0.5054 | 7 (.1719) | 7/10 |
| 6 | 2, 3, 4, 5, 8, 15 | 0.6967 | 0.4793 | 2 (.9893) | 3/10 |
| 7 | 2, 3, 4, 6, 8, 10, 15 | 0.7814 | 0.4940 | 2 (.9893) | 8/10 |
| 8 | 2, 3, 4, 5, 6, 8, 10, 15 | 0.8598 | 0.4808 | 2 (.9893) | 9/10 |
| 9 | 2, 3, 4, 5, 6, 8, 10, 13, 14 | 0.9034 | 0.4658 | 1 (.9990) | 5/10 |
| 10 | 2, 3, 4, 5, 6, 7, 8, 10, 13, 14 | 0.9343 | 0.4639 | 2 (.9893) | 8/10 |
| 11 | 2, 3, 4, 5, 6, 7, 8, 10, 12, 13, 14 | 0.9547 | 0.5043 | 7 (.1719) | 10/10 |
| 12 | 2, 3, 4, 5, 6, 7, 8, 9, 10, 12, 13, 14 | 0.9661 | 0.4751 | 4 (.8281) | 8/10 |
| 13 | 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 12, 14, 16 | 0.9743 | 0.5423 | 8 (.0547) | 8/10 |
| 14 | 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 12, 14, 15, 16 | 0.9796 | 0.5047 | 6 (.3770) | 7/10 |
| 15 | 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 14, 15, 16 | 0.9817 | 0.5044 | 6 (.3770) | 7/10 |
| 16 | 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16 | 0.9830 | 0.5081 | 7 (.1719) | 10/10 |
Numbers 1–16 indicate the following studied variations: 1 = PNTR of ApoA1, 2 = MTHFR 677C>T, 3 = VEGFA rs2010963, 4 = VEGFA rs1570360, 5 = VEGFA rs833061, 6 = KDR rs2071559, 7 = KDR rs2305948, 8 = KDR rs1870377, 9 = VKORC1 rs2359612, 10 = ALOX5AP rs10507391, 11 = ALOX5AP rs4769874, 12 = NOTCH3 rs1043994, 13 = Chr.9p21.3 rs2383206, 14 = Chr.9p21.3 rs2383207, 15 = Chr.9p21.3 rs10757278, 16 = Chr.9p21.3 rs10757274.
p value was obtained from the GMDR analysis which adjusted for age, gender, BMI, fasting glucose, triglycerides, total cholesterol, HDL‐cholesterol, LDL‐cholesterol, family history of stroke, systolic and diastolic blood pressure, smoking, and drinking status.
In the hemorrhagic stroke model of GMDR, the best gene–gene interaction was found between VEGFA rs833061T>C and KDR rs2305948C>T, which scored 6/10 for cross‐validation consistency and 10 for sign test (p = .001; Table S4). The one‐locus model of KDR rs2305948 was also identified for hemorrhagic stroke, with score of 10 for cross‐validation consistency and sign test (p = .001; Table S4). It was reported in our previous study that subjects carrying the rs2305948T allele of KDR had remarkably higher risks for intracerebral hemorrhage (OR 2.20, 95% CI 1.70–2.84, p = 1.5 × 10−9) and for stroke recurrence (relative risk 1.40, 95% CI 1.12–1.75, p < .003; Zhang et al., 2009). Significant high‐order interactions were not detected for atherothrombotic stroke or lacunar infarction (Tables S5 and S6) by the GMDR analysis.
Since our main purpose was to assess the genetic interactions for stroke risk, the best two‐locus model was mainly analyzed. The associations of the two identified gene–gene interactions with stroke and its subtypes were further validated by the traditional logistic regression model after adjustment for vascular risk factors and correction for multiple comparisons.
For the joint effect of the two genes VKORC1 and Chr.9p21.3, the score distributions by GMDR analysis showed that the genotype combinations of VKORC1 rs2359612TC/CC and Chr.9p21.3 rs10757274AG/GG were high‐risk interactive variables (Figure S2), and thus, individuals with VKORC1 rs2359612TC/CC and Chr.9p21.3 rs10757274 AG/GG were grouped as "risk carriers" in the logistic regression models. The results showed that when compared to individuals with wild‐type genotypes of VKORC1 rs2359612TT and Chr.9p21.3 rs10757274AA, subjects carrying both VKORC1 rs2359612C and Chr.9p21.3 rs10757274G had a higher risk for overall stroke (ORs 1.83, 95% CI 1.32–2.52, p < .001, q < 0.016) and atherothrombotic stroke (ORs 1.78, 95% CI 1.20–2.65, p = .004, q = 0.02; Table 4). However, the two genes VKORC1 and Chr.9p21.3 had no significant joint effects on the risk of hemorrhagic stroke and lacunar stroke after correction for multiple comparisons (Table S7).
Table 4.
The joint effects of VKORC1 and Chr.9p21.3 on stroke risk validated by the logistic regression model
| VKORC1 | Chr.9p21.3 | Risk of overall stroke | q‐value# | Risk of atherothrombotic stroke | |||||
|---|---|---|---|---|---|---|---|---|---|
|
rs2359612 T>C |
rs10757274 A>G |
Controls/cases, n (%) | ORs (95% CI)* | p * | Controls/cases, n (%) | ORs (95% CI)* | p * | q‐value# | |
| TT | AA | 314 (24.9%)/312 (22.2%) | 1.00 | 314 (24.9%)/136 (22.4%) | 1.00 | ||||
| TT | AG | 555 (43.9%)/529 (37.7%) | 0.86 (0.68–1.08) | .20 | 0.32 | 555 (43.9%)/228 (37.6%) | 0.89 (0.66–1.19) | .41 | 0.47 |
| TT | GG | 230 (18.2%)/288 (20.5%) | 1.18 (0.90–1.56) | .23 | 0.33 | 230 (18.2%)/132 (21.7%) | 1.33 (0.95–1.86) | .10 | 0.23 |
| TC | AA | 49 (3.9%)/65 (4.6%) | 1.56 (0.98–2.48) | .06 | 0.16 | 49 (3.9%)/22 (3.6%) | 1.22 (0.67–2.23) | .52 | 0.52 |
| TC+CC | AG+GG | 114 (9.0%)/210 (14.9%) | 1.83 (1.32–2.52) | <.001 | <0.016 | 114 (9.0%)/89 (14.7%) | 1.78 (1.20–2.65) | .004 | 0.02 |
Abbreviations: Chr.9p21.3, chromosome 9p21.3; CI, confidential interval; ORs, odds ratio; VKORC1, vitamin K epoxide reductase complex subunit 1.
ORs (95% CI) and p values were obtained by the logistic regression analysis after adjustment for conventional risk factors as in Table 3.
q‐value (false discovery rate) was obtained with the use of Benjamini–Hochberg procedure for controlling the false positive rate in multiple comparisons.
For the interactions of VEGFA and KDR, the genotype combinations of VEGFA rs833061 TC/CC and KDR rs2305948CT/TT were considered to be high‐risk interactive variables in the score distributions by GMDR analysis (Figure S3). Thus, individuals with VEGFA rs833061TC/CC and KDR rs2305948CT/TT were grouped as "risk carriers" in further logistic regression analysis. When compared to individuals with wild‐type genotypes of VEGFA rs833061TT and KDR rs2305948CC, subjects carrying the risk alleles of both VEGFA rs833061C and KDR rs2305948T had a higher risk for hemorrhagic stroke (ORs 2.30, 95% CI 1.50–3.55, p < .001, q < 0.02; Table S8). However, the two genes, VEGFA and KDR, were not found to have significant joint effects on the risk of overall stroke, atherothrombotic stroke, or lacunar infarction (Table S9). These validations were consistent with the results from GMDR analysis which use the data‐reduction strategy to reduce the dimensionality from multidimensional to one‐dimensional.
3.3. Association of the significant gene–gene interaction model with carotid IMT
In the association analysis with carotid IMT, the combination of risk alleles of VKORC1 rs2359612C and Chr.9p21.3 rs10757274G was positively associated with the increased IMT (γ = .31, p = 2.0 × 10−4) after adjustment for age, gender, and vascular risk factors with multiple linear regression analysis. The IMT was markedly higher in the subjects carrying both VKORC1 rs2359612C and Chr.9p21.3 rs10757274G risk alleles when compared with those having the wild‐type genotypes of VKORC1 rs2359612TT and Chr.9p21.3 rs10757274AA (Figure S4). On the other hand, no association was found between the IMT and variants combination of VEGFA rs833061C and KDR rs2305948T which increased the risk of hemorrhagic stroke, supporting that stroke subtypes have significantly different pathogenesis and different genetic risk factor profiles.
3.4. Combined effects of environmental/lifestyle and genetic factors on stroke risk
Since stroke is a multifactorial disorder, the extent to which genetic risk can be affected by environmental factors remains undetermined. In this study, we further assessed whether environmental factors (including hypertension, smoking, drinking, physical inactivity, and overweight/obesity) could affect the occurrence of stroke among people at high genetic risks. After adjustment for conventional vascular risk factors and correction for multiple comparisons, we found that hypertension conferred significantly higher risk for stroke regardless of the genetic risk profiles; moreover, hypertensive subjects who carried the combined at‐risk alleles of VKORC1 rs2359612C and Chr.9p21.3 rs10757274G would have a remarkably increased risk for stroke (ORs 10.61, 95% CI 6.85–16.44, q < 0.002; p < .001; P interaction = 0.01), compared with those having wild‐type genotypes and without hypertension (Figure 1a). For those who lack regular exercise (less than once weekly), the combined at‐risk alleles of VKORC1 rs2359612C and Chr.9p21.3 rs10757274G added an additional risk to stroke, with ORs of 6.15 (95% CI 3.60–10.50, q < 0.002; p for trend < 0.001; P interaction = 0.01; Figure 1b). The statistical data are shown in Tables S10 and S11. We did not find that lifestyle factors such as smoking, drinking, and overweight/obesity had significant interactions with the two genes VKORC1 and Chr.9p21.3 (P interaction > 0.05), and the risk effects of VKORC1 rs2359612C and Chr.9p21.3 rs10757274G on stroke were independent of smoking, drinking, and overweight/obesity after adjustment for conventional risk factors and multiple comparisons (Tables S12 and S13).
Figure 1.
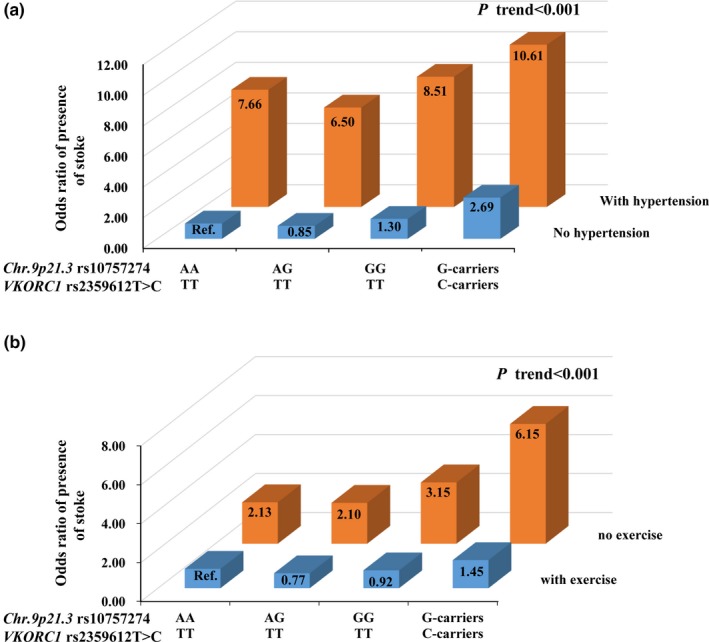
Jointed effects of the vitamin K epoxide reductase complex subunit 1 (VKORC1) and Chr.9p21.3 interactions with environmental factors on stroke risk. Abbreviations: Ref, reference group. The odds ratio (ORs) and p values were calculated by the logistic regression analysis after adjustment for age, gender, and conventional vascular risk factors, and correction for multiple comparisons. (a) Joint effects of the VKORC1 rs2359612 and Chr.9p21.3 rs10757274 with hypertension on stroke risk, and the reference group was defined as subjects without hypertension and carrying wild‐type genotypes of rs2359612TT and rs10757274AA; (b) Joint effects of the VKORC1 rs2359612 and Chr.9p21.3 rs10757274 with physical inactivity on stroke risk, and the reference group was defined as subjects with regular exercise and carrying wild‐type genotypes of rs2359612TT and rs10757274AA
4. ASSOCIATION OF HIGH‐RISK GENOTYPE COMBINATIONS WITH THE PROGNOSIS OF POSTSTROKE
During a median follow‐up period of 4.5 years for stroke patients, we found that the combined at‐risk alleles of VKORC1 rs2359612C and Chr.9p21.3 rs10757274G conferred almost twice the risk for cardiovascular and cerebrovascular diseases, including stroke recurrence (adjusted HR 1.84, 95% CI 1.24–2.73; p = .003), CVD events (adjusted HR 1.65, 95% CI 1.15–2.38; p = .007), and cardiovascular mortality (adjusted HR, 2.16, 95% CI 1.24–3.79; p = .007) in the Cox regression models after adjustment for age, gender, and conventional vascular risk factors (Table 5). Next, the Kaplan–Meier curves revealed a significantly higher cumulative hazard rate of stroke recurrence, CVD events, and cardiovascular mortality among patients carrying the combined risk alleles of VKORC1 rs2359612C and Chr.9p21.3 rs10757274G as compared with those having wild‐type genotypes of VKORC1 rs2359612TT and Chr.9p21.3 rs10757274AA (Figure 2).
Table 5.
Relative risk of cardiovascular outcomes by the VKORC1 and Chr.9p21.3 interactions during a longitudinal follow‐up of stroke patients
| VKORC1 rs2359612 | Chr.9p21.3 rs10757274 | Person‐years | Outcomes, n (%) | Adjusted Hazards ratio (95% CI)* | p value* |
|---|---|---|---|---|---|
| Stroke recurrence | |||||
| TT | AA | 1,293.48 | 49 (16.4%) | 1.00 | |
| TT | AG+GG | 3,272.89 | 169 (21.7%) | 1.45 (1.05–2.00) | .02 |
| CT | AA | 256.46 | 12 (19.7%) | 1.41 (0.75–2.66) | .29 |
| CT+CC | AG+GG | 789.97 | 50 (25.9%) | 1.84 (1.24–2.73) | .003 |
| CVD events | |||||
| TT | AA | 1,231.65 | 61 (20.5%) | 1.00 | |
| TT | AG+GG | 3,089.48 | 194 (24.9%) | 1.32 (0.99–1.76) | .06 |
| CT | AA | 253.16 | 13 (21.3%) | 1.18 (0.65–2.16) | .59 |
| CT+CC | AG+GG | 751.52 | 57 (29.5%) | 1.65 (1.15–2.38) | .007 |
| CVD mortality | |||||
| TT | AA | 1,290.37 | 23 (7.7%) | 1.00 | |
| TT | AG+GG | 3,269.28 | 93 (11.9%) | 1.64 (1.04–2.59) | .04 |
| CT | AA | 271.70 | 3 (4.9%) | 0.74 (0.22–2.48) | .63 |
| CT+CC | AG+GG | 814.07 | 27 (14.0%) | 2.16 (1.24–3.79) | .007 |
Abbreviations: Chr.9p21.3, Chromosome 9p21.3; CVD, cardiovascular diseases; VKORC1, vitamin K epoxide reductase complex subunit 1.
Adjusted Hazards ratio (95% CI) and p values were obtained by the Cox regression analysis after adjustment for age, gender, BMI, fasting glucose, triglycerides, total cholesterol, HDL‐cholesterol, LDL‐cholesterol, family history of stroke, systolic and diastolic blood pressure, smoking, and drinking status.
Figure 2.
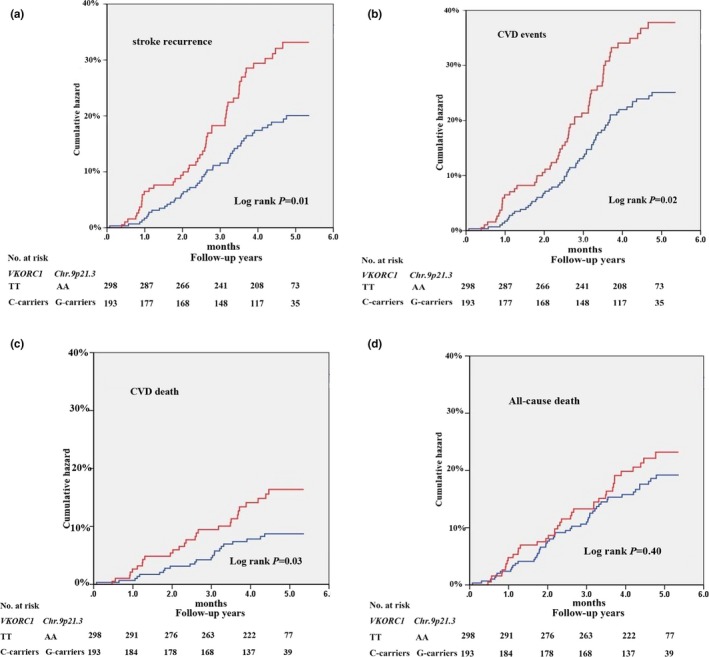
Kaplan–Meier curves of stroke prognosis among patients who had high‐risk allele combinations of VKORC1 rs2359612C and Chr.9p21.3 rs10757274G. CVD, cardiovascular diseases. The red line represented the cumulative hazards of patients who had the combined risk alleles of VKORC1 rs2359612C and Chr.9p21.3 rs10757274G, and the blue line as the cumulative hazards of patients who had the wild‐type genotypes of rs2359612TT and rs10757274AA. Log‐rank p values were obtained from the Kaplan–Meier analysis after adjustment for conventional risk factors as same as the Cox regression analysis in the Table 5. (a) stroke recurrence events; (b) CVD events; (c) Cardiovascular death; (d) All‐cause death
5. DISCUSSION
In the present study, we evaluated the multi‐locus interaction on stroke risk by analyzing a total of 16 variants in eight susceptibility genes including APOA1, MTHFR, VKORC1, ALOX5AP, NOTCH3, VEGFA, and its receptor KDR, and Chr.9p21.3 locus. These risk genes play critical roles in inflammation, lipid metabolism, endothelial dysfunction, and abnormal vascular remodeling during the development of atherosclerosis. We identified for the first time that the genetic interaction of VKORC1 rs2359612T>C and Chr.9p21.3 rs10757274A>G not only contributed to a higher susceptibility to overall stroke and atherothrombotic stroke, but also a worsen prognosis of poststroke such as stroke recurrence and CVD‐related mortality, after adjustment for age, gender, and conventional risk factors. In addition, the gene–gene interaction of VEGFA rs833061C>T and KDR rs2305948T>C, which are related to abnormal vascular remodeling, was identified to confer a higher risk for hemorrhagic stroke, indicating the different pathogenic mechanisms from atherothrombotic stroke. Furthermore, the gene–environmental interaction analysis showed that hypertension or lack of exercise (less than once weekly) can remarkably increase this risk effect among individuals at high genetic basis combining both VKORC1 rs2359612C and Chr.9p21.3 rs10757274G. Lifestyle factors such as smoking, drinking, and overweight/obesity status were not found to have significant interactions with VKORC1 and Chr.9p21.3.
Atherosclerosis is the major pathophysiological mechanism of stroke. The two‐factor genetic interaction between VKORC1 and Chr.9p21.3 on stroke risk can be potentially explained by their common biological roles in the inflammatory process and formation of thrombosis. VKORC1 is a key protein in the recycling of vitamin K and plays a vital role in activating the coagulation cascade and maintaining integrity of the vasculature (Suh et al., 2009). The genetic variants of VKORC1 gene have been reported to affect warfarin dose response and blood clotting through effects on the circle of vitamin K (Schwarz et al., 2008), and persons with the risk genotype of rs2359612TT had higher levels of serum undercarboxylated vitamin K‐dependent proteins, which support the association of VKORC1 gene with coronary heart disease and stroke (Wang et al., 2013). ANRIL, a newly annotated gene encoding a long antisense noncoding RNA on the Chr.9p21.3 risk region, regulates the atherosclerotic process such as in thrombogenesis, vascular remodeling and/or repair, and plaque stability (Holdt et al., 2010; Jarinova et al., 2009). Supporting, our data also provide consistent evidence that the combination of risk alleles in VKORC1 and Chr.9p21.3 loci was not only prospectively associated with worse prognosis of poststroke but also correlated with increased IMT, a useful marker of preclinical atherosclerosis.
On the other hand, the two genes VKORC1 and Chr.9p21.3 were not identified to have joint effects on the risk of hemorrhagic stroke and lacunar stroke after adjustment for conventional risk factors and multiple comparisons. The pathogenesis of atherothrombotic stroke is mainly involved in the formation and rupture of atherosclerotic plaque in large cerebral vessels, such as anterior, middle and posterior cerebral artery, internal and external carotid artery, vertebral and basilar artery, etc. Obviously, hemorrhagic stroke and lacunar infarction are different from atherothrombotic stroke. Intracerebral hemorrhage often results from long‐term arterial stiffness and arteriolosclerosis, microaneurysms, lipohyalinosis, and cerebral amyloid angiopathy occurring in older people (Qureshi et al., 2001). As for lacunar infarction, lacunas result from the occlusion of those small deep penetrating cerebral arteries (with a diameter of <15 mm).
In our findings, increased risk of hemorrhagic stroke was found to be related with genetic interaction of VEGFA rs833061 and KDR rs2305948, which may be attributed to the critical role of VEGFA and its receptor KDR signaling pathway in the development and maintenance of brain vasculature (Greenberg & Jin, 2005; Moulton, 2006). The lack of sufficient VEGFA signaling can result in vascular degeneration and the weak, thin‐walled vessel formation (Shalaby et al., 1995), which may reduce vessel compliance and increase the risk of spontaneous vessel wall rupture and hemorrhagic stroke under stress (Qureshi et al., 2001). Exonic variant rs2305948, located at the KDR binding domain, has been shown to affect the efficiency of VEGFA binding to KDR (Wang et al., 2007); rs833061, located at the promoter region of VEGFA gene, can promote the gene transcription and expression level (Zhang et al., 2009). Our findings showed that the interaction of these genetic variants further amplified the risk of hemorrhagic stroke, which may be helpful for clarifying the etiology of stroke.
Interestingly, our data showed that hypertension or lack of regular exercise can significantly increase the risk effect among individuals at high genetic basis with both VKORC1 and Chr.9p21.3 risk genes. Stroke is a multifactorial disease affected by both genetic and environmental factors, therefore, the extent to which genetic risk can be affected by environmental factors needs to be clarified. Unhealthy lifestyle accompanying with industrialization and urbanization has become increasingly prevalent in China, which may increase the risk of chronic disease including stroke. Our findings regarding the gene–environmental interaction analysis provide additional evidence that blood pressure control and regular exercise are likely to be important strategies for stroke prevention.
The strengths of the study are two aspects. First, the large number of stroke patients allowed us to examine the genetic interactions in different subtypes. In previous studies, the number of hemorrhagic strokes was too small to allow for conclusions as to whether genetic interactions have any effects on this particular subtype of stroke. The second strength included its prospective design, high follow‐up rate, and long‐term follow‐up period. It is the first study to assess whether genetic interactions are associated with the outcome after stroke.
However, some limitations in this study should be mentioned. First, we did not assess all candidate risk genes that have been identified in the Caucasian population by recent genome‐wide association studies. Considering the potential population stratification and difference in ethnicity, we examined the effects of multiple genetic markers based on our previous data in this stroke population, in which no unequal genetic admixture or population stratification was found to affect an association between a genetic variant and disease risk (Wang et al., 2006). Second, this study did not repeatedly assess the genetic interactions in an independent stroke cohort, thus more validations are needed in other stroke population. Third, among hemorrhagic stroke patients carrying risk alleles of VEGFA and KDR, no gene–environmental interactions were observed due to the insufficient sample size, and further studies are needed in larger hemorrhagic stroke cohorts.
In summary, our findings provided novel evidence that people with multiple genetic risk factors can be targets for stroke prevention. The interaction of VKORC1 and Chr.9p21.3 was identified to be associated with increased risk of first stroke and its worse prognosis during a longitudinal follow‐up of stroke complications in a Chinese case‐control study. The interaction of VEGFA and KDR was found to be associated with hemorrhagic stroke. Stroke is highly prevalent in China; therefore, the population‐attributable fraction may be high even if genetic variants confer a modestly relative risk.
CONFLICT OF INTEREST
None declared.
AUTHORS' CONTRIBUTIONS
CF and WZ developed the study design. CF and WZ performed the data analysis and interpretation and prepared the manuscript. XT, YY, RH, YW, YS, and SY collected the samples and interpreted the data. All authors critically reviewed drafts of the manuscript. All authors read and approved the final manuscript.
Supporting information
ACKNOWLEDGMENTS
We thank the technicians and staffs at the State Key Laboratory of Cardiovascular Disease at Fuwai Hospital and at collaborating hospitals for data collection and management.
Feng C, Yang Y, Yang S, et al. Effect of gene–gene and gene–environment interaction on the risk of first‐ever stroke and poststroke death. Mol Genet Genomic Med. 2019;7:e846 10.1002/mgg3.846
Funding information
This work is supported by the grants from the Ministry of Science and Technology, China (2018YFC1312405 and 2011BAI11B04) and the grant from the Chinese Academy of Medical Sciences Innovation Fund for Medical Sciences (2016‐I2M‐1‐006).
REFERENCES
- Adams, H. P. Jr , Bendixen, B. H. , Kappelle, L. J. , Biller, J. , Love, B. B. , Gordon, D. L. , & Marsh, E. E. 3rd (1993). Classification of subtype of acute ischemic stroke. Definitions for use in a multicenter clinical trial. TOAST. Trial of Org 10172 in acute stroke treatment. Stroke, 24(1), 35–41. [DOI] [PubMed] [Google Scholar]
- Benjamini, Y. , & Hochberg, Y. (2000). On the adaptive control of the false discovery rate in multiple testing with independent statistics. Journal of Educational and Behavioral Statistics, 25(1), 60–83. 10.3102/10769986025001060 [DOI] [Google Scholar]
- Chambless, L. E. , Folsom, A. R. , Clegg, L. X. , Sharrett, A. R. , Shahar, E. , Nieto, F. J. , … Evans, G. (2000). Carotid wall thickness is predictive of incident clinical stroke: The atherosclerosis risk in communities (ARIC) study. American Journal of Epidemiology, 151(5), 478–487. 10.1093/oxfordjournals.aje.a010233 [DOI] [PubMed] [Google Scholar]
- Culverhouse, R. , Suarez, B. K. , Lin, J. , & Reich, T. (2002). A perspective on epistasis: Limits of models displaying no main effect. American Journal of Human Genetics, 70(2), 461–471. 10.1086/338759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichmann, A. , & Simons, M. (2012). VEGF signaling inside vascular endothelial cells and beyond. Current Opinion in Cell Biology, 24(2), 188–193. 10.1016/j.ceb.2012.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg, D. A. , & Jin, K. (2005). From angiogenesis to neuropathology. Nature, 438(7070), 954–959. 10.1038/nature04481 [DOI] [PubMed] [Google Scholar]
- He, Y. , Lam, T. H. , Jiang, B. , Li, L. S. , Sun, D. L. , Wu, L. , … Hu, F. B. (2014). Changes in BMI before and during economic development and subsequent risk of cardiovascular disease and total mortality: A 35‐year follow‐up study in china. Diabetes Care, 37(9), 2540–2547. 10.2337/dc14-0243 [DOI] [PubMed] [Google Scholar]
- Helgadottir, A. , Manolescu, A. , Thorleifsson, G. , Gretarsdottir, S. , Jonsdottir, H. , Thorsteinsdottir, U. , … Stefansson, K. (2004). The gene encoding 5‐lipoxygenase activating protein confers risk of myocardial infarction and stroke. Nature Genetics, 36(3), 233–239. 10.1038/ng1311 [DOI] [PubMed] [Google Scholar]
- Hoh, J. , & Ott, J. (2003). Mathematical multi‐locus approaches to localizing complex human trait genes. Nature Reviews Genetics, 4(9), 701–709. 10.1038/nrg1155 [DOI] [PubMed] [Google Scholar]
- Holdt, L. M. , Beutner, F. , Scholz, M. , Gielen, S. , Gabel, G. , Bergert, H. , … Teupser, D. (2010). ANRIL expression is associated with atherosclerosis risk at chromosome 9p21. Arteriosclerosis Thrombosis and Vascular Biology, 30(3), 620–627. 10.1161/atvbaha.109.196832 [DOI] [PubMed] [Google Scholar]
- Hou, T. T. , Lin, F. , Bai, S. , Cleves, M. A. , Xu, H. M. , & Lou, X. Y. (2019). Generalized multifactor dimensionality reduction approaches to identification of genetic interactions underlying ordinal traits. Genetic Epidemiology, 43(1), 24–36. 10.1002/gepi.22169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarinova, O. , Stewart, A. F. R. , Roberts, R. , Wells, G. , Lau, P. , Naing, T. , … McPherson, R. (2009). Functional analysis of the chromosome 9p21.3 coronary artery disease risk locus. Arteriosclerosis Thrombosis and Vascular Biology, 29(10), 1671–1677 10.1161/atvbaha.109.189522 [DOI] [PubMed] [Google Scholar]
- Johnson, A. D. , Hwang, S.‐J. , Voorman, A. , Morrison, A. , Peloso, G. M. , Hsu, Y.‐H. , … O'Donnell, C. J. (2013). Resequencing and clinical associations of the 9p21.3 region: A comprehensive investigation in the Framingham heart study. Circulation, 127(7), 799–810 10.1161/circulationaha.112.111559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jürgens, Günther , Taddei‐Peters, W. C. , Költringer, P. , Petek, W. , Chen, Q. I. , Greilberger, J. , … Ransom, J. H. (1995). Lipoprotein(a) serum concentration and apolipoprotein(a) phenotype correlate with severity and presence of ischemic cerebrovascular disease. Stroke, 26(10), 1841–1848. 10.1161/01.STR.26.10.1841 [DOI] [PubMed] [Google Scholar]
- Khera, A. V. , Emdin, C. A. , Drake, I. , Natarajan, P. , Bick, A. G. , Cook, N. R. , … Kathiresan, S. (2016). Genetic risk, adherence to a healthy lifestyle, and coronary disease. New England Journal of Medicine, 375(24), 2349–2358. 10.1056/NEJMoa1605086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiechl, S. , & Willeit, J. (1999). The natural course of atherosclerosis. Part I: Incidence and progression. Arteriosclerosis Thrombosis and Vascular Biology, 19(6), 1484–1490. [DOI] [PubMed] [Google Scholar]
- Li, Z. , Sun, L. I. , Zhang, H. , Liao, Y. , Wang, D. , Zhao, B. , … Hui, R. (2003). Elevated plasma homocysteine was associated with hemorrhagic and ischemic stroke, but methylenetetrahydrofolate reductase gene C677T polymorphism was a risk factor for thrombotic stroke: A Multicenter Case‐Control Study in China. Stroke, 34(9), 2085–2090. 10.1161/01.STR.0000086753.00555.0D [DOI] [PubMed] [Google Scholar]
- Liu, L. S. (2011). Chinese guidelines for the management of hypertension. Zhonghua Xin Xue Guan Bing Za Zhi, 39(7), 579–615. 10.3760/cma.j.issn.0253-3758.2011.07.002 [DOI] [PubMed] [Google Scholar]
- Lloyd‐Jones, D. M. , Hong, Y. , Labarthe, D. , Mozaffarian, D. , Appel, L. J. , Van Horn, L. , … Rosamond, W. D. (2010). Defining and setting national goals for cardiovascular health promotion and disease reduction: The American Heart Association's strategic Impact Goal through 2020 and beyond. Circulation, 121(4), 586–613. 10.1161/CIRCULATIONAHA.109.192703 [DOI] [PubMed] [Google Scholar]
- Lorenz, M. W. , Markus, H. S. , Bots, M. L. , Rosvall, M. , & Sitzer, M. (2007). Prediction of clinical cardiovascular events with carotid intima‐media thickness: A systematic review and meta‐analysis. Circulation, 115(4), 459–467. 10.1161/CIRCULATIONAHA.106.628875 [DOI] [PubMed] [Google Scholar]
- Lou, X. Y. , Chen, G. B. , Yan, L. , Ma, J. Z. , Zhu, J. , Elston, R. C. , & Li, M. D. (2007). A generalized combinatorial approach for detecting gene‐by‐gene and gene‐by‐environment interactions with application to nicotine dependence. American Journal of Human Genetics, 80(6), 1125–1137. 10.1086/518312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markus, H. S. (2011). Stroke genetics. Human Molecular Genetics, 20(R2), R124–R131. 10.1093/hmg/ddr345 [DOI] [PubMed] [Google Scholar]
- Moulton, K. S. (2006). Angiogenesis in atherosclerosis: Gathering evidence beyond speculation. Current Opinion in Lipidology, 17(5), 548–555. 10.1097/01.mol.0000245261.71129.f0 [DOI] [PubMed] [Google Scholar]
- Polak, J. F. , Pencina, M. J. , O'Leary, D. H. , & D'Agostino, R. B. (2011). Common carotid artery intima‐media thickness progression as a predictor of stroke in multi‐ethnic study of atherosclerosis. Stroke, 42(11), 3017–3021. 10.1161/STROKEAHA.111.625186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qureshi, A. I. , Tuhrim, S. , Broderick, J. P. , Batjer, H. H. , Hondo, H. , & Hanley, D. F. (2001). Spontaneous intracerebral hemorrhage. New England Journal of Medicine, 344(19), 1450–1460. 10.1056/NEJM200105103441907 [DOI] [PubMed] [Google Scholar]
- Ross, O. A. , Soto‐Ortolaza, A. I. , Heckman, M. G. , Verbeeck, C. , Serie, D. J. , Rayaprolu, S. , … Meschia, J. F. (2013). NOTCH3 variants and risk of ischemic stroke. PLoS ONE, 8(9), e75035 10.1371/journal.pone.0075035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz, U. I. , Ritchie, M. D. , Bradford, Y. , Li, C. , Dudek, S. M. , Frye‐Anderson, A. , … Stein, C. M. (2008). Genetic determinants of response to warfarin during initial anticoagulation. New England Journal of Medicine, 358(10), 999–1008. 10.1056/NEJMoa0708078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalaby, F. , Rossant, J. , Yamaguchi, T. P. , Gertsenstein, M. , Wu, X. F. , Breitman, M. L. , & Schuh, A. C. (1995). Failure of blood‐island formation and vasculogenesis in Flk‐1‐deficient mice. Nature, 376(6535), 62–66. 10.1038/376062a0 [DOI] [PubMed] [Google Scholar]
- Smith, J. G. , Melander, O. , Lövkvist, Håkan , Hedblad, B. O. , Engström, G. , Nilsson, P. , … Lindgren, A. (2009). Common genetic variants on chromosome 9p21 confers risk of ischemic stroke: A large‐scale genetic association study. Circulation Cardiovascular Genetics, 2(2), 159–164. 10.1161/CIRCGENETICS.108.835173 [DOI] [PubMed] [Google Scholar]
- Suh, J.‐W. , Baek, S.‐H. , Park, J.‐S. , Kang, H.‐J. , Chae, I.‐H. , Choi, D.‐J. , … Kim, H.‐S. (2009). Vitamin K epoxide reductase complex subunit 1 gene polymorphism is associated with atherothrombotic complication after drug‐eluting stent implantation: 2‐Center prospective cohort study. American Heart Journal, 157(5), 908–912. 10.1016/j.ahj.2009.02.001 [DOI] [PubMed] [Google Scholar]
- Sun, L. I. , Li, Z. , Zhang, H. , Ma, A. , Liao, Y. , Wang, D. , … Hui, R. (2003). Pentanucleotide TTTTA repeat polymorphism of apolipoprotein(a) gene and plasma lipoprotein(a) are associated with ischemic and hemorrhagic stroke in Chinese: A multicenter case‐control study in China. Stroke, 34(7), 1617–1622. 10.1161/01.STR.0000078370.12085.02 [DOI] [PubMed] [Google Scholar]
- Wang, Y. , Chen, J. , Zhang, Y. U. , Bin, L. V. , Sun, K. , Yu, W. , … Hui, R. (2013). VKORC1 rs2359612C allele is associated with increased risk of coronary artery disease in the presence of coronary calcification. Human Genetics, 132(1), 29–37. 10.1007/s00439-012-1222-y [DOI] [PubMed] [Google Scholar]
- Wang, Y. , Zhang, W. , Zhang, Y. , Yang, Y. , Sun, L. , Hu, S. , … Hui, R. (2006). VKORC1 haplotypes are associated with arterial vascular diseases (stroke, coronary heart disease, and aortic dissection). Circulation, 113(12), 1615–1621. 10.1161/circulationaha.105.580167 [DOI] [PubMed] [Google Scholar]
- Wang, Y. , Zheng, Y. I. , Zhang, W. , Yu, H. , Lou, K. , Zhang, Y. U. , … Hui, R. (2007). Polymorphisms of KDR gene are associated with coronary heart disease. Journal of the American College of Cardiology, 50(8), 760–767. 10.1016/j.jacc.2007.04.074 [DOI] [PubMed] [Google Scholar]
- Wang, Z. , Hu, S. , Sang, S. , Luo, L. , & Yu, C. (2017). Age‐period‐cohort analysis of stroke mortality in China: Data from the global burden of disease study 2013. Stroke, 48(2), 271–275. 10.1161/STROKEAHA.116.015031 [DOI] [PubMed] [Google Scholar]
- Zhang, W. , Chen, Y. , Liu, P. , Chen, J. , Song, L. , Tang, Y. , … Hui, R. (2012). Variants on chromosome 9p21.3 correlated with ANRIL expression contribute to stroke risk and recurrence in a large prospective stroke population. Stroke, 43(1), 14–21 10.1161/strokeaha.111.625442 [DOI] [PubMed] [Google Scholar]
- Zhang, W. , Sun, K. , Zhen, Y. , Wang, D. , Wang, Y. , Chen, J. , … Hui, R. (2009). VEGF receptor‐2 variants are associated with susceptibility to stroke and recurrence. Stroke, 40(8), 2720–2726. 10.1161/STROKEAHA.109.554394 [DOI] [PubMed] [Google Scholar]
- Zhang, W. L. , Yang, X. M. , Shi, J. , Sun, K. , & Hui, R. T. (2006). Polymorphism of SG13S114T/A in the ALOX5AP gene and the risk for stroke in a large Chinese cohort. Yi Chuan Xue Bao, 33(8), 678–684. 10.1016/S0379-4172(06)60099-1 [DOI] [PubMed] [Google Scholar]
- Zhao, M. , Wang, X. , He, M. , Qin, X. , Tang, G. , Huo, Y. , … Cai, Y. (2017). Homocysteine and stroke risk: Modifying effect of methylenetetrahydrofolate reductase C677T polymorphism and folic acid intervention. Stroke, 48(5), 1183–1190. 10.1161/STROKEAHA.116.015324 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


