Abstract
Chemotherapy resistance remains a challenge in the clinical management of metastatic colorectal cancer (mCRC). Here, early changes in cell-free circulating tumour DNA (ctDNA) levels were explored as a marker of therapeutic efficacy. Twenty-four mCRC patients were enrolled and treated with FOLFIRI based first-line therapy. Blood samples collected pre-treatment, at day 7, 14, 21, 60 and at progression were analysed for cell-free DNA (cfDNA) and ctDNA levels using digital droplet PCR. A subset of samples were additionally analysed by targeted sequencing. Patients with high pre-treatment ctDNA or cfDNA levels (≥75th centile) had significantly shorter progression free survival (PFS) than patients with lower levels. Despite an overall decline in ctDNA levels from pre-treatment to first CT-scan, serial analysis identified seven patients with temporary increases in ctDNA consistent with growth of resistant cells. These patients had shorter PFS and shorter overall survival. Targeted sequencing analyses of cfDNA revealed dramatic changes in the clonal composition in response to treatment. Our study suggests that increasing ctDNA levels during the first cycles of first-line FOLFIRI treatment is a predictor of incipient progressive disease and poorer survival. Thus, we demonstrate the importance of monitoring ctDNA levels as early as one week after treatment onset to enable early detection of treatment failure.
Subject terms: Tumour biomarkers, Chemotherapy
Introduction
Colorectal cancer (CRC) is one of the most frequently diagnosed cancers worldwide, and approximately 20% of all patients present with metastatic disease1. The majority of these have incurable spread of disease that requires treatment with palliative chemotherapy2,3. Treatment of metastatic CRC (mCRC) is improving, and the median overall survival (OS) for patients presenting with mCRC is currently approaching thirty months in clinical trials3. 5-Fluorouracil and leucovorin administered together with Irinotecan (FOLFIRI) has contributed to the improved clinical outcomes of mCRC4 and is commonly used as a first- or second-line regimen. Despite treatment improvements, intrinsic and acquired resistance mechanisms ultimately lead to disease progression5. At present, the gold standard for assessing treatment effects is the imaging-based Response Evaluation Criteria in Solid Tumours (RECIST)6. Although RECIST is instrumental for standardised assessment and communication of radiological management of cancer patients, its current implementation has some limitations. These include lack of reproducibility, difficulties with classification of patients that have multiple or “non-measurable” metastases, and some studies have reported poor correlation with important clinical endpoints, such as progression free survival (PFS) and OS7. Hence, new approaches enabling early assessment of chemotherapy efficacy could conceivably lead to early adaptation and hence optimized treatment of mCRC.
Recently, the quantificaion of total cell-free DNA (cfDNA) and circulating tumour DNA (ctDNA) has been proposed as a way to monitor changes in tumour burden8–12.
Therefore, the primary aim of this phase II study of mCRC patients treated with FOLFIRI based first-line therapy, was to assess whether changes in plasma cfDNA and ctDNA, during the first months of treatment, could be used as an early marker of FOLFIRI effectiveness.
Results
Study pipeline
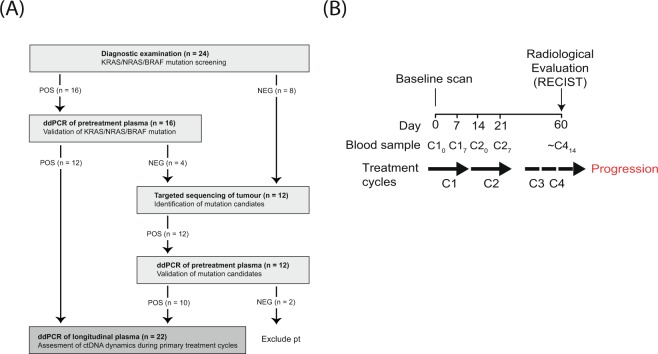
In order to apply a clinically applicable approach we used the following strategy to select patient-specific mutations as ctDNA markers (patient enrolment and study pipeline are presented in Fig. 1A). Tumour samples from all patients were screened for known hotspot mutations in KRAS, BRAF and NRAS as part of routine diagnostic examination. The pre-treatment plasma sample from patients with a tumour hotspot mutation was screened for that mutation by droplet digital polymerase chain reaction (ddPCR) and, if confirmed, the mutation was chosen as a ddPCR marker to monitor plasma ctDNA in longitudinal samples. ctDNA specific ddPCR assays for patients without KRAS, BRAF and NRAS hotspot mutations were developed to match mutation calls from next generation targeted sequencing of biopsies from primary and metastatic lesions.
Figure 1.
Study design and overview of collected samples. (A) Study design and pipeline for selection of patient-specific mutations for ddPCR analysis. Boxes stipulate the method applied and objective at individual analysis steps with POS and NEG indicating a positive or negative result, respectively. The number of patients are shown in parenthesis. (B) Overview of collected samples and treatment cycles. ddPCR, digital droplet PCR; CX, treatment cycle X; CXY, treatment cycle X at day Y.
Pre-treatment total cfDNA and ctDNA levels in relation to clinicopathological parameters
Twenty-four patients with mCRC were included (Table 1 and Supplementary Table 1). KRAS, NRAS or BRAF mutations had been identified in tumour samples from sixteen of the twenty-four patients (75%), and in twelve of these (75%) the mutation could be verified in the pre-treatment plasma sample by ddPCR. Tumour DNA from the four patients, where the hotspot mutation was not detected, and from the eight patients without known tumour hotspot mutation, were submitted to targeted sequencing. Sequencing revealed a median of eight mutations eligible for ddPCR assay development per tumour. One or two patient-specific ddPCR assays were designed for each patient, and finally, a total of sixteen mutation assays were applied to the pre-treatment plasma samples (Supplementary Table 3). Eleven were detectable, which corresponded to ten of the twelve patients. In total, ddPCR detected mutated DNA in pre-treatment plasma of twenty-two patients (12 with KRAS, BRAF mutations and 10 patient specific). The two patients without detectable mutated DNA were excluded from all ddPCR analyses.
Table 1.
Pre-treatment patient characteristics and cfDNA/ctDNA levels in plasma.
| Baseline Characteristics | Patients available for total DNA analysis (n = 24) | Patients available for ctDNA analysis (n = 22) | ||||
|---|---|---|---|---|---|---|
| No of patients | Alleles per mL plasma (CHR3/gCYC assay) Median (range) | p-value | Patients | Mutated alleles per mL plasma. Median (range) | p-value | |
| Age (median/range) | 64.5 (29–81) | 64.5 (45–81) | ||||
| <Median | 11 | 5437 (876–72286) | 0.73 | 11 | 69 (8,2–15400) | 0.12 |
| >Median | 11 | 4733 (833–43029) | 11 | 513 (44–24231) | ||
| Gender | ||||||
| Female | 11 | 5400 (833–72286) | 0.62 | 10 | 94 (23–15400) | 0.77 |
| Male | 13 | 5457 (876–42783) | 12 | 179 (8–24231) | ||
| ECOG Performance status | ||||||
| 0 | 14 | 5428 (876–72286) | 14 | 108 (8–15400) | ||
| 1 | 7 | 5400 (833–16733) | 0.37 | 5 | 1120 (81–3086) | 0.55 |
| 2 | 1 | 3217 | 0.67 | 1 | 59 | 0.67 |
| Unknown | 2 | 21386 (7696–35074) | 0.5 | 2 | 12127 (23–24231) | 0.93 |
| Sites of metastatic disease | ||||||
| Liver metastasis | 12 | 19149 (876–72286) | 0.04* | 12 | 684 (23–23231) | 0.16 |
| No liver metastasis | 12 | 4508 (833–16733) | 10 | 91 (8–3086) | ||
| Lung metastasis | 11 | 5457 (833–43029) | 0.50 | 10 | 381 (8–24231) | 0.58 |
| No lung metastasis | 13 | 5400 (933–72286) | 12 | 104 (23–15400) | ||
| Mutation status of tumour | ||||||
| WT | 7 | 3617 (833–30600) | 5 | 69 (27–513) | ||
| KRAS mut | 12 | 6268 (1857–72286) | 0.05* | 12 | 1803 (8–24231) | 0.33 |
| NRAS mut | 3a | 16733 (4067–42783) | 0.17 | 3a | 107 (101–10199) | 0.39 |
| BRAF mut | 3a | 2108 (933–4067) | 0.65 | 3a | 107 (81–109) | 0.79 |
| Number of metastatic sites | ||||||
| 1 | 12 | 4742 (876–72286) | 12 | 91 (23–15400) | ||
| 2 | 10 | 5400 (833–40629) | 0.77 | 6 | 817 (8–24231) | 0.21 |
| >3 | 2 | 36814 (30600–43029) | 0.20 | 4 | 1652 (59–11242) | 0.45 |
| Treatment efficacy, 1. RECIST | ||||||
| Stabil disease (SD) | 19 | 5457 (833–72286) | 0.52 | 17 | 248 (8–24231) | 0.16 |
| Partial/complete response (PR/CR) | 5 | 4067 (2610–5620) | 5 | 44 (27–2486) | ||
| Progression disease (PD) | 0 | 0 | ||||
Wilcoxon rank sum test was applied to test for correlations between patient characteristics and cfDNA and ctDNA. aTwo patients were excluded (see Fig. 1A) and hence data from n = 22 patients is shown. bOne patient (pt109) had a colon tumour with a BRAF mutation and a rectum tumour with a NRAS mutation. p-values < 0.05 are indicated in bold and with *.
ctDNA, circulating tumour DNA; cfDNA, cell-free DNA, ECOG, Eastern Cooperative Oncology Group; RECIST, Response Evaluation Criteria In Solid Tumours.
At last follow up (January 2018) progressive disease had occurred in twenty-one of the twenty-two patients, and twelve patients had died. The median PFS and OS of the cohort (n = 22) was 218 days (95%CI = 197–265) and 315 days (95%CI = 274–373), respectively.
Pre-treatment cfDNA and ctDNA levels are associated with treatment outcome
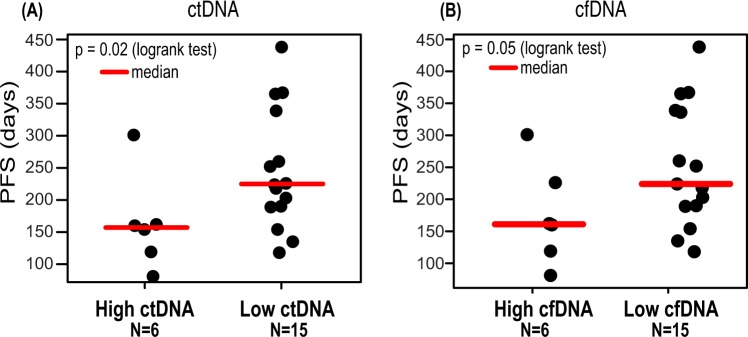
The median pre-treatment levels of ctDNA and cfDNA were 108 (range 8–24231) and 5408 (range 833–72286) GEs per mL plasma, respectively. Patients with high ctDNA levels (≥75th centile) at pre-treatment (C10, see Fig. 1B) had a shorter PFS (157 days versus 226 days) than patients with low pre-treatment ctDNA levels (HR = 3.34, 95%CI = 1.20–9.29, p = 0.02) (Fig. 2A). The same was evident for cfDNA (HR = 2.68, 95%CI = 0.96–7.48, p = 0.05) (Fig. 2B).
Figure 2.
Progression free survival for patients with or without a high pre-treatment level of ctDNA/cfDNA. Patients with a high pre-treatment ctDNA level (A) (75th centile) or cfDNA level (B) have a shorter PFS than patients with a low level. ctDNA, circulating tumour DNA; cfDNA, cell-free DNA.
ctDNA levels decline from pre-treatment to first response evaluation
Plasma samples were analysed prior to the first treatment cycle (C10), at day seven (C17) during the first cycle, at day zero and seven during the second cycle (C20 and C27, respectively), at first radiological status evaluation (C414) and at the time of clinical validated progression (Fig. 1B). A ctDNA decline from pre-treatment to first response evaluation was observed for all patients (p = 0.001, Wilcoxon signed rank test) (Supplementary Fig. 1.) This was in agreement with RECIST evaluations, which indicated either stable disease (n = 17) or partial or complete response (n = 5) (Table 1). We investigated whether patients with at least a 10-fold reduction in the level of ctDNA from pre-treatment to pre-cycle-two (C20) had a longer PFS, as recently proposed by Tie et al.13 (Supplementary Fig. 2). We observed a shorter PFS in patients with a ≥ 10-fold reduction in ctDNA (n = 6, HR = 3.20, 95%CI = 1.10–9.20, p = 0.02). As this appeared to conflict with the general assumption that a reduction in ctDNA is reflecting a reduced tumour burden and response to the therapy, we searched for confounders. Thereby, it was revealed that all six patients with ≥10-fold ctDNA reductions had higher pre-treatment ctDNA levels than the median, while this was only the case for 5/15 of the patients with <10-fold ctDNA reductions (Fisher’s Exact Test, p-value = 0.06, odds ratio 8.89).
Multiple increases in early ctDNA level are associated with decreased PFS
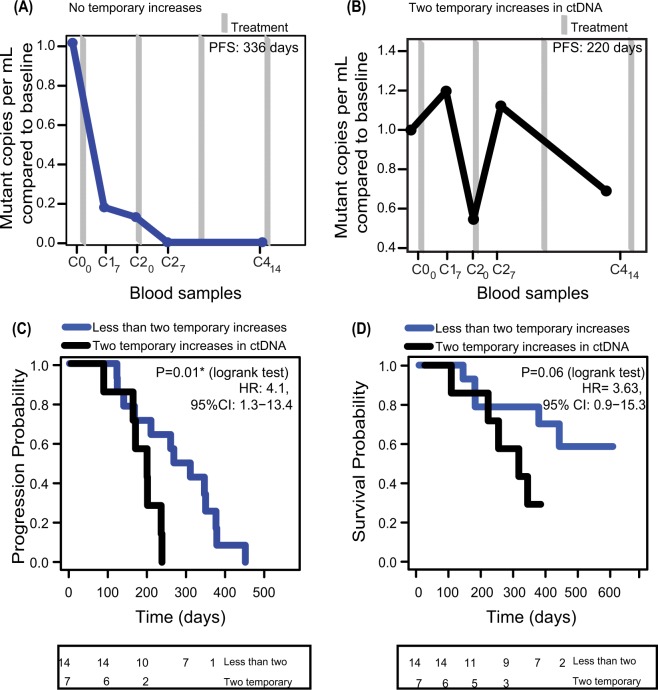
Despite the overall trend of declining ctDNA levels towards the first response evaluation (Supplementary Fig. 1), ctDNA levels nevertheless fluctuated during or between treatment cycles. Longitudinal ctDNA levels for all patients are given in Supplementary Fig. 3. In seven patients, we observed two occurrences of temporary ctDNA level increases, i.e. a higher ctDNA level compared to the previous drawn blood sample, during the first 60 days of treatment. We speculated if these temporary raises could indicate growth of resistant cells and potentially would be associated with a shorter time to progression. In agreement with this hypothesis the mean PFS of the seven patients was significantly shorter than for the rest of the patients (p = 0.01, HR = 4.11, 95%CI = 1.27–13.36) (Fig. 3C). Their OS were correspondingly shorter (p = 0.06, HR = 3.63, 95%CI = 0.87–15.25) (Fig. 3D).
Figure 3.
Time to progression for patients with or without two occurrences of temporary increasing ctDNA levels. Patients with multiple occurrences of temporary ctDNA increases have a shorter PFS and OS than patients with stable/decreasing ctDNA levels. For one patient, only C10 and C414 samples were accessible and therefore this patient was excluded from this analysis. (A) Representative patient with one temporary ctDNA increase during the first 60 days of treatment. (B) Representative patient with two instances of temporary ctDNA increases. (C) Progression free survival (PFS). Boxed numbers represents the number of patients in each group not having clinical progression at the given timepoint. (D) Overall survival (OS). Boxed numbers represents the number of patients in each group alive at the indicated timepoint.
Targeted sequencing of pre-treatment cfDNA samples identifies mutations present at time of treatment initiation
In order to determine if alterations selected on basis of results from the diagnostic examination were as informative as those selected from tumour sequencing, we investigated if the chosen markers were increasing at time of clinical progression (n = 10 samples available). Only four of seven diagnostic work-up ctDNA markers increased at progression, while all three mutations identified by sequencing increased (Table 2). We speculated whether sequencing of the cfDNA directly would be an even better approach for identifying mutations suited for monitoring of treatment response. To address this, the pre-treatment plasma remnants after the former analyses were sequenced using a custom gene panel (Nugen Target Enrichment) targeting the genomic regions most frequently mutated in CRC (Supplementary Table 5). Pre-treatment plasma was available for 17 of the 24 patients, two of which no ctDNA marker had previously been identified. Often only small plasma volumes were left and therefore the average target coverage was limited for some samples – median 246x (range 100–419x). Matching germline samples were also subjected to sequencing using the same gene panel. Genomic variants were identified in all of the pre-treatment samples. On average 59 variants were identified per sample (range 2–153, Supplementary Table 6), including the two patients where no ctDNA marker previously had been identified (pt106 and pt108). Generally, a good concordance was observed between the ddPCR results and the targeted sequencing results for the 14 ddPCR markers covered by the plasma sequencing panel (Table 3). Four ddPCR-detected markers were not detected by plasma-sequencing. For three of these, ddPCR indicated minor allele frequencies (MAF) below 2%. At the position of these mutations the sequencing coverage was only 57x, 5x and 125x (mean for whole panel was 112x, 195x, and 310x), potentially explaining why these variants were not detected. One mutation (ctDNA marker for pt117) was not identified by sequencing (coverage 109x) even though ddPCR analysis indicated a MAF of 37%. Importantly, other mutations were identified by plasma sequencing in these four patients. Furthermore, often the mutation selected for ddPCR was not the most abundantly observed by plasma sequencing. In conclusion, sequencing of plasma could in all cases have been used for analyses of changes in ctDNA levels in response to therapy.
Table 2.
Evaluation of whether mutations applied as ctDNA markers for ddPCR are informative or non-informative at progression.
| Patient | ctDNA fold changea | Informativeb | Days to progressionc | Mutation detected by ddPCR at progression | Source of selection of ctDNA marker for ddPCR |
|---|---|---|---|---|---|
| pt101 | 46 | Yes | 6 (252) | Yes | Diagnostic examination |
| pt103 | 0.04 | No | 35 (154) | Yes | Diagnostic examination |
| pt104 | 12 | Yes | 15 (301) | Yes | Sequencing of primary tumor |
| pt105 | 38 | Yes | 10 (135) | Yes | Diagnostic examination |
| pt109 | 164 | Yes | 5 (365) | Yes | Sequencing of liver metastasis |
| pt113 | 8 | Yes | 0 (260) | Yes | Sequencing of lung metastasis |
| pt115 | 16 | Yes | 19 (189) | Yes | Diagnostic examination |
| pt116 | 30 | Yes | 38 (119) | Yes | Diagnostic examination |
| pt118 | 0.8 | No | 1 (160) | Yes | Diagnostic examination |
| pt119 | 0.9 | No | 9 (162) | Yes | Diagnostic examination |
Informative versus non-informative mutations applied as ctDNA markers for ddPCR. ctDNA fold change is measured as ctDNA change from first status evaluation to sample nearest progression. An informative mutation is defined as a mutation increasing towards progression.
actDNA change from first status evaluation to sample nearest progression.
bSample was considered informative if an increase in ctDNA levels were observed towards progression.
cSamples collected up to 38 days before progression were considered progression samples.
Table 3.
Concordance between ddPCR and plasma-sequencing analysis of pre-treatment plasma samples.
| Pta | Mutation for ddPCR analysis | Mutation present in sequencing of pre-treatment sample | MAF% detected by sequencing | Mean coverage of cfDNA sequencing | MAF% detected by ddPCR |
|---|---|---|---|---|---|
| pt101 | KRAS G12D | no | Not available | 112 | 1.7 |
| pt102 | TP53–221T < TTC | yes | 0.3 | 406 | 3.8 |
| pt103 | KRAS G12D | yes | 28 | 306 | 36 |
| pt104 | APC-507C > T | yes | 40 | 275 | 49 |
| pt105 | BRAF V600E | yes | 9 | 419 | 2.7 |
| pt107 | KRAS G12D | yes | 30 | 238 | 39 |
| pt109 | PARK2-214G > A | Not on panel | Not on panel | 211 | 17 |
| pt110 | KRAS G12C | yes | 17 | 152 | 79 |
| pt111 | KRAS G12V | no | Not available | 195 | 0.7 |
| pt112 | ERBB2-392A > G | yes | 4 | 327 | 7.4 |
| pt113 | TP53-003G > A | no | Not available | 310 | 0.8 |
| pt114 | Tp53-533G > A | yes | 13 | 253 | 1.7 |
| pt116 | KRAS G12A | yes | 33 | 310 | 36 |
| pt117 | KRAS G12V | no | Not available | 109 | 37 |
| pt118 | KRAS G13D | yes | 36 | 170 | 38 |
Targeted sequencing of pre-treatment plasma sample. For 17 out of 24 patients plasma DNA were available for targeted sequencing. NA means that no sample within two months of progression was available.
aNo mutated DNA in patients pt106 and pt108 were detected.
Therapy induced selection pressure dramatically changes the clonal composition of ctDNA
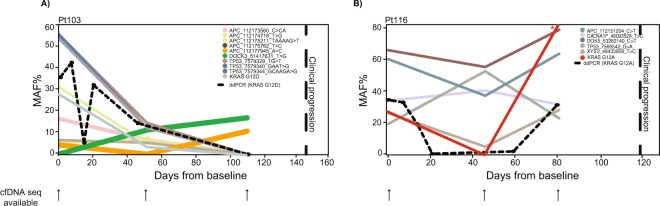
In this study we used a one marker strategy for monitoring therapy response. As it can be seen from Table 2, the risk of this strategy is that the selected marker may turn out to be subclonal, and while informative of the initial response, it may be non-informative of progression. Therefore, we hypothesized that a strategy based on monitoring multiple mutations would be better because this would increase the chance of including an informative marker. To address this, we compared our one-marker ddPCR and our targeted cfDNA sequencing strategies. For patients 103 and 116 sufficient plasma cfDNA from pre-treatment, first status evaluation and progression samples were available for analysis with both ddPCR and targeted sequencing (Fig. 4). It was evident from sequencing of cfDNA from patient 103 that treatment onset changed the composition of ctDNA dramatically (Fig. 4A). Sequencing of cfDNA from patient 116 revealed that even though the KRAS mutation chosen for ddPCR analysis was informative of progression (Table 3), it was not the most optimal mutation to monitor (Fig. 4B). The TP53 G > A mutation was increasing already after the first time point indicating that the treatment was not effective already at the first status scan evaluation. This knowledge could have been used to change treatment prior than at clinical progression.
Figure 4.
Targeted sequencing of longitudinal cfDNA from two patients. (A,B) Targeted sequencing was performed on cfDNA from patient 103 and 116 (pre-treatment, first status evaluation and a progression sample - collected within 40 days of clinical progression). Several mutations were detected in all samples. Shown are mutations detected in two or more samples. *Due to low coverage (<5x) in the specified sample the reported MAF may be an overestimate.
In summary, targeted sequencing of cfDNA gives a panel of mutations that can be monitored, thus giving a better indication of which mutations that are clonal and therefore, believed to be informative, in contrast to ddPCR where prior knowledge of mutational status often is a request. Consequently, direct sequencing of cfDNA will probably be the best approach for the future.
Discussion
Identification of patients that do not benefit from chemotherapy is of great importance, both to reduce side effects of ineffective treatments, and to facilitate changes in the treatment strategy. In this study, we aimed to investigate whether changes in plasma ctDNA during the first three weeks of treatment were predictive of FOLFIRI efficacy. Our results indicate that FOLFIRI efficacy can be predicted prior to the first response evaluation and opens for rapid revision of the treatment strategy.
Patient and disease characteristics were as expected from other studies on this patient group14. However, OS was shorter than reported in recent randomized trials involving mCRC, but comparable to survival in retrospective series on patients receiving combination chemotherapy15, as well as to population based cohorts with incurable disease were treatment is merely palliative as is the case for the patients in the current study16.
We found that high pre-treatment ctDNA levels were negatively correlated to outcome. This is in line with previous observations13,17. Equally high total cfDNA at diagnosis correlated to shorter PFS, as previously reported18,19. We did not find any correlation between the number or type of metastasis and the ctDNA levels (data not shown).
Recently, it has been suggested that a large reduction in the ctDNA level shortly after initiation of treatment is a potential marker of treatment effect13,17. Tie et al. investigated the ctDNA levels in a cohort consisting of 47 patients treated with 5-fuorouracil and oxaliplatin and four treated with FOLFIRI and found a trend towards longer PFS for patients with a 10-fold, or higher, ctDNA reduction (p = 0.27, 95%CI = 0.6–5.6)13. We could not confirm this trend in our study, neither with a ten-fold reduction threshold nor other thresholds tested. It should be noticed that both studies include small cohorts, and therefore the results should be taken with caution. Moreover, Tie et al. investigate samples collected at different time points than in the current study.
In our study, we analysed 103 serial plasma samples collected between pre-treatment and first RECIST evaluation for twenty-two patients (overview of all analysed plasma samples are given in Suppl. Table 7). Uniquely, we measured ctDNA levels as early as one, two and three weeks after treatment start. The ctDNA levels were surprisingly dynamic in this window, with several temporary increases in ctDNA (Supplementary Fig. 3). Seven patients even showed two temporary increases in ctDNA. Speculating if the increasing ctDNA levels could reflect growth of resistant cells, we compared PFS in these patients relative to the remaining patients and found that they had a significantly shorter PFS, and shortened OS. In agreement with these findings, Garlan et al., recently reported that mCRC patients with increasing ctDNA levels between the pre-treatment sample and the second or third treatment cycle had significant shorter PFS and OS20. Thus, by early serial ctDNA analysis it appears to be possible to predict which patients have minimal benefit of the given therapy, potentially providing an early opportunity to revise the treatment strategy.
In the current work, a single mutation was used for ctDNA monitoring of each patient. We chose this strategy in order to make it clinically applicable, robust and affordable. A weakness of the one-marker strategy is that it is essential to select an ancestral mutation representative of all tumour cells. If subclonal, the results may potentially lead to incorrect conclusions. Our analyses, from ten patients with samples available close to progression, revealed that sequencing of the tumour samples facilitated selection of more informative mutations as compared to the diagnostic examination. In the clinic, RAS/RAF mutational status is determined in order to direct the choice of treatment, and do not involve quantification. Consequently, we could not determine the likelihood of a mutation being subclonal or clonal on basis of the results from the diagnostic examination. From the tumour sequencing on the other hand, we selected mutations on basis of the minor allele frequency and driver potential, and thus increased the chance of selecting a clonal mutation for ddPCR analysis.
We hypothesized that applying a multi-target strategy would be better. Through targeted plasma sequencing we identified mutations in all patients – and further confirmed the ctDNA levels determined by ddPCR. We next hypothesised that targeted sequencing of longitudinally samples would give a better indication of progression than mutations selected on basis of tumour sequencing of diagnostic examination. To investigate this, we applied targeted sequencing directly to longitudinal cfDNA samples from a small number of patients. These analyses indicated the major effects of chemotherapeutics on clonal composition. From sequencing of samples from patient 103 it was evident that some mutations, including the one chosen for ddPCR analysis, were not informative of progression, as these clones apparently were eliminated by the applied therapy. However, an APC (12177945_A > C) mutation appeared subclonal at baseline and increased towards progression. Additionally, a DOCK3 mutation that was not detected in the pre-treatment sample emerged during the first cycles of therapy, and seemed to be part of one of the clones driving resistance to therapy and thus progression. Thus, mutations expected to be clonal, based upon sequencing of the pre-treatment sample, proved to be non-informative of progression as other clones evolved during the applied selection pressure. This stresses the need to not only monitor more than one mutation, but also to do it unbiased, e.g. without prior knowledge, as new mutations do arise during treatment.
The targeted sequencing analysis of patient 103 indicated the possibility of there being more than one tumour lesion present in this patient as the mutations emerging towards progression (APC A > C and DOCK3) did not have any ancestral clones in common with the rest of the identified mutations, which seemed to respond to the applied therapy.
Our study has some limitations. Firstly, the modest cohort size inherently makes the confidence intervals of our findings wide. Secondly, the use of only one single patient-specific ctDNA marker may limit the sensitivity towards detecting ctDNA and impose the risk that the selected marker is non-ancestral and hence may turn out non-informative of future resistant clone(s). Thirdly, the chosen method for cfDNA sequencing analysis yielded relatively low sequencing coverage levels, thus making it difficult to detect low frequency alterations. In future studies, multiplexing strategies, e.g. based on targeted massive parallel sequencing may limit such a risk as both indicated by the findings presented in this article and by others21–23. Theoretically, multiplexing will also increase the sensitivity for detecting ctDNA24,25.
In conclusion, very early evaluation of ctDNA changes during therapy may be beneficial for patients through rapid detection of treatment failure. Additional studies are needed to thoroughly assess the potential clinical benefit.
Materials and Methods
Patients and collection of blood samples
This prospective, observational phase II study recruited patients undergoing treatment for mCRC at the Department of Oncology, Aarhus University Hospital, Denmark. Inclusion criteria were age ≥18, non-resectable, histopathological verified mCRC, with indication for first-line FOLFIRI-based treatment, and measurable disease according to RECIST version 1.16. Patients were treated according to Danish National Guidelines. Chemotherapy included: Leucovorin (400 mg/m2 intravenous infusion) and Fluoruracil (400 mg/m2 bolus infusion or 2400 mg/m2 continuous infusion/46 hours), Irinotecan 180 mg/m2 and some RAS/RAF wildtype (WT) patients got additionally Panitumumab 6 mg/kg or Cetuximab 500 mg/kg, every two weeks (overview of treatment and patient characteristics is given in Supplementary Table 1). Treatment decisions were made after each four treatment cycles based on CT-scanning of the chest and abdomen/pelvic areas and evaluation according to RECIST version 1.1. The study was approved by the The Central Denmark Region Commitees on Health Research Ethics (M-2014-298-14) and the Danish Data Protection Agency. Written informed consents were obtained from the patients.
PFS and OS were defined as time from first day of treatment until the date of clinical progression (RECIST evaluation) or death from any cause, respectively. Patients who did not experience an event were censored by the last date of observation.
Isolation and quantification of DNA
Within two hours of the blood draw, EDTA Plasma (median 3.5 mL, range 1.5–4.0) was isolated from 10 mL of blood by centrifugation at 1400 g for 20 min at 20 °C. DNA was extracted from formalin-fixed, paraffin-embedded (FFPE) tumour tissue using QiAamp DNA FFPE tissue kit (Qiagen). Tumour DNA was quantified using Qubit (Thermofischer) and stored at −80 °C. Leucocyte DNA was extracted with the QIAamp DNA Blood Midi Kit (control DNA). Cell free plasma DNA (cfDNA) was extracted using the QIAamp Circulating Nucleic Acid Kit (Qiagen). The isolated cfDNA was eluted in 60 μL elution buffer and stored at −80 °C. Quantification of cfDNA was done using ddPCR as previously described8, and is reported as copies per mL plasma or genomic equivalents (GE). We used a B-cell immunoglobulin rearrangement specific ddPCR assay26 to identify cfDNA samples with contaminating lymphocyte DNA. These samples were excluded as contamination is an indication of undesired cell lysis.
DNA quantification by ddPCR
ctDNA was detected and quantified using ddPCR. The experiments were conducted according to the ddPCR MIQE-guidelines (Supplementary Table 2)27. Serial plasma DNA samples were analysed on a QX200 AutoDG Droplet Digital PCR System according to the manufacturer’s instructions (Bio-Rad). Each analysis included a positive, a negative (matched leucocyte DNA) and a non-template control (H2O). The positive control sample was either a matched tumour DNA or a ~150 bp template DNA oligo of the mutated sequence. The ddPCR reaction volume was 22 µL, consisting of 13 µL mastermix (18 mM forward and reverse primer, 0.05 mM probe, 2x Supermix for probes (no UTP) (Cat. No. 186–3023; Bio-Rad)) and 9 µL cfDNA. Data were analysed using QuantaSoft v1.6.6 software (Bio-Rad). Thresholds for separating positive and negative droplets were determined based on the positive and negative control samples on each plate. Mutated copies per mL plasma were calculated and used as a measure of concentration. Both previously reported and novel mutation specific ddPCR assays were used (Supplementary Table 3). The novel assays were designed using Primer328 and included two probes to quantify both the mutated and the WT alleles. Amplicon length was, when possible, kept below 100 nucleotides. Limit of detection (LOD) and limit of blank (LOB) for each assay (Supplementary Table 3) were determined using a panel of 24 negative control samples from healthy donors as described previously29. For all plasma samples reported to be negative for the assessed patient-specific mutation, a minimum cfDNA input of 700 GEs were analysed (median 1303 GEs).
Targeted sequencing of tumour DNA
DNA from tumours without known hotspot mutations in KRAS, BRAF, or NRAS were subjected to targeted sequencing using a previously reported hybridization capture panel (Roche Nimblegen) covering 410 cancer genes30,31. An overview of the sequenced tumour samples is presented in Supplementary Table 4. Tumour and matched normal DNAs were fragmented (Covaris E220) and subjected to library preparation using the KAPA Hyper Library Preparation Kit (KAPA Biosystems). The indexed libraries (twelve tumour and normal DNA pairs) were pooled and paired-end sequenced using Illumina® NextSeq Medium v2, 300 bp flow cells. Adapter sequences were trimmed using Trim Galore (Babraham Bioinformatics), and mapped to the hg19 reference genome using BWA MEM (v. 0.7.5)32. Picard MarkDuplicates was used to inspect the alignment and remove duplicates. Mapping in areas with INDELs were refined using GATK IndelRealigner33 and systematic errors in base quality scores were identified and adjusted using GATK BaseRecalibrator33. Somatic SNVs and INDELs were called using a combination of GATK MuTect234 and VarScan235.
Patient specific somatic point mutations to be used for ddPCR assay development were selected based on following criteria: Minimum 30 reference reads in the tumour sample (median tumour coverage was 526x), should be among the most clonal mutations in the tumour as indicated by a high allele frequency, and the alternative allele frequency should be below 0.002 in the germline sample. Additionally, mutations in known CRC driver genes were preferred (such as APC, TP53).
Targeted sequencing of circulating-free DNA
We performed targeted sequencing using a commercial version of anchored multiplex PCR (AMP) (NuGEN). Libraries were prepared from 3–129 ng (median 49 ng) DNA according to manufacturer’s instructions. A custom Nugen Target Enrichment panel was designed (NuGEN). The panel covers 28622 bases including: (1) the 127 most frequently mutated regions in CRC patients identified from TCGA data36, (2) five most frequently mutated regions identified by Giannakis et al.37 and not identified by TCGA, (3) all of the coding regions of TP53 and APC, and 4) 18 ID-SNPs (Supplementary Table 5). Unique identifiers (UIDs) are included as part of the adapter design for error correction. Libraries were subjected to 21 cyclers of PCR amplification. Pools were single-end sequenced using Illumina® NextSeq Medium v2, 150 bp flow cells, with 15 base index reads to ensure sequencing of the region containing the UIDs. Data was handled and processed as previously described38. A random 6 nucleotide unique molecular identifier (UMI) were identified in each read, before the reads were mapped to the human hg19 reference genome using BWA MEM (v. 0.7.5)32. Barcodes that had mutated during PCR amplification, or contained sequencing errors, were rescued using UMI-tools group (v. 0.4.0)39 with the directional method and edit distance set to 1. Rescue of erroneous UMIs increases both sensitivity and specificity, as well as hinders inflation of the estimated number of unique molecules sequenced. Consensus families were obtained using a custom script by collapsing reads with the same rescued UMI barcode and genomic position. Consensus families with less than three reads were discarded. For each position a consensus base was obtained by requiring 55% similarity across all reads in a family, otherwise an N was inserted. Consensus sequencing quality scores were generated using the mean of individual quality scores per base in each family. Consensus reads were mapped to hg19 using BWA MEM and variants identified using MuTect2 (GATK v. 3.7) using default settings. All reported mutations were accepted as confident by Mutect2 (PASS variants). The median unique coverage were 253x (range: 109–419x).
Statistical analysis
Wilcoxon rank sum test was applied to test association between patient characteristics and cfDNA and ctDNA. Survival data were analysed by the Kaplan–Meier method, and differences between the groups were estimated by the log-rank test. Analyses were performed using R statistical software40 and R packages survival and rms41,42. A p-value ≤ 0.05 was considered statistically significant. Effect sizes are indicated by 95% confidence intervals.
Supplementary information
Acknowledgements
We thank Iver Kristiansen Nordentoft and Pia Kjølhede Andersen for setting up sequencing experiments. We acknowledge the patients included in this study. K.S. and this work was supported by the Danish Cancer Society (R99-A6323) and the Novo Nordisk Foundation. I. Lyskjær was supported by the Lundbeck Foundation (R180-2014-3998). M. H. Rasmussen was funded by The Danish Council for Strategic Research (DFF – 4183-00619). C. L. Andersen was funded by The Danish Cancer Society (R146-A9466-16-S2) and The Novo Nordisk Foundation (ID: 12747).
Author Contributions
K.S. planned the clinical study design. K.S. and C.K. collected the samples. Samples was analysed with ddPCR by I.L., C.D. and B.S. I.L. performed the sequencing. M.K. and S.V. made the pipelines for mutational calling. I.L., M.R., C.A., C.K. and K.S. analysed the data. S.K. and M.R. conducted the radiological and histological analysis. I.L., M.R., A.F.B.J. and C.A. drafted the manuscript, which was revised by all authors.
Data Availability
The data from findings of this study are available from the corresponding author upon reasonable request.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Iben Lyskjær and Camilla Skovhus Kronborg contributed equally.
Karen-Lise Garm Spindler and Claus Lindbjerg Andersen jointly supervised this work.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-47708-1.
References
- 1.van der Geest LG, et al. Nationwide trends in incidence, treatment and survival of colorectal cancer patients with synchronous metastases. Clin Exp Metastasis. 2015;32:457–465. doi: 10.1007/s10585-015-9719-0. [DOI] [PubMed] [Google Scholar]
- 2.Schmoll HJ, et al. ESMO Consensus Guidelines for management of patients with colon and rectal cancer. a personalized approach to clinical decision making. Ann Oncol. 2012;23:2479–2516. doi: 10.1093/annonc/mds236. [DOI] [PubMed] [Google Scholar]
- 3.Van Cutsem E, et al. ESMO consensus guidelines for the management of patients with metastatic colorectal cancer. Ann Oncol. 2016;27:1386–1422. doi: 10.1093/annonc/mdw235. [DOI] [PubMed] [Google Scholar]
- 4.Douillard JY, Group VS. Irinotecan and high-dose fluorouracil/leucovorin for metastatic colorectal cancer. Oncology (Williston Park) 2000;14:51–55. [PubMed] [Google Scholar]
- 5.Housman G, et al. Drug resistance in cancer: an overview. Cancers (Basel) 2014;6:1769–1792. doi: 10.3390/cancers6031769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eisenhauer EA, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1) Eur J Cancer. 2009;45:228–247. doi: 10.1016/j.ejca.2008.10.026. [DOI] [PubMed] [Google Scholar]
- 7.Sharma M. R., Maitland M. L., Ratain M. J. RECIST: No Longer the Sharpest Tool in the Oncology Clinical Trials Toolbox--Point. Cancer Research. 2012;72(20):5145–5149. doi: 10.1158/0008-5472.CAN-12-0058. [DOI] [PubMed] [Google Scholar]
- 8.Reinert T, et al. Analysis of circulating tumour DNA to monitor disease burden following colorectal cancer surgery. Gut. 2016;65:625–634. doi: 10.1136/gutjnl-2014-308859. [DOI] [PubMed] [Google Scholar]
- 9.Bettegowda C, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med. 2014;6:224ra224. doi: 10.1126/scitranslmed.3007094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dawson SJ, et al. Analysis of circulating tumor DNA to monitor metastatic breast cancer. N Engl J Med. 2013;368:1199–1209. doi: 10.1056/NEJMoa1213261. [DOI] [PubMed] [Google Scholar]
- 11.Spindler KL, et al. Clinical utility of KRAS status in circulating plasma DNA compared to archival tumour tissue from patients with metastatic colorectal cancer treated with anti-epidermal growth factor receptor therapy. Eur J Cancer. 2015;51:2678–2685. doi: 10.1016/j.ejca.2015.06.118. [DOI] [PubMed] [Google Scholar]
- 12.Scholer LV, et al. Clinical Implications of Monitoring Circulating Tumor DNA in Patients with Colorectal Cancer. Clin Cancer Res. 2017;23:5437–5445. doi: 10.1158/1078-0432.CCR-17-0510. [DOI] [PubMed] [Google Scholar]
- 13.Tie J, et al. Circulating tumor DNA as an early marker of therapeutic response in patients with metastatic colorectal cancer. Ann Oncol. 2015;26:1715–1722. doi: 10.1093/annonc/mdv177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Labianca R, et al. Intermittent versus continuous chemotherapy in advanced colorectal cancer: a randomised ‘GISCAD’ trial. Ann Oncol. 2011;22:1236–1242. doi: 10.1093/annonc/mdq580. [DOI] [PubMed] [Google Scholar]
- 15.Zacharakis M, et al. Predictors of survival in stage IV metastatic colorectal cancer. Anticancer Res. 2010;30:653–660. [PubMed] [Google Scholar]
- 16.Engstrand J, Nilsson H, Stromberg C, Jonas E, Freedman J. Colorectal cancer liver metastases - a population-based study on incidence, management and survival. BMC Cancer. 2018;18:78. doi: 10.1186/s12885-017-3925-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gray ES, et al. Circulating tumor DNA to monitor treatment response and detect acquired resistance in patients with metastatic melanoma. Oncotarget. 2015;6:42008–42018. doi: 10.18632/oncotarget.5788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Spindler KL, Pallisgaard N, Andersen RF, Brandslund I, Jakobsen A. Circulating free DNA as biomarker and source for mutation detection in metastatic colorectal cancer. PLoS One. 2015;10:e0108247. doi: 10.1371/journal.pone.0108247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boysen AK, et al. Cell-free DNA levels and correlation to stage and outcome following treatment of locally advanced rectal cancer. Tumour Biol. 2017;39:1010428317730976. doi: 10.1177/1010428317730976. [DOI] [PubMed] [Google Scholar]
- 20.Garlan F, et al. Early Evaluation of Circulating Tumor DNA as Marker of Therapeutic Efficacy in Metastatic Colorectal Cancer Patients (PLACOL Study) Clin Cancer Res. 2017;23:5416–5425. doi: 10.1158/1078-0432.CCR-16-3155. [DOI] [PubMed] [Google Scholar]
- 21.Shu Y, et al. Circulating Tumor DNA Mutation Profiling by Targeted Next Generation Sequencing Provides Guidance for Personalized Treatments in Multiple Cancer Types. Sci Rep. 2017;7:583. doi: 10.1038/s41598-017-00520-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murtaza M, et al. Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature. 2013;497:108–112. doi: 10.1038/nature12065. [DOI] [PubMed] [Google Scholar]
- 23.Phallen Jillian, Sausen Mark, Adleff Vilmos, Leal Alessandro, Hruban Carolyn, White James, Anagnostou Valsamo, Fiksel Jacob, Cristiano Stephen, Papp Eniko, Speir Savannah, Reinert Thomas, Orntoft Mai-Britt Worm, Woodward Brian D., Murphy Derek, Parpart-Li Sonya, Riley David, Nesselbush Monica, Sengamalay Naomi, Georgiadis Andrew, Li Qing Kay, Madsen Mogens Rørbæk, Mortensen Frank Viborg, Huiskens Joost, Punt Cornelis, van Grieken Nicole, Fijneman Remond, Meijer Gerrit, Husain Hatim, Scharpf Robert B., Diaz Luis A., Jones Siân, Angiuoli Sam, Ørntoft Torben, Nielsen Hans Jørgen, Andersen Claus Lindbjerg, Velculescu Victor E. Direct detection of early-stage cancers using circulating tumor DNA. Science Translational Medicine. 2017;9(403):eaan2415. doi: 10.1126/scitranslmed.aan2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Taly V, et al. Multiplex picodroplet digital PCR to detect KRAS mutations in circulating DNA from the plasma of colorectal cancer patients. Clin Chem. 2013;59:1722–1731. doi: 10.1373/clinchem.2013.206359. [DOI] [PubMed] [Google Scholar]
- 25.Jackson JB, et al. Multiplex Preamplification of Serum DNA to Facilitate Reliable Detection of Extremely Rare Cancer Mutations in Circulating DNA by Digital PCR. J Mol Diagn. 2016;18:235–243. doi: 10.1016/j.jmoldx.2015.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pallisgaard N, Spindler KL, Andersen RF, Brandslund I, Jakobsen A. Controls to validate plasma samples for cell free DNA quantification. Clin Chim Acta. 2015;446:141–146. doi: 10.1016/j.cca.2015.04.015. [DOI] [PubMed] [Google Scholar]
- 27.Huggett JF, et al. The digital MIQE guidelines: Minimum Information for Publication of Quantitative Digital PCR Experiments. Clin Chem. 2013;59:892–902. doi: 10.1373/clinchem.2013.206375. [DOI] [PubMed] [Google Scholar]
- 28.Untergasser A, et al. Primer3–new capabilities and interfaces. Nucleic Acids Res. 2012;40:e115. doi: 10.1093/nar/gks596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Milbury CA, et al. Determining lower limits of detection of digital PCR assays for cancer-related gene mutations. Biomol Detect Quantif. 2014;1:8–22. doi: 10.1016/j.bdq.2014.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cheng DT, et al. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): A Hybridization Capture-Based Next-Generation Sequencing Clinical Assay for Solid Tumor Molecular Oncology. J Mol Diagn. 2015;17:251–264. doi: 10.1016/j.jmoldx.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hyman DM, et al. Precision medicine at Memorial Sloan Kettering Cancer Center: clinical next-generation sequencing enabling next-generation targeted therapy trials. Drug Discov Today. 2015;20:1422–1428. doi: 10.1016/j.drudis.2015.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McKenna A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.GATK. Mutect2.
- 35.Koboldt DC, et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012;22:568–576. doi: 10.1101/gr.129684.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Network, T. R. http://cancergenome.nih.gov/.
- 37.Giannakis M, et al. Genomic Correlates of Immune-Cell Infiltrates in Colorectal Carcinoma. Cell Rep. 2016;15:857–865. doi: 10.1016/j.celrep.2016.03.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Christensen E, et al. Optimized targeted sequencing of cell-free plasma DNA from bladder cancer patients. Sci Rep. 2018;8:1917. doi: 10.1038/s41598-018-20282-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smith T, Heger A, Sudbery I. UMI-tools: modeling sequencing errors in Unique Molecular Identifiers to improve quantification accuracy. Genome Res. 2017;27:491–499. doi: 10.1101/gr.209601.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.R_Core_Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria (2013).
- 41.Jr FE. H. rms: Regression Modeling Strategies. R package version. 2017;5:1–1. [Google Scholar]
- 42.T, T. A Package for Survival Analysis in S_. version 2.38. (2015).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data from findings of this study are available from the corresponding author upon reasonable request.