Abstract
Advances in the molecular characteristics of cancers have facilitated the classification system from morphology to molecular characteristic-based subtypes. Cancer profiling has expanded in its focus from protein-coding genes to noncoding RNAs, with advances in the depth and quality of transcriptome sequencing. Here, we examined the profiles of long noncoding RNAs (lncRNAs) according to breast cancer subtype categories in The Cancer Genome Atlas (TCGA) database to identify a cohort of breast cancer- and oestrogen receptor (ER)-negative-associated lncRNAs. According to the prioritization of variation in ER-negative-associated lncRNAs, we identified and investigated the role of LINC00511 in breast cancer. We determined that high LINC00511 expression was an unfavourable prognostic factor for patients with breast cancer. Furthermore, LINC00511 promoted tumour growth by accelerating the G1/S transition and inhibiting apoptosis. At the transcriptional level, ER deficiency directly affected the expression of LINC00511 activated by transcription factor AP-2 (TFAP-2) in breast cancer cells. Moreover, mechanistic investigations demonstrated that ER-negative-associated LINC00511 interacted with enhancer of zeste homologue 2 (EZH2, the catalytic subunit of polycomb repressive complex 2, PRC2) and recruited PRC2 to mediate histone methylation, contributing to the repression of CDKN1B in the nucleus. This process resulted in altered ER-negative breast cancer cell biology. By highlighting the oncogenic function of LINC00511, we revealed the role of lncRNAs in regulating the network of cell cycle control in ER-negative breast cancer and suggested the exploitation of LINC00511 as an anticancer therapy in the future.
Subject terms: Breast cancer, Long non-coding RNAs
Introduction
Breast cancer is one of the most commonly diagnosed cancers and the leading cause of death in women worldwide1. With advances in the recognition of the molecular characteristics of cancers, breast cancer has been categorized into four major subtypes: luminal A, luminal B, human epidermal growth factor receptor 2 (HER2)-positive and basal-like2,3. Among the major molecular characteristics of breast cancer, oestrogen receptor (ER) is crucial to the classification of breast cancer subtypes and tailored individualized therapy4,5. Aberrant ER transcriptional activity is involved in the endocrine response and cell cycle progression6,7. Patients with ER-negative breast cancer tend to have a poorer prognosis than those with ER-positive breast cancer8. However, the network of underlying regulatory mechanisms in the initiation and progression of ER-negative breast cancer remains poorly understood.
Long noncoding RNAs (lncRNAs) are heterogeneous categories of transcripts more than 200 base pairs in length. Because they lack the ability to translate into proteins, they were once considered “transcriptional noise”9,10. In recent years, the “transcriptional noise” has no longer been considered irrelevant, and lncRNAs were confirmed to be involved in multiple pathological conditions such as carcinogenesis and cancer progression11–13. In our previous study, we reported that the lncRNA EGOT, a downregulated lncRNA in cancer tissues, enhanced cellular autophagosome accumulation and sensitized cells to paclitaxel in breast cancer9,14. With advances in scientific research, the old, traditional dogmas of prominent RNA functions were overthrown and a supplement or modification to traditional understanding was gradually established10,13,15.
Our cognition of cancer is an evolving process. Douglas Hanahan and Robert A. Weinberg provided an excellent overview summarizing ten hallmarks of cancer in 201116. Among these hallmarks, epigenetic regulation, identified as a heritable variation, plays a significant role in tumourigenesis. Aberrant epigenetic regulation contributes to the chaos of gene expression, chromatin organization, cell differentiation, etc17,18. Some lncRNAs have been reported to serve as epigenetic regulators influencing the expression of target genes, such as MALAT1, PVT1 and HOTAIR18–21. Moreover, sustained cell proliferation, which ensures the continuity of heritable variations in cancer, such as genetic modifications, was identified as another important hallmark of cancer. Uncontrolled cell proliferation results from defects in the control of the cell cycle, particularly at the G1/S transition22. To date, some lncRNAs have been confirmed to regulate the G1/S transition to affect tumour growth22–24. Often, the hallmarks of cancer are interwoven rather than isolated, similar to the regulatory mechanisms of lncRNAs in cancers. Some lncRNAs, such as SNHG1, have been reported to epigenetically repress the expression of cyclin-dependent kinase inhibitors (CDKIs) to promote the cell cycle25.
Here, we repurposed and integrated multi-RNA-sequencing (RNA-seq) analyses of lncRNA expression profiles among entries in the breast tissue cohort from The Cancer Genome Atlas (TCGA) database to identify the lncRNAs that are enriched in both breast cancer (cancer versus normal) and ER-negative breast cancer (ER-negative versus ER-positive)11,13. We identified and investigated the role of LINC00511 in breast cancer according to the prioritization of variation in ER-negative-associated lncRNAs. In previous studies, LINC00511 was reported to exert an oncogenic function in many cancers, such as breast cancer, non-small cell lung cancer, ovarian cancer and glioma26–29. We found that LINC00511 promoted tumour growth by accelerating the G1/S transition and inhibiting apoptosis in breast cancer. We demonstrated that ER deficiency directly affected the expression of LINC00511 activated by TFAP-2 and identified EZH2, which is involved in histone methylation, as a protein that interacts with LINC00511 and contributes to the repression of CDKN1B in ER-negative breast cancer. We aimed to provide detailed knowledge of LINC00511 in breast cancer progression. By integrating these data with the TCGA clinical data, we aimed to identify and validate the carcinogenic mechanism of LINC00511 in ER-negative breast cancer. In the future, it may be exploited for anticancer therapy.
Results
Identification of breast cancer- and ER-negative-associated lncRNAs
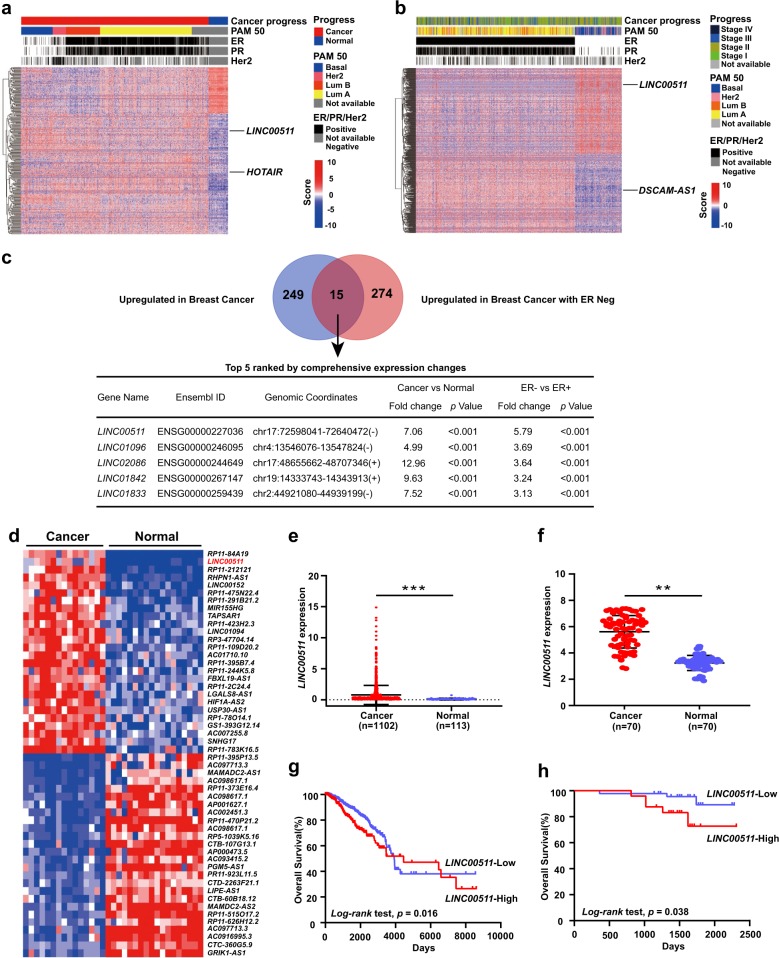
Utilizing the large-scale cancer genome RNA-seq expression data from the TCGA database, we focused on the potential carcinogenesis of lncRNAs that were differentially expressed in breast cancer tissues compared with adjacent normal tissues. We identified 264 upregulated lncRNAs in breast cancer tissues (Fig. 1a). HOTAIR, a well-known lncRNA that promotes breast cancer aggression, was included30. We also identified differentially expressed lncRNAs in ER-positive breast cancer tissues compared with ER-negative tissues within the lncRNA expression profiles in cancer tissues. We identified 289 upregulated lncRNAs in ER-negative breast cancer tissues (Fig. 1b). Among these lncRNAs, DSCAM-AS1 was affirmed to be responsible for the oncogenicity in ER-positive breast cancer11. Because of a poor prognosis encountered by patients with ER-negative breast cancer (relative to patients with ER-positive breast cancer), we investigated the potential oncogenic function of ER-negative-associated lncRNAs that were upregulated in both breast cancer (cancer versus normal) (Fig. 1a) and ER-negative breast cancer (ER-negative versus ER-positive) (Fig. 1b). We examined the intersection of these two lncRNA profiles, and 15 lncRNAs were upregulated in both the cancer versus normal analysis and in the ER-negative versus ER-positive analysis, as shown in the Venn diagram (Fig. 1c and Table S1). According to the prioritization of variation in ER-negative-associated lncRNAs, we selected LINC00511 for further investigation.
Fig. 1. Identification of breast cancer- and ER-negative-associated lncRNAs.
a Hierarchical clustering heat map of the differentially expressed lncRNAs in breast cancer tissues and adjacent normal tissues generated from RNA-seq data from the TCGA database. A total of 342 lncRNAs were differentially expressed, with a p value < 1E-20 and a fold change >2. Among these lncRNAs, 264 were upregulated in breast cancer tissues, and 78 were upregulated in adjacent normal tissues. The cancer progression, PAM50, ER, PR and HER2 statuses of each sample are shown above the heat map. HOTAIR was employed as a positive control. b Hierarchical clustering heat map of the differentially expressed lncRNAs in ER-positive and ER-negative breast cancer tissues generated from RNA-seq data from the TCGA database. A total of 551 lncRNAs were differentially expressed, with a p value < 1E-4 and a fold change >2. Among these lncRNAs, 262 were upregulated in ER-positive breast cancer tissues, and 289 were upregulated in ER-negative breast cancer tissues. The cancer progression, PAM50, ER, PR and HER2 statuses of each sample are shown above the heat map. DSCAM-AS1 was employed as a positive control. c Venn diagram depiction of the intersection of the lncRNAs upregulated in both breast cancer and ER-negative breast cancer tissues (264 versus 289, respectively). The top five lncRNAs are listed according to the prioritization of variation in ER-negative-associated lncRNAs. d Heat map of the differentially expressed lncRNAs in breast cancer tissues, adjacent normal tissues and non-breast cancer patient tissues generated from RNA-seq data of the whole-transcriptome sequencing of 33 breast cancer specimens (RNA-seq data of this study has been deposited into the NCBI GEO database under accession number GSE71651). e Scatter diagram depiction of LINC00511 expression in breast cancer tissues and adjacent normal tissues generated from the TCGA database. f qRT-PCR analysis of LINC00511 expression in 70 pairs of breast cancer and adjacent normal tissues. g Kaplan–Meier survival analysis of OS in breast cancer patients based on LINC00511 expression generated from the TCGA database (n = 1086, high = 379, low = 707, p < 0.05). h Kaplan–Meier survival analysis of OS in breast cancer patients based on LINC00511 expression in our cohort (n = 70, high = 24, low = 46, p < 0.05). Data are shown as the mean ± standard deviation (SD). Student’s t test was used for the statistical analysis: *p < 0.05; **p < 0.01; ***p < 0.001. Data represent three independent experiments
We also examined the protein-coding potential of LINC00511 through the CPAT database (http://lilab.research.bcm.edu/cpat/) and the CPC database (http://cpc.cbi.pku.edu.cn/) (Fig. S1a, b). Moreover, an RNA pull-down assay showed no interaction/binding between LINC00511 and ribosomal protein LP0, a component of the 60S ribosomal subunit (Fig. S1c)31,32. These data suggest that LINC00511 has no protein-coding potential.
LINC00511 expression is increased in breast cancer and correlates with a poor prognosis
The whole-transcriptome sequencing of 33 breast cancer specimens, including breast cancer tissue (N = 15), adjacent normal tissue (N = 15) and non-breast cancer patient tissue (N = 3), showed that LINC00511 was highly expressed in cancer tissues compared with adjacent normal tissues (Fig. 1d). The expression of LINC00511 was further validated by analysing 70 pairs of breast cancer and adjacent normal tissues from the Harbin Medical University Cancer Centre (HMUCC) and RNA-seq data from the TCGA database (Fig. 1e, f). To explore the relationship between LINC00511 expression and the prognosis of breast cancer patients, we performed a Kaplan–Meier analysis and a log-rank test to assess the effects of LINC00511 expression and clinical outcomes on overall survival in the TCGA database and 70 paired breast cancer tissues, respectively. As shown in Fig. 1g, h, high LINC00511 expression indicated a remarkably poorer prognosis than low LINC00511 expression did in patients. Next, we examined the correlation of LINC00511 expression with patients’ clinicopathological characteristics in breast cancer. LINC00511 expression was positively correlated with tumour size, ER, progesterone receptor (PR), Ki-67 and p53 status (p < 0.05). No significant association was found between LINC00511 expression and age, tumour-node-metastasis (TNM) stages, histological grade, lymph-node-metastasis (LNM) or HER2 status (Table 1).
Table 1.
Correlation between LINC00511 expression and clinicopathological characteristics of breast cancer patients
| Clinical parameter test p-value | No. cases (n = 70) | LINC00511 expression | Chi-squared | |
|---|---|---|---|---|
| High cases | Low cases | |||
| Age (years) | 0.75 | |||
| ≤50 | 39 | 14 | 25 | |
| >50 | 31 | 10 | 21 | |
| Tumour size (cm) | 0.03 | |||
| ≤2 | 33 | 7 | 26 | |
| >2 | 37 | 17 | 20 | |
| TNM stages | 0.08 | |||
| I–II | 65 | 20 | 45 | |
| III–IV | 5 | 4 | 1 | |
| Histological grade | 0.77 | |||
| I–II | 51 | 18 | 33 | |
| III | 19 | 6 | 13 | |
| LNM | 0.74 | |||
| Positive | 36 | 13 | 23 | |
| Negative | 34 | 11 | 23 | |
| ER status | <0.01 | |||
| Positive | 19 | 1 | 18 | |
| Negative | 51 | 23 | 28 | |
| PR status | 0.04 | |||
| Positive | 14 | 1 | 13 | |
| Negative | 56 | 23 | 33 | |
| Her2 status | 0.06 | |||
| Positive | 37 | 9 | 28 | |
| Negative | 33 | 15 | 18 | |
| Ki-67 status | 0.02 | |||
| ≤14% | 54 | 23 | 31 | |
| >14% | 16 | 1 | 15 | |
| P53 status | <0.01 | |||
| Positive | 29 | 16 | 13 | |
| Negative | 41 | 8 | 33 | |
LNM lymph node metastasis
Characteristics of LINC00511 in ER-negative breast cancer
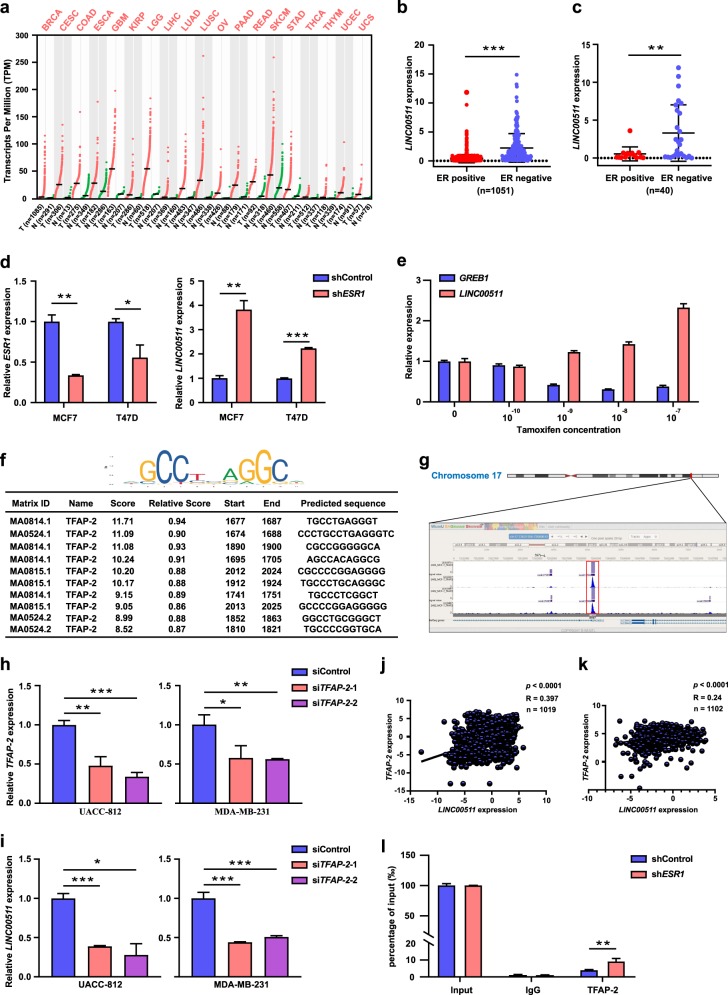
In addition to breast cancer, LINC00511 exhibited a highly cancer-specific expression pattern in multiple cancers according to an analysis of the Gene Expression Profiling Interactive Analysis database (http://gepia.cancer-pku.cn/) (Fig. 2a). As shown in Fig. 2b, c, LINC00511 expression was upregulated in the ER-negative breast cancer cohort from the TCGA database and in ER-negative breast cancer cells from the Cancer Cell Line Encyclopedia (CCLE) database (http://portals.broadinstitute.org/ccle/about). To investigate the relationship between LINC00511 and ER deficiency, we knocked down ER expression in MCF7 and T47D cells and found that LINC00511 expression was upregulated (Fig. 2d). Furthermore, LINC00511 expression was induced in MCF7 cells after stimulation with the anti-oestrogen agent tamoxifen, and tamoxifen induced LINC00511 expression in a dose-dependent manner (Fig. 2e). We deduced that certain regulatory mechanisms involved in LINC00511 expression may be involved when blocking ER signalling.
Fig. 2. Characteristics of LINC00511 in ER-negative breast cancer.
a Scatter diagram depiction of LINC00511 expression in multiple cancer tissues and adjacent normal tissues generated from the GEPIA database. b Scatter diagram depiction of LINC00511 expression in ER-positive and ER-negative breast cancer tissues generated from the TCGA database. c Scatter diagram depiction of LINC00511 expression in ER-positive and ER-negative breast cancer cells generated from the CCLE database. d qRT-PCR analysis of LINC00511 expression following the knockdown of ESR1 expression in MCF7 and T47D cell lines. e qRT-PCR analysis of LINC00511 expression in MCF7 cells treated with different doses of tamoxifen. f The upper corner of the picture showed the TFAP-2 binding motif, and the lower table showed the prediction of TFAP-2 binding sites within the promoter region of LINC00511 provided by the JASPAR database. g Analysis of TFAP-2 ChIP-seq data from MCF7 cells at the LINC00511 promoter locus generated from data from the ENCODE and Cistrome Data Browser databases. h qRT-PCR analysis of the knockdown efficiency of TFAP-2 expression in UACC-812 and MDA-MB-231 cells. i qRT-PCR analysis of LINC00511 expression in UACC-812 and MDA-MB-231 cell lines following the knockdown of TFAP-2 expression. j Scatter diagram analysis of the correlation between TFAP-2 and LINC00511 generated from the CCLE database. k Scatter diagram analysis of the correlation between TFAP-2 and LINC00511 generated from the TCGA database. l ChIP and qRT-PCR analysis of TFAP-2 occupancy at the LINC00511 promoter region and the altered occupancy efficiency following the knockdown of ER expression in MCF7 cells. IgG was used as a negative control. Enrichment was quantified relative to input controls. Data are shown as the mean ± SD. Student’s t test was used for the statistical analysis: *p < 0.05; **p < 0.01; ***p < 0.001. Data represent three independent experiments
We utilized bioinformatics to analyse the potential regulation of LINC00511 expression. Using the University of California at Santa Cruz (UCSC) database (http://genome.ucsc.edu/cgi-bin/hgGateway), we obtained the location information around the transcriptional start site (TSS, −2000 bases to +200 bases, “−” indicates “upstream of the TSS” and “+” indicates “downstream of the TSS”). Then, we used the JASPAR database (http://jaspar.genereg.net/) to scan this segment and performed a promoter binding analysis. We predicted that LINC00511 was the potential target gene of TFAP-2 (Fig. 2f). We validated this interaction in the ENCODE database (https://www.encodeproject.org/) and in the Cistrome Data Browser database (http://cistrome.org/db/#/) (Fig. 2g). We ascertained the basic expression levels of LINC00511 in breast cancer cells through qRT-PCR assays, and UACC-812 and MDA-MB-231 cells, which express higher levels of LINC00511 than other breast cancer cells, were selected for subsequent investigation (Fig. S2). We knocked down TFAP-2 expression in UACC-812 and MDA-MB-231 cells and observed that LINC00511 expression was impaired (Fig. 2h, i). We found that LINC00511 expression was positively correlated with TFAP-2 in cancer cells and breast cancer tissues according to the CCLE and TCGA databases (Fig. 2j, k). The occupancy of TFAP-2 at the promoter region of LINC00511 was also confirmed through chromatin immunoprecipitation (ChIP) assays (Fig. 2l). These results illustrated that TFAP-2 promoted LINC00511 expression at the transcriptional level. Next, we wanted to determine whether there was a correlation between ER deficiency and TFAP-2 activity in regulating LINC00511 expression. ChIP assays confirmed that the occupancy efficiency of TFAP-2 at the specific promoter region was enhanced after knocking down ER expression in MCF7 cells (Fig. 2l). These data indicated that ER deficiency promoted LINC00511 expression by enhancing the occupancy efficiency of TFAP-2 at specific promoter regions, resulting in increased transcriptional activity.
LINC00511 impacts ER-negative breast cancer cell proliferation and apoptosis in vitro
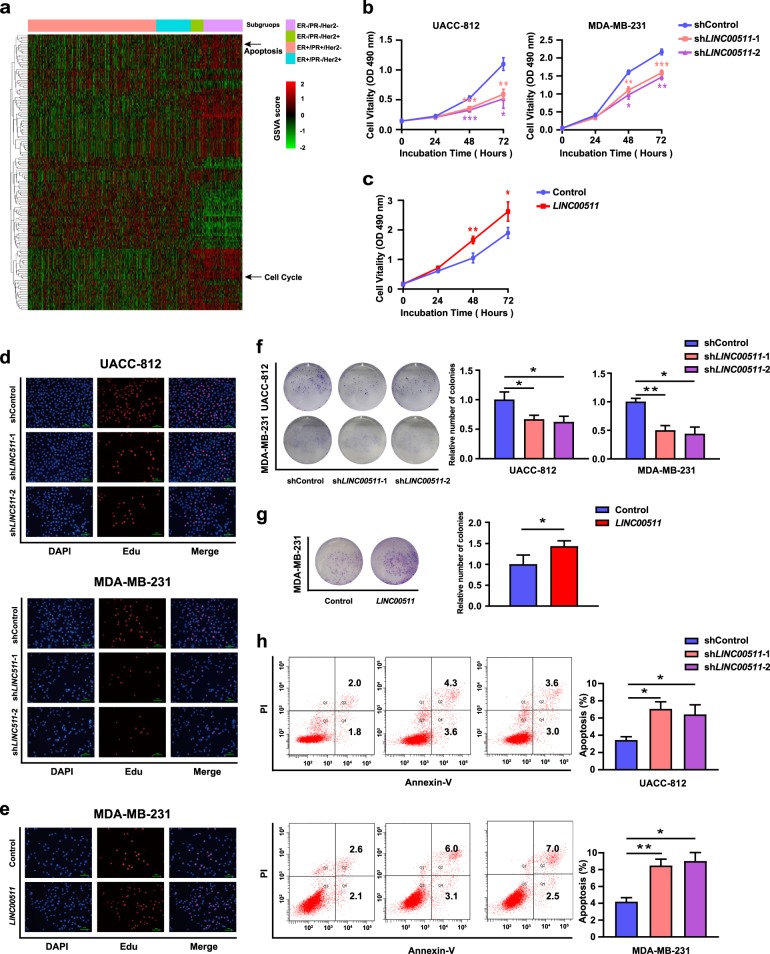
To gain insight into the functional role of LINC00511 in ER-negative breast cancer, we performed a gene set variation analysis (GSVA) to determine whether the cancer phenotypes regulated by LINC00511 were shared across breast cancer patients from the TCGA database33 (Fig. 3a). We focused on a set of cancer-specific signatures that are shown in Table S2. Among these cancer-specific signatures, we focused our attention on cell proliferation and cell apoptosis pathways, which were enriched more in ER-negative breast cancer patients than in ER-positive breast cancer patients. LINC00511 expression was knocked down in UACC-812 and MDA-MB-231 cells and overexpressed in MDA-MB-231 cells, as shown in Fig. S3a–c. Cell Counting Kit-8 assays showed that the knockdown of LINC00511 expression significantly inhibited the viability and proliferation of UACC-812 and MDA-MB-231 cells relative to shcontrol cells, while the overexpression of LINC00511 substantially promoted the viability and proliferation of MDA-MB-231 cells (Fig. 3b, c). Ethynyldeoxyuridine (EdU) assays also supported the conclusions in UACC-812 and MDA-MB-231 cells (Fig. 3d, e). The results of colony-forming growth assays revealed that the knockdown of LINC00511 expression greatly attenuated the colony-forming ability of UACC-812 and MDA-MB-231 cells, but the overexpression of LINC00511 enhanced the colony-forming ability of MDA-MB-231 cells (Fig. 3f, g). Furthermore, flow cytometry (cell apoptosis) analysis demonstrated a significantly increased proportion of apoptotic cells following the knockdown of LINC00511 expression (Fig. 3h). Taken together, these results indicated that LINC00511 promoted ER-negative breast cancer cell growth by affecting cell proliferation and apoptosis.
Fig. 3. LINC00511 impacts ER-negative breast cancer cell proliferation and apoptosis in vitro.
a Cancer gene signature analyses and clustering of GSVA scores for LINC00511 in breast cancer patients obtained from the TCGA database. b CCK-8 analysis of the viability and proliferation of UACC-812 and MDA-MB-231 cells following the knockdown of LINC00511 expression. c CCK-8 analysis of the viability and proliferation of MDA-MB-231 cells following the overexpression of LINC00511 expression. d EdU analysis of the proliferation ability of UACC-812 and MDA-MB-231 cells following the knockdown of LINC00511 expression. e EdU analysis of the proliferation ability of MDA-MB-231 cells following the overexpression of LINC00511 expression. f Analysis of the cell colony formation ability of UACC-812 and MDA-MB-231 cells following the knockdown of LINC00511 expression. The number of colonies was counted on the 14th day after seeding. g Analysis of the cell colony formation ability of MDA-MB-231 cells following the overexpression of LINC00511 expression. The number of colonies was counted on the 14th day after seeding. h Flow cytometry (cell apoptosis) analysis of the apoptosis of UACC-812 and MDA-MB-231 cells following the knockdown of LINC0051 expression. Data are shown as the mean ± SD. Student’s t test was used for the statistical analysis: *p < 0.05; **p < 0.01; ***p < 0.001. Data represent three independent experiments
LINC00511 accelerates the G1/S transition, in part, by regulating CDKN1B expression
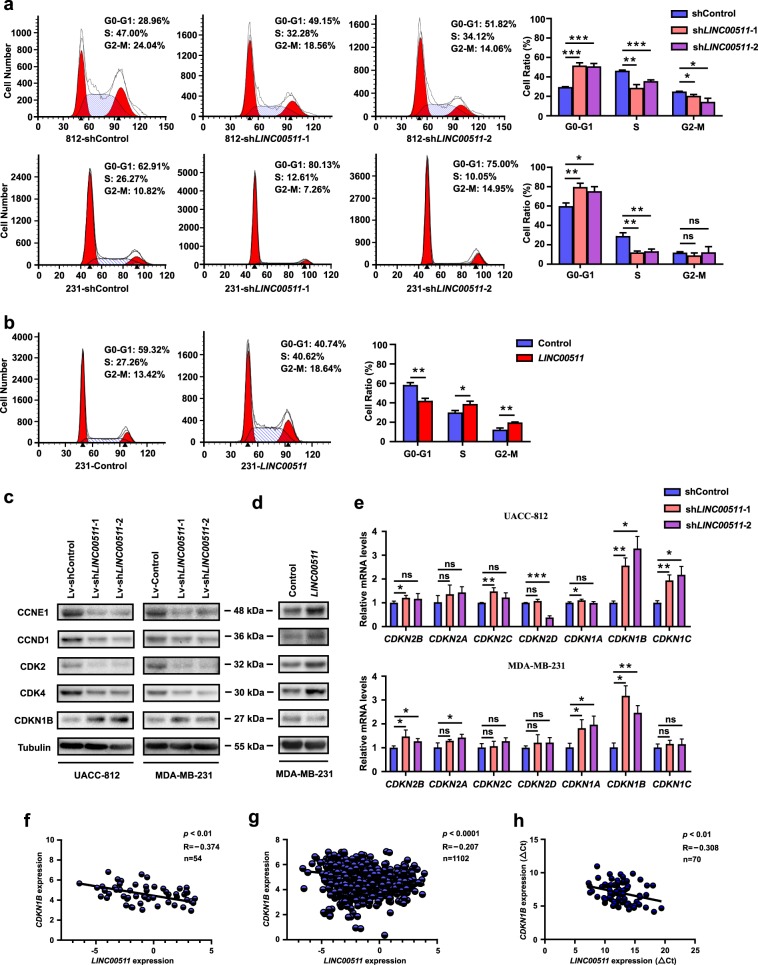
To investigate the specific regulatory mechanisms of LINC00511 in promoting cell proliferation, flow cytometry (cell cycle distribution) analysis revealed that the knockdown of LINC00511 expression significantly decreased the proportion of UACC-812 and MDA-MB-231 cells in S phase. In contrast, the overexpression of LINC00511 increased the proportion of MDA-MB-231 cells in S phase (Fig. 4a, b). These results indicate that LINC00511 may play an important role in the G1/S transition of the cell cycle. We also detected key cell cycle-related proteins that play irreplaceable roles in G1 phase. The results revealed that CCNE1, CCND1, CDK4 and CDK2 expression was impaired in UACC-812 and MDA-MB-231 cells following the knockdown of LINC00511 expression, and the overexpression of LINC00511 increased the expression of these proteins in MDA-MB-231 cells (Fig. 4c, d).
Fig. 4. LINC00511 accelerates the G1/S transition, in part, by regulating CDKN1B expression.
a Flow cytometry (cell cycle distribution) analysis of the proportion of UACC-812 and MDA-MB-231 cells in the G0/G1, S and G2/M phases following the knockdown of LINC00511 expression. b Flow cytometry (cell cycle distribution) analysis of the proportion of MDA-MB-231 cells in the G0/G1, S and G2/M phases following the overexpression of LINC00511 expression. c Western blot analysis of the key cell cycle-related proteins (CCNE1, CCND1, CDK4, CDK2 and CDKN1B) of UACC-812 and MDA-MB-231 cells following the knockdown of LINC00511 expression. d Western blot analysis of the key cell cycle-related proteins (CCNE1, CCND1, CDK4, CDK2 and CDKN1B) of MDA-MB-231 cells following the overexpression of LINC00511 expression. e qRT-PCR analysis of CDKI mRNAs (the INK4 and Cip/Kip families) of UACC-812 and MDA-MB-231 cells following the knockdown of LINC00511 expression. f Scatter diagram analysis of the correlation between LINC00511 and CDKN1B expression obtained from the CCLE database. g Scatter diagram analysis of the correlation between LINC00511 and CDKN1B expression generated from the TCGA database. h Scatter diagram analysis of the correlation between LINC00511 and CDKN1B expression in 70 breast cancer tissues in our cohort. Data are shown as the mean ± SD. Student’s t test was used for the statistical analysis: *p < 0.05; **p < 0.01; ***p < 0.001. Data represent three independent experiments
CDKIs, which are classified into the INK4 and Cip/Kip families, act as brakes to halt cell cycle progression34. We suspected that LINC00511 played a role through CDKIs. qRT-PCR assays were performed to examine the correlation between LINC00511 and CDKI mRNAs. CDKN1B was the only mRNA that was increased in both UACC-812 and MDA-MB-231 cells following the knockdown of LINC00511 expression (Fig. 4e). Furthermore, the observation was validated at the protein level in UACC-812 and MDA-MB-231 cells following the knockdown or overexpression of LINC00511 expression (Fig. 4c, d). Some lncRNAs are involved in the regulation of protein synthesis or protein stability in mammalian cells9. We found that neither CDKN1B protein synthesis nor stability was affected using the protein synthesis inhibitor cycloheximide (CHX) and the proteasome inhibitor MG132 following the knockdown or overexpression of LINC00511 expression (Fig. S4a–c). After reviewing the public databases, we aimed to confirm the correlation between LINC00511 and CDKN1B by analysing the RNA-seq data in the CCLE and TCGA databases. The results revealed a negative correlation between LINC00511 and CDKN1B (Fig. 4f, g). The correlation was further validated in 70 breast cancer tissues in the HMUCC cohort (Fig. 4h). The above results suggested that LINC00511 contributed to breast cancer cell growth by accelerating the G1/S transition in the cell cycle, in part, by regulating CDKN1B expression at transcriptional level.
LINC00511 is required for the epigenetic repression of CDKN1B by interacting with EZH2
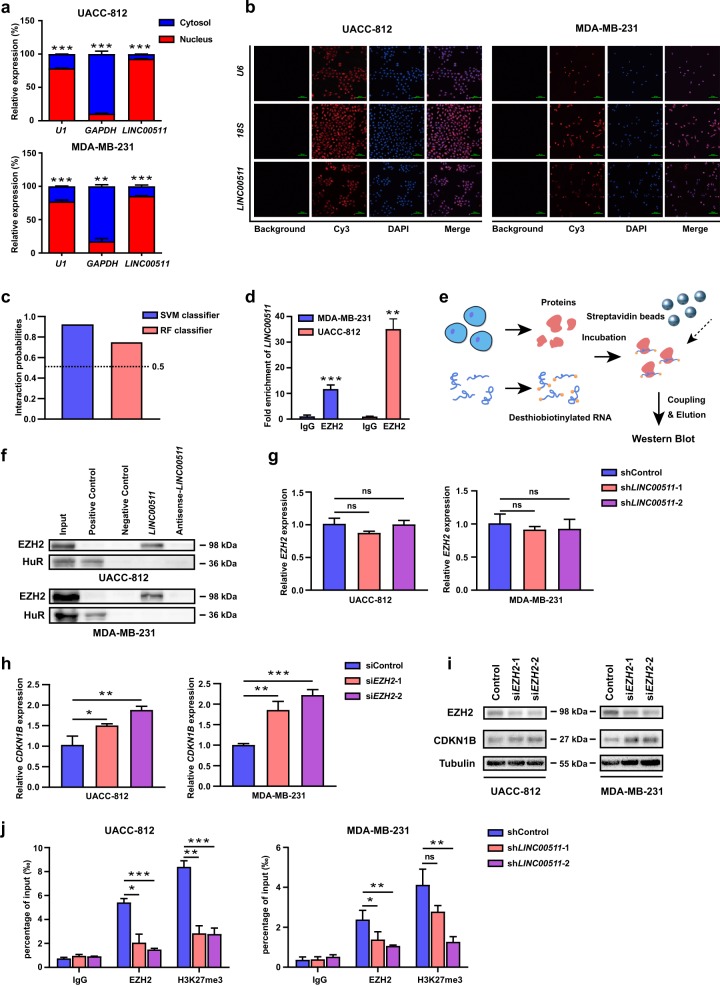
To explore the specific mechanisms of LINC00511, we conducted a fractionated nuclear and cytoplasmic RNA analysis and RNA fluorescence in situ hybridization (RNA-FISH) assays to ensure the subcellular location of LINC00511 in UACC-812 and MDA-MB-231 cells. LINC00511 was mainly located in the nucleus (Fig. 5a, b). Previous studies have demonstrated that some lncRNAs in the nucleus recruit the PRC2 complex for epigenetic regulation19,35. The methyltransferase EZH2 is the core subunit of the PRC2 complex, and several RNA immunoprecipitation (RIP) experiments have shown a massive number of lncRNAs that bind to EZH236. Thus, we predicted the interaction probabilities between LINC00511 and EZH2 with the protein-RNA prediction software RPISeq (http://pridb.gdcb.iastate.edu/RPISeq.html) (Fig. 5c). To verify our prediction, we performed RNA immunoprecipitation assays with EZH2 antibodies. The results revealed that LINC00511 could bind to EZH2 in UACC-812 and MDA-MB-231 cells (Fig. 5d). As shown in Fig. 5e, RNA pull-down assays also confirmed that LINC00511 could directly interact with EZH2 in UACC-812 and MDA-MB-231 cells (Fig. 5f).
Fig. 5. LINC00511 is required for the epigenetic repression of CDKN1B by interacting with EZH2.
a qRT-PCR analysis of the relative RNA expression levels after nuclear and cytoplasmic RNA separation. GAPDH was used as a cytoplasmic marker, and U1 was used as a nuclear marker. b Representative RNA-FISH images of the subcellular location of LINC00511 in UACC-812 and MDA-MB-231 cells (red). Nuclei were stained with DAPI (blue). 18S rRNA was used as a cytoplasmic marker, and U6 was used as a nuclear marker. c Analysis of the interaction probabilities of LINC00511 and EZH2 with the prediction software RPISeq. d RNA immunoprecipitation and qRT-PCR analysis of endogenous EZH2 binding to LINC00511 in UACC-812 and MDA-MB-231 cells with the anti-EZH2 antibody. IgG was used as the control. e Schematic of the RNA pull-down assays for the identification of LINC00511-associated proteins. f Western blot analysis of EZH2 following the pull-down of LINC00511 or antisense-LINC00511 in UACC-812 and MDA-MB-231 cells. HuR was used as a positive control. g qRT-PCR analysis of the EZH2 expression of UACC-812 cells and MDA-MB-231 cells following the knockdown of LINC00511 expression. h qRT-PCR analysis of the CDKN1B expression of UACC-812 and MDA-MB-231 cells following the knockdown of EZH2 expression. i Western blot analysis of the CDKN1B expression of UACC-812 and MDA-MB-231 cells following the knockdown of EZH2 expression. j ChIP and qRT-PCR analysis of EZH2 and H3K27me3 occupancy at the CDKN1B promoter region in UACC-812 and MDA-MB-231 cells following the knockdown of LINC00511 expression. IgG was used as a negative control. Enrichment was quantified relative to input controls. Data are shown as the mean ± SD. Student’s t test was used for the statistical analysis: *p < 0.05; **p < 0.01; ***p < 0.001. Data represent three independent experiments
Subsequently, we wanted to determine whether EZH2 was involved in the repression of CDKN1B. We found no effect on EZH2 expression following the knockdown of LINC00511 expression in UACC-812 and MDA-MB-231 cells (Fig. 5g). Simultaneously, the knockdown efficiency of EZH2 expression in UACC-812 and MDA-MB-231 cells was shown in Fig. S5a, b. qRT-PCR and western blot analyses confirmed that the expression of CKDN1B was increased following the knockdown of EZH2 expression in UACC-812 and MDA-MB-231 cells (Fig. 5h, i). So far, we hypothesized that LINC00511 regulated CDKN1B expression through EZH2-mediated H3K27me3 trimethylation at the promoter region of CDKN1B. As shown in Fig. 5j, ChIP assays were performed to confirm that the occupancy capacity of EZH2 and H3K27me3 at the specific promoter region of CDKN1B was impaired after knocking down LINC00511 in UACC-812 and MDA-MB-231 cells.
Knockdown of LINC00511 expression represses tumour growth in vivo
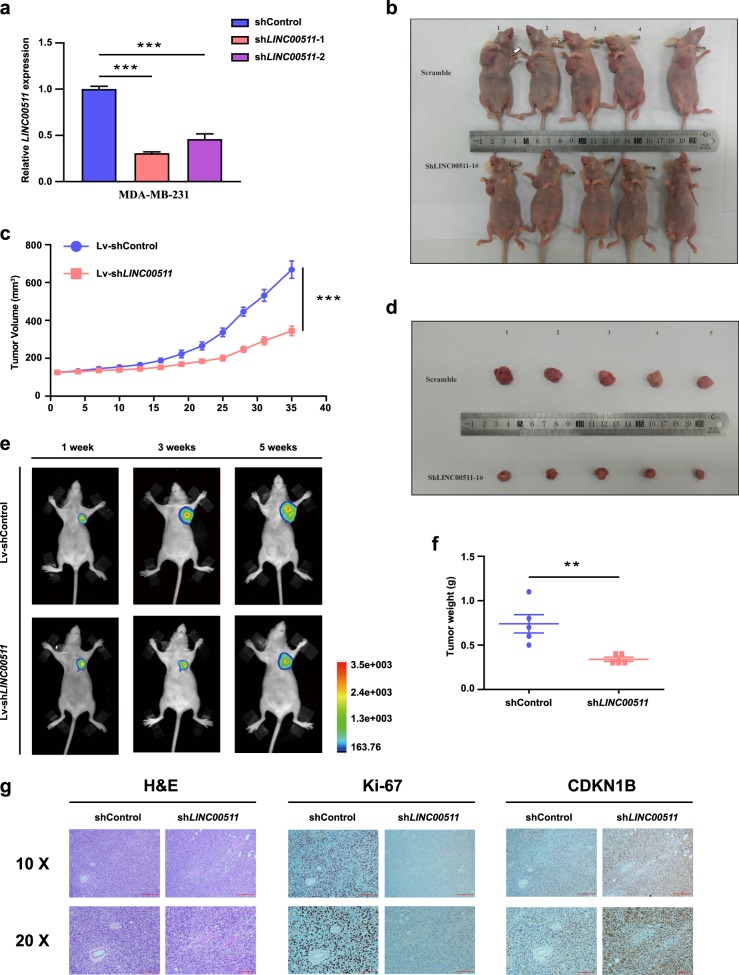
For the purpose of further confirming that LINC00511 affected tumour growth in vivo, luciferase-labelled MDA-MB-231 cells transfected with a shcontrol vector or a short hairpin RNA (shRNA) against LINC00511 (shLINC00511) were injected into nude mice. We chose luciferase-labelled MDA-MB-231 cells transfected with shLINC00511–1 as the experimental cohort because of the higher knockdown efficiency of LINC00511 expression in MDA-MB-231 cells, as shown in Fig. 6a. On the 35th day following injection, the tumours that developed in the shLINC00511 cohort were remarkably smaller than those that developed in the shcontrol cohort (Fig. 6b and d). Moreover, the average tumour volumes and respective weights were remarkably lower in the shLINC00511 cohort than the shcontrol cohort (Fig. 6c and f). Bioluminescent imaging was utilized to detect tumour growth dynamically every other week. As shown in Fig. 6e, the xenograft growth ability of MDA-MB-231 cells in the shLINC00511 cohort was weaker than that in the shcontrol cohort.
Fig. 6. Knockdown of LINC00511 expression represses tumour growth in vivo.
a qRT-PCR analysis of the knockdown efficiency of LINC00511 expression in luciferase-labelled MDA-MB-231 cells. b Representative images of nude mice bearing tumours from the shcontrol and shLINC00511 cohorts. c The growth curves of tumour volumes calculated every 3 days after injection in the shcontrol and shLINC00511 cohorts. d Representative images of tumours from the shcontrol and shLINC00511 cohorts. e Representative bioluminescent images of nude mice bearing tumours of luciferase-labelled MDA-MB-231 cells from the shcontrol and shLINC00511 cohorts. Image capture occurred during the 1st, 3rd and 5th weeks. f Scatter diagram depiction of the final tumour weights in shcontrol and shLINC00511 cohorts. g Representative images of haematoxylin and eosin (H&E) staining and Ki-67 and CDKN1B immunostaining of tumours from the shcontrol and shLINC00511 cohorts. Data are shown as the mean ± SD. Student’s t test was used for the statistical analysis: *p < 0.05; **p < 0.01; ***p < 0.001. Data represent three independent experiments
Moreover, the microscopic observation of tumours revealed that tumours that developed in the shcontrol cohort showed stronger Ki-67 expression than those in the shLINC00511 cohort, and the tumours that developed in the shLINC00511 cohort showed stronger CDKN1B expression than those that developed in the shcontrol cohort, as detected by immunohistochemistry (IHC) analysis (Fig. 6g). These results supported a role for LINC00511 in promoting breast cancer tumour growth in vivo.
Discussion
With the large population of breast cancer patients worldwide, the heterogeneity of breast cancer is still an obstacle in the assessment and treatment of breast cancer. The emergence of an evaluation system for molecular classification promoted the development of tailored individualized therapy37,38. Among the major molecular receptors, ER status separated breast cancer into distinct subgroups of which 70% express ER39. While ER, which is regarded as the decisive hormone receptor in breast cancer, is essential in the algorithm for treatment decision making, a poor prognosis can be observed in ER-negative patients40. Further investigation and study of the mechanisms through which ER-negative breast cancers become aggressive and eventually evade traditional therapy is of clinical importance.
In previous studies of ER-positive breast cancer, the activation of ER signalling was found to be involved in crosstalk with multiple signalling pathways, such as PI3K/AKT/mTOR or ER-CCND1-CDK4/6-RB, and to promote cell cycle progression41–43. Compared with ER-positive breast cancer, the specific oncogenic molecular and regulatory mechanisms remain poorly understood in ER-negative breast cancer. In this study, we identified several potential ER-negative-associated lncRNAs in breast cancer and mechanistically elaborated the oncogenic function of the one of the most intriguing candidates in ER-negative breast cancer. We firmly believed that the functional interpretation of other candidates will contribute to our understanding of ER-negative biology. lncRNAs function through multiple mechanisms, such as participating in epigenetic regulation, endogenous competition regulation and endogenous transport regulation44–46. According to the prioritization of variation in ER-negative-associated lncRNAs, we identified and investigated the oncogenic function of LINC00511 in ER-negative breast cancer. At the transcriptional level, ER deficiency directly affected the expression of LINC00511 activated by TFAP-2 in breast cancer cells. We determined that LINC00511 promotes tumour growth and inhibits apoptosis. The study of the interaction between LINC00511 and EZH2, the catalytic subunit of PRC2, was a crucial step towards understanding the mechanisms through which LINC00511 exerted its oncogenic function in breast cancer. Through the epigenetic silencing of CDKN1B, which acts as a brake to halt cell cycle progression, LINC00511 accelerated the G1/S transition to sustain cell proliferation (Fig. 7). Further investigation of LINC00511 in oncogenesis and cancer progression is necessary.
Fig. 7. Schematic illustration of the possible regulatory mechanisms in this study.
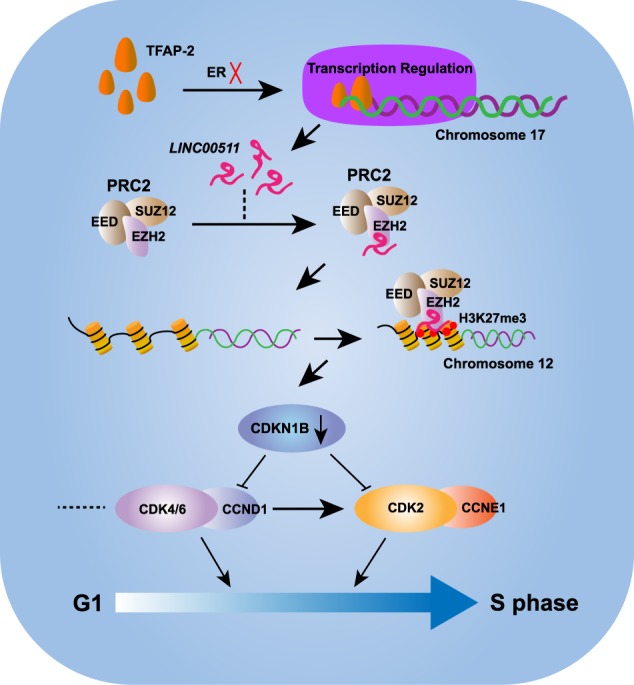
ER deficiency promoted TFAP-2-inducible LINC00511 expression by enhancing the occupancy efficiency of TFAP-2 at specific promoter regions. The ER-negative-associated LINC00511, which was mainly located in the nucleus, repressed the expression of CDKN1B via interacting with EZH2 to recruit PRC2 to mediate histone methylation, contributing to the G1/S transition to sustain cell proliferation
Along with the deeper understanding of molecule-mediated oncogenic mechanisms in ER-negative breast cancer, we expect improvements in treatment strategies in ER-negative breast cancer. It is necessary to prolong survival and alter the poor prognosis of ER-negative breast cancer patients. In recent years, small molecule compound drugs targeting novel tumour mediators have brought us hope, but the treatment of ER-negative breast cancer remains a great clinical challenge47–49. Ultimately, our study provides insight into the oncogenic function of LINC00511 in promoting ER-negative tumourigenesis, and further investigation of other candidates is likely to yield a greater understanding of ER-negative breast cancer biology. Moreover, these lncRNAs may be exploited as potential anticancer treatments in the future.
Materials and methods
Cell lines, cell culture and treatment
All the breast cancer cell lines were obtained from the Institute of Biochemistry and Cell Biology of the Chinese Academy of Sciences (Shanghai, China) and periodically authenticated (Cellbio). Unless otherwise specified, cells were maintained in Roswell Park Memorial Institute (RPMI) 1640 medium (Gibco, Carlsbad, CA, USA), Dulbecco’s modified Eagle’s medium (DMEM) plus GlutaMAX (Gibco, Carlsbad, CA, USA) or Leibovitz’s L15 medium (PYG0038, Boster, China) supplemented with 10% foetal bovine serum (FBS; 0500, ScienCell, USA) and 1% penicillin-streptomycin in a humidified incubator at 37 °C with 5% CO2 or air. For the ER deprivation experiments, MCF7 cells were treated with the ER antagonist tamoxifen (T5648–1G, Sigma, USA), for 24 h. For the ubiquitination assay, UACC-812 and MDA-MB-231 cells were treated with MG132 (HY-13259, MCE, USA) for 40 min. For the CHX chase assay, MDA-MB-231 cells were incubated with 50 μg/ml CHX (HY-12320, MCE, USA) for the indicated durations (0, 15, 30, 60, 120 and 180 min), as previously described9.
Knockdown and overexpression studies
The stable knockdown of LINC00511 and ESR1 was accomplished by lentiviral constructs containing two different LINC00511 and ESR1 shRNAs or no targeting shRNA (Umibio (Shanghai) Co.,Ltd, China) in the presence of polybrene (107689, Sigma). The transduced cells were cultured in culture media containing 1 µg ml−1 puromycin (Catalogue Number 540411, Calbiochem, USA) for 2 weeks. The knockdown of TFAP-2 and EZH2 was accomplished with a small interfering RNA (siRNA; HANBIO, China). The transfections were performed with INTERFERin® (Polyplus-transfection® SA) according to the manufacturer’s instructions. The target sequences used for the shRNAs or siRNAs are listed in Table S3.
The overexpression of LINC00511 was accomplished with a plasmid containing full-length LINC00511 cloned into the pcDNA3.1 vector between theEcoRI and KpnI sites (Umibio (Shanghai) Co.,Ltd, China). The transfections were performed with jetPRIME® (Polyplus-transfection® SA) according to the manufacturer’s instructions. Cells were collected at 48 h post transfection.
The knockdown and overexpressing cell lines were identified by a qRT-PCR assay or western blot analysis.
qRT-PCR assay
The E.Z.N.A.® Total RNA Kit I (Catalogue Number R6834–01, Omega Bio-Tek, USA) was utilized to isolate RNA from cell lysates. From 1 µg of isolated RNA, a Transcriptor First Strand cDNA Synthesis Kit and Random Primer (Catalogue Number 04897030001, Roche, USA) were used to generate cDNA according to the manufacturer’s protocol. The 7500 Fast Real-Time PCR system (Applied Biosystems, USA) was utilized for qRT-PCR. A relative quantification method was used to analyse the qRT-PCR data, and actin was used as a reference for the mRNAs or lncRNAs. Each sample was analysed in triplicate. The primer sequences synthesized by Shanghai Generay Biotech Co.,Ltd, are listed in Table S3.
Western blot assay and antibodies
Western blot assays were performed by running cell lysates on 8–10% SDS polyacrylamide gels (Solarbio) to separate proteins. The separated proteins were then transferred to a polyvinylidene fluoride membrane via wet transfer at 300 mA for 60–90 min. After incubating with blocking buffer (Becton Dickinson, USA) for 1 h, the indicated antibodies were added to the membrane and incubated at 4 °C overnight. The blots were incubated with goat anti-rabbit IgG H&L (HRP) or goat anti-mouse IgG H&L (HRP) for 1 h at room temperature. FluorChem HD2 (Protein Sample, USA) was used to detect the proteins via enhanced chemiluminescence. All antibodies used in this study are described in Table S4.
Subcellular fractionation
Cellular fractionation was performed using NE-PERTM Nuclear and Cytoplasmic Extraction Reagents (Catalogue Number 78835, Thermo Fisher) according to the manufacturer’s instructions. qRT-PCR was performed to detect the isolated RNA, with GAPDH and U1 used as the reference for cytoplasmic and nuclear RNA, respectively.
Fluorescence in situ hybridization
RNA-FISH was performed with a RiboTM Fluorescence In Situ Hybridization Immobilized Kit (Catalogue Number 10910, RiboBio Co., Ltd, China) according to the manufacturer’s instructions. LINC00511, U6 and 18S (referencesfor the nucleus and cytoplasm) hybridized with cy3 oligonucleotide probes were observed with a confocal laser scanning microscope (FV1200, Olympus, Japan).
Chromatin immunoprecipitation (ChIP)
An EZ-ChIPTM ChIP Kit (Catalogue Number #17–371, Millipore, USA) was utilized to perform ChIP assays according to the manufacturer’s instructions. Briefly, ∼2 × 107 shcontrol MCF7 cells or MCF7 cells in which the stable knockdown of ER was validated were used for each ChIP assay. Cells were crosslinked using 1% formaldehyde for 20 min, and cross-linking was quenched for 10 min at room temperature using a 1/10 volume of 1.25 M glycine. The cells treated with enzyme lysis and sonication yielded an average chromatin fragment size of 300–500 bp. The DNAs bound to the antibody against TFAP-2 (sc-12762, Santa Cruz, USA) via overnight incubation at 4 °C were purified with a Universal DNA Purification Kit (DP214, Tiangen, China) according to the manufacturer’s instructions. ChIP DNA was subjected to quantitative PCR and is reported as % input ± standard error of the mean (S.E.M.). The primers used for PCR are listed in Table S3.
RNA immunoprecipitation (RIP)
Magna RIPTM RNA-Binding Protein Immunoprecipitation Kits (Catalogue Number #17–700, Millipore, USA) were used according to the manufacturer’s instructions. The abundance of LINC00511 was detected by quantitative PCR using total RNA as an input control. The antibody used for RIP is listed in Table S4.
RNA pull-down assay
The T7 RiboMAXTM Express Large Scale RNA Production System (Catalogue Number P1320, Promega, USA) was used to produce abundant LINC00511, including sense and antisense RNAs, in vitro according to the manufacturer’s instructions. The Pierce™ RNA 3′ End Desthiobiotinylation Kit (Catalogue Number 20163, Thermo Fisher, USA) was applied to label the 3′ ends of the LINC00511 RNAs (including sense and antisense) with a desthiobiotin tag according to the manufacturer’s instructions. Protein-RNA interactions were determined using a Pierce™ Magnetic RNA-Protein Pull-Down Kit (Catalogue Number 20164, Thermo Fisher, USA) with lysates from MDA-MB-231 and UACC-812 cells. Then, western blot assays were used to detect the precipitated proteins.
Cell proliferation and colony formation assays
MDA-MB-231 and UACC-812 cell proliferation and colony formation abilities were assessed as previously described9.
EdU proliferation assay
A Cell-LightTM EdU Apollo567 In Vitro Kit (Catalogue Number C10310–1, RiboBio, China) was used to perform the EdU proliferation assay according to the manufacturer’s instructions as previously described50.
Apoptosis and cell cycle analysis
MDA-MB-231 and UACC-812 cells expressing the indicated constructs were treated with the Cell Cycle Staining Kit (Catalogue Number 70-CCS012, Multiscience, China) and an Annexin V, FITC Apoptosis Detection Kit (Catalogue Number AD10, Dojindo, Japan) according to the manufacturer’s instructions and then analysed by flow cytometry (BD FACSCalibur, USA). The results are presented as the percentage of cells in each phase.
Xenograft analysis
All experimental procedures were approved by the Institutional Animal Care and Use Committee of the Center of Harbin Medical University and conformed to all regulatory standards. A total of 5 × 106 luciferase-labelled MDA-MB-231 control cells or shLINC00511–1cells suspended in 0.2 ml of PBS with phenol-red-free Matrigel (Catalogue Number 356234, Corning, USA) (1:1) were injected into the axilla of 5-week-old pathogen-free female athymic BALB/c mice obtained from Shanghai SLAC Laboratory Animal Co., Ltd (www.slaccas.com, China). Bioluminescence imaging was carried out using a Carestream Image Station System (Multimodal Pro Light Source, Carestream Health, Inc., CA). When the tumours became palpable, tumour volume was assessed by digital calliper measurements using the formula (width2 × length)/2 (mm3), and the whole body weight was measured once every 3 days. All mice were euthanized at the end of the experiment.
IHC assay
The tumours were stained for H&E and immunostained for Ki-67 and CDKN1B as previously described9.
Statistical analysis
Statistical analysis was performed using SPSS 17.0 software (SPSS Software, USA) and GraphPad Prism 8 (GraphPad Software, USA). Student’s t test and the chi-square test were used to determine significant differences where appropriate. Survival was calculated by the Kaplan–Meier method, with the log-rank test applied for comparison. All statistical tests were two-sided, and a probability level of 0.05 indicated statistical significance.
Supplementary information
Acknowledgements
This work was supported by funding from the Project Nn10 of Harbin Medical University Cancer Hospital (Grant numberNn102017–02), the National Natural Science Foundation of China (Grant numbers 81602323, 81872149 and 81802649), the Outstanding Youth Project of Heilongjiang Provincial Natural Science Foundation (Grant numberYQ2019H027), the Wu Lien-teh Science Foundation of Harbin Medical University (Grant number WLD-QN1706), the Distinguished Young Scholars of Harbin Medical University Cancer Hospital (Grant number JCQN2018–03), the Yong Elite Training Foundation Grant of Harbin Medical University Cancer Hospital (Grant number JY2016–02), and the BaJian QingNian Grant of Harbin Medical University Cancer Hospital (Grant number BJQN2019–09).
Author contributions
Conception and design: J.Z. Data acquisition and analysis (acquired and managed patient information and statistical analysis): J.Z. and J.Z. Molecular biology experiments: J.Z., S.S. and H.W. Experimental operation and recording of tumours in vivo: J.Z. and X.Z. S.X. and D.P. contributed to the study design and supervision. All authors contributed to the paper. All authors read and approved the final paper.
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Edited by A. Stephanou
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Shouping Xu, Email: shoupingxu@hrbmu.edu.cn.
Da Pang, Email: pangda@ems.hrbmu.edu.cn.
Supplementary information
Supplementary Information accompanies this paper at (10.1038/s41419-019-1835-3).
References
- 1.Zhang H, et al. Epigenetic regulation of NAMPT by NAMPT-AS drives metastatic progression in triple-negative breast cancer. Cancer Res. 2019 doi: 10.1158/0008-5472.CAN-18-3418. [DOI] [PubMed] [Google Scholar]
- 2.Polyak K. Heterogeneity in breast cancer. J. Clin. Investig. 2011;121:3786–3788. doi: 10.1172/JCI60534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sorlie T, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc. Natl. Acad. Sci. USA. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harbeck N, Gnant M. Breast cancer. Lancet. 2017;389:1134–1150. doi: 10.1016/S0140-6736(16)31891-8. [DOI] [PubMed] [Google Scholar]
- 5.Nagini S. Breast cancer: current molecular therapeutic targets and new players. Anticancer Agents Med. Chem. 2017;17:152–163. doi: 10.2174/1871520616666160502122724. [DOI] [PubMed] [Google Scholar]
- 6.Doisneau-Sixou SF, et al. Estrogen and antiestrogen regulation of cell cycle progression in breast cancer cells. Endocr. Relat. Cancer. 2003;10:179–186. doi: 10.1677/erc.0.0100179. [DOI] [PubMed] [Google Scholar]
- 7.Shang Y, Hu X, DiRenzo J, Lazar MA, Brown M. Cofactor dynamics and sufficiency in estrogen receptor-regulated transcription. Cell. 2000;103:843–852. doi: 10.1016/S0092-8674(00)00188-4. [DOI] [PubMed] [Google Scholar]
- 8.Sotiriou C, et al. Breast cancer classification and prognosis based on gene expression profiles from a population-based study. Proc. Natl Acad. Sci. USA. 2003;100:10393–10398. doi: 10.1073/pnas.1732912100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu S, et al. Ai-lncRNA EGOT enhancing autophagy sensitizes paclitaxel cytotoxicity via upregulation of ITPR1 expression by RNA-RNA and RNA-protein interactions in human cancer. Mol. Cancer. 2019;18:89. doi: 10.1186/s12943-019-1017-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang J, Wang P, Wan L, Xu S, Pang D. The emergence of noncoding RNAs as Heracles in autophagy. Autophagy. 2017;13:1004–1024. doi: 10.1080/15548627.2017.1312041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Niknafs YS, et al. The lncRNA landscape of breast cancer reveals a role for DSCAM-AS1 in breast cancer progression. Nat. Commun. 2016;7:12791. doi: 10.1038/ncomms12791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang Z, et al. lncRNA epigenetic landscape analysis identifies EPIC1 as an oncogenic lncRNA that interacts with MYC and promotes cell-cycle progression in cancer. Cancer Cell. 2018;33:706–720 e709. doi: 10.1016/j.ccell.2018.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Iyer MK, et al. The landscape of long noncoding RNAs in the human transcriptome. Nat. Genet. 2015;47:199–208. doi: 10.1038/ng.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu SP, et al. Downregulation of the long noncoding RNA EGOT correlates with malignant status and poor prognosis in breast cancer. Tumour Biol. 2015;36:9807–9812. doi: 10.1007/s13277-015-3746-y. [DOI] [PubMed] [Google Scholar]
- 15.Ulitsky I, Bartel DP. P. lincRNAs: genomics, evolution, and mechanisms. Cell. 2013;154:26–46. doi: 10.1016/j.cell.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 17.Shen H, Laird PW. Interplay between the cancer genome and epigenome. Cell. 2013;153:38–55. doi: 10.1016/j.cell.2013.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qu D, et al. Long noncoding RNA MALAT1 releases epigenetic silencing of HIV-1 replication by displacing the polycomb repressive complex 2 from binding to the LTR promoter. Nucleic Acids Res. 2019;47:3013–3027. doi: 10.1093/nar/gkz117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gupta RA, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tsai MC, et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu Y, et al. Long non-coding RNA PVT1 promotes cell proliferation and migration by silencing ANGPTL4 expression in cholangiocarcinoma. Mol. Ther.Nucleic Acids. 2018;13:503–513. doi: 10.1016/j.omtn.2018.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang YL, et al. Lnc-UCID promotes G1/S transition and hepatoma growth by preventing DHX9-mediated CDK6 down-regulation. Hepatology. 2019 doi: 10.1002/hep.30613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu Y, et al. MYC targeted long noncoding RNA DANCR promotes cancer in part by reducing p21 Levels. Cancer Res. 2018;78:64–74. doi: 10.1158/0008-5472.CAN-17-0815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tang J, et al. Bidirectional transcription of Linc00441 and RB1 via H3K27 modification-dependent way promotes hepatocellular carcinoma. Cell Death Dis. 2017;8:e2675. doi: 10.1038/cddis.2017.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu Y, et al. Epigenetic silencing of tumor suppressor gene CDKN1A by oncogenic long non-coding RNA SNHG1 in cholangiocarcinoma. Cell Death Dis. 2018;9:746. doi: 10.1038/s41419-018-0768-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sun CC, et al. Long intergenic noncoding RNA 00511 acts as an oncogene in non-small-cell lung cancer by binding to EZH2 and suppressingp57. Mol. Ther. Nucleic Acids. 2016;5:e385. doi: 10.1038/mtna.2016.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang, J., Tian, Y., Zheng, H., Ding, Y. & Wang, X. An integrated analysis reveals the oncogenic function of lncRNA LINC00511 in human ovarian cancer. Cancer Med., 10.1002/cam4.2171 (2019). [DOI] [PMC free article] [PubMed]
- 28.Li Chen, Liu Hongjiang, Yang Jipeng, Yang Jiankai, Yang Liang, Wang Yuanyu, Yan Zhongjie, Sun Yuchen, Sun Xiaofeng, Jiao Baohua. Long noncoding RNA LINC00511 induced by SP1 accelerates the glioma progression through targeting miR‐124‐3p/CCND2 axis. Journal of Cellular and Molecular Medicine. 2019;23(6):4386–4394. doi: 10.1111/jcmm.14331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu S, Kong D, Chen Q, Ping Y, Pang D. Oncogenic long noncoding RNA landscape in breast cancer. Mol. Cancer. 2017;16:129. doi: 10.1186/s12943-017-0696-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu, L. C. et al. Long noncoding RNA HOTAIR promotes invasion of breast cancer cells through chondroitin sulfotransferase CHST15. Int. J. Cancer, 10.1002/ijc.32319 (2019). [DOI] [PMC free article] [PubMed]
- 31.Francisco-Velilla R, Fernandez-Chamorro J, Ramajo J, Martinez-Salas E. The RNA-binding protein Gemin5 binds directly to the ribosome and regulates global translation. Nucleic Acids Res. 2016;44:8335–8351. doi: 10.1093/nar/gkw702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xie JJ, et al. Super-enhancer-driven long non-coding RNA LINC01503, regulated by TP63, is over-expressed and oncogenic in squamous cell carcinoma. Gastroenterology. 2018;154:2137–2151 e2131. doi: 10.1053/j.gastro.2018.02.018. [DOI] [PubMed] [Google Scholar]
- 33.Hanzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinform. 2013;14:7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tesio M, Trumpp A. Breaking the cell cycle of HSCs by p57 and friends. Cell Stem Cell. 2011;9:187–192. doi: 10.1016/j.stem.2011.08.005. [DOI] [PubMed] [Google Scholar]
- 35.Khalil AM, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc. Natl Acad. Sci. USA. 2009;106:11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Laugesen A, Hojfeldt JW, Helin K. Molecular mechanisms Directing PRC2 Recruitment and H3K27 Methylation. Mol. Cell. 2019;74:8–18. doi: 10.1016/j.molcel.2019.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rivenbark AG, O’Connor SM, Coleman WB. Molecular and cellular heterogeneity in breast cancer: challenges for personalized medicine. Am. J. Pathol. 2013;183:1113–1124. doi: 10.1016/j.ajpath.2013.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ellsworth RE, Blackburn HL, Shriver CD, Soon-Shiong P, Ellsworth DL. Molecular heterogeneity in breast cancer: state of the science and implications for patient care. Semin. Cell Dev. Biol. 2017;64:65–72. doi: 10.1016/j.semcdb.2016.08.025. [DOI] [PubMed] [Google Scholar]
- 39.Petrossian K, et al. ERalpha-mediated cell cycle progression is an important requisite for CDK4/6 inhibitor response in HR+ breast cancer. Oncotarget. 2018;9:27736–27751. doi: 10.18632/oncotarget.25552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yip CH, Rhodes A. Estrogen and progesterone receptors in breast cancer. Future Oncol. 2014;10:2293–2301. doi: 10.2217/fon.14.110. [DOI] [PubMed] [Google Scholar]
- 41.Chan HJ, Petrossian K, Chen S. Structural and functional characterization of aromatase, estrogen receptor, and their genes in endocrine-responsive and -resistant breast cancer cells. J. Steroid Biochem. Mol. Biol. 2016;161:73–83. doi: 10.1016/j.jsbmb.2015.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Foster JS, Henley DC, Ahamed S, Wimalasena J. Estrogens and cell-cycle regulation in breast cancer. Trends Endocrinol. Metab. 2001;12:320–327. doi: 10.1016/S1043-2760(01)00436-2. [DOI] [PubMed] [Google Scholar]
- 43.O’Leary B, Finn RS, Turner NC. Treating cancer with selective CDK4/6 inhibitors. Nat. Rev. Clin. Oncol. 2016;13:417–430. doi: 10.1038/nrclinonc.2016.26. [DOI] [PubMed] [Google Scholar]
- 44.Miao H, et al. A long noncoding RNA distributed in both nucleus and cytoplasm operates in the PYCARD-regulated apoptosis by coordinating the epigenetic and translational regulation. PLoS Genet. 2019;15:e1008144. doi: 10.1371/journal.pgen.1008144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.He F, et al. Long noncoding RNA PVT1-214 promotes proliferation and invasion of colorectal cancer by stabilizing Lin28 and interacting with miR-128. Oncogene. 2019;38:164–179. doi: 10.1038/s41388-018-0432-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang L, et al. Long non-coding RNA HOTAIR promotes exosome secretion by regulating RAB35 and SNAP23 in hepatocellular carcinoma. Mol. Cancer. 2019;18:78. doi: 10.1186/s12943-019-0990-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Costales MG, et al. A designed small molecule inhibitor of a non-coding RNA sensitizes HER2 negative cancers to herceptin. J. Am. Chem. Soc. 2019;141:2960–2974. doi: 10.1021/jacs.8b10558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vellanki SH, et al. Natural compound tetrocarcin-A downregulates junctional adhesion molecule-A in conjunction with HER2 and inhibitor of apoptosis proteins and inhibits tumor cell growth. Cancer Lett. 2019;440-441:23–34. doi: 10.1016/j.canlet.2018.09.032. [DOI] [PubMed] [Google Scholar]
- 49.Aviles P, et al. MI130004, a novel antibody-drug conjugate combining trastuzumab with a molecule of marine origin, shows outstanding in vivo activity against HER2-expressing tumors. Mol. Cancer Ther. 2018;17:786–794. doi: 10.1158/1535-7163.MCT-17-0795. [DOI] [PubMed] [Google Scholar]
- 50.Kong F, et al. ZFPM2-AS1, a novel lncRNA, attenuates the p53 pathway and promotes gastric carcinogenesis by stabilizing MIF. Oncogene. 2018;37:5982–5996. doi: 10.1038/s41388-018-0387-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.