Abstract
Background: Esophageal squamous cell carcinoma (ESCC), one of the most aggressive cancers, is endemic in Sub-Saharan Africa, constituting a major health burden. It has the most divergence in cancer incidence globally, with high prevalence reported in East Asia, Southern Europe, and in East and Southern Africa. Its etiology is multifactorial, with lifestyle, environmental, and genetic risk factors. Very little is known about the role of genetic factors in ESCC development and progression among African populations. The study aimed to systematically assess the evidence on genetic variants associated with ESCC in African populations.
Methods: We carried out a comprehensive search of all African published studies up to April 2019, using PubMed, Embase, Scopus, and African Index Medicus databases. Quality assessment and data extraction were carried out by two investigators. The strength of the associations was measured by odds ratios and 95% confidence intervals.
Results: Twenty-three genetic studies on ESCC in African populations were included in the systematic review. They were carried out on Black and admixed South African populations, as well as on Malawian, Sudanese, and Kenyan populations. Most studies were candidate gene studies and included DNA sequence variants in 58 different genes. Only one study carried out whole-exome sequencing of 59 ESCC patients. Sample sizes varied from 18 to 880 cases and 88 to 939 controls. Altogether, over 100 variants in 37 genes were part of 17 case-control genetic association studies to identify susceptibility loci for ESCC. In these studies, 25 variants in 20 genes were reported to have a statistically significant association. In addition, eight studies investigated changes in cancer tissues and identified somatic alterations in 17 genes and evidence of loss of heterozygosity, copy number variation, and microsatellite instability. Two genes were assessed for both genetic association and somatic mutation.
Conclusions: Comprehensive large-scale studies on the genetic basis of ESCC are still lacking in Africa. Sample sizes in existing studies are too small to draw definitive conclusions about ESCC etiology. Only a small number of African populations have been analyzed, and replication and validation studies are missing. The genetic etiology of ESCC in Africa is, therefore, still poorly defined.
Keywords: esophageal squamous cell carcinoma, genetic association, somatic variant, germline mutation, sequence variants, systematic review, African populations
Introduction
Esophageal cancer is an aggressive and fatal cancer of the 18digestive tract. It accounts for an estimated 455,800 new cases and 400,200 deaths per year globally, making it the eighth most common cancer in the world (Murphy et al., 2017). The malignant tumors are characterized by two major subtypes: esophageal squamous cell carcinoma (ESCC), which is the more common type and contributes 90%, and esophageal adenocarcinoma (EAC) (Kaz and Grady, 2014; Abnet et al., 2017). ESCC presents with poor prognosis and low survival rate (<5%) in low resource settings (Yazbeck et al., 2016; Murphy et al., 2017). The asymptomatic development of ESCC results in diagnosis at late stage for patients and is characterized by dysphagia. At this stage, treatment is limited to palliative care.
ESCC is endemic in specific geographic locations worldwide and has the most divergence in cancer incidence globally, with high prevalence reported in East Asia, Southern Europe, as well as in Eastern and Southern Africa (Abnet et al., 2017). This peculiar distribution draws questions on the specificity of certain risk factors to particular populations. The African ESCC corridor, which includes Ethiopia, Rwanda, Burundi, Malawi, Kenya, Uganda, Tanzania, and South Africa, is an ESCC hotspot region (Munishi et al., 2015; Schaafsma et al., 2015). It has also been reported that in Sub-Saharan Africa, ESCC develops in younger patients than in other regions (Kayamba et al., 2015).
The etiology of esophageal carcinoma is multifactorial. The risk factors reported worldwide comprise several lifestyle and environmental and genetic factors (Pink et al., 2011; Sewram et al., 2014; Chen et al., 2015; Sewram et al., 2016; Huang and Yu, 2018). Growing evidence supports the hypothesis that genomic alterations and epigenetic modifications contribute to tumor development (Baba et al., 2017). ESCC has both an inherited and cellular genetic basis (Abnet et al., 2017; Coleman et al., 2018). Familial syndromes associated with increased risk of malignancy include tylosis and Fanconi anemia (Abnet et al., 2017). The majority of genetic studies on ESCC have been case-control association studies analyzing single-nucleotide polymorphisms (SNPs) in various candidate genes. However, the reproducibility of these studies has been low. Some of the more common SNPs associated with ESCC have been identified in the aldehyde dehydrogenase 2 family gene (ALDH2) and an acetaldehyde dehydrogenase gene (ADH1B) (Abnet et al., 2017). Variants in these genes have been shown to increase susceptibility to ESCC development, and they are also associated with alcohol consumption (Abnet et al., 2017). Two meta-analyses published in 2018 reported associations between the genes MTHFR and GSTT1 and esophageal cancer development (He et al., 2018; Kumar and Rai, 2018). However, the meta-analyses were done on predominantly Asian and Western populations. In recent years, the focus of ESCC research in the Western and Asian countries has shifted from candidate gene studies to genome-wide association studies (GWAS) and whole-exome sequencing (WES) to identify variants associated with ESCC. Combined analysis of different study designs has provided a better understanding of ESCC etiology in Asian populations (Abnet et al., 2017). Genes with variants implicated in the development of ESCC in these populations include phospholipase c epsilon 1 (PLCE1), caspase 8 (CAP8), tumor protein 53 (TP53), and human leukocyte antigen (HLA) (Abnet et al., 2017).
The genetic etiology of ESCC in Africa is not well understood, since there have been very few studies on ESCC in African populations. This is in part due to the unavailability of adequate research infrastructure. A lack of comprehensive assessment and validation of existing evidence through systematic reviews has also contributed to this knowledge gap. A number of small studies on African populations have yielded varied associations between genetic variants and ESCC. There is, therefore, a need to systematically assess the current evidence in order to map out the contribution of genetic factors in the development of ESCC in African populations using critically appraised data.
The aim of the current systematic review was to assess all genetic (cross-sectional, case-control, and cohort) studies reporting on germline and somatic variants where risk factor estimates were calculated. This was achieved through the following: 1) critical appraisal of African literature on association of genetic factors to ESCC development; 2) comprehensive analysis of genetic (germline and somatic) variants in the reported studies; 3) data synthesis through pooled analysis, if feasible; and 4) comparison of genetic variants identified in African populations to those reported in other geographic regions.
Materials and Methods
We followed the Preferred Reporting Items for Systematic Reviews and Meta-Analyses guidelines (PRISMA) (Little et al., 2009). However, because PRISMA is not a quality assessment tool, other instruments were used to assess quality control.
Data Sources and Search Strategy
We carried out a literature search on all published African ESCC studies up to April 2019. We developed a comprehensive set of search terms subjectively and iteratively. We searched the following electronic bibliographic databases without time or language limits: Medline (PubMed), Embase (OViD), Scopus, African Index Medicus, and Africa-wide information (EbsCOHost). We also checked the reference lists of potentially relevant articles for additional citations and used the “related citations” search key in PubMed to identify similar papers.
We checked Medline (PubMed) to identify controlled vocabulary (MeSH) terms related to esophageal cancer and also identified text keywords based on our knowledge of the field ( Table 1 ). Medline search terms were modified for other electronic databases to conform to their search functions.
Table 1.
Medline (PubMed) search strategy to identify published African ESCC literature.
| #1 | Search cancer or carcinoma or neoplasm* Field: Title/Abstract |
| #2 | Search Esophageal or oesophageal Field: Title/Abstract |
| #3 | #1 and #2 |
| #4 | Search “Esophageal cancer” Field: Title/Abstract |
| #5 | Search “oesophageal cancer” or “oesophageal neoplasm*” Field: Title/Abstract |
| #6 | Search “Esophageal Neoplasms”[Mesh] |
| #7 | Search “Esophageal Neoplasms” Field: Title/Abstract |
| #8 | Search “Esophageal squamous cell carcinoma” or “oesophageal squamous cell carcinoma” or ESCC Field: Title/Abstract |
| #9 | Search ((((#3) OR #4) OR #5) OR #6) OR #7 OR #8 |
| #10 | Search “Africa”[Mesh] |
| #11 | Search algeria OR angola OR benin OR botswana OR burkina faso OR burundi OR cameroon OR cape verde OR central african republic OR chad OR comoros OR congo OR “Democratic Republic of Congo” OR DRC OR djibouti OR equatorial guinea OR egypt OR eritrea OR ethiopia OR gabon OR gambia OR ghana OR guinea OR bissau OR ivory coast OR (Côte d’ Ivoire) OR jamahiriya OR kenya OR lesotho OR liberia OR Libya OR madagascar OR malawi OR mali OR mauritania OR mauritius OR mayotte OR morocco OR mozambique OR namibia OR niger OR nigeria OR principe OR reunion OR rwanda OR “Sao Tome” OR senegal OR seychelles OR “Sierra Leone” OR somalia OR “South Africa” OR st helena OR sudan OR swaziland OR tanzania OR togo OR tunisia OR uganda OR zaire OR zambia OR zimbabwe OR “Central Africa” OR “West Africa” OR “East Africa” OR “Southern Africa” OR “South Africa” Field: Title/Abstract |
| #12 | Search (#10) or #11 |
| #13 | Search (#9) AND #12 |
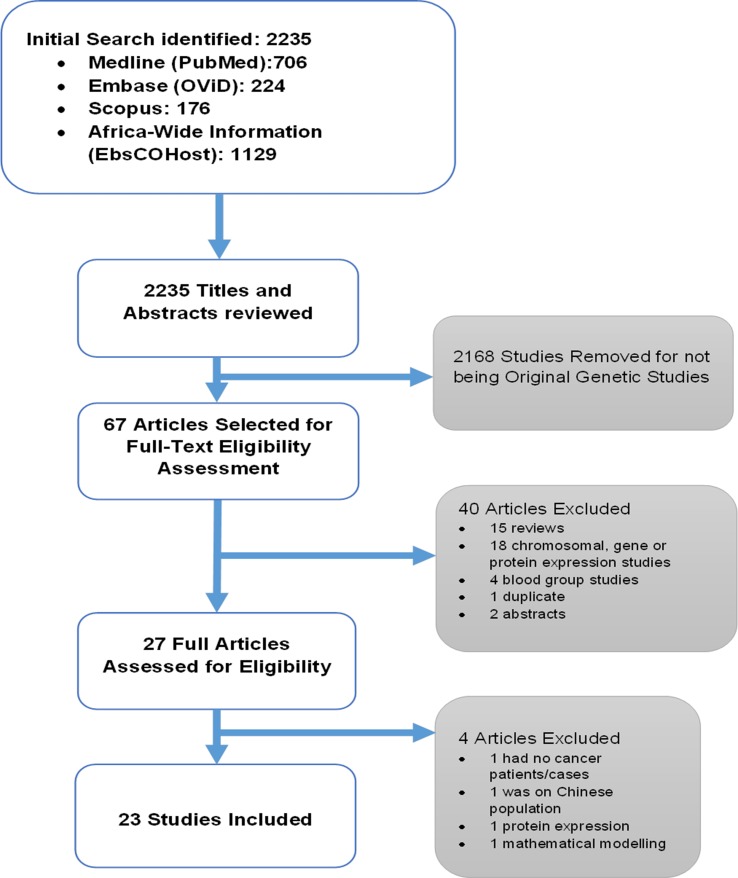
Screening for eligible studies was carried out by two authors (HS and HK). First, the two authors read the titles and abstracts independently and then met to finalize an initial list. Full articles of the studies selected based on the initial screening were read and assessed for inclusion to the systematic review. Figure 1 shows the outline for selection of eligible studies.
Figure 1.
Outline of the systematic review.
Quality Control and Data Extraction
Quality of the methodology used in the published studies was assessed using a quality assessment tool adapted from the STrengthening the REporting of Genetic Association studies (STREGA) statement (Little et al., 2009). The quality assessment for genetic association studies to identify ESCC susceptibility loci included reporting on power calculations, detailed population characteristics for cases, description of ESCC diagnosis, screening of cases and controls, reporting a measure of association using odds ratios, adjustment of population stratification, assessment of genotyping error, reporting the Hardy–Weinberg equilibrium, correction for multiple testing, and reporting of National Center for Biotechnology Information (NCBI) rs numbers for variants ( Table S1 ).
For somatic mutation studies, quality assessment included the following: description of ESCC diagnosis, reporting of tissues used [cancerous (Ca) and normal neighboring tissue (NET)], detailed population characteristics, variant classification and type, confirmation of variants identified, reporting of amino acid change, and use of pathogenicity scoring ( Table S2 ).
Data extraction was carried out by two authors (HS and HK) using data extraction forms. Two separate extraction forms were prepared for the germline (genetic susceptibility) and somatic mutation studies. The data extraction form for the genetic susceptibility studies included the following: description of the population (age, sex, sample size, smoking, and alcohol use for cases and controls separately), genotyping method, statistical analysis test, minor allele frequency (MAF), genotype frequency, haplotype frequency, and environmental association frequency. The somatic mutation study extraction form had the same variables excluding gene–environment interaction frequency and haplotype frequency.
The South African Admixed Population is reported as mixed ancestry in the tables according to how it was reported in the articles.
Data Analysis
A meta-analysis could not be performed as there were only two SNPs analyzed in more than one study and even those were analyzed in only two independent studies. For a meta-analysis to be carried out, SNPs have to be assessed in at least three separate case-control studies. TP53 in the somatic variant studies was analyzed in four separate studies, but two of the studies had cases only with no controls, and the remaining two assessed different parts of the gene. The results of this systematic review will, therefore, be reported in a descriptive manner.
We were able to find rs numbers for most of the variants even if the authors of the original studies did not report them and have included them in the tables of this systematic review. We used the canonical SNP identifier (rs number) and dbSNP (version 152; April 2019) database at NCBI (https://www.ncbi.nlm.nih.gov/snp/) for this. We also determined the locus positions of the microsatellite markers reported in a study by Naidoo et al. (2005) using the primer-BLAST database at NCBI (https://www-ncbi-nlm-nih-gov.ez.sun.ac.za/tools/primer-blast).
To determine the linkage disequilibrium (LD) measures between the SNPs reported in the same genes, we obtained the imputed data set from the Thousand Genomes project (1000 Genomes Release Phase 3 2013-05-02) and used bcftools to extract all individuals from African populations, not including African Americans, and the 77 SNPs discussed here using all synonyms (alternative rs IDs) for SNPs (Auton et al., 2015). We obtained a dataset of 504 individuals and 67 SNPs. We computed all pair-wise r2-values using PLINK (v1.09) (Danecek et al., 2011; Chang et al., 2015).
Results
Systematic Review Outline
The selection process for all the included studies is shown in Figure 1 . The initial database search identified 2,235 articles. Titles and abstracts of these articles were reviewed, and 2,168 studies were removed for not being original genetic studies. The 67 articles that remained were selected for full-text eligibility assessment. This process resulted in the removal of 40 articles: 15 review articles, 18 chromosomal, gene or protein expression studies, 4 blood group studies, 1 duplicate, and 2 abstracts. A total of 27 full articles were then assessed for eligibility, and four articles were removed for not meeting the criteria, as follows: one study had no cancer patients/cases (Adams et al., 2003), one focused on the Chinese population (Li et al., 2016), while one focused on protein expression (Jaskiewicz and De Groot, 1994; Huang and Yu, 2018), and the other was a mathematical model study (Uys and Van Helden, 2003). In the end, 23 studies were included and analyzed in the systematic review.
Study Characteristics
The characteristics of all the genetic susceptibility and somatic variant studies included are shown in Tables 2 and 3 , respectively. The 23 studies included in the study were published between 1990 and 2019. There were 17 genetic susceptibility and eight somatic variant studies. Two studies reported on both genetic susceptibility and somatic variants.
Table 2.
Characteristics of genetic susceptibility studies for ESCC in African populations.
| Study (PMID) |
Location | Year | Population | Age, y (SD) | Sample size | Sex, cases n (%) | Sex, ctrl n (%) | Clinical assessment | Analysis method | Smoking n (%) | Alcohol n (%) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cases | Ctrl | Cases | Ctrl | Male | Female | Male | Female | Cases | Ctrl | Cases | Ctrl | Cases | Ctrl | |||||
|
Bye et al., 2011
(21926110) |
South Africa | 2011 | Black | 59.8 (11.3) | – | 358 | 477 | 182 (50.8) | 176 (49.2) | – | – | Histology | – | TaqMan Assay | 228 (63.7) | – | 228 (63.7) | – |
| Mixed ancestry | 60.5 (10.6) | – | 201 | 427 | 131 (65.2) | 70 (34.8) | – | – | Histology | – | TaqMan Assay | 189 (94.1) | – | 163 (81.1) | – | |||
| Bye et al., 2012 (22865593) | South Africa | 2012 | Black | 59.8 (11.3) | 48.8 (16.7) | 407 | 849 | 199 (48.9) | 208 (51.1) | 335 (39.5) | 511 (60.2) | Histology | – | TaqMan Assay and KASP | 242 (59.5) | 333 (39.2) | 253 (62.2) | 452 (53.2) |
| Mixed ancestry | 60.6 (10.6) | 46.7 (16.8) | 257 | 860 | 165 (64.2) | 91 (35.4) | 309 (35.9) | 551 (64.1) | Histology | – | TaqMan Assay and KASP | 240 (93.4) | 597 (69.4) | 212 (82.5) | 419 (48.7) | |||
| Chelule et al., 2006 (17264406) | South Africa | 2006 | Black | 18–741 | 18–74 | 70 | 261 | – | – | – | – | Histology | – | PCR-RFLP | – | – | – | – |
| Chen et al., 2019 (30753320) | South Africa | 2019 | Black7 | 60.2 (11.3) | 48.9 (16.8) | 591 | 852 | 284 (48.1) | 307 (51.9) | 342 (40.1) | 507 (59.5) | Histology | – | TaqMan Assay | 364 (61.6) | 338 (39.7) | 370 (62.6) | 458 (53.7) |
| Black8 | 58.2 (10.2) | 50.0 (15.5) | 880 | 939 | 545 (61.9) | 332 (37.7) | 240 (25.6) | 698 (74.3) | Histology | iPLEX and TaqMan Assays | 598 (68.0) | 333 (35.5) | 473 (53.8) | 633 (67.4) | ||||
| Dandara et al., 2005 (15978331) | South Africa | 2005 | Black | – | – | 142 | 178 | – | – | – | – | Histology | – | PCR-RFLP | 179 | 162 | 171 | 160 |
| Mixed ancestry | – | – | 99 | 94 | Histology | PCR-RFLP | ||||||||||||
| Dandara et al., 2006 (16272171) | South Africa | 2006 | Black | 61.23 | 61.85 | 145 | 194 | 85 (59) | 60 (41) | 111 (57) | 83 (43) | Histology | – | PCR-RFLP | 95 (65) | 123 (63) | 98 (68) | 127 (65) |
| Mixed ancestry | 61.49 | 69.53 | 100 | 94 | 78 (78) | 22 (22) | 45 (48) | 49 (52) | Histology | – | PCR-RFLP | 93 (93) | 74 (79) | 73 (73) | 45 (48) | |||
| Dietzsch et al., 2003 (12925954) | South Africa | 2003 | Black and mixed ancestry | 59.6 | 58.7 | 582 | 226 | 44 | 14 | 167 | 59 | – | – | PCR and PAGE |
||||
| Eltahir et al., 2012 (23053979) | Sudan | 2012 | 18 | 235 | Histology | PCR-RFLP | ||||||||||||
| Li et al., 2005 (15899651) | South Africa | 2005 | Black and mixed ancestry | 61.1 (10.5) | 65.7 (10.2) | 189 | 198 | – | – | – | – | Histology | – | PCR-SSCP and DNA sequencing | 144 (76) | 122 (62) | 133 (70) | 114 (58) |
| Li et al., 2008 (18254707) | South Africa | 2008 | Black3 | – | – | 142 | 178 | – | – | – | – | Histology | – | PCR- RLFP | 179 | 162 | 71 | 160 |
| Mixed3 ancestry | 101 | 100 | Histology | PCR-RFLP | ||||||||||||||
| Li et al., 2010 (20540773) | South Africa | 2010 | Black3 | 61.23 | 61.85 | 145 | 194 | 85 (59) | 60 (41) | 111 (57) | 83 (43) | Histology | – | PCR-RFLP | 95 (65) | 123 (63) | 98 (68) | 127 (65) |
| Mixed3 ancestry | 61.49 | 69.53 | 100 | 94 | 78 (78) | 22 (22) | 45 (48) | 49 (52) | Histology | – | PCR- RFLP | 93 (93) | 74 (79) | 73 (73) | 45 (48) | |||
| Matejcic et al., 2011 (22216261) | South Africa | 2011 | Black | – | – | 330 | 479 | – | – | – | – | Histology | – | TaqMan assay and gel electrophoresis | 210 | – | 204 | – |
| Mixed ancestry | – | – | 232 | 428 | – | – | – | – | Histology | – | TaqMan assay and gel electrophoresis | 216 | – | 189 | – | |||
| Matejcic et al., 2015 (26447020) | South Africa | 2015 | Black | 59.6 (10.7) |
56.7 (15.0) |
463 | 480 | 229 (49) | 234 (51) | 235 (49) | 245 (51) | Histology | – | TaqMan assay | 280 (60) | 222 (46) | 286 (62) | 278 (58) |
| Mixed ancestry | 60.7 (10.3) | 57.7 (14.3) | 269 | 288 | 177 (66) | 92 (34) | 178 (62) | 110 (38) | Histology | – | TaqMan Assay | 250 (93) | 226 (78) | 215 (80) | 172 (60) | |||
| Strickland et al., 2012 (21901748) | South Africa | 2012 | Black | 59/664 | – | 96 | 88 | 48 | 48 | – | – | Histology | Brush biopsy |
HEX SSCP and DNA sequencing | 58 | – | 58 | – |
| Vogelsang et al., 2012 (22623965) | South Africa | 2012 | Black | 59.8 (11.3) | 56.1 (16.2) | 3455 | 344 | 166 (48.1) | 179 (51.9) | 120 (34.9) | 224 (65.1) | Histology | – | Allele-specific quantitative PCR | 209 (60.6) | 117 (34.0) | 160 (46.4) | 92 (26.7) |
| Mixed ancestry | 60.7 (10.2) | 56.8 (16.5) | 2056 | 266 | 136 (66.3) | 69 (33.7) | 82 (30.8) | 184 (69.2) | Histology | – | Allele-specific quantitative PCR | 189 (92.2) | 162 (60.9) | 118 (57.6) | 38 (14.3) | |||
| Vos et al., 2003 (12550754) | South Africa | 2003 | Black | 57 (11) | 57 (11) | 74 | 118 | – | – | – | – | Histology | – | SSCP and DNA sequencing | – | – | – | – |
| Zaahl et al., 2005 (15860357) | South Africa | 2005 | Mixed ancestry | – | – | 105 | 110 | 82 | 23 | 43 | 67 | Histology | – | SSCP and DNA sequencing | – | – | – | – |
1Only range of age was reported for the combined group of cases and controls.
257 had ESCC.
3Same population as in Dandara et al. (2005) study.
459+/–13 for male (n = 48) and 66+/– (n = 48) for female patients.
5326 had ESCC.
6182 had ESCC.
7Western and Eastern Cape Province Black Population.
8Gauteng Province Black Population.
Ctrl, controls; ESCC, esophageal squamous cell carcinoma; HEX, heteroduplex; KASP, competitive allele specific PCR; PAGE, polyacrylamide gel electrophoresis; PCR, polymerase chain reaction; RFLP, restriction fragment length polymorphism; SD, Standard deviation; SSCP, single-strand conformation polymorphism.
Table 3.
Characteristics of studies on somatic changes in ESCC in African populations.
| Study (PMID) | Country | Year | Population | Sample size | Age, y (SD) | Sex n (%) | Clinical assessment | Analysis method | Smoking n (%) | Alcohol n (%) | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ca | NET | Blood | Cases | Male | Female | Ca | NET | |||||||
| Dietzsch and Parker, 2002 (12435113) | South Africa | 2002 | Black | 33 | 33 | – | 57.4 | 23 (70) | 10 (30) | Histology | – | PCR and DNA sequencing analysis | – | – |
| Dietzsch et al., 2003 (12925954) | South Africa | 2003 | Black and mixed ancestry | 581 | 58 | – | 59.6 | 29 (67) | 14 (33) | – | – | PCR and PAGE | – | – |
| Gamieldien et al.,1998 (9808520) | South Africa | 1998 | Black | 76 | 9 | 50 | 57 (11) | 49 (65) | 27 (35) | Histology | Histology | PCR and HEX-SSCP | – | – |
| Liu et al., 2016 (29148985) | Malawi | 2016 | Malawian | 59 | – | 59 | 56 | 27 (45.8) | 31 (52.5) | Histology | - | WES | 24 (40.7) | 14 (23.7) |
| Naidoo et al., 2005 (15735161) | South Africa | 2005 | South African | 100 | 100 | – | 56 | 53 (54) | 45 (46) | Histology | Histology | PCR | – | – |
| Patel et al., 2011 (22040862) | Kenya | 2011 | Kenyan | 28 | – | – | 56.03 (12.30) | 13 (46) | 15 (54) | – | – | PCR and DNA sequencing | 6 (21) | 10 (36) |
| Victor et al., 1990 (2199031) | South Africa | 1990 | Black and mixed ancestry | 27 | – | – | – | – | – | – | – | PCR and dot blot hybridization | – | – |
| Vos et al., 2003 (12550754) | South Africa | 2003 | South African | 74 | – | 37 | – | – | – | Histology | – | SSCP and DNA sequencing | – | – |
Ca, cancer tissue; HEX-SSCP, heteroduplex single-strand conformation polymorphism; NET, neighboring tissue; PAGE, polyacrylamide gel electrophoresis; PCR, polymerase chain reaction; WES, whole exome sequencing.
157 had ESCC and 1 had adenocarcinoma.
Genetic Susceptibility Studies
The 17 genetic susceptibility studies ( Table 2 ) were all case-control studies (Dietzsch et al., 2003; Vos et al., 2003; Dandara et al., 2005; Li et al., 2005; Zaahl et al., 2005; Chelule et al., 2006; Dandara et al., 2006; Li et al., 2008; Li et al., 2010; Bye et al., 2011; Matejcic et al., 2011; Bye et al., 2012; Eltahir et al., 2012; Strickland et al., 2012; Vogelsang et al., 2012; Matejcic et al., 2015; Chen et al., 2019) published between 2003 and 2019. Sixteen articles reported on the South African population and one article on the Sudanese population. The majority (13/17; 76%) of the studies reported on the main subject characteristics (ethnicity, sex, age, and type of clinical assessment). Sample sizes for ESCC patients ranged from 18 to 880 with six of the studies having over 200 patient samples. Sample sizes for controls ranged from 88 to 939 with nine of the studies having over 200 control samples. It is difficult to estimate the total number of patients analyzed in these 17 studies, since it appears that the same authors used the same sample set for different SNPs in different publications. Our assessment showed that Bye et al. (2011) and Bye et al. (2012) used the same participants. In addition, studies by Li et al. (2005)and Li et al. (2008) used the same participants as Dandara et al. (2005). The remaining 12 studies do not seem to have any obvious sample overlap.
Altogether, 16 out of 17 studies clinically assessed for ESCC through histology. None of the studies clinically assessed controls for ESCC with the exception of one study (Strickland et al., 2012), which assessed controls using a brush biopsy. Nine studies reported on smoking and alcohol consumption status for all participants (Dandara et al., 2005; Li et al., 2005; Dandara et al., 2006; Li et al., 2008 Li et al., 2010; Bye et al., 2012; Vogelsang et al., 2012; Matejcic et al., 2015; Chen et al., 2019), while three (Bye et al., 2011; Matejcic et al., 2011; Strickland et al., 2012) reported those risk factors for only the ESCC patients.
The Hardy–Weinberg equilibrium deviation was assessed in 11 (65%) studies; however, only six (35%) of the studies reported power calculations, and three (18%) studies reported the evaluation of a genotyping error. Detailed characteristics of the study population were reported in 12 of the studies for cases and 10 for controls. Correction for multiple testing was reported in only seven (41%) studies. NCBI rs numbers were reported in eight (47%) studies. Our quality assessment scoring had 11 items ( Table S1 ), and each item had a weight of 1 point; therefore, total maximum quality score was 11. Overall, only seven of the 17 (41%) studies scored half or above half (5.5). The highest score was 9 (Vogelsang et al., 2012; Chen et al., 2019), and the lowest score was 1 (Vos et al., 2003; Zaahl et al., 2005).
Somatic Variant Studies
Somatic variant studies ( Table 3 ) constituted of eight studies published between 1990 and 2016 (Victor et al., 1990; Gamieldien et al., 1998; Dietzsch and Parker, 2002; Dietzsch et al., 2003; Vos et al., 2003; Naidoo et al., 2005; Patel et al., 2011; Liu et al., 2016). A total of 455 patients were assessed, with the control group comprising 200 NET and 146 blood samples. Of the 455 patient samples, one was reported to be an adenocarcinoma from one study; therefore, the exact ESCC patient population was 454. The study populations were from South Africa, Kenya, and Malawi.
Clinical diagnosis of ESCC was determined by histology in five (75%) studies, and the remaining three did not report on how clinical assessment was done. Four (50%) studies reported using both cancer tissue and NET for assessment. Three of these studies had an equal number of cancer tissue and NET samples. Two (25%) studies did not have any control samples, and the remaining two (25%) studies collected blood samples only as controls. Only two studies reported on smoking and alcohol consumption status. On patient characteristics, age and sex were reported in six (75%) of the studies. Variant classification and type were reported in all of the studies, but confirmation of results was reported in only two studies. No studies used pathogenicity scoring. Amino acid change was also reported in only two of the studies. Our quality assessment score had seven items ( Table S2 ), and each item had a weight of 1 point; therefore, total maximum score for the quality assessment was 7. Overall, six of the eight (75%) studies scored half or above half (3.5). The highest score was 6 (Gamieldien et al., 1998), and the lowest score was 0 (Victor et al., 1990).
Description of Genes Studied
A total of 58 genes were investigated in the 23 studies, which were selected for the systematic review, with 37 genes studied in the genetic susceptibility studies and 23 in the somatic variant studies. Two genes were investigated in both studies. In addition, the somatic studies investigated six genetic loci without specific gene names. A summary of SNPs analyzed in the genetic susceptibility studies is shown in Table 4 . Over 100 SNPs were analyzed, and 25 SNPs were reported to be associated with ESCC (four SNPs using p values only, and 21 SNPs using p values and odds ratios). The 25 SNPs were in 20 genes: ADH1B, ADH3, ALDH2, AR, CASP8, CHEK2, CP, CYP2E1, CYP3A5, GSTT2B, MGMT, MLH3, MSH3, NAT2, PTGS2 (also known as COX-2), PLCE1, PMS1, RUNX1, SLC11A1, and TP53. The associations with all 25 SNPs were identified in South African populations, while none were found in the Sudanese population.
Table 4.
Summary of studies investigating genetic susceptibility of ESCC in African populations.
| Gene | Variant (rs number) | Study (PMID) | Population | ESCC | Controls | Effect allele | Findings and Comments2 | ||
|---|---|---|---|---|---|---|---|---|---|
| n | MAF | n | MAF | ||||||
| ADH1B | rs1229984 (Arg48His) | Bye et al., 2011 (21926110) | Black South African | 358 | 0 | 477 | 0 | Not informative | |
| Bye et al., 2011 (21926110) | Mixed ancestry South African | 201 | 0.054 | 427 | 0.098 | A | OR = 0.52 (0.32–0.86) p = 0.009 | ||
| ADH2 | ADH2*1/*2/*3 | Li et al., 2008 (18254707) | Black South African | 142 | 0.01 | 174 | 0.01 | Not informative | |
| Li et al., 2008 (18254707) | Mixed ancestry South African | 96 | 0.03 | 94 | 0.03 | Not informative | |||
| ADH3 | ADH3*1/*2 | Li et al., 2008 (18254707) | Black South African | 141 | 0.46 | 174 | 0.32 | NS | |
| Li et al., 2008 (18254707) | Mixed ancestry South African | 96 | 0.38 | 94 | 0.31 | *2 | OR = 1.80; p = 0.0004 | ||
| ADH7 | rs1573496 (Gly92Ala) | Bye et al., 2011 (21926110) | Black South African | 358 | 0 | 477 | 0.001 | Not informative | |
| Bye et al., 2011 (21926110) | Mixed ancestry South African | 201 | 0.014 | 427 | 0.02 | NS | |||
| ALDH2 | rs671 (Glu504Lys) | Bye et al., 2011 (21926110) | Black South African | 358 | 0 | 477 | 0 | Not informative | |
| Bye et al., 2011 (21926110) | Mixed ancestry South African | 201 | 0 | 427 | 0 | Not informative | |||
| rs441 (-261 C/T) |
Bye et al., 2011 (21926110) | Black South African | 358 | 0.154 | 477 | 0.145 | NS | ||
| Bye et al., 2011 | Mixed ancestry South African | 201 | 0.18 | 427 | 0.194 | NS | |||
| rs886205 (+82 A/G) | Bye et al., 2011 (21926110) | Black South African | 358 | 0.247 | 477 | 0.252 | NS | ||
| Bye et al., 2011 (21926110) | Mixed ancestry South African | 201 | 0.402 | 427 | 0.489 | G | OR = 0.70 (0.55–0.89); p = 0.004 | ||
| ALDH2*1/*2 | Li et al., 2008 (18254707) | Black South African | 142 | 0.10 | 174 | 0.04 | *2 | OR = 2.35; p = 0.008 | |
| Li et al., 2008 (18254707) | Mixed ancestry South African | 101 | 0.03 | 1004 | 0.04 | Not informative | |||
| rs4767364 (A/G) | Chen et al., 2019 (30753320) | Black South African5 | 880 | 0.12 | 939 | 0.11 | NS | ||
| ALS2CR12 | rs13016963 (G/A) | Chen et al., 2019 (30753320) | Black South African4 | 591 | 0.35 | 852 | 0.35 | NS | |
| Chen et al., 2019 (30753320) | Black South African5 | 880 | 0.39 | 939 | 0.38 | NS | |||
| rs10201587 (A/G) | Chen et al., 2019 (30753320) | Black South African5 | 880 | 0.38 | 939 | 0.39 | NS | ||
| AR | CAG-repeat in exon 1 | Dietzsch et al., 2003 (12925954) | Black South African males | 29 | 109 | NS | |||
| Dietzsch et al., 2003 (12925954) | Mixed ancestry South African males | 15 | 58 | NS | |||||
| GGC-repeat in exon 1 | Dietzsch et al., 2003 (12925954) | Black South African males | 29 | 109 | (GGC)≤16 | OR = 2.7 (1.14–6.36); p = 0.018 | |||
| Dietzsch et al., 2003 (12925954) | Mixed ancestry South African males | 15 | 58 | NS | |||||
| ATP1B2/TP53 | rs1642764 (C/T) | Chen et al., 2019 (30753320) | Black South African4 | 591 | 0.21 | 852 | 0.20 | NS | |
| Chen et al., 2019 (30753320) | Black South African5 | 880 | 0.18 | 939 | 0.18 | NS | |||
| rs1641511 (A/G) | Chen et al., 2019 (30753320) | Black South African5 | 880 | 0.39 | 939 | 0.42 | NS | ||
| C20orf54 | rs13042395 | Bye et al., 2012 (22865593) | Black South African | 407 | 0.002 | 849 | 0.005 | Not informative | |
| Bye et al., 2012 (22865593) | Mixed ancestry South African | 257 | 0.067 | 860 | 0.068 | NS | |||
| CASP8 | rs1045485 (Asp302His) | Bye et al., 2011 (21926110) | Black South African | 358 | 0.154 | 477 | 0.152 | NS | |
| Bye et al., 2011 (21926110) | Mixed ancestry South African | 201 | 0.169 | 427 | 0.126 | C | OR = 1.42 (1.01–1.98); p = 0.040 | ||
| rs3834129 (-652 6N ins/del) |
Bye et al., 2011 (21926110) | Black South African | 358 | 0.518 | 477 | 0.502 | NS | ||
| Bye et al., 2011 (21926110) | Mixed ancestry South African | 201 | 0.385 | 427 | 0.386 | NS | |||
| rs10931936 (C/T) | Chen et al., 2019 (30753320) | Black South African4 | 591 | 0.19 | 852 | 0.20 | NS | ||
| Chen et al., 2019 (30753320) | Black South African5 | 880 | 0.22 | 939 | 0.20 | NS | |||
| CHEK2 | rs4822983 (C/T) | Chen et al., 2019 (30753320) | Black South African4 | 591 | 0.46 | 852 | 0.39 | T | OR = 1.32 (1.12–1.56); p = 0.001 |
| Chen et al., 2019 (30753320) | Black South African5 | 880 | 0.43 | 939 | 0.42 | NS | |||
| rs1033667 (C/T) | Chen et al., 2019 (30753320) | Black South African4 | 591 | 0.44 | 852 | 0.38 | T | OR = 1.30 (1.10–1.53) P = 0.002 |
|
| Chen et al., 2019 (30753320) | Black South African5 | 880 | 0.42 | 939 | 0.39 | NS | |||
| CP | rs34053109 (C/G) | Strickland et al., 2012 (21901748) | Black South African | 84 | 0 | 85 | 0.01 | Not informative | |
| rs17838834 (T/C) | Strickland et al., 2012 (21901748) | Black South African | 90 | 0.33 | 85 | 0.23 | NS | ||
| rs701749 (C/T) | Strickland et al., 2012 (21901748) | Black South African | 79 | 0.01 | 78 | 0.02 | Not informative | ||
| rs17838833 (delT) | Strickland et al., 2012 (21901748) | Black South African | 79 | 0.01 | 78 | 0 | Not informative | ||
| rs17838832 (T/C) | Strickland et al., 2012 (21901748) | Black South African | 80 | 0.33 | 78 | 0.3 | NS | ||
| rs34334174 (C/T) | Strickland et al., 2012 (21901748) | Black South African | 80 | 0.14 | 78 | 0.08 | NS | ||
| 5’UTR-308G/A | Strickland et al., 2012 (21901748) | Black South African | 52 | 0.05 | 64 | 0 | A | p = 0.012; sample size very small | |
| rs17838831 (A/G) | Strickland et al., 2012 (21901748) | Black South African | 53 | 0.21 | 64 | 0.22 | NS | ||
| rs138512757 (Thr83) | Strickland et al., 2012 (21901748) | Black South African | 92 | 0.02 | 84 | 0.01 | Not informative | ||
| rs35438054 (Val223) | Strickland et al., 2012 (21901748) | Black South African | 95 | 0.01 | 85 | 0.01 | Not informative | ||
| rs797045480 (Val246Ala) | Strickland et al., 2012 (21901748) | Black South African | 95 | 0.01 | 85 | 0 | Not informative | ||
| rs34067682 (IVS4-14C/T) |
Strickland et al., 2012 (21901748) | Black South African | 84 | 0.12 | 83 | 0.12 | NS | ||
| rs34624984 (Arg367Cys) |
Strickland et al., 2012 (21901748) | Black South African | 94 | 0.02 | 86 | 0.01 | Not informative | ||
| rs34237139 (Tyr425) |
Strickland et al., 2012 (21901748) | Black South African | 91 | 0.01 | 87 | 0 | Not informative | ||
| rs35272481 (IVS7+9T/C) | Strickland et al., 2012 (21901748) | Black South African | 91 | 0.01 | 87 | 0 | Not informative | ||
| rs701753 (D544E) |
Strickland et al., 2012 (21901748) | Black South African | 95 | 0.23 | 81 | 0.27 | NS | ||
| rs147192657 (Gly633 T/C) |
Strickland et al., 2012 (21901748) | Black South African | 88 | 0.07 | 84 | 0 | C | p = 0.0004 | |
| rs16861582 (IVS15-12T/C) |
Strickland et al., 2012 (21901748) | Black South African | 93 | 0,44 | 88 | 0.41 | NS | ||
| CYP2E1 | CYP2E1*1 (c1)/CYP2E1*5 (c2) | Chelule et al., 2006 (17264406) | Black South African | 30 | 0.04 | 331 | 0.06 | Limited power | |
| -1053C/T | Li et al., 2005 (15899651) | Black and Mixed ancestry South African | 189 | 0.01 | 198 | 0.02 | NS | ||
| -1293G/A | Li et al., 2005 (15899651) | Black and Mixed ancestry South African | 189 | 0.01 | 198 | 0.03 | NS | ||
| 7632T/A | Li et al., 2005 (15899651) | Black and Mixed ancestry South African | 189 | 0.18 | 198 | 0.07 | A | OR = 5.90 (3.25–10.7); p = 0.001 for genotype distribution | |
| CYP3A5 | CYP3A5*1 | Dandara et al., 2005 (15978331) | Black South African | 142 | 0.627 | 178 | 0.638 | NS | |
| Dandara et al., 2005 (15978331) | Mixed ancestry South African | 99 | 0.384 | 94 | 0.287 | NS | |||
| CYP3A5*3 (6986A/G) | Dandara et al., 2005 (15978331) | Black South African | 142 | 0.155 | 178 | 0.138 | NS | ||
| Dandara et al., 2005 (15978331) | Mixed ancestry South African | 99 | 0.475 | 94 | 0.590 | G | OR = 0.60 (0.39–0.94); p = 0.025 | ||
| CYP3A5*6 (1490G/A) | Dandara et al., 2005 (15978331) | Black South African | 142 | 0.190 | 178 | 0.213 | NS | ||
| Dandara et al., 2005 (15978331) | Mixed ancestry South African | 99 | 0.136 | 94 | 0.122 | NS | |||
| CYP3A5*7 (27131-32insT; frameshift) | Dandara et al., 2005 (15978331) | Black South African | 142 | 0.028 | 178 | 0.011 | NS | ||
| Dandara et al., 2005 (15978331) | Mixed ancestry South African | 99 | 0.005 | 94 | 0 | Not informative | |||
| CYP3A5 all variants | Dandara et al., 2005 (15978331) | Black South African | 142 | 0.373 | 178 | 0.441 | NS | ||
| Dandara et al., 2005 (15978331) | Mixed ancestry South African | 99 | 0.616 | 94 | 0.713 | OR = 0.65 (0.42–0.99); p = 0.045 | |||
| FAS | rs1800682 (-670 G > A) |
Bye et al., 2011 (21926110) | Black South African | 358 | 0.219 | 477 | 0.225 | NS | |
| Bye et al., 2011 (21926110) | Mixed ancestry South African | 201 | 0.356 | 427 | 0.406 | NS | |||
| rs2234767 (-1377 G > A) |
Bye et al., 2011 (21926110) | Black South African | 358 | 0.096 | 477 | 0.072 | NS | ||
| Bye et al., 2011 (21926110) | Mixed ancestry South African | 201 | 0.139 | 427 | 0.183 | NS | |||
| FASL | rs763110 (-844 T > C) |
Bye et al., 2011 (21926110) | Black South African | 358 | 0.192 | 477 | 0.189 | NS | |
| Bye et al., 2011 (21926110) | Mixed ancestry South African | 201 | 0.416 | 427 | 0.386 | NS | |||
| GSTP1 | rs1695 (Ile105Val) |
Matejcic et al., 2011 | Black South African | 325 | 0.518 | 474 | 0.534 | NS | |
| rs1695 (Ile105Val) |
Matejcic et al., 2011 | Mixed ancestry South African | 229 | 0.454 | 428 | 0.438 | NS | ||
| rs1695 (Ile105Val) |
Li et al., 2010 (20540773) | Black South African | 0.39 | 0.37 | NS | ||||
| rs1695 (Ile105Val) |
Li et al., 2010 (20540773) | Mixed ancestry South African | 0.38 | 0.41 | NS | ||||
| rs1138272 (Ala114Val) |
Li et al., 2010 (20540773) | Black South African | 0.22 | 0.07 | NS | ||||
| rs1138272 (Ala114Val) |
Li et al., 2010 (20540773) | Mixed ancestry South African | 0.19 | 0.03 | NS | ||||
| GSTT1 | Deletion allele | Matejcic et al., 2011 (22216261) | Black South African | 311 | 0.574 | 462 | 0.554 | NS | |
| Matejcic et al., 2011 (22216261) | Mixed ancestry South African | 217 | 0.493 | 414 | 0.495 | NS | |||
| GSTT2B | Deletion allele | Matejcic et al., 2011 (22216261) | Black South African | 320 | 0.336 | 461 | 0.371 | NS | |
| Matejcic et al., 2011 (22216261) | Mixed ancestry South African | 226 | 0.418 | 425 | 0.501 | OR = 0.71 (0.57–0.90); p = 0.004 | |||
| MGMT | rs12917 (Leu84Phe) |
Bye et al., 2011 (21926110) | Black South African | 358 | 0.189 | 477 | 0.195 | NS | |
| Bye et al., 2011 (21926110) | Mixed ancestry South African | 201 | 0.222 | 427 | 0.168 | OR = 1.41 (1.05–1.91); p = 0.023 | |||
| MLH1 | rs13320360 (c.546-191T/C) |
Vogelsang et al., 2012 (22623965) | Black South African | 343 | 0.15 | 340 | 0.17 | NS | |
| Vogelsang et al., 2012 (22623965) | Mixed ancestry South African | 203 | 0.07 | 264 | 0.06 | NS | |||
| MLH3 | rs28756991 (Arg797His) |
Vogelsang et al., 2012 (22623965) | Black South African | 345 | 0.11 | 342 | 0.12 | NS | |
| Vogelsang et al., 2012 (22623965) | Mixed ancestry South African | 205 | 0.09 | 264 | 0.4 | G | OR = 2.07 (1.04–4.12); p = 0.038 | ||
| MSH2 | rs17217772 (Asn127Ser) |
Vogelsang et al., 2012 (22623965) | Black South African | 341 | 0.06 | 343 | 0.06 | NS | |
| Vogelsang et al., 2012 (22623965) | Mixed ancestry South African | 204 | 0.03 | 264 | 0.03 | NS | |||
| rs10188090 (c.2635-765G/A) |
Vogelsang et al., 2012 (22623965) | Black South African | 343 | 0.09 | 342 | 0.10 | NS | ||
| Vogelsang et al., 2012 (22623965) | Mixed ancestry South African | 205 | 0.31 | 265 | 0.33 | NS | |||
| rs3771280 (c.1510+118T/C) |
Vogelsang et al., 2012 (22623965) | Black South African | 344 | 0.11 | 339 | 0.12 | NS | ||
| Vogelsang et al., 2012 (22623965) | Mixed ancestry South African | 202 | 0.35 | 266 | 0.37 | NS | |||
| MSH3 | rs26279 (Ala1045Thr) |
Vogelsang et al., 2012 (22623965) | Black South African | 341 | 0.40 | 344 | 0.43 | NS | |
| Vogelsang et al., 2012 (22623965) | Mixed ancestry South African | 204 | 0.38 | 263 | 0.32 | A | OR = 2.71 (1.34–5.50); p = 5.71×10-3 | ||
| rs1428030 (c.1341-12568A/G) |
Vogelsang et al., 2012 (22623965) | Black South African | 342 | 0.29 | 342 | 0.27 | NS | ||
| Vogelsang et al., 2012 (22623965) | Mixed ancestry South African | 201 | 0.23 | 264 | 0.20 | NS | |||
| rs1805355 (Pro231Pro) |
Vogelsang et al., 2012 (22623965) | Black South African | 343 | 0.28 | 339 | 0.29 | NS | ||
| Vogelsang et al., 2012 (22623965) | Mixed ancestry South African | 203 | 0.24 | 265 | 0.22 | NS | |||
| NAT1 | rs1057126 (1088T > A NAT1*10) |
Matejcic et al., 2015 (26447020) | Black South African | 463 | 54.8 | 480 | 57.7 | NS | |
| Matejcic et al., 2015 (26447020) | Mixed ancestry South African | 269 | 43.4 | 288 | 40.1 | NS | |||
| rs15561 (1095C > A NAT1*10, NAT1*3) | Matejcic et al., 2015 (26447020) | Black South African | 463 | 55.7 | 480 | 57.7 | NS | ||
| Matejcic et al., 2015 (26447020) | Mixed ancestry South African | 269 | 46.5 | 288 | 43 | NS | |||
| NAT2 | rs1799930 (590G/A NAT2*6) |
Matejcic et al., 2015 (26447020) | Black South African | 463 | 24.7 | 480 | 21.4 | NS | |
| Matejcic et al., 2015 (26447020) | Mixed ancestry South African | 269 | 22.4 | 288 | 22 | NS | |||
| rs1801280 (341T/C NAT2*5) |
Matejcic et al., 2015 (26447020) | Black South African | 463 | 27.1 | 480 | 29 | NS | ||
| Matejcic et al., 2015 (26447020) | Mixed ancestry South African | 269 | 25.2 | 288 | 33.2 | C | 0R = 0.57 (0.38–0.87) p = 0.01 | ||
| rs1799931 (857G/A NAT2*7) |
Matejcic et al., 2015 (26447020) | Black South African | 463 | 0.01 | 480 | 0.05 | Not informative | ||
| Matejcic et al., 2015 (26447020) | Mixed ancestry South African | 269 | 0.05 | 288 | 0.04 | NS | |||
| rs1801279 (191G/A NAT2*14) |
Matejcic et al., 2015 (26447020) | Black South African | 463 | 0.053 | 480 | 0.063 | NS | ||
| Matejcic et al., 2015 (26447020) | Mixed ancestry South African | 269 | 0.038 | 288 | 0.023 | NS | |||
| UNC5CL | rs10484761 (G/A) | Bye et al., 2012 (22865593) | Black South African | 407 | 0.467 | 849 | 0.477 | NS | |
| Bye et al., 2012 (22865593) | Mixed ancestry South African | 257 | 0.354 | 860 | 0.314 | NS | |||
| PTGS2 | rs20417 (-765 G/C) |
Bye et al., 2011 (21926110) | Black South African | 358 | 0.471 | 477 | 0.513 | NS | |
| Bye et al., 2011 (21926110) | Mixed ancestry South African | 201 | 0.376 | 427 | 0.321 | NS | |||
| rs689466 (-1195 A/G) |
Bye et al., 2011 (21926110) | Black South African | 358 | 0.064 | 477 | 0.053 | NS | ||
| Bye et al., 2011 (21926110) | Mixed ancestry South African | 201 | 0.103 | 427 | 0.155 | G | OR = 0.63 (0.43–0.91); p = 0.014 | ||
| PDE4D | rs10052657 (C/A) |
Bye et al., 2012 (22865593) | Black South African | 407 | 0.137 | 849 | 0.128 | NS | |
| Bye et al., 2012 (22865593) | Mixed ancestry South African | 257 | 0.175 | 860 | 0.155 | NS | |||
| PLCE1 | rs2274223 (His1927Arg) |
Bye et al., 2012 (22865593) | Black South African | 407 | 0.416 | 849 | 0.403 | NS | |
| Bye et al., 2012 (22865593) | Mixed ancestry South African | 257 | 0.437 | 860 | 0.40 | NS | |||
| rs17417407 (Arg548Leu) |
Bye et al., 2012 (22865593) | Black South African | 407 | 0.166 | 849 | 0.211 | T | OR = 0.74 (0.60–0.93); p = 0.008 | |
| Bye et al., 2012 (22865593) | Mixed ancestry South African | 257 | 0.174 | 860 | 0.18 | NS | |||
| rs1438095332 (5’UTR 14 bp indel) |
Bye et al., 2012 (22865593) | Black South African | 321 | 0.234 | 456 | 0.242 | NS | ||
| rs199781223 (Gly1199Ser) | Bye et al., 2012 (22865593) | Black South African | 321 | 0.053 | 449 | 0.045 | NS | ||
| rs37655253
(Ile1777Thr) |
Bye et al., 2012 (22865593) | Black South African | 316 | 0.472 | 452 | 0.463 | NS | ||
| rs58539480 (Pro1890Leu) |
Bye et al., 2012 (22865593) | Black South African | 307 | 0.073 | 429 | 0.064 | NS | ||
| rs17417407 (G/T) | Chen et al., 2019 (30753320) | Black South African4 | 591 | 0.17 | 852 | 0.21 | T | OR = 0.76 (0.60–0.95); p = 0.014 | |
| Chen et al., 2019 (30753320) | Black South African5 | 880 | 0.19 | 939 | 0.19 | NS | |||
| rs7084339 (G/A) | Chen et al., 2019 (30753320) | Black South African5 | 880 | 0.48 | 939 | 0.46 | NS | ||
| rs3765524 (T/C) | Chen et al., 2019 (30753320) | Black South African4 | 591 | 0.47 | 852 | 0.47 | NS | ||
| Chen et al., 2019 (30753320) | Black South African5 | 880 | 0.48 | 939 | 0.46 | NS | |||
| rs2274223 (A/G) | Chen et al., 2019 (30753320) | Black South African4 | 591 | 0.42 | 852 | 0.40 | NS | ||
| Chen et al., 2019 (30753320) | Black South African5 | 880 | 0.41 | 939 | 0.43 | NS | |||
| rs11187850 (A/G) | Chen et al., 2019 (30753320) | Black South African5 | 880 | 0.21 | 939 | 0.19 | NS | ||
| PMS1 | rs5742938 (c.-21+639G/A) |
Vogelsang et al., 2012 (22623965) | Black South African | 345 | 0.18 | 344 | 0.15 | NS | |
| Vogelsang et al., 2012 (22623965) | Mixed ancestry South African | 203 | 0.43 | 266 | 0.48 | A | OR = 1.73 (1.07–2.79); p = 0.027 | ||
| rs13404927 (c.699+3331G/A) |
Vogelsang et al., 2012 (22623965) | Black South African | 342 | 0.18 | 339 | 0.19 | NS | ||
| Vogelsang et al., 2012 (22623965) | Mixed ancestry South African | 204 | 0.14 | 264 | 0.12 | NS | |||
| RUNX1 | rs2014300 (A/G) | Bye et al., 2012 (22865593) | Black South African | 407 | 0.378 | 849 | 0.403 | NS | |
| Bye et al., 2012 (22865593) | Mixed ancestry South African | 257 | 0.438 | 860 | 0.370 | G | OR = 1.33 (1.09–1.63); p = 0.0055 | ||
| rs2014300 (A/G) | Chen et al., 2019 (30753320) | Black South African4 | 591 | 0.38 | 852 | 0.40 | NS | ||
| Chen et al., 2019 (30753320) | Black South African5 | 880 | 0.36 | 939 | 0.36 | NS | |||
| rs2834718 (T/A) | Chen et al., 2019 (30753320) | Black South African5 | 880 | 0.33 | 939 | 0.33 | NS | ||
| SLC11A1 | -237C/T | Zaahl et al., 2005 (15860357) | Mixed ancestry South African | 105 | 0.029 | 110 | 0.1 | p < 0.004 | |
| -8G/A | Zaahl et al., 2005 (15860357) | Mixed ancestry South African | 105 | 0.004 | 110 | 0.009 | NS | ||
| IVSI-28C/T | Zaahl et al., 2005 (15860357) | Mixed ancestry South African | 105 | 0.028 | 110 | 0.0004 | p < 0.05 | ||
| GT-repeat | Zaahl et al., 2005 (15860357) | Mixed ancestry South African | 0.171 | 0.191 | NS | ||||
| SULT1A1 | 638G/A in Exon 7 | Dandara et al., 2006 (16272171) | Black South African | 145 | 0.42 | 194 | 0.37 | NS1 | |
| Dandara et al., 2006 (16272171) | Mixed ancestry South African | 100 | 0.40 | 94 | 0.29 | NS | |||
| TMEM173 | rs13181561 (A/G) | Chen et al., 2019 (30753320) | Black South African5 | 880 | 0.48 | 939 | 0.49 | NS | |
| rs13153461 (G/A) | Chen et al., 2019 (30753320) | Black South African4 | 591 | 0.04 | 852 | 0.05 | NS | ||
| TP53 | 16-bp insertion in intron 3 | Vos et al., 2003 (12550754) | Black South African | 74 | 0.108 | 118 | 0.364 | ||
| rs200073907 (Exon 4 codon 34) |
Vos et al., 2003 (12550754) | Black South African | 74 | 0.115 | 118 | 0.102 | NS | ||
| rs750578863 (Exon 4 codon 36) |
Vos et al., 2003 (12550754) | Black South African | 73 | 0.089 | 115 | 0.143 | NS | ||
| Arg72Pro | Vos et al., 2003 (12550754) | Black South African | 73 | 0.356 | 115 | 0.409 | p < 0.05 | ||
| Arg72Pro | Eltahir et al., 2012 (23053979) | Sudanese | 25 | 0.49 | 235 | 0.51 | NS | ||
| rs1800371 (G/A) | Chen et al., 2019 (30753320) | Black South African4 | 591 | 0.02 | 852 | 0.03 | NS | ||
| Chen et al., 2019 (30753320) | Black South African5 | 880 | 0.03 | 939 | 0.02 | NS | |||
| XBP1 | rs2239815 (C/T) | Chen et al., 2019 (30753320) | Black South African4 | 591 | 0.21 | 852 | 0.16 | T | OR = 1.41 (1.15–1.74) p = 0.001 |
| Chen et al., 2019 (30753320) | Black South African5 | 880 | 939 | NS | |||||
|
1
Increased risk among smokers with SULT1A1*2/*2 genotype, but sample size was small.
2 When OR > 1, effect allele = increased risk; when OR < 1, effect allele = protective effect. 3 rs3765525 has been merged into rs959421. 4 Western and Eastern Cape Province Black Population. 5 Gauteng Province Black Population. | |||||||||
Table 5 shows a summary of the pathways for the 20 genes. All the genes encode for proteins. Three of the genes, ADH1B, ADH3, and ALDH2, are involved in alcohol metabolism (Li et al., 2008; Bye et al., 2011). Three mismatch repair genes, MLH3, MSH3, and PMS1, play a role in genomic integrity (Vogelsang et al., 2012). They are reported to also play a role in carcinogenesis. MGMT is involved in cell defense against mutagens, and mutations in the gene are reported to be associated with cancer formation (Bye et al., 2011). NAT2 and GSTT2B play a role in the activation and deactivation of drugs and carcinogens, with reports of mutations being associated with carcinogenesis (Matejcic et al., 2015). Genes regulating cell apoptosis are TP5, CHEK2, and CASP8 (Vos et al., 2003; Bye et al., 2011; Eltahir et al., 2012; Chen et al., 2019). TP53 and CHEK2 are also involved in gene expression and DNA repair. Regulation of gene expression is facilitated by PLCE1 and SLC11A1 (Zaahl et al., 2005; Bye et al., 2012). The AR gene regulates the sex hormones, androgens (Dietzsch et al., 2003), while CYP2E1 and CYP3A5 are involved in steroid, cholesterol, and lipid synthesis (Dandara et al., 2005; Li et al., 2005; Chelule et al., 2006). CYP2E1 also metabolizes drugs and has been implicated in carcinogenesis. CP facilitates transportation of iron from organs into the blood cells; RUNX1 plays a role in hematopoiesis and PTGS2 in inflammation and mitogenesis (Bye et al., 2011; Bye et al., 2012; Strickland et al., 2012).
Table 5.
Biological pathways for genetic susceptibility studies showing putative association with ESCC in African populations.
| Gene | Full name | Pathway |
|---|---|---|
| ADH1B | Alcohol dehydrogenase 1B (class I), beta polypeptide | Ethanol metabolism |
| ADH3 | Alcohol dehydrogenase ADH3 | Metabolizes ethanol into acetaldehyde |
| ALDH2 | Aldehyde dehydrogenase 2 family member | Alcohol metabolism. Implicated in increased susceptibility for cancer |
| AR | Androgen receptor | Regulates binding of androgens on androgen receptor |
| CASP8 | Caspase 8 | Cell apoptosis |
| CHEK2 | Checkpoint kinase 2 | Tumor suppressor gene. Mutations associated with predisposition to carcinogenesis |
| CP | Ceruloplasmin | Peroxidation of iron through its transportation from organs and tissue into blood |
| CYP2E1 | Cytochrome P450 family 2 subfamily E member 1 | Drug metabolism and catalysis and synthesis of cholesterol, steroids, and other lipids. Implicated in cancer development |
| CYP3A5 | cytochrome P450 family 3 subfamily A member 5 | Involved in drug metabolism and in the synthesis of cholesterol, steroids, and other lipids |
| GSTT2B | Glutathione S-transferase theta 2B (gene/pseudogene) | Conjugation of glutathione to electrophilic and hydrophobic compounds. Plays a role in carcinogenesis |
| MGMT | O-6-methylguanine-DNA methyltransferase | DNA repair and defense from alkylating agents which cause mutagenesis and toxicity. Implicated in several cancers. |
| MLH3 | MutL homolog 3 | Maintenance of genomic integrity following cell division and DNA replication. Germline mutations implicated in cancer and somatic mutations implicated in microsatellite instability |
| MSH3 | MutS homolog 3 | Forms heterodimers with MSH2. Involved in mismatch repair and implicated in cancer development. |
| NAT2 | N-acetyltransferase 2 | Activation and deactivation of arylamine and hydrazine drugs and carcinogens. Implicated in high cancer incidence and drug toxicity. |
| PTGS2 | Prostaglandin-endoperoxide synthase 2 | A dioxygenase and a peroxidase involved in both inflammation and mitogenesis |
| PLCE1 | Phospholipase C epsilon 1 | Regulation of cell growth, differentiation, and gene expression. |
| PMS1 | PMS1 homolog 1, mismatch repair system component | Mismatch repair gene. Mutations implicated in cancer development. |
| RUNX1 | Runt related transcription factor 1 | Development of hematopoiesis |
| SLC11A1 | Solute carrier family 11 (proton-coupled divalent metal ion transporter), member 1 | Regulation of gene expression. |
| TMEM173 | Transmembrane protein 173 | Regulation of the innate immune response to viral and bacterial infections. Role in tumorigenesis still inadequate |
| TP53 | Tumor protein 53 | Regulation of gene expression, cell cycle, apoptosis, and DNA repair. |
| XBP1 | X-box binding protein 1 | Regulation of genes involved in endoplasmic reticulum protein synthesis, folding, glycosylation, redox metabolism, autophagy, lipid biogenesis, and vesicular trafficking. Associated with development of cancer. |
Nine of the 25 associated SNPs were from small studies with fewer than 150 cases and controls. These SNPs are in the following six genes: ADH3, AR, CP, CYP3A5, SLC11A1, and TP53. Because of the small sample size, the reliability and replicability of these results are uncertain. Sixteen of the SNPs came from studies with at least 150 cases and controls, and one study with 142 cases. These sample sizes could potentially give reliable and replicable results. The 16 SNPs were from the following genes: ADH1B, ALDH2, CASP8, CHEK2, CYP2E1, GSTT2B, MGMT, MLH3, MSH3, NAT2, PLCE1, PMS1, PTGS2, and RUNX1.
Two of the 16 SNPs are in the ALDH2 geneand were analyzed in two different studies. However, it is not clear whether these two SNPs are the same because, while one study reported the NCBI rs number (rs886205) (Bye et al., 2011), the other study did not (Li et al., 2008).The two SNPs reported very different MAF, and opposite odds ratios of 2.35 and 0.70 demonstrating increased risk and a protective effect, respectively.
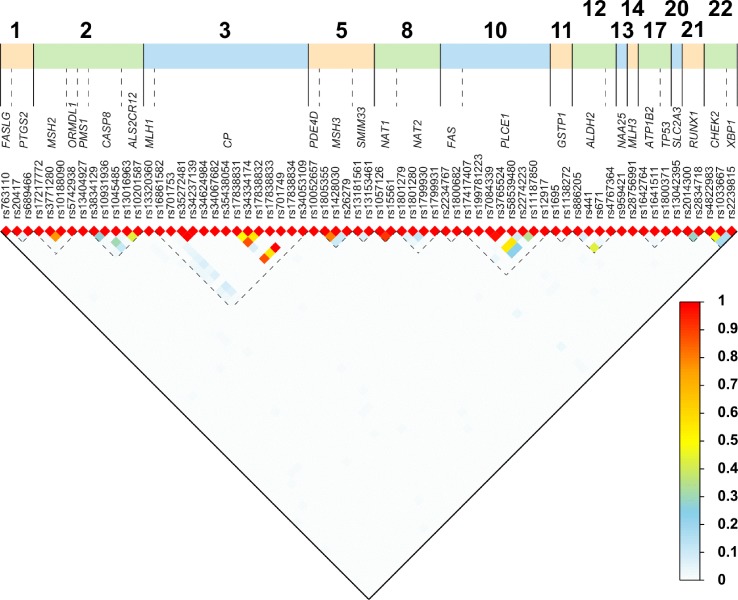
Six of the 16 SNPs were reported to reduce the risk of ESCC, and they are the following: ADH1B (Arg48His; rs1229984), ALDH2 (+82 A > G; rs886205), GSTT2B (deletion allele), NAT2 (341T > C; rs1801280), PTGS2 (-1195 A > G; rs689466), and PLCE1 (Arg548Leu; rs17417407). The remaining 10 SNPs were reported to increase the risk of ESCC: ALDH2 (ALDH2*1/*2), CASP8 (Asp302His; rs1045485), CHEK2 (rs4822983 C > T, and rs1033667, C > T), CYP2E1 (7632T > A), MGMT (Leu84Phe; rs12917), MLH3 (Arg797His; rs28756991), MSH3 (Ala1045Thr; rs26279), PMS1 (c.-21+639G > A; rs5742938), and RUNX1 (rs2014300). Eleven of the 16 SNPs showed association in the South African Admixed population, while only four showed association in the Black South African population and one in a combined South African population. All the studies used PCR-based methods for genotyping. Using the 1000 Genomes Database, r2 analysis was carried out on SNPs reported in the same gene, to assess the LD between the SNPs. Thirteen pairs of SNPs in MHS2, CP, MSH3, PLCE1,CHEK2, and NAT1 genes had r2 > 0.45, shown in Figure 2 and Table S3 .
Figure 2.
Linkage disequilibrium (LD) plot for paired SNPs. We obtained the rs numbers of the variants from dbSNP (version 152; April 2019; (https://www.ncbi.nlm.nih.gov/snp/)) and used the canonical SNP identifier. To determine the LD between the SNPs, we obtained the imputed data set from the Thousand Genomes project (1000 Genomes Release Phase 3 2013-05-02) and used bcftools to extract all individuals from African populations not including African Americans, and the 77 SNPs discussed here using all synonyms (alternative rs IDs) for SNPs (Auton et al., 2015). We obtained a dataset of 504 individuals and 67 SNPs. We computed all pair-wise r2 using PLINK (v1.09) (Danecek et al., 2011; Chang et al., 2015).
Altogether 44 somatic changes were reported in the following 22 genes: AR, CCND1, CDKN2A, COL1A2, EFGR, EP300, FAT1, FAT2, FAT3, FAT4, FBXW7, JAG1, KMT2C (MLL3), KMT2D (MLL2), MUC2, NFE2L2, NOTCH1, NOTCH3, PIK3CA, SERPINB4, TP53, and TP63, and six genetic loci without specific gene names ( Table 6 ). The specific locus positions with the corresponding microsatellite markers are as follows: 2p (D2S123), 3p13 (D3S659), 3p24.2-25 (D3S1255), 4q12 (Bat 25), 2p21-p16.3 (Bat 26), and 1p12-13.3 (Bat 40). These variants were reported in the South African (20 variants), Kenyan (three variants), and Malawian (21 variants) populations. While the majority of the studies used PCR-based methods, a more recent study used WES as the analysis method (Liu et al., 2016). A total of 18 of the 22 genes with somatic variants in cancer tissue were discovered using WES. Statistical significance was not reported for any of the 44 variants. The most common type of somatic variants was missense mutations, reported in 14 of the 22 genes (64%) (Patel et al., 2011; Liu et al., 2016). Other somatic changes included copy number gains (14%), copy number losses (5%), deletions (14%), insertions (14%), and frameshift mutations (14%). In three studies (Dietzsch and Parker, 2002; Dietzsch et al., 2003; Naidoo et al., 2005), microsatellite instability and loss of heterozygosity (LOH) were reported (14%).
Table 6.
Summary of studies investigating somatic changes linked to ESCC in African patients.
| Gene | Study (PMID) | Population | Findings |
|---|---|---|---|
| AR | Dietzsch et al., 2003 (12925954) | Black and mixed ancestry South African | LOH at CAG locus |
| CCND1 | Liu et al., 2016 (29148985) | Malawian | Enriched copy number gains |
| CDKN2A | Gamieldien et al., 1998 (9808520) | Black South African | Insertions Deletions Frameshift mutations |
| Liu et al., 2016 (29148985) | Malawian | Copy number losses | |
| COL1A2 | Dietzsch and Parker, 2002 (12435113) | Black South African | LOH (promoter and 1st intron) No evidence of MSI or allelic amplification |
| EFGR | Liu et al., 2016 (29148985) | Malawian | Copy number gains |
| EP300 | Liu et al., 2016 (29148985) | Malawian | Missense mutations |
| FAT1 | Liu et al., 2016 (29148985) | Malawian | Nonsense mutations |
| FAT2 | Liu et al., 2016 (29148985) | Malawian | Missense mutations |
| FAT3 | Liu et al., 2016 (29148985) | Malawian | Missense mutations |
| FAT4 | Liu et al., 2016 (29148985) | Malawian | Missense mutations |
| FBXW7 | Liu et al., 2016 (29148985) | Malawian | Frameshift mutations |
| JAG1 | Liu et al., 2016 (29148985) | Malawian | Missense mutations |
| KMT2C (MLL3) | Liu et al., 2016 (29148985) | Malawian | Missense mutations |
| KMT2D (MLL2) | Liu et al., 2016 (29148985) | Malawian | Nonsense mutations |
| Mismatch repair genes | Naidoo et al., 2005 (15735161) | South African | LOH and MSI at: D2S123 (2p) D3S659 (3p13) D3S1255 (3p3p24.2-25) Bat 25 (4q12) Bat 26 (2p2p21-p16.3) Bat 40 (1p12-13.3) |
| MUC2 | Liu et al., 2016 (29148985) | Malawian | Missense mutations |
| NFE2L2 | Liu et al., 2016 (29148985) | Malawian | Missense mutations |
| NOTCH1 | Liu et al., 2016 (29148985) | Malawian | Missense mutations |
| NOTCH3 | Liu et al., 2016 (29148985) | Malawian | Missense mutations |
| PIK3CA | Liu et al., 2016 (29148985) | Malawian | Missense mutations |
| Ras genes | Victor et al., 1990 (2199031) | South African | No mutations found in codon 12, 13 or 61 |
| SERPINB4 | Liu et al., 2016 (29148985) | Malawian | Missense mutations |
| TP53 | Liu et al., 2016 (29148985) | Malawian | Missense and nonsense mutations |
| Gamieldien et al., 1998 (9808520) | Black South African | Exon 5–8 frameshift mutations: point mutations, deletions and insertions |
|
| Patel et al., 2011 (22040862) | Kenyan | Exon 5–8 mutations: missense, nonsense and deletions | |
| Vos et al., 2003 (12550754) | South African | 16-bp insertion in intron 3 | |
| Vos et al., 2003 (12550754) | South African | Exon 4 polymorphism in codons 34, 36 and 72 LOH (16-bp repeat locus) |
|
| TP63 | Liu et al., 2016 (29148985) | Malawian | Copy number gains |
LOH, loss of heterozygosity; MSI, microsatellite instability.
Table 7 shows a summary of the pathways in the 22 genes reporting somatic changes. Five genes, AR, EP300, KMT2D, KMT2C, and TP53, play a role in the regulation of transcription (Gamieldien et al., 1998; Dietzsch et al., 2003; Vos et al., 2003; Patel et al., 2011; Liu et al., 2016). The encoded protein for the AR gene functions as a steroid hormone activated transcription factor, while KMT2D has a role in methylation. Both TP53 and EP300 have been implicated in a number of cancers (Gamieldien et al., 1998; Vos et al., 2003; Patel et al., 2011; Liu et al., 2016). TP53 additionally functions in DNA repair, gene expression, and apoptosis. The mismatch repair genes also facilitate DNA repair (Naidoo et al., 2005). CCND1, CDKN2A, FAT1/2/3/4, and Ras genes are all reported to be involved in cell cycle pathways including regulation of mitotic events, cell proliferation, and cell growth and death (Victor et al., 1990; Gamieldien et al., 1998; Liu et al., 2016). NOTCH1 and NOTCH3 both facilitate cell and tissue development (Liu et al., 2016). JAG1 plays a role in hematopoiesis while NFE2L2 is involved in response to inflammation including production of free radicals (Liu et al., 2016). PIK3CA is an oncogene implicated in tumor development while SERPINB4 modulates response against tumor cells (Liu et al., 2016). EGFR and COL1A2 genes encode for epidermal growth factor and type 1 collagen, respectively (Dietzsch and Parker, 2002; Liu et al., 2016). FBXW7 is a tumor suppressor involved in ubiquitin degradation (Liu et al., 2016). MUC2 facilitates the formation of a mucous barrier that protects the gut lumen (Liu et al., 2016). TP63 gene is involved in tissue and organ development including skin and heart, and in adult stem cell regulation (Liu et al., 2016).
Table 7.
Biological pathways for somatic changes studies showing putative association with ESCC in African populations.
| Gene | Full name | Pathway |
|---|---|---|
| AR | Androgen receptor gene | Regulation of gene expression and the protein functions as a steroid-hormone activated transcription factor. |
| CCND1 | Cyclin D1 | Regulators of CDK kinases and mitotic events. Mutations and overexpression of the gene has been associated with cancer development. |
| CDKN2A | Cyclin dependent kinase inhibitor 2A | A tumor suppressor gene which regulates the cell cycle. Commonly inactivated in a variety of tumors. |
| CHEK2 | ||
| COL1A2 | Collagen type I, alpha 2 chain | Encodes for type I collagen, which is an abundant connective tissue protein and part of extracellular matrix. |
| EFGR | Epidermal growth factor receptor | Encodes for the growth factor epidermal growth factor receptor. |
| EP300 | E1A binding protein p300 | Encodes the adenovirus E1A-associated cellular p300 transcriptional co-activator protein which functions in transcription regulation. Mutations have been implicated in tumorigenesis. |
| FAT1/2/3/4 | FAT atypical cadherin 1/2/3/4 | Human homologues of the Drosophila FAT genes. Putative tumor suppressor involved in cell proliferation during Drosophila development. |
| FBXW7 | F-box and WD repeat domain containing 7 | Encodes an F-Box protein which binds directly to cyclin E and potentially targets cyclin E for ubiquitin-mediated degradation. |
| JAG1 | Jagged 1 | Encodes for the human homolog of the Drosophila jagged 1 protein which is involved in hematopoiesis. |
| KMT2C (MLL3) | Lysine methyltransferase 2C | The gene is member of the myeloid/lymphoid or mixed-lineage leukemia (MLL) family. It encodes a nuclear protein involved in transcriptional regulation. |
| KMT2D (MLL2) | Lysine methyltransferase 2D | Methylation of histones and transcriptional regulation. |
| Mismatch repair genes | Mismatch repair genes | DNA repair. Mutations have been implicated in cancer. |
| MUC2 | Mucin 2, oligomeric mucus/gel-forming | Formation of insoluble mucous barrier that protects the gut lumen. |
| NFE2L2 | Nuclear factor, erythroid 2 like 2 | Encodes for proteins involved in response to inflammation including free radical production. |
| NOTCH1 | NOTCH1 | Development of cell and tissue. Mutations have been reported to be linked with tumorigenesis. |
| NOTCH3 | NOTCH3 | The third discovered human homologue of the Drosophila melanogaster type I membrane protein notch. Involved in intercellular signaling pathways in neural development. |
| PIK3CA | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha | Oncogenic and implicated in cancer development. |
| Ras genes | Rat sarcoma | Regulation of cell signaling pathways, and cell growth and death. |
| SERPINB4 | Serpin family B member 4 | Inactivation of granzyme M, an enzyme that kills tumor cells. Highly expressed in tumor cells. |
| TP53 | Tumor protein p53 | Regulates transcription, expression of target genes, thereby inducing cell cycle arrest, apoptosis, senescence, DNA repair, or changes in metabolism. Implicated in a number of cancers. |
| TP63 | Tumor protein p63 | Involved in the following processes in skin development and maintenance, adult stem/progenitor cell regulation, heart development, and premature aging. |
Interaction Studies
Combinations of specific genotypes with environmental factors were also reported to be associated with ESCC in a number of studies ( Table 2 ). The main two environmental factors studied were smoking and alcohol consumption. The interaction between smoking and alcohol status and specific genotypes was measured and reported as frequency (percentage) and assessed using p values and odds ratios in nine genetic susceptibility studies (Dandara et al., 2005; Li et al., 2005; Li et al., 2010; Dandara et al., 2006; Li et al., 2008; Li et al., 2010; Bye et al., 2011; Matejcic et al., 2011; Vogelsang et al., 2012; Matejcic et al., 2015). Four studies showed statistically significant associations between both alcohol and smoking status and variants in the CYP3A5, CYP2E1, GST, and NAT2 genes (Dandara et al., 2005; Li et al., 2005; Matejcic et al., 2015). SULT1A1 variants were associated with smoking status only (Dandara et al., 2006). Other interaction studies included wood/charcoal use and mutations in the GST genes (Li et al., 2010), as well as red and white meat intake and SNPs in NAT1/2 genes (Matejcic et al., 2015).
Discussion
General Systematic Review Findings
In this study, we systematically evaluated the genetic variants reported to be associated with ESCC in African populations providing the first systematic review on genetic factors of ESCC in this region. Of all studies that have been published on genetic association to ESCC in the African populations, only 23 fit our selection criteria. It was clear from the beginning that there is a dearth of information on this topic. Our analysis showed that 25 germline SNPs were reported to be associated with ESCC in the South African population. However, none of these SNPs were repeated in three or more independent studies; hence, a meta-analysis was not possible. Additionally, only three (ALDH2, PLCE and CYP2E1) of the 20 genes were analyzed in two independent studies, but testing for different SNPs. We determined that it was unlikely that the two ALDH2 SNPs analyzed were the same SNPs. This is because the MAFs were significantly different and, while one SNP had a protective effect (reduced risk), the other increased risk. The lack of studies re-assessing the same genetic variants poses a major hurdle in validating existing evidence on the association between genetic variants and ESCC development. This makes resolving the genetic etiology of ESCC in African populations difficult.
Genetic Susceptibility to ESCC
Of the 25 SNPs from the genetic susceptibility studies that showed an association to ESCC, we concluded that results on 16 SNPs had the potential to be reliable and reproducible due to the larger sample sizes. Ten of the SNPs were reported to increase the risk of ESCC, while six were reported to reduce the risk. However, it was noted that the majority (11) of these SNPs showed association in the South African Admixed population and the studies did not report controlling for population stratification. This is a highly admixed population (Chimusa et al., 2013), in which the predominant ancestral lines are Khoesan (32–43%), Bantu-speaking Africans (20–36%), European (21–28%), and Asian (9–11%) (De Wit et al., 2010). This diverse population is a result of South Africa’s colonial and trade history, and constitutes 9% of the total South African population (De Wit et al., 2010). Genetic variability can also be seen in the Black South African population (Chimusa et al., 2013). Without controlling for population stratification, the reproducibility of these results is questionable. It is, however, important to note that the majority of these studies were carried out several years ago, and information on population stratification and methods to detect it may not have been available as yet.
Re-examination of common SNPs from the Chinese population was done in three of the studies (Bye et al., 2011; Bye et al., 2012; Chen et al., 2019), but the findings were not conclusive. It is possible that there may be population-specific differences influencing the genetic etiology of ESCC in the African populations. This may also point to the role of environmental factors contributing to the genetic susceptibility to ESCC through gene-environment interactions.
Somatic Changes in ESCC
Forty-four somatic variants were reported, but only two were significantly associated with ESCC. The paucity of information was also evident in the somatic variant studies. There were significantly fewer studies (8) on somatic variants than on genetic susceptibility (17). The molecular profiling of tumors is of great importance as it is relevant in the development of targeted cellular therapeutics. One gene (CDKN2A) was analyzed in two studies, but these studies focused on a different variant. Another gene, TP53, was analyzed in four studies, but two studies analyzed different parts of the gene, and two had no control data. It was evident, however, that the WES study provided with a wider variety of genetic variants associated with ESCC (Liu et al., 2016). The WES study overall had the largest number of genetic variants of all the 23 studies and was able to identify variants in an unbiased manner.
Common Limitations Among the African Studies
There were no GWAS among the studies we analyzed, but reports from the Chinese and European studies demonstrated that GWAS are able to successfully identify common genetic variants associated with ESCC (Abnet et al., 2017). To date, GWAS has successfully identified more than 700 loci for cancer risk. However, these studies have been predominantly done in populations of European ancestry (80%), with African and Latin American populations contributing less than 1% (Van Loon et al., 2018). A shift to WES and GWAS on the African populations might, therefore, yield better results in identifying variants that play a role in ESCC development. The African Esophageal Cancer Consortium, which was initiated in 2016 by African investigators and International partners, released a call to action to, among other priority activities, increase molecular research on esophageal cancer in Africa, particularly GWAS and genomic profiling (Van Loon et al., 2018).
One of the main deficiencies in the studies was that the majority of the genetic susceptibility studies did not report a power calculation, or a genotyping error, and this may have resulted in studies being underpowered and with increased type II error. Few studies reported correction for multiple testing; however, many of the studies were not analyzing multiple variants at the same time. The lack of correction for multiple testing, therefore, is not a reflection on the methodological quality. Very few studies reported NCBI rs numbers. In most studies, the diagnosis of ESCC in patients was adequately defined with no ambiguity on the number of patients with ESCC. There were, however, three studies that combined samples from patients with squamous cell and adenocarcinoma into one case group, which could introduce bias (Dietzsch et al., 2003; Eltahir et al., 2012; Vogelsang et al., 2012).
It is important to note that rs numbers were poorly documented in the majority of the studies assessed in this systematic review. Additionally, in many of these studies, the positions of the SNPs using genome coordinates were not reported, hence making it difficult to locate the SNPs. In the absence of an rs number, we recommend that authors report the position using genome coordinates and the version of the genome used as a reference.
The somatic variant studies also had adequately defined ESCC diagnosis for the majority of the studies. While the variant classification and type were reported by most studies, there was no confirmation of the results (except for two studies). Overall, for both the germline and somatic variant studies, the quality of reporting for the majority of the studies was not adequate. Other important limitations and biases are the lack of controlling for population stratification and small sample sizes in the study populations, which may have led to unreliable results.
Limitations of the Systematic Review
While we did a comprehensive search in four of the main literature databases, it is possible that we could have missed some non-English studies on African populations. Because of the lack of replication and validation studies, we could not carry out a meta-analysis in the current study. Furthermore, we did not re-analyze the data and relied on reported p values and odds ratios for descriptive analysis.
Conclusions
While this review has highlighted a number of genes that may be potentially associated with ESCC in the African populations, limitations such as lack of reproducibility, quality of reporting, and quality of assessment remain a major concern. The implications of having these inconsistencies and lack of reproducibility are that the genetic etiology of ESCC in Africa will continue to be unclear. The region lags behind in contributing to genetic knowledge and literature on ESCC. Importantly, any preventative, diagnostic, or therapeutic interventions cannot be effectively identified or applied in these populations.
The identification of genetic markers of esophageal cancer susceptibility has clear translational benefits to African populations in understanding the underlying disease risk and heritability. Benefits include the utilization of genetic information to improve risk prediction, which can be translated into prevention and screening programs relevant and specific to the African population. These studies also play a role in identifying and quantifying the interactions of modifiable environmental risk factors, which interact with these genetic variants, and hence provide a platform for better targeted interventions. The ability to sufficiently translate genetic research on the African population is dependent on more genetic studies done on the population.
Our recommendations are that more and larger genetic studies be done on the African populations, particularly focusing on WES and GWAS approaches. This will require multinational collaborations between the African countries.
Ethics Statement
The study was approved by the Stellenbosch University Health Research Ethics Committee as part of the Doctoral Studies of HS (HREC Reference #: S18/10/250).
Author Contributions
VL, VS, and HS carried out literature searches. HS, VS, and HK appraised the articles, summarized the results, prepared the tables and figures, and drafted the manuscript. VS and VL reviewed the articles and edited the manuscript. VS and HK conceptualized the idea for the research, obtained funding, supervised the project, and wrote sections of the manuscript. VL provided specialist expertise and knowledge, and critically reviewed the manuscript. GT carried out the r2 analyses, prepared the r2 figure and table, and critically reviewed and revised the manuscript. All authors approved the final version of the manuscript.
Funding
This work was supported by the African Cancer Institute, Faculty of Medicine and Health Sciences, Stellenbosch University. HS acknowledges the Beit Trust Hardship Fund for providing a Doctoral Scholarship in part aid of tuition and registration fees and the Collaboration for Evidence-based Healthcare and Public Health in Africa (CEBHA+), as part of the Research Networks for Health Innovation in Sub-Saharan Africa Funding Initiative of the German Federal Ministry of Education and Research. GT was supported by the South African Tuberculosis Bioinformatics Initiative (SATBBI), a Strategic Health Innovation Partnership grant from the South African Medical Research Council and South African Department of Science and Technology.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2019.00642/full#supplementary-material
References
- Abnet C. C., Arnold M., Wei W. Q. (2017). Epidemiology of esophageal squamous cell carcinoma. Gastroenterology 154, 360–373. 10.1053/j.gastro.2017.08.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams C. H., Werely C. J., Victor T. C., Hoal E. G., Rossouw G., Van Helden P. D. (2003). Allele frequencies for glutathione S-transferase and N-acetyltransferase 2 differ in African population groups and may be associated with oesophageal cancer or tuberculosis incidence. Clin. Chem. Lab. Med. 41, 600–605. 10.1515/CCLM.2003.090 [DOI] [PubMed] [Google Scholar]
- Auton A., Brooks L. D., Durbin R. M., Garrison E. P., Kang H. M., Korbel J. O., et al. (2015). A global reference for human genetic variation. Nature 526, 68–74. 10.1038/nature15393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba Y., Yamamura K., Nakagawa S., Mima K., Ishimoto T., Iwatsuki M., et al. (2017). Abstract 4930: genetic and epigenetic characteristics of esophageal cancer tissues with microbiome fusobacterium nucleatum. Cancer Res. 77, 4930–4930. 10.1158/1538-7445.AM2017-4930 [DOI] [Google Scholar]
- Bye H., Prescott N. J., Lewis C. M., Matejcic M., Moodley L., Robertson B., et al. (2012). Distinct genetic association at the PLCE1 locus with oesophageal squamous cell carcinoma in the South African population. Carcinogenesis 33, 2155–2161. 10.1093/carcin/bgs262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bye H., Prescott N. J., Matejcic M., Rose E., Lewis C. M., Parker M. I., et al. (2011). Population-specific genetic associations with oesophageal squamous cell carcinoma in South Africa. Carcinogenesis 32, 1855–1861. 10.1093/carcin/bgr211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C. C., Chow C. C., Tellier L. C., Vattikuti S., Purcell S. M., Lee J. J. (2015). Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience 4, 7. 10.1186/s13742-015-0047-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chelule P. K., Pegoraro R. J., Gqaleni N., Dutton M. F. (2006). The frequency of cytochrome P450 2E1 polymorphisms in Black South Africans. Dis. Markers 22, 351–354. 10.1155/2006/980392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W. C., Bye H., Matejcic M., Amar A., Govender D., Khew Y. W., et al. (2019). Association of genetic variants in CHEK2 with oesophageal squamous cell carcinoma in the South African Black population. Carcinogenesis 40, 513–520. 10.1093/carcin/bgz026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Winckler B., Lu M., Cheng H., Yuan Z., Yang Y., et al. (2015). Oral microbiota and risk for esophageal squamous cell carcinoma in a high-risk area of China. PLoS One 10, e0143603. 10.1371/journal.pone.0143603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chimusa E. R., Daya M., Moller M., Ramesar R., Henn B. M., Van Helden P. D., et al. (2013). Determining ancestry proportions in complex admixture scenarios in South Africa using a novel proxy ancestry selection method. PLoS One 8, e73971. 10.1371/journal.pone.0073971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman H. G., Xie S. H., Lagergren J. (2018). The Epidemiology of Esophageal Adenocarcinoma. Gastroenterology 154, 390–405. 10.1053/j.gastro.2017.07.046 [DOI] [PubMed] [Google Scholar]
- Dandara C., Ballo R., Parker M. I. (2005). CYP3A5 genotypes and risk of oesophageal cancer in two South African populations. Cancer Lett. 225, 275–282. 10.1016/j.canlet.2004.11.004 [DOI] [PubMed] [Google Scholar]
- Dandara C., Li D. P., Walther G., Parker M. I. (2006). Gene-environment interaction: the role of SULT1A1 and CYP3A5 polymorphisms as risk modifiers for squamous cell carcinoma of the oesophagus. Carcinogenesis 27, 791–797. 10.1093/carcin/bgi257 [DOI] [PubMed] [Google Scholar]
- Danecek P., Auton A., Abecasis G., Albers C. A., Banks E., Depristo M. A., et al. (2011). The variant call format and VCFtools. Bioinformatics 27, 2156–2158. 10.1093/bioinformatics/btr330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Wit E., Delport W., Rugamika C. E., Meintjes A., Moller M., Van Helden P. D., et al. (2010). Genome-wide analysis of the structure of the South African Coloured Population in the Western Cape. Hum. Genet. 128, 145–153. 10.1007/s00439-010-0836-1 [DOI] [PubMed] [Google Scholar]
- Dietzsch E., Laubscher R., Parker M. I. (2003). Esophageal cancer risk in relation to GGC and CAG trinucleotide repeat lengths in the androgen receptor gene. Int. J. Cancer 107, 38–45. 10.1002/ijc.11314 [DOI] [PubMed] [Google Scholar]
- Dietzsch E., Parker M. I. (2002). Infrequent somatic deletion of the 5’region of the COL1A2 gene in oesophageal squamous cell cancer patients. Clin. Chem. Lab. Med. 40, 941–945. 10.1515/CCLM.2002.165 [DOI] [PubMed] [Google Scholar]
- Eltahir H. A., Adam A. A., Yahia Z. A., Ali N. F., Mursi D. M., Higazi A. M., et al. (2012). p53 Codon 72 arginine/proline polymorphism and cancer in Sudan. Mol. Biol. Rep. 39, 10833–10836. 10.1007/s11033-012-1978-0 [DOI] [PubMed] [Google Scholar]
- Gamieldien W., Victor T. C., Mugwanya D., Stepien A., Gelderblom W. C., Marasas W. F., et al. (1998). p53 and p16/CDKN2 gene mutations in esophageal tumors from a high-incidence area in South Africa. Int. J. Cancer 78, 544–549. [DOI] [PubMed] [Google Scholar]
- He F., Liu C., Zhang R., Hao Z., Li Y., Zhang N., et al. (2018). Association between the Glutathione-S-transferase T1 null genotype and esophageal cancer susceptibility: a meta-analysis involving 11,163 subjects. Oncotarget 9, 15111–15121. 10.18632/oncotarget.24534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang F. L., Yu S. J. (2018). Esophageal cancer: risk factors, genetic association, and treatment. Asian J. Surg. 41, 210–215. 10.1016/j.asjsur.2016.10.005 [DOI] [PubMed] [Google Scholar]
- Jaskiewicz K., De Groot K. M. (1994). p53 gene mutants expression, cellular proliferation and differentiation in oesophageal carcinoma and non-cancerous epithelium. Anticancer Res. 14, 137–140. [PubMed] [Google Scholar]
- Kayamba V., Bateman A. C., Asombang A. W., Shibemba A., Zyambo K., Banda T., et al. (2015). HIV infection and domestic smoke exposure, but not human papillomavirus, are risk factors for esophageal squamous cell carcinoma in Zambia: a case-control study. Cancer Med. 4, 588–595. 10.1002/cam4.434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaz A. M., Grady W. M. (2014). Epigenetic biomarkers in esophageal cancer. Cancer Lett. 342, 193–199. 10.1016/j.canlet.2012.02.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P., Rai V. (2018). MTHFR C677T polymorphism and risk of esophageal cancer: an updated meta-analysis. Egypt. J. Med. Hum. Genet. 19, 273–284. 10.1016/j.ejmhg.2018.04.003 [DOI] [Google Scholar]
- Li D.-P., Dandara C., Walther G., Parker M. I. (2008). Genetic polymorphisms of alcohol metabolising enzymes: their role in susceptibility to oesophageal cancer. Clin. Chem. Lab. Med. 46, 323–328. 10.1515/CCLM.2008.073 [DOI] [PubMed] [Google Scholar]
- Li D., Dandara C., Parker M. I. (2005). Association of cytochrome P450 2E1 genetic polymorphisms with squamous cell carcinoma of the oesophagus. Clin. Chem. Lab. Med. 43, 370–375. 10.1515/CCLM.2005.067 [DOI] [PubMed] [Google Scholar]
- Li D., Dandara C., Parker M. I. (2010). The 341C/T polymorphism in the GSTP1 gene is associated with increased risk of oesophageal cancer. BMC Genet. 11, 47. 10.1186/1471-2156-11-47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Yu X., Zhang Z. Y., Wu C. L., Xu H. L. (2016). Interaction of XRCC1 Arg399Gln polymorphism and alcohol consumption influences susceptibility of esophageal cancer. Gastroenterol. Res. Pract. 2016, 9495417. 10.1155/2016/9495417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little J., Higgins J. P., Ioannidis J. P., Moher D., Gagnon F., Von Elm E., et al. (2009). STrengthening the REporting of Genetic Association Studies (STREGA)–an extension of the STROBE statement. Genet. Epidemiol. 33, 581–598. 10.1002/gepi.20410 [DOI] [PubMed] [Google Scholar]
- Liu W., Snell J. M., Jeck W. R., Hoadley K. A., Wilkerson M. D., Parker J. S., et al. (2016). Subtyping sub-Saharan esophageal squamous cell carcinoma by comprehensive molecular analysis. JCI Insight 1, e88755. 10.1172/jci.insight.88755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matejcic M., Li D., Prescott N. J., Lewis C. M., Mathew C. G., Parker M. I. (2011). Association of a deletion of GSTT2B with an altered risk of oesophageal squamous cell carcinoma in a South African population: a case-control study. PLoS One 6, e29366. 10.1371/journal.pone.0029366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matejcic M., Vogelsang M., Wang Y., Parker M. I. (2015). Erratum to: NAT1 and NAT2 genetic polymorphisms and environmental exposure as risk factors for oesophageal squamous cell carcinoma: a case-control study. BMC Cancer 15, 658. 10.1186/s12885-015-1681-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munishi M. O., Hanisch R., Mapunda O., Ndyetabura T., Ndaro A., Schüz J., et al. (2015). Africa’s oesophageal cancer corridor: do hot beverages contribute? Cancer Causes Control 26, 1477–1486. 10.1007/s10552-015-0646-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy G., Mccormack V., Abedi-Ardekani B., Arnold M., Camargo M. C., Dar N. A., et al. (2017). International cancer seminars: a focus on esophageal squamous cell carcinoma. Ann Oncol. 28, 2086–2093. 10.1093/annonc/mdx279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naidoo R., Ramburan A., Reddi A., Chetty R. (2005). Aberrations in the mismatch repair genes and the clinical impact on oesophageal squamous carcinomas from a high incidence area in South Africa. J. Clin. Pathol. 58, 281–284. 10.1136/jcp.2003.014290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel K., Mining S., Wakhisi J., Gheit T., Tommasino M., Martel-Planche G., et al. (2011). TP53 mutations, human papilloma virus DNA and inflammation markers in esophageal squamous cell carcinoma from the Rift Valley, a high-incidence area in Kenya. BMC Res. Notes 4, 469. 10.1186/1756-0500-4-469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pink R. C., Bailey T. A., Iputo J. E., Sammon A. M., Woodman A. C., Carter D. R. (2011). Molecular basis for maize as a risk factor for esophageal cancer in a South African population via a prostaglandin E2 positive feedback mechanism. Nutr. Cancer 63, 714–721. 10.1080/01635581.2011.570893 [DOI] [PubMed] [Google Scholar]
- Schaafsma T., Wakefield J., Hanisch R., Bray F., Schüz J., Joy E. J. M., et al. (2015). Africa’s oesophageal cancer corridor: geographic variations in incidence correlate with certain micronutrient deficiencies. PloS One 10, e0140107. 10.1371/journal.pone.0140107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sewram V., Sitas F., O’connell D., Myers J. (2014). Diet and esophageal cancer risk in the Eastern Cape Province of South Africa. Nutr. Cancer 66, 791–799. 10.1080/01635581.2014.916321 [DOI] [PubMed] [Google Scholar]
- Sewram V., Sitas F., O’connell D., Myers J. (2016). Tobacco and alcohol as risk factors for oesophageal cancer in a high incidence area in South Africa. Cancer Epidemiol. 41, 113–121. 10.1016/j.canep.2016.02.001 [DOI] [PubMed] [Google Scholar]
- Strickland N. J., Matsha T., Erasmus R. T., Zaahl M. G. (2012). Molecular analysis of ceruloplasmin in a South African cohort presenting with oesophageal cancer. Int. J. Cancer 131, 623–632. 10.1002/ijc.26418 [DOI] [PubMed] [Google Scholar]
- Uys P., Van Helden P. D. (2003). On the nature of genetic changes required for the development of esophageal cancer. Mol. Carcinog. 36, 82–89. 10.1002/mc.10100 [DOI] [PubMed] [Google Scholar]
- Van Loon K., Mwachiro M. M., Abnet C. C., Akoko L., Assefa M., Burgert S.L., et al. (2018). The African esophageal cancer consortium: a call to action. J. Glob. Oncol. 4, 1–9. 10.1200/JGO.17.00163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Victor T., Du Toit R., Jordaan A. M., Bester A. J., Van Helden P. D. (1990). No evidence for point mutations in codons 12, 13, and 61 of the ras gene in a high-incidence area for esophageal and gastric cancers. Cancer Res. 50, 4911–4914. [PubMed] [Google Scholar]
- Vogelsang M., Wang Y., Veber N., Mwapagha L. M., Parker M. I. (2012). The cumulative effects of polymorphisms in the DNA mismatch repair genes and tobacco smoking in oesophageal cancer risk. PLoS One 7, e36962. 10.1371/journal.pone.0036962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vos M., Adams C. H., Victor T. C., Van Helden P. D. (2003). Polymorphisms and mutations found in the regions flanking exons 5 to 8 of the TP53 gene in a population at high risk for esophageal cancer in South Africa. Cancer Genet. Cytogenet. 140, 23–30. 10.1016/S0165-4608(02)00638-6 [DOI] [PubMed] [Google Scholar]
- Yazbeck R., Jaenisch S. E., Watson D. I. (2016). From blood to breath: new horizons for esophageal cancer biomarkers. World J. Gastroenterol. 22, 10077–10083. 10.3748/wjg.v22.i46.10077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaahl M. G., Warnich L., Victor T. C., Kotze M. J. (2005). Association of functional polymorphisms of SLC11A1 with risk of esophageal cancer in the South African Colored population. Cancer Genet. Cytogenet. 159, 48–52. 10.1016/j.cancergencyto.2004.09.017 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.