Abstract
To date, 29 distinct microRNAs (miRNAs) have been reported to be expressed during herpes simplex virus infections. Sequence analysis of mature herpes simplex virus-1 (HSV-1) miRNAs revealed five sets of miRNAs that are complementary to each other: miR-H6-5p/H1-3p, miR-H6-3p/H1-5p, H2-5p/H14-3p, miR-H2-3p/H14-5p, and miR-H7/H27. However, the roles of individual miRNAs and consequences of this complementarity remain unclear. Here, we focus on two of these complementary miRNAs, miR-H6-5p and miR-H1-3p, using loss-of-function experiments in vitro and in a mouse model of infection using an miRNA sponge approach, including tandem multiplex artificial miRNA-binding sequences that do not match perfectly to the target miRNA inserted downstream of a green fluorescent protein reporter gene. Infection with recombinant virus expressing the miR-H6-5p sponge reduced viral protein levels and virus yield. Decreased accumulation of viral proteins was also observed at early stages of infection in the presence of both an miR-H6-5p inhibitor and plasmid-expressed miR-H1-3p. Moreover, establishment of latency and reactivation did not differ between the recombinant virus expressing the miR-H6-5p sponge and wild-type HSV-1. Taken together, these data suggest that miR-H6-5p has an as-yet-unidentified role in the early stages of viral infection, and its complement miR-H1-3p suppresses this role in later stages of infection. This report extends understanding of the roles of miRNAs in infection by herpes simplex viruses, supporting a model of infection in which the production of virus and its virulent effects are tightly controlled to maximize persistence in the host and population.
Electronic supplementary material
The online version of this article (10.1007/s12250-019-00111-6) contains supplementary material, which is available to authorized users.
Keywords: miR-H6-5p, miR-H1-3p, Sponge, Replication, Herpes simplex virus-1 (HSV-1)
Introduction
Herpes simplex viruses (HSV) are common human pathogens that establish lytic infection in cells of the oral mucosa and result in lifelong latent infection in the trigeminal ganglia. Following initial lytic infection, the viruses are transported to the sensory or autonomic neurons where they establish latent infection (Roizman and Whitley 2013; Yu and He 2016). Virus reactivation occurs throughout life in response to environmental stress, and may lead to anterograde transport of infectious virions from the latently infected ganglia to a site at or near the place of initial infection (i.e., the oral mucosa) where the virus is transmissible by physical contact between infected tissues of the donor and recipient. During lytic infection, viral genes are expressed in three temporally distinct classes (α, β, and γ). By contrast, only the latency associated transcript (LAT) is detected in latently infected neurons (Roizman and Whitley 2013; Aranda and Epstein 2015; Bloom 2016). Recent evidence has also pointed to an association of microRNAs (miRNAs) in mediating the infection of HSVs at different stages (Munson and Burch 2012; Jurak et al. 2014); however, the precise roles and mechanisms remain unclear.
Upon entry into epithelial cells, HSV initiates lytic infection during which more than 80 gene products are expressed in a tightly regulated manner. Infection culminates in the production and release of infectious progeny. Recent evidence has revealed that both HSV-1 and HSV-2 encode miRNAs during lytic infection. In particular, herpes simplex virus-1 (HSV-1) encodes at least 29 miRNAs that are dispersed throughout the genome with enrichment in or near the LAT gene (Cui et al. 2006; Sun and Li 2012; Flores et al. 2013; Du et al. 2015; Han et al. 2016). Several pairs of these miRNAs are encoded on the sense and antisense strands of the genome and, as such, are complementary. Analysis of the mature sequence of HSV-1 miRNAs revealed five sets of complementary miRNAs: H6-5p/H1-3p, H6-3p/H1-5p, H2-5p/H14-3p, H2-3p/H14-5p, and H7/H27 (Umbach et al. 2009; Jurak et al. 2010; Kramer et al. 2011; Sun and Li, 2012; Wu et al. 2013). HSV-1-encoded miRNAs show different expression patterns in productive infection, latent infection, and reactivation from latency (Umbach et al. 2009; Kramer et al. 2011; Sun and Li 2012; Du et al. 2015). Although the exact function of most virally encoded miRNAs remains undiscovered, several lines of evidence support a role in the regulation of both viral and host gene expression. To date, the viral gene targets and corresponding miRNAs identified include ICP0, ICP34.5, and ICP4 and their respective miRNAs miR-H2, miR-H4-5p, and miR-H6-3p (Flores et al. 2013; Jiang et al. 2016; Zhao et al. 2015; Umbach et al. 2008). Host targets of miR-H4-5p, miR-H6-3p, and miR-H8 have been identified as the p16 and PI3 K-Akt pathways, IL6 regulation, the cellular transcriptional repressor KLHL24, and the immune regulator PIGT, respectively (Flores et al. 2013; Zhao et al. 2015; Enk et al. 2016; Kim et al. 2017; Wu et al. 2013). HSV-1 encodes the five pairs of miRNAs complementary to each other, while HSV-2 encodes three complementary miRNA sets (HSV-2-miR-H7-5p/-H20, -H4-3p/-H19, and -H11-5p/-H11-3p). Complementary miRNAs are common among herpes virus families but their role in virus infection has not been precisely determined. miR-H6, encoded by both HSV-1 and HSV-2, is located upstream of the LAT promoter regions but the sequence differs slightly between the two viruses (Jurak et al. 2010; Sun and Li 2012). Interestingly, HSV-1, but not HSV-2, encodes a miRNA (miR-H6) that is complementary to miR-H1. This observation prompted us to explore the functions of miR-H6/miR-H1 miRNAs during HSV-1 lytic infection or latent infection. miR-H6 (both miR-H6-5p and miR-H6-3p) is located upstream of the LAT promoter region on the strand opposite the LAT, while miR-H1 (miR-H1-3p and miR-H1-5p) is complementary in sequence to miR-H6. HSV-1 miR-H1 and miR-H6 are abundantly expressed in lytically infected cells and may be detected by quantitative polymerase chain reaction (qPCR) and northern blotting as reported by several groups (Cui et al. 2006; Jurak et al. 2010). miR-H6-3p was reported to inhibit the expression of the transfected essential immediate early protein ICP4, and miR-H6-3p has extended seed complementary to ICP4; however, its role in viral replication from lytic and latent infection remains unclear.
Specifically, we hypothesized that miR-H6-5p is involved in the control of viral gene expression during the early stages of infection. Toward this end, we conducted a loss-of-function analysis of the effect of miR-H6-5p on infection and reactivation. At least three methods are commonly used for miRNA loss-of-function studies: genetic knockouts (Kurata and Lin 2018), miRNA inhibitors (Horwich and Zamore 2008; Liu et al. 2008), and sponge technologies (Ebert and Sharp 2010; Kluiver et al. 2012; Tay et al. 2015). We constructed miRNA sponges as an effective method to study the effects of miR-H6-5p on viral replication. miRNA sponges are tandem multiplex artificial miRNA-binding sequences that are typically imperfectly matched to the target miRNA, with a reporter gene such as green fluorescent protein (GFP) inserted upstream. This method is particularly useful to study a single miRNA of a complementary pair in both in vivo and in vitro settings. For comparison, we also conducted loss-of-function analyses using an H6-5p miRNA inhibitor and H1-3p miRNA. We first examined the pattern of accumulation of miRNAs encoding H6-5p, H6-3p, H1-5p, and H1-3p during the course of HSV-1(F) productive infection. Then we evaluated the effects of loss of miR-H6-5p on infection in HEp-2 cells in vitro and in a murine model of HSV-1 infection in vivo. One subset of miRNAs accumulates in the murine trigeminal ganglia harboring latent virus and decreases in amount during reactivation (Du et al. 2011). A distinct subset of viral miRNAs is detectable only after the latent virus undergoes reactivation (Du et al. 2015). Thus, we designed experiments to determine whether miR-H6-5P is essential to HSV latent infection or reactivation. These results can help to extend the roles of miRNAs in HSV infection and establishment, and highlight new targets for treatment.
Materials and Methods
Cell Lines and Virus
HEp-2, HEK293T, and Vero cells were obtained from the American Type Culture Collection. HEp-2 and HEK293T cells were cultured in high-glucose Dulbecco’s modified Eagle medium (DMEM) supplemented with 5% and 10% (vol/vol) fetal bovine serum, respectively. Vero cells were cultured in high-glucose DMEM supplemented with 5% (vol/vol) newborn calf serum. The prototype HSV-1 strain HSV-1(F) (Ejercito et al. 1968) was gifted from Roizman’s lab (The University of Chicago) and amplified and titrated on Vero cells.
Antibodies, miRNA Mimic, and miRNA Inhibitor
Antibodies against ICP0, ICP8 (Rumbaugh-Goodwin Institute for Cancer Research, Inc.), ICP27 (Rumbaugh-Goodwin Institute for Cancer Research, Inc.; Ackermann et al. 1984), VP16 (from the University of Chicago; McKnight et al. 1987), and US11 (from the University of Chicago; Roller and Roizman 1992) have been validated and described elsewhere. In addition, we used anti-GFP monoclonal antibody (Sungene Biotech) and anti-GAPDH (Cell Signaling Technology) as the reporter protein and loading control, respectively, for our study. The miRNA non-target (NT) mimic, miRNA H6-5p mimic, non-target (NT) inhibitor, and miRNA H6-5p inhibitor were designed and purchased from GenePharma. The sequences are as follows: NT mimic, sense: 5′-UUCUCCGAACGUGUCACGUUU-3′, and antisense: 5′-ACGUGACACGUUCGGAGAAUU-3′; H6-5p mimic, sense: 5′-GGUGGAAGGCAGGGGGGUGUA-3′, and antisense: 5′-UUCCACCUUCCGUCCCCCCAC-3′; NT inhibitor: 5′-CAGUACUUUUGUGUAGUACAA-3′; H6-5p inhibitor: 5′-UACACCCCCCUGCCUUCCACC-3′.
miR-H6-5p Sponge Design and Plasmid Construction
We constructed an H6-5p sponge consisting of a decoy vector expressing tandem repeats of the H6-5p binding sites. The binding sites for H6-5p were perfectly complementary in the seed region, with a bulge at positions 8–12 to prevent RNA interference-type cleavage and degradation of the sponge RNA. Repeats were separated by an AATT sequence. To validate the designed sponge targets, specifically miR-H6-5p, we subcloned the sponge sequence into a psiCHECK™-2 Luciferase vector, which was co-transfected with either miR-H6-5p mimic or non-target miRNA mimic (NT). Thus, if the miR-H6-5p sponge soaks up H6-5p mimic, the expression of luciferase will decrease.
The miRNA sponge sequence complementary to miR-H6-5p and the Scramble sequence were designed using Life Technologies’ BLOCK-iT™ RNAi Designer and synthesized by Ige Biotechnology (Guangzhou, China). The dual luciferase plasmid (luc-H6-5p sponge) used to test the biological function of the miR-H6-5p sponge was constructed as follows: miR-H6-5p sponge (for sequences see supplementary Table S1) was subcloned into plasmid psiCHECK™-2 (Promega, Mannheim, Germany) and a Xho I-Not I site was inserted downstream of the Renilla luciferase gene.
Sponge expression plasmids included the sponge-deficient P1000, P1066 containing a scramble sequence, and P1065 containing the miR-H6-5p sponge, which were constructed as follows: UL3 (Afl II-Spe I), Egr-1 promoter (Spe I-Bst XI), EGFP (EcoR I-Hind III), BGH poly(A) (Xho I-Not I), and UL4 (Apa I-Xba I) fragments were amplified by PCR using primers encoding respective restriction enzyme sites, and then cloned into the corresponding sites of pBluescript II KS(+) (Stratagene) along with the synthesized miRNA sponge Hind III-Xho I fragments. Primers were used for plasmid construction and SYBR Green qPCR are summarized on Supplementary Table S2. The miR-H1-3p expression plasmid (AmiRH1-3P) was constructed as follows. Synthesized miRNA fragments (5′-TACACCCCCCTGCCTTCCACCCTGTTTTGGCCACTGACTGACGCAGAAGGTTCCTGCTGAC-3′, underlined sequences indicate the mature miRH1-3p sequence) were digested with BamH I and Xho I restriction enzymes sites and cloned into the corresponding sites of the pcDNA6.2-GW/EmGFP-miR-neg control plasmid (Invitrogen).
Construction of Recombinant Virus
The recombinant virus was designed to express the miR-H6-5p sponge. In brief, the tandem repeated H6-5p sponge fused at 3′-end of the EGFP-coding sequence under control of the Egr-1 promoter was inserted between the UL3 and UL4 genes. The resulting virus was designated R1065. The cassette containing UL3-Egr-1 promoter-EGFP-miR-H6-5p sponge-BGH poly(A)-UL4 was digested with Bgl II and Pac I from P1065, and cloned into the corresponding sites of pKO5 to generate pKO-1065. Construction of BAC-1065, encoding Egr-1 promoter-EGFP-miR-H6-5p sponge-BGH poly(A), was reported previously (Gu and Roizman 2007). Plasmid DNAs isolated from BAC-1065 were transfected into Vero cells. Plaques were established and purified three times on Vero cells. Viral DNAs were isolated from individual plaques, and recombinant virus R1065 was identified by PCR and sequenced to verify that no additional sequences were introduced during assembly. The TK gene was rescued by co-transfection with a plasmid encoding the gene.
RNA Isolation and Real-time Quantitative PCR (qPCR)
Replicate cultures of HEp-2 cells seeded in 6-well plates were exposed to 10 plaque-forming units (PFU) of HSV-1(F) per cell, or HEp-2 cells in 12-well plates were exposed to 10 PFU of HSV-1(F) per cell after transfection of 1 μg of P1065, P1066, or P1000 for 24 h. The cells were harvested at various times after infection (1, 3, 6, 12, and 24 h). Total RNA was isolated using TRIzol reagent (Invitrogen) according to the manufacturer’s instructions as described previously with minor modifications (Han et al. 2016). The miRNAs tested in this study were reverse-transcribed from 5 ng total RNA in duplicate by specific stem-loop primers as described in the TaqMan miRNA reverse transcription kit (Applied Biosystems, Inc.). The expression of miRNAs was determined by real-time PCR using TaqMan Universal Master Mix II kit (Applied Biosystems, Inc). For quantification, miRNA copy numbers were normalized by the levels of 18s rRNA. The primers and TaqMan MGB probes are listed in Supplementary Table S2.
Reporter Gene Assays
To determine the biological function of the miR-H6-5p sponge, duplicate HEK293T cells in 12-well plates were co-transfected with 1 μg of pluc-H6-5p sponge with either mock, 20 or 40 nmol/L of non-target (NT) miRNA mimic, or 20 or 40 nmol/L of miR-H6-5p mimic using Lipofectamine 2000 (Life Technologies). Firefly and Renilla luciferase activities were determined at 24 h post-transfection according to the manufacturer’s instructions (Promega). Relative luciferase activity was determined by calculating the ratio of Renilla to firefly luciferase activity.
Western Blot Analysis
Cells in 12-well plates were harvested as described above and lysed with a RIPA lysis buffer (Beyotime) supplemented with 1 mmol/L protease inhibitor phenylmethylsulfonyl fluoride (Beyotime) and phosphatase inhibitor (Beyotime). Cell lysates were heat-denatured and separated by sodium dodecyl sulfate–sulfate–polyacrylamide gel electrophoresis, and transferred to polyvinylidene difluoride membranes (Millipore). EGFP served as a marker of infection for R1065 and GAPDH served as a loading control. The proteins were detected by incubation with the appropriate primary antibody, followed by horseradish peroxidase-conjugated secondary antibody (Pierce) and the ECL reagent (Pierce), and exposed to a film. The densities of corresponding bands were quantified using ImageJ software.
Virus Titration
Cells were seeded in a 6-well plate at densities of 5 × 105 cells per well for 24 h and then exposed to 0.1 PFU per cell of HSV-1(F) or R1065. The cells were harvested at 3, 6, 12, 24, and 48 h post-infection. The virus was titrated on Vero cells after three freeze–thaw cycles and brief sonication.
Murine Model of HSV-1 Latent Infection and Reactivation
Five-week-old inbred female BALB/c mice (Guangdong Medical Lab Animal Center) were infected with 105 PFU of HSV-1 (F) or recombinant virus R1065 through the corneal route as reported previously (Du et al. 2011). At 30 days after infection, the trigeminal ganglia were either analyzed immediately for viral gene expression or incubated for 24 h in medium containing 199 V with anti-NGF antibody (Abcam) at 37 °C, 5% CO2 for 24 h to induce reactivation from latency. The harvested ganglia in groups of six were analyzed for the amounts of mRNAs encoding representative α (ICP27), β (ICP8), γ1 (VP16), and LAT.
Ganglia RNA Isolation and Real-time qPCR
RNAs were extracted from the trigeminal ganglia with mirVana miRNA isolation kit (Ambion) according to the manufacturer’s instructions. The miRNAs tested in this study were reverse-transcribed from 5 ng total RNA in duplicate by specific stem-loop primers as described in the TaqMan miRNA reverse transcription kit (Applied Biosystems, Inc.). The expression of miRNAs was determined by real-time PCR using TaqMan Universal Master Mix II kit (Applied Biosystems, Inc); miRNA copy numbers were normalized by U6 snRNA. The primers and TaqMan MGB probes are listed in Supplementary Table S2. RNA was reverse-transcribed with ReverTra Ace® qPCR RT Kit (TOYOBO) according to the manufacturer’s instructions. Real-time PCR was performed using SYBR Green PCR Master Mix Kit (TOYOBO). The primers used for amplification of ICP27, ICP8, VP16, and LAT genes have been reported elsewhere (Du et al. 2011), and were synthesized by Ige Biotechnology. The probes for detecting viral ICP0, ICP27, VP16, LAT and cellular 18s are listed in Supplementary Table S2. The viral mRNA copy numbers were normalized to 20 ng RNA by comparison with cellular 18s rRNA.
Statistical Analysis
Unless stated otherwise, data represent mean ± standard deviation. All statistical analyses, including t-tests and one-way analysis of variance (ANOVA), were performed using the software GraphPad Prism 6. P < 0.05 or P < 0.01 was considered to be statistically significant or highly significant, respectively.
Results
Accumulation of miR-H6-5p, H6-3p, H1-5p, and H1-3p during the Course of HSV-1 Productive and Latent Infection
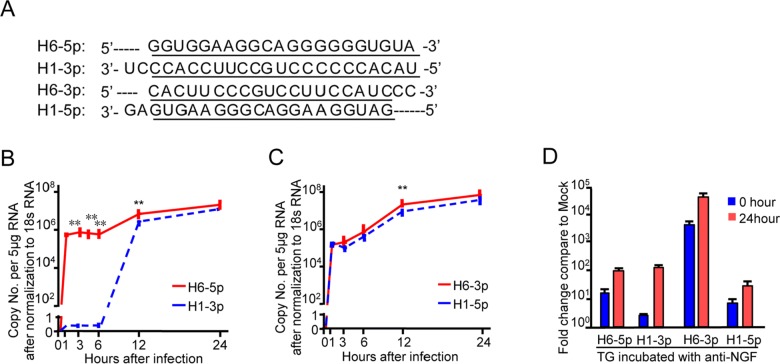
As mentioned in Introduction, sequence analysis of HSV-1 revealed two pairs of complementary miRNAs H6-5p and H1-3p as one pair and H6-3p and H1-5p as another (Fig. 1A). During productive infection in HEp-2 cells, H6-5p (Fig. 1B, red line) and H6-3p (Fig. 1C, red line) were detected 1 h after infection, reached a peak level of expression at 12 h post-infection, and remained at high levels up to the final 24-h collection time point. H1-3p (Fig. 1B, blue dotted line), which has the complementary sequence to H6-5p, started to accumulate after 6 h and remained at high levels from 12 to 24 h. However, the overall expression level of H1-3p was lower than that of H6-5p. The pattern and levels of H1-5p accumulation (Fig. 1C, blue dotted line) matched those of H6-3p.
Fig. 1.
Pattern of accumulation of miR-H6 and miR-H1 during the time course of HSV-1 productive infection. A Underlined sequences indicate regions of complementarity between mature miR-H6-5p and H1-3p, H6-3p, and H1-5p. B, C Levels of miR-H6-5p, H1-3p, H6-3p, and H1-5p in HEp-2 cells exposed to 10 PFU of HSV-1(F) per cell with qPCR. Data shown are normalized to 18s rRNA. The two independent experiments were performed in triplicate. **P < 0.01, considered highly statistically significant. D Accumulation of miR-H6-5p, H6-3p, H1-5p, and H1-3p in the murine trigeminal ganglia during latency and upon reactivation from the latent state. miRNAs quantified and normalized with respect to the U6 cellular small RNA are shown as the fold change compared with miRNAs present in ganglia excised from mock-infected mice.
In vivo, during the latent infection stage in mice, H6-3p and H6-5p were expressed at higher levels than H1-5p and H1-3p (Fig. 1D, 0 h), and all four miRNAs accumulated upon latency reactivation (Fig. 1D, 24 h).
Transfection of H6-5p Sponge in HEp-2 Cells Leads to Decreased Accumulation of Viral miRNA H6-5p at 3 h Post-infection
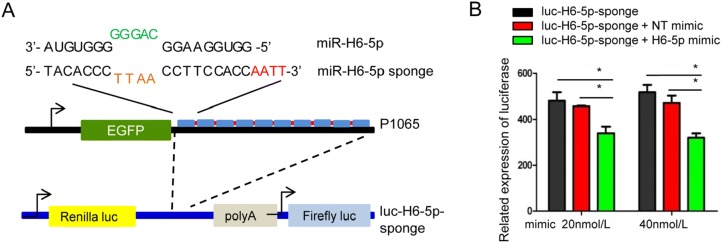
A schematic of the designed H6-5p sponge is shown in Fig. 2A. As shown in Fig. 2B, there was a greater than 20% decrease in luciferase activity when the sponge vector was co-transfected with 20 nmol/L of the H6-5p mimic and a greater than 40% decrease in luciferase activity was observed when co-transfected with 40 nmol/L miR-H6-5p mimic compared with non-target miRNA mimic (NT) or sponge alone. These results confirmed successful design of the H6-5p sponge.
Fig. 2.
Design of an artificial miR-H6-5p sponge. A Schematic representation and sequence of the designed miR-H6-5p sponge to miR-H6-5p. Complementary sequences are indicated. The nucleotides in green and light brown indicate the mismatch region and nucleotides in red indicate linker bridges between the 10 repeats of the sponge. The miR-H6-5p sponge was subcloned into the 3′ UTR of the Renilla gene of psiCHECK™-2 Luciferase vector to make the luc-H6-5p sponge plasmid. B Validation of biological function of the miR-H6-5p sponge with plasmid psiCHECK™-2. Co-transfection of luc-H6-5p sponge plasmid with either mock, or 20 or 40 nmol/L of non-target (NT) miRNA mimic or with miR-H6-5p mimic for 24 h. Decreases in luciferase activity correlate with the ability of the miR-H6-5p sponge to sequester miR-H6-5p. The two independent experiments performed in triplicate are shown. *P < 0.05, one-way ANOVA.
There was no difference in the overall pattern of timing of accumulation of all four miRNAs in cells transfected with control sponge plasmids (P1000, P1066) from those of mock-treated but HSV-1(F)-infected cells. There was a moderate decrease in miRNA accumulation, which is likely due to non-specific suppression of HSV-1 gene expression by the plasmid (Mallon et al. 2012). Notably, at 3 h post-infection, the levels of miR-H6-5p decreased by more than tenfold in P1065-transfected cells. The relative decrease was maintained until the final time point of 24 h post-infection. No other miRNA tested had reduced levels in P1065-transfected cells at this point. This observation supports the notion that P1065 effectively binds H6-5p miRNA (Fig. 3).
Fig. 3.
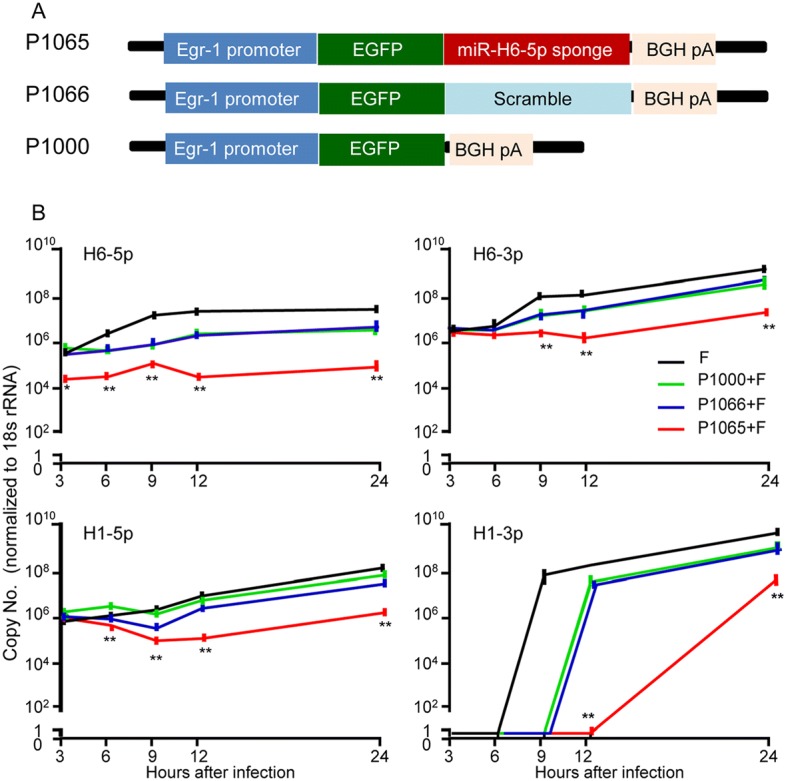
Accumulation of viral miRNAs H6-5p, H6-3p, H1-5p, and H1-3p in HEp-2 cells transfected with the plasmid encoding miR-H6-5p sponge. A Schematic representation of plasmids designed to express EGFP-fused miR-H6-5p sponge (P1065), a scrambled sequence (P1066), and no-sponge (P1000) under the Egr-1 promoter. B Expression levels of miRNAs in HEp-2 cells in 12-well plates after transfection with 1 μg of P1065, P1066, or P1000 for 24 h normalized to 18s rRNA. The two independent experiments performed in triplicate are shown. *P < 0.05; **P < 0.01, one-way ANOVA comparing the P1065 group to the other groups at the same time point.
Interestingly, transfection of P1065 resulted in a 6-h delay in the accumulation of H1-3p and more than tenfold decrease in its levels. Since the mature sequence of H6-5p is complementary to the sequence of H1-3p, this finding suggested that the H6-5p sponge could effectively interfere with the accumulation of H1-3p.
Infection of Recombinant Virus Expressing miR-H6-5p Decreased Accumulation of Viral Protein and Yields in the Lytic but not Latent Infection Stage
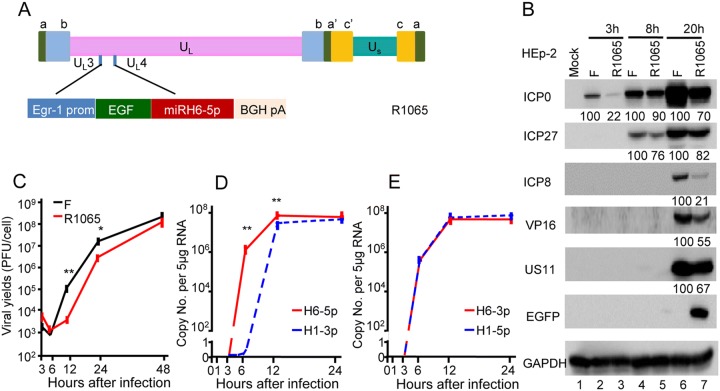
After construction and exposure to the recombinant virus R1065 (Fig. 4A), Western blotting demonstrated the reduced accumulation of proteins that are not specific to a kinetic class (Fig. 4B, lanes 3, 5, 7). The viral yield of R1065 was tenfold lower than that of the wild-type at 12 and 24 h post-infection, but became similar at 48 h post-infection (Fig. 4C). Moreover, cells infected with R1065 (Fig. 4D, 4E) showed reduced expression levels of miR-H6-5p, H6-3p, and H1-5p at 3 h after infection, while the expression of miR-H1-3p was unchanged (Fig. 1B, 1C).
Fig. 4.
Decreased viral yield and protein expression following infection with recombinant HSV (R1065)-expressing miR-H6-5p sponge. A Schematic representation of the cassette of the Egr-1 promoter, EGFP-fused miR-H6-5p sponge inserted into the wild-type viral genome between the UL3 and UL4 genes. B Accumulation of viral proteins in cells infected with wild-type HSV-1(F) and the recombinant R1065. The result is representative of two independent experiments, and the numbers under the blots indicate the relative expression calculated by ImageJ software. C Virus yields of R1065. The two independent experiments were performed in triplicate. Student’s t test was performed to compare groups at the same time point (*P < 0.05). D and E Expression levels of miRNAs in HEp-2 cells exposed to 10 PFU of R1065 normalized to 18s rRNA. The two independent experiments (each experiment was performed in triplicate) are shown. Student’s t-test was performed to compare groups at the same time point (**P < 0.01).
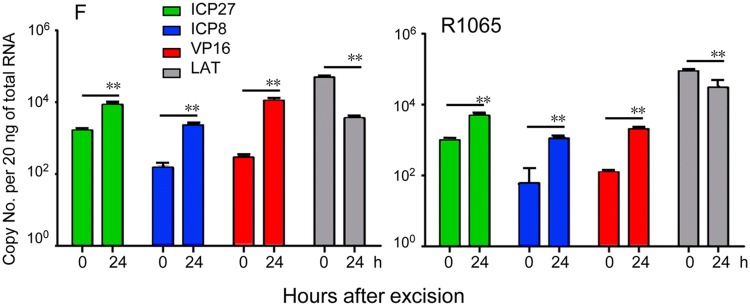
In the mouse model, recombinant virus carrying the H6-5p sponge induced the expression of all representative viral gene groups to levels indistinguishable from those of ganglia harboring latent WT virus. There was significantly less of a decrease in the amounts of LAT in ganglia harboring latent R1065 virus or induced for reactivation (Fig. 5).
Fig. 5.
Viral gene expression during virus reactivation from R1065 latently infected ganglia. Trigeminal ganglia were excised from six mice 30 days after corneal inoculation of 105 PFU of HSV-1(F) or R1065, and incubated in 199 V medium containing anti-NGF antibody for 0 or 24 h. The geometric mean of amounts of viral mRNAs encoding ICP27, ICP8, VP16, and LAT per groups of six ganglia selected at random are shown. The two independent experiments of qPCR using cDNAs as templates were performed in triplicate (**P < 0.01).
Both miR-H6-5p Inhibitor and Plasmid-expressed miR-H1-3p Decreased the Accumulation of Viral Proteins and Yield to a Degree Similar to the miR-H6-5p Sponge
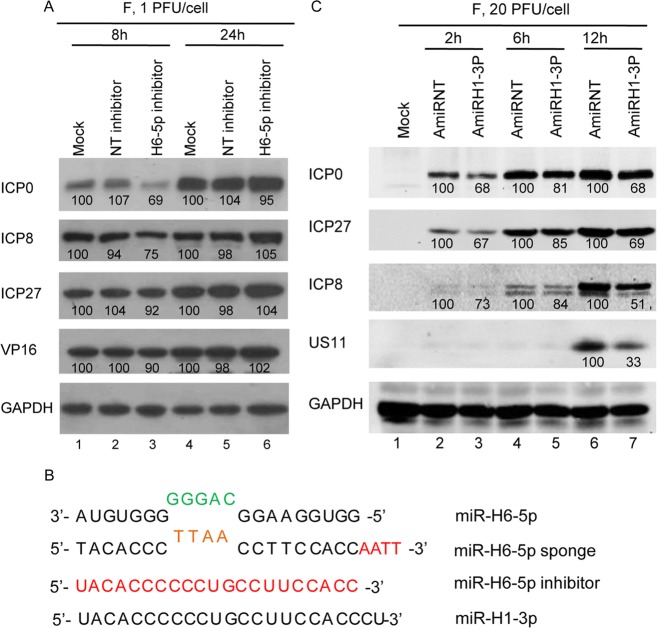
To confirm the efficacy of the sponge method for depleting the selected miRNAs, we compared the effects to those of specific inhibitors (Fig. 6A). In cells transfected with the H6-5p inhibitor, the amounts of ICP0 and ICP8 were lower than those in mock-transfected or in NT inhibitor-transfected cells at 8 h post-infection (lane 3) but not at 24 h post-infection (lane 6).
Fig. 6.
Accumulation of viral proteins in HEp-2 cells transfected with miR-H6-5p inhibitor or plasmid encoding miR-H1-3p. A Protein analysis of HEp-2 cells mock-treated or transfected with 100 nmol/L of miR-H6-5p inhibitor or non-target miRNA inhibitor (NT-inhibitor) for 24 h then exposed to 1 PFU of HSV-1(F) per cell. B Protein analysis of HEp-2 cells transfected with plasmids expressing miR-H1-3p (AmiRH1-3P) and non-target miRNA (AmiRNT). The result is representative of two independent experiments shown, and the numbers under the blots indicate the relative expression level calculated by ImageJ software. C Sequence alignment of miR-H6-5p, miR-H6-5p sponge, miR-H6-5p inhibitor, and miR-H1-3p.
Given that H1-3p expression occurs sometime after 6 h post-infection and is complementary to H6-5p, it is reasonable to assume that H1-3p plays a role in regulating H6-5p levels during HSV infection. To test this hypothesis, we constructed the plasmid AmiRH1-3p expressing miR-H1-3p to test whether miR-H1-3p reduces viral protein levels to a similar degree as the miR-H6-5p sponge. The sequence of miR-H1-3p is shown in Fig. 6B. Monolayers of HEp-2 cells were transfected with 0.5 μg of non-target miRNA (AmiRNT) or AmiRH1-3P for 24 h, exposed to 20 PFU of HSV-1(F) per cell, and harvested at 2, 6, and 12 h after infection. Compared with plasmid transfection of AmiRNT, transfection of AmiRH1-3P readily reduced the accumulation of ICP0, ICP27, ICP8, and US11 as of 2, 6, and 12 h (Fig. 6C, lanes 3, 5, 7). Thus, plasmid-expressed miR-H1-3p decreased the accumulation of viral proteins by two mechanisms: (1) direct suppression of miR-H6-5p in a manner similar to the miR-H6-5p sponge, and (2) interference of miR-H6-5p at the beginning of viral replication.
Discussion
Accumulating evidence supports the hypothesis that miRNAs play a role in the maintenance of the latent state of infection and reactivation of HSV (Umbach et al. 2008, 2009; Sun and Li 2012; Du et al. 2015). In support of this hypothesis, we observed differential expression patterns of miRNAs during latent infection and reactivation. Specifically, miR-H6-5p, miR-H6-3p, and miR-H1-5p accumulation began at early stages of infection, while miR-H1-3p accumulated at later stages of infection. Viruses expressing individual miRNAs should be precisely regulated and benefit their own development. Considering that miR-H6-3p and miR-H1-5p, and miR-H6-5p and miR-H1-3p are complementary pairs, we hypothesized that there may be functional regulation between the complementary pairs to counter each other. Umbach et al. (2008) reported that miR-H6-3p might target ICP4, an important early gene of HSV-1. miR-H1-5p accumulates in the same pattern as H6-3p, which may help to tightly regulate the function of miR-H6-3p. Specifically, miR-H6-5p accumulation starts at the early stages of infection, but the complementary miR-H1-3p accumulates at later stages of infection, suggesting that miR-H6-5p exerts functions to facilitate early virus infection. To test this hypothesis and to explore the function of H6-5p during viral productive infection, latency, and reactivation, an artificial miRNA sponge was designed against miR-H6-5p and used in loss-of-function studies. We evaluated the kinetics of expression of HSV-1 miR-H6-5p, H6-3p, H1-5p, and H1-3p in HEp-2 infected cells. H6-5p accumulated at 1 h after infection, and its expression was maintained at a high level throughout the course of infection. miR-H1-3p started to accumulate after 6 h, and remained at approximately constant levels through the 12- and 24-h time points.
Given that H1-3p is complementary in sequence to H6-5p, based on the expression patterns of complementary miRNAs, it is reasonable to consider that miR-H6-5p plays a beneficial role during the early stages of infection and has an inimical role at the later stages. To confirm this idea, we inhibited miR-H6-5p by three means: (1) H6-5p sponge, (2) H6-5p miRNA inhibitor, and (3) H1-3p miRNA. The H6-5p sponge encoding repeated miRNA antisense sequences effectively sequestered H6-5p as a valuable tool for loss-of-function studies. Indeed, miR-H6-5p levels were significantly decreased following transfection of the sponge into cells, suggesting that H6-5p produced by HSV-1 is effectively soaked by an H6-5p sponge. The recombinant virus R1065 expressing the H6-5p sponge showed a decrease of viral protein accumulation as well as virus growth curve compared to control virus. Importantly, these changes were more pronounced at early stages after infection in support of our hypothesis that the function of H6-5p is required during early infection. Transfection of the H6-5p miRNA inhibitor reduced viral yields albeit with less efficiency than the H6-5p sponge. H6-5p miRNA inhibitor was designed to block H6-5p regulation of target gene expression by competitive binding to miR-H6-5p. Thus, this reduced efficiency may be explained by either excess miR-H6-5p or insufficient binding of the miRNA inhibitor.
The rationale behind the experiments testing overexpression of H1-3p miRNA was twofold: first, to determine if H1-3p miRNA inhibits the function of H6-5p, and second, to determine if the early expression of miR-H1-3p affects viral protein production or virus yield. Our results demonstrated that miR-H1-3p inhibits the function of miR-H6-5p, and, if transfected prior to infection, can reduce both viral protein accumulation and overall yield. Notably, the inhibitory effect of transfected miR-H1-3p was greater than that of the H6-5p miRNA inhibitor.
Given that miR-H6-5p was produced early post-infection, and that the transcription and translation of viral genes were repressed, along with decreased virus yield when blocking the function of the miR-H6-5p using either the miR-H6-5p sponge or inhibitor, miR-H6-5p is likely important for replication during the early stage of infection. miR-H1-3p, which is complementary to miR-H6-5p, accumulates at a later stage of infection. Interestingly, when miR-H1-3p was expressed by an expression plasmid at the early stage of infection, the translation of viral genes was also repressed and the viral yield decreased (data not shown). This suggests that miR-H1-3p should counter the function of miR-H6-5p and prevent over-replication at the late stages of infection, so as to contribute to completing the replication cycle and establish long-term infection. Thus, when blocking the function of the miR-H6-5p using a miR-H6-5p sponge, miR-H1-3p was expressed with a 6-h delay as a result of lacking sufficient miR-H6-5p, and excessive miR-H1-3p will be detrimental to viral replication.
In conclusion, our data demonstrate that miR-H6-5p is important for the early stages of HSV-1 infection and its loss causes a reduction in viral yield. An additional observation was that miR-H1-3p, with a complementary sequence to miR-H6-5p, accumulates in the middle and late infection stages at a time when the miR-H6-5p function is expected to be detrimental to virus infection. The implication of our findings is that miR-H1-3p counters the role of miRNA H6-5p to ensure more efficient infection. This fits with an emerging model that HSV limits or controls its replication to ensure more productive infection over the long term in the infected cell, host, and population.
Lastly, it is interesting to predict the potential targeted genes of miR-H6-5p. Unsurprisingly, bioinformatics analysis identified a long list of targeted genes (Supplementary Table S3). A few of these genes have previously been reported to be associated with HSV productive infection (Supplementary Table S3, genes marked with asterisks), such as phosphatidylinositol binding clathrin assembly protein (PICALM) (Carter 2011), zinc finger DHHC-type containing 3 (ZDHHC3) (Wang et al. 2018), aquaporin 4 (AQP4) (Martinez-Torres et al. 2007), SUMO1/sentrin specific peptidase 7 (SENP7) (Cui et al. 2017), and inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase beta (IKBKB) (Gregory et al. 2004). These candidate targets offer opportunities for further studies to understand the detailed mechanisms of the molecular mechanisms underlying HSV infection and establishment toward developing new treatment strategies.
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Acknowledgements
These studies were supported by grants from Shenzhen Science and Innovation Commission Project Grants JCYJ20170411094933148 and Dapeng Research Project Grants KY20160301 to Shenzhen International Institute for Biomedical Research; Guangzhou Science and Innovation Commission Project Grants 2016070100039; Guangzhou Education Bureau Project Grants 1201620034 to Guangzhou Medical University.
Author Contributions
RH, XZ, ZH, and GZ designed research; RH, XZ, and SR XL performed research; RH, XZ, ZH, and GZ analyzed data and wrote the paper.
Compliance with Ethical Standards
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Animal and Human Rights Statement
All animal experiments were performed in accordance with protocols approved by the Institutional Animal Use Committee.
Contributor Information
Zhiyuan Han, Phone: +86-20-37105269, Email: zac_zhiyuan0420@126.com.
Grace Guoying Zhou, Phone: +86-755-28291816, Email: zhoug@siitm.org.cn.
References
- Ackermann M, Braun DK, Pereira L, Roizman B. Characterization of herpes simplex virus 1 alpha proteins 0, 4, and 27 with monoclonal antibodies. J Virol. 1984;52:108–118. doi: 10.1128/jvi.52.1.108-118.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aranda AM, Epstein AL. Herpes simplex virus type 1 latency and reactivation: an update. Med Sci (Paris) 2015;31:506–514. doi: 10.1051/medsci/20153105012. [DOI] [PubMed] [Google Scholar]
- Bloom DC. Alphaherpesvirus latency: a dynamic state of transcription and reactivation. Adv Virus Res. 2016;94:53–80. doi: 10.1016/bs.aivir.2015.10.001. [DOI] [PubMed] [Google Scholar]
- Carter CJ. Alzheimer's disease plaques and tangles: cemeteries of a pyrrhic victory of the immune defence network against herpes simplex infection at the expense of complement and inflammation-mediated neuronal destruction. Neurochem Int. 2011;58:301–320. doi: 10.1016/j.neuint.2010.12.003. [DOI] [PubMed] [Google Scholar]
- Cui C, Griffiths A, Li G, Silva LM, Kramer MF, Gaasterland T. Prediction and identification of herpes simplex virus 1-encoded microRNAs. J Virol. 2006;80:5499–5508. doi: 10.1128/JVI.00200-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Y, Yu H, Zheng X, Peng R, Wang Q, Zhou Y, Wang R, Wang J, Qu B, Shen N, Guo Q, Liu X, Wang C. SENP7 potentiates cGAS activation by relieving SUMO-mediated inhibition of cytosolic DNA sensing. PLoS Pathog. 2017;13:e1006156. doi: 10.1371/journal.ppat.1006156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du T, Zhou G, Roizman B. HSV-1 gene expression from reactivated ganglia is disordered and concurrent with suppression of latency-associated transcript and miRNAs. Proc Natl Acad Sci USA. 2011;108:18820–18824. doi: 10.1073/pnas.1117203108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du T, Han Z, Zhou G, Roizman B. Patterns of accumulation of miRNAs encoded by herpes simplex virus during productive infection, latency, and on reactivation. Proc Natl Acad Sci USA. 2015;112:E49–55. doi: 10.1073/pnas.1422657112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert MS, Sharp PA. MicroRNA sponges: progress and possibilities. RNA. 2010;16:2043–2050. doi: 10.1261/rna.2414110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ejercito PM, Kieff ED, Roizman B. Characterization of herpes simplex virus strains differing in their effects on social behaviour of infected cells. J Gen Virol. 1968;2:357–364. doi: 10.1099/0022-1317-2-3-357. [DOI] [PubMed] [Google Scholar]
- Enk J, Levi A, Weisblum Y, Yamin R, Charpak-Amikam Y, Wolf DG. HSV1 MicroRNA modulation of GPI anchoring and downstream immune evasion. Cell Rep. 2016;17:949–956. doi: 10.1016/j.celrep.2016.09.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores O, Nakayama S, Whisnant AW, Javanbakht H, Cullen BR, Bloom DC. Mutational inactivation of herpes simplex virus 1 microRNAs identifies viral mRNA targets and reveals phenotypic effects in culture. J Virol. 2013;87:6589–6603. doi: 10.1128/JVI.00504-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory Y, Hargett D, Holmes D, Money E, Bachenheimer SL. Efficient replication by herpes simplex virus type 1 involves activation of the IkappaB kinase-IkappaB-p65 pathway. J Virol. 2004;78:13582–13590. doi: 10.1128/JVI.78.24.13582-13590.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu H, Roizman B. Herpes simplex virus-infected cell protein 0 blocks the silencing of viral DNA by dissociating histone deacetylases from the CoREST-REST complex. Proc Natl Acad Sci USA. 2007;104:17134–17139. doi: 10.1073/pnas.0707266104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Z, Liu X, Chen X, Zhou X, Du T, Roizman B. miR-H28 and miR-H29 expressed late in productive infection are exported and restrict HSV-1 replication and spread in recipient cells. Proc Natl Acad Sci USA. 2016;113:E894–901. doi: 10.1073/pnas.1525674113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horwich MD, Zamore PD. Design and delivery of antisense oligonucleotides to block microRNA function in cultured drosophila and human cells. Nat Protoc. 2008;3:1537–1549. doi: 10.1038/nprot.2008.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X, Brown D, Osorio N, Hsiang C, BenMohamed L, Wechsler SL. Increased neurovirulence and reactivation of the herpes simplex virus type 1 latency-associated transcript (LAT)-negative mutant dLAT2903 with a disrupted LAT miR-H2. J Neurovirol. 2016;22:38–49. doi: 10.1007/s13365-015-0362-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurak I, Kramer MF, Mellor JC, van Lint AL, Roth FP, Knipe DM. Numerous conserved and divergent microRNAs expressed by herpes simplex viruses 1 and 2. J Virol. 2010;84:4659–4672. doi: 10.1128/JVI.02725-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurak I, Hackenberg M, Kim JY, Pesola JM, Everett RD, Preston CM. Expression of herpes simplex virus 1 microRNAs in cell culture models of quiescent and latent infection. J Virol. 2014;88:2337–2339. doi: 10.1128/JVI.03486-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Iizasa H, Kanehiro Y, Fekadu S, Yoshiyama H. Herpesviral microRNAs in cellular metabolism and immune responses. Front Microbiol. 2017;8:1318. doi: 10.3389/fmicb.2017.01318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kluiver J, Slezak-Prochazka I, Smigielska-Czepiel K, Halsema N, Kroesen BJ, van den Berg A. Generation of miRNA sponge constructs. Methods. 2012;58:113–117. doi: 10.1016/j.ymeth.2012.07.019. [DOI] [PubMed] [Google Scholar]
- Kramer MF, Jurak I, Pesola JM, Boissel S, Knipe DM, Coen DM. Herpes simplex virus 1 microRNAs expressed abundantly during latent infection are not essential for latency in mouse trigeminal ganglia. Virology. 2011;417:239–247. doi: 10.1016/j.virol.2011.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurata JS, Lin RJ. MicroRNA-focused CRISPR-Cas9 library screen reveals fitness-associated miRNAs. RNA. 2018;24:966–981. doi: 10.1261/rna.066282.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Sall A, Yang D. MicroRNA: an emerging therapeutic target and intervention tool. Int J Mol Sci. 2008;9:978–999. doi: 10.3390/ijms9060978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallon S, Wakim BT, Roizman B. Use of biotinylated plasmid DNA as a surrogate for HSV DNA to identify proteins that repress or activate viral gene expression. Proc Natl Acad Sci USA. 2012;109:E3549–3557. doi: 10.1073/pnas.1218783109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Torres FJ, Völcker D, Dörner N, Lenhard T, Nielsen S, Haas J, Kiening K, Meyding-Lamadé U. Aquaporin 4 regulation during acute and long-term experimental Herpes simplex virus encephalitis. J Neurovirol. 2007;13:38–46. doi: 10.1080/13550280601145340. [DOI] [PubMed] [Google Scholar]
- McKnight JL, Kristie TM, Roizman B. Binding of the virion protein mediating alpha gene induction in herpes simplex virus 1-infected cells to its cis site requires cellular proteins. Proc Natl Acad Sci USA. 1987;84:7061–7065. doi: 10.1073/pnas.84.20.7061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munson DJ, Burch AD. A novel miRNA produced during lytic HSV-1 infection is important for efficient replication in tissue culture. Arch Virol. 2012;157:1677–1688. doi: 10.1007/s00705-012-1345-4. [DOI] [PubMed] [Google Scholar]
- Roizman B, Whitley RJ. An inquiry into the molecular basis of HSV latency and reactivation. Annu Rev Microbiol. 2013;67:355–374. doi: 10.1146/annurev-micro-092412-155654. [DOI] [PubMed] [Google Scholar]
- Roller RJ, Roizman B. The herpes simplex virus 1 RNA binding protein US11 is a virion component and associates with ribosomal 60S subunits. J Virol. 1992;66:3624–3632. doi: 10.1128/jvi.66.6.3624-3632.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Li Q. The miRNAs of herpes simplex virus (HSV) Virol Sin. 2012;27:333–338. doi: 10.1007/s12250-012-3266-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay FC, Lim JK, Zhu H, Hin LC, Wang S. Using artificial microRNA sponges to achieve microRNA loss-of-function in cancer cells. Adv Drug Deliv Rev. 2015;81:117–127. doi: 10.1016/j.addr.2014.05.010. [DOI] [PubMed] [Google Scholar]
- Umbach JL, Kramer MF, Jurak I, Karnowski HW, Coen DM, Cullen BR. MicroRNAs expressed by herpes simplex virus 1 during latent infection regulate viral mRNAs. Nature. 2008;454:780–783. doi: 10.1038/nature07103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umbach JL, Nagel MA, Cohrs RJ, Gilden DH, Cullen BR. Analysis of human alphaherpesvirus microRNA expression in latently infected human trigeminal ganglia. J Virol. 2009;83:10677–10683. doi: 10.1128/JVI.01185-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Mott KR, Cilluffo M, Kilpatrick CL, Murakami S, Ljubimov AV, Kousoulas KG, Awasthi S, Luscher B, Ghiasi H. The absence of DHHC3 affects primary and latent herpes simplex virus 1 infection. J Virol. 2018;92:e01599–17. doi: 10.1128/JVI.01599-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W, Guo Z, Zhang X, Guo L, Liu L, Liao Y. A microRNA encoded by HSV-1 inhibits a cellular transcriptional repressor of viral immediate early and early genes. Sci China Life Sci. 2013;56:373–383. doi: 10.1007/s11427-013-4458-4. [DOI] [PubMed] [Google Scholar]
- Yu X, He S. The interplay between human herpes simplex virus infection and the apoptosis and necroptosis cell death pathways. Virol J. 2016;13:77. doi: 10.1186/s12985-016-0528-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Zhang C, Hou G, Song J. MicroRNA-H4-5p encoded by HSV-1 latency-associated transcript promotes cell proliferation, invasion and cell cycle progression via p16-mediated PI3 K-Akt signaling pathway in SHSY5Y cells. Int J Clin Exp Med. 2015;8:7526–7534. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.