Abstract
Fire blight is a devastating disease of apple and pear caused by the bacterium Erwinia amylovora. One of its main symptoms is canker formation on perennial tissues which may lead to the death of limbs and/or the entire tree. E. amylovora overwinters in cankers which play an important role in initiating fire blight epidemics. However, knowledge of pathogen biology in cankers is scarce, in part due to limitations of classical microbiology methods and the inability of most molecular techniques to distinguish live from dead cells. In this work, a viability digital PCR (v-dPCR) protocol using propidium monoazide (PMA) was developed, allowing for the first time the selective detection and absolute quantification of E. amylovora live cells in apple and pear cankers collected in two time periods. Some key factors affecting the v-dPCR performance were the maceration buffer composition, the target DNA amplicon length, the thermal cycle number and the use of sodium dodecyl sulfate or PMA enhancer for Gram-negative bacteria to improve the effect of PMA. In the future, this methodology could shed light on E. amylovora population dynamics in cankers and provide clues on the effect of management practices, host cultivar, host water/nutritional status, etc., on bacterial survival.
Subject terms: Microbiology techniques, Bacterial techniques and applications
Introduction
Erwinia amylovora is the etiological agent of fire blight of rosaceous plants, a devastating plant disease affecting economically important pome fruit crops like apple (Malus pumila Mill.) and pear (Pyrus communis L.), as well as ornamental and wild species1,2. Fire blight is a systemic disease attacking almost every plant organ, causing necrosis and characteristic exudates in actively growing tissues, and formation of cankers in the perennial ones, mainly on branches, the trunk, and/or the rootstock. E. amylovora overwinters in cankers until spring, when favorable environmental conditions break the host’s winter dormancy. With the host’s growth renewal, the pathogen cells multiply and emerge on the surface of some cankers, serving as the primary inoculum source for new disease outbreaks1,3.
While a role of other reservoirs in fire blight epidemics has been discussed4–6, cankers are widely considered one of the main sources of E. amylovora cells for the spread of the disease. However, knowledge of E. amylovora population dynamics in cankers through time and the impact of environmental and/or host-specific factors on E. amylovora survival in cankers is scarce, partially due to limitations of classical microbiology detection methods employed in plant disease diagnostics. Most attempts to determine the presence of E. amylovora in cankers have focused on the isolation on culture media and/or classical PCR7–11. Culture-dependent methods can underestimate the number of viable bacteria due to the impaired growth of stressed cells, growth inhibition by competitive microbiota, and/or the existence of pathogen cell populations in the viable but nonculturable (VBNC) state, which involves the inability of live bacteria to form colonies on solid media12. On the other hand, classical PCR detection neither discriminates between the live and dead E. amylovora cells nor allows their quantification. Improvement of molecular methods for pathogen quantification and/or selective detection of viable cells have been two important research topics in the last two decades13–18.
Digital PCR (dPCR) is a technology gaining importance in the field of plant pathology19–22. This technique builds on traditional PCR amplification and fluorescent probe–based detection methods such as quantitative PCR (qPCR), while enabling the absolute quantification of nucleic acids without requiring standard curves. This makes interlaboratory comparison of quantification data easier and less laborious. The main feature distinguishing dPCR from other PCR variants is the partition of samples into thousands of independent PCR sub-reactions, so that each partition receives either one or no target DNA sequences. End-point PCRs occur in parallel in each individual partition. The positive and negative amplification reactions are detected and quantified by means of fluorescence and the final concentration of target DNA copies in the sample is determined by Poisson distribution statistics23,24.
Similar to qPCR, dPCR allows the detection and quantification of specific DNA targets, but it is unable to determine if the amplified genetic material comes from live or dead cells. Many works have attempted to use the viability PCR dye propidium monoazide (PMA) for selective amplification of live bacterial DNA25–28. PMA is a DNA intercalating agent able to penetrate only compromised dead cell membranes. After photo-activation, PMA binds covalently to DNA and inhibits amplification by polymerases. Hence, only DNA originating from live cells can be detected by PCR29. However, despite the promising uses of PMA for molecular detection of live cells, several studies have highlighted major drawbacks of the technique leading to detection of false positives30.
In this study we developed a viability dPCR (v-dPCR) protocol for E. amylovora combining the chip-based QuantStudio 3D (QS3D) dPCR system and PMA. After optimization, v-dPCR allowed selective detection and absolute quantification of live E. amylovora cells in natural apple and pear cankers. This newly developed methodology will allow investigation of unknown aspects of E. amylovora biology including host interactions, pathogen population dynamics during canker formation and maturation, detection of nonculturable cells in plant samples, and assessment of the effect of environmental and/or host-related factors on E. amylovora survival in plant tissues.
Results and Discussion
Direct transfer of a qPCR protocol to the QS3D dPCR system
A known qPCR protocol for E. amylovora31 was transferred to the QS3D dPCR platform, using the same primer and probe concentrations and thermal cycling conditions as a first step to test the QS3D dPCR technology.
Analysis of no-template negative controls and DNA samples from E. amylovora-free plant material extracts resulted in detection of a background fluorescence signal easily distinguished from that corresponding to the positive amplification of target DNA (i.e. positive fluorescence calls) (Supplementary Fig. S1). However, a variable number of positive calls (0–5) was detected in some of the negative controls prepared with E. amylovora-free apple and pear plant macerates, with fluorescence intensity values similar to those in positive controls (Supplementary Fig. S2). These calls might be produced by cross-reaction with DNA of the host or common microorganisms present in samples, as reported previously19. Based on these results, only samples containing more than 5 positive calls per chip were considered positive for E. amylovora detection in further assays.
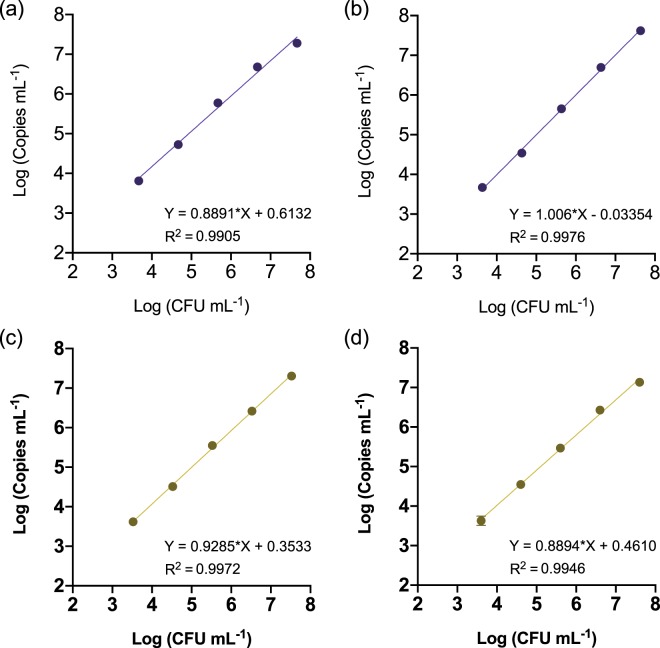
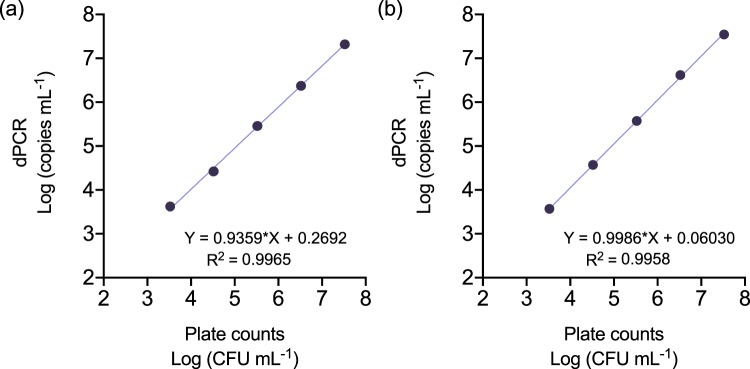
Correlation assays showed a linear relationship with a high correlation index (R2 > 0.99) between colony numbers and target DNA copy counts by dPCR in the range of 103 to 107 CFU mL−1 (Fig. 1). Pathogen detection in samples containing bacterial concentrations below 103 CFU mL−1 or above 107 CFU mL−1 led to inconsistent results or inaccurate quantification values due to chip saturation, respectively (Supplementary Tables S1 and S2).
Figure 1.
Correlation between the Log-transformed values of plate counts and QS3D dPCR absolute quantification values. Serial tenfold dilutions of E. amylovora ATCC 49946 (a,b) and CFBP 1430 (c,d) were performed in apple (a,c) and pear (b,d) branch macerates prepared in AMB, followed by DNA extraction and dPCR using a protocol transferred from Pirc et al.31. Dots and error bars are average values of three biological repeats of the experiment ± SD. Absent error bars indicate SD values smaller than the represented symbols.
The results in this section were similar for both E. amylovora strains (ATCC 49946 and CFBP 1430) and hosts (apple and pear), suggesting the QS3D dPCR system as a robust technology potentially applicable for analysis of host plant material contaminated with different strains of the pathogen.
Live-cell numbers are overestimated by v-dPCR using standard PMA treatments and the dPCR protocol transferred from a qPCR assay
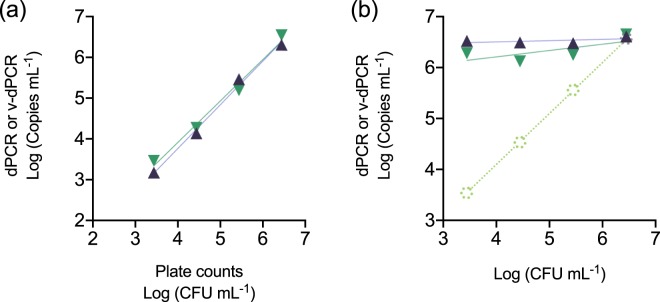
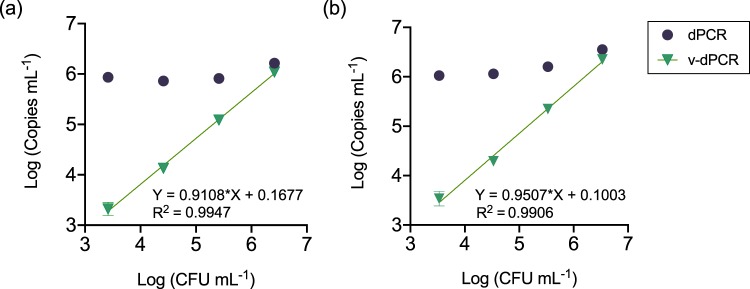
To evaluate the use of PMA for selective detection of live cells, apple plant macerates were artificially inoculated either with increasing live E. amylovora cell concentrations, or with the same live cell concentrations in a background of dead E. amylovora cells. Aliquots of the same samples were analyzed by dPCR and v-dPCR to calculate total and viable cell numbers, respectively. A good correlation between the inoculated CFU mL−1 and the total (R2 > 0.99) and live (R2 > 0.98) cell concentrations was observed in samples inoculated with the increasing live cell concentrations (Fig. 2a). However, the analysis of mixed live/dead samples revealed the failure of v-dPCR to distinguish live from dead cells, with live cell concentration values being close to those of total cell counts (i.e. most of the dead cells in mixed samples were quantified as live ones by v-dPCR) (Fig. 2b).
Figure 2.
Correlation between plate counts and v-dPCR values in the presence of dead cells. Apple macerates prepared in AMB were inoculated with E. amylovora live cells ranging from 103 to 106 CFU mL−1 (control) (a), or the same range of live cell concentrations, but with a constant background of 106 dead cell mL−1 (b). Each dilution was subjected to dPCR to quantify total target DNA copies mL−1 (purple), and v-dPCR, to quantify the copies mL−1 coming from live cells (green), using primers and probe of Pirc et al.31. The linear regression equations and coefficients of correlation corresponding to dPCR and v-dPCR values in (a) were Y = 1.070*X–0.5165 (R2 = 0.9925) and Y = 1.014*X–0.1432 (R2 = 0.9825), respectively. Dashed lines and symbols in (b) represent the expected live cell concentration values (pale green), calculated based on plate counts of the live cell stock used to prepare samples. The slopes of the regression lines corresponding to dPCR and v-dPCR values in (b) did not statistically differ from 0 (p > 0.05). In both graphs, each value is the mean of three independent repeats. Absent error bars indicate SD values smaller than the represented symbols.
The overestimation of live cells in PMA-treated samples has also been documented in previous works32,33. Factors conditioning PCR inhibition by PMA are the dye capacity to penetrate dead cell membranes and a concentration ensuring its binding to all the target DNA copies in the sample. Accordingly, insufficient PMA concentrations and/or the use of killing methods causing insufficient bacterial membrane damage might lead to the detection and quantification of dead cells as live ones.
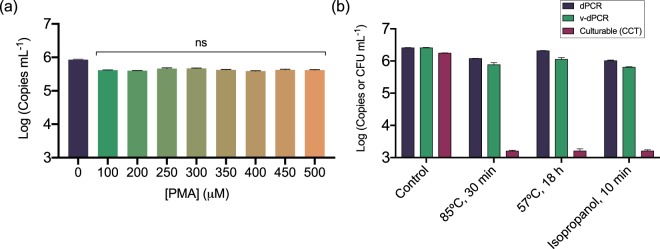
The PMA concentration and photo-activation conditions used in this work were based on other works analyzing plant material and/or more complex samples26,27. The use of high PMA concentrations, up to 500 μM, did not have an apparent effect on false-positive signal suppression in samples containing only heat-killed E. amylovora cells (Fig. 3a). This indicated that almost all the dead cells were detected and quantified as live cells regardless of the PMA concentration. Similarly, the PMA treatment of mixtures of E. amylovora live and dead cells obtained by different killing methods (85 °C, 30 min; 57 °C, 18 h; 70% isopropanol, 10 min) revealed no apparent effects of the killing method on the suppression of false-positive signals coming from the dead cells (Fig. 3b). Hence, based on these results, the overestimation of E. amylovora live cells by v-dPCR did not seem to be particularly associated with an insufficient PMA concentration or an inappropriate killing method.
Figure 3.
Effect of PMA concentration and the E. amylovora killing method on the exclusion of false-positive signals by v-dPCR. Apple branch macerates prepared in AMB were inoculated with 106 E. amylovora heat-killed cells mL−1 (85 °C for 30 min), and treated with PMA concentrations ranging from 0–500 μM before DNA extraction and dPCR (a). E. amylovora live cells (103 CFU mL−1) were mixed with dead cells (106 cell mL−1) obtained either by heating at 85 °C for 30 min, heating at 57 °C for 18 h, or exposure to 70% isopropanol for 10 min. Samples were then analyzed by dPCR, v-dPCR (treatment with 100 μM PMA) and plate counts (b). D-PCRs were performed using the same primers and probe as Pirc et al.31. Represented data are mean values of experiments performed in triplicate. Error bars show the SD.
Improvement of v-dPCR false-positive suppression by targeting larger DNA sequences
Another important parameter conditioning the potential of v-dPCR to discriminate between live and dead cells is the length of the target DNA determined by primers30,34. Similar to propidium iodide, PMA does not have a preference for specific nucleotides or DNA sequences, so the dye binding to DNA follows a random distribution. The employed PMA concentrations for viability-PCR are far from saturation34. Therefore, the probability of PMA to bind the target DNA and inhibit PCR amplification increases with larger amplicon sizes. Likewise, greater numbers of thermal cycles might increase the probability to amplify target DNA from a dead cell and/or to detect the fluorescence generated by false positive reactions.
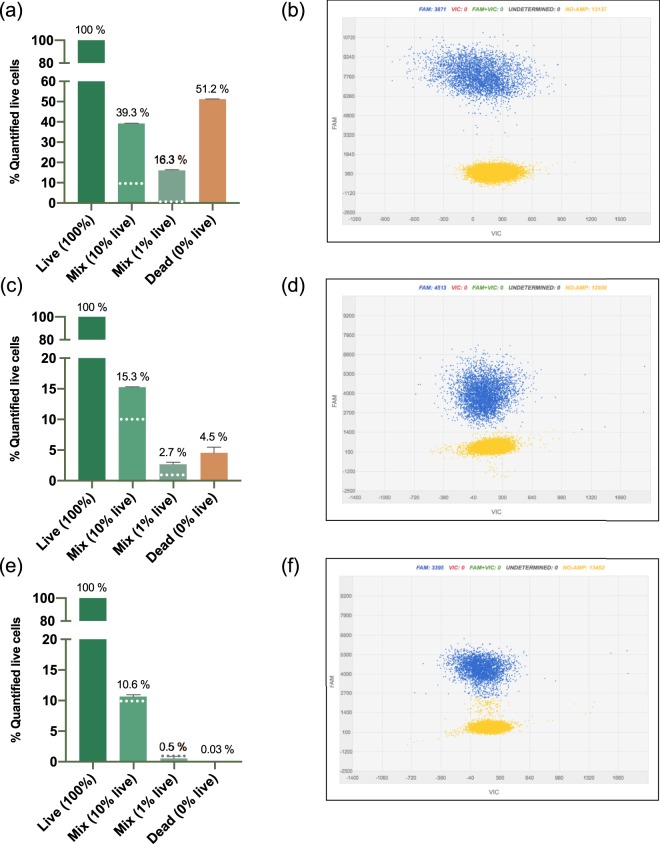
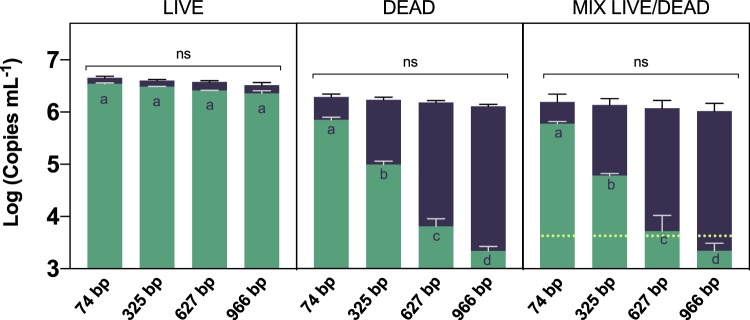
The effects of a drastic reduction of thermal cycles from 40 to 30 and the use of a large amplicon size of 966 bp versus 74 bp were tested to determine the next steps for v-dPCR optimization (Fig. 4). Compared to the original protocol (74 bp amplicon; 40 thermal cycles) (Fig. 4a,b), both the reduction of thermal cycles (Fig. 4c,d) and the increase of the amplicon size (Fig. 4e,f) had a positive effect on the suppression of false positive signals by v-dPCR in samples containing dead E. amylovora cells, either mixed or not with live cells. However, the reduction of cycle numbers had a strong impact on fluorescence intensity values, which on average, became closer to the ones corresponding to no-template reactions (Fig. 4d).
Figure 4.
Effect of the thermal cycle number and the amplicon size on the reduction of false-positive signals by v-dPCR. Apple branch macerates prepared in AMB were inoculated with either E. amylovora at 7 × 106 CFU mL−1 (100% live cells), 7 × 106 dead cells mL−1 (0% live cells), or live/dead cell mixtures containing a 10% and a 1% of live cells and a 90% and a 99% of dead cells, respectively. After PMA treatment and DNA extraction, dPCR was conducted using: i) same primers and probe as Pirc et al.31, with 40 amplification cycles (a,b); ii), reducing the number of thermal cycles to 30 (c,d); iii) using a primers targeting a 966 bp DNA sequence, and 40 amplification cycles (e,f). Each column shows mean values of two independent experiments. Error bars are the SD. Dashed horizontal lines indicate the theoretical percentage of live cells in samples containing live/dead cell mixtures (10% and 1%). Representative dPCR 2D scatter plots showing fluorescence values of the FAM labelled probe in wells with positive calls (blue) and fluorescent values corresponding to negative amplifications (yellow) (b,d,f).
Based on these results, new primers targeting DNA sequences with lengths ranging between 74 bp and 966 bp (Table 1) were designed and their effect on false-positive signal suppression analyzed. The new primers were specific against a collection of 30 E. amylovora strains, including reference strains and natural isolates. They provided negative amplification results with 16 other bacterial species, including bacterial phytopathogens of the genera Dickeya, Pectobacterium, Pantoea and Pseudomonas, as well as unidentified saprophytic bacteria usually isolated together with E. amylovora from apple cankers on CCT medium (Supplementary Table S3).
Table 1.
Primer and probe combinations used in this study.
| Primer/Probe | Sequence (5′ –> 3′) | Reference | Amplicon size in combination with Ams189R |
|---|---|---|---|
| Primers | |||
| Ams189R† | GGG TAT TTG CGC TAA TTT TAT TCG | Pirc et al., 2009 | |
| Ams116F | TCC CAC ATA CTG TGA ATC ATC CA | Pirc et al., 2009 | 74 bp |
| Ams03KbF | ATA TCC TTG TTG ATA AAC AGG TGC G | This study | 325 bp |
| Ams06KbF† | AAT TGG TTC CGC TAT AAC TTG CAG | This study | 627 bp |
| Ams09KbF | TAA TAG ACT GAG CTG AAA GTA TCG TGT | This study | 966 bp |
| Probe | |||
| Ams141T | FAM-CCA GAA TCT GGC CCG CGT ATA CCG-TAMRA | Pirc et al., 2009 | |
†Primer combination used in the final optimized protocol for dPCR and v-dPCR.
The amplicon size had no effect on dPCR total cell quantification, regardless of the cell viability status (p > 0.05) (Fig. 5). Similarly, the increase of the amplicon length did not affect the v-dPCR outcome during the analysis of samples composed of only live cells (p > 0.05), but was linked to a significant reduction of false-positive signals in samples containing dead cells (p < 0.0001) (Fig. 5).
Figure 5.
Effect of the amplicon length on the suppression of false positive signals by v-dPCR. Apple branch macerates prepared in AMB were inoculated with E. amylovora live (106 CFU mL−1), dead (106 dead cells mL−1) or a defined ratio of live (103 CFU mL−1) and dead cells (106 dead cell mL−1). Samples were sub-aliquoted for total (purple) and live (green) cell quantification by dPCR and v-dPCR, respectively, using primer combinations targeting a 74, 325, 627 and 966 bp DNA sequence. Data are mean values of three biological repeats of the same experiment. Error bars show the SD. Different letters denote statistically significant differences between live cell counts of samples treated with PMA. Non-statistically significant (ns) differences were detected between dPCR total cell counts regardless of the amplicon length.
The v-dPCR analysis of defined mixtures of live and dead cells using the primer combination targeting a 966 bp DNA sequence underestimated the number of live cells with respect to the CFUs inoculated per sample (p < 0.05). Cell number underestimation associated with large amplicon lengths for qPCR is usually attributed to accumulation of DNA damage that, similarly to PMA, inhibits or reduces the efficiency of amplification by the DNA polymerase34. Based on these results, the 966 bp amplicon size was discarded for further assays. The v-dPCR analysis of the same samples with the primer combination targeting a 627 bp DNA sequence provided viability values slightly above, but non-significantly different to, the inoculated CFUs (p < 0.05). However, the use of smaller amplicon lengths, led to a clear overestimation of viable cell counts with respect to the inoculated CFUs (p < 0.05).
Sample processing in diluted AMB and the addition of a membrane destabilizer during PMA treatments enhance false-positive suppression by v-dPCR
The sample matrix may interfere with v-dPCR by complexing or reacting with PMA molecules, thus impeding the ability of this viability dye to penetrate dead cell membranes and/or bind to DNA, as well as by hampering PMA photo-activation. In other studies, PMA concentrations and photo-activation protocols similar to the ones used in this work have been used to analyze plant material27,35,36 and more complex samples with different degrees of turbidity and particle size26.
Sample processing is usually conducted in water or a buffered solution. We used anti-oxidant maceration buffer (AMB)37 as recommended by European and American plant protection organizations38,39 for E. amylovora diagnostics. This solution buffers pH around neutral and contains a variety of antioxidants which improve the isolation of stressed E. amylovora cells on culture media and remove PCR inhibitors during DNA purification37–39. However, some of the AMB components might also interact with PMA, DNA, cell membranes or all of them.
Several works have reported an additional improvement of viability qPCR by complementing the viability dye treatment with a membrane destabilizer40,41. Similarly, a commercial product, PMA Enhancer for Gram-negative bacteria, improving PMA affinity to dead-cell DNA can be combined with PMA to improve the selective detection of live cells.
We determined the effect of the maceration buffer and the addition of the membrane destabilizer sodium dodecyl sulfate (SDS) or PMA Enhancer during PMA treatments. Suspensions of E. amylovora live, dead, and a mixture of live and dead cells were prepared in plant macerates obtained in either AMB or tenfold diluted AMB (0.1xAMB). Additionally, v-dPCR was performed using either PMA, PMA plus 0.0025% SDS (w/v) (PMA + SDS), or PMA plus PMA Enhancer (PMA + E) (Fig. 6). The SDS assay concentration was chosen based on a minimum inhibitory concentration assay (Supplementary Fig. S3). The dPCR reactions were performed using the primer combination Ams06KbF/Ams189R (627 bp amplicon size), which in a previous experiment (Fig. 5) significantly reduced false positive detection providing v-dPCR quantification values similar to the inoculated CFU mL−1.
Figure 6.
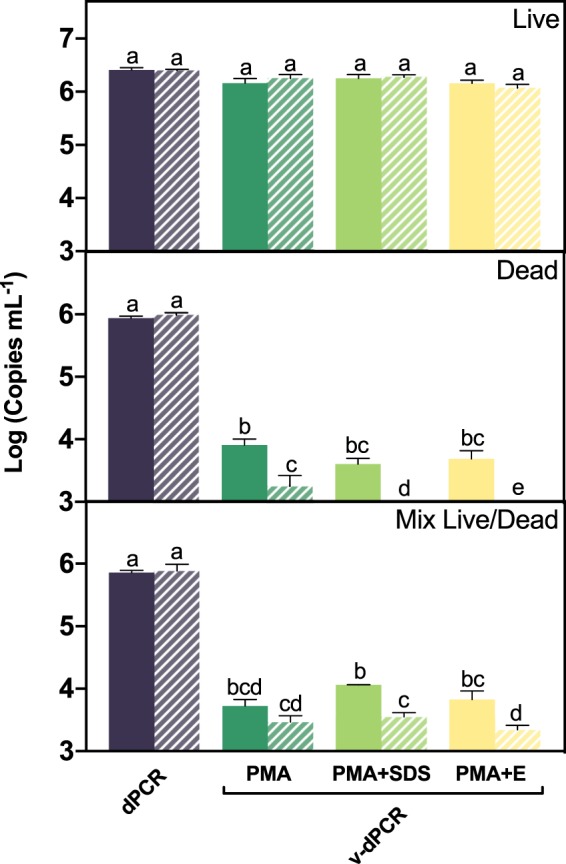
Effect of the maceration buffer, SDS and PMA Enhancer for Gram negative bacteria on the suppression of E. amylovora dead cell signals by v-dPCR. Apple branch macerates prepared either with AMB (solid columns) or 0.1xAMB (dashed columns) were inoculated with E. amylovora live (106 CFU mL−1), dead (106 dead cells mL−1) or a mixture of live (103 CFU mL−1) and dead cells (106 dead cell mL−1). Total cell numbers were quantified by dPCR. The effect of SDS and PMA Enhancer on viable cell quantification was assessed comparing v-dPCR viable cell counts of samples (i) only treated with PMA (100 μM); (ii) PMA plus 0.0025% (w/v) SDS (PMA + SDS); (iii) PMA plus PMA Enhancer (PMA + E). For both, total and viable cell quantification, primers targeting a 627 bp DNA sequence were used. Columns represent mean values of three independent experiments in duplicate. Error bars show the SD. Different letters denote statistically significant differences.
Total cell quantification by dPCR was similar (p > 0.05) in samples processed in AMB and in 0.1xAMB, regardless of the type of sample analyzed (live, dead, live/dead mix) (Fig. 6). Viable cell counts (v-dPCR) in samples composed of only live cells were also similar (p > 0.05) regardless of the maceration buffer or the PMA treatment employed to inhibit dead-cell DNA amplification (Fig. 6). However, the two-way ANOVA analysis revealed an effect of both the maceration buffer (p < 0.001) and the PMA treatment (p < 0.05) in samples containing dead cells, alone and mixed with live cells (Fig. 6). In samples containing only heat-killed cells, a total exclusion of dead-cell signals was achieved by using 0.1xAMB and PMA treatments combined with SDS or PMA Enhancer (Fig. 6). In samples containing mixed live/dead cells, the effect of the maceration buffer concentration was only significant in samples treated with PMA + SDS or PMA + E (p < 0.01), but not in the samples treated only with PMA (p > 0.05).
The same samples were analyzed using the primer pair targeting the 325 bp DNA sequence. However, the false-positive signals were consistently detected regardless of the maceration buffer concentration and the type of PMA treatment, confirming that a larger amplicon size is required to efficiently remove signals coming from dead cells (Supplementary Fig. S4).
These results show that, while AMB does not seem to have a detrimental effect on total cell quantification by dPCR, one or more AMB components are probably involved in the failure of PMA treatments to suppress false-positive signals. This hypothesis is also supported by the fact that extremely high PMA concentrations of up to 500 μM in samples processed with AMB did not inhibit the DNA polymerase (Fig. 3a), as reported by other authors using lower concentrations of PMA33,42. The most concentrated AMB component is polyvinylpyrrolidone (PVP) (2% w/v), a polymer used in DNA extraction protocols due to its ability to remove PCR inhibitors present in plant material. However, PVP also interacts with DNA43, with the PVP-DNA complex possessing less negative charge density and being more hydrophobic than DNA alone43. Although PVP interactions with DNA do not affect PCR amplification, it could negatively impact the PMA capacity to bind DNA. Further studies are required to determine which of the AMB compound/s negatively affect v-dPCR and by what mechanism.
In vitro performance of the dPCR and the optimized v-dPCR in samples with increasing live cell concentrations
The previous assay revealed the conditions allowing a complete dead-cell signal suppression by v-dPCR: (i) sample processing in 0.1xAMB, (ii) treatment with PMA + E (or SDS), (iii) and dPCR amplification using the primers Ams06KbF/Ams189R (Fig. 6).
Before testing these conditions with natural samples, we first characterized the dPCR performance with primers Ams06KbF/Ams189R, by analyzing increasing E. amylovora live cell concentrations in apple and pear plant macerates prepared in 0.1xAMB (Fig. 7). Afterwards, we also tested the efficiency of the new v-dPCR conditions (sample processing with 0.1xAMB, treatment with PMA + E, dPCR with primers Ams06KbF/Ams189R) in quantifying increasing E. amylovora live cell concentrations in the presence of dead cells (Fig. 8).
Figure 7.
Correlation between the Log-transformed values of plate counts and QS3D dPCR absolute quantification values, using primers Ams06KbF and Ams189R. Serial tenfold dilutions of E. amylovora were performed in apple (a) and pear (b) branch macerates prepared in 0.1xAMB, and subjected to DNA extraction and dPCR. Each dot represents the average value of two biological repeats performed in triplicate. Absent error bars indicate SD values smaller than the represented symbols.
Figure 8.
Correlation between plate counts and v-dPCR values in the presence of dead cells, using improved methods for plant material processing and dPCR. Apple and pear branch macerates prepared in 0.1xAMB were inoculated with increasing live cell concentrations (from 103 to 106 CFU mL−1) in a constant background of 106 dead cell mL−1. Total cell counts were performed by dPCR. For viable cell counts, samples were treated with PMA plus 1x PMA Enhancer before dPCR. In all cases, dPCR was performed using primers targeting a 627 bp DNA sequence. Each dot corresponds to mean values of two independent experiments performed in triplicate. Error bars show the SD. Absent error bars indicate SD values smaller than the represented symbols. Solid lines represent the linear regression best fit through all the v-dPCR dataset points. The corresponding linear regression equations and coefficients of correlation (R2) are also indicated.
The dPCR quantification range using primers Ams06KbF and Ams189R (Fig. 7; Table 2) was 103 to 107 CFU mL−1, similar to the one with primers Ams116F/Ams189R (74 bp amplicon)31 in this work (Fig. 1) and in a previous one using a different dPCR platform19. Other quantitative techniques provide wider ranges of detection and quantification. However, the lack of requirement of calibration controls makes dPCR a good option to simplify the analysis of natural samples and allows their comparability23. The mean coefficients of variation (%CV) of two independent experiments calculated for each of the assayed bacterial concentrations (from 103 to 107 CFU mL−1), ranged from 1.23 to 23.36% in apple, and 1.26–24.78% in pear. The highest %CV were observed in samples containing 103 CFU mL−1, and the lowest ones in samples containing 106 CFU mL−1. Bacterial concentrations below 103 CFU mL−1 and higher than 107 CFU mL−1 led to unsuccessful detection of E. amylovora cells in most of the replicates and to chip saturation, respectively, similar to preliminary experiments using the primers designed by Pirc et al.31 (Table 2). Therefore, the limit of detection and quantification for our newly developed dPCR protocol had the same value regardless of the analyzed plant material, ca. 103 CFU mL−1 (i.e. the pathogen was detected in all the analyzed replicates, with a %CV below 25%).
Table 2.
Characterization of the QS3D dPCR performance using the primer combination Ams06KbF/Ams189R, targeting a 627 bp DNA sequence.
| Host | Log CFU mL−1 b | Log Copies mL−1 (±SD)c,d | Copies rxn−1 (±SD)d,e | # Qualified by QT (± SD)d,f | # Pos (±SD)d,g |
|---|---|---|---|---|---|
| Apple cv. Honeycrisp a | 7.33 | 7.32 ± 0.06 | 4.406 ± 0.623 | 17353 ± 279 | 17057 ± 190 |
| 6.33 | 6.37 ± 0.01 | 0.494 ± 0.014 | 17537 ± 195 | 6824 ± 183 | |
| 5.33 | 5.46 ± 0.06 | 0.061 ± 0.009 | 17205 ± 812 | 1021 ± 183 | |
| 4.33 | 4.42 ± 0.08 | 0.005 ± 0.001 | 17656 ± 381 | 93 ± 20 | |
| 3.33 | 3.62 ± 0.10 | 0.001 ± 0.000 | 17388 ± 339 | 14 ± 5 | |
| Pear cv. Bosc a | 7.33 | 7.54 ± 0.10 | 4.328 ± 0.988 | 17725 ± 630 | 17351 ± 423 |
| 6.33 | 6.62 ± 0.11 | 0.518 ± 0.126 | 15655 ± 602 | 7019 ± 1101 | |
| 5.33 | 5.57 ± 0.10 | 0.047 ± 0.010 | 17752 ± 829 | 802 ± 139 | |
| 4.33 | 4.57 ± 0.08 | 0.005 ± 0.001 | 17490 ± 393 | 80 ± 16 | |
| 3.33 | 3.62 ± 0.09 | 0.000 ± 0.000 | 17707 ± 705 | 8 ± 2 |
aE. amylovora-free plant material was processed in a plastic bag containing 0.1xAMB in a ratio 1:50 (w/v), by hammering.
bData shown in this column correspond to average bacterial concentrations of two different E. amylovora stocks, employed to inoculate samples in two independent assays performed in triplicate. Values in each dilution were calculated based on the stock concentrations. Hence, the SD in all cases was the same, ±0.24.
cLog Copies mL−1 = Log [copies μL−1rxn × 1/Drxn tube × 1/EDNA Extr × 1/VolPl.Macerates]; i.e. Log (copies μL−1rxn × 1.79 × 103) for Apple, and Log (copies μL−1rxn × 4.81 × 103) for pear.
dAverage values of two independent experiments performed in triplicate.
eQuantity of target DNA copies, expressed in copies per reaction well.
fTotal number of reaction wells in the chip fitting the selected quality threshold.
gNumber of positive calls for FAM dye in the chip, as determined from the Review Calls scatter plot. Based on the no-template control and plant material negative control analysis, chips with 5 or less than 5 positive calls were considered negative for E. amylovora detection, and the provided dPCR values were not considered for quantification.
Apple and pear plant macerates containing increasing live E. amylovora cell concentrations with a background of dead cells were used to determine the ability of the optimized sample processing protocol and v-dPCR conditions to suppress false-positive signals during live cell quantification (Fig. 8). Regardless of the analyzed plant material (apple or pear), dPCR analysis of samples provided total cell numbers coinciding with the inoculated bacterial concentrations, around 106 mixed live/dead cells mL−1. Similarly, v-dPCR live cell values correlated well (R2 > 0.99) with the inoculated CFU mL−1 despite the assayed ratio of live:dead cells ranging from 1:1 in samples containing 106 live cells mL−1 and 106 dead cells mL−1, to 1:999 in samples containing 103 live cells mL−1 plus 106 dead cells mL−1 (Fig. 8). Live:dead cell ratios higher than 1:999 were also tested, but led to an overestimation of live cells, similar to that reported by other authors32,33. Hence, the method seems suitable for the analysis of samples containing relevant concentrations of live bacterial cells without an excessive load of dead bacteria.
Absolute quantification of E. amylovora live cells in natural cankers
Although many works have addressed the characterization of cankers8,44–46, accurate estimations of live E. amylovora populations in these structures are limited and usually based on culture dependent methods. We tested the sample processing with 0.1xAMB, primers targeting a 625 bp DNA sequence, and different PMA treatments for v-dPCR to analyze apple and pear cankers originating from fire blight infections occurring in early spring and harvested in mid-summer and mid-winter. The assayed PMA treatments consisted of PMA, PMA + SDS, PMA + E, and equivalent treatments using PMAxx, which is an improved commercial version of PMA. Total and culturable cell numbers were also quantified by dPCR (using the same primers) and plate counts on SNAN and CCT media, respectively.
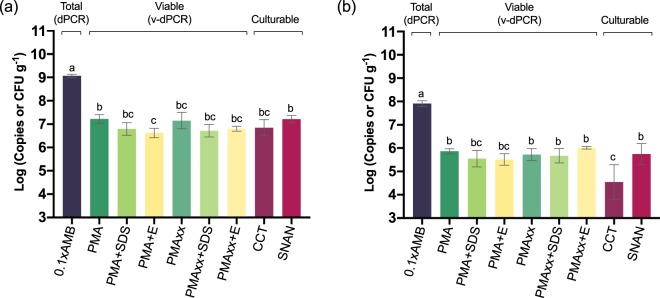
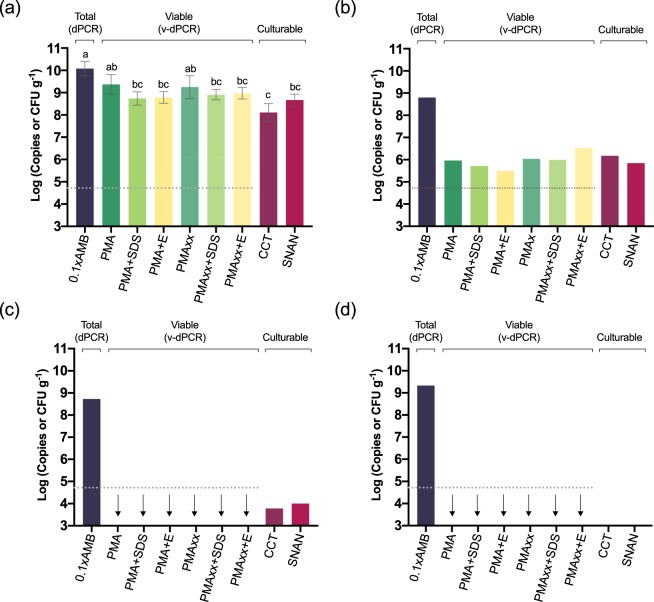
In apple cankers, E. amylovora total cell numbers in mid-summer (Fig. 9a) were about one log unit higher than in mid-winter (p < 0.0001) (Fig. 9b). Live cell counts were about 2 log units lower than the total ones, regardless of the time period or the viability-staining treatment assayed, with differences between total and viable or culturable cell quantification values being very significant (p < 0.0001) (Fig. 9). No differences were detected between the v-dPCR viable cell counts performed using PMA and PMAxx, regardless of the time period assayed or their combination with SDS or PMA Enhancer (p > 0.05). In most cases, the addition of SDS or PMA Enhancer reduced viable cell counts compared to the control treatments with PMA or PMAxx alone (Fig. 9a,b), although these differences were statistically significant only in the case of mid-summer apple cankers treated with PMA + E (p < 0.05) (Fig. 9a).
Figure 9.
E. amylovora total, live and culturable cell populations in fire blight cankers on apple (cv. Honeycrisp). Fire blight infections started in early-spring of 2016, and cankers were collected in mid-summer (July 2016) (a) and mid-winter (January 2017) (b). Samples were homogenized with 0.1xAMB. Total cell counts were performed by dPCR, using primers Ams06KbF and Ams189R. For v-dPCR, sample sub-aliquots were treated with PMA, PMAxx, and combinations of both dyes with SDS (+SDS) or PMA Enhancer (+E). Culturable cell counts were performed by spread plate on SNAN and CCT media. Three cankers were analyzed per time period. Each column represents mean values of total, viable or culturable cell counts of three different cankers, with error bars indicating the SD.
E. amylovora culturable counts on CCT and SNAN were similar in mid-summer cankers (p > 0.05), but in the mid-winter cankers, a lower E. amylovora recovery on CCT compared to SNAN was observed (p < 0.01) (Fig. 9b). CCT contains crystal violet and other selective compounds47 which show low toxicity for non-stressed E. amylovora cells. Poor recovery efficiencies on selective media as compared to general media are usually linked to injured bacteria which become more sensitive to the selective compounds of the medium48. Accordingly, another advantage of v-dPCR is the detection of live cells regardless of their ability to form colonies on solid media. This might be especially important for detecting cells in the VBNC state. This survival strategy has been reported in more than 100 bacterial species12 including E. amylovora and other important plant pathogens, which represents a challenge for their management.
The E. amylovora cell counts in pear cankers harvested in mid-summer (Fig. 10a) were also higher than in those sampled in mid-winter (p < 0.0001) (Fig. 10b–d). The differences between total and viable cell numbers in pear cankers collected in mid-summer (about 0.5 log units) were smaller than those in apple at the same time period (ca. 2 log units) (Fig. 9). This reflects a greater number of E. amylovora dead cells in apple than in pear cankers, which might be linked to the higher resistance of apple to fire blight in comparison to pear. V-dPCR data revealed no differences between PMA- and PMAxx-treated samples (p > 0.05), or between the samples treated with SDS or PMA Enhancer compared to their counterparts treated only with PMA or PMAxx (p > 0.05) (Fig. 10a). Similarly, no statistically significant differences were detected between plate counts carried out on CCT and SNAN (p > 0.05) (Fig. 10a).
Figure 10.
E. amylovora total, live and culturable cell populations in fire blight cankers on pear. Fire blight infections originated in early-spring of 2016, and cankers collected in mid-summer (July 2016) (a) and mid-winter (January 2017) (b–d). For sample analysis, cankers were homogenized with 0.1xAMB. Total cell counts were performed by DNA extraction and dPCR. For v-dPCR, samples were treated with PMA, PMAxx, or any of these dyes combined with SDS (+SDS) or PMA Enhancer (+E). In both cases, dPCR was performed using primers Ams06KbF/Ams189R and the probe Ams141T. Culturable cell counts were performed by spread plate on SNAN and CCT media. In cankers collected in mid-summer, E. amylovora quantification values were similar, so each column represents mean values of total, viable and culturable cell counts of three different cankers, with error bars indicating the SD (a). In cankers collected in mid-winter, substantial differences among cankers in E. amylovora populations were observed. Separate graphs were used to highlight differences among cankers (b,c,d). Dashed lines represent the limit of detection and quantification of dPCR. Arrows represent viable cell counts equal to 0 or below the dPCR detection limit. The absence of columns corresponding to plate counts represent values below the detection limit of the technique, which is shown in graphs as the minimum value at the Y axis.
In pear cankers harvested in mid-winter, total cell numbers also showed similar values in all of the analyzed samples, although culturable and viable cell counts significantly differed among cankers (Fig. 10b–d). In only one of the three cankers collected in mid-winter (Fig. 10b) were E. amylovora viable cell populations at the levels similar to those in apple cankers collected in the same time period (Fig. 9b). In the remaining cankers (Fig. 10c,d), the number of viable cells fell below the detection limit of the technique under our experimental conditions (ca. 103 CFU mL−1 of plant macerates, or about 5 × 104 CFU g−1 of canker). This coincided with E. amylovora culturable cell counts below the v-dPCR detection limit (ca. 104 CFU g−1) (Fig. 10c,d) or no detection of culturable bacteria on any of the assayed media (Fig. 10d) (limit of detection of plate counts: 20 CFU mL−1 of plant macerates or 103 CFU g−1 of canker).
In both apple and pear cankers, v-dPCR provided viable cell counts similar to those on SNAN with most of the assayed PMA treatments (Fig. 9a,b), indicating that most of the viable cells in cankers are in a culturable state. In mid-summer apple samples that were treated with PMA + E, a small decrease of viable versus culturable cell counts was observed (p < 0.05) (Fig. 9a), although these differences were not detected in apple cankers collected in winter or in pear cankers in any of the assayed periods (Figs 9 and 10).
Unlike in previous assays where the use of PMA Enhancer or SDS was required to eliminate false-positive signals (Fig. 6), the analysis of natural cankers revealed a good suppression of dead-cell DNA amplification, even in samples treated with PMA or PMAxx alone (i.e. v-dPCR live cell counts were similar to those of culturable cells in most of the analyzed samples) (Figs 9 and 10). The differences between in vitro and natural sample analysis are probably related to conditions in cankers favoring a more efficient degradation of dead cell membranes compared to the killing method employed in this work. Moreover, the analysis of pear cankers harvested in mid-winter revealed the complete elimination of false-positive signals in samples containing at least 100,000 times more dead cells than live ones (i.e. ca. 109 total cells g−1 from which only about 104 cells or less were culturable) (Fig. 10c,d). These results show the efficiency of most of the tested treatments to analyze E. amylovora viable cell populations in cankers. Accordingly, based on our data, the suggested protocol to perform E. amylovora viable-cell analysis of cankers by v-dPCR consists of the following steps: i) Sample processing with 0.1xAMB, in a ratio 1:50 (w/v); ii) treatment of samples with PMAxx, PMAxx + E or PMAxx + SDS; iii) dPCR using primers Ams06KbF/Ams189R. Detailed information on each step can be found in the Methods section and a detailed protocol for the analysis of E. amylovora total, viable, and culturable cell populations in cankers is provided as supplementary information (Fig. S5).
Overall, our results reveal the valuable potential of v-dPCR and the combined analysis of total, viable, and culturable E. amylovora populations in cankers to unveil cryptic stages of the E. amylovora life cycle or specific aspects linked to host-pathogen interactions. Although preliminary, canker analysis revealed that winter weather conditions and/or the cessation of the host’s vegetative growth are probable factors inducing the general decline of E. amylovora live cell populations in winter, particularly on pear. Our results also correlate well with the hypothesis that the pathogen overwinters in only a certain percentage of cankers1,3. Differences between bacterial cell numbers in apple versus pear cankers might indicate a host- and/or bacterial strain-dependent survival of E. amylovora. More research on these topics will provide more information on E. amylovora cell population dynamics in cankers and its relationship with environmental and/or host-related factors, as well as on establishing links between host resistance and E. amylovora survival in cankers. Our protocol might also be useful for data generation to improve the accuracy of fire blight forecasting systems, which currently assume abundance of primary bacterial inoculum every year for disease renewal49.
Methods
Bacterial strains and culture media
Unless stated otherwise, the E. amylovora strain ATCC 49946 was used in all the experiments. Other E. amylovora strains and/or plant pathogenic bacterial species employed in this work are summarized in the Supplementary Table S3.
Bacterial cultures were cryopreserved at −80 °C in 20% glycerol (v/v). All the strains were grown at 28 °C in/on the general media Luria-Bertani50 (LB) and/or Sucrose Nutrient Agar51 (SNA). E. amylovora isolation and plate counts from natural samples were carried out on SNA amended with 21.6 mg L−1 natamycin (Amtech Biotech Co. Ltd., Qiqihar, China) (SNAN) and/or on Crystal violet-Cycloheximide-Thallium nitrate (CCT) medium48.
E. amylovora-free plant material, sources and obtaining of plant macerates
All the E. amylovora-free plant material was collected from orchards at the Cornell University’s Hudson Valley Research Laboratory (HVRL) in Highland (NY, USA). 1-year-old apple ‘Honeycrisp’ and pear ‘Bosc’ branches were harvested, cut in 15-cm-long pieces and stored at −80 °C after fast freezing in liquid N2 until use.
To obtain E. amylovora free plant macerates, branch pieces were left to thaw at room temperature for 10 min. Slices from the branch surface containing bark and vascular cambium were excised with a sterile scalpel, weighed, placed into a Ziploc plastic bag containing either ice-cold AMB38,39 or 0.1xAMB in 10 mM phosphate buffered saline (PBS) pH 7.4, and homogenized by crushing with a hammer. The volume of maceration buffer in each bag was proportional to the sample weight, in a ratio 1:50 (w/v). Plant macerates were freshly prepared before each experiment, and chilled on ice until use. The absence of E. amylovora in this plant material was confirmed by microbiological and molecular methods, according the International Standard for Phytosanitary Measures 27 (ISPM 27, Annex 13) for E. amylovora detection in asymptomatic plant material39.
Preparation of E. amylovora live and/or dead cell stocks
In most experiments, E. amylovora-free plant macerates were inoculated with either E. amylovora live, dead or a mixture of live and dead cells. Live cell suspensions were prepared with overnight cultures (16 h) in LB, by centrifugation and resuspension of the pellets in sterile distilled water (diH2O) thrice.
Unless otherwise specified, dead cells were obtained by heat-treatment, exposing 1 mL of E. amylovora live cell suspensions in diH2O to 85 °C for 30 min. In both cases, the OD600 nm of the cell stocks was adjusted to 1 or 0.1 (109 or 108 CFU mL−1, respectively) and employed to inoculate plant macerates at the desired concentrations, either with the stocks or with their serial dilutions in diH2O.
Live cell concentrations in stocks were confirmed by drop-plate on LB agar, as described elsewhere52. The absence of live cells in heat-treated bacterial suspensions was confirmed by negative growth on LB agar after spread-plating 0.1 mL of the challenged cell suspension in triplicate and incubation for 72 h at 28 °C.
All the experiments including E. amylovora-free plant macerates included negative controls, to ensure the absence of E. amylovora cells prior to sample inoculation.
DNA extractions
Before dPCR, 0.5 mL aliquots from plant macerates, treated or not with the viability-PCR dye PMA (see below), were subjected to DNA extraction using DNeasy Plant Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer instructions and resuspending DNA in a final volume of 200 μL. When required, the same kit was also used for bacterial genomic DNA extractions, starting from 0.5 mL overnight cultures in LB. Samples were stored at −20 °C until use.
dPCR conditions
dPCR was performed with the QS3D dPCR System (ThermoFisher Scientific). Each dPCR reaction was prepared in a final volume of 16 μL, composed of 8 μL of QS3D dPCR Master Mix v2 (Applied Biosystems, Frederick, MD, USA), 0.8 μL of 20x primers/probe mix (final concentrations, 0.9 and 0.2 μM, respectively) (Tables 1), 6.4 μL of direct or diluted DNA sample and 0.8 μL of nuclease free water. The QS3D dPCR chips were loaded with 15 μL reaction mix by using the QS3D dPCR automatic loader, and the dPCR amplification was conducted with a GeneAmp 9700 PCR thermocycler (Applied Biosciences, Foster City, CA, USA). Amplification conditions were: 10 min at 95 °C for DNA polymerase activation, followed by 39 two-step cycles of 1 min at 60 °C and 15 sec. 98 °C, and a final extension at 60 °C for 2 min. The annealing temperature, and primer/probe concentrations were chosen based on previous works19,31. One-off assays using higher and lower annealing temperatures led to a smaller separation between background and positive fluorescence signals, so we continued using 60 °C with all primer combinations.
After DNA amplification, the chips were transferred to the QS3D Instrument for imaging, and end-point fluorescence data were collected and analyzed with the QS3D AnalysisSuite Cloud Software (version 3.1.2-PRC-build-03, Thermo Fisher Scientific) under the Absolute Quantification module, maintaining automatic settings. The global threshold was determined automatically by the software, and adjusted manually when required using the signals observed in no-template controls as a reference. Output data provided by the software included the mean number of copies per reaction, the number of copies per μL, the number of partitions qualified for quantification, the number of partitions negative for target DNA amplification, along with other parameters related to quantification and the quality assessment of the analysis. Only chips with more than 15,000 out of 20,000 partitions qualifying for quantification were used for analysis.
Discrimination between positive and negative fluorescence calls was based on the QS3D Analysis Suite output data (Thermo Fisher Scientific). Signals of no-template controls and those from DNA extractions corresponding to E. amylovora-free plant material, from both apple and pear, as well as positive controls with different E. amylovora DNA concentrations were used to discriminate background fluorescence from positive calls. Based on these experiments, only chips with a number of positive calls higher than 5 were considered positive for E. amylovora detection.
Quantitative data provided by the QS3D Analysis Suite Cloud Software was used to calculate the number of target copies per g of canker tissue, with the formula:
where Copies μL−1Rxn are the copies of target DNA per μL in the reaction tube, provided by the QS3D Analysis Suite software; DRxn tube is the dilution of sample DNA in the dPCR reaction tube (e.g. 6.4 μL sample DNA in 16 μL of dPCR mastermix); DDNA is an optional dilution of sample DNA before dPCR (e.g. typically, 1/50–1/100 dilutions of sample DNA were required for the analysis of total cell quantification in natural canker samples); EDNA extr is the efficiency of the DNA extraction with the kit (in our case, a 27.9% for apple, and a 10.4% for pear tissues); VolElution (μL) is the final elution volume of sample DNA after extraction with the kit (in our case, 200 μL); VolPl. Macerates (mL) is the volume of plant macerates used for DNA extraction (usually, 0.5 mL); the RatioCanker:AMB was taken into consideration to calculate the concentration of E. amylovora cells per gram of analyzed canker (ratio canker:AMB, 1:50).
Other parameters related to the dPCR performance such as the quantity of target DNA copies per reaction well, the total number of reaction wells in the chip that exceed the selected quality threshold, or the number of positive calls for FAM dye in the chip were also provided by the software, and are specified in tables within the text or as supplementary information (Supplementary Tables S1, S2, S5), according to the digital MIQE guidelines53.
Digital PCR using a transferred protocol for qPCR
A preliminary evaluation of the QS3D dPCR performance and robustness (capacity of the method to remain unaffected by variations in sample composition) was performed before testing the technique for the analysis of E. amylovora live cell populations in cankers.
E. amylovora-free apple and pear macerates obtained using AMB were inoculated with strains ATCC 49946 and CFBP 1430 at final concentrations ranging from 102 to 108 CFU mL−1. After DNA extraction, samples were analyzed by dPCR using the same primers, probe and annealing temperature of an already existing qPCR protocol31 which had previously successfully been used with a different dPCR platform19. Data from three independent experiments were log-transformed and the correlation between dPCR counts of target DNA copies and the inoculated CFUs mL−1 was analyzed by linear regression analysis, using GraphPad Prism 8 (version 8.1.2). The same software was used for the statistical analysis of the remaining experiments in this work.
PMA treatment
The PMA treatment was designed based on Fittipaldi et al.30 and works analyzing plant material27 or more complex samples26. Unless otherwise stated, 0.4–0.5 mL plant macerates were treated with PMA (Biotium Inc., CA, USA) at a final concentration of 100 μM, in 2 mL tubes and incubated in the dark for 5 min with shaking (150 rpm). Tubes were then placed horizontally on ice, and exposed to light from two halogen bulbs (500 W each) at a distance of 20 cm from the light source, as described elsewhere26,27. In other works, the usual times for PMA photolysis range from 2 to 20 min30. In our case, light exposure periods above 5 min were enough to provide consistent results among sample replicates. Hence, a standard 10-min PMA photo-activation was applied in all the experiments. For natural canker analysis, a commercial, improved version of PMA, known as PMAxx (Biotium Inc., CA, USA), was also used the same as described for PMA. After the PMA (or PMAxx) treatment, samples were centrifuged at 13,000 rpm for 10 min, the supernatant discarded and pellets stored at −80 °C until use.
Correlation between v-dPCR and culturable cell counts
Apple macerates were prepared in AMB and inoculated with live cells ranging from 103 to 106 CFU mL−1. Samples were sub-aliquoted and subjected to v-dPCR after PMA treatment. Aliquots of the same samples non-treated with PMA were used as controls, to quantify total target DNA copies by dPCR. The experiment was repeated in three independent assays. Correlation between v-dPCR and the inoculated CFUs was determined by linear regression analysis.
Correlation between v-dPCR and culturable cell counts in the presence of dead cells
E. amylovora-free plant macerates were prepared with AMB (in initial analyses) or 0.1xAMB, and inoculated with 106 E. amylovora dead cells mL−1. Aliquots of these dead cell suspensions were additionally inoculated with increasing live cell concentrations ranging from 103 to 106 CFU mL−1. Samples were sub-aliquoted in two parts. One part was treated with PMA, to calculate viable cell concentrations, and the other part was directly subjected to dPCR, to determine total cell counts. The experiment was repeated in three independent assays, and coefficients of correlation between v-dPCR and culturable cell counts were determined by linear regression analysis.
Effect of PMA concentration and the E. amylovora killing method on the exclusion of dead-cell signals by v-dPCR
PMA concentrations ranging 100–500 μM were used to stain aliquots of apple plant macerates prepared in AMB and inoculated with 106 E. amylovora dead cells mL−1. Samples non-treated with PMA were used as controls. The assay was performed in triplicate. Mean differences between v-dPCR values for each PMA concentration were calculated after log-transformation of data, by ordinary one-way ANOVA and a Tukey’s multiple comparison test.
To detect a possible effect of the killing method on the suppression of false-positive signals by v-dPCR, apple branch macerates prepared in AMB were inoculated with E. amylovora live cells (103 CFU mL−1) mixed with dead cells (106 cell mL−1) obtained either by heating at 85 °C for 30 min (standard killing method used in this work), heating at 57 °C for 18 h, or exposure to 70% isopropanol for 10 min. Samples were then sub-aliquoted and analyzed in parallel by dPCR, v-dPCR (regular treatment with 100 μM PMA) and plate counts. The experiment was performed in triplicate. The differences between log-transformed viable (v-dPCR) and culturable cell counts for each killing method were calculated, and compared to the control by a Kruskal-Wallis with Dunn’s post hoc test.
Effect of the thermal cycle number and the amplicon length on false-positive signal suppression and live cell population discrimination by v-dPCR
Apple branch macerates prepared in AMB were inoculated either with E. amylovora at 7 × 106 CFU mL−1 (live cell control), or live/dead cell mixtures containing a 10%, a 1%, or a 0% of live cells with respect to the live cell control, and a constant background of 7 × 106 dead cells mL−1. For v-dPCR, samples were treated with PMA and dPCR was performed under different conditions: (i) using the same primers and probe as Pirc et al.31, with 40 amplification cycles; (ii) Pirc et al.31 primers, but reducing the number of thermal cycles to 30; (iii) or a newly designed primer combination targeting a 966 bp DNA sequence (Ams09KbF/Ams189R) (Table 1), and 40 amplification cycles. The assay was performed in two biological repeats. For an easier comparison of theoretical (100%, 10%, 1%, 0%) and obtained live cell percentage values, viable cell concentrations in the control and in live/dead cell mixtures were expressed as percentages with respect to the live cell control. Differences between fluorescence peaks of negative (background signal) and positive calls (positive detection of target DNA) by dPCR were monitored through the QS3D Analysis Suite.
Primer design and effect of increasing amplicon lengths on the exclusion of dead-cell signals by v-dPCR
To obtain different amplicon lengths, the reverse primer Ams189R, from a previous publication31 was combined either with the original primer Ams116F31 or with newly designed primers binding DNA upstream of the reverse primer sequence (Table 1). Primers were designed with Primer-BLAST (https://www.ncbi.nlm.nih.gov/tools/primer-blast/), using the amsC gene sequence of E. amylovora ATCC 49946 plus additional 500 bp upstream of the reverse primer sequence as template. All primers were designed using the web application default settings, including an optimal melting temperature of 60 ± 3 °C, a G + C content between 20–80% and primer sizes of 20 ± 5 nucleotides.
Prior to primer utilization for dPCR, the size of the amplicons with each primer pair was confirmed by classical PCR. Although signal generation by dPCR is less dependent on highly efficient assays than qPCR53, a gradient PCR was performed with all primer pairs to identify possible differences in the efficiency of PCR amplification at different temperatures. All primers showed similar optimal annealing temperatures. Also, preliminary assays on dPCR showed 60 °C as the optimal temperature for both the original primers from the work of Pirc et al.31, and the primers designed in this work. Temperatures above and below 60 °C led to a worse separation between background fluorescence signals and the ones corresponding to positive detection of target DNA. None of the assayed primers showed signs of unspecific amplification either by classical PCR (no unspecific bands) or by dPCR (no more than 2 clusters of fluorescence signals, corresponding to negative and positive amplification) after 40 DNA amplification cycles.
Primer specificity for the E. amylovora target DNA was tested using genomic DNA extractions of a collection of E. amylovora strains, including reference strains, natural isolates, and other plant pathogenic and saprophytic bacterial species listed in the Supplementary Table S3.
To determine the effect of the amplicon length on v-dPCR live/dead cell discrimination, apple branch macerates prepared in AMB were artificially inoculated with either E. amylovora live (106 CFU mL−1), dead (106 dead cells mL−1) or a defined ratio of live:dead cells (103:106 cells mL−1). Each sample was sub-aliquoted for total and live cell quantification. The dPCR reactions were performed with primer combinations targeting a 74, 325, 627 and 966 bp DNA sequence, respectively (Table 1). The assay was repeated in three independent experiments. The effect of the amplicon size on total (dPCR) or live (v-dPCR) cell quantification in samples composed of live, dead or a live/dead cell mixture was analyzed by two-way ANOVA with Tukey’s multiple comparisons test.
Effect of the maceration buffer and the addition of SDS or PMA Enhancer on dead-cell signal suppression by v-dPCR
Apple branch macerates prepared either in AMB or in 0.1xAMB were inoculated with either E. amylovora live (106 CFU mL−1), dead (106 dead cell mL−1), or mixed live (103 CFU mL−1) and dead (106 dead cell mL−1) cells as mentioned above. Each sample was sub-aliquoted and subjected to: (i) dPCR to quantify total cells; (ii) treatment with PMA; (iii) PMA plus 0.0025% SDS (w/v) (PMA + SDS); (iv) PMA plus 1x PMA Enhancer for Gram-negative bacteria (Biotium Inc., CA, USA) (PMA + E). Based on the results from the previous experiment, samples were subjected to DNA extraction and dPCR, using the primers targeting the 627 bp DNA sequence.
The SDS concentration was selected based on a minimal inhibitory concentration (MIC) assay performed in fresh LB broth inoculated with a 1/100 dilution of an overnight culture of E. amylovora ATCC 49946 in the same medium, and amended with increasing two-fold concentrations of SDS ranging from 0 to 0.32% (w/v). The greatest SDS concentration not having a measurable effect on the OD600 nm after a 16-h incubation period was chosen to determine the effect of SDS on the selective discrimination of live cells during PMA treatments.
These assays were performed in triplicate, in two independent experiments. After log-transformation of data, a two-way ANOVA with Sidak’s multiple comparisons tests was conducted to determine the main effects of the maceration buffer and the PMA treatment.
dPCR performance using primers Ams06KbF and Ams189R
Similar to the preliminary assay using the primers of Pirc et al.31, the dPCR performance with the primers Ams06KbF/Ams189R was characterized using apple and pear plant macerates prepared in 0.1xAMB and inoculated with increasing E. amylovora concentrations ranging from 103 to 107 CFU mL−1. Data from two independent experiments performed in triplicate were used to determine the correlation between dPCR values and the inoculated CFUs mL−1 by linear regression analysis. The assay variability (%CV) for each of the tested bacterial concentrations was calculated by dividing the SD by the mean of the replicates, multiplied by 100. The dynamic range of the technique, as well as the linear relationship and correlation coefficient (R2) for dPCR values and culturable cell counts were determined after log-transformation of data. The limits of detection (the lowest concentration at which all replicates are positive for detection) and quantification (the lowest concentration showing positive detection in all the replicates with a %CV equal or below 25%) were defined based on Pavšič et al.54.
Selective live cell discrimination in mixed live/dead cell populations by optimized plant material processing and v-dPCR conditions
The v-dPCR selective detection of E. amylovora viable cells in the presence of dead cells, as well as the correlation of v-dPCR values to the inoculated CFUs mL−1 was tested in apple and pear plant macerates homogenized in 0.1xAMB and inoculated with increasing live cell concentrations (from 103 to 106 CFU mL−1) with a constant background of 106 dead cell mL−1. Total cell counts were performed by dPCR. For viable cell counts, samples were treated with PMA plus 1x PMA Enhancer before dPCR. In all cases, dPCR was performed using primers Ams06KbF and Ams189R. Assays were performed in triplicate and repeated twice, and the linear regression best fit through all the v-dPCR dataset points was calculated with GraphPad Prism 8.1.2 (227).
Canker sampling and analysis of E. amylovora total, live and culturable cell populations
A total of 12 natural fire blight apple (cv. Honeycrisp) and pear (cv. Bosc) cankers were collected from orchards in Peru (NY, USA) and at the HVRL experimental station in Highland (NY, USA), respectively. In both cases, fire blight infections started in early spring of 2016, canker formation was monitored through time, and samples were collected in mid-summer (July 2016) and mid-winter (January 2017). All the sampled cankers had defined margins. Branch diameters and other data related to canker characteristics are detailed in the Supplementary Table S4.
For canker analysis, tissue slices containing the bark and the vascular cambium below the bark were excised from the canker area plus a 4 mm wide ring surrounding the canker margin, using a sterile scalpel. The sliced samples were weighed, transferred to individual plastic bags, mixed in a ratio 1:50 (w/v) with 0.1xAMB, and homogenized with a hammer, as described above.
Afterward, canker macerates were aliquoted to perform (i) plate counts on SNAN and CCT by spread plating 50 μL after serial tenfold dilutions of samples in 10 mM PBS pH 7.4; (ii) total cell quantification by dPCR; (iii) and v-dPCR after treatment with PMA, PMAxx, or combinations of any of these two viability PCR dyes with 0.0025% SDS (final concentration) or 1x PMA Enhancer for Gram-negative bacteria, according to the manufacturer’s instructions.
All dPCR reactions were run using the primer pair and probe Ams06KbF/Ams189R and Ams141T, respectively (Table 1). When required, samples for dPCR and v-dPCR were diluted from tenfold up to 500-fold to reach target copy concentrations inside of the dPCR quantification range.
In apple cankers, a strong consistency between the total, viable and culturable cell counts was observed among the samples collected in the same time period. A similar phenomenon was observed in pear cankers collected in mid-summer. However, the mid-winter cankers revealed very different E. amylovora population patterns depending on the analyzed sample. For a more accurate statistical analysis to assess differences among viability-staining treatments, consistent data among samples harvested in the same time period were grouped together (Figs 9a,b and 10a), while mid-winter cankers differing in the E. amylovora population content were represented separately to illustrate specific aspects of the v-dPCR efficiency as well as other important insights tied to pathogen biology.
Supplementary information
Acknowledgements
We thank Alan Collmer, Kerik Cox, Awais Khan, and Melanie J. Filiatrault (Cornell University) as well as George Sundin (Michigan State University) for kindly providing some of the bacterial strains employed in this work. We also acknowledge Peter Schweitzer at the Institute of Biotechnology, Cornell University, for training and Jerry Calvin and David Esteban (Vassar College) for allowing us to use their facilities at Biology Department in preliminary stages of this work. This material is based upon work supported by the National Institute of Food and Agriculture, U.S. Department of Agriculture, Hatch/Multistate Research Program projects NYG-625835 and NYG-625857 under 1009897 and 1014444, the NY State Farm Viability Institute grant number 81927/A001-FVI 17 006, and the NY State Specialty Crop Block Grant Program grant number SCG 82535/A001-SCG 17 005 to SGA and SGA’s unrestricted laboratory program funds. We thank the faculty and staff of Cornell University’s HVRL and the owners of Northern Orchard Co Inc for help in this research and maintenance of HVRL experiment plots.
Author Contributions
R.D.S., C.M. and S.G.A. contributed to the study design. R.D.S. and C.M. performed all the experiments. R.D.S. carried out the analysis of results, data interpretation and article drafting. C.M. and S.G.A. reviewed and edited the article. All authors approved the final version of the manuscript. S.G.A. secured all the project funding.
Data Availability
Data supporting the findings of this study are available within the publication, as supplementary material and/or from the corresponding authors on request.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
10/29/2019
An amendment to this paper has been published and can be accessed via a link at the top of the paper.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-47976-x.
References
- 1.van der Zwet, T., Orolaza-Halbrendt, N. & Zeller, W. Fire Blight: History, Biology, and Management (APS Press, 2012).
- 2.Marco-Noales E, et al. Iberian wild pear (Pyrus bourgaeana) is a new host of Erwinia amylovora, the causal agent of fire blight. Plant Dis. 2017;101:502. [Google Scholar]
- 3.Thomson, S. V. Epidemiology Of Fire Blight. In Fire blight: The Disease and Its Causative Agent, Erwinia amylovora (ed. Vanneste, J. L.) 9–36 (CAB International, 2000).
- 4.Billing E. Fire blight. Why do views on host invasion by Erwinia amylovora differ? Plant Pathol. 2011;60:178–189. [Google Scholar]
- 5.Weißhaupt S, et al. Alternative inoculum sources for fire blight: the potential role of fruit mummies and non-host plants. Plant Pathol. 2016;65:470–483. [Google Scholar]
- 6.Sobiczewski P, Iakimova ET, Mikiciński A, Węgrzynowicz-Lesiak E, Dyki B. Necrotrophic behaviour of Erwinia amylovora in apple and tobacco leaf tissue. Plant Pathol. 2017;66:842–855. [Google Scholar]
- 7.Beer SV, Opgenorth DC. Erwinia amylovora on fire blight canker surfaces and blossoms in relation to disease occurrence. Phytopathology. 1976;66:317–322. [Google Scholar]
- 8.Beer S, Norelli J. Fire Blight epidemiology: Factors affecting release of Erwinia amylovora by cankers. Phytopathology. 1977;67:1119–1125. [Google Scholar]
- 9.Paulin JP. Overwintering of Erwinia amylovora: sources of inoculum in spring. Acta Hort. 1981;117:49–54. [Google Scholar]
- 10.Sobiczewski P, Pulawska J, Berczynski S, Konicka M. Fire blight detection and control in Poland. Acta Hortic. 1999;489:115–120. [Google Scholar]
- 11.Aćimović SG, Balaž JS, Aćimović DD, Reeb PD. High magnitude of fire blight symptom development and canker formation from July onwards on two apple cultivars under severe natural infections. J. Plant Pathol. 2014;96:159–168. [Google Scholar]
- 12.Oliver JD. The viable but nonculturable state for bacteria: Status update. Microbe Mag. 2016;11:159–164. [Google Scholar]
- 13.Schaad NW, Frederick RD. Real-time PCR and its application for rapid plant disease diagnostics. Can. J. Plant Pathol. 2002;24:250–258. [Google Scholar]
- 14.Rudi, K., Moen, B., Drømtorp, S. M. & Holck, A. L. Use of ethidium monoazide and PCR in combination for quantification of viable and dead cells in complex samples. 71, 1018–1024 (2005). [DOI] [PMC free article] [PubMed]
- 15.Schaad NW, Berthier-Schaad Y, Knorr D. A high throughput membrane BIO-PCR technique for ultra-sensitive detection of Pseudomonas syringae pv. phaseolicola. Plant Pathol. 2007;56:1–8. [Google Scholar]
- 16.Scuderi G, et al. Development of a simplified NASBA protocol for detecting viable cells of the citrus pathogen Xanthomonas citri subsp. citri under different treatments. Plant Pathol. 2010;59:764–772. [Google Scholar]
- 17.Lau HY, et al. Specific and sensitive isothermal electrochemical biosensor for plant pathogen DNA detection with colloidal gold nanoparticles as probes. Sci. Rep. 2017;7:38896. doi: 10.1038/srep38896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Daranas N, et al. Monitoring viable cells of the biological control agent Lactobacillus plantarum PM411 in aerial plant surfaces by means of a strain-specific viability quantitative PCR method. Appl. Environ. Microbiol. 2018;84:e00107–18. doi: 10.1128/AEM.00107-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dreo T, et al. Optimising droplet digital PCR analysis approaches for detection and quantification of bacteria: a case study of fire blight and potato brown rot. Anal. Bioanal. Chem. 2014;406:6513–6528. doi: 10.1007/s00216-014-8084-1. [DOI] [PubMed] [Google Scholar]
- 20.Rački N, Dreo T, Gutierrez-Aguirre I, Blejec A, Ravnikar M. Reverse transcriptase droplet digital PCR shows high resilience to PCR inhibitors from plant, soil and water samples. Plant Methods. 2014;10:42. doi: 10.1186/s13007-014-0042-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gutiérrez-Aguirre I, Rački N, Dreo T, Ravnikar M. Droplet digital PCR for absolute quantification of pathogens. Met. Mol. Biol. 2015;1302:331–347. doi: 10.1007/978-1-4939-2620-6_24. [DOI] [PubMed] [Google Scholar]
- 22.Bahder BW, Helmick EE, Mou D, Harrison NA, Davis R. Digital PCR technology for detection of palm-infecting phytoplasmas belonging to group 16SrIV that occur in Florida. Plant Dis. 2018;102:1008–1014. doi: 10.1094/PDIS-06-17-0787-RE. [DOI] [PubMed] [Google Scholar]
- 23.Huggett JF, Whale A. Digital PCR as a novel technology and its potential implications for molecular diagnostics. Clin. Chem. 2013;59:1691–1693. doi: 10.1373/clinchem.2013.214742. [DOI] [PubMed] [Google Scholar]
- 24.Quan P, Sauzade M, Brouzes E. dPCR: A Technology Review. Sensors. 2018;18:1271. doi: 10.3390/s18041271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cawthorn D-M, Witthuhn RC. Selective PCR detection of viable Enterobacter sakazakii cells utilizing propidium monoazide or ethidium bromide monoazide. J. Appl. Microbiol. 2008;105:1178–1185. doi: 10.1111/j.1365-2672.2008.03851.x. [DOI] [PubMed] [Google Scholar]
- 26.Bae S, Wuertz S. Discrimination of viable and dead fecal Bacteroidales bacteria by quantitative PCR with Propidium monoazide. Appl. Environ. Microbiol. 2009;75:2940–2944. doi: 10.1128/AEM.01333-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bertolini E, et al. Transmission of ‘Candidatus Liberibacter solanacearum’ in carrot seeds. Plant Pathol. 2015;64:276–285. [Google Scholar]
- 28.Duarte-Guevara P, Duarte-Guevara C, Ornob A, Bashir R. On-chip PMA labeling of foodborne pathogenic bacteria for viable qPCR and qLAMP detection. Microfluid. Nanofluidics. 2016;20:114. [Google Scholar]
- 29.Nocker A, Cheung C-Y, Camper AK. Comparison of propidium monoazide with ethidium monoazide for differentiation of live vs. dead bacteria by selective removal of DNA from dead cells. J. Microbiol. Methods. 2006;67:310–320. doi: 10.1016/j.mimet.2006.04.015. [DOI] [PubMed] [Google Scholar]
- 30.Fittipaldi M, Nocker A, Codony F. Progress in understanding preferential detection of live cells using viability dyes in combination with DNA amplification. J. Microbiol. Methods. 2012;91:276–289. doi: 10.1016/j.mimet.2012.08.007. [DOI] [PubMed] [Google Scholar]
- 31.Pirc M, Ravnikar M, Tomlinson J, Dreo T. Improved fireblight diagnostics using quantitative real-time PCR detection of Erwinia amylovora chromosomal DNA. Plant Pathol. 2009;58:872–881. [Google Scholar]
- 32.Løvdal T, Hovda MB, Björkblom B, Møller SG. Propidium monoazide combined with real-time quantitative PCR underestimates heat-killed Listeria innocua. J. Microbiol. Methods. 2011;85:164–169. doi: 10.1016/j.mimet.2011.01.027. [DOI] [PubMed] [Google Scholar]
- 33.Slimani S, et al. Evaluation of propidium monoazide (PMA) treatment directly on membrane filter for the enumeration of viable but non cultivable Legionella by qPCR. J. Microbiol. Methods. 2012;88:319–321. doi: 10.1016/j.mimet.2011.12.010. [DOI] [PubMed] [Google Scholar]
- 34.Contreras PJ, Urrutia H, Sossa K, Nocker A. Effect of PCR amplicon length on suppressing signals from membrane-compromised cells by propidium monoazide treatment. J. Microbiol. Methods. 2011;87:89–95. doi: 10.1016/j.mimet.2011.07.016. [DOI] [PubMed] [Google Scholar]
- 35.Meng X, et al. Rapid detection and quantification of viable Pseudomonas syringae pv. lachrymans cells in contaminated cucumber seeds using propidium monoazide and a real-time PCR assay. Can. J. Plant Pathol. 2016;38:296–306. [Google Scholar]
- 36.Han S, et al. Detection of Clavibacter michiganensis subsp. michiganensis in viable but nonculturable state from tomato seed using improved qPCR. PLoS One. 2018;13:e0196525. doi: 10.1371/journal.pone.0196525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gorris MT, et al. A sensitive and specific detection of Erwinia amylovora based on ELISA-DASI enrichment method with monoclonal antibodies. Acta Hort. 1996;411:41–46. [Google Scholar]
- 38.EPPO standard PM 7/20 (2): Erwinia amylovora. OEPP/EPPO Bull. 43, 21–45 (2013).
- 39.ISPM 27. Annex 13. Erwinia amylovora Rome, IPPC, FAO, https://www.ippc.int/en/publications/83443/ (2016).
- 40.Nkuipou-Kenfack E, Engel H, Fakih S, Nocker A. Improving efficiency of viability-PCR for selective detection of live cells. J. Microbiol. Methods. 2013;93:20–24. doi: 10.1016/j.mimet.2013.01.018. [DOI] [PubMed] [Google Scholar]
- 41.Lee JL, Levin RE. Discrimination of viable and dead Vibrio vulnificus after refrigerated and frozen storage using EMA, sodium deoxycholate and real-time PCR. J. Microbiol. Methods. 2009;79:184–188. doi: 10.1016/j.mimet.2009.08.014. [DOI] [PubMed] [Google Scholar]
- 42.Yáñez MA, et al. Quantification of viable Legionella pneumophila cells using propidium monoazide combined with quantitative PCR. J. Microbiol. Methods. 2011;85:124–130. doi: 10.1016/j.mimet.2011.02.004. [DOI] [PubMed] [Google Scholar]
- 43.Li, S. Y. Polymers for DNA binding. PhD thesis, Seton Hall University, South Orange, NJ (Seton Hall University Dissertations and Theses, ETDs, 2013).
- 44.Steiner PW. Predicting apple blossom infections by Erwinia amylovora using the Maryblyt model. Acta Hortic. 1990;273:139–148. [Google Scholar]
- 45.Biggs AR. Characteristics of fire blight cankers following shoot inoculations of three apple cultivars. HortScience. 1994;29:795–797. [Google Scholar]
- 46.Kielak K, Sobiczewski P, Pulawska J. Overwintering of Erwinia amylovora in naturally and artificially infected appIe shoots. Acta Hort. 2002;590:157–162. [Google Scholar]
- 47.Ishimaru C, Klos EJ. New medium for detecting Erwinia amylovora and its use in epidemiological studies. Phytopathology. 1984;74:1342–1345. [Google Scholar]
- 48.Apajalahti JHA, Kettunen A, Nurminen PH, Jatila H, Holben WE. Selective plating underestimates abundance and shows differential recovery of bifidobacterial species from human feces. Appl. Environ. Microbiol. 2003;69:5731–5735. doi: 10.1128/AEM.69.9.5731-5735.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Biggs AR, Turechek WW. Fire blight of apples and pears: Epidemiological concepts comprising the Maryblyt forecasting program. Plant Heal. Prog. 2010;11:23. [Google Scholar]
- 50.Bertani G. Studies on Lysogenesis. I. The mode of phage liberation by lysogenic. Escherichia coli. J. Bacteriol. 1952;62:293–300. doi: 10.1128/jb.62.3.293-300.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dye DW. A taxonomic study of the genus Erwinia. I. The ‘amylocora’ group. N. Z. J. Sci. 1968;11:590–607. [Google Scholar]
- 52.Santander RD, Català-Senent JF, Marco-Noales E, Biosca EG. In planta recovery of Erwinia amylovora viable but nonculturable cells. Trees. 2012;26:75–82. [Google Scholar]
- 53.Huggett JF, et al. The Digital MIQE Guidelines: Minimum Information for Publication of Quantitative Digital PCR Experiments. Clin. Chem. 2013;59:892–902. doi: 10.1373/clinchem.2013.206375. [DOI] [PubMed] [Google Scholar]
- 54.Pavšič J, Žel J, Milavec M. Assessment of the real-time PCR and different digital PCR platforms for DNA quantification. Anal. Bioanal. Chem. 2016;408:107–121. doi: 10.1007/s00216-015-9107-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data supporting the findings of this study are available within the publication, as supplementary material and/or from the corresponding authors on request.