Abstract
Background
MicroRNAs are small regulatory RNAs with important roles in carcinogenesis. Genetic variants in these regulatory molecules may contribute to disease. We aim to identify allelic variants in microRNAs as susceptibility factors to gastric cancer using association studies and functional approaches.
Methods
Twenty‐one single nucleotide variants potentially functional, because of their location in either the seed, mature or precursor region of 22 microRNAs, were selected for association studies. Genetic association with gastric cancer in 365 cases and 1,284 matched controls (European Prospective Investigation into Cancer and Nutrition Cohort) was analysed using logistic regression. MicroRNA overexpression, transcriptome analysis, and target gene validation experiments were performed for functional studies.
Results
rs3746444:T>C, in the seed of MIR499A and mature MIR499B, associated with the cardia adenocarcinoma location; rs12416605:C>T, in the seed of MIR938, associated with the diffuse subtype; and rs2114358:T>C, in the precursor MIR1206, associated with the noncardia phenotype. In all cases, the association was inverse, indicating a protective affect against gastric cancer of the three minor allelic variants. MIR499 rs3746444:T>C and MIR1206 rs2114358:T>C are reported to affect the expression of these miRNAs, but the effect of MIR938 rs12416605:C>T is unknown yet. Functional approaches showed that the expression of MIR938 is affected by rs12416605:C>T and revealed that MIR938 could regulate a subset of cancer‐related genes in an allele‐specific fashion. Furthermore, we demonstrated that CXCL12, a chemokine participating in gastric cancer metastasis, is specifically regulated by only one of the rs12416605:C>T alleles.
Conclusion
rs12416605 appears to be involved in gastric cancer by affecting the regulatory function of MIR938 on genes related to this cancer type, particularly on CXCL12 posttranscriptional regulation.
Keywords: functional SNV, gastric cancer, gene regulation, genetic susceptibility, microRNA
1. INTRODUCTION
Gastric Cancer (GC) or stomach cancer is the fifth most common cancer in the world in terms of incidence and the third leading cause of cancer death in both sexes worldwide (Ferlay et al., 2015; Globocan, 2018). There are two main locations of gastric adenocarcinoma: proximal (cardia) and distal (noncardia); histologically there are two subtypes of the disease, the intestinal‐type, which presents clearly defined glandular structures, and the undifferentiated diffuse‐type, which consists of individually infiltrating neoplastic cells (Crew & Neugut, 2006). GC is a multifactorial disease, whose major risk factors are environmental, such as Helicobacter pylori infection, diet or tobacco, as well as genetic (González, Sala, & Rokkas, 2013). Several GC susceptibility genes have already been identified by both candidate gene analysis and genome‐wide association studies, among which there are genes involved in immunoinflammatory response, DNA repair, cellular adherence, proliferative processes, mucosa protection, and Helicobacter pylori's cellular signaling pathways (Loh et al., 2009; McLean & El‐Omar, 2014; Saeki, Ono, Sakamoto, & Yoshida, 2013; Sala et al., 2012). A multistep model of carcinogenesis involving the effects of common low penetrance and rare disease‐causing variants is currently accepted for GC (Correa, 1992; Fletcher & Houlston, 2010). Once the gastric adenocarcinoma has appeared, its prognosis is highly dependent on the progression and metastasis of the tumor, which are intricate processes partially controlled by oncogenes and tumor suppressor genes, whose expression is further regulated by microRNAs (miRNAs) (Esquela‐Kerscher & Slack, 2006).
MiRNAs are nonprotein‐coding small RNAs that negatively regulate the gene expression posttranscriptionally (Bartel, 2009). The action of miRNAs as gene repressors is performed by means of partial complementarity to miRNA‐binding sites at their target messenger RNAs (mRNAs) resulting in either degradation of the target mRNAs or inhibition of translation (Filipowicz, Bhattacharyya, & Sonenberg, 2008). Each mRNA can be repressed by multiple miRNAs, as well as each miRNA is a potential regulator of hundreds of transcripts (Friedman, Farh, Burge, & Bartel, 2009; Lewis, Shih, Jones‐Rhoades, Bartel, & Burge, 2003) accordingly, it has been estimated that more than half of the total human protein‐coding genes are regulated by miRNAs through complex regulatory networks that control almost every cellular processes, including development, differentiation, proliferation and apoptosis, and having important roles in carcinogenesis (Friedman et al., 2009; Kloosterman & Plasterk, 2006; ). Aberrant miRNA expression has been related to the diagnosis and stage of many types of cancer and numerous studies validate their prognostic and predictive value (Lu et al., 2005; Shen, Stass, & Jiang, 2013). In GC various miRNAs have been found differentially expressed between tumor and normal tissues and some of them are associated with the progression and prognosis of GC, being proposed as potential GC biomarkers (Katada et al., 2009; Lu et al., 2005; Shen et al., 2013; Zhou et al., 2015).
Genetic association studies have pointed to several miRNA allelic variants as susceptibility factors to GC (Espinosa‐Parrilla et al., 2014; Peng et al., 2010; Xia et al., 2016; Yang et al., 2014). Nevertheless, very little is known about the real contribution of these genetic variants to GC pathology. One of the few exceptions is a report showing that the A allele of rs11671784 in the MIR27A reduces the expression of the miRNA and is associated with a reduction in the risk of GC (Yang et al., 2014). A better understanding of the role of single‐nucleotide variants (SNVs) in miRNAs in the regulation of gastric carcinogenesis could be useful to assess individual susceptibility to GC and to evaluate their functional relevance and potential use as diagnostic and therapy tools.
In this work, we analysed the genetic association between GC and 21 SNVs selected because of their location in functional miRNA regions in a cohort of 365 cases and 1,284 healthy controls from the European Prospective Investigation into Cancer and Nutrition (EPIC) cohort. We found associations between different GC subtypes and three SNVs: rs3746444:T>C in the seed of MIR499A and mature MIR499B, rs12416605:C>T in the seed of MIR938, and rs2114358:T>C in the precursor sequence of MIR1206. Moreover, we performed functional analyses based on miRNA overexpression experiments, transcriptome analyses and target gene‐validation tests, and concluded that rs12416605:T>C alters the regulatory action of MIR938 specifically by affecting the regulation of CXCL12, a chemokine gene involved in the etiology of GC.
2. MATERIALS AND METHODS
2.1. Ethical compliance
Patient recruitment, consent, and DNA extraction were carried out as previously described (Riboli et al., 2002). Signed informed consent was obtained from all the individuals prior to their participation in the study. The study was approved by ethics committees at the International Agency for Research on Cancer and in each of the EPIC recruitment centers. This study conforms to the Declaration of Helsinki standards for Medical research involving human subjects.
2.2. Subjects
Study subjects were selected from the EPIC cohort according to a nested case–control design aimed at assessing the genetic and environmental risk factors for GC (EurGast study). The EPIC cohort has been extensively presented (Riboli et al., 2002). Briefly, it includes 521,457 individuals recruited between 1992 and 2000 in 23 centers from 10 North to South European countries. As previously reported (Espinosa‐Parrilla et al., 2014), GC Cases were subjects with adenocarcinoma of the stomach (code C16 of the International Classification of Diseases, 10th Revision) having blood collected and diagnosed during the follow‐up. An independent panel of pathologists confirmed and validated the diagnosis, tumor subsite, and morphology (Carneiro et al., 2007). Prevalent tumors, cancer located in gastric stump, as well as tumors other than adenocarcinoma were excluded. For each case, up to four control subjects matched by center, gender, age (±2.5 years), and date of blood collection (±45 days) were randomly selected among cohort members who were alive and free of cancer at the time of diagnosis. The initial study population consisted of 373 GC cases and 1,332 controls. After exclusion of eight cases and 48 controls whose DNA was not amplified or had a genotyping call rate lower than 80%, the final analysis was performed on 365 GC cases and 1,284 matched controls (Espinosa‐Parrilla et al., 2014) (Table 1). Regarding the localization of tumors, 29.3% were in the cardia or gastroesophageal junction, 49.6% were noncardia, 1.6% presented a mixed localization, and 19.5% were unspecified; as for the histological type, 34.5% were intestinal, 35.1% were diffuse, 2.2% were mixed, and 28.2% were unspecified (Table 1).
Table 1.
Main characteristics of the gastric cancer cases and controls analyzed
| Cases | Controls | ||||
|---|---|---|---|---|---|
| N | (%) | N | (%) | ||
| Sex | Male | 214 | (58.6%) | 759 | (59.1%) |
| Female | 151 | (41.4%) | 525 | (40.9%) | |
| Anatomical subtype of gastric cancer | Cardia | 107 | (29.3%) | – | – |
| Non‐cardia | 181 | (49.6%) | – | – | |
| Mixed | 6 | (1.6%) | – | – | |
| Unknown | 71 | (19.5%) | – | – | |
| Histological subtype of gastric cancer | Intestinal | 126 | (34.5%) | – | – |
| Diffuse | 128 | (35.1%) | – | – | |
| Mixed | 8 | (2.2%) | – | – | |
| Unknown | 103 | (28.2%) | – | – | |
| Mean | (Sd) | Mean | (Sd) | ||
|---|---|---|---|---|---|
| Age at recruitment (years) | 58.4 | (7.9) | 58.4 | (7.69) |
2.3. SNV selection, genotyping, quality control, and data filtering
We selected a panel of 29 non‐monomorphic SNVs located in different functional regions of 30 human miRNAs including all SNVs (dbSNP version 126) located on miRNA genes (miRBase release 13.0) that were known at the moment of the design of the study (Table S1). Genotyping of these SNVs was included in a panel of 1,536 SNVs described in detail elsewhere (Sala et al., 2012). Briefly, genomic DNA was extracted from buffy coat and genotyped at the Spanish National Genotyping Centre (CEGEN) by use of the Illumina BeadStation Platform and GoldenGate Technology (Illumina, San Diego, CA), according to the manufacturer's protocols. In addition to the internal genotyping controls, ~5% of the samples (n = 100) were genotyped in duplicate with overall agreement of 99.2% (Sala et al., 2012). Among the miRNA SNVs selected for the study, eight were excluded from the analysis because of technical genotyping problems (Table S1). The total number of SNVs successfully genotyped and available for analysis was therefore 21 (Table 2). As previously described (Espinosa‐Parrilla et al., 2014), deviation from Hardy–Weinberg equilibrium (HWE) was established at p < 10−4 (Fisher exact test). According to this criterion, none of the analyzed SNVs deviated from HWE among controls (Table S1). In all cases significance from deviation was above 0.05 and only one SNP (rs10505168 in MIR2053), which was not associated with GC, showed a p ≤ 0.05 (p = 0.008).
Table 2.
Main characteristics of the miRNA SNVs analyzed
| SNV | miRNA Gene | Chromosome | MAF Controls | MAF Cases | Location | Alleles (major/minor) | Global Fst |
|---|---|---|---|---|---|---|---|
| rs41291179 | MIR216A | 2 | 0.053 | 0.047 | precursor | A/T | 0.1068 |
| rs13186787 | MIR1294 | 5 | 0.002 | 0.003 | precursor | A/G | 0.0188 |
| rs2910164 | MIR146A | 5 | 0.240 | 0.229 | seed | C/G | 0.1012 |
| rs41274239 | MIR96 | 7 | 0.004 | 0.003 | precursor | A/G | 0.0010 |
| rs2114358 | MIR1206 | 8 | 0.395 | 0.358 | precursor | T/C | 0.0302 |
| rs10505168 | MIR2053 | 8 | 0.280 | 0.274 | precursor | A/G | 0.0768 |
| rs11259096 | MIR1265 | 10 | 0.048 | 0.054 | precursor | T/C | 0.2416 |
| rs17091403 | MIR2110 | 10 | 0.102 | 0.084 | precursor | C/T | 0.031 |
| rs4919510 | MIR608 | 10 | 0.191 | 0.178 | mature | C/G | 0.1406 |
| rs12416605 | MIR938 | 10 | 0.258 | 0.248 | seed | C/T | 0.2635 |
| rs11020790 | MIR548L | 11 | 0.013 | 0.007 | precursor | C/T | 0.1503 |
| rs7311975 | MIR1178 | 12 | 0.028 | 0.026 | seed | T/C | 0.3701 |
| rs11614913 | MIR196A2 | 12 | 0.394 | 0.410 | mature | C/T | 0.2026 |
| rs2289030 | MIR492 | 12 | 0.058 | 0.047 | precursor | C/G | 0.1107 |
| rs11844707 | MIR1185‐2 | 14 | 0.001 | 0.000 | precursor | G/A | 0.2655 |
| rs6505162 | MIR423 | 17 | 0.461 | 0.452 | precursor | A/C | 0.3750 |
| rs17759989 | MIR633 | 17 | 0.029 | 0.022 | precursor | A/G | 0.0410 |
| rs895819 | MIR27A | 19 | 0.320 | 0.310 | precursor | T/C | 0.0412 |
| rs3746444 | MIR499A | 20 | 0.192 | 0.185 | seed | T/C | 0.0000 |
| rs3746444 | MIR499B | 20 | 0.192 | 0.185 | mature | A/G | 0.0000 |
| rs4822739 | MIR548J | 22 | 0.051 | 0.049 | precursor | C/G | 0.0613 |
| rs5965660 | MIR888 | X | 0.187 | 0.218 | precursor | T/G | NA |
Abbreviations: Fst, fixation index; MAF, minor allele frequency.
2.4. Statistical analyses
As previously described, odds ratios (ORs) and 95% confidence intervals (95% CIs) were estimated for general GC as well as for its anatomical and histological subtypes using unconditional logistic regression adjusted by the matching variables gender, age, and country using the SNVassoc R package and applying a gene‐based 1000‐permutation test on the log‐additive model to account for multiple testing of the different SNPs analyzed in each miRNA gene region (Espinosa‐Parrilla et al., 2014). Other risk factors for GC such as Helicobacter pylori infection, smoking and some other lifestyle and dietary factors independent of the genetic variants were not included in the models to retain maximal statistical power. The possibility of population stratification was considered and excluded as previously detailed (Sala et al., 2012).
2.5. miRNA cloning
A genomic region of 521 bp including the 83 bp of the precursor molecule of MIR938 (RefSeq: NR_030634.1) and at least 100 bp at the 5′ and 100 bp at the 3′ flanking regions was amplified from human DNA samples using the forward primer: 5′ cacacacaAGATCTCCAAATCATTCTGGCAGTGA3′ and reverse primer: 5′‐cacacacaCTGCAGTTCATTGCTTGTTGGGATCA‐3′. PCR fragments were cloned into the pmR‐ZsGreen1 vector (Clontech; Mountain View, CA) through PstI and BglIl restriction sites.
2.6. miRNA transfection experiments
For RT‐qPCR experiments, HeLa cells were grown as previously described and scaled to 1.8 × 105 cells/well in 12‐well plates (Lopez‐Valenzuela et al., 2012). After 24 hr, cells were double‐transfected with Lipofectamine 2000 (Invitrogen; Carlsbad, CA) and 0.75 µg of DNA with the plasmid constructions carrying either one or the other allelic variant of the studied miRNAs, plus a nonhuman miRNA that was used as a control for normalization of transfection. For transcriptome analyses 4.5 × 105 cells were grown in 6‐well plates and after 24 hr, scaled amount of DNA was single‐transfected with only one of the above described constructs. Transfections were stopped at 24 hr for both RT‐qPCR expression and transcriptome analysis, and 1 ml/well of QIAzol Lysis Reagent (QIAGEN; Venlo, Netherlands) was used for total RNA extraction using miRNeasy Mini Kit (QIAGEN; Venlo, Netherlands). Three and four independent experiments were performed for transcriptome and RT‐qPCR experiments, respectively. Each experiment included three technical replicates in all cases.
2.7. Expression analysis by real‐time quantitative reverse transcription polymerase chain reaction (RT‐qPCR)
MiRNA expression analyses were performed by RT‐qPCR starting from 300 ng of total RNA from transfected cells by using the specific MIR938 forward primer: 5′‐ CAGTGCCCTTAAAGGTGA‐3′ and MIR938 reverse primer 5′‐CAGTTTTTTTTTTTTTTTACTGGGT‐3′. Primers design and RT‐qPCR protocol were performed as previously described (Balcells, Cirera, & Busk, 2011; Gallego et al., 2016), using LightCycler® 480 SYBR Green I Master (Roche Diagnostics; Rotkreuz, Switzerland) following manufacturer's protocol. Standard curves were calculated by pooling samples from transfection experiments. Comparison of the expression levels between the two MIR938 variants was performed from RT templates (30x) by quantifying the relative expression ratio of the studied miRNAs based on the efficiency and on the quantification of cycle deviation (ΔCp) between the two variants, and comparing versus the reference miRNA MIR25 as previously described (Gallego et al., 2016).
2.8. Whole‐Genome expression analyses in HeLa cells using beadchip microarrays
RNA samples from three independent transfection experiments in HeLa cells were used for microarray expression experiments in Agilent SurePrint G3 Human Gene Expression microarrays (8×60k), starting from 300 ng of total RNA. Data were analyzed using the Array File Maker (AFM) 4.0 software package (Array File Maker; Stanford, CA). We considered genes deregulated when comparing each miRNA allelic variant versus the empty construct and differential regulation between alleles when comparing each miRNA allelic variant versus the other [nominal p < 0.05 with at least 1.2 fold change (FC)]. Analyses on the biological functions associated with deregulated genes were performed using the Ingenuity Pathway Analysis software (IPA, www.ingenuity.com) and DAVIDGO (http://david.abcc.ncifcrf.gov/home.jsp).
2.9. 3′ Untranslated regions (UTR) cloning and luciferase activity assay in HeLa cells
A fragment of 1,106 bp of the 3′ UTR of CXCL12 (RefSeq: NM_000609.6) was amplified by PCR using forward primer 5′‐acacacgctagcTGTGTTACCTGAAAACACTGTGC‐3′ and reverse primer 5′‐acacactctagaAAGGGACAATTTTTGTTGATGG‐3′ and cloned into PGL4.13 Promega vector containing a Firefly reporter gene, after digestion with XbaI enzyme. Co‐transfection experiments have been previously described (Lopez‐Valenzuela et al., 2012). Briefly, HeLa cells were seeded at 1.3 × 104 cells per well in 96‐well plates and co‐transfected 24 hr later with the Firefly reporter constructs described above or the empty pGL4.13 vector (24 ng), the Renilla reporter plasmid pGL4.75 (3 ng), and 10 nM miRNA mimic for the mature miRNAs containing either the T or C alleles of rs12416605:C>T, and the negative control C2 (Dharmacon; Lafayette, CO), using Lipofectamine 2000 (Invitrogen; Carlsbad, CA). Activities of Firefly and Renilla luciferases were measured 24 hr after transfection using Dual‐Glo Luciferase assay system (Promega; Fitchburg, WI). Relative reporter firefly luciferase activity was obtained by normalization to the Renilla luciferase activity. To correct for vector‐dependent unspecific effects, each relative reporter activity was normalized to the empty vector co‐transfected with the corresponding miRNA. Results were then compared with the mean of the C2 negative control. Each experiment was done in triplicate and six independent experiments were performed for each miRNA. Statistical significance was determined using Student's t test (p < 0.05).
2.10. Target gene prediction analysis
Potential target genes for MIR938 allelic variants were predicted using the PITA (Probability of Interaction by Target Accessibility) algorithm (Kertesz, Iovino, Unnerstall, Gaul, & Segal, 2007). Minimum accessibility energy score (ΔG) and the energy‐based score for microRNA‐target interactions (ΔΔG) were calculated for all described 3′ UTRs of human RefSeq genes and either the MIR938 C or T alleles. A transcript was considered as a predicted target when ΔΔG ≤ −8.
3. RESULTS
3.1. Association of miRNA SNVs with GC risk and tumor subtype
To analyze the possible association between gastric adenocarcinoma and putative functional SNVs located in miRNAs, we genotyped a set of 21 selected SNVs: 15 located in the precursor region, outside of the mature miRNA; two in the mature region, outside the seed miRNA; three in the seed miRNA region; and one SNV, rs3746444:T>C, that was located in the seed of MIR499A and in the mature MIR499B, since both miRNA genes overlap in sense and antisense strands of the same genomic region (Table 2).
Next, we investigated if there were miRNA SNVs significantly associated with GC and its subtypes in 365 incident GC cases with different histological and adenocarcinoma location phenotypes matched to 1,284 controls from de EPIC cohort (Table 1). Results of the association analyses under the log‐additive model are shown in Table 3. The strongest association was found for rs2114358:T>C in the precursor MIR1206, which appeared inversely associated with the risk of the non‐cardia localization of the adenocarcinoma (per allele OR = 0.73, 95% CI = 0.58–0.93, p = 0.0093) but not for the cardia localization. Additionally, rs3746444:T>C in the mature MIR499A and seed of MIR499B was found inversely associated only with the risk of the cardia localization of the adenocarcinoma (per allele OR = 0.64, 95% CI = 0.42–0.98, p = 0.0308). As for the histological subtypes, rs12416605:C>T in the seed of MIR938, appeared inversely associated with the risk for the diffuse subtype of GC (per allele OR = 0.7, 95% CI = 0.51–0.97, p = 0.0281) but not for the intestinal one (Table 3). These results are suggestive of a protective effect against GC for the minor alleles of all three SNVs (C rs2114358, C rs3746444, and T rs12416605).
Table 3.
Results of the association analyses of miRNA SNVs with general gastric cancer and its histological and anatomical subtypes under the log‐additive model
| SNV | miRNA gene | General GC | Intestinal | Diffuse | Cardia | Non‐Cardia | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| OR (95% CI) | p‐value | OR (95% CI) | p‐value | OR (95% CI) | p‐value | OR (95% CI) | p‐value | OR (95% CI) | p‐value | ||
| rs41291179 | MIR216A | 0.87 (0.59–1.28) | 0.4657 | 1.03 (0.58–1.84) | 0.9165 | 0.96 (0.53–1.72) | 0.8786 | 0.69 (0.33–1.45) | 0.3036 | 0.98 (0.6–1.61) | 0.9343 |
| rs13186787 | MIR1294 | 1.76 (0.32–9.81) | 0.5308 | 0 (0–NA) | 0.4122 | 5.38 (0.93–31.27) | 0.0937 | 0 (0–NA) | 0.3872 | 4.16 (0.73–23.89) | 0.1452 |
| rs2910164 | MIR146A | 0.95 (0.78–1.15) | 0.5930 | 1.11 (0.82–1.49) | 0.4988 | 0.8 (0.58–1.11) | 0.1723 | 1.18 (0.85–1.64) | 0.3135 | 0.93 (0.71–1.21) | 0.5839 |
| rs41274239 | MIR96 | 0.66 (0.14–3.01) | 0.5720 | 0 (0–NA) | 0.1467 | 0.83 (0.1–6.67) | 0.8590 | 0 (0–NA) | 0.2888 | 0.55 (0.07–4.35) | 0.5363 |
| rs2114358 | MIR1206 | 0.85 (0.71–1.01) | 0.0600 | 0.8 (0.61–1.06) | 0.1145 | 0.81 (0.62–1.06) | 0.1267 | 1.04 (0.77–1.4) | 0.8092 | 0.73 (0.58–0.93) | 0.0093 |
| rs10505168 | MIR2053 | 0.97 (0.81–1.17) | 0.7663 | 0.93 (0.7–1.24) | 0.6348 | 0.88 (0.66–1.18) | 0.3960 | 0.83 (0.6–1.14) | 0.2486 | 1.07 (0.84–1.37) | 0.5666 |
| rs11259096 | MIR1265 | 1.11 (0.77–1.61) | 0.5721 | 1.36 (0.8–2.33) | 0.2728 | 0.87 (0.45–1.69) | 0.6764 | 1.48 (0.83–2.62) | 0.1975 | 0.99 (0.58–1.69) | 0.9816 |
| rs17091403 | MIR2110 | 0.81 (0.6–1.09) | 0.1505 | 0.79 (0.49–1.27) | 0.3074 | 0.87 (0.55–1.37) | 0.5374 | 0.67 (0.39–1.16) | 0.1345 | 0.85 (0.57–1.26) | 0.4017 |
| rs4919510 | MIR608 | 0.92 (0.74–1.15) | 0.4724 | 1.15 (0.83–1.58) | 0.4140 | 0.81 (0.56–1.16) | 0.2426 | 1.18 (0.83–1.69) | 0.3594 | 0.87 (0.64–1.17) | 0.3449 |
| rs12416605 | MIR938 | 0.95 (0.78–1.15) | 0.5733 | 0.96 (0.71–1.31) | 0.8152 | 0.7 (0.51–0.97) | 0.0281 | 0.88 (0.63–1.24) | 0.4604 | 1.07 (0.83–1.38) | 0.5896 |
| rs11020790 | MIR548L | 0.55 (0.21–1.42) | 0.1846 | 0.59 (0.14–2.53) | 0.4467 | 0.28 (0.04–2.09) | 0.1300 | 0 (0–NA) | 0.0493 | 0.6 (0.18–2) | 0.3731 |
| rs7311975 | MIR1178 | 0.95 (0.56–1.6) | 0.8504 | 1.33 (0.65–2.75) | 0.4514 | 0.86 (0.36–2.03) | 0.7197 | 1.2 (0.53–2.72) | 0.6743 | 0.68 (0.3–1.5) | 0.3144 |
| rs11614913 | MIR196A2 | 1.07 (0.9–1.27) | 0.4543 | 1.12 (0.85–1.46) | 0.4189 | 1.21 (0.92–1.59) | 0.1649 | 0.9 (0.67–1.21) | 0.4830 | 1.18 (0.93–1.48) | 0.1702 |
| rs2289030 | MIR492 | 0.79 (0.54–1.16) | 0.2166 | 0.6 (0.3–1.19) | 0.1192 | 0.98 (0.56–1.71) | 0.9371 | 0.62 (0.3–1.29) | 0.1739 | 1.11 (0.7–1.76) | 0.6630 |
| rs11844707 | MIR1185−2 | 0 (0–NA) | 0.3216 | 0 (0–NA) | 0.4983 | 0 (0–NA) | 0.6115 | 0 (0–NA) | 0.4749 | 0 (0–NA) | 0.5279 |
| rs6505162 | MIR423 | 0.96 (0.8–1.15) | 0.6746 | 0.81 (0.6–1.09) | 0.1627 | 1.06 (0.8–1.42) | 0.6662 | 1.04 (0.76–1.42) | 0.8249 | 0.88 (0.69–1.13) | 0.3195 |
| rs17759989 | MIR633 | 0.71 (0.41–1.25) | 0.2212 | 0.28 (0.07–1.16) | 0.0305a | 0.77 (0.33–1.8) | 0.5299 | 1.07 (0.45–s2.54) | 0.8757 | 0.46 (0.18–1.15) | 0.0632 |
| rs895819 | MIR27A | 0.96 (0.8–1.16) | 0.6801 | 1.08 (0.81–1.45) | 0.5919 | 0.92 (0.69–1.25) | 0.6048 | 1.02 (0.74–1.41) | 0.8912 | 0.91 (0.7–1.17) | 0.4577 |
| rs3746444 | MIR499A/B | 0.95 (0.77–1.18) | 0.6560 | 0.95 (0.68–1.32) | 0.7490 | 1.07 (0.77–1.48) | 0.6977 | 0.64 (0.42–0.98) | 0.0308 | 1.1 (0.83–1.46) | 0.5004 |
| rs4822739 | MIR548J | 0.96 (0.65–1.41) | 0.8297 | 1.57 (0.93–2.65) | 0.1063 | 0.81 (0.43–1.53) | 0.5027 | 1.3 (0.7–2.4) | 0.4172 | 0.77 (0.43–1.35) | 0.3401 |
| rs5965660 | MIR888 | 1.12 (0.96–1.32) | 0.1625 | 1.13 (0.88–1.45) | 0.3360 | 1.02 (0.77–1.35) | 0.8729 | 1.16 (0.89–1.51) | 0.2716 | 1.24 (1–1.54) | 0.0524 |
Significant associations are shown in bold.
This genetic association is not supported by the confidence interval, which is most likely due to the low frequency of the variant (see Table S1).
Analyses by other inheritance models (codominant, dominant, and recessive) indicated that the only association that was stronger with a model different from the log‐additive was that of rs3746444:T>C with the cardia phenotype, which was better explained by the dominant model (OR = 0.58, 95% CI = 0.36–0.94, p = 0.0210).
3.2. rs12416605:C>T affects the expression of MIR938 and several gene regulatory pathways
rs2114358 in MIR1206, and rs3746444:T>C in MIR499 have already been shown to affect the dosage of the corresponding miRNAs by altering either their biogenesis or stability (Kim, Prokunina‐Olsson, & Chanock, 2012; Torruella‐Loran et al., 2016). Conversely, no studies have been conducted to analyse the functional effect of rs12416605:C>T in MIR938. We therefore investigated if there were differences in miRNA expression or in gene regulation between both rs12416605:C>T MIR938 alleles that could explain their genetic association with GC.
To test the effect of rs12416605:C>T in MIR938 expression, we cloned both miRNA alleles and co‐transfected them in HeLa cells (after confirming that these cells had undetectable levels of endogenous MIR938) together with a control miRNA for normalization of transfection (Figure 1a). After measuring MIR938 expression by RT‐qPCR, we observed that the rs12416605 C allele was 1.49 times more expressed than the T allele (Student t test, p < 0.05) indicating that this variant in MIR938 may be affecting the final expression of the miRNA (Figure 1b).
Figure 1.
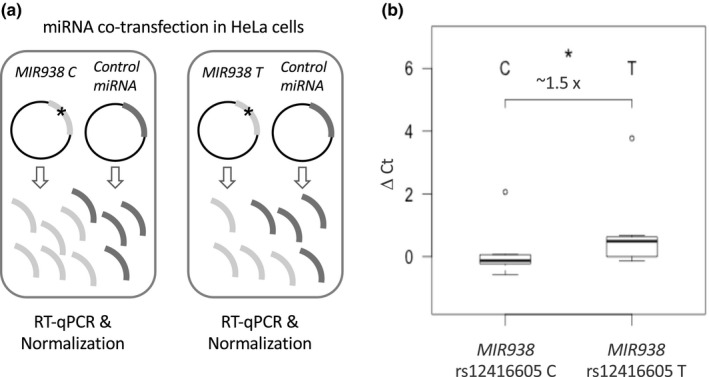
Effect of rs12416605:C>T on MIR938 expression. (a) Outline of the miRNA co‐transfection experiments in HeLa cells using the pmR‐ZsGreen1 vector (Clontech) containing either one of the two MIR938 rs12416605 alleles or a nonhuman control miRNA used for normalization of transfection. (b) Box‐plot showing differences in the expression levels (ΔCt), measured by qPCR for the C and T MIR938 rs12416605 alleles after co‐transfection experiments, related to the control reference miRNA. Each experiment was done in triplicate and four independent experiments were performed. Data reported here are the means ± SEM of all experiments performed. A significant reduction between the expression of the C and T alleles was found (p < 0.05, Student's t test)
In addition to differences in the expression levels between the rs12416605 C and T alleles, this nucleotide substitution could also change the spectrum of miRNA target genes; therefore, we investigated which genes could be regulated by each miRNA allelic variant through miRNA overexpression experiments and transcriptome analyses. After transfection of both miRNA alleles in HeLa cells, we selected those transcripts statistically significant up‐ or downregulated with respect to the control miRNA, using a relaxed cutoff value (p < 0.05, over a 1.2 FC). The MIR938 C allele deregulated 2,045 transcripts, whereas the T allele deregulated 1,825 transcripts. Among those, 829 were deregulated by both the alleles (this represents 40.5% and 45.5% of the transcripts deregulated by the C and T allele, respectively), indicating that about 50% of the transcripts were exclusively regulated by each rs12416605:C>T allele. Next, we used Ingenuity Pathway Analysis software to analyse the functions associated with the genes deregulated after overexpression of each MIR938 allelic variant. Both the alleles presented cancer as their top disease associated with their deregulated genes. Interestingly, variant C but not variant T presented gastrointestinal disease among the top five diseases to which their deregulated genes were associated (Table S2). According to cancer as the top associated disease and among the top 10 upregulated and downregulated genes, there were several genes that are implicated in the etiology of gastric cancer, such as the protein tyrosine phosphatase nonreceptor type 11 (PTPN11) (Liu et al., 2015) or the Fibroblast Growth Factor 1 (FGF1) (Zhuo et al., 2016); (Table 4). Remarkably, most of the top 10 downregulated transcripts (8 out of 10) but none of the top 10 upregulated transcripts were common deregulated genes between the two rs12416605:C>T alleles (Table 4).
Table 4.
Top 10 deregulated genes by MIR938 rs12416605:C>T alleles compared to the empty vector control and ordered from the highest to the lowest fold change (FC)
| MIR938 C | MIR938 T | |||||
|---|---|---|---|---|---|---|
| Gene | FC | p values | Gene | FC | p values | |
| Top 10 up‐regulated genes | TSPAN19 | 4.44 | 3.36E‐02 | HPGDS | 4.39 | 1.30E‐03 |
| C18orf34 | 3.74 | 5.17E‐03s | TRPA1 | 3.60 | 8.12E‐04 | |
| SLC4A5 | 3.56 | 4.79E‐03 | SORBS1 | 3.51 | 8.07E‐04 | |
| CHD6 | 3.33 | 4.65E‐02 | PEX5L‐AS2 | 3.39 | 4.82E‐06 | |
| BC015129 | 3.14 | 5.30E‐04 | OR1D5 | 3.31 | 2.58E‐03 | |
| IL34 | 3.07 | 9.48E‐04 | SLC23A2 | 3.26 | 1.46E‐03 | |
| OR5B2 | 2.93 | 3.85E‐03 | FLJ21408 | 3.16 | 2.03E‐07 | |
| GRIP1 | 2.77 | 3.19E‐03 | PRR15L | 3.09 | 3.36E‐03 | |
| SRRM4 | 2.73 | 2.92E‐02 | RBMS3 | 2.99 | 2.02E‐03 | |
| ABCA8 | 2.71 | 2.00E‐02 | RNF157 | 2.95 | 1.60E‐05 | |
| Top 10 down‐regulated genes | NKX2‐8 | −6.65 | 1.72E‐03 | NKX2‐8 | −5.95 | 3.02E‐03 |
| ZNF730 | −5.10 | 2.00E‐03 | ZNF730 | −5.00 | 2.22E‐03 | |
| THPO | −4.71 | 1.45E‐03 | THPO | −4.68 | 1.51E‐03 | |
| MFSD4 | −4.27 | 3.79E‐04 | DNAJC3 | −3.93 | 2.53E‐03 | |
| PTPN11 | −4.13 | 8.18E‐03 | HNF1A | −3.85 | 7.25E‐03 | |
| DPCR1 | −3.99 | 1.13E‐02 | MFSD4 | −3.79 | 9.88E‐04 | |
| HNF1A | −3.92 | 6.52E‐03 | FGF1 | −3.78 | 3.40E‐02 | |
| MDM1 | −3.67 | 1.02E‐02 | PTPN11 | −3.74 | 1.35E‐02 | |
| C1orf210 | −3.66 | 1.91E‐03 | DPCR1 | −3.72 | 1.58E‐02 | |
| CCDC103 | −3.62 | 2.32E‐03 | C1orf210 | −3.63 | 2.00E‐03 | |
In grey, common deregulated transcripts to both the alleles.
3.3. The chemokine CXCL12 is regulated by MIR938 in an allele‐dependent manner
We next investigated if there were cancer‐related genes differentially regulated by the MIR938 alleles that could be exclusive direct target genes of only one of the rs12416605:C>T allelic variants of MIR938. We used PITA algorithm to predict MIR938 C and MIR938 T targets among the whole set of differentially regulated transcripts that were related to cancer according to DAVIDGO (Table S3). Three GC‐related genes were predicted to be exclusively regulated by one of the MIR938 allelic variants: the chemokine receptor 5 CCR5, the Leptin receptor LEPR, and the chemokine CXCL12 (Table 5). Among these genes, the one that presented the highest difference in the PITA prediction between alleles was the CXCL12 chemokine, which showed a predicted target site for the rs12416605 C allele (ΔΔG = −9.08) whereas any good prediction was found for the T allele (ΔΔG = −0.93) (Table 5, Figure 2a). These differences between MIR938 alleles and the fact that CXCL12 is reported to be involved in GC through its participation in cell migration and invasion (Cheng et al., 2017; Izumi et al., 2016; Rubie, Kauffels, Kölsch, Glanemann, & Justinger, 2016) led us to investigate the possible interaction between CXCL12 and the rs12416605:C>T alleles by means of a dual‐luciferase assay in HeLa cells. A luciferase reporter pGL4.13 construct, either empty or carrying the 3′UTR of the CXCL12, was co‐transfected with the corresponding miRNA mimic: MIR938 T, MIR938 C, or a control miRNA. As shown in Figure 2b, a statistically significant reduction in the luciferase activity was observed for the rs12416605 C allele of MIR938 when co‐transfected with the 3′ UTR of the CXCL12 compared to both, the control miRNA and the T allelic variant (p < 0.01, Student's t test). The observed reduction in the luciferase activity would be compatible with a 40% repression of CXCL12 exclusively by the rs12416605 C allele of MIR938, suggesting that differences in the regulation of CXCL12 between both alleles might be underlying the genetic association of GC with the rs12416605 SNV in MIR938 and the protective effect found for the T allele against diffuse GC.
Table 5.
Cancer‐related genes predicted to be differentially regulated by MIR938 rs12416605:C>T alleles according to PITA target site predictions
| Gene | Alleles |
Best Pita Score ΔΔG |
|---|---|---|
| CXCL12 (Chemokine C‐X‐C motif ligand 12) | C | −9.08 |
| T | −0.93 | |
| CFLAR (CASP8 and FADD‐Like Apoptosis Regulator) | C | −15.51 |
| T | −11.4 | |
| CCR5 (C‐C chemokine receptor 5) | C | −5.79 |
| T | −12.81 | |
| LEPR (Leptin receptor) | C | −4.77 |
| T | −11.81 |
The best Pita score (energy for microRNA‐target interaction, ΔΔG) for each allele is shown. Relevant Pita score scores are indicated in bold face.
Figure 2.
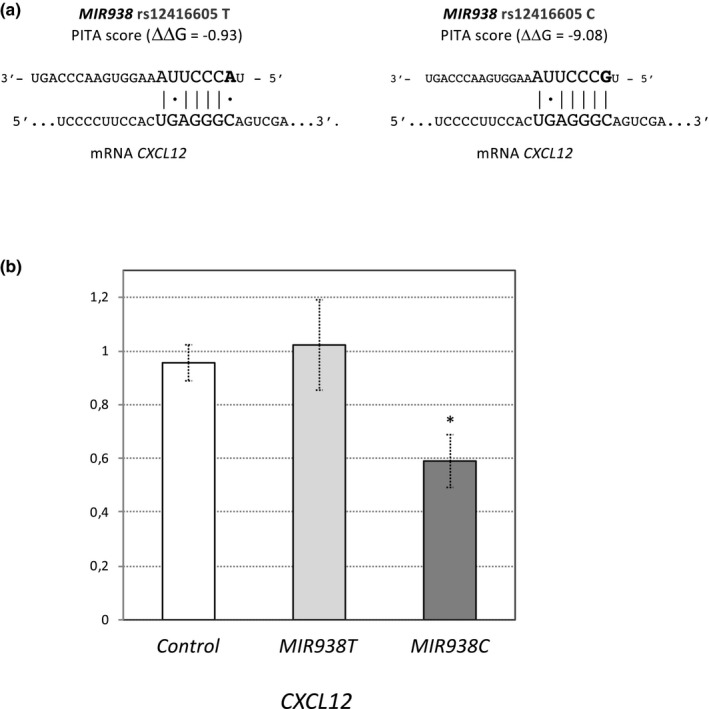
Allele‐specific regulation of the chemokine CXCL12 by MIR938. (a) Sequence alignment between the MIR938 rs12416605 C and T alleles and the CXCL12 mRNA showing the PITA predicted target site and ΔΔG PITA scores for both alleles on this site. (b) Results of the luciferase‐reporter assay testing the interaction between MIR938 rs12416605 C and T alleles and the 3′UTR of the CXCL12 in HeLa cells. Ratios of the Firefly and Renilla luciferase luminescence are presented after normalization to the empty plasmid pGL4.13. Each experiment sswas done in triplicate and six independent experiments were performed. Data reported here are the means ± SEM of all experiments performed. A significant reduction in the luciferase activity between both the alleles and between the C allele and the control miRNA was found (*, p < 0.01, Student's t‐test)
4. DISCUSSION
Even though changes in the expression of miRNAs have been largely associated with GC and impaired miRNA expression may be related to genetic variation in their sequences, still there is lack of evidence explaining the real contribution of miRNA genetic variants to GC pathology. In the present study, we evaluated several putative allelic variants in functional miRNA regions as candidate susceptibility factors to GC by means of association studies and demonstrated a possible causative effect for at least one of these variants using a functional approach.
We identified three common genetic variants in miRNA genes that associated with GC. One of them, rs3746444:T>C in MIR499, has largely been reported to be associated with cancer and has further been suggested as having a role in the GC etiology (Ahn et al., 2013; Chen, Yang, Chaugai, Wang, & Wang, 2014). In the present work, we identified the C allele of this SNV as a protective factor against the cardia GC subtype under the log‐additive and dominant models, which is partially in agreement with a recent meta‐analysis study in which rs3746444:T>C was reported to be associated with GC in Asian but not European populations (Chen et al., 2014). Even though this last study did not find association in Europeans, another study supports our finding by reporting association of this SNV with another cancer type in populations of European origin (Nikolić et al., 2015). Furthermore, we have recently shown that the two alleles of rs3746444:T>C may influence the processing and expression of MIR499A and MIR499B, which ultimately may be affecting the regulation of their target genes in a dosage and allele‐dependent manner (Torruella‐Loran et al., 2016). More importantly, we showed that the T allele of MIR499A caused a significant repression of the cadherin CDH1 and the cell adhesion molecule CLH1, which could be associated with a malignant behavior of the T allele and, consequently, with the protective effect found for the C allele against GC.
Another of the SNVs that we found associated with GC was rs2114358:T>C, in the precursor molecule of MIR1206. Even though this miRNA is in the cancer risk loci 8q24.2, this is the first report of a genetic association of MIR1206 with cancer. Significantly, a previous report demonstrated an effect of rs2114358:T>C in the biogenesis of the mature miRNA forms (Kim, Prokunina‐Olsson, et al., 2012), and several genes involved in GC such us FGF2 and TP53INP1 appear as targets of MIR1206 in miRTarBase. Given the location of this variant in the precursor region outside the mature miRNA molecule, the functional effect of rs2114358:T>C might be through allele‐specific variation of miRNA expression rather than alteration in the spectrum of target genes.
The third SNV associated with GC was rs12416605:C>T in the seed of MIR938. Interestingly, a previous study reported the association of GC with another SNV (rs2505901) in the MIR938 primary precursor molecule as well as another variant (rs2275913) in IL17A, a MIR938‐targeted gene (Arisawa et al., 2012). To investigate the possible contribution of rs12416605:C>T to GC, we analyzed the effect of both the alleles in the expression levels of MIR938. Remarkably, statistically significant differences were observed between the expression levels of each miRNA variant: The C variant of MIR938 was 1.49 times more expressed than the T variant. Besides the alteration in the expression levels of MIR938, this MIR938 variant was also predicted to regulate several gastric cancer‐related genes in an allele‐dependent manner. For instance, the CASP8 and FADD‐Like Apoptosis Regulator (CFLAR) gene has strong ΔΔG PITA scores, particularly for the C rs12416605 allele (−15.51 and −11.4 for the C and T alleles, respectively, Table 5). CFLAR is a gene that encodes the FLICE‐like inhibitory protein (c‐FLIP) that acts as an inhibitor of the Fas‐mediated apoptosis and helps tumor cells to escape from TRAIL‐mediated apoptosis, hence promoting metastasis and tumor progression (Olsson & Zhivotovsky, 2011; Zhang et al., 2004; Zhou et al., 2004). Several SNVs in the CFLAR gene have been associated with GC in Chinese populations (Hyland et al., 2004) reinforcing a role for CFLAR in GC. Moreover, CFLAR mRNA and its protein were found highly expressed in gastric adenocarcinomas compared with normal gastric mucosa tissue in patients of Chinese origin stressing the importance of CFLAR expression regulation in the pathophysiology of GC (Zhou et al., 2004), which may be mediated by MIR938. In addition to CFLAR, which might be regulated by both allelic variants of MIR938, we found at least three other genes that were predicted as exclusive targets of only one of the MIR938 variants and that are known to play important roles in GC (Table 5). For instance, the C‐C chemokine receptor 5 (CCR5) and the Leptin receptor (LEPR) are related to GC (Chang, Du, Zhao, Ma, & Cao, 2014; Kim, Chin, et al., 2012; Shi et al., 2014), and were predicted to be regulated exclusively by the T allele. Leptin and its receptor are known to play a role in Helicobacter pylori infection (Azuma et al., 2001), and a polymorphism in LEPR has been associated with GC risk in Korean populations (Kim, Chin, et al., 2012). Similarly, CCR5 was predicted as targeted solely by the T MIR938 allele. In this context, the expression of CCR5 has been found to favor metastasis in GC patients (Cao et al., 2011) and thereby, repression of the CCL5/CCR5 axis has been suggested as a possible therapeutic strategy against GC progression (Aldinucci & Casagrande, 2018), which would be compatible with a protective effect of the T MIR938 allele mediated by its putative repression of CCR5. Finally, the chemokine CXCL12 was predicted to be regulated exclusively by the MIR938 C allelic variant with the best ΔΔG PITA score of −9.08. CXCL12 has been shown to have a critical role in GC promoting cell migration and invasion, although a controversial participation for CXCL12 in metastasis has been reported: several authors show that absence of CXCL12 expression might lead to GC metastasis whereas some others suggest that CXCL12 overexpression promotes cell invasion (Cheng et al., 2017; Izumi et al., 2016; Rubie et al., 2016; Zhi et al., 2012). In this context, the CXCL12/CXCR4 axis has been proposed as a promising therapeutic target for advanced GC (Xue, Mao, Ren, & Chu, 2017). The here observed repression of about 40% of this chemokine by the C but not the T allele, together with the increased expression of MIR938 related to the rs12416605 C allele, might cause an amplified downregulation of CXCL12 in C carriers that could be associated with a promotion of GC metastasis. This would agree with our finding of the T allele as a protective factor and with the finding of a higher frequency of the T allele in European compared with Asian populations (29% vs. 2%, 1,000 genomes data), which is reflected by a high‐global fixation index (Fst) for this SNV (0.2635 for the global Fst, Table 2) and in agreement with Asian populations having a higher predisposition to GC.
In conclusion, we replicated a previously described association of MIR499 rs3746444:T>C with GC in European populations and identified two new unreported associations of miRNA SNVs with GC: rs12416605:T>C in MIR938 and rs2114358:T>C in MIR1206. We showed a significant difference in MIR938 expression between rs12416605 alleles and suggested that the protective effect of the T MIR938 allele against diffuse GC could be mediated by the lack of repression of the chemokine CXCL12. These findings strongly suggest a role for CXCL12 posttranscriptional regulation in the development of GC and as a candidate regulatory pathway for anticancer therapies.
Supporting information
ACKNOWLEDGMENTS
This work was funded by the “Ministerio de Educación, Gobierno de Chile, Comisión Nacional de Investigación Científica y Tecnológica (CONICYT) ‐ Fondo de Fomento al Desarrollo Científico y Tecnológico (FONDECYT regular)” [grant Nº 1170446]; the Spanish “Ministerio de Ciencia e Innovación” [grant BFU2010‐18477]; “the Spanish Ministerio de Economía y Competitividad ‐ Instituto de Salud Carlos III”; the European Regional Development Funds (ERDF/FEDER) ‘A way to build Europe' [grants PI070130 and PI12/01187] and “LaCaixa” Foundation [grant BM06‐130‐0]. We also thank CERCA Program / Generalitat de Catalunya for their institutional support. MRV and DZC were employed by the Fondecyt regular grant 1170446.
Torruella‐Loran I, Ramirez Viña MK, Zapata‐Contreras D, et al; on behalf of the EPIC gastric cancer working group . rs12416605:C>T in MIR938 associates with gastric cancer through affecting the regulation of the CXCL12 chemokine gene. Mol Genet Genomic Med. 2019;7:e832 10.1002/mgg3.832
REFERENCES
- Ahn, D. H. , Rah, H. , Choi, Y. K. , Jeon, Y. J. , Min, K. T. , Kwack, K. , … Kim, N. K. (2013). Association of the miR‐146aC>G, miR‐149T>C, miR‐196a2T>C, and miR‐499A>G polymorphisms with gastric cancer risk and survival in the Korean population. Molecular Carcinogenesis, 52(Suppl 1), E39–E51. 10.1002/mc.21962 [DOI] [PubMed] [Google Scholar]
- Aldinucci, D. , & Casagrande, N. (2018). Inhibition of the CCL5/CCR2 Axis against the progression of gastric cancer. International Journal of Molecular Sciences, 19(5), E1477 10.3390/ijms19051477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arisawa, T. , Tahara, T. , Shiroeda, H. , Matsue, Y. , Minato, T. , Nomura, T. , … Shibata, T. (2012). Genetic polymorphisms of IL17A and pri‐microRNA‐938, targeting IL17A 3’‐UTR, influence susceptibility to gastric cancer. Human Immunology, 73(7), 747–752. 10.1016/j.humimm.2012.04.011 [DOI] [PubMed] [Google Scholar]
- Azuma, T. , Suto, H. , Ito, Y. , Ohtani, M. , Dojo, M. , Kuriyama, M. , & Kato, T. (2001). Gastric leptin and Helicobacter pylori infection. Gut, 49(3), 324–329. 10.1136/gut.49.3.324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balcells, I. , Cirera, S. , & Busk, P. K. (2011). Specific and sensitive quantitative RT‐PCR of miRNAs with DNA primers. BMC Biotechnology, 11, 70 10.1186/1472-6750-11-70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel, D. P. (2009). MicroRNAs: Target recognition and regulatory functions. Cell, 136(2), 215–233. 10.1016/j.cell.2009.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, Z. , Xu, X. , Luo, X. , Li, L. I. , Huang, B. , Li, X. , … Gong, J. (2011). Role of RANTES and its receptor in gastric cancer metastasis. Huazhong University of Science and Technology, 31(3), 342–347. 10.1007/s11596-011-0378-3 [DOI] [PubMed] [Google Scholar]
- Carneiro, F. , Moutinho, C. , Pera, G. , Caldas, C. , Fenger, C. , Offerhaus, J. , … Gonzalez, C. A. (2007). Pathology findings and validation of gastric and esophageal cancer cases in a European cohort (EPIC/EURGAST). Scandinavian Journal of Gastroenterology, 42(5), 618–627. 10.1080/00365520601101641 [DOI] [PubMed] [Google Scholar]
- Chang, W. J. , Du, Y. , Zhao, X. , Ma, L. Y. , & Cao, G. W. (2014). Inflammation‐related factors predicting prognosis of gastric cancer. World Journal of Gastroenterology, 20(16), 4586–4596. 10.3748/wjg.v20.i16.4586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, C. , Yang, S. , Chaugai, S. , Wang, Y. , & Wang, D. W. (2014). Meta‐analysis of Hsa‐mir‐499 polymorphism (rs3746444) for cancer risk: Evidence from 31 case‐control studies. BMC Medical Genetics, 15, 1–11. 10.1186/s12881-014-0126-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, Y. U. , Qu, J. , Che, X. , Xu, L. , Song, N. A. , Ma, Y. , … Liu, Y. (2017). CXCL12/SDF‐1α induces migration via SRC‐mediated CXCR11‐EGFR cross‐talk in gastric cancer cells. Oncology Letters, 14(2), 2103–2110. 10.3892/ol.2017.6389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correa, P. (1992). Human gastric carcinogenesis: A multistep and multifactorial process–First American Cancer Society Award Lecture on Cancer Epidemiology and Prevention. Cancer Research, 52(24), 6735–6740. [PubMed] [Google Scholar]
- Crew, K. D. , & Neugut, A. I. (2006). Epidemiology of gastric cancer. World Journal of Gastroenterology, 12(3), 354–362. 10.3748/wjg.v12.i3.354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinosa‐Parrilla, Y. , Muñoz, X. , Bonet, C. , Garcia, N. , Venceslá, A. , Yiannakouris, N. , … Sala, N. (2014). Genetic association of gastric cancer with miRNA clusters including the cancer‐related genes MIR29, MIR25, MIR93 and MIR106: Results from the EPIC‐EURGAST study. International Journal of Cancer, 135(9), 2065–2076. 10.1002/ijc.28850 [DOI] [PubMed] [Google Scholar]
- Esquela‐Kerscher, A. , & Slack, F. J. (2006). Oncomirs—microRNAs with a role in cancer. Nature Reviews Cancer, 6(4), 259–269. 10.1038/nrc1840 [DOI] [PubMed] [Google Scholar]
- Ferlay, J. , Soerjomataram, I. , Dikshit, R. , Eser, S. , Mathers, C. , Rebelo, M. , … Bray, F. (2015). Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. International Journal of Cancer, 136(5), E359–E386. 10.1002/ijc.29210 [DOI] [PubMed] [Google Scholar]
- Filipowicz, W. , Bhattacharyya, S. N. , & Sonenberg, N. (2008). Mechanisms of post‐transcriptional regulation by microRNAs: Are the answers in sight? Nature Reviews Genetics, 9(2), 102–114. 10.1038/nrg2290 [DOI] [PubMed] [Google Scholar]
- Fletcher, O. , & Houlston, R. S. (2010). Architecture of inherited susceptibility to common cancer. Nature Reviews Cancer, 10(5), 353–361. 10.1038/nrc2840 [DOI] [PubMed] [Google Scholar]
- Friedman, R. C. , Farh, K. K. , Burge, C. B. , & Bartel, D. P. (2009). Most mammalian mRNAs are conserved targets of microRNAs. Genome Research, 19(1), 92–105. 10.1101/gr.082701.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego, A. , Melé, M. , Balcells, I. , García‐Ramallo, E. , Torruella‐Loran, I. , Fernández‐Bellon, H. , … Espinosa‐Parrilla, Y. (2016). Functional implications of human‐specific changes in great ape microRNAs. PLoS ONE, 11(4), e0154194 10.1371/journal.pone.0154194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- GLOBOCAN . (2018). IARC global cancer observatory web site. Retrieved from http://gco.iarc.fr/ [Google Scholar]
- González, C. A. , Sala, N. , & Rokkas, T. (2013). Gastric cancer: Epidemiologic aspects. Helicobacter, 18(Suppl 1), 34–38. 10.1111/hel.12082 [DOI] [PubMed] [Google Scholar]
- Hyland, P. L. , Lin, S.‐W. , Hu, N. , Zhang, H. , Wang, L. , Su, H. , … Taylor, P. R. (2004). Genetic variants in fas signaling pathway genes and risk of gastric cancer. International Journal of Cancer, 134(4), 822–831. 10.1002/ijc.28415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumi, D. , Ishimoto, T. , Miyake, K. , Sugihara, H. , Eto, K. , Sawayama, H. , … Baba, H. (2016). CXCL12/CXCR24 activation by cancer‐associated fibroblasts promotes integrin β1 clustering and invasiveness in gastric cancer. International Journal of Cancer, 138(5), 1207–1219. 10.1002/ijc.29864 [DOI] [PubMed] [Google Scholar]
- Katada, T. , Ishiguro, H. , Kuwabara, Y. , Kimura, M. , Mitui, A. , Mori, Y. , … Fujii, Y. (2009). microRNA expresion profile in undifferentiated gastric cancer. International Journal of Oncology, 34(2), 537–542. [PubMed] [Google Scholar]
- Kertesz, M. , Iovino, N. , Unnerstall, U. , Gaul, U. , & Segal, E. (2007). The role of site accessibility in microRNA target recognition. Nature Genetics, 39(10), 1278–1284. 10.1038/ng2135 [DOI] [PubMed] [Google Scholar]
- Kim, E. Y. , Chin, H. M. , Park, S. M. , Jeon, H. M. , Chung, W. C. , Paik, C. N. , & Jun, K. H. (2012). Susceptibility of gastric cancer according to leptin and leptin receptor gene polymorphisms in Korea. Journal of the Korean Surgical Society, 83(1), 7–13. 10.4174/jkss.2012.83.1.7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, H. K. , Prokunina‐Olsson, L. , & Chanock, S. J. (2012). Common genetic variants in miR‐1206 (8q24.2) and miR‐612 (11q13.3) affect biogenesis of mature miRNA forms. PLoS ONE, 7(10), e47454 10.1371/journal.pone.0047454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloosterman, W. P. , & Plasterk, R. H. (2006). The diverse functions of microRNAs in animal development and disease. Developmental Cell, 11(4), 441–450. 10.1016/j.devcel.2006.09.009 [DOI] [PubMed] [Google Scholar]
- Lewis, B. P. , Shih, I. , Jones‐Rhoades, M. W. , Bartel, D. P. , & Burge, C. B. (2003). Prediction of mammalian microRNA targets. Cell, 115(7), 787–798. 10.1016/S0092-8674(03)01018-3 [DOI] [PubMed] [Google Scholar]
- Liu, N. , Zhang, J. , Sun, S. , Yang, L. , Zhou, Z. , Sun, Q. , & Niu, J. (2015). Expression and clinical significance of fibroblast growth factor 1 in gastric adenocarcinoma. OncoTargets and Therapy, 8, 615–621. 10.2147/OTT.S79204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh, M. , Koh, K. X. , Yeo, B. H. , Song, C. M. , Chia, K. S. , Zhu, F. , … Soong, R. (2009). Meta‐analysis of genetic polymorphisms and gastric cancer risk: Variability in associations according to race. European Journal of Cancer, 45(14), 2562–2568. 10.1016/j.ejca.2009.03.017 [DOI] [PubMed] [Google Scholar]
- Lopez‐Valenzuela, M. , Ramírez, O. , Rosas, A. , García‐Vargas, S. , de la Rasilla, M. , Lalueza‐Fox, C. , & Espinosa‐Parrilla, Y. (2012). An ancestral miR‐1304 allele present in neanderthals regulates genes involved in enamel formation and could explain dental differences with modern humans. Molecular Biology and Evolution, 29(7), 1797–1806. 10.1093/molbev/mss023 [DOI] [PubMed] [Google Scholar]
- Lu, J. , Getz, G. , Miska, E. A. , Alvarez‐Saavedra, E. , Lamb, J. , Peck, D. , … Golub, T. R. (2005). MicroRNA expression profiles classify human cancers. Nature, 435(7043), 834–838. 10.1038/nature03702 [DOI] [PubMed] [Google Scholar]
- McLean, M. H. , & El‐Omar, E. M. (2014). Genetics of gastric cancer. Nature Reviews Gastroenterology & Hepatology, 11(11), 664–674. 10.1038/nrgastro.2014.143 [DOI] [PubMed] [Google Scholar]
- Nikolić, Z. , Savić Pavićević, D. , Vučić, N. , Cidilko, S. , Filipović, N. , Cerović, S. , … Brajušković, G. (2015). Assessment of association between genetic variants in microRNA genes hsa‐miR‐499, hsa‐miR‐196a2 and hsa‐miR‐27a and prostate cancer risk in Serbian population. Experimental and Molecular Pathology, 99(1), 145–150. 10.1016/j.yexmp.2015.06.009 [DOI] [PubMed] [Google Scholar]
- Olsson, M. , & Zhivotovsky, B. (2011). Caspases and cancer. Cell Death & Differentiation, 18(9), 1441–1449. 10.1038/cdd.2011.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, S. , Kuang, Z. , Sheng, C. , Zhang, Y. , Xu, H. , & Cheng, Q. (2010). Association of MicroRNA‐196a‐2 gene polymorphism with gastric cancer risk in a Chinese population. Digestive Diseases and Sciences, 55(8), 2288–2293. 10.1007/s10620-009-1007-x [DOI] [PubMed] [Google Scholar]
- Riboli, E. , Hunt, K. J. , Slimani, N. , Ferrari, P. , Norat, T. , Fahey, M. , … Saracci, R. (2002). European Prospective Investigation into Cancer and Nutrition (EPIC): Study populations and data collection. Public Health Nutrition, 5(6B), 1113–1124. 10.1079/PHN2002394 [DOI] [PubMed] [Google Scholar]
- Rubie, C. , Kauffels, A. , Kölsch, K. , Glanemann, M. , & Justinger, C. (2016). CXCL12/CXCR40 display an inverse mRNA expression profile in gastric carcinoma that correlates with tumor progression. Oncology Letters, 11(1), 360–364. 10.3892/ol.2015.3850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeki, N. , Ono, H. , Sakamoto, H. , & Yoshida, T. (2013). Genetic factors related to gastric cancer susceptibility identified using a genome‐wide association study. Cancer Science, 104(1), 1–8. 10.1111/cas.12042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sala, N. , Muñoz, X. , Travier, N. , Agudo, A. , Duell, E. J. , Moreno, V. , … González, C. A. (2012). Prostate stem cell antigen gene is associated with diffuse and intestinal gastric cancer in Caucasians: Results from the EPIC‐Eurgast study. International Journal of Cancer, 130(10), 2417–2427. 10.1002/ijc.26243 [DOI] [PubMed] [Google Scholar]
- Shen, J. , Stass, S. , & Jiang, F. (2013). MicroRNAs as potential biomarkers in human solid tumors. Cancer Letters, 329(2), 125–136. 10.1016/j.canlet.2012.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, H. , Shu, H. , Huang, C. , Gong, J. , Yang, Y. , Liu, R. , … Liu, P. (2014). Association of LEPR K109R polymorphisms with cancer risk: A systematic review and pooled analysis. Journal of BUON, 19(3), 847–854. [PubMed] [Google Scholar]
- Torruella‐Loran, I. , Laayouni, H. , Dobon, B. , Gallego, A. , Balcells, I. , Garcia‐Ramallo, E. , & Espinosa‐Parrilla, Y. (2016). MicroRNA genetic variation: From population analysis to functional implications of three allele variants associated with cancer. Human Mutation, 37(10), 1060–1073. 10.1002/humu.23045 [DOI] [PubMed] [Google Scholar]
- Xia, Z. G. , Yin, H. F. , Long, Y. , Cheng, L. , Yu, L. J. , Guo, W. J. , … Wei, Y. (2016). Genetic variant of miR‐146a rs2910164 C>G and gastric cancer susceptibility. Oncotarget, 7(23), 34316–34321. 10.18632/oncotarget.8814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue, L. J. , Mao, X. B. , Ren, L. L. , & Chu, X. Y. (2017). Inhibition of CXCL12/CXCR47 axis as a potential targeted therapy of advanced gastric carcinoma. Cancer Medicine, 6(6), 1424–1436. 10.1002/cam4.1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Q. , Jie, Z. , Ye, S. , Li, Z. , Han, Z. , Wu, J. , … Jiang, Y. (2014). Genetic variations in miR‐27a gene decrease mature miR‐27a level and reduce gastric cancer susceptibility. Oncogene, 33(2), 193–202. 10.1038/onc.2012.569 [DOI] [PubMed] [Google Scholar]
- Zhang, X. , Jin, T. , Yang, H. , DeWolf, W. C. , Khosravi‐Far, R. , & Olumi, A. F. (2004). Persistent c‐FLIP ( L ) expression is necessary and sufficient to maintain resistance to tumor necrosis factor‐related apoptosis‐inducing ligand–mediated apoptosis in prostate cancer. Cancer Research, 64(19), 7086–7091. 10.1158/0008-5472.CAN-04-1498 [DOI] [PubMed] [Google Scholar]
- Zhi, Y. , Chen, J. , Zhang, S. , Zhang, S. , Chang, X. , Ma, J. , & Dai, D. (2012). Down‐regulation of CXCL12 by DNA hypermethylation and its involvement in gastric cancer metastatic progression. Digestive Diseases and Sciences, 57(3), 650–659. 10.1007/s10620-011-1922-5 [DOI] [PubMed] [Google Scholar]
- Zhou, X. D. , Yu, J. P. , Liu, J. , Luo, H. S. , Chen, H. X. , & Yu, H. G. (2004). Overexpression of cellular FLICE‐inhibitory protein (FLIP) in gastric adenocarcinoma. Clinical Science, 106(4), 397–405. 10.1042/CS20030238 [DOI] [PubMed] [Google Scholar]
- Zhou, X. , Zhu, W. , Li, H. , Wen, W. , Cheng, W. , Wang, F. , … Liu, P. (2015). Diagnostic value of a plasma microRNA signature in gastric cancer: A microRNA expression analysis. Scientific Reports, 5(22), 11251 10.1038/srep11251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuo, C. , Shao, M. , Chen, C. , Lin, C. , Jiang, D. , Chen, G. , … Lin, X. (2016). Chemotherapy effectiveness and prognosis of gastric cancer influenced by PTPN11 polymorphisms. Cellular Physiology and Biochemistry, 39(4), 1537–1552. 10.1159/000447856 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


