Antimicrobial resistance (AMR) represents an existential threat to the function of modern medicine. Genomics and machine learning methods are being increasingly used to analyze and predict AMR. This type of surveillance is very important to try to reduce the impact of AMR. Machine learning models are typically trained using genomic data, but the aspects of the genomes that they use to make predictions are rarely analyzed. In this work, we showed how, by using different types of machine learning models and performing this analysis, it is possible to identify the key genes underlying AMR in nontyphoidal Salmonella (NTS). NTS is among the leading cause of foodborne illness globally; however, AMR in NTS has not been heavily studied within the food chain itself. Therefore, in this work we performed a broad-scale analysis of the AMR in NTS isolates from commercial chicken farms and identified some priority AMR genes for surveillance.
KEYWORDS: AMR prediction, Salmonella, antimicrobial resistance, food chain, genomics, machine learning
ABSTRACT
Nontyphoidal Salmonella (NTS) is a leading global cause of bacterial foodborne morbidity and mortality. Our ability to treat severe NTS infections has been impaired by increasing antimicrobial resistance (AMR). To understand and mitigate the global health crisis AMR represents, we need to link the observed resistance phenotypes with their underlying genomic mechanisms. Broiler chickens represent a key reservoir and vector for NTS infections, but isolates from this setting have been characterized in only very low numbers relative to clinical isolates. In this study, we sequenced and assembled 97 genomes encompassing 7 serotypes isolated from broiler chicken in farms in British Columbia between 2005 and 2008. Through application of machine learning (ML) models to predict the observed AMR phenotype from this genomic data, we were able to generate highly (0.92 to 0.99) precise logistic regression models using known AMR gene annotations as features for 7 antibiotics (amoxicillin-clavulanic acid, ampicillin, cefoxitin, ceftiofur, ceftriaxone, streptomycin, and tetracycline). Similarly, we also trained “reference-free” k-mer-based set-covering machine phenotypic prediction models (0.91 to 1.0 precision) for these antibiotics. By combining the inferred k-mers and logistic regression weights, we identified the primary drivers of AMR for the 7 studied antibiotics in these isolates. With our research representing one of the largest studies of a diverse set of NTS isolates from broiler chicken, we can thus confirm that the AmpC-like CMY-2 β-lactamase is a primary driver of β-lactam resistance and that the phosphotransferases APH(6)-Id and APH(3″-Ib) are the principal drivers of streptomycin resistance in this important ecosystem.
IMPORTANCE Antimicrobial resistance (AMR) represents an existential threat to the function of modern medicine. Genomics and machine learning methods are being increasingly used to analyze and predict AMR. This type of surveillance is very important to try to reduce the impact of AMR. Machine learning models are typically trained using genomic data, but the aspects of the genomes that they use to make predictions are rarely analyzed. In this work, we showed how, by using different types of machine learning models and performing this analysis, it is possible to identify the key genes underlying AMR in nontyphoidal Salmonella (NTS). NTS is among the leading cause of foodborne illness globally; however, AMR in NTS has not been heavily studied within the food chain itself. Therefore, in this work we performed a broad-scale analysis of the AMR in NTS isolates from commercial chicken farms and identified some priority AMR genes for surveillance.
INTRODUCTION
Genomic methods are being increasingly established as key tools in rapid continuous surveillance, tracking, and control strategy development for infectious diseases (1). They are critical to our ability to study the evolution and spread of antimicrobial resistance (AMR), especially as we adopt a broader One Health (2) approach that integrates clinical, food production, and environmental settings. AMR is a current and growing global health crisis with soaring levels of observed multidrug resistance in a broad range of pathogens (3) combined with record low levels of novel drug discovery (4). There is a global consensus that AMR poses a severe and growing threat to human and animal health (3).
Currently, phenotypic antibiotic susceptibility testing (AST) is the principal method for the identification of AMR in treatment protocol determination and surveillance programs. Unfortunately, AST is highly variable between laboratories and can take days to weeks longer than genomic approaches (5). Despite the development of high-quality curated databases such as the Comprehensive Antibiotic Resistance Database (CARD) (6), we still observe a high level of variability in our ability to predict the phenotypic AMR profile from purely genomic data (7, 8). This disconnect can be attributed to fundamental limitations in the genomic methods used to describe phenotype (i.e., representing genetic capacity but not necessarily gene expression) as well as gaps in our knowledge of resistance determinants. Therefore, despite these expression-related limitations, AST prediction from genomics data is still a highly useful tool for the identification of novel mechanisms and key resistance drivers as well as for determination of the propensity for a given driver to be transmitted. This is vital for prioritizing research and surveillance efforts for the determinants driving AMR.
There have been several approaches used for predicting AST from genomic data sets; these can be divided into AMR gene-centered and gene-free k-mer-based models. The simplest of the approaches in the first category is that of annotation of known AMR genes within the genome and the direct tallying of their associated resistances; for example, in cases in which the genome contained a broad-spectrum β-lactamase such as New Delhi metallo-β-lactamase 1 (NDM-1), the isolate would be considered resistant to β-lactam antibiotics (7, 9). Alternatively, the presence and absence of AMR genes can be used as features to train machine learning (ML) classification models (10). These models learn to determine the weights across the genes that best explain the observed pattern of resistance to a given antibiotic. Such approaches are likely to perform best when organisms are well studied and the AMR mechanisms are relatively well characterized.
The approaches classified into the second category, consisting of the gene-free k-mer-based models, provide an alternative approach. This approach, while more data intensive, attempts to identify the parts of the genome that correlate best to the resistance pattern (11). Gene-free approaches do not include a priori assumptions about the AMR determinants in the genome and allow discovery of new genomic features (11–13). However, in certain data sets with limited diversity, such approaches may identify non-AMR-related k-mers that are shared by the resistant organisms only incidentally.
Salmonella is a broadly distributed Gram-negative bacterium found in a range of environments throughout the food chain, including in many prominent food-producing animals, e.g., poultry, pigs, and cattle (14). Additionally, it is known to have numerous inter- and intraspecific and environmental transmission routes (15). Nontyphoidal Salmonella (NTS) serovars represent the leading global cause of foodborne-related lost years of life (2.4 to 6.2 million years) and lost disability-adjusted life years (2.5 to 6.3 million) (15). Additionally, NTS serovars are conservatively (16) estimated to cause 31.8 to 211.2 million infections globally and 36.3 to 89.1 million deaths annually (15). An increased prevalence of resistant NTS is of great concern due to the association of AMR with worse clinical outcomes (17, 18). Therefore, due to the heavy global burden of NTS, understanding AMR dynamics in this system is highly important.
Previous studies have investigated the utility of genomic methods for AMR prediction of Salmonella infections by certain clinical isolates and serovars (7, 19). However, isolates of Salmonella from one of the most important vectors of foodborne salmonellosis, chicken, have been previously characterized genomically in only relatively low numbers (20). Therefore, this study aims both to expand this AMR genomic characterization to a broader sampling of NTS serovars isolated from commercial chicken farms and to train prediction models for AMR phenotypes. These models are then used to identify the most important drivers of the observed resistance patterns in this ecological context.
RESULTS
Assembly and annotation.
All 97 de novo assemblies resulted in genomes comprising between 19 and 105 contigs (mean = 43.56) of average length of between 45.72 kb and 247.03 kb (mean = 122.18 kb). The distribution of N50 values ranged from a minimum of 68.01 kb to a maximum of 735.38 kb (mean = 341.91 kb), and total genome sizes ranged from 4.64 Mb to 5.05 Mb (mean = 4.84 Mb). G+C% content displayed a tight range over the assembled genomes, with a mean of 52.11% and a standard deviation of 0.11%. All assemblies were deposited in GenBank, and all accession numbers and assembly metrics can be found in Table S1 in the supplemental material.
NCBI GenBank accession numbers and assembly metrics for all genomes (BioProject identifier PRJNA521409). Download Table S1, CSV file, 0.01 MB (14KB, csv) .
© Crown copyright 2019.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Phylogeny.
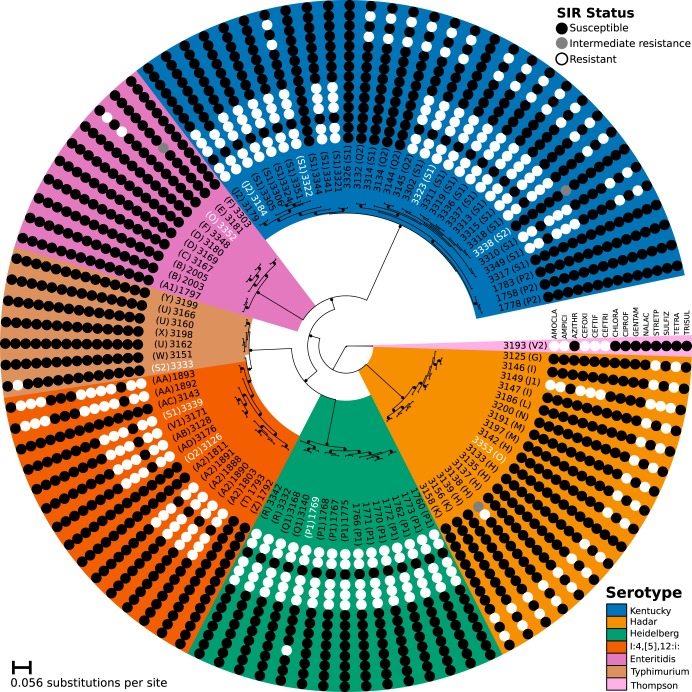
Phylogenetic analysis of a core genome single nucleotide polymorphism (SNP) alignment shows a relatively well-supported monophyletic distribution of serotypes (see Fig. 1). Isolate 3333 was the one exception to this monophyly, branching with 100% ultrafast bootstrap support as sister to the well (100%)-supported Salmonella enterica serovar S. I:4,[5],12:i: clade.
FIG 1.
Core genome SNP phylogeny. The figure shows the IQTree maximum likelihood phylogeny generated from core genome SNP alignment. Internal tree nodes with ≥90% ultrafast bootstrap support are noted by black circles. Correspondences of serotype clades to the lowest common ancestor of each are highlighted according to the following color scheme (as indicated by the legend): a blue background indicates S. Kentucky serovars, orange S. Hadar, green S. Heidelberg, red-orange S. I:4,[5],12:i:, purple S. Enteritidis, brown S. Typhimurium, and light pink S. Thompson (outgroup). A randomly chosen name representing the farm from which a sample was isolated is indicated in parentheses. AST results are indicated using circles, with resistance indicated by a white circle, intermediate resistance by a gray circle, and susceptibility by a black circle. Antibiotics are abbreviated per standard shorthand from the taxon label outward as follows: amoxicillin-clavulanic acid (AMOCLA), ampicillin (AMPICI), azithromycin (AZITHR), cefoxitin (CEFOXI), ceftiofur (CEFTIF), ceftriaxone (CEFTRI), chloramphenicol (CHLORA), ciprofloxacin (CIPROF), gentamicin (GENTAM), nalidixic acid (NALAC), streptomycin (STREPT), sulfamethoxazole (SULFIZ), tetracycline (TETRA), and trimethoprim-sulfamethoxazole (TRISUL). Taxa for which the AST was systematically predicted incorrectly are indicated in white characters.
Antibiotic susceptibility testing.
In contrast to the largely monophyletic serotype patterns observed in the phylogenetic analysis, the observed complements of resistances to the antibiotics tested showed variation both within and between the serovars (Fig. 1; see also Table S2). S. Kentucky serotypes, in particular, displayed the greatest range of observed AMR phenotypes, with 6 distinct patterns of resistance. S. Hadar serotypes had 4 resistance sets, and S. I:4,[5],12:i: serotypes displayed 3 different sets. The remaining isolates with more than a single exemplar, namely, S. Typhimurium, S. Enteritidis, and S. Heidelberg, were more consistent, showing only 2 different resistance patterns each.
Full MIC results for all isolates and their CLSI breakpoint SIR status. Download Table S2, XLSX file, 0.01 MB (14.1KB, xlsx) .
© Crown copyright 2019.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The most commonly observed set of cooccurring resistances (see Fig. S1G in the supplemental material) to a group of antibiotics consisted of the 48.4% (47/97) of isolates resistant to all the β-lactam antibiotics tested, i.e., the aminopenicillins (amoxicillin-clavulanic acid [AMOCLA] and ampicillin [AMPICI]) and the cephalosporins (cefoxitin [CEFOXI], ceftiofur [CEFTIF], and ceftriaxone [CEFTRI]). Only 2 isolates were susceptible to some but not all β-lactam antibiotics: a single S. Kentucky isolate (3184) that was susceptible to CEFTRI while being resistant to the other β-lactams and a single S. Hadar isolate (3138) that showed intermediate resistance and full resistance to AMOCLA and AMPICI, respectively, but susceptibility to the 3 cephalosporins. In terms of serotypes, this group of β-lactam-resistant isolates included all of the S. Heidelberg isolates, 20/32 (65.63%) of the S. Kentucky isolates, 10/15 (66.6%) of the S. I:4,[5],12:i: isolates, 1/17 (5.9%) of the S. Hadar isolates, and the lone S. Thompson isolate.
(A to G) Observed number of resistant isolates by serotype. Antimicrobials are presented in the same order for all panels as indicated by axis label on the bottommost panels. (H) Cooccurrence pattern of resistance across all serotypes. This UpSet plot shows the different sets of resistances observed across the isolates. The top bar plot indicates how many isolates were resistant to each specific group of antibiotics. The corresponding set of antibiotics is indicated below the top bar plot by the dark colored dots. The bar plot to the left shows how frequently resistance to each antibiotic occurred regardless of which set of resistances it appeared within. Download FIG S1, PDF file, 0.1 MB (147.6KB, pdf) .
© Crown copyright 2019.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The other most commonly observed group of shared resistances (see Fig. S1H) was that consisting of the 36.1% (35/97) of isolates resistant to the aminoglycoside streptomycin (STREPT) and tetracycline (TETRA). This pattern of shared resistances included 16/17 (94.1%) of the S. Hadar isolates, 16/32 (50%) of the S. Kentucky isolates, 2/15 (13.3%) of the S. I:4,[5],12:i: isolates, and 1/10 (10%) of the S. Enteritidis isolates. There were only 2 exceptions to the pattern of isolates being resistant to either of STREPT or TETRA (implying resistance to the other): S. Typhimurium isolate 3333 and S. Heidelberg isolate 1769.
Constituting a set apart from those resistant to the β-lactams, streptomycin, and tetracycline, only 5 isolates were resistant to any other antibiotic tested: 2/15 (13.3%) of the S. I:4,[5],12:i: isolates (1892 and 1893) showed resistance to chloramphenicol (CHLORA) and sulfamethoxazole (SULFIZ), a single (1/32, 3.13%) S. Kentucky isolate (3338) and a single (1/10, 10%) S. Enteritidis isolate (3181) had intermediate resistance to CHLORA (see Fig. S2), and, finally, a single S. Hadar isolate (1/17 5.9%) was also resistant to SULFIZ.
Hierarchical clustering of observed phenotypic AST results. Black cells indicate susceptibility, while red and cream cells represent intermediate susceptibility and resistance, respectively. The serotypes are indicated along the left axis per the legend. Each row represents a single isolate and each column a single antibiotic tested. Isolates and antibiotics are ordered according to their similarity to one another as inferred by a pair of hierarchical clustering inferences as shown by the pair of annotated dendrograms. The antibiotics tested were amoxicillin-clavulanic acid (AMOCLA), ampicillin (AMPICI), azithromycin (AZITHR), cefoxitin (CEFOXI), ceftiofur (CEFTIF), ceftriaxone (CEFTRI), chloramphenicol (CHLORA), ciprofloxacin (CIPROF), gentamicin (GENTAM), nalidixic acid (NALAC), streptomycin (STREPT), sulfamethoxazole (SULFIZ), tetracycline (TETRA), and trimethoprim-sulfamethoxazole (TRISUL). Download FIG S2, PDF file, 0.04 MB (41.9KB, pdf) .
© Crown copyright 2019.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The isolates showing resistance to the greatest numbers of antibiotics (i.e., the most multiresistant isolates) were S. I:4,[5],12:i: isolates (1893 and 1892) and the S. Hadar isolate (3149), which were resistant to chloramphenicol and sulfamethoxazole, respectively, as well as to the β-lactams and to the aminoglycoside and tetracycline tested. A total of 12/32 (37.5%) S. Kentucky isolates were resistant to all of these antibiotics apart from chloramphenicol and sulfamethoxazole. At the other end of the spectrum, S. Enteritidis was the most susceptible serotype with 9/10 (90%) of isolates showing susceptibility (or intermediate susceptibility) to all antibiotics. Similarly, 6/7 (85.7%) of the S. Typhimurium isolates, 7/32 (21.88%) of the S. Kentucky isolates, 5/15 (33.3%) of the S. I:4,[5],12:i: isolates, and 1/17 (5.9%) of the S. Hadar isolates also showed susceptibility (or intermediate susceptibility) to all tested antibiotics.
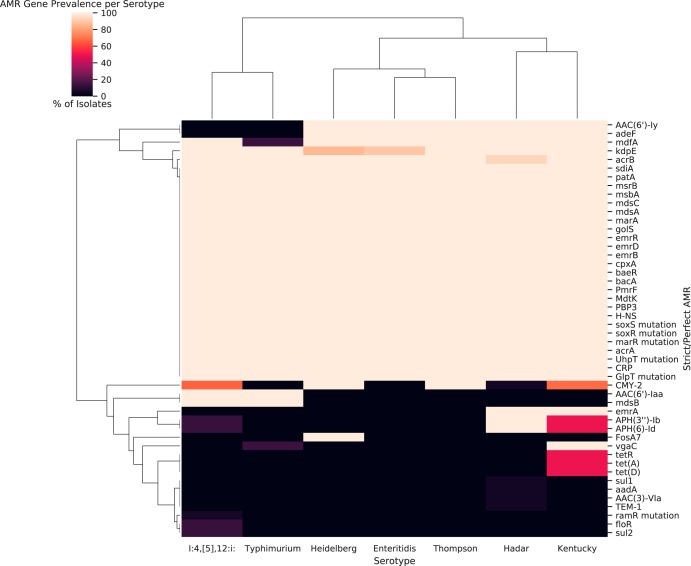
Antimicrobial resistance gene analysis.
Twenty-five putative AMR genes were identified across all isolates and serotypes by CARD’s “strict” or “perfect” match criteria (see Fig. 2). Every isolate had a pair of fosfomycin resistance-related mutations in the glpT and uhpT transporters (21) as well as a pmrF gene associated with resistance to polymyxin antibiotics (22), a Penicillin-binding protein (PBP) 3 gene (23), and a bacA gene (associated with low-level resistance to bacitracin) (24). Additionally, there were 16 efflux components and determinants associated with their expression, including general efflux regulators such as H-NS histone-like protein (25) and cpxA (26). Genes implicated in specific efflux systems included genes associated with the acrAB system: sdiA (27), marR and marA (28), and soxR and soxS (29). Interestingly, while acrB was detected in all but a single S. Hadar strain, acrA was not detected in any isolate. The other ubiquitous efflux pump-related systems included mdsABC-associated genes golS, mdsA, and mdsC (30); MATE efflux system mdt-associated efflux components CRP (31), baeR (32), and mdtK (33); and emrAB-TolC components and regulators emrB, emrD, and emrR (34, 35). Interestingly, only the S. Hadar and S. Kentucky isolates had emrA and only the S. Typhimurium and S. I:4,[5],12:i: isolates had mdsB. Finally, there were patA and msbA transporters found in all isolates despite no detection of patB (36). kdpE, a regulator of potassium transport associated with aminoglycoside resistance (37), was nearly ubiquitous (absent in 2/15 S. Heidelberg and 1/10 S. Enteritidis isolates).
FIG 2.
All AMR genes detected by serotype under CARD’s “Strict” and “Perfect” criteria (including efflux system components). Each cell indicates the percentage of isolates belonging to the relevant serotype (column) that contained each detected determinant (rows). Black blocks indicate that no isolates of that serotype had that AMR gene, and cream blocks indicate that 100% of the isolates had that AMR gene. Serotypes and genes are each ordered via hierarchical clustering as indicated by the dendrograms.
Some predicted AMR genes had more varied serotype distributions. The chloramphenicol exporter mdfA (38) was ubiquitous in all serotypes apart from S. Typhimurium, where it was present in a single isolate (1/7). Similarly, the aminoglycoside resistance gene AAC(6’)-Iy (39) and the adeFGH efflux component adeF (40) were present in all genomes apart from those of the S. Typhimurium and S. I:4,[5],12:i: isolates. The β-lactamase CMY-2 gene (41) was present in all of the S. Heidelberg and S. Thompson strains but absent from the S. Enteritidis and S. Typhimurium isolates. CMY-2 was more unevenly distributed in other serotypes, being detected in 5.88% of S. Hadar isolates, 66.67% of I:4,[5],12:I isolates, and 68.75% of S. Kentucky isolates. All S. Typhimurium and S. I:4,[5],12:i: isolates but no isolates of other serotypes contained AAC(6’)-Iaa. Similarly, APH(6)-Id was found in all S. Hadar isolates but in only 13.33% of the S. I:4,[5],12:i: isolates and 50% of the S. Kentucky isolates. Every S. Kentucky isolate also had the streptogramin A resistance-related vgaC gene (42), but it was otherwise found only in a single S. Typhimurium isolate (3333).
AMR genes detected within only a single serotype included fosA7 in every S. Heidelberg isolate sequenced, and tet genes corresponding to efflux pump-related proteins, namely, tetR, tetD, and tetA (43), were found exclusively in 16/32 (50%) of S. Kentucky isolates. The rarest AMR genes in our data set, floR (phenicol resistance) (44) and sul2 (sulfonamide resistance) (45), were found in only 2 (13.3%) of S. I:4,[5],12:i: isolates (1892 and 1893) collected from the same farm. Another S. I:4,[5],12:i: isolate was the lone carrier of ramR mutations related to upregulation of acrAB (46). Finally, the TEM-1 β-lactamase gene (47) was found in only a single S. Hadar isolate (3142), with aadA, sul1, and AAC(3)-VIa found in only a single different S. Hadar isolate (3186).
Predicting phenotype from genotype.
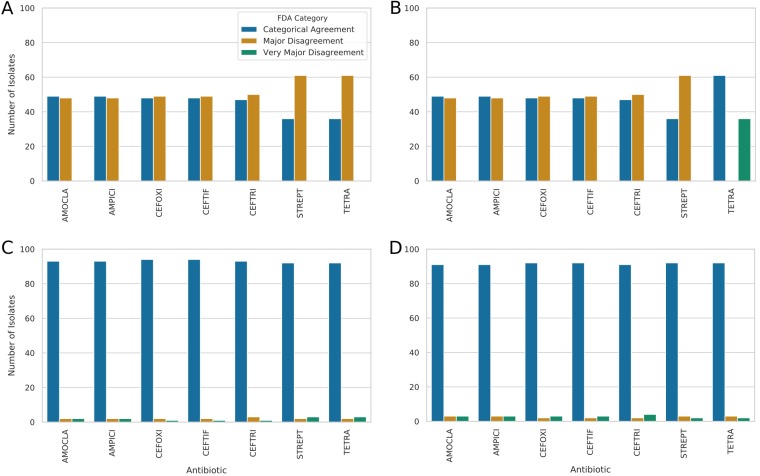
The relationship between the observed AST phenotype and the AMR determinants detected within the sequenced genomes was assessed according to standard FDA categories. If the predictions from the genome or models trained using the genome matched the phenotypic data, then the result was classified as representing “categorical agreement.” However, if the prediction was of resistance but the phenotype showed susceptibility, then the result was classified as representing a “major disagreement,” and if the prediction was of susceptibility but the phenotype showed resistance, then the result was classified as representing a “very major disagreement.”
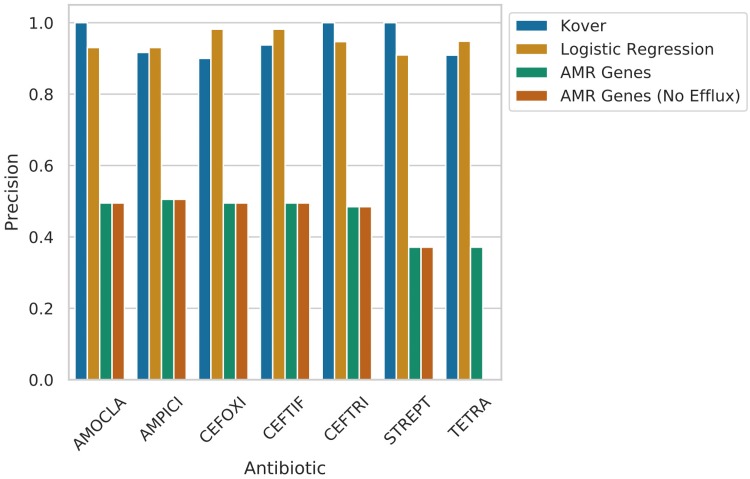
By using the antibiotic resistance ontology (ARO) that CARD is built upon (48), we were able to directly tally the associated resistances with the detected AMR determinants. As can be seen in the high levels of major disagreement (48% to 62% of isolates; Fig. 3) and poor precision (0 to 0.5; Fig. 4), directly tallying resulted in a massive overprediction of resistance. As this was initially believed to be due to the presence of the efflux pump components, we also performed this direct tallying without efflux pumps but observed the same overprediction of resistance. The one exception to this pattern was tetracycline resistance, which was consistently underpredicted in the absence of efflux (i.e., all isolates were classified as “susceptible with very major disagreement”). Overall, the level of precision seen from direct tallying of ranges is low, at approximately 0.5 for β-lactams and 0.38 for streptomycin and tetracycline (0.0 precision with no efflux genes for tetracycline). This represents the underlying proportion of resistant isolates in our data set.
FIG 3.
FDA categorization of AST prediction performance across the antibiotics with sufficient numbers of susceptible and resistant isolates for assessment. “Categorical Agreement” represents the cases in which the prediction matched the observed phenotype, “Major Disagreement” corresponds to a prediction of resistance but a determination of susceptibility by the AST, and “Very Major Disagreement” indicates a prediction of susceptibility but a determination of resistance by the AST. (A) Performance of direct tallying of the presence of AMR genes as detected by RGI. (B) The same procedure was performed but with exclusion of efflux determinants. (C) Accuracy of prediction of resistance patterns by the use of binary logistic regression models trained using the AMR genes as features. (D) Accuracy of prediction of resistance directly from the genome by the use of a k-mer-based set-covering machine model.
FIG 4.
Observed precision for direct tallying with and without efflux pumps and test set average classifier precision for set-covering machine and logistic regression models. These results clearly show that both machine learning approaches created far more precise predictions of AST (>0.9) than direct tallying of the AMR determinants.
Logistic regression.
A simple set of binary logistic regression models using detected AMR genes as features were able to predict the AST. On a held-out test set, average precision ranged from 0.91 for streptomycin to 0.98 for ceftiofur and cefoxitin (see Fig. 4). This meant that, overall, there were a maximum of only 3 major disagreements and a maximum of 4 very major disagreements among the 97 isolates.
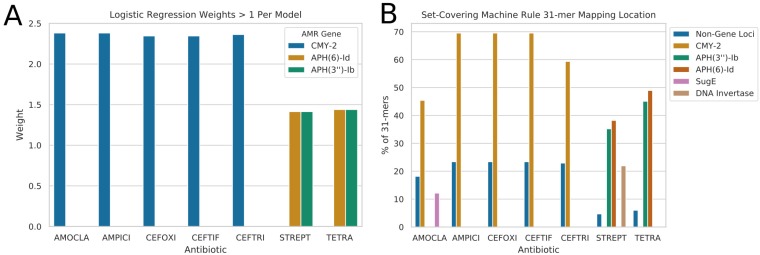
Inspection of the mostly highly weighted AMR determinants for each trained logistic regression model revealed that the β-lactamase AmpC-like CMY-2 gene was most important for prediction of resistance to β-lactam antibiotics (see Fig. 5). No other determinant had a weighting of greater than 25% of that of CMY-2 for the β-lactam models. For the streptomycin and tetracycline models, the phosphotransferase genes APH(6)-Id and APH(3”)-Ib were the most highly weighted determinants by a factor of 2 or greater.
FIG 5.
A plot of the most important features and their identity for the machine learning models. (A) Learnt coefficients/weights on the AMR gene presence/absence matrix by the logistic regression models. Only weights greater than 1 are displayed on this plot. (B) Top genomic origins of the 31-mers learnt by the set-covering machines. Non-Gene Loci are the 31-mers mapping outside any of the identified genes.
Set-covering machine.
Similarly, the k-mer-based set-covering machine classifiers also greatly outperformed direct tallying. Set-covering machines (implemented using the Kover algorithm as described previously [13, 49]) represent a type of machine learning model that learns a set of Boolean rules, e.g., presence/absence (and higher-order conjunctions) of specific features (k-mers in our case) which predict the resistance label (50).
Performance for all 7 antibiotics represented greater than 0.9 precision and was only slightly poorer than that of logistic regression despite having only the genomic 31-mers as features (see Fig. 3; see also Fig. 4). Overall, under the FDA categorization metrics, the set-covering machines performed similarly to logistic regression and considerably better than direct tallying.
When the 31-mers identified by the trained set-covering machines as the most highly equivalently important rules to predict AST were mapped back to the underlying genomes, the majority mapped to the same AMR genes that were most highly weighted by logistic regression, i.e., CMY-2 for β-lactam antibiotics and APH(3”)-Ib and APH(6)-Id for streptomycin and tetracycline (see Fig. 5). However, there were a few additionally weighted non-AMR-specific genes that had significant numbers of mapping k-mers. For example, the amoxicillin-clavulanic acid model included 12.1% of k-mers mapping to the gene sugE, and the streptomycin model had a reasonably high 21.9% rate of mapping to a hin DNA invertase gene.
DISCUSSION
Overall, the serotypes isolated from broiler chicken in this study included the top 5 serovars implicated in human salmonellosis in Canada (51). The most common clinical isolate serovar, S. Enteritidis (56% of cases) (51), was relatively less common in the chicken isolates studied here (10.31%) than in the clinical data. This might reflect the earlier sampling date of these isolates, as the most recent Canadian FoodNet report indicated that 41% of Salmonella isolates from broiler chicken manure at sentinel sites were S. Enteritidis (51). As S. Enteritidis has become more common in Canadian salmonellosis cases, it is perhaps reassuring that all broiler chicken S. Enteritidis isolates in this study were totally susceptible to all 14 tested antibiotics. Other serotypes either were found in our isolates in proportions similar to those of the Canadian clinical isolates (for S. Typhimurium, 7.2% versus 7%) or were more common in our data set, including S. Heidelberg (31.9% versus 3%) and S. I:4,[5],12:i: (15.4% versus 3%) (51). Rare clinical isolates such as S. Kentucky (33%) and S. Hadar (17.5%) were far more common in our isolates, but S. Infantis, which is the 5th most common cause of salmonellosis (3%) (51), was totally absent. These observed overlaps and differences with respect to the diversity of human clinical and broiler chicken NTS isolates underscore the utility and relevance of this type of genomic surveillance work.
The Salmonella in silico typing resource (SISTR) in-silico serotyping conducted is supported by the largely monophyletic distribution of serotypes within the core genome phylogeny (Fig. 1). There was only one exception to this pattern, namely, an S. Typhimurium isolate (3333) which branched within the I:4,[5]:12:i clade. As this is a monophasic S. Typhimurium variant, we would expect this to branch within the phylogenetically adjacent S. Typhimurium clade. This suggests that this (well-supported) branching location might reflect some form of phylogenetic reconstruction artifact. Otherwise, the inferred relationships between serotype clades largely agree with those previously inferred in dedicated Salmonella phylogenomic analyses (52).
There were limitations to this data set for the purposes of predicting phenotypic resistance from genomic data. Tests of several antibiotics identified no or very few resistant isolates, meaning that it was possible to train models for only a subset of antibiotics (e.g., AMOCLA, AMPICI, CEFOXI, CEFTIF, CEFTRI, STREPT, and TETRA) due to the problem of label imbalance. The treatment of phenotypes showing intermediate resistance as resistant is another potential cause of distortion in the trained models. However, only a single model (AMOCLA) used any isolates with intermediate phenotypes (a single isolate, representing 1.03% of the data). This means that, even if it were misleading to treat this isolate as resistant, doing so would have a correspondingly small effect on the learnt weights and k-mer mappings of the AMOCLA models. Finally, we do lose potentially interesting granularity in our models by performing only binary predictions of susceptibility or resistance. Ideally, we would train regression models that directly predict MIC values. Unfortunately, this is a difficult problem due to this data set being somewhat small for this level of prediction and due to the nature of MIC measurement. MICs are generally measured to within an accuracy of only a 2-fold dilution, meaning that the amount of measurement error is greater for higher MIC values than for lower ones. The other problem is that MICs at the extremes of the standardized measured range are presented only as inequalities (i.e., >256 mg/liter or <0.5 mg/liter) which is difficult to handle mathematically. However, future work incorporating larger numbers of genomes and measurements could use approaches such as maximum margin interval trees to address these issues (53).
This being said, by using the learnt weights and k-mer mapping locations (see Fig. 5) in the high-precision (see Fig. 4) AMR gene-based and gene-free resistance prediction models generated in this study, it was possible to identify key drivers of observed resistance patterns for the subset of antibiotics with a reasonable balance of resistant and susceptible isolates in our data set. Therefore, we attempted to identify key AMR drivers for β-lactam resistance (specifically, amoxicillin-clavulanic acid, ampicillin, cefoxitin, ceftiofur, and ceftriaxone) as well as for streptomycin resistance and tetracycline resistance. It should be noted that these results are specific to the isolates and serovars within our data set; while there are some highly encouraging functional connections, expanding this approach to incorporate new serotypes would be best served by retraining the models.
The β-lactam models were largely identical, pinpointing the β-lactamase CMY-2 gene (or k-mers derived from this gene) as the most important feature in all individual models. Only the amoxicillin-clavulanic acid k-mer model featured less than 50% k-mers deriving from a CMY-2 gene. This model featured a low but notable proportion of k-mers (12.1%) deriving from a sugE gene. sugE is associated with resistance to quaternary ammonium compounds (forming part of an efflux pump), specifically, cetylpyridinium, cetyldimethylethyl ammonium, and cetrimide, cations commonly used as disinfectants (54). This gene has previously been detected on the same plasmid as CMY-2 (55) and may play a coselection role. Every single isolate with CMY-2, with the exception of 3186, had a directly adjacent lipocalin gene (blc) on the same strand followed by a sugE gene in the opposite orientation (see Fig. S4 in the supplemental material). This suggests that sugE was likely learnt to be predictive purely due to its colocation/linkage instead of due to any specific antimicrobial resistance-related function. The presence of this adjacent blc may further potentiate the resistance, as these have been reported in other bacteria to increase MICs of β-lactam antibiotics by binding the antibiotic in the medium (56, 57).
This result is supported by previous work identifying CMY-2 as a key driver of extended-spectrum-β-lactam resistance in Escherichia coli derived from broiler chickens (58). CMY-2 has been established as displaying relatively broad distribution in samples derived from a range of bacteria and food production animals in Canada (59) as well as globally (60–62). Importantly, evidence of the direct connections of NTS with CMY-2-related extended-spectrum-β-lactam resistance in human clinical infections has been established (if somewhat poorly understood) (63). Previous work in poultry E. coli isolates has shown that without active selection from antibiotic usage, this gene is rapidly lost in chicken samples (58). However, CMY-2 does persist in poultry farm environmental samples (58), suggesting that further work is needed to elucidate the selective forces determining persistence and transmission of this critical resistance gene. One potential avenue for this work, underlined by the k-mers derived from the sugE gene, is that of looking at the role which the use of quaternary ammonium-based disinfectants plays in this process. Similarly, there is a need to experimentally investigate whether the presence of blc can lead to increased β-lactam resistance in Salmonella such as they have been shown to do in other bacteria (56, 57) and what impact this has on the retention of CMY-2.
For both the logistic regression and k-mer set-covering machine models, there were a subset of genomes that were consistently mispredicted (whether they were in the training set or the test set). Specifically, for the β-lactam antimicrobials (AMPICI, AMOCLA, CEFOXI, CEFTRI, and CEFTIF), every individual model failed to correctly predict the phenotype for isolates 3338, 3126, and 3339, with one additional misprediction in just the CEFTRI models for 3184. These taxa are highlighted in Fig. 1 with white taxon names and do not exhibit any clear monophyletic phylogenetic groupings, serotype-based trend, or sampling location. It should be noted, however, that these taxa displayed resistance phenotypes that were different from those of their closest relatives despite similar predicted AMR gene complements; e.g., 3333 is the only S. Typhimurium isolate with any observed resistance, and 3352 is the only S. Enteritidis isolate with clear resistance phenotypes.
Some isolates that were predicted to be resistant to β-lactams but were found to be susceptible in the AST showed a perfect hit for the CMY-2 β-lactamase (see Fig. 2). Specifically, isolates 3186, 3126, and 3338 all had CMY-2 genes clearly present but were found to be susceptible to β-lactams, with the observed MICs at the lower end of the measured range (see Table S2 in the supplemental material). These inferred susceptibilities were consistent even following multiple replications of the AST results. This likely suggests an uncharacterized context-related expression determinant for CMY-2 (i.e., the gene is present but not expressed) to explain why these strains had the capacity for β-lactam resistance but did not display that phenotype during testing. This could be tested in future work via approaches that directly measure the presence of specific proteins such as matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometry (64). On the other hand, S. I:4,[5],12:i isolate 3339 and S. Hadar isolate 3149 were both resistant to the β-lactam tested but had no detectable CMY-2 gene. Additionally, with the exception of genes of efflux pump components, there were no AMR genes or mutations clearly associated with β-lactam resistance detected in these isolates. However, previous work has shown that Salmonella can display resistance to β-lactam antibiotics without any detectable β-lactamase genes being present (65). This suggests the presence of an undetected or uncharacterized β-lactam resistance mechanism in these isolates. It is possible that the 18% to 23.3% of k-mers mapping to intergenic regions for these models may play a role in this resistance mechanism.
For the streptomycin resistance models, the most important predictor was the presence of the phosphotransferase genes APH(6)-Id and APH(3”)-Ib. These are two among a large number of known aminoglycoside resistance genes detected in Salmonella isolates (66). They are also frequently found on mobile genetic elements such as transposons and plasmids (66). Additionally, they have previously been associated with resistance to streptomycin in animal isolates (67). Interestingly, there were also a number of k-mers (21.9%) associated with a hin DNA invertase gene in the set-covering machine model for streptomycin. This gene has been previously associated with phase variation in Salmonella (68).
Similarly to the β-lactam models, there was a subset of isolates what were consistently mispredicted for streptomycin (3322, 3323, 3352, 3353, and 1769). Among the isolates where there was a failure to successfully predict streptomycin resistance, it was found that these had no detectable APH(6)-Id or APH(3″)-Ib genes. However, several did show evidence of a cryptic aminoglycoside N-acetyltransferase enzyme gene, ACC(6′)-Iy, in their genome (isolates 3322, 3352, and 1769) but so did a large number of isolates that displayed susceptibility to streptomycin (e.g., every S. Enteritidis, S. Heidelberg, S. Thompson, S. Hadar, and S. Kentucky isolate). All 3 of the isolates with the cryptic aminoglycoside N-acetyltransferase enzyme gene ACC(6′)-Iy were the lone examples of a resistant isolate within clades that otherwise contained only susceptible isolates. In the other direction, isolate 3323 was predicted to be resistant due to the presence of both the APH(6)-Id gene and the APH(3″)-Ib gene but was found to be susceptible to streptomycin in the AST. Similarly, this isolate branched within a resistant clade that was found to be totally resistant to these antibiotics. Isolate 3353 displayed a pattern similar to that shown by the lone S. Hadar isolate and thus was found not to be resistant to streptomycin despite having a strict full-length hit with respect to APH(6)-Id and 100% identity and a partial (52%) hit to APH(3″)-Ib. These prediction failures were not attributable to a failure to detect AMR genes due to poor assembly quality. All the consistently mispredicted genomes had assembly quality metrics that were largely representative of the assemblies overall (see Fig. S3).
Visualization of the N50 distribution of assembled genomes. This figure shows that the assemblies of genomes that were consistently mispredicted in terms of AMR for TETRA/STREPT and the β-lactam antibiotics tested are not notably more fragmented than the assemblies as a whole. Download FIG S3, PDF file, 0.03 MB (29.9KB, pdf) .
© Crown copyright 2019.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
CMY-2. The blaCMY-2 genes were typically flanked by ISEcp1 downstream and lipocalin (blc) on the same strand followed by a quaternary ammonium compound(s) (sugE) on the opposite strand. Download FIG S4, PDF file, 0.08 MB (79.2KB, pdf) .
© Crown copyright 2019.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The data corresponding to the tetracycline resistance model weights and k-mer locations were more perplexing. Specifically, the learnt weights and k-mers in that model were nearly identical to those seen with the streptomycin models despite the differences in the drug classes and known resistance mechanisms. These phosphotransferase genes [APH(6)-Id and APH(3″)-Ib] have never been associated with a mechanism through which they could convey tetracycline resistance directly. This result renders the tetracycline model somewhat suspect and could represent overfitting to the training data due to the strong cooccurrence of streptomycin resistance and tetracycline resistance among isolates in our data set. While these aminoglycoside resistance genes are frequently detected on chromosomal fragments containing the tet(B) tetracycline resistance gene (69), this particular tetracycline gene was totally absent in these isolates, so there is little evidence to support any hypothesis involving colocalization. In terms of mispredicted isolates, they was largely the same isolates as were seen in the streptomycin testing (3322, 3323, 3352, and 3353), with the exception of 1769 and the addition of isolate 3333.
Overall, this work shows the potential utility and pitfalls of the effective use of genomic data for the surveillance of AMR. We demonstrate the propensity of overprediction of resistance that occurs in tallying resistance directly from AMR gene predictions. Additionally, we show the utility of comparing the learnt parameters of simple machine learning models to help identify key drivers of antimicrobial resistance. Specifically, we identify the AmpC-like β-lactamase CMY-2 gene as the primary driver of resistance to aminopenicillins and to second- and third-generation cephalosporins in broiler chicken nontyphoidal Salmonella enterica serovars (see Fig. 5; see also Table S4). As this β-lactamase has been reported in human NTS isolates (63), this underscores the importance of monitoring of CMY-2 β-lactamase prevalence and transmission in food production both within Canada (59) and globally (60–62). This work also revealed that APH(6)-Id and APH(3″)-Ib genes are key determinants driving streptomycin resistance in Canadian chicken NTS isolates (see Fig. 5; see also Table S4). Reassuringly, the most commonly clinically relevant serotype, S. Enteritidis, was shown to be susceptible to all common antimicrobials in Canadian poultry sources. Additionally, despite previous detection of colistin resistance genes in CMY-2-bearing poultry isolates (70) there was no evidence of colistin resistance genes present in the genomes of these isolates based on the Resistance Gene Identifier (RGI) analyses (see Fig. 2).
MATERIALS AND METHODS
Isolation.
A total of 97 Salmonella serovar isolates obtained from 23 broiler chicken farms in British Columbia, Canada, were sequenced in this study. Isolates were selected based on their prevalence, pulsotype, and antibiotic susceptibility profiles as outlined previously (71).
Sequencing.
Genomic DNA was extracted from overnight cultures in 5 ml of brain heart infusion (BHI) broth (BD, NJ, USA) using DNeasy blood and tissue kits (Qiagen) as specified in the protocols (71).
The extracted DNA was stored in 10 mM Tris-HCl buffer (pH 8.0) and quantified by the use of an Invitrogen Qubit 2.0 Fluorometer (Life Technologies). The quality of DNA was visualized by electrophoresis on a 1% agarose gel, and the DNA was stored at −20°C until construction of the genomic libraries. The libraries were then sequenced using a MiSeq v3 sequencer in paired-end mode to generate 2 × 250-bp reads.
Assembly and annotation.
Genomes were assembled using a standardized MiSeq assembly pipeline implemented within the Integrated Rapid Infectious Disease Analysis (IRIDA) platform of the Public Health Agency of Canada (72). This workflow trims reads to remove low-quality sequences and then merges overlapping paired reads with Fast Length Adjustment of SHort (FLASH) (v1.2.9) reads (73). The merged and remaining unmerged reads were then used to generate a de novo assembly with SPAdes (v3.9.0) (74). The resultant assembly was then filtered to remove short (>1,000-bp) contigs, repetitive (1.75 repeat cutoff ratio) contigs, and low-coverage (0.33 coverage cutoff ratio) contigs.
Assembly metrics were calculated for the final assemblies using QUAST (v5.0.2) (75) (see Table S1 in the supplemental material). The final assemblies were annotated with Prokka (v1.13) (76) using the “Salmonella” genus, “enterica” species, and Gram-negative options and a 1E−5 minimum expectation value. Additionally, assemblies were screened for plasmids using abricate (v0.8.7) (77) and the plasmidfinder database (27 August 2018) (78). Plasmid screening results were then summarized and visualized using the Pandas (v0.22.0) (79) and Seaborn (v0.8.1) Python libraries (80) (see Fig. S5 in the supplemental material; see also Table S3).
Visualization of the relative levels of coverage of plasmids within the genome assemblies as detected using abricate and the plasmidfinder database. Download FIG S5, PDF file, 0.04 MB (46.8KB, pdf) .
© Crown copyright 2019.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Plasmid prediction results from the genomes studied (determined via the plasmidFinder database and abricate). Download Table S3, CSV file, 0.01 MB (5.9KB, csv) .
© Crown copyright 2019.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Relative risk calculations (determined using SAS-implemented chi-square tests) for the subset of AMR genes identified in the predictive modeling and literature as key drivers of resistance. Download Table S4, CSV file, 0.01 MB (585B, csv) .
© Crown copyright 2019.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Serotyping.
Serotyping was performed using the assembled genomic contigs and the Salmonella In Silico Typing Resource (SISTR) tool (v1.0.2) (81).
Code for this and all further analyses in this paper can be found in the associated Jupyter Notebook (82) and in the relevant folder under “analyses” in the git repository (83) (i.e., in this case, “analyses/serotyping”). This repository can be found at https://github.com/fmaguire/salmonella_ast_prediction/.
Phylogenetics.
A pangenome analysis was conducted using the Prokka annotations and Roary (v3.12.0) (84). From the identified 3,743 core genes present in ≥99% of genomes, an alignment was inferred in Roary. SNPs were then extracted from this alignment using “snp-sites” (v2.4.0) (85). This resulted in an alignment consisting of 77,795 sites.
A maximum likelihood phylogeny was then inferred in IQTree (v1.6.5) (86, 87) with UltraFast Bootstrap support. The inference was performed under the General Time Reversible Model with ascertainment bias correction and nucleotide frequencies (GTR+ASC+F), as this was the consensus best-fitting model for all information criteria as determined by ModelFinder (88). Phylogeny was then visualized and annotated with SISTR serotyping, AST results, and encoded origin using ETE3 (v3.1.1) (89).
Code used to perform this can be found in the notebook under “analyses/phylogeny.”
Antibiotic susceptibility testing.
Phenotypic antibiotic susceptibility testing (AST) was conducted for a panel of 14 standard antibiotics as described previously (71). In brief, a Sensititre automated system (Trek Diagnostic Systems, Cleveland, OH) was used to determine MICs (Table S2), and the results were analyzed according to Clinical and Laboratory Standards Institute guidelines for the following antibiotics: amoxicillin-clavulanic acid (AMOCLA), ampicillin (AMPICI), azithromycin (AZITHR), cefoxitin (CEFOXI), ceftiofur (CEFTIF), ceftriaxone (CEFTRI), chloramphenicol (CHLORA), ciprofloxacin (CIPROF), gentamicin (GENTAM), nalidixic acid (NALAC), streptomycin (STREPT), sulfamethoxazole (SULFIZ), tetracycline (TETRA), and trimethoprim-sulfamethoxazole (TRISUL).
This phenotypic testing was fully repeated for all isolates to confirm the resistance status. The code used to visualize and explore these results is available in the Jupyter Notebook under “analyses/ast.”
AMR gene identification.
AMR gene identification was performed using Resistance Gene Identifier (RGI) v4.0.3 (6) on assembled contigs. This involved Prodigal v2.6.3 (90) open reading frame (ORF) calling and DIAMOND v0.8.36 (91)-based homology searches. Loose hits were excluded from the results, and the reference database used was CARD (v2.0.1 release) (6). Results were further separated using RGI’s “perfect” (the predicted gene matches a known curated resistance gene completely at the amino acid level [including SNPs]) and “strict” (above a gene-specific threshold of bitscore-based similarity) credibility levels. Predictions were then grouped and analyzed using CARD’s in-built antibiotic resistance ontology (ARO) and the pandas (v0.22.0) (79) and seaborn (v0.8.1) (80) Python libraries.
The code used to perform this can be found in the notebook and under “analyses/rgi.”
Comparison of phenotype to genotype.
AST results were compared with the “strict” and “perfect” RGI predictions with and without efflux pump inclusion separately. This was done by using the ARO to identify the class of antibiotics associated with resistance shown by a given detected AMR determinant. These classes were then cross-referenced to the individual antibiotics tested in the AST. If an AMR determinant was detected in a given genome, it was considered to represent a prediction of resistance to the pertinent antibiotics tested. As there were so few isolates with intermediate resistances, all intermediate resistances in the AST were classified as resistant for this and subsequent analyses.
Standard FDA criteria were then used to tally how effective the ‘perfect’ and ‘strict+perfect’ AMR determinants were in predicting the AST as binary presence/absence indicators. These results fell into 3 types (as specific MICs were not being predicted): categorical agreement (CA; the genomic data and AST both predicted susceptibility or resistance); major disagreement (maj; the genomic data predicted resistance but the AST showed susceptibility); and very major disagreement (vmaj; the genomic data predicted susceptible but the AST showed resistance).
Code used to perform this can be found in the notebook and under “analyses/prediction/direct_tallying.”
Logistic regression.
Antibiotics with either no resistant isolates (azithromycin, ciprofloxacin, gentamicin, nalidixic acid, and trimethoprim-sulfamethoxazole) or an extreme imbalance of resistant/susceptible isolates (chloramphenicol and sulfamethoxazole), defined as the minority class consisting of <5% of isolates, were excluded from the machine learning analyses.
For each of the remaining antibiotics, a simple binary logistic regression model was fitted using the RGI-detected AMR determinants as the input features and “susceptible” and “resistant” as the output labels. Any AMR gene that was found in every isolate was removed from the data matrix. This was performed using scikit-learn v0.20.1 (92). Each model was trained on 80% of the training data (using a stratified test:train split) after resampling was performed using the Synthetic Minority Oversampling Technique (SMOTE) (via imblearn v0.4.3 [93]) to improve label balance. Logistic regression models were tuned using 3-fold cross-validation over the training set, with test-set performance evaluated using precision-recall curves. Performance was then compared across the whole data set using the FDA criteria as described above.
All code used to perform this analysis can be found in the “logistic_regression” folder under “analyses/prediction.”
Set-covering machine.
In order to assess whether other genomic factors not detected by RGI were likely to contribute to AMR, a k-mer-based set-covering machine approach was applied to the whole genomes for the balanced subset of antibiotics (with exclusion criteria used as described above for logistic regression). This was performed using Kover v2.0.0 (49), and individual rule sets were inferred using 10-fold cross-validation for each of the same antibiotic resistances as were used as described for the logistic regression. The trade-off hyperparameter (p) was selected using cross-validation across a range of possible values from 0.1 to 9999999. A maximum of 10 k-mers were allowed to be included in each rule, and a maximum of 10,000 equivalent rules were output. To assess where the inferred k-mers derived from in the genomes, the inferred equivalent conjunction k-mers were mapped to the genomes using BWA-MEM (94). The resulting individual model SAM files were then analyzed using the PySAM (v0.15.0) library to tally their mapping locations.
All code used to perform this analysis can be found in the “set_covering_machines” folder under “analyses/prediction” in the associated git repository.
CMY-2 locus analysis.
The contigs bearing CMY-2 were annotated using Rapid Annotation using Subsystem Technology (RAST) (95), and the 20-kb regions flanking the CMY-2 gene were visualized in SEED Viewer version 2.0 (96).
Relative risk calculation.
Relative risk was calculated from chi-square tests using the SAS software package and the whole data set. The subset of genes selected was determined using the results of the predictive models as well as the Tet and AAC(6’)-Iy genes that have previously been associated with resistance in the literature.
Data availability.
All data used in this study are available in GenBank (BioProject identifier PRJNA521409). The full list of accession numbers per genome can be found in Table S1. The code used to perform all analyses is available in the git repository https://github.com/fmaguire/salmonella_ast_prediction/.
ACKNOWLEDGMENTS
We are grateful to the Animal Health Centre (British Columbia Ministry of Agriculture, Abbotsford, British Columbia, Canada) and the participating broiler chicken farmers for assistance. We also acknowledge Shane Thiessen and Aaron Petkau at Public Health Agency of Canada (PHAC) for assistance with our account and for Virtual Private Network (VPN) access to PHAC’s IRIDA platform.
This work was funded by support from Agriculture and Agri-Food Canada through the Federal Genomics Research and Development Initiative to mitigate antimicrobial resistance (GRDI-AMR) project (PSS 1858) of Government of Canada provided to M.S.D. F.M. is supported by a Donald Hill Family Fellowship in Computer Science, a Genome Canada grant, and a Canadian Institutes of Health Research grant.
REFERENCES
- 1.Gardy JL, Loman NJ. 2018. Towards a genomics-informed, real-time, global pathogen surveillance system. Nat Rev Genet 19:9. doi: 10.1038/nrg.2017.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Robinson T, Bu D, Carrique-Mas J, Fèvre E, Gilbert M, Grace D, Hay S, Jiwakanon J, Kakkar M, Kariuki S, Laxminarayan R, Lubroth J, Magnusson U, Thi Ngoc P, van Boeckel TP, Woolhouse M. 2016. Antibiotic resistance is the quintessential One Health issue. Trans R Soc Trop Med Hyg 110:377–380. doi: 10.1093/trstmh/trw048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.World Health Organization. 2015. Global action plan on antimicrobial resistance. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 4.Brown ED, Wright GD. 2016. Antibacterial drug discovery in the resistance era. Nature 529:336. doi: 10.1038/nature17042. [DOI] [PubMed] [Google Scholar]
- 5.Bradley P, Gordon NC, Walker TM, Dunn L, Heys S, Huang B, Earle S, Pankhurst LJ, Anson L, De Cesare M, Piazza P, Votintseva A, Golubchik T, Wilson D, Wyllie D, Diel R, Niemann S, Feuerriegel S, Kohl T, Ismail N, Omar S, Smith E, Buck D, McVean G, Walker A, Peto T, Crook D, Iqbal Z. 2015. Rapid antibiotic-resistance predictions from genome sequence data for Staphylococcus aureus and Mycobacterium tuberculosis. Nat Commun 6:10063. doi: 10.1038/ncomms10063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jia B, Raphenya AR, Alcock B, Waglechner N, Guo P, Tsang KK, Lago BA, Dave BM, Pereira S, Sharma AN, Doshi S, Courtot M, Lo R, Williams LE, Frye JG, Elsayegh T, Sardar D, Westman EL, Pawlowski AC, Johnson TA, Brinkman FSL, Wright GD, McArthur AG. 28 October 2016, posting date. CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res doi: 10.1093/nar/gkw1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Neuert S, Nair S, Day MR, Doumith M, Ashton PM, Mellor KC, Jenkins C, Hopkins KL, Woodford N, de Pinna E, Godbole G, Dallman TJ. 2018. Prediction of phenotypic antimicrobial resistance profiles from whole genome sequences of non-typhoidal Salmonella enterica. Front Microbiol 9:592. doi: 10.3389/fmicb.2018.00592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Anderson DJ, Miller B, Marfatia R, Drew R. 2012. Ability of an antibiogram to predict Pseudomonas aeruginosa susceptibility to targeted antimicrobials based on hospital day of isolation. Infect Control Hosp Epidemiol 33:589–593. doi: 10.1086/665721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McDermott PF, Tyson GH, Kabera C, Chen Y, Li C, Folster JP, Ayers SL, Lam C, Tate HP, Zhao S. 22 August 2016, posting date. The use of whole genome sequencing for detecting antimicrobial resistance in nontyphoidal Salmonella. Antimicrob Agents Chemother doi: 10.1128/AAC.01030-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pesesky MW, Hussain T, Wallace M, Patel S, Andleeb S, Burnham CAD, Dantas G. 2016. Evaluation of machine learning and rules-based approaches for predicting antimicrobial resistance profiles in Gram-negative Bacilli from whole genome sequence data. Front Microbiol 7:1887. doi: 10.3389/fmicb.2016.01887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nguyen M, Long SW, McDermott PF, Olsen RJ, Olson R, Stevens RL, Tyson GH, Zhao S, Davis JJ. 2018. Using machine learning to predict antimicrobial minimum inhibitory concentrations and associated genomic features for nontyphoidal Salmonella. bioRxiv https://www.biorxiv.org/content/10.1101/380782v2. [DOI] [PMC free article] [PubMed]
- 12.Davis JJ, Boisvert S, Brettin T, Kenyon RW, Mao C, Olson R, Overbeek R, Santerre J, Shukla M, Wattam AR, Will R, Xia F, Stevens R. 2016. Antimicrobial resistance prediction in PATRIC and RAST. Sci Rep 6:27930. doi: 10.1038/srep27930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Drouin A, Giguère S, Déraspe M, Marchand M, Tyers M, Loo VG, Bourgault AM, Laviolette F, Corbeil J. 2016. Predictive computational phenotyping and biomarker discovery using reference-free genome comparisons. BMC Genomics 17:754. doi: 10.1186/s12864-016-2889-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.World Health Organization. 2018. Salmonella (non-typhoidal) fact sheet. http://www.who.int/news-room/fact-sheets/detail/salmonella-(non-typhoidal).
- 15.World Health Organization. 2015. WHO estimates of the global burden of foodborne diseases: foodborne disease burden epidemiology reference group 2007-2015. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 16.Ao TT, Feasey NA, Gordon MA, Keddy KH, Angulo FJ, Crump JA. 2015. Global burden of invasive nontyphoidal Salmonella disease, 2010. Emerg Infect Dis 21:941. doi: 10.3201/eid2106.140999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Angulo FJ, Mølbak K. 2005. Human health consequences of antimicrobial drug–resistant Salmonella and other foodborne pathogens. Clin Infect Dis 41:1613–1620. doi: 10.1086/497599. [DOI] [PubMed] [Google Scholar]
- 18.Krueger AL, Greene SA, Barzilay EJ, Henao O, Vugia D, Hanna S, Meyer S, Smith K, Pecic G, Hoefer D, Griffin P. 2014. Clinical outcomes of nalidixic acid, ceftriaxone, and multidrug-resistant nontyphoidal Salmonella infections compared with pansusceptible infections in FoodNet sites, 2006–2008. Foodborne Pathog Dis 11:335–341. doi: 10.1089/fpd.2013.1642. [DOI] [PubMed] [Google Scholar]
- 19.Tyson GH, Zhao S, Li C, Ayers S, Sabo JL, Lam C, Miller RA, McDermott PF. 2017. Establishing genotypic cutoff values to measure antimicrobial resistance in Salmonella. Antimicrob Agents Chemother 61:e02140-16. doi: 10.1128/AAC.02140-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dhanani AS, Block G, Dewar K, Forgetta V, Topp E, Beiko RG, Diarra MS. 2015. Genomic comparison of non-typhoidal Salmonella enterica serovars Typhimurium, Enteritidis, Heidelberg, Hadar and Kentucky isolates from broiler chickens. PLoS One 10:e0128773. doi: 10.1371/journal.pone.0128773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Takahata S, Ida T, Hiraishi T, Sakakibara S, Maebashi K, Terada S, Muratani T, Matsumoto T, Nakahama C, Tomono K. 2010. Molecular mechanisms of fosfomycin resistance in clinical isolates of Escherichia coli. Int J Antimicrob Agents 35:333–337. doi: 10.1016/j.ijantimicag.2009.11.011. [DOI] [PubMed] [Google Scholar]
- 22.Gunn JS, Lim KB, Krueger J, Kim K, Guo L, Hackett M, Miller SI. 1998. PmrA–PmrB-regulated genes necessary for 4-aminoarabinose lipid A modification and polymyxin resistance. Mol Microbiol 27:1171–1182. doi: 10.1046/j.1365-2958.1998.00757.x. [DOI] [PubMed] [Google Scholar]
- 23.Misawa K, Tarumoto N, Tamura S, Osa M, Hamamoto T, Yuki A, Kouzaki Y, Imai K, Ronald RL, Yamaguchi T, Murakami T, Maesaki S, Suzuki Y, Kawana A, Maeda T. 2018. Single nucleotide polymorphisms in genes encoding penicillin-binding proteins in β-lactamase-negative ampicillin-resistant Haemophilus influenzae in Japan. BMC Res Notes 11:53. doi: 10.1186/s13104-018-3169-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shaaly A, Kalamorz F, Gebhard S, Cook GM. 2013. Undecaprenyl pyrophosphate phosphatase confers low-level resistance to bacitracin in Enterococcus faecalis. J Antimicrob Chemother 68:1583–1593. doi: 10.1093/jac/dkt048. [DOI] [PubMed] [Google Scholar]
- 25.Nishino K, Yamaguchi A. 2004. Role of histone-like protein H-NS in multidrug resistance of Escherichia coli. J Bacteriol 186:1423–1429. doi: 10.1128/jb.186.5.1423-1429.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Srinivasan VB, Vaidyanathan V, Mondal A, Rajamohan G. 2012. Role of the two component signal transduction system CpxAR in conferring cefepime and chloramphenicol resistance in Klebsiella pneumoniae NTUH-K2044. PLoS One 7:e33777. doi: 10.1371/journal.pone.0033777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rahmati S, Yang S, Davidson AL, Zechiedrich EL. 2002. Control of the AcrAB multidrug efflux pump by quorum-sensing regulator SdiA. Mol Microbiol 43:677–685. doi: 10.1046/j.1365-2958.2002.02773.x. [DOI] [PubMed] [Google Scholar]
- 28.Alekshun MN, Levy SB. 1997. Regulation of chromosomally mediated multiple antibiotic resistance: the mar regulon. Antimicrob Agents Chemother 41:2067–2075. doi: 10.1128/AAC.41.10.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O’Regan E, Quinn T, Pagès JM, McCusker M, Piddock L, Fanning S. 2009. Multiple regulatory pathways associated with high-level ciprofloxacin and multidrug resistance in Salmonella enterica serovar enteritidis: involvement of RamA and other global regulators. Antimicrob Agents Chemother 53:1080–1087. doi: 10.1128/AAC.01005-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pontel LB, Audero MEP, Espariz M, Checa SK, Soncini FC. 2007. GolS controls the response to gold by the hierarchical induction of Salmonella-specific genes that include a CBA efflux-coding operon. Mol Microbiol 66:814–825. doi: 10.1111/j.1365-2958.2007.05963.x. [DOI] [PubMed] [Google Scholar]
- 31.Nishino K, Senda Y, Yamaguchi A. 2008. CRP regulator modulates multidrug resistance of Escherichia coli by repressing the mdtEF multidrug efflux genes. J Antibiot 61:120. doi: 10.1038/ja.2008.120. [DOI] [PubMed] [Google Scholar]
- 32.Nagakubo S, Nishino K, Hirata T, Yamaguchi A. 2002. The putative response regulator BaeR stimulates multidrug resistance of Escherichia coli via a novel multidrug exporter system, MdtABC. J Bacteriol 184:4161–4167. doi: 10.1128/JB.184.15.4161-4167.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nishino K, Latifi T, Groisman EA. 2006. Virulence and drug resistance roles of multidrug efflux systems of Salmonella enterica serovar Typhimurium. Mol Microbiol 59:126–141. doi: 10.1111/j.1365-2958.2005.04940.x. [DOI] [PubMed] [Google Scholar]
- 34.Lomovskaya O, Lewis K. 1992. emr, an Escherichia coli locus for multidrug resistance. Proc Natl Acad Sci U S A 89:8938–8942. doi: 10.1073/pnas.89.19.8938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lomovskaya O, Lewis K, Matin A. 1995. emrR is a negative regulator of the Escherichia coli multidrug resistance pump emrAB. J Bacteriol 177:2328–2334. doi: 10.1128/jb.177.9.2328-2334.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Garvey MI, Baylay AJ, Wong RL, Piddock LJ. 2011. Overexpression of patA and patB, which encode ABC transporters, is associated with fluoroquinolone resistance in clinical isolates of Streptococcus pneumoniae. Antimicrob Agents Chemother 55:190–196. doi: 10.1128/AAC.00672-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hirakawa H, Nishino K, Hirata T, Yamaguchi A. 2003. Comprehensive studies of drug resistance mediated by overexpression of response regulators of two-component signal transduction systems in Escherichia coli. J Bacteriol 185:1851–1856. doi: 10.1128/JB.185.6.1851-1856.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bohn C, Bouloc P. 1998. The Escherichia coli cmlA gene encodes the multidrug efflux pump Cmr/MdfA and is responsible for isopropyl-β-D-thiogalactopyranoside exclusion and spectinomycin sensitivity. J Bacteriol 180:6072–6075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Magnet S, Courvalin P, Lambert T. 1999. Activation of the cryptic AAC(6’)-Iy aminoglycoside resistance gene of Salmonella by a chromosomal deletion generating a transcriptional fusion. J Bacteriol 181:6650–6655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Coyne S, Rosenfeld N, Lambert T, Courvalin P, Périchon B. 2010. Overexpression of resistance-nodulation-cell division pump AdeFGH confers multidrug resistance in Acinetobacter baumannii. Antimicrob Agents Chemother 54:4389–4393. doi: 10.1128/AAC.00155-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bauernfeind A, Stemplinger I, Jungwirth R, Giamarellou H. 1996. Characterization of the plasmidic beta-lactamase CMY-2, which is responsible for cephamycin resistance. Antimicrob Agents Chemother 40:221–224. doi: 10.1128/AAC.40.1.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Novotna G, Janata J. 2006. A new evolutionary variant of the streptogramin A resistance protein, Vga(A) LC, from Staphylococcus haemolyticus with shifted substrate specificity towards lincosamides. Antimicrob Agents Chemother 50:4070–4076. doi: 10.1128/AAC.00799-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roberts MC. 2005. Update on acquired tetracycline resistance genes. FEMS Microbiol Lett 245:195–203. doi: 10.1016/j.femsle.2005.02.034. [DOI] [PubMed] [Google Scholar]
- 44.Arcangioli MA, Leroy-Sétrin S, Martel JL, Chaslus-Dancla E. 1999. A new chloramphenicol and florfenicol resistance gene flanked by two integron structures in Salmonella typhimurium DT104. FEMS Microbiol Lett 174:327–332. doi: 10.1111/j.1574-6968.1999.tb13586.x. [DOI] [PubMed] [Google Scholar]
- 45.Daly M, Villa L, Pezzella C, Fanning S, Carattoli A. 2005. Comparison of multidrug resistance gene regions between two geographically unrelated Salmonella serotypes. J Antimicrob Chemother 55:558–561. doi: 10.1093/jac/dki015. [DOI] [PubMed] [Google Scholar]
- 46.Abouzeed YM, Baucheron S, Cloeckaert A. 2008. ramR mutations involved in efflux-mediated multidrug resistance in Salmonella enterica serovar Typhimurium. Antimicrob Agents Chemother 52:2428–2434. doi: 10.1128/AAC.00084-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Salverda ML, De Visser JAG, Barlow M. 2010. Natural evolution of TEM-1 β-lactamase: experimental reconstruction and clinical relevance. FEMS Microbiol Rev 34:1015–1036. doi: 10.1111/j.1574-6976.2010.00222.x. [DOI] [PubMed] [Google Scholar]
- 48.McArthur AG, Waglechner N, Nizam F, Yan A, Azad MA, Baylay AJ, Bhullar K, Canova MJ, De Pascale G, Ejim L, Kalan L, King A, Koteva K, Morar M, Mulvey M, O’Brien J, Pawlowski A, Piddock L, Spanogiannopoulos P, Sutherland A, Tang I, Taylor P, Thaker M, Wang W, Yan M, T Y, Wright G. 6 May 2013, posting date. The comprehensive antibiotic resistance database. Antimicrob Agents Chemother doi: 10.1128/AAC.00419-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Drouin A, Giguère S, Sagatovich V, Déraspe M, Laviolette F, Marchand M, Corbeil J. 2014. Learning interpretable models of phenotypes from whole genome sequences with the Set Covering Machine. arXiv 1412.1074 [q-bio.GN]. https://arxiv.org/abs/1412.1074.
- 50.Marchand M, Shawe-Taylor J. 2002. The set covering machine. J Mach Learn Res 3:723–746. http://www.jmlr.org/papers/volume3/marchand02a/marchand02a.pdf. [Google Scholar]
- 51.Public Health Agency of Canada. 2018. FoodNet Canada annual report 2017. Public Health Agency of Canada, Ottawa, Canada. [Google Scholar]
- 52.Timme RE, Pettengill JB, Allard MW, Strain E, Barrangou R, Wehnes C, Van Kessel JS, Karns JS, Musser SM, Brown EW. 2013. Phylogenetic diversity of the enteric pathogen Salmonella enterica subsp. enterica inferred from genome-wide reference-free SNP characters. Genome Biol Evol 5:2109–2123. doi: 10.1093/gbe/evt159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Drouin A, Hocking T, Laviolette F. 2017. Maximum margin interval trees, p 4947–4956. In Guyon I, Luxburg UV, Bengio S, Wallach H, Fergus R, Vishwanathan S, Garnett R (ed), Proceedings of Advances in Neural Information Processing Systems 30 (NIPS 2017). Neural Information Processing Systems, San Diego, CA. [Google Scholar]
- 54.Chung YJ, Saier MH. 2002. Overexpression of the Escherichia coli sugE gene confers resistance to a narrow range of quaternary ammonium compounds. J Bacteriol 184:2543–2545. doi: 10.1128/JB.184.9.2543-2545.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Giles W, Benson AK, Olson M, Hutkins RW, Whichard J, Winokur P, Fey PD. 2004. DNA sequence analysis of regions surrounding blaCMY-2 from multiple Salmonella plasmid backbones. Antimicrob Agents Chemother 48:2845–2852. doi: 10.1128/AAC.48.8.2845-2852.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.El-Halfawy OM, Klett J, Ingram RJ, Loutet SA, Murphy ME, Martín-Santamaría S, Valvano MA. 2017. Antibiotic capture by bacterial lipocalins uncovers an extracellular mechanism of intrinsic antibiotic resistance. mBio 8:e00225-17. doi: 10.1128/mBio.00225-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Naguib MM, Valvano MA. 2018. Vitamin E increases antimicrobial sensitivity by inhibiting bacterial lipocalin antibiotic binding. mSphere 3:e00564-18. doi: 10.1128/mSphere.00564-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dame-Korevaar A, Fischer EA, Stegeman A, Mevius D, van Essen-Zandbergen A, Velkers F, van der Goot J. 2017. Dynamics of CMY-2 producing E. coli in a broiler parent flock. Vet Microbiol 203:211–214. doi: 10.1016/j.vetmic.2017.03.024. [DOI] [PubMed] [Google Scholar]
- 59.Martin LC, Weir EK, Poppe C, Reid-Smith RJ, Boerlin P. 2012. Characterization of bla CMY-2 plasmids in Salmonella and Escherichia coli isolates from food animals in Canada. Appl Environ Microbiol 78:1285–1287. doi: 10.1128/AEM.06498-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Campos J, Mourão J, Silveira L, Saraiva M, Belo Correia C, Maçãs AP, Peixe L, Antunes P. 2017. P-262-extended-spectrum cephalosporin-resistant CMY-2-producing Salmonella Heidelberg and S. Minnesota in poultry meat imported into the European Union. In Congress of Microbiology and Biotechnology (MICROBIOTEC 2017), Escola Superior de Biotecnologia da Universidade Católica do Porto, 7–9 December 2017. http://hdl.handle.net/10400.18/4894.
- 61.Campos J, Mourão J, Silveira L, Saraiva M, Correia CB, Maçãs AP, Peixe L, Antunes P. 2018. Imported poultry meat as a source of extended-spectrum cephalosporin-resistant CMY-2-producing Salmonella Heidelberg and Salmonella Minnesota in the European Union, 2014–2015. Int J Antimicrob Agents 51:151–154. doi: 10.1016/j.ijantimicag.2017.09.006. [DOI] [PubMed] [Google Scholar]
- 62.Tiba-Casas MR, Camargo CH, Soares FB, Doi Y, Fernandes SA. 25 September 2018, posting date. Emergence of CMY-2-producing Salmonella Heidelberg associated with IncI1 plasmids isolated from poultry in Brazil. Microb Drug Resist doi: 10.1089/mdr.2018.0044. [DOI] [PubMed] [Google Scholar]
- 63.Madec JY, Haenni M, Nordmann P, Poirel L. 2017. Extended-spectrum β-lactamase/AmpC-and carbapenemase-producing Enterobacteriaceae in animals: a threat for humans? Clin Microbiol Infect 23:826–833. doi: 10.1016/j.cmi.2017.01.013. [DOI] [PubMed] [Google Scholar]
- 64.Espinosa RF, Rumi V, Marchisio M, Cejas D, Radice M, Vay C, Barrios R, Gutkind G, Di Conza J. 2018. Fast and easy detection of CMY-2 in Escherichia coli by direct MALDI-TOF mass spectrometry. J Microbiol Methods 148:22–28. doi: 10.1016/j.mimet.2018.04.001. [DOI] [PubMed] [Google Scholar]
- 65.Radford D, Strange P, Lepp D, Hernandez M, Rehman MA, Diarra MS, Balamurugan S. 2018. Genomic and proteomic analyses of Salmonella enterica serovar Enteritidis identifying mechanisms of induced de novo tolerance to ceftiofur. Front Microbiol 9:2123. doi: 10.3389/fmicb.2018.02123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Michael G, Schwarz S. 2016. Antimicrobial resistance in zoonotic nontyphoidal Salmonella: an alarming trend? Clin Microbiol Infect 22:968–974. doi: 10.1016/j.cmi.2016.07.033. [DOI] [PubMed] [Google Scholar]
- 67.Pezzella C, Ricci A, DiGiannatale E, Luzzi I, Carattoli A. 2004. Tetracycline and streptomycin resistance genes, transposons, and plasmids in Salmonella enterica isolates from animals in Italy. Antimicrob Agents Chemother 48:903–908. doi: 10.1128/AAC.48.3.903-908.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Silverman M, Zieg J, Hilmen M, Simon M. 1979. Phase variation in Salmonella: genetic analysis of a recombinational switch. Proc Natl Acad Sci U S A 76:391–395. doi: 10.1073/pnas.76.1.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lucarelli C, Dionisi AM, Filetici E, Owczarek S, Luzzi I, Villa L. 2012. Nucleotide sequence of the chromosomal region conferring multidrug resistance (R-type ASSuT) in Salmonella Typhimurium and monophasic Salmonella Typhimurium strains. J Antimicrob Chemother 67:111–114. doi: 10.1093/jac/dkr391. [DOI] [PubMed] [Google Scholar]
- 70.Maamar E, Alonso CA, Hamzaoui Z, Dakhli N, Abbassi MS, Ferjani S, Saidani M, Boubaker IBB, Torres C. 2018. Emergence of plasmid-mediated colistin-resistance in CMY-2-producing Escherichia coli of lineage ST2197 in a Tunisian poultry farm. Int J Food Microbiol 269:60–63. doi: 10.1016/j.ijfoodmicro.2018.01.017. [DOI] [PubMed] [Google Scholar]
- 71.Diarra MS, Delaquis P, Rempel H, Bach S, Harlton C, Aslam M, Pritchard J, Topp E. 2014. Antibiotic resistance and diversity of Salmonella enterica serovars associated with broiler chickens. J Food Protect 77:40–49. doi: 10.4315/0362-028.JFP-13-251. [DOI] [PubMed] [Google Scholar]
- 72.Matthews TC, Bristow FR, Griffiths EJ, Petkau A, Adam J, Dooley D, Kruczkiewicz P, Curatcha J, Cabral J, Fornika D, Winsor G, Courtot M, Bertelli C, Roudgar A, Feijao P, Mabon P, Enns E, Thiessen J, Keddy A, Isaac-Renton J, Gardy JL, Tang P, Consortium I, Carriço JA, Chindelevitch L, Chauve C, Graham MR, McArthur AG, Taboada EN, Beiko RG, Brinkman FS, Hsiao WW, Van Domselaar G. 2018. The Integrated Rapid Infectious Disease Analysis (IRIDA) platform. bioRxiv https://www.biorxiv.org/content/10.1101/381830v1.
- 73.Magoč T, Salzberg SL. 2011. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27:2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gurevich A, Saveliev V, Vyahhi N, Tesler G. 2013. QUAST: quality assessment tool for genome assemblies. Bioinformatics 29:1072–1075. doi: 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 77.Seemann T. 2017. ABRicate. Mass screening of contigs for antimicrobial resistance or virulence genes. https://github.com/tseemann/abricate.
- 78.Carattoli A, Zankari E, García-Fernández A, Larsen MV, Lund O, Villa L, Aarestrup FM, Hasman H. 2014. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother 58:3895–3903. doi: 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.McKinney W. 2011. pandas: a foundational Python library for data analysis and statistics. https://www.researchgate.net/publication/265194455_pandas_a_Foundational_Python_Library_for_Data_Analysis_and_Statistics.
- 80.Waskom M, Botvinnik O, O’Kane D, Hobson P, Lukauskas S, Gemperline DC, Augspurger T, Halchenko Y, Cole JB, Warmenhoven J, de Ruiter J, Pye C, Hoyer S, Vanderplas J, Villalba S, Kunter G, Quintero E, Bachant P, Martin M, Meyer K, Miles A, Ram Y, Yarkoni T, Williams ML, Evans C, Fitzgerald C, Brian Fonnesbeck C, Lee A, Qalieh A. 2017. mwaskom/seaborn: v0.8.1 (September 2017). doi: 10.5281/zenodo.883859. [DOI]
- 81.Yoshida CE, Kruczkiewicz P, Laing CR, Lingohr EJ, Gannon VP, Nash JH, Taboada EN. 2016. The Salmonella in silico typing resource (SISTR): an open Web-accessible tool for rapidly typing and subtyping draft Salmonella genome assemblies. PLoS One 11:e0147101. doi: 10.1371/journal.pone.0147101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kluyver T, Ragan-Kelley B, Pérez F, Granger BE, Bussonnier M, Frederic J, Kelley K, Hamrick JB, Grout J, Corlay S, Ivanov P, Avila D, Abdalla S, Willing C, Jupyter development team. 2016. Jupyter Notebooks—a publishing format for reproducible computational workflows. https://eprints.soton.ac.uk/403913/. [Google Scholar]
- 83.Torvalds L, Hamano J. 2010. Git: Fast version control system. http://git-scm.com.
- 84.Page AJ, Cummins CA, Hunt M, Wong VK, Reuter S, Holden MT, Fookes M, Falush D, Keane JA, Parkhill J. 2015. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics 31:3691–3693. doi: 10.1093/bioinformatics/btv421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Page AJ, Taylor B, Delaney AJ, Soares J, Seemann T, Keane JA, Harris SR. 29 April 2016, posting date. SNP-sites: rapid efficient extraction of SNPs from multi-FASTA alignments. Microb Genom doi: 10.1099/mgen.0.000056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol 32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Trifinopoulos J, Nguyen LT, von Haeseler A, Minh BQ. 2016. W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res 44:W232–W235. doi: 10.1093/nar/gkw256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kalyaanamoorthy S, Minh BQ, Wong TK, von Haeseler A, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods 14:587. doi: 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Huerta-Cepas J, Serra F, Bork P. 2016. ETE 3: reconstruction, analysis, and visualization of phylogenomic data. Mol Biol Evol 33:1635–1638. doi: 10.1093/molbev/msw046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hyatt D, Chen GL, LoCascio PF, Land ML, Larimer FW, Hauser LJ. 2010. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Buchfink B, Xie C, Huson DH. 2015. Fast and sensitive protein alignment using DIAMOND. Nat Methods 12:59. doi: 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- 92.Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, Blondel M, Prettenhofer P, Weiss R, Dubourg V, Vanderplas J, Passos A, Cournapeau D, Brucher M, Perrot M, Duchesnay É. 2011. Scikit-learn: machine learning in Python. J Mach Learn Res 12:2825–2830. [Google Scholar]
- 93.Lemaître G, Nogueira F, Aridas CK. 2017. Imbalanced-learn: a Python toolbox to tackle the curse of imbalanced datasets in machine learning. J Mach Learn Res 18:559–563. [Google Scholar]
- 94.Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv 1303.3997 [q-bio.GN]. https://arxiv.org/abs/1303.3997.
- 95.Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen G, Olson R, Osterman A, Overbeek R, McNeil L, Paarmann D, Paczian T, Parrello B, Pusch G, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O. 2008. The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Overbeek R, Olson R, Pusch GD, Olsen GJ, Davis JJ, Disz T, Edwards RA, Gerdes S, Parrello B, Shukla M, Vonstein V, Wattam A, Xia F, Stevens R. 2014. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Res 42:D206–D214. doi: 10.1093/nar/gkt1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
NCBI GenBank accession numbers and assembly metrics for all genomes (BioProject identifier PRJNA521409). Download Table S1, CSV file, 0.01 MB (14KB, csv) .
© Crown copyright 2019.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Full MIC results for all isolates and their CLSI breakpoint SIR status. Download Table S2, XLSX file, 0.01 MB (14.1KB, xlsx) .
© Crown copyright 2019.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
(A to G) Observed number of resistant isolates by serotype. Antimicrobials are presented in the same order for all panels as indicated by axis label on the bottommost panels. (H) Cooccurrence pattern of resistance across all serotypes. This UpSet plot shows the different sets of resistances observed across the isolates. The top bar plot indicates how many isolates were resistant to each specific group of antibiotics. The corresponding set of antibiotics is indicated below the top bar plot by the dark colored dots. The bar plot to the left shows how frequently resistance to each antibiotic occurred regardless of which set of resistances it appeared within. Download FIG S1, PDF file, 0.1 MB (147.6KB, pdf) .
© Crown copyright 2019.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Hierarchical clustering of observed phenotypic AST results. Black cells indicate susceptibility, while red and cream cells represent intermediate susceptibility and resistance, respectively. The serotypes are indicated along the left axis per the legend. Each row represents a single isolate and each column a single antibiotic tested. Isolates and antibiotics are ordered according to their similarity to one another as inferred by a pair of hierarchical clustering inferences as shown by the pair of annotated dendrograms. The antibiotics tested were amoxicillin-clavulanic acid (AMOCLA), ampicillin (AMPICI), azithromycin (AZITHR), cefoxitin (CEFOXI), ceftiofur (CEFTIF), ceftriaxone (CEFTRI), chloramphenicol (CHLORA), ciprofloxacin (CIPROF), gentamicin (GENTAM), nalidixic acid (NALAC), streptomycin (STREPT), sulfamethoxazole (SULFIZ), tetracycline (TETRA), and trimethoprim-sulfamethoxazole (TRISUL). Download FIG S2, PDF file, 0.04 MB (41.9KB, pdf) .
© Crown copyright 2019.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Visualization of the N50 distribution of assembled genomes. This figure shows that the assemblies of genomes that were consistently mispredicted in terms of AMR for TETRA/STREPT and the β-lactam antibiotics tested are not notably more fragmented than the assemblies as a whole. Download FIG S3, PDF file, 0.03 MB (29.9KB, pdf) .
© Crown copyright 2019.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
CMY-2. The blaCMY-2 genes were typically flanked by ISEcp1 downstream and lipocalin (blc) on the same strand followed by a quaternary ammonium compound(s) (sugE) on the opposite strand. Download FIG S4, PDF file, 0.08 MB (79.2KB, pdf) .
© Crown copyright 2019.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Visualization of the relative levels of coverage of plasmids within the genome assemblies as detected using abricate and the plasmidfinder database. Download FIG S5, PDF file, 0.04 MB (46.8KB, pdf) .
© Crown copyright 2019.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Plasmid prediction results from the genomes studied (determined via the plasmidFinder database and abricate). Download Table S3, CSV file, 0.01 MB (5.9KB, csv) .
© Crown copyright 2019.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Relative risk calculations (determined using SAS-implemented chi-square tests) for the subset of AMR genes identified in the predictive modeling and literature as key drivers of resistance. Download Table S4, CSV file, 0.01 MB (585B, csv) .
© Crown copyright 2019.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data Availability Statement
All data used in this study are available in GenBank (BioProject identifier PRJNA521409). The full list of accession numbers per genome can be found in Table S1. The code used to perform all analyses is available in the git repository https://github.com/fmaguire/salmonella_ast_prediction/.