Abstract
DNA methylation acts at the interface of genetic and environmental factors relevant for autism spectrum disorder (ASD). Placenta, normally discarded at birth, is a potentially rich source of DNA methylation patterns predictive of ASD in the child. Here, we performed whole methylome analyses of placentas from a prospective study MARBLES (Markers of Autism Risk in Babies—Learning Early Signs) of high-risk pregnancies. A total of 400 differentially methylated regions (DMRs) discriminated placentas stored from children later diagnosed with ASD compared to typically developing controls. These ASD DMRs were significantly enriched at promoters, mapped to 596 genes functionally enriched in neuronal development, and overlapped genetic ASD risk. ASD DMRs at CYP2E1 and IRS2 reached genome-wide significance, replicated by pyrosequencing and correlated with expression differences in brain. Methylation at CYP2E1 associated with both ASD diagnosis and genotype within the DMR. In contrast, methylation at IRS2 was unaffected by within DMR genotype but modified by preconceptional maternal prenatal vitamin use. This study therefore identified two potentially useful early epigenetic markers for ASD in placenta.
Introduction
Autism spectrum disorder (ASD) is a heterogeneous neurodevelopmental disorder diagnosed by a combination of behavioral features including restricted interests, repetitive behaviors, language deficits and impairments in social communication (1). In the United States, 1 in 59 children are diagnosed with ASD, at a mean age of 4.2 years (1). ASD is currently diagnosed by clinicians trained on the Autism Diagnostic Observation Schedule (ADOS) and the Autism Diagnostic Interview—Revised (ADI-R) according to the Statistical Manual of Mental Disorders (DSM-5), which is most accurate at or after 36 months (1). However, an early assessment of ASD risk could identify infants and toddlers who would benefit from behavioral interventions that improve cognitive, social and language skills.
Monozygotic versus dizygotic twin and sibling studies suggest a strong genetic basis for ASD (2–4). However, mutations in any individual gene account for less than 1% of ASD cases (2,5). Genetic sequencing analyses can only identify a potentially causative genetic abnormality in ~ 25% of clinical ASD diagnoses (2,5,6). While genome-wide association studies (GWAS) also support common genetic variants in ASD, the complexity and heterogeneity of ASD have been a major challenge (7–9). Evidence for environmental risk factors in ASD points to in utero maternal exposures such as air pollution, fever or asthma and nutrients, specifically the absence of preconceptional prenatal vitamin intake (10–13). Maternal prenatal vitamins, which contain high levels of folate and other additional B vitamins, protect offspring by up to 70% for neural tube defects (14–19) and correlate with an overall 40% reduction in ASD risk if taken during the first month of pregnancy (P1) (12,13,20). This finding was replicated with a large prospective study in Norway including over 80 000 pregnancies (20), and more recently in the MARBLES high risk cohort (21).
DNA methylation shows dynamic changes during fetal development (22–24) and contains the molecular memory of in utero experiences such as maternal nutrition (15,25). The term placenta is an accessible fetal tissue that maintains the distinctive embryonic bimodal DNA methylation pattern, in which expressed genes are marked by higher methylation levels (24,26). The placenta therefore offers a unique window to study DNA methylation patterns that may reflect altered fetal development relevant to ASD genetic risk (24,27,28). Specifically, a recent study of polygenic risk scores for schizophrenia demonstrated a significant interaction of genetic risk with maternal perinatal environmental factors that affected placental gene expression (29). Previous analyses of DNA methylation patterns in placenta samples from a high-risk ASD cohort also identified an association between self-reported use of lawn and garden pesticides and large-scale changes in DNA methylation patterns and identified a putative enhancer of the DLL1 gene as differentially methylated in ASD (30,30).
Here, we continue the epigenetic investigation of ASD risk through the novel approach of identifying differentially methylated regions (DMRs) in whole methylomes from placenta samples from male children later diagnosed with ASD compared to children with typical development (TD) controls. Two genome-wide significant ASD-associated DMRs at CYP2E1 and IRS2 were further validated and investigated for effects of genotype, RNA expression and protein levels as well as interactions with preconception prenatal vitamin use. Understanding the epigenetic patterns of ASD associated with maternal prenatal vitamin use in placenta could lead to the development of preventative and therapeutic early interventions for high-risk children with ASD.
Results
Placenta ASD DMRs discriminate ASD from TD samples
To identify novel differentially methylated gene loci between ASD and TD, a DMR bioinformatic analysis was performed on placenta whole-genome bisulfite sequencing (WGBS) data (30,31). A total of 400 DMRs were identified with a threshold of >10% methylation difference between ASD and TD groups, and these were associated with 596 genes using the Genomics Regions Enrichment of Annotations Tool (GREAT) on the default association settings (Fig. 1A, Supplementary Material, Table 1). There was no bias for gene length in the ASD DMR associated genes compared to all human genes (Supplementary Material, Fig. 1). Hypermethylated were 296 DMRs, while 104 DMRs were hypomethylated in ASD compared to TD placenta (Fig. 1A). Principal component analysis (PCA) using methylation levels for each sample over the 400 DMRs demonstrated a clear separation of placental samples by child outcome of ASD versus TD (Fig. 1B). In addition, most ASD DMRs showed a highly significant association with Mullen Scales of Early Learning (MSEL) scores and autism severity score from ADOS but not with potential confounding variables (Supplementary Material, Table 2, Supplementary Material, Fig. 2). Demographic and clinical variables of children and their mothers in the MARBLES (Markers of Autism Risk in Babies—Learning Early Signs) study were not significantly associated with child diagnosis (Supplementary Material, Table 3). Furthermore, there was no significant enrichment between placental cell type-specific genes from single-cell RNA-seq data (31) and genes identified associated with ASD DMRs in placenta, suggesting that the differences were not due to major shifts in cell-type composition (Supplementary Material, Fig. 3, Supplementary Material, Table 4).
Figure 1.
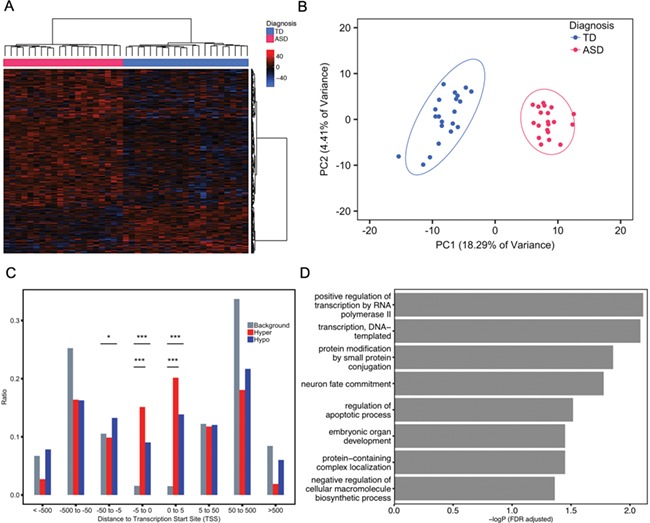
DMRs in placenta distinguished ASD from TD child outcomes. (A) Heatmap and hierarchical clustering of 20 ASD versus 21 TD placental samples using methylation levels in the 400 identified ASD DMRs. Percent methylation for each sample relative to the mean methylation at each ASD DMR was plotted as a heatmap, with black representing no difference, hyper-methylation (red) and hypo-methylation (blue). Columns were clustered by child outcome, ASD (red) or TD (blue), while rows were clustered by methylation direction. (B) PCA of TD versus ASD placental samples on the basis of methylation at 400 ASD DMRs. Ellipses represent the 95% confidence interval for each group. The non-overlapping ellipses showed a significant difference between ASD and TD for these DMRs’ methylation level (P < 0.05). (C) Location relative to genes for hypermethylated (red) or hypomethylated (blue) ASD DMRs compared to background (grey). Distributions of locations relative to transcription start sites (TSS) are shown on the x-axis. The ratio plotted on the y-axis is calculated by the number of genes at each binned location divided by the total number of genes (Supplementary Material, Table 5). *P < 0.05, **P < 0.01, ***P < 0.001 by Fisher’s exact test. (D) Bar graph represents the significant results from gene ontology and pathway enrichment analysis of ASD DMRs associated genes compared to background by Fisher’s exact test (FDR adjusted -log P-value, x-axis).
Placenta ASD DMRs were enriched for transcription start sites and genes that function in transcriptional regulation and neuronal fate
To further study the location and function of ASD DMRs in placenta, we calculated the location of each ASD DMR relative to the assigned gene’s transcription start site (TSS) (Supplementary Material, Table 5). Both hyper- and hypomethylated ASD DMRs were enriched within 5 kb on either side of TSS compared to background regions (Fig. 1C). Gene ontology (GO) analysis of ASD DMRs genes revealed significant enrichment for functions in transcription, protein modification, embryonic organ development and neuron fate commitment by Fisher’s exact test after false discovery rate (FDR) multiple test correction (Fig. 1D,Supplementary Material, Table 6).
Placenta DMR genes were enriched in ASD but not ID risk genes
To test a hypothesized overlap between epigenetic and genetic ASD risk loci observed previously in ASD and neurodevelopmental disorder brain tissues (33), we investigated the possible overlap of placenta ASD DMR genes with identified genetic risk factors for ASD and other types of intellectual disability (ID). First, the curated Simons Foundation Autism Research Initiative (SFARI) gene list was separated into six categories based on SFARI ASD gene scores (34). The entire list of SFARI genes as well as the high-confidence gene list both showed significant overlap with placenta ASD DMR genes (Fig. 2A, Supplementary Material, Table 7). The 39 genes in common between the SFARI gene list and placenta ASD DMRs were significantly enriched for functions in positive regulation of histone H3K4 methylation, multicellular organ development and system development. Second, high-risk ASD genes from Sanders et al. (8) and likely gene-disrupting (LGD) recurrent mutations and missense mutation on de novo mutations to ASD gene lists from whole-genome exome sequencing (9) were also significantly enriched for placental ASD DMRs. In contrast, no significant enrichment was observed for placental ASD DMRs with ID, Alzheimer’s disease or lung cancer genetic risk (35) or a random set of 400 genomic regions mapped to 600 genes (Fig. 2A, Supplementary Material, Table 7). When placental ASD DMRs were separated by direction of change, hypomethylated ASD DMRs exhibited more categories of significant enrichment with ASD genetic risk compared to hypermethylated ASD DMRs (Supplementary Material, Fig. 4). Differentially expressed genes in ASD postmortem brain were extracted from multiple studies (36–39). Placenta ASD DMR genes significantly overlapped with ASD cortex differentially expressed genes in the largest of these studies (39) (Supplementary Material, Fig. 5, Supplementary Material, Table 8).
Figure 2.
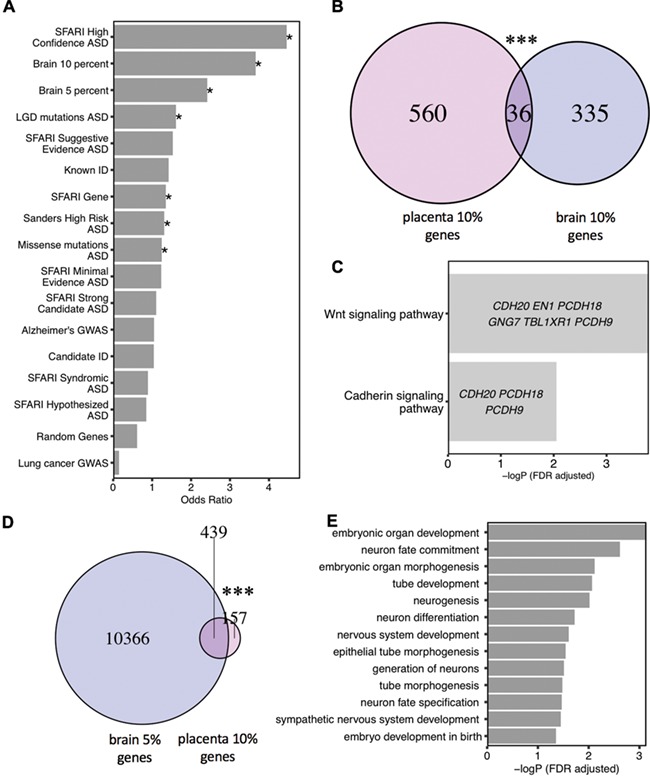
Placenta ASD DMR genes overlapped with ASD DMR associated genes from postmortem brain and known genetic risk for ASD but not for other disorders. (A) Placenta ASD DMR associated genes were compared for significant overlap with ASD DMR genes identified from ASD postmortem brain (33) (based on 10% or 5% methylation difference cutoffs), as well as multiple curated gene lists of ASD, ID or unrelated disorder genetic risks or a randomly generated gene list (*P < 0.05 FDR corrected two-tailed Fisher’s exact test, ranked by odds ratio). SFARI: Simons Foundation Autism Research Initiative (34), LGD: likely gene-disrupting mutation, ASD: autism spectrum disorder, Alzheimer: Alzheimer’s Disease, ID: intellectual disability. (B) Venn diagram represents the significant overlap of 36 genes associated with placenta ASD DMRs and brain ASD DMRs based on 10% methylation differences in brain between ASD versus TD by Fisher’s exact test (P < 0.001***) (Supplementary Material, Table 7). Methylation data from human postmortem brain was obtained from previous published data sets, GSE8154 (ASD and TD) (33). (C) GO and pathway analysis on the 36 genes in common between placenta ASD DMRs and brain ASD DMRs associated genes. Enrichment tests were done on Fisher’s exact test with FDR 0.05 correction. Genes in each GO term are shown within each bar. (D) Venn diagram represents significant overlap of 439 genes between placenta ASD DMRs and DMRs from brain with 5% methylation difference between ASD and TD (Fisher’s exact test, P < 0.001***) (Supplementary Material, Table 7). (E) Multiple developmental pathways were significantly enriched on the overlapped 439 genes with Fisher’s exact test after FDR 0.05 correction.
Placenta ASD DMR genes significantly overlapped with brain ASD DMRs that were enriched at Wnt and cadherin signaling pathways and developmental functions
From a prior methylation analysis in brain frontal cortex, 210 ASD discriminating DMRs were identified (10% methylation difference between ASD and TD), which mapped to 371 genes (33). A significant overlap of ASD DMR genes was observed between placenta and brain (Fisher’s exact test, P < 0.001), with 36 genes in common at a 10% difference in ASD versus TD brain (Fig. 2B, Supplementary Material, Table 7). Those 36 genes were significantly enriched for functions in the Wnt signaling and cadherin signaling pathways (Fig. 2C). Of these shared genes, four overlapped with SFARI genetic risk: GADD45B, MC4R, PCDH9 and TBL1XR1. With a more inclusive 5% methylation difference in brain that has been described previously (33,40), 74% of the placental ASD DMR genes significantly overlapped with those identified in brain (Fisher’s exact test, P < 0.001) (Fig. 2D). Pathways significantly associated with this more extended list included neuronal fate commitment, neurogenesis and neuronal differentiation, as well as embryonic developmental terms such as tube development (Fig. 2E). Among shared 439 genes, 32 genes were also identified as SFARI ASD genetics risk genes, including AUTS2, KMT2A, SETBP1 and TBR1 (Supplementary Material, Table 7).
Placenta ASD DMRs were enriched for placental and brain active promoter H3K4me3 peaks, promoter flanking regions and CpG shores
To functionally annotate the ASD DMRs identified by placenta WGBS, multiple histone modification ChIP-seq peaks and chromatin state predictions from multiple tissue types in the Roadmap Epigenomics Projects were compared to ASD DMR chromosomal locations for enrichment compared to background regions (41). Placental ASD DMRs were significantly enriched at epigenomic regulatory regions, specifically H3K4me3 and H3K4me1 marks across multiple tissues, although placental H3K4me3 marks showed the strongest (odds ratio = 17.08, FDR q < 1.8E-42), and brain H3K4me3 marks showed the second strongest enrichment (odds ratio = 13.75, FDR q < 3.55E-31) (Fig. 3A). Importantly, we also found a significant overlap between genes we identified by placenta ASD DMR and those identified as differential in ASD prefrontal neuron H3K4me3 peaks (Fisher’s exact test, P < 0.05) (Supplementary Material, Fig. 6) (41). Next, we overlapped ASD DMRs with published chromatin state predictions that use histone modification ChIP-seq data to annotate the genome into 15 functional states (chromHMM) (43). Placental ASD DMRs showed significant enrichment in regions flanking TSS (TssAFlnk) and active transcription start site (TssA) compared to background over multiple tissues (Fig. 3B). When separated by directional change in ASD, both hyper- and hypomethylated ASD DMRs were significantly enriched for H3K4me3 peaks, transcription start sites and their flanking regions, as well as enhancers (Supplementary Material, Fig. 7).
Figure 3.
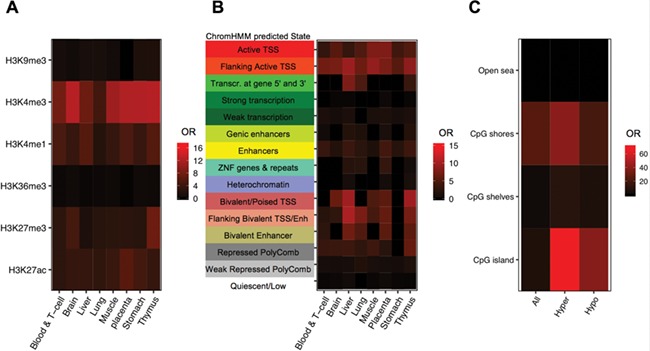
Placenta ASD DMRs were enriched at H3K4me3 regions, flanking promoter regions and CpG shores. (A) Placenta ASD DMRs were examined for enrichment with histone modification ChIP-seq peaks from the Epigenome Roadmap using the LOLA package. Enrichments are plotted as the odds ratio in a heat map for each of eight different tissue types and six types of modified histone marks (107). (B) Enrichment tests on chromatin states from chromHMM categories in the Epigenome Roadmap and placental ASD DMRs from this study were performed using LOLA, with each row representing a different ChromHMM predicted state and each column a single tissue type. (C) Placenta ASD DMRs (categorized as all, hypermethylated or hypomethylated in ASD) were tested for enrichment based on CpG island location. The human genome was separated into CpG islands, CpG shores, CpG shelves and open sea.
We also separated the genome into four parts relative to CpG island location (44–46). CpG shores were defined as the region within 2 kb on both sides of CpG islands, while another 2 kb extension from the shores were defined as CpG shelves. The remaining regions were defined as ‘open sea’. Placental ASD DMRs showed significant enrichment at CpG shores, and hypermethylated ASD DMRs more significantly overlapped CpG islands compared to hypomethylated DMRs (Fig. 3C).
Two genome-wide significant placental ASD DMRs at CYP2E1 and IRS2 validate by pyrosequencing and correlated with gene expression
Of the 400 ASD DMRs identified in ASD placenta, 2 reached genome-wide significance by family-wide error rate (FWER), including chr10: 133527713–133529507, located inside CYP2E1 (cytochrome P450 2E1), and chr13: 109781111–109782389, located inside IRS2 (insulin receptor substrate 2) (Fig. 4). The CYP2E1 DMR was located after the first exon, included the first intron and part of the second exon, and was hypomethylated in ASD versus TD (Fig. 4A). The IRS2 DMR spanned the end of the first exon and the beginning of first intron and was hypermethylated in ASD versus TD (Fig. 4B). Both CYP2E1 and IRS2 were also present in the gene lists overlapping with brain ASD DMR related genes and high-risk ASD genes (Fig. 2A, Supplementary Material, Table 7) (8).
Figure 4.
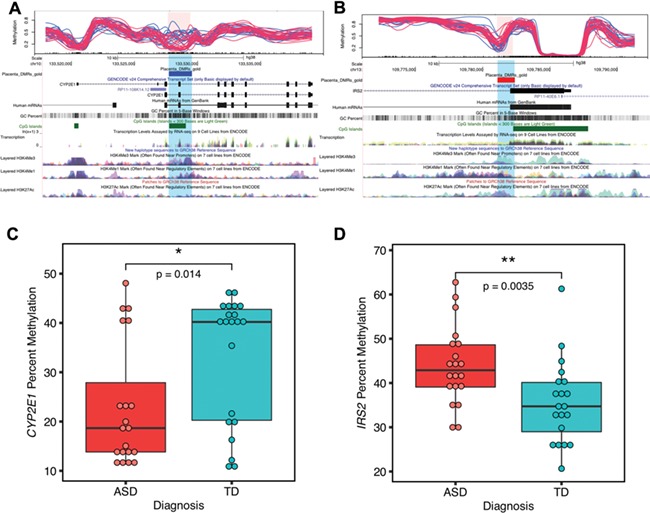
Two genome-wide significant placental DMRs located at CYP2E1 and IRS2 were validated by pyrosequencing. (A) and (B) show the location relative to genes and CpG islands of the two genome-wide significant DMRs (highlighted in pink and blue) in the UCSC Genome Browser. In the upper tracks, each line represents percent methylation (y-axis) of a single individual by WGBS analysis. Blue lines represent TD, and red lines represent ASD samples. (A) Hypomethylated DMR at CYP2E1 with 10 kb upstream and 10 kb downstream. (B) Hypermethylated DMR at IRS2 with 10 kb upstream and 10 kb downstream. (C) The CYP2E1 DMR percent methylation was significantly associated with child outcome and verified by pyrosequencing (two-tailed t-test, P = 0.014). The y-axis represents the average percent DNA methylation across the DMR regions from pyrosequencing. Each dot represented one sample. (D) Pyrosequencing validation on IRS2 DMR’s methylation with child outcome (two-tailed t-test, P = 0.0035). *P < 0.05, **P < 0.01.
Pyrosequencing was performed as an independent method to verify methylation differences between ASD and TD placental samples at CYP2E1 and IRS2 DMRs (Supplementary Material, Table 9). For the CYP2E1 DMR, there was a significant difference in average percent methylation detected by pyrosequencing between ASD and TD samples (Fig. 4C). A total of 13 CpG sites were included in the CYP2E1 DMR pyrosequencing test, and all but 2 also showed individually significant differences between ASD and TD after FDR correction (Supplementary Material, Table 9, Supplementary Material, Fig. 8A). Pyrosequencing also confirmed a significant difference between ASD and TD average percent methylation at the IRS2 DMR (Fig. 4D), and all of the 11 CpG sites individually assayed at IRS2 (Supplementary Material, Table 9, Supplementary Material, Fig. 8B). There were no significance differences in the percent methylation variances of ASD and TD samples on both CYP2E1 DMR (two-tailed F-test, P = 0.8) and IRS2 DMR (two-tailed F-test, P = 0.7). To test the relevance of these loci to females, 10 female placental samples were also verified by pyrosequencing and showed a significance difference between ASD and TD in percent methylation at CYP2E1 and IRS2 DMRs in the same direction as that observed in males (Supplementary Material, Fig. 9).
While MARBLES placenta samples were not collected in a manner conducive to RNA stability for gene expression analyses, we were able to examine expression level of both CYP2E1 and IRS2 in MARBLES umbilical cord blood during the same time period from an Affymetrix Human Gene 2.0 array analysis in a related study (47). No significant differences by ASD diagnosis were observed in cord blood CYP2E1 or IRS2 expression or placental IRS2 protein levels, although trends were consistent with the direction of methylation differences (Supplementary Materials, Figs 10–11). Furthermore, both CYP2E1 and IRS2 were previously identified as differentially expressed genes in ASD brain from three previous studies (36–38) as well as in a meta-analysis of ASD compared to TD in human male cortex (dbMDEGA database) (48) (Supplementary Material, Fig. 5, Supplementary Material, Table 8). Together, these results suggest that methylation differences identified in ASD placenta are also relevant to differential gene expression patterns identified in brain.
CYP2E1 but not IRS2 DMR methylation levels were associated with common variants within each DMR
We performed Sanger sequencing within the CYP2E1 and IRS2 ASD DMRs to identify single-nucleotide polymorphisms (SNPs) located inside each DMR that could explain some of the methylation differences. Two SNPs (rs943975, rs1536828) were identified within the boundaries of the CYP2E1 DMR in the 41 placenta samples (Supplementary Material, Table 10). A significant association between rs1536828 (but not rs943975) genotype and CYP2E1 DMR percent methylation was observed, with samples homozygous for the minor allele (G/G) showing the lowest methylation (Fig. 5A). This significant genotype–methylation association remained when the minor (GG) and heterozygous (CG) CYP2E1 genotypes were combined and compared to the major (CC) samples (Supplementary Material, Fig. 12). Further confirming the effect of CYP2E1 genotype on methylation, five placental samples of each rs1536828 genotype showed a similar significant association of genotype with CYP2E1 DMR percent methylation by pyrosequencing (Supplementary Material, Fig. 13). A single informative SNP (rs9301411) was also identified within the IRS2 DMR (Supplementary Material, Table 10) but was not significantly associated with methylation level (Fig. 5B). However, genotypes at rs943975 (minor allele frequency: 0.4), rs1536828 (minor allele frequency: 0.32) and rs9301411 (minor allele frequency: 0.08) were not significantly associated with diagnosis. In addition, these SNPs were not located inside any known transcription factor motifs identified by MEME (Multiple Expectation maximization for Motif Elicitation) (49), and there were no differences on the MEME motif structure on the samples with and without SNPs (Supplementary Material, Fig. 14).
Figure 5.
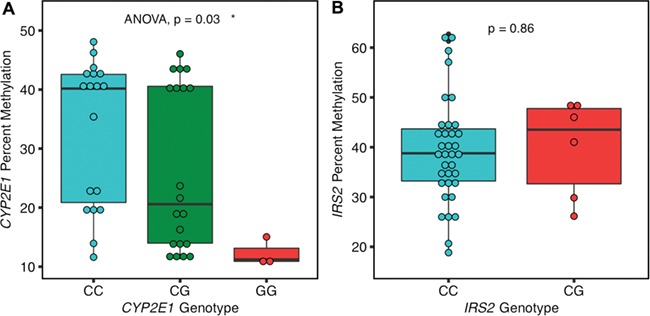
Cis genotype was significantly associated with CYP2E1 but not IRS2 DMR methylation levels.( A) CYP2E1 genotype at rs1536828 within the ASD DMR was significantly associated with CYP2E1 DMR average percent methylation tested by ANOVA (P = 0.03). (B) IRS2 genotype at rs9301411 within the ASD DMR was not significantly associated with IRS2 DMR methylation by two-tailed t-test (P = 0.86).
Preconception prenatal vitamin use corresponded to protective placental DNA methylation patterns at CYP2E1, IRS2 and genome wide
Placental samples from mothers who took prenatal vitamins during the first month of pregnancy showed a trend for higher CYP2E1 DMR methylation that was not significant but in the same direction expected for protection from ASD (Fig. 6A). At the IRS2 DMR, however, there was a significant association with maternal prenatal vitamin use and lower methylation, also in ASD-protective direction (Fig. 6B). The first month of pregnancy was chosen given it was associated with reduced risk for ASD in this MARBLES cohort (21); examination of the association with methylation at the IRS2 DMR for prenatal vitamin intake at other months indicated that the months near conception were most significant (Supplementary Material, Fig. 15).
Figure 6.
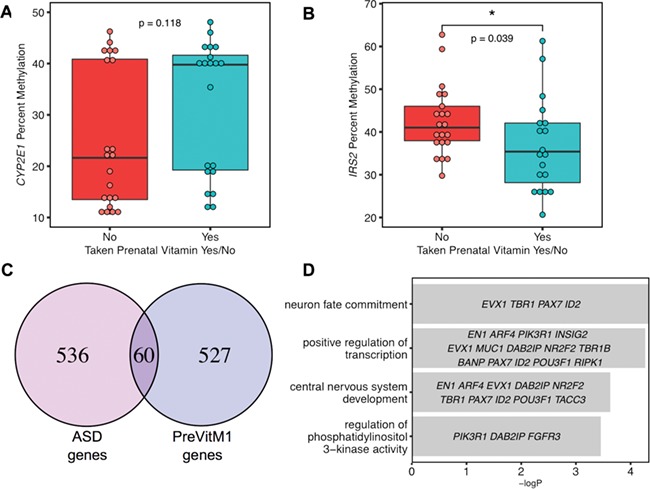
Preconception prenatal vitamin use was a significant modifier of IRS2 methylation and associated DMRs overlapped ASD DMRs in placenta. For (A) and (B), the x-axis represents maternal prenatal vitamins use during the first month of pregnancy, while the y-axis shows the percent methylation. (A) CYP2E1 DMR methylation was not significantly altered by P1 prenatal vitamin use. (B) Higher percent methylation at IRS2 DMR was significantly associated with not taking prenatal vitamins at P1 (two-tailed t-test, P = 0.039), which is in the same direction as ASD risk. (C) DMRs identified based on P1 prenatal vitamins use were associated with 587 genes, which showed a significant overlap with ASD DMR associated genes (Fisher’s exact test, P < 2.528e-16). (D) GO and pathway analysis were performed on the overlapped gene list (60 genes) (Supplementary Material, Table 11) between placenta ASD DMR and P1 prenatal vitamin DMR-associated genes for enrichment by Fisher’s exact test with -log (P-value) represented on the x-axis. Genes in each GO term are shown within each bar.
To further explore the relationship between placental methylation patterns influenced by prenatal vitamin use in the first month of pregnancy, placental methylomes were analyzed for DMRs by prenatal vitamin use in the first month of pregnancy (PreVitM1) with more than 10% methylation difference, and 376 DMRs were identified in over 587 genes (Supplementary Material, Table 11). A total of 60 genes overlapped between PreVitM1 DMRs and ASD DMRs in placenta (Supplementary Material, Table 11, Fig. 6C). GO analysis showed that genes common to PreVitM1 and ASD DMRs were significantly enriched for functions in neuron fate commitment, transcription regulation, central nervous system development and regulation of phosphatidylinositol 3-kinase activity (Fig. 6D).
To further investigate the potential inter-relatedness of diagnosis, prenatal vitamin use and cis genotype on methylation at CYP2E1 and IRS2 DMRs, we calculated associations between each factor and methylation separately by two-tailed t-test or analysis of variance (ANOVA), as well as two-way diagnosis and PreVitM1; diagnosis and genotype; genotype and PreVitM1 by Pearson’s chi-squared test. No significant association was found among each of the three factors and each DMR methylation level by ANOVA (Supplementary Material, Table 12).
Discussion
This study utilized whole methylome analysis of prospectively stored placenta samples in a high-risk ASD cohort to bioinformatically identify novel gene loci that may be useful for understanding and predicting ASD risk. This unbiased analysis of ASD DMRs in placenta tissue resulted in several novel findings. First, the 596 genes identified from 400 placental ASD DMRs significantly overlapped with genetic risk for ASD from curated databases and gene functions in neurons. Second, two genome-wide significant placental ASD DMRs at CYP2E1 and IRS2 were discovered that were validated by pyrosequencing and also overlapped with ASD-associated genetic variation and gene expression changes. Lastly, we investigated genotype and nutrient factors correlating with methylation at CYP2E1 and IRS2, demonstrating specific effects for cis genotype and diagnosis at CYP2E1 and prenatal vitamin use at IRS2. These results therefore suggest that DNA methylation patterns in placenta provide a link between genetics, environment and fetal epigenetic programming, which could reflect early development relevant to the complex etiology of ASD. The epigenomic signature of ASD in placenta also provides important insights into gene functions, pathways, gene–environment interactions and potential biomarkers that may be useful in improving early detection of ASD.
This study is the first to identify 400 potential ASD DMRs that distinguish between ASD and TD placenta samples and highlights specific locations and gene functions of differential methylation in placental samples from children with ASD. First, these placental ASD DMRs were highly enriched around transcription start sites and H3K4me3 marks that are clear marks of gene regulatory functions (50,51). Furthermore, GO analysis of the 596 genes mapped to placental ASD DMRs pointed to enriched gene functions in transcription, neuron fate and embryonic development, which were expected based on previous studies (31,52,53).
Genes with ASD DMRs in both placenta and brain were enriched for Wnt and cadherin signaling pathways. Wnt signaling is important in embryogenesis, tissue regeneration and neurodevelopment (54), while cadherin signaling plays a vital role in connecting major intracellular signaling pathways with adhesion protein complexes (55,55). Our results therefore complement previous studies that have shown the importance of Wnt and cadherin pathways in the etiology of ASD (57–59). We also replicated our previous finding of differential methylation at DLL1 in ASD placenta (31) (Supplementary Materials, Tables 1 and 7). DLL1 encodes a ligand of Notch, activated by Wnt signaling (60).
When overlapped with data sets of genetic risk for neurodevelopmental disorders including ASD and ID (8,9,34), placenta ASD DMRs were significantly enriched for ASD but not for ID genetic risk, illustrating the specificity of the ASD DMRs identified in our study. The highest overlap of ASD DMR associated genes was with the SFARI high-confidence genes, including KMT2A, MYT1L and TBR1 (34).
Our study identified novel methylation differences at CYP2E1 and IRS2, which exhibited genome-wide significant differences between ASD and TD. Both CYP2E1 and IRS2 are identified as ASD genetic risk genes in multiple databases related to ASD genetic risk across different tissues and populations (8,33). Both CYP2E1 and IRS2 DMRs are located close to the TSS site at CpG shore intragenic regions, which is also consistent with the enrichment for TSS flanking regions and H3K4me3 promoter marks in the 400 ASD DMRs. In our prior studies of differential methylation associated with UBE3A duplication in Dup15q syndrome, a genetic cause of ASD, both CYP2E1 and IRS2 were identified with differential methylation and H3K4me3 peaks in a Dup15q cell line model and differential methylation in Dup15q brain (40,61). Furthermore, CYP2E1 and IRS2 both showed differential H3K4me3 marks at their promoters in ASD versus control in the prefrontal cortex (42). Structural variants and SNPs in cis-regulatory elements also showed significant contribution to ASD (62,63). In contrast, neither CYP2E1 nor IRS2 were included as significant differential splicing events for ASD (36,39).
CYP2E1 encodes a member of the cytochrome P450 superfamily that is involved in the metabolism of drugs (64). Previous studies showed those proteins essential for embryonic development in human, rat and zebrafish (65–69). We observed a significant association between methylation and cis genotype at the CYP2E1 DMR, a finding that is consistent with the identification of this locus in a screen for human metastable epialleles variability between individuals (70). In addition, in immune models of ASD, maternal interleukin-6 (IL6) crosses the placenta, disrupting development of hippocampal spatial learning (71–73). Previous studies showed that IL6 inhibits CYP1A1, CYP1A2 and CYP2E1 expression (74–77), consistent with the lower methylation and expression levels in ASD versus TD observed in our study. In addition, CYP2E1 expression is transcriptionally regulated by the JAK2/STAT3 pathway, providing a potential convergent pathway with IRS2 (Fig. 7) (74).
Figure 7.
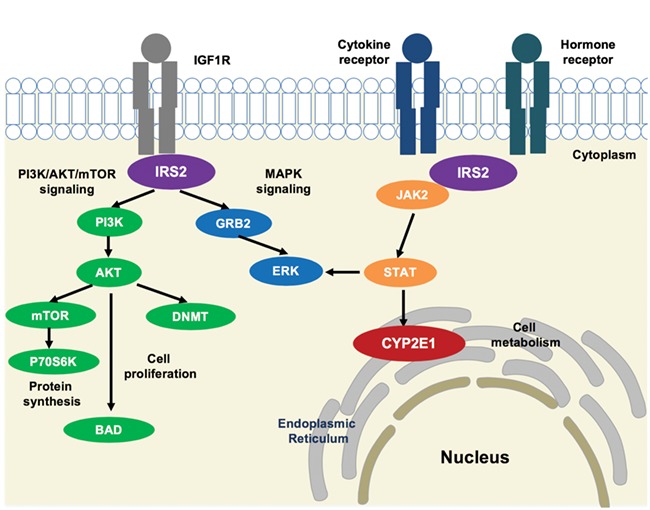
Potential pathway convergence of proteins encoded by both ASD DMRs. IRS2 interacts with transmembrane protein IGF1R at the intracellular membrane, resulting in activation of the PI3K/AKT/mTOR and MAPK signaling pathways involved in protein synthesis, cell proliferation and gene expression (81). An AKT-mediated ubiquitin pathway leads to de novo DNA methylation changes by DNMT (84). IRS2 also interacts with cytokine and hormone receptors and induces JAK2/STAT3 signaling (81,83). STAT activation leads to CYP2E1 localization at the endoplasmic reticulum, changing cellular metabolism (74).
IRS2 encodes for insulin receptor substrate 2, a cytoplasmic signaling molecule that mediates the effects of insulin and insulin-like growth factor 1 (IGF1) (78) and cytokine receptors (79). A GWAS noise reduction method to correct for false–positive association with ASD identified cadherin and signaling transduction pathways that included IRS2 as high-confidence ASD genes (80). IRS2 has a phosphotyrosine-binding domain that contributes to the intracellular affinity to cell membrane receptors (Fig. 7) (79). PI3K/AKT/mTOR and MAPK signaling pathways are linked with IRS2 in the regulation of protein synthesis and cell proliferation (81). Furthermore, a previous study using a reverse pathway genetic approach identified the MAPK signaling pathway associated with ASD (82). When activated by cytokine and hormone receptors, IRS2 stimulates JAK2, leading to STAT and MAPK signaling activation (81,83). The IGF1 pathway, which includes IRS2, also mediates de novo DNA methylation by DNA methyltransferase (DNMT) through AKT (84). This pathway may explain why methylation at the IRS2 DMR was sensitive to maternal prenatal vitamin intake, since IRS2 stimulates the mTOR (mechanistic target of rapamycin) pathway, which responds to nutrients and growth factors signaling to regulate protein synthesis (83). The link between epigenetic alterations in IRS2 and risk for ASD is particularly intriguing given a growing body of epidemiologic evidence demonstrating higher ASD risk in offspring born to mothers who experienced diabetes during pregnancy, including some very large and methodologically sound studies with clinical diagnoses of both maternal diabetes and child ASD (85).
We did not observe any significant associations between other potential cofounders such as maternal age, pregnancy BMI (Body Mass Index), gestational age or pre-eclampsia with ASD diagnosis in the MARBLES study (30). Cell-type heterogeneity in the placenta may complicate the interpretation of our results; however, placental ASD DMRs were not significantly associated with cell-type markers, and our previous study did not detect differences in methylation levels by placental region at specific gene loci (31). This study serves as a proof-of-principle that placenta methylation patterns detected by WGBS may be informative in ASD. Replication with additional samples, on female samples, and other similar prospective cohorts, and improved sequencing and bioinformatic strategies will be important in future studies. Approaches such as methylation quantitative trait loci analysis will be important in future studies with larger sample sizes for identifying possible common genetic variants associated with placental ASD DMRs. In addition, functional analyses on JAK, mTOR and MAPK signaling pathways in ASD placenta and brain would be important to provide mechanical insight.
In conclusion, we identified two high-confidence genes differentially methylated in ASD from an unbiased analysis of DNA methylation in placenta from high-risk pregnancies and investigated possible genetic and environmental modifiers of methylation at both loci. Methylation levels at the CYP2E1 DMR were associated with genotype, while the methylation levels at the IRS2 DMR were associated with prenatal vitamin use. Our results are consistent with a previous study using the Illumina 450 K array, which showed that both genetic and environmental effects influence DNA methylation levels (4). Placenta reflects the essential interface between the fetus and mother, mediating the impacts of endocrine and growth factors in the maternal environment on fetal development (86,87). Both CYP2E1 and IRS2 are related to protein synthesis, cell proliferation and cell metabolism, consistent with previous studies of convergent gene pathways in ASD (8,33,36,48,88). These results therefore provide evidence that placental methylation levels reflect the intersection of genetic and environmental risk and protective factors that are expected to be useful for early intervention and prevention of ASD.
Materials and Methods
MARBLES study design, sample selection and DNA isolation
The MARBLES study design was described in a previous publication (89). In MARBLES, mothers of at least one child with confirmed ASD who were pregnant or planning a pregnancy were recruited in the Northern California area. Inclusion criteria for the study were 1) mother or father has one or more biological child(ren) with ASD; 2) mother is 18 years or older; 3) mother is pregnant; 4) mother speaks, reads and understands English sufficiently to complete the protocol, and the younger sibling will be taught to speak English; and 5) mother lives within 2.5 hours of the Davis/Sacramento, California region at time of enrollment. With shared genetics, the next child has a 13-fold higher risk for developing ASD compared to the general population (89). Demographic, diet, lifestyle, environmental and medical information were prospectively collected through telephone-assisted interviews and mailed questionnaires throughout pregnancy and the postnatal period. Infants received standardized neurodevelopmental assessments beginning at 6 months and concluding at 3 years of age (89). Diagnostic assessments at 3 years included the gold standard ADOS (90), the ADI-R (91) and the MSEL (92). Participants were classified into outcome groups including ASD and TD, based on a previously published algorithm that uses ADOS and MSEL scores (93,94). Children with ASD outcomes have scores over the ADOS cutoff and meet DSM-5 criteria for ASD. Children with TD outcomes have all MSEL scores within 2.0 SD (Standard Deviation) and no more than one MSEL subscale that is 1.5 SD below the normative mean and scores on the ADOS at least three more points below the ASD cutoff. This study utilized 41 male MARBLES placenta samples, including 20 samples from children later diagnosed with ASD and 21 children determined to have TD, matched for enrollment time frame and date of birth. DNA was isolated from 50–100 mg frozen placental tissues (20 ASD and 21 TD) using the Gentra Puregene tissue kit (Qiagen, Germantown, MD).
WGBS
Raw sequencing data (fastq files) were published previously (31). Briefly, WGBS libraries were made with the sonicated genomic DNA (around 300 bp) and ligated with methylated Illumina (San Diego, CA) adapters using NEB’s NEBNext DNA library prep kit (26,31). The library was bisulfite converted using EZ DNA Methylation lighting kit (Zymo, Irvine, CA), amplified for 12 cycles using PfuTurbo Cx Hotstart DNA Polymerase (Agilent, Santa Clara, CA) and purified with Agencourt AMPure XP Beads (Beckman Coulter, Brea, CA). The quality and quantity of libraries were measured on a Bioanalyzer (Agilent, Santa Clara, CA) and sequenced on Illumina HiSeq 2000 with each sample per single lane. Reads after trimming were uniquely mapped to human reference genome (hg38) as described previously using BS-Seeker2 on average 1.6X genome converge with 99.3% bisulfite conversion efficiency (measured through the percentage of non-CpG cytosines that were unconverted) (26,40,95).
ASD DMRs and genome-wide significant DMRs
DMRs were called as described in previous publications (40,96) using the default settings. In this case, each ASD DMR contained greater than 10% methylation difference between ASD and TD samples at least three CpGs within 300 base pairs and a P-value of <0.05. Background regions were defined using the same conditions as DMRs but without any percent methylation filters to identify all possible DMR locations based on CpG density and sample sequencing coverage. Hypermethylated ASD DMRs were defined as higher percent methylation in ASD versus TD, while hypomethylated ASD DMRs were defined as lower percent methylation in ASD versus TD samples. Genome-wide, significant DMRs were identified based on a FWER < 0.05, determined by permuting the samples 1000× by chromosome and counting the number of null permutations with equal or better DMRs ranked by number of CpGs and areaStat (97).
Hierarchical clustering and PCA
Methylation was extracted at each ASD DMR for every sample. Percent methylation of each sample was normalized to the mean methylation of each ASD DMR. ASD DMRs were grouped by Ward’s method of hierarchical clustering (98). PCA was performed on methylation at all ASD DMRs across all samples using the prcomp function and ggbiplot package in R. The ellipses for each group were illustrated as the 95% confidence interval. The lack of overlapping ellipses for ASD and TD samples indicated significant methylation difference in ASD DMRs between groups (P < 0.05).
Placental cell type-specific genes identification
Cell type-specific genes were extracted from Vento-Tormo et al. (32) on 38 different types of cells with the top 30 cell type-specific signature genes. Genes for each cell types were filtered as unique genes that were only expressed in specific cell types. A test for significance of overlapping genes was done using Fisher’s exact test.
Assignment of DMRs to genes and relative location to TSS
Genes were assigned to DMRs using the GREAT on the default association settings (5.0 kb upstream and 1.0 kb downstream, up to 1000.0 kb max extension) (99). The distance (kb) was calculated from the ASD DMRs, hypermethylated ASD DMRs, hypomethylated ASD DMRs and background regions to TSS of the GREAT assigned gene. The gene length was calculated for both placental ASD DMR genes and all genes in human genome and tested for potential distribution differences by Pearson’s chi-squared test.
GO term and pathway enrichment analysis
GO analysis was done using Protein Analysis Through Evolutionary Relationships overrepresentation test, with the GO Ontology database (100,101) and Fisher’s exact test with FDR multiple test correction. GO term enrichments were presented by the hierarchical terms rather than specific subclass functional classes, as described previously (102,103).
Tests for ASD DMR enrichments
All tests of enrichment for ASD DMRs were compared to a set of all possible background regions that are calculated in the DMR analysis pipeline. Enrichment tests for placenta ASD DMRs associated genes and published gene lists were done using the GeneOverlap R package that implements Fisher’s exact test and adjusted for FDR correction (104). *P < 0.05, **P < 0.01, ***P < 0.001 by Fisher’s exact test with FDR corrected. Brain cortex (BA9) ASD DMRs were defined as either a 5% or 10% methylation difference between ASD and TD and were described previously using the same method as placenta ASD DMRs (33). The SFARI database was used for the five categories of ASD risk genes (34). High-effect ASD risk gene lists were also identified from Sanders et al. (8). LGD recurrent ASD mutations and missense mutation on de novo mutations were obtained from Iossifov et al. (9). Gene lists on ID were obtained from Gilissen et al. (35). Alzheimer’s disease GWAS gene lists were extracted from SNPs showing association with Alzheimer’s disease (P ≤ 1 × 10−3) (105). Lung cancer GWAS gene lists were acquired from Landi et al. (106). The random genes category contains the same number of regions as the placenta ASD DMRs to serve as a specificity control. ASD DMRs were examined for enrichment with known chromatin marks compared to the background using LOLA R package with two-tailed Fisher’s exact test after FDR correction (107). Placenta histone marks H3K4me1, H3K4me3, H4K9me3, H3K36me3, H3K27me3 and H3K27ac were extracted from Encyclopedia of DNA Elements placenta ChIP-seq data set (108,109). ASD DMRs were also analyzed for overlap with chromatin states predicted by chromHMM, which use histone modification ChIP-seq data to separate the genome into 15 functional states in the Roadmap Epigenomics Project using a Hidden Markov Model (41,110). For promoters, chromHMM separates active TSS (TssA), TSS flank (TssAFlnk), bivalent TSS (TssBiv) and bivalent TSS flank (BivFlnk) states. For enhancers, genic enhancer (EnhG), enhancer (Enh) and bivalent enhancer (EnhBiv) are the different states. Human CpG island locations were extracted from UCSC Genome Browser (111). CpG island shores were defined as 2 kb flanking regions on both sides of CpG island. CpG island shelf was characterized as 2 kb flanking regions on both sides of CpG island shore, not including CpG island or CpG island shore. CpG island ‘open sea’ includes all genomic regions except CpG island, CpG island shore and CpG island shelf. A custom R script was used to generate the locations of CpG islands (https://github.com/Yihui-Zhu/AutismPlacentaEpigenome).
Pyrosequencing
Genomic DNA (500 ng) was bisulfite converted using the EZ DNA Methylation kit (Zymo, Irvine, CA). Amplification and sequencing primers were designed using the PyroMark Assay Design Software 2.0 (Qiagen, Germantown, MD). DMRs were amplified using the PyroMark PCR (Polymerase Chain Reaction) kit (Qiagen, Germantown, MD). Pyrosequencing of 13 CpG sites at CYP2E1 gene and 11 CpG sites in human IRS2 gene was performed in triplicate. Pyrosequencing was performed on a Pyromark Q24 Pyrosequencer (Qiagen, Germantown, MD) with the manufacturer’s recommended protocol. Enzyme, substrate and dNTPs (deoxyribonucleotide triphosphate) were from the Pyromark Gold Q24 Reagents (Qiagen, Germantown, MD), and the methylation levels were analyzed using Pyromark Q24 software.
CYP2E1-related DMR pyrosequencing primers:
Forward: GGTGTTTTGTTTTGGGGTTGA
Reverse: ACCCATTCAATATTCACAACAATC (5′ Biotin)
Sequencing: GGTTGATGATGGGGA
Amplification region: chr10: 133527817–133527938 (hg38).
IRS2-related DMR pyrosequencing primers:
Forward: TTAGGAATATAGGGAAAGGTGAAAGT
Reverse: CCACCCATTCACCCATTCTA (5′ Biotin)
Sequencing: GGGAAAGGTGAAAGTT
Amplification region: chr13: 109781623–109781794 (hg38).
Gene expression in umbilical cord blood
Gene expression data assayed by Affymetrix Human Gene 2.0 array were extracted from a previous publication on umbilical cord blood from subjects in the MARBLES study (GEO ID: GSE123302) (47). Placenta and cord blood were collected at the same time period in the same study. Raw intensity values from cord blood samples were normalized by RMA (Robust Multi-array Average), and data from 70 male samples were extracted, including 30 ASD and 40 TD samples. Normalized expression was examined at the only probe annotated to CYP2E1 (16711001) and the only probe annotated to IRS2 (16780917). Analysis was done on those two probes with 70 samples on the normalized matrix data.
Western blot
In western blot experiments, placental proteins were isolated with RIPA (Radioimmunoprecipitation assay) buffer containing 10 mm Tris-Cl (pH 8.0), 1 mm EDTA, 1% Triton X-100, 0.1% sodium deozydholate, 0.1% SDS, 140 mm NaCl, 1 mm PMSF and complete protease inhibitors (ThermoFisher, Waltham, MA), incubated at 37°C for 30 min, sonicated and heated at 95°C for 5 min. Bicinchoninic acid protein assay (ThermoFisher, Waltham, MA) was used to determine protein concentration. Protein samples (20–30 ug) were resolved on 4–20% tris-gylcine polyacrylamide gels (Biorad, Hercules, CA). Proteins were separated and transferred to nitrocellulose membranes for 60 min at a constant voltage of 100. The membranes were blocked in Odyssey Blocking Buffer [PBS (phosphate-buffered saline)] (Licor, Lincoln, NE, 927–40 000) for 40 min. Anti-IRS2 (1:5000, Cell Signaling, Danvers, MA, 3089S) and anti-GAPDH (Glyceraldehyde 3-phosphate dehydrogenase) (1:10000, Advanced Immunochemical, Inc., Long Beach, CA, 2-RGM2) were incubated with the membrane with Odyssey Blocking Buffer containing 0.2% Tween overnight at 4°C. Membranes were washed with 1× PBS containing 0.2% Tween and then incubated with secondary antibodies, IRDye 800CW Donkey anti-Mouse IgG (1:50000, Licor, Lincoln, NE, 926–32 212) and IRDye 680RD Donkey anti-Rabbit IgG (1:50000, Licor, Lincoln, NE, 926–68 073) for 1 h. Membranes were scanned using a Licor(Lincoln, NE) Odyssey infrared imaging system based on the manufacturer’s guidance (with resolution: 84; quality: medium, 600-channel: 6; 800-channel: 5). Relative protein quantification was done using the ImageJ software program (112) in densitometry mode. IRS2 signals were normalized to GAPDH for each sample.
Sanger sequencing
PCR amplification was performed on each sample using PCR 10× buffer, 25 mm MgCl2, 5 m betaine, 10 mm dNTPs, DMSO, and HotStart Taq (Qiagen, Germantown, MD). Each PCR program was unique to the region being amplified with specific primers. The PCR product was then resolved by gel electrophoresis using a 1% Agarose gel in 1× TE (Tris and EDTA) to later be extracted using the gel extraction kit (Qiagen, Germantown, MD) based on the default protocol. After DNA quantitation by NanoDrop, the samples were sent to the UC Davis Sequencing Facility for sequencing on the 3730 Genetic Analyzer (ThermoFisher, Waltham, MA) with DNA Sequencing Analysis software v.5.2 (ThermoFisher, Waltham, MA). The sequencing results were assembled and analyzed using CodonCode Aligner version 7.0 (CodonCode).
CYP2E1-related SNP (rs943975, rs1536828) primers:
Forward: CTACAAGGCGGTGAAGGAAG
Reverse: CCCATCCCCATAAACTCTCC.
IRS2-related SNP (rs943975) primers:
Forward: TTAGGAATATAGGGAAAGGTGAAAGT
Reverse: CCACCCATTCACCCATTCTA.
Code availability
Custom scripts for WGBS analysis are available at https://github.com/kwdunaway/WGBS_Tools with the instructions. Custom Scripts for DMR finder are available at https://github.com/cemordaunt/DMRfinder with the instructions. The rest of code and scripts for each figure and tables are available at https://github.com/Yihui-Zhu/AutismPlacentaEpigenome.
Data availability
WGBS data were previously published, Gene Expression Omnibus (GEO) accession number GSE67615 (31). The rest of the relevant data and information are included in supplementary tables.
Supplementary Material
Acknowledgements
We would like to thank members of the LaSalle lab and UCD Children’s Center for Environmental Health for helpful discussions and the MARBLES study participants.
Conflict of Interest statement. The authors declare no competing interests.
Funding
National Institute of Health (R01 ES025574, R01 ES029213, P01 ES011269, R01ES020392, NIH-UL1-TR000002); Environmental Protection Agency (83543201); Department of Defense (AR110194); Intellectual and Developmental Disability Research Centers (U54HD079125); Environmental Health Support Center (P30ES023513). This work used the Vincent J. Coates Genomics Sequencing Laboratory at UC Berkeley, supported by National Institute of Health S10 Instrumentation Grants (S10RR029668, S10RR027303). STAR grant RD-83329201 from the US Environmental Protection Agency (EPA); and grants R24ES028533, R01ES028089, R01ES020392, R01ES025574, P01ES011269.
Reference
- 1. Baio J., Wiggins L., Christensen D.L., Maenner M.J., Daniels J., Warren Z., Kurzius-Spencer M., Zahorodny W., Robinson Rosenberg C., White T. et al. (2018) Prevalence of autism spectrum disorder among children aged 8 years—autism and developmental disabilities monitoring network, 11 sites, United States, 2014. MMWR. Surveill. Summ., 67,–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Tsai P.C. and Bell J.T. (2015) Power and sample size estimation for epigenome-wide association scans to detect differential DNA methylation. Int. J. Epidemiol., 44,1429–1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wessels W.H. and Pompe van Meerdervoort M. (1979) Monozygotic twins with early infantile autism. A case report. S. Afr. Med. J., 55, 955–957. [PubMed] [Google Scholar]
- 4. Hannon E., Knox O., Sugden K., Burrage J., Wong C.C.Y., Belsky D.W., Corcoran D.L., Arseneault L., Moffitt T.E., Caspi A. et al. (2018) Characterizing genetic and environmental influences on variable DNA methylation using monozygotic and dizygotic twins. PLoS Genet., 14, e1007544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bourgeron T. (2015) From the genetic architecture to synaptic plasticity in autism spectrum disorder. Nat. Rev. Neurosci., 16, 551–563. [DOI] [PubMed] [Google Scholar]
- 6. Turner T.N., Hormozdiari F., Duyzend M.H., McClymont S.A., Hook P.W., Iossifov I., Raja A., Baker C., Hoekzema K., Stessman H.A. et al. (2016) Genome sequencing of autism-affected families reveals disruption of putative noncoding regulatory DNA. Am. J. Hum. Genet., 98, 58–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Grove J., Ripke S., Als T.D., Mattheisen M., Walters R., Won H., Pallesen J., Agerbo E., Andreassen O.A., et al. (2019) Identification of common genetic risk variants for autism spectrum disorder. Nature Genetics, 51, 431–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sanders S.J., He X., Willsey A.J., Ercan-Sencicek A.G., Samocha K.E., Cicek A.E., Murtha M.T., Bal V.H., Bishop S.L., Dong S. et al. (2015) Insights into autism spectrum disorder genomic architecture and biology from 71 risk loci. Neuron, 87, 1215–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Iossifov I., O’Roak B.J., Sanders S.J., Ronemus M., Krumm N., Levy D., Stessman H.A., Witherspoon K.T., Vives L., Patterson K.E. et al. (2014) The contribution of de novo coding mutations to autism spectrum disorder. Nature, 515, 216–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Raz R., Roberts A.L., Lyall K., Hart J.E., Just A.C., Laden F. and Weisskopf M.G. (2015) Autism spectrum disorder and particulate matter air pollution before, during, and after pregnancy: a nested case–control analysis within the Nurses’ Health Study II Cohort. Environ. Health Perspect., 123, 264–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zerbo O., Iosif A.-M., Walker C., Ozonoff S., Hansen R.L. and Hertz-Picciotto I. (2013) Is maternal influenza or fever during pregnancy associated with autism or developmental delays? Results from the CHARGE (CHildhood Autism Risks from Genetics and Environment) study. J. Autism Dev. Disord., 43, 25–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Schmidt R.J., Hansen R.L., Hartiala J., Allayee H., Schmidt L.C., Tancredi D.J., Tassone F. and Hertz-Picciotto I. (2011) Prenatal vitamins, one-carbon metabolism gene variants, and risk for autism. Epidemiology, 22, 476–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Schmidt R.J., Tancredi D.J., Ozonoff S., Hansen R.L., Hartiala J., Allayee H., Schmidt L.C., Tassone F. and Hertz-Picciotto I. (2012) Maternal periconceptional folic acid intake and risk of autism spectrum disorders and developmental delay in the CHARGE (CHildhood Autism Risks from Genetics and Environment) case–control study. Am. J. Clin. Nutr., 96, 80–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Relton C.L., Wilding C.S., Laffling A.J., Jonas P.A., Burgess T., Binks K., Tawn E.J. and Burn J. (2004) Low erythrocyte folate status and polymorphic variation in folate-related genes are associated with risk of neural tube defect pregnancy. Mol. Genet. Metab., 81, 273–281. [DOI] [PubMed] [Google Scholar]
- 15. Howsmon D.P., Kruger U., Melnyk S., James S.J. and Hahn J. (2017) Classification and adaptive behavior prediction of children with autism spectrum disorder based upon multivariate data analysis of markers of oxidative stress and DNA methylation. PLoS Comput. Biol., 13,–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rush E.C., Katre P. and Yajnik C.S. (2014) Vitamin B12: one carbon metabolism, fetal growth and programming for chronic disease. Eur. J. Clin. Nutr., 68, 2–7. [DOI] [PubMed] [Google Scholar]
- 17. Kalkbrenner A.E., Schmidt R.J. and Penlesky A.C. (2014) Environmental chemical exposures and autism spectrum disorders: a review of the epidemiological evidence. Curr. Probl. Pediatr. Adolesc. Health Care, 44, 277–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Caramaschi D., Sharp G.C., Nohr E.A., Berryman K., Lewis S.J., Davey Smith G. and Relton C.L. (2017) Exploring a causal role of DNA methylation in the relationship between maternal vitamin B12 during pregnancy and child’s IQ at age 8, cognitive performance and educational attainment: a two-step Mendelian randomization study. Hum. Mol. Genet., 26, 3001–3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zeisel S.H. (2009) Importance of methyl donors during reproduction. Am. J. Clin. Nutr., 89, 673S–677S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Suren P., Roth C., Bresnahan M., Haugen M., Hornig M., Hirtz D., Lie K.K., Lipkin W.I., Manus P., Reichborn-Kjennerud T. et al. (2013) Association between maternal use of folic acid supplements and risk of autism spectrum disorders in children. J. Am. Med. Assoc., 309, 570–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Schmidt R.J., Iosif A.-M., Guerrero Angel E., Ozonoff S. (2019) Association of maternal prenatal vitamin use with risk for autism spectrum disorder recurrence in young siblings. JAMA psychiatry. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Vogel Ciernia A. and LaSalle J. (2016) The landscape of DNA methylation amid a perfect storm of autism aetiologies. Nat. Rev. Neurosci., 17, 411–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Crawley J.N., Heyer W.D. and LaSalle J.M. (2016) Autism and cancer share risk genes, pathways, and drug targets. Trends Genet., 32, 139–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Smallwood S.A. and Kelsey G. (2012) De novoDNA methylation: a germ cell perspective. Trends Genet., 28, 33–42. [DOI] [PubMed] [Google Scholar]
- 25. Teh A.L., Pan H., Chen L., Ong M.L., Dogra S., Wong J., MacIsaac J.L., Mah S.M., McEwen L.M., Saw S.M. et al. (2014) The effect of genotype and in utero environment on interindividual variation in neonate DNA methylomes. Genome Res., 24, 1064–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Schroeder D.I., Blair J.D., Lott P., Yu H.O.K., Hong D., Crary F., Ashwood P., Walker C., Korf I., Robinson W.P. et al. (2013) The human placenta methylome. Proc. Natl. Acad. Sci. U. S. A., 110, 6037–6042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Schroeder D.I., Jayashankar K., Douglas K.C., Thirkill T.L., York D., Dickinson P.J., Williams L.E., Samollow P.B., Ross P.J., Bannasch D.L. et al. (2015) Early developmental and evolutionary origins of gene body DNA methylation patterns in mammalian placentas. PLoS Genet., 11,–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Watson E.D. and Cross J.C. (2005) Development of structures and transport functions in the mouse placenta. Physiology, 20, 180–193. [DOI] [PubMed] [Google Scholar]
- 29. Ursini G., Punzi G., Chen Q., Marenco S., Robinson J.F., Porcelli A., Hamilton E.G., Mitjans M., Maddalena G., Begemann M. et al. (2018) Convergence of placenta biology and genetic risk for schizophrenia. Nat. Med., 24, 792–801. [DOI] [PubMed] [Google Scholar]
- 30. Schmidt R.J., Schroeder D.I., Crary-Dooley F.K., Barkoski J.M., Tancredi D.J., Walker C.K., Ozonoff S., Hertz-Picciotto I. and LaSalle J.M. (2016) Self-reported pregnancy exposures and placental DNA methylation in the MARBLES prospective autism sibling study. Environ. Epigenet., 2,dvw024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Schroeder D.I., Schmidt R.J., Crary-Dooley F.K., Walker C.K., Ozonoff S., Tancredi D.J., Hertz-Picciotto I. and LaSalle J.M. (2016) Placental methylome analysis from a prospective autism study. Mol. Autism, 7, 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Vento-Tormo R., Efremova M., Botting R.A., Turco M.Y., Vento-Tormo M., Meyer K.B., Park J.-E., Stephenson E., Polański K., Goncalves A. et al. (2018) Single-cell reconstruction of the early maternal–fetal interface in humans. Nature, 563, 347–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Vogel Ciernia A., Laufer B.I., Dunaway K.W., Mordaunt C.E., Coulson R.L., Yasui D.H. and LaSalle J.M. (2018) Epigenomic convergence of genetic and immune risk factors in autism brain. bioRxiv , 10.1101/270827. [DOI] [PMC free article] [PubMed]
- 34. Abrahams B.S., Arking D.E., Campbell D.B., Mefford H.C., Morrow E.M., Weiss L.A., Menashe I., Wadkins T., Banerjee-Basu S. and Packer A. (2013) SFARI Gene 2.0: a community-driven knowledgebase for the autism spectrum disorders (ASDs). Mol. Autism, 4, 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gilissen C., Hehir-Kwa J.Y., Thung D.T., van de Vorst M., van Bon B.W.M., Willemsen M.H., Kwint M., Janssen I.M., Hoischen A., Schenck A. et al. (2014) Genome sequencing identifies major causes of severe intellectual disability. Nature, 511, 344–347. [DOI] [PubMed] [Google Scholar]
- 36. Voineagu I., Wang X., Johnston P., Lowe J.K., Tian Y., Horvath S., Mill J., Cantor R.M., Blencowe B.J. and Geschwind D.H. (2011) Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature, 474, 380–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gupta S., Ellis S.E., Ashar F.N., Moes A., Bader J.S., Zhan J., West A.B. and Arking D.E. (2014) Transcriptome analysis reveals dysregulation of innate immune response genes and neuronal activity-dependent genes in autism. Nat. Commun., 5, 5748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gandal M.J., Haney J.R., Parikshak N.N., Leppa V., Ramaswami G., Hartl C., Schork A.J., Appadurai V., Buil A., Werge T.M. et al. (2018) Shared molecular neuropathology across major psychiatric disorders parallels polygenic overlap. Science, 359, 693–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Parikshak N.N., Swarup V., Belgard T.G., Irimia M., Ramaswami G., Gandal M.J., Hartl C., Leppa V., Ubieta L.d.l.T., Huang J. et al. (2016) Genome-wide changes in lncRNA, splicing and regional gene expression patterns in autism. Nature, 540, 423–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Dunaway K.W., Islam M.S., Coulson R.L., Lopez S.J., Vogel Ciernia A., Chu R.G., Yasui D.H., Pessah I.N., Lott P., Mordaunt C. et al. (2016) Cumulative impact of polychlorinated biphenyl and large chromosomal duplications on DNA methylation, chromatin, and expression of autism candidate genes. Cell Rep., 17, 3035–3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kundaje A., Meuleman W., Ernst J., Bilenky M., Yen A., Heravi-Moussavi A., Kheradpour P., Zhang Z., Wang J., Ziller M.J. et al. (2015) Integrative analysis of 111 reference human epigenomes. Nature, 518,317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Shulha H.P., Cheung I., Whittle C., Wang J., Virgil D., Lin C.L., Guo Y., Lessard A., Akbarian S. and Weng Z. (2012) Epigenetic signatures of autism. Arch. Gen. Psychiatry, 69, 314. [DOI] [PubMed] [Google Scholar]
- 43. Ernst J. and Kellis M. (2012) ChromHMM: automating chromatin-state discovery and characterization. Nat. Methods, 9, 215–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Timp W., Bravo H.C., McDonald O.G., Goggins M., Umbricht C., Zeiger M., Feinberg A.P. and Irizarry R.A. (2014) Large hypomethylated blocks as a universal defining epigenetic alteration in human solid tumors. Genome Med., 6, 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sandoval J., Heyn H., Moran S., Serra-Musach J., Pujana M.A., Bibikova M. and Esteller M. (2011) Validation of a DNA methylation microarray for 450,000 CpG sites in the human genome. Epigenetics, 6, 692–702. [DOI] [PubMed] [Google Scholar]
- 46. Aryee M.J., Jaffe A.E., Corrada-Bravo H., Ladd-Acosta C., Feinberg A.P., Hansen K.D. and Irizarry R.A. (2014) Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics, 30, 1363–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Mordaunt C.E., Park B.Y., Bakulski K.M., Feinberg J.I., Croen L.A., Ladd-Acosta C., Newschaffer C.J., Volk H.E., Ozonoff S., Hertz-Picciotto I. et al. (2018) A meta-analysis of two high-risk prospective cohort studies reveals autism-specific transcriptional changes to chromatin, autoimmune, and environmental response genes in umbilical cord blood. bioRxiv, 10.1101/486498. [DOI] [PMC free article] [PubMed]
- 48. Zhang S., Deng L., Jia Q., Huang S., Gu J., Zhou F., Gao M., Sun X., Feng C. and Fan G. (2017) dbMDEGA: a database for meta-analysis of differentially expressed genes in autism spectrum disorder. BMC Bioinformatics, 18,494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Bailey T.L., Boden M., Buske F.A., Frith M., Grant C.E., Clementi L., Ren J., Li W.W. and Noble W.S. (2009) MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res., 37, W202–W208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Koudritsky M. and Domany E. (2008) Positional distribution of human transcription factor binding sites. Nucleic Acids Res., 36, 6795–6805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Carninci P., Sandelin A., Lenhard B., Katayama S., Shimokawa K., Ponjavic J., Semple C.A.M., Taylor M.S., Engström P.G., Frith M.C. et al. (2006) Genome-wide analysis of mammalian promoter architecture and evolution. Nat. Genet., 38, 626–635. [DOI] [PubMed] [Google Scholar]
- 52. Geschwind D.H. and Levitt P. (2007) Autism spectrum disorders: developmental disconnection syndromes. Curr. Opin. Neurobiol., 17, 103–111. [DOI] [PubMed] [Google Scholar]
- 53. Dapretto M., Davies M.S., Pfeifer J.H., Scott A.A., Sigman M., Bookheimer S.Y. and Iacoboni M. (2006) Understanding emotions in others: mirror neuron dysfunction in children with autism spectrum disorders. Nat. Neurosci., 9, 28–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Katoh M. and Katoh M. (2006) Notch ligand, JAG1, is evolutionarily conserved target of canonical WNT signaling pathway in progenitor cells. Int. J. Mol. Med., 17, 681–685. [PubMed] [Google Scholar]
- 55. Klezovitch O. and Vasioukhin V. (2015) Cadherin signaling: keeping cells in touch. F1000Res., 4, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Yap A.S. and Kovacs E.M. (2003) Direct cadherin-activated cell signaling: a view from the plasma membrane. J. Cell Biol., 160, 11–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kalkman H. (2012) A review of the evidence for the canonical Wnt pathway in autism spectrum disorders. Mol. Autism, 3, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Krey J.F. and Dolmetsch R.E. (2007) Molecular mechanisms of autism: a possible role for Ca2+ signaling. Curr. Opin. Neurobiol., 17, 112–119. [DOI] [PubMed] [Google Scholar]
- 59. Betancur C., Sakurai T. and Buxbaum J.D. (2009) The emerging role of synaptic cell-adhesion pathways in the pathogenesis of autism spectrum disorders. Trends Neurosci., 32, 402–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Hofmann M., Schuster-Gossler K., Watabe-Rudolph M., Aulehla A., Herrmann B.G. and Gossler A. (2004) WNT signaling, in synergy with T/TBX6, controls Notch signaling by regulating Dll1 expression in the presomitic mesoderm of mouse embryos. Genes Dev., 18, 2712–2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Lopez S.J., Dunaway K., Islam M.S., Mordaunt C., Vogel Ciernia A., Meguro-Horike M., Horike S., Segal D.J. and LaSalle J.M. (2017) UBE3A-mediated regulation of imprinted genes and epigenome-wide marks in human neurons. Epigenetics, 12, 982–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Brandler W.M., Antaki D., Gujral M., Kleiber M.L., Whitney J., Maile M.S., Hong O., Chapman T.R., Tan S., Tandon P. et al. (2018) Paternally inherited cis-regulatory structural variants are associated with autism. Science, 360, 327–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Sun W., Poschmann J., Cruz-Herrera del Rosario R., Parikshak N.N., Hajan H.S., Kumar V., Ramasamy R., Belgard T.G., Elanggovan B., Wong C.C.Y. et al. (2016) Histone acetylome-wide association study of autism spectrum disorder. Cell, 167, 1385–1397e11. [DOI] [PubMed] [Google Scholar]
- 64. Gonzalez F.J. (1988) The molecular biology of cytochrome P450s. Pharmacol. Rev., 40, 91–92. [PubMed] [Google Scholar]
- 65. Jones S.M., Boobis A.R., Moore G.E. and Stanier P.M. (1992) Expression of CYP2E1 during human fetal development: methylation of the CYP2E1 gene in human fetal and adult liver samples. Biochem. Pharmacol., 43, 1876–1879. [DOI] [PubMed] [Google Scholar]
- 66. Kishida M. and Callard G.V. (2001) Distinct cytochrome P450 aromatase isoforms in zebrafish (Danio rerio) brain and ovary are differentially programmed and estrogen regulated during early development 1. Endocrinology, 142, 740–750. [DOI] [PubMed] [Google Scholar]
- 67. Ko Y., Choi I., Green M.L., Simmen F.A. and Simmen R.C.M. (1994) Transient expression of the cytochrome P450 aromatase gene in elongating porcine blastocysts is correlated with uterine insulin-like growth factor levels during peri-implantation development. Mol. Reprod. Dev., 37,–11. [DOI] [PubMed] [Google Scholar]
- 68. Majdic G., Sharpe R.M., O’Shaughnessy P.J. and Saunders P.T. (1996) Expression of cytochrome P450 17alpha-hydroxylase/C17-20 lyase in the fetal rat testis is reduced by maternal exposure to exogenous estrogens. Endocrinology, 137, 1063–1070. [DOI] [PubMed] [Google Scholar]
- 69. Hakkola J., Raunio H., Purkunen R., Pelkonen O., Saarikoski S., Cresteil T. and Pasanen M. (1996) Detection of cytochrome P450 gene expression in human placenta in first trimester of pregnancy. Biochem. Pharmacol., 52, 379–383. [DOI] [PubMed] [Google Scholar]
- 70. Silver M.J., Kessler N.J., Hennig B.J., Dominguez-Salas P., Laritsky E., Baker M.S., Coarfa C., Hernandez-Vargas H., Castelino J.M., Routledge M.N. et al. (2015) Independent genomewide screens identify the tumor suppressor VTRNA2-1 as a human epiallele responsive to periconceptional environment. Genome Biol., 16, 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Jonakait G.M. (2007) The effects of maternal inflammation on neuronal development: possible mechanisms. Int. J. Dev. Neurosci., 25, 415–425. [DOI] [PubMed] [Google Scholar]
- 72. Boksa P. (2010) Effects of prenatal infection on brain development and behavior: a review of findings from animal models. Brain. Behav. Immun., 24, 881–897. [DOI] [PubMed] [Google Scholar]
- 73. Krakowiak P., Walker C.K., Bremer A.A., Baker A.S., Ozonoff S., Hansen R.L. and Hertz-Picciotto I. (2012) Maternal metabolic conditions and risk for autism and other neurodevelopmental disorders. Pediatrics, 129,e1121–e1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Patel S.A.A., Bhambra U., Charalambous M.P., David R.M., Edwards R.J., Lightfoot T., Boobis A.R. and Gooderham N.J. (2014) Interleukin-6 mediated upregulation of CYP1B1 and CYP2E1 in colorectal cancer involves DNA methylation, miR27b and STAT3. Br. J. Cancer, 111, 2287–2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Jover R., Bort R., Gómez-Lechón M.J. and Castell J.V. (2002) Down-regulation of human CYP3A4 by the inflammatory signal interleukin-6: molecular mechanism and transcription factors involved. FASEB J., 16, 1799–1801. [DOI] [PubMed] [Google Scholar]
- 76. Abdel-Razzak Z., Loyer P., Fautrel A., Gautier J.C., Corcos L., Turlin B., Beaune P. and Guillouzo A. (1993) Cytokines down-regulate expression of major cytochrome P-450 enzymes in adult human hepatocytes in primary culture. Mol. Pharmacol., 44, 707–715. [PubMed] [Google Scholar]
- 77. Hakkola J., Hu Y. and Ingelman-Sundberg M. (2002) Mechanisms of down-regulation of CYP2E1 expression by inflammatory cytokines in rat hepatoma cells. J. Pharmacol. Exp. Ther., 304, 1048–1054. [DOI] [PubMed] [Google Scholar]
- 78. Park H.J., Kim S.K., Kang W.S., Park J.K., Kim Y.J., Nam M., Kim J.W. and Chung J.H. (2016) Association between IRS1 gene polymorphism and autism spectrum disorder: a pilot case–control study in Korean males. Int. J. Mol. Sci., 17,–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Sun X.J., Wang L.-M., Zhang Y., Yenush L., Myers M.G. Jr., Glasheen E., Lane W.S., Pierce J.H. and White M.F. (1995) Role of IRS-2 in insulin and cytokine signalling. Nature, 377, 173–177. [DOI] [PubMed] [Google Scholar]
- 80. Hussman J.P., Chung R.-H., Griswold A.J., Jaworski J.M., Salyakina D., Ma D., Konidari I., Whitehead P.L., Vance J.M., Martin E.R. et al. (2011) A noise-reduction GWAS analysis implicates altered regulation of neurite outgrowth and guidance in autism. Mol. Autism, 2,. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Machado-Neto J.A., de Melo Campos P. and Traina F. (2017) IRS2 (insulin receptor substrate 2). Atlas Genet. Cytogenet. Oncol. Haematol., 21, 35–43. [Google Scholar]
- 82. Mitra I., Lavillaureix A., Yeh E., Traglia M., Tsang K., Bearden C.E., Rauen K.A. and Weiss L.A. (2017) Reverse pathway genetic approach identifies epistasis in autism Spectrum disorders. PLoS Genet., 13, e1006516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Carvalheira J.B.C., Ribeiro E.B., Folli F., Velloso L.A. and Saad M.J.A. (2003) Interaction between leptin and insulin signaling pathways differentially affects JAK-STAT and PI 3-kinase-mediated signaling in rat liver. Biol. Chem., 384, 151–159. [DOI] [PubMed] [Google Scholar]
- 84. Fang Q.L., Yin Y.R., Xie C.R., Zhang S., Zhao W.X., Pan C., Wang X.M. and Yin Z.Y. (2015) Mechanistic and biological significance of DNA methyltransferase 1 upregulated by growth factors in human hepatocellular carcinoma. Int. J. Oncol., 46, 782–790. [DOI] [PubMed] [Google Scholar]
- 85. Xu G., Jing J., Bowers K., Liu B. and Bao W. (2014) Maternal diabetes and the risk of autism spectrum disorders in the offspring: a systematic review and meta-analysis. J. Autism Dev. Disord., 44, 766–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Koukoura O., Sifakis S. and Spandidos D.A. (2012) DNA methylation in the human placenta and fetal growth (review). Mol. Med. Rep., 5, 883–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Zeltser L.M. and Leibel R.L. (2011) Roles of the placenta in fetal brain development. Proc. Natl. Acad. Sci. U. S. A., 108, 15667–15668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Xu L.-M., Li J.-R., Huang Y., Zhao M., Tang X. and Wei L. (2012) AutismKB: an evidence-based knowledgebase of autism genetics. Nucleic Acids Res., 40, D1016–D1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Hertz-Picciotto I., Schmidt R.J., Walker C.K., Bennett D.H., Oliver M., Shedd-Wise K.M., LaSalle J.M., Giulivi C., Puschner B., Thomas J. et al. (2018) A prospective study of environmental exposures and early biomarkers in autism spectrum disorder: design, protocols, and preliminary data from the MARBLES study. Environ. Health Perspect., 126, 117004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Lord C., Risi S., Lambrecht L., Cook E.H., Leventhal B.L., DiLavore P.C., Pickles A. and Rutter M.L. (2000) Autism Diagnostic Observation Schedule (ADOS). J. Autism Dev. Disord., 30, 205–223. [PubMed] [Google Scholar]
- 91. Lord C., Rutter M. and Le Couteur A. (1994) Autism Diagnostic Interview—revised: a revised version of a diagnostic interview for caregivers of individuals with possible pervasive developmental disorders. J. Autism Dev. Disord., 24, 659–685. [DOI] [PubMed] [Google Scholar]
- 92. Mullen E.M. (1995) Mullen Scales of Early Learning. (AGS ed.).Circle Pines, MN: American Guidance Service Inc. [Google Scholar]
- 93. Chawarska K., Shic F., Macari S., Campbell D.J., Brian J., Landa R., Hutman T., Nelson C.A., Ozonoff S., Tager-Flusberg H. et al. (2014) 18-month predictors of later outcomes in younger siblings of children with autism spectrum disorder: a baby siblings research consortium study. J. Am. Acad. Child Adolesc. Psychiatry, 53, 1317–1327e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Ozonoff S., Young G.S., Belding A., Hill M., Hill A., Hutman T., Johnson S., Miller M., Rogers S.J., Schwichtenberg A.J. et al. (2014) The broader autism phenotype in infancy: when does it emerge? J. Am. Acad. Child Adolesc. Psychiatry, 53, 398–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Guo W., Fiziev P., Yan W., Cokus S., Sun X., Zhang M.Q., Chen P.-Y. and Pellegrini M. (2013) BS-Seeker2: a versatile aligning pipeline for bisulfite sequencing data. BMC Genomics, 14, 774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Coulson R.L., Yasui D.H., Dunaway K., Laufer B.I., Vogel Ciernia A., Zhu Y., Mordaunt C.E., Totah T.S. and Lasalle J.M. (2018) Snord116-dependent diurnal rhythm of DNA methylation in mouse cortex. Nat Commun., 9, 1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Box J.F. (1980) R. A. Fisher and the Design of Experiments, 1922–1926. Am. Stat., 34,. [Google Scholar]
- 98. Wilks D.S. (2011) Cluster Analysis. Int. Geophys., 100, 603–616. [Google Scholar]
- 99. McLean C.Y., Bristor D., Hiller M., Clarke S.L., Schaar B.T., Lowe C.B., Wenger A.M. and Bejerano G. (2010) GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol., 28, 495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T. et al. (2000) Gene ontology: tool for the unification of biology. Nat. Genet., 25, 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. The Gene Ontology Consortium (2017) Expansion of the Gene Ontology knowledgebase and resources. Nucleic Acids Res., 45, D331–D338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Thomas P.D., Campbell M.J., Kejariwal A., Mi H., Karlak B., Daverman R., Diemer K., Muruganujan A. and Narechania A. (2003) PANTHER: a library of protein families and subfamilies indexed by function. Genome Res., 13, 2129–2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Mi H., Muruganujan A. and Thomas P.D. (2012) PANTHER in 2013: modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucleic Acids Res., 41, D377–D386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Shen L., Shao N.-Y., Liu X., Maze I., Feng J. and Nestler E.J. (2013) diffReps: detecting differential chromatin modification sites from ChIP-seq data with biological replicates. PLoS One, 8, e65598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Harold D., Abraham R., Hollingworth P., Sims R., Gerrish A., Hamshere M.L., Pahwa J.S., Moskvina V., Dowzell K., Williams A. et al. (2009) Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer’s disease. Nat. Genet., 41, 1088–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Landi M.T., Chatterjee N., Yu K., Goldin L.R., Goldstein A.M., Rotunno M., Mirabello L., Jacobs K., Wheeler W., Yeager M. et al. (2009) A genome-wide association study of lung cancer identifies a region of chromosome 5p15 associated with risk for adenocarcinoma. Am. J. Hum. Genet., 85, 679–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Sheffield N.C. and Bock C. (2016) LOLA: enrichment analysis for genomic region sets and regulatory elements in R and Bioconductor. Bioinformatics, 32,587–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Sloan C.A., Chan E.T., Davidson J.M., Malladi V.S., Strattan J.S., Hitz B.C., Gabdank I., Narayanan A.K., Ho M., Lee B.T. et al. (2016) ENCODE data at the ENCODE portal. Nucleic Acids Res., 44, D726–D732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. ENCODE Project Consortium, T.E.P (2012) An integrated encyclopedia of DNA elements in the human genome. Nature, 489, 57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Ernst J. and Kellis M. (2017) Chromatin-state discovery and genome annotation with ChromHMM. Nat. Protoc., 12, 2478–2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Kent W.J., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M. and Haussler D. (2002) The human genome browser at UCSC. Genome Res., 12,996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Schneider C.A., Rasband W.S. and Eliceiri K.W. (2012) NIH Image to ImageJ: 25 years of image analysis. Nat. Methods, 9, 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
WGBS data were previously published, Gene Expression Omnibus (GEO) accession number GSE67615 (31). The rest of the relevant data and information are included in supplementary tables.


