Abstract
Background
Inflammation is an important priming event in the pathogenesis of pulmonary diseases, including chronic obstructive pulmonary disease (COPD). Increasing evidence indicates that microRNAs (miRNAs) contribute to the pathogenesis of COPD by regulating inflammatory response. Therefore, it is necessary to investigate novel molecular targets in COPD without any validation in COPD samples in airway inflammation. The aim of our study is to reveal novel miRNAs that can influence molecular targets for COPD and to examine the underlying mechanism in airway inflammation.
Methods
We identified the downregulation of miR-181a-2-3p in the serum of COPD patients and further investigated the role of miR-181a-2-3p in cadmium (Cd)-induced inflammation of a human bronchial epithelial cell line (BEAS-2B) and normal human bronchial epithelial (NHBE) cells.
Results
Our results showed that expression of miR-181a-2-3p was significantly decreased in Cd-treated cells and silencing of miR-181a-2-3p enhanced Cd-induced inflammatory responses and inflammasome activation. This negative regulatory effect of miR-181a-2-3p on inflammation is partly mediated by the calcium signaling pathway. Furthermore, global gene expression profiling revealed that Toll-like receptor 4 or sequestosome 1 genes were identified as potential targets of miR-181a-2-3p, which were significantly upregulated by knockdown of miR-181a-2-3p in Cd-treated cells.
Conclusions
Our results strongly suggest that miR-181a-2-3p has a critical role in Cd-induced inflammation of airway by regulating its potential target genes, which could be molecular targets for COPD.
Keywords: Chronic obstructive pulmonary disease (COPD), miR-181a-2-3p, cadmium (Cd), inflammation, inflammasome
Introduction
Chronic respiratory diseases are provoked by inflammatory responses to external toxicants, such as smoke, air pollution, and heavy metals (1). Inflammatory signals activate a range of responses in the multiple cell types of airway epithelium (2). As a first-line defense, damaged epithelial cells recruit inflammatory cells to the airway by producing inflammatory cytokines and mediators and increasing mucus secretion and other signals that promote generating reactive oxygen species (ROS). Chronic obstructive pulmonary disease (COPD) is characterized by progressive airflow limitation because of airway inflammation and destruction of lung parenchyma. However, the mechanisms underlying COPD remain unclear. Although its pathogenesis is uncertain, cigarette smoking remains the main environmental risk factor for COPD (3,4), with its involved aberrant inflammatory and dysregulated cellular responses, where airways and lung tissue are affected. However, according to the World Health Organization, only 20% of the smokers suffer from COPD, suggesting that environmental factors and genetic susceptibility may also contribute to the pathogenesis of this disease (5-7).
As an environmental risk factor, cadmium (Cd) is a harmful component of cigarette smoke and particulate matter. Emerging evidence demonstrates that Cd exposure causes its excessive accumulation in the lungs which leads to the airway inflammation and lung dysfunction observed in COPD (8-11). Although the underlying mechanisms associated with Cd exposure have not been addressed in COPD, recent studies have suggested that Cd is closely associated with the severity and progression of COPD (8-10). Furthermore, exposure to Cd leads to epigenetic modulations, including aberrant methylation of DNA repair genes, histone modification, or microRNAs (miRNAs) (11-13).
miRNAs are small noncoding RNAs. These gene regulatory networks are rather complex, because a single miRNA can bind to the 3'-untranslated region (3'-UTR) of multiple target mRNAs, typically resulting in transcriptional or translational regulation of gene expression (14,15). By doing so, miRNAs are involved in various biological processes, such as cellular development, differentiation, proliferation, and apoptosis. In particular, recent studies have reported that several miRNAs have an important role in positive and negative regulation of the inflammatory response and participate in various regulatory network motifs in respiratory disease (16-19). Despite the critical role of miRNAs in inflammatory response, limited studies have focused on its role in inflammation-induced COPD. Numerous studies have revealed the dysregulation of miRNAs and their pathogenic roles in COPD. For instance, miR-26a acts as a regulator of the nuclear factor-κB (NF-κB) pathway (20) by regulating its target gene and activating signal cointegrator 1 complex subunit 3 (ASCC3) (21). In addition, miR-146a is significantly downregulated in lung fibroblasts of COPD patients and correlated positively with cytokine activation (22). Other studies suggested that miR-218-5p is downregulated in recruiting inflammatory cells toward the sites of inflammation through targeting chemokine (C-C motif) ligand 20 and interleukin (IL)-8, thereby assisting in sustaining the inflammation (23). However, knowledge of this state has remained limited to establish their role as biomarkers for early prevention, prognosis, and possible therapy (24). Therefore, it is necessary to develop novel therapeutic methods and targets through understanding the mechanism of COPD pathogenesis mediated by miRNA.
We identified the downregulation of miR-181a-2-3p in serum and lung tissues of COPD patients and further revealed its negative regulatory role in enhancing Cd-induced inflammatory responses of a human bronchial epithelial cell line (BEAS-2B) and normal human primary bronchial epithelial (NHBE) cells. Moreover, silencing of miR-181a-2-3p revealed its potential target genes, which could be implicated in the pathological events of COPD.
Methods
Preparation of serum samples
Serum samples were obtained from 58 COPD patients and 38 subjects with normal lung function registered in the COPD cohort study. The demographic characteristics of donors in each group are shown in Table 1. Methods for recruiting the cohort population have been reported previously (25,26). COPD was defined as a post-bronchodilator forced expiratory volume in 1 s/forced vital capacity ratio less than 0.7. The study was approved by the institutional review board of Kangwon National University Hospital (2012-06-007). All participants provided written informed consent.
Table 1. Characteristics of COPD and control subjects used for expression profiling of miRNAs.
| Characteristics | COPD (n=58) | Control (n=38) | P value |
|---|---|---|---|
| Male (%) | 40 (69.0) | 22 (57.9) | 0.27 |
| Age (years) | 73.9±5.2 | 72.0±7.0 | 0.05 |
| Smoking (pack-years) | 20.0±21.7 | 16.0±22.8 | 0.78 |
| FEV1 (% of predicted) | 75.3±18.7 | 97.5±21.2 | <0.001 |
| FEV1/FVC | 57.7±6.5 | 75.4±3.9 | <0.001 |
Unless otherwise stated, data are represented as mean ± standard deviation. FEV1, forced expiratory volume in 1 s; FVC, forced vital capacity; COPD, chronic obstructive pulmonary disease.
Cell culture
BEAS-2B was kindly provided by the Biomedical Research Institute at Seoul National University Hospital. BEAS-2B cells were maintained in defined keratinocyte serum-free medium containing epidermal growth factor, 100 U/mL penicillin, and 100 µg/mL streptomycin (Thermo Fisher Scientific, Waltham, MA, USA). NHBE cells (CC-2540; Lonza Group, Allendale, NJ, USA) isolated from the epithelial lining of airways above the bifurcation of a normal human donor lung were cultured in bronchial epithelial growth medium (BulletKit medium, CC-3171, Lonza Group) and used before passage five in all experiments. Cell cultures were incubated at 37 °C in a humidified atmosphere containing 5% CO2.
In vitro transfection experiments of miR-181a-2-3p mimetics or inhibitors
Unless otherwise indicated, all materials for miRNA study were purchased from Qiagen (Hilden, Germany). For a transient transfection approach with the aim to inhibit or enhance miR-181a-2-3p function, cells were transfected using the fast-forward HiPerFect Transfection Reagent protocol according to the manufacturer’s instructions. A specific miR-181a-2-3p mimic, inhibitors or negative inhibitors were purchased commercially. For the reference to normalize the findings, we used the miScript inhibitor negative control under the same concentrations and conditions used for the mimic/inhibitor (50 nM). Transfected bronchial epithelial cells were incubated under their normal growth conditions, and the effects of miR-181a-2-3p manipulations on changes in gene expression levels were measured by quantitative reverse transcription-polymerase chain reaction (RT-PCR) after 24 h as described above.
Cd treatment
BEAS-2B and NHBE cells were seeded at a density of 2.0×105 cells per well in 6-well plates and incubated until they reach 70–80% confluency. Cells were then transfected with miR-181a-2-3p mimics and/or inhibitors, followed by treatment with or without Cd for 24 h (27,28).
ELISA assay
Secreted IL-1β was measured in BEAS-2B cell culture supernatants using the human IL-1β/IL-1F2 Quantikine ELISA Kit (R&D Systems, MN, USA). The ELISA plates were read using a microplate reader (Molecular Devices, Sunnyvale, CA, USA).
Inflammasome activation
THP-1 cells (1×106 cells/well) in 12-well plates were differentiated into macrophage-like cells by incubating with phorbol 12-myristate 13-acetate (100 nM, PMA; InvivoGen, San Diego, CA, USA) for 72 h. THP-1-derived macrophages were primed for 3 h with lipopolysaccharide (LPS) (1 µg/mL), and then cells were stimulated as follows by the NLRP3, NLRC4, or AIM2 activators ATP (2 mM), flagellin (500 µg/mL), or dsDNA (2 µg/mL), respectively, with miR-181a-2-3p mimic/inhibitor (50–100 nM), and then the IL-1β (p17, active form) secretion was assayed.
Pharmacological reagents treatment
Unless otherwise indicated, all pharmacological reagents were obtained from Tocris Bioscience (Bristol, UK) or Cayman Chemical (Ann Arbor, MI, USA) and used at the following working concentrations: Toll-like receptor 4 (TLR4) inhibitor (TAK242, 2.5 µM; Cayman Chemical), phospholipase C (PLC) inhibitor (U73122, 10 µM; Tocris Bioscience), IP3/TRP channel inhibitor [2-aminoethoxydiphenyl borate (2-APB), 10 µM; Tocris], and nicotinamide adenine dinucleotide phosphate oxidase (NOX) inhibitor [diphenyleneiodonium (DPI), 10 µM; Tocris]. Dimethyl sulfoxide at 0.1% concentration was used as the vehicle control. Cells were pretreated with the concerned pharmacological reagents for 1 h followed by transfection with miRNA transfection protocol.
Western blotting analysis
An equal amount of proteins from control and each treatment sample were loaded by sodium dodecyl sulfate-polyacrylamide gel electrophoresis gel (10% or 15%) and transferred onto polyvinyl difluoride membranes (Millipore, Billerica, MA, USA). Nonspecific binding proteins were blocked with 3% skim milk in 1× phosphate-buffered saline with 0.05% Tween 20 for 60 min. Membranes were incubated with primary antibodies against antihuman IL-1β antibody (AF-201-NA, R&D Systems, Minneapolis, MN, USA), NF-κB sampler kit antibodies (#9936; Cell Signaling Technology; Danvers, MA, USA), and CEBP Antibody Sampler Kit (#12814, Cell Signaling Technology), PI3 Kinase Antibody Sampler Kit (#9655, Cell Signaling Technology), or anti-β-actin antibody (sc4778, Santa Cruz Biotechnology, Inc., Dallas, TX, USA) overnight at 4 °C. The membranes were probed further with horseradish peroxidase-conjugated secondary anti-sera (A9917, A6667, or A5420; Sigma-Aldrich Corp., St. Louis, MO, USA) and visualized with PierceFast Western blot kit (Thermo Fisher Scientific) and a cooled CCD camera system (Bio-Rad Laboratories, Hercules, CA, USA).
miRNA and mRNA extraction and real-time PCR
According to the manufacturer’s instructions, total miRNA was isolated from the serum using the miRNeasy Serum/Plasma Kit (Qiagen) and reverse transcription was performed using the miScript II RT Kit (Qiagen). Subsequently, complementary DNAs (cDNAs) were amplified from the following 20 miRNAs using the custom miScript miRNA PCR Array (CMIHS02261C; Qiagen): hsa-miR-101-3p, hsa-miR-125b-1-3p, hsa-miR-143-3p, hsa-miR-148a-3p, hsa-miR-151a-3p, hsa-miR-181a-2-3p, hsa-miR-181b-5p, hsa-miR-181c-5p, hsa-miR-181d-5p, hsa-miR-23b-3p, hsa-miR-26a-5p, hsa-miR-28-3p, hsa-miR-30a-5p, hsa-miR-30c-2-3p, hsa-miR-30e-3p, hsa-miR-361-5p, hsa-miR-501-3p, hsa-miR-660-5p, hsa-miR-769-5p, and hsa-mir-191-5p. The following thermal cycling conditions were used: 95 °C for 15 min, followed by 40 cycles of 95, 55, and 70 °C for 15, 30, and 30 s, respectively. Quantitation was normalized to hsa-miR-RNU6 (reference gene). Data were analyzed using PCR array data analysis tools (Qiagen). For mRNA analysis, total RNA was extracted from cells by TRIzol (Invitrogen) according to the manufacturer’s protocol and reverse-transcribed into cDNA using the QuantiTect® reverse transcription kit. cDNA was quantitated using Power SYBR Green PCR (Applied Biosystems by Life Technologies, Warrington, UK) and the QuantStudio 6 Flex Real-Time PCR system (Applied Biosystems). Primer sequences are listed in Table S1.
Table S1. Primer sequences used for quantitative RT-PCR.
| Gene | Primer | Sequence 5' to 3' | Product size (bp) | Tm (°C) |
|---|---|---|---|---|
| IL-1α | F | ATC AGT ACC TCA CGG CTG CT | 179 | 60.5 |
| R | TGG GTA TCT CAG GCA TCT CC | |||
| IL-1β | F | CTG TCC TGC GTG TTG AAA GA | 161 | 58.4 |
| R | TTC TGC TTG AGA GGT GCT GA | |||
| IL-6 | F | TAC CCC CAG GAG AAG ATT CC | 164 | 60.5 |
| R | TTT TCT GCC AGT GCC TCT TT | |||
| IL-8 | F | GTG CAG TTT TGC CAA GGA GT | 186 | 60.5 |
| R | CTC TGC ACC CAG TTT TCC TT | |||
| TNFα | F | AAC CTC CTC TCT GCC ATC AA | 162 | 58.4 |
| R | CCA AAG TAG ACC TGC CCA GA | |||
| COX2 | F | TGA GCA TCTACG GTT TGC TG | 159 | 58.4 |
| R | TGC TTG TCT GGA ACA ACT GC | |||
| SQSTM1 | F | GAG TTC CAG CAC AGA GGA GA | 210 | 58.4 |
| R | AAG ACA GAT GGG TCC AGT CA | |||
| TLR4 | F | GAC AAC CAG CCT AAA GTA TT | 206 | 62.1 |
| R | TGC CAT TGA AAG CAA CTC TG | |||
| PPP3CA | F | GCG CAT CTT ATG AAG GAG GGA | 115 | 61.3 |
| R | TGA CTG GCG CAT CAA TAT CCA | |||
| PPP3CB | F | CCC CAA CAC ATC GCT TGA CAT | 140 | 61.3 |
| R | GGC AGC ACC CTC ATT GAT AAT TC | |||
| PPP3CC | F | ACC GCG TCA TCA AAG CTG T | 126 | 57.3 |
| R | CTT CCA GTC GTC CTT CCT TTA C | |||
| PPP3R1 | F | CCT TTG GAA ATG TGC TCA CAC T | 137 | 60.3 |
| R | GGA TTC TGT TGT AAC TCA GGC AG | |||
| PPP3R2 | F | GCA GAG CAC GGA CAG CTA TC | 213 | 62.5 |
| R | GGG CTT TCT CCA CGA AAA TGA | |||
| PPP4R2 | F | ACG AAG GCC CTG TAA GTA GT | 129 | 58.4 |
| R | CTT CTG TGG CTT CGT CAT CA | |||
| NAFTC1 | F | GCA GAG CAC GGA CAG CTA TC | 101 | 62.5 |
| R | GGG CTT TCT CCA CGA AAA TGA | |||
| NAFTC2 | F | GCT GGT TCC GGT GTA CTC G | 172 | 61.7 |
| R | AGA GAC CAC TCG AAT CTG CCA | |||
| NAFTC3 | F | GCT CGA CTT CAA ACT CGT CTT | 96 | 59.4 |
| R | GAT GCA CAA TCA TCT GGC TCA | |||
| NAFTC4 | F | CTT CTC CGA TGC CTC TGA CG | 173 | 62.5 |
| R | CGG GGC TTG GAC CAT ACA G | |||
| 18S rRNA | F | GTA ACC CGT TGA ACC CCA TT | 152 | 58.4 |
| R | CCA TCC AAT CGG TAG TAG CG |
Identification of miRNAs targets and network analysis
Following incubation with/without miR-181a-2-3p inhibitor (50 nM) for 24 h, Cd-treated BEAS-2B cells were collected as described previously and total RNA was extracted using TRIzol. Affymetrix GeneChip Human 2.0 ST Array (Affymetrix, Santa Clara, CA, USA) was hybridized with complementary RNA (cRNA) probes at the Macrogen core facility (Seoul, Korea). The expression value and detection calls were computed from the raw data and gene set enrichment analysis (GSEA) version 4.0 (Broad Institute, Cambridge, MA, USA) to interpret expression profiles from microarrays. GSEA originally was developed to identify cohorts of genes whose functions are integrated into certain biological processes and/or specific signaling pathways. Pathways were ranked according to the significance of the enrichment, and the validation mode measure of significance was used to identify pathways of greatest enrichment. Significance was tested by comparing the observed enrichment with the enrichment seen in data sets in which sample labels were permutated randomly. All data analysis and visualization of differentially expressed genes was conducted using Cytoscape.
Statistical analyses
Statistical analyses were performed with Student’s t-test and two-way analysis of variance for multiple groups using GraphPad Prism (GraphPad Software, San Diego, CA, USA). P values are indicated in the figures. Moreover, Fisher’s exact test was performed to estimate the significance of differential expressions of miRNAs in serum samples using R program (version 3.5). P<0.05 was considered as statistically significant, which is indicated in the figures.
Results
miR-181a-2-3p is downregulated in lung tissues and serum of COPD patients compared with normal subjects
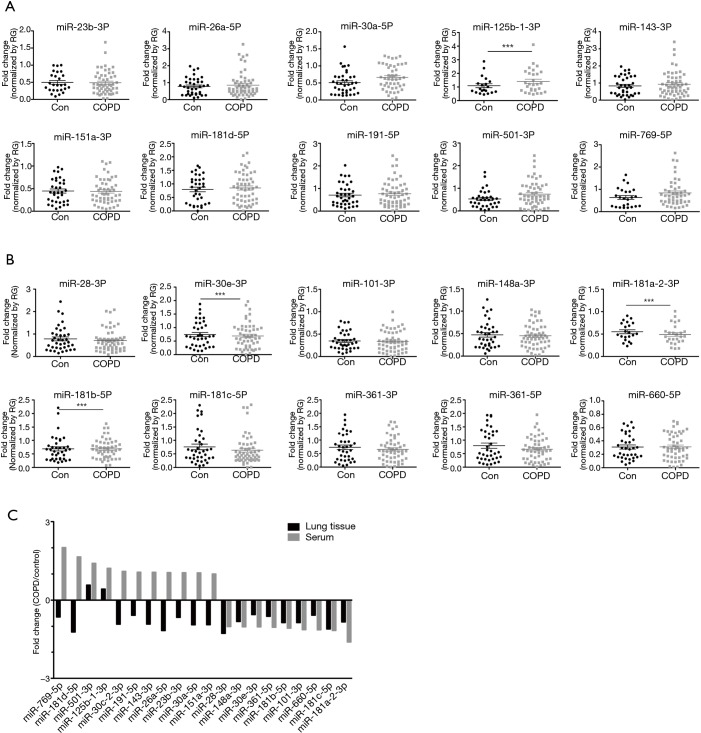
Profiling miRNA expression patterns in lung tissues of COPD patients and determining regulatory mechanisms of specific miRNAs enhance our understanding of molecular mechanisms of COPD. On the basis of our previous report of such profiling in COPD patients and normal subjects (29), we selected several miRNAs (10 upregulated and 10 downregulated) to determine their differential expressions in serum samples from COPD patients and normal subjects (Table 2). To draw further conclusions about the correlation between selected miRNAs in lung tissues and body fluid, we compared their expression levels in the serum of 58 smokers with COPD and 38 healthy smokers without COPD. Among these miRNAs, miR-181a-2-3p was significantly downregulated in the serum samples of COPD patients, whereas miR-181d-5p, miR-501-3p, miR-769-5p, and miR-191-5p were upregulated (Figure 1A,B). Also, miR-181a-2-3p and miR-501-3p were similarly dysregulated between lung tissues and serum of the COPD patients (Figure 1C).
Table 2. Expression levels of miRNAs in the COPD and control group.
| miRNAs | COPD vs. control | ||
|---|---|---|---|
| Fold regulation | 95% CI | P value | |
| hsa-miR-769-5p | 2.0108 | 0.77, 3.26 | 1.000 |
| hsa-miR-181d-5p | 1.6559 | 0.81, 2.50 | 1.000 |
| hsa-miR-501-3p | 1.4094 | 0.61, 2.21 | 0.502 |
| hsa-miR-125b-1-3p | 1.2192 | 0.60, 1.83 | <0.0001 |
| hsa-miR-191-5p | 1.0708 | 0.95, 1.19 | 0.835 |
| hsa-miR-143-3p | 1.0675 | 0.68, 1.46 | 0.536 |
| hsa-miR-26a-5p | 1.0577 | 0.94, 1.18 | 0.835 |
| hsa-miR-23b-3p | 1.0504 | 0.90, 1.20 | 0.522 |
| hsa-miR-30a-5p | 1.0456 | 0.97, 1.12 | 1.000 |
| hsa-miR-151a-3p | 1.0011 | (0.89, 1.12) | 0.537 |
| hsa-miR-361-3p | −1.0044 | (0.65, 1.34) | 0.835 |
| hsa-miR-28-3p | −1.011 | (0.86, 1.12) | 1.000 |
| hsa-miR-148a-3p | −1.0189 | (0.83, 1.14) | 0.679 |
| hsa-miR-30e-3p | −1.0218 | (0.74, 1.21) | <0.0001 |
| hsa-miR-361-5p | −1.0354 | (0.58, 1.35) | 0.302 |
| hsa-miR-181b-5p | −1.0709 | (0.83, 1.04) | 0.659 |
| hsa-miR-101-3p | −1.1287 | (0.74, 1.03) | 0.679 |
| hsa-miR-660-5p | −1.1339 | (0.62, 1.15) | 1.000 |
| hsa-miR-181c-5p | −1.1594 | (0.61, 1.11) | 0.831 |
| hsa-miR-181a-2-3p | −1.5998 | (0.28, 0.97) | 0.008 |
Fold change (2−ÄÄCt) is normalized gene expression. (2−ÄCt) in test sample divided the normalized gene expression (2−ÄCt) in the control sample. Quantitation was normalized to reference control. COPD, chronic obstructive pulmonary disease; CI, confidence interval.
Figure 1.
Selection of candidate miRNAs. Differentially expressed miRNAs in the serum from subjects with COPD compared with healthy smoker. (A) Ten miRNAs (miR-23b-3p, miR-26a-5p, miR-30a-5p, miR-125b-1-3p, miR-143-3p, miR-151a-3p, miR-181d-5p, miR-191-5p, miR-501-3p, and miR-769-5p) were substantially upregulated; (B) 10 miRNAs (miR-28-3p, miR-30e-3p, miR-101-3p, miR-148a-3p, miR-181a-2-3p, miR-181b-5p, miR-181c-5p, miR-361-3p, miR-361-5p, and miR-660-5p) were downregulated in COPD patients; (C) analysis of differential miRNA expression profiles from serum and lung tissues (published miRNA profile) of COPD patients, represented by fold-change. ***, P<0.001 were considered significant. COPD, chronic obstructive pulmonary disease.
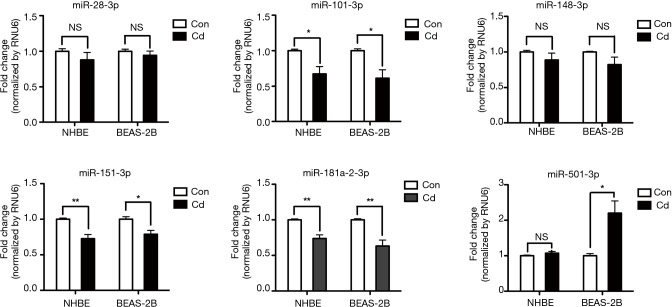
Cd, a major component of cigarette smoke and particulate matter, induces inflammatory responses and endoplasmic reticulum stress in human bronchial epithelial cells via the C/EBP-DDIT3 signaling pathway (28). Thus, we investigated whether expression of miRNAs is influenced by Cd treatment in BEAS-2B and NHBE cells. We found that expression of miR-28-3p, miR-101-3p, miR-148-3p, miR-151-3p, miR-181a-2-3p, and miR-501-3p was influenced by Cd treatment in BEAS-2B and/or NHBE cells (Figure 2). Interestingly, miR-181a-2-3p was downregulated in Cd-treated bronchial epithelial cells as well as lung tissue and serum of COPD patients, suggesting that miR-181a-2-3p may have an important role in promoting inflammatory response associated with the pathogenesis of pulmonary diseases.
Figure 2.
Analysis of dysregulated miRNAs expression of Cd-treated cells. Bronchial epithelial cells (BEAS-2B and primary NHBE cells) were stimulated with 10 µM Cd for 24 h. The miRNA levels were analyzed using SYBR green-based quantitative real-time PCR assays. All data are representative of at least three independent experiments. *, P<0.05; **, P<0.01 were considered significant. Cd, cadmium; NS, not significant; NHBE, normal human bronchial epithelial.
Knockdown of miR-181α-2-3p enhances inflammatory responses and inflammasome activation in human bronchial epithelial cells
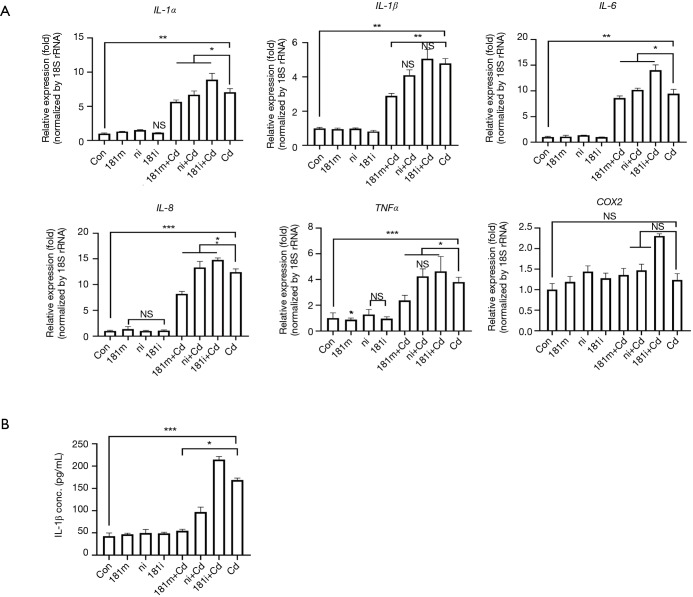
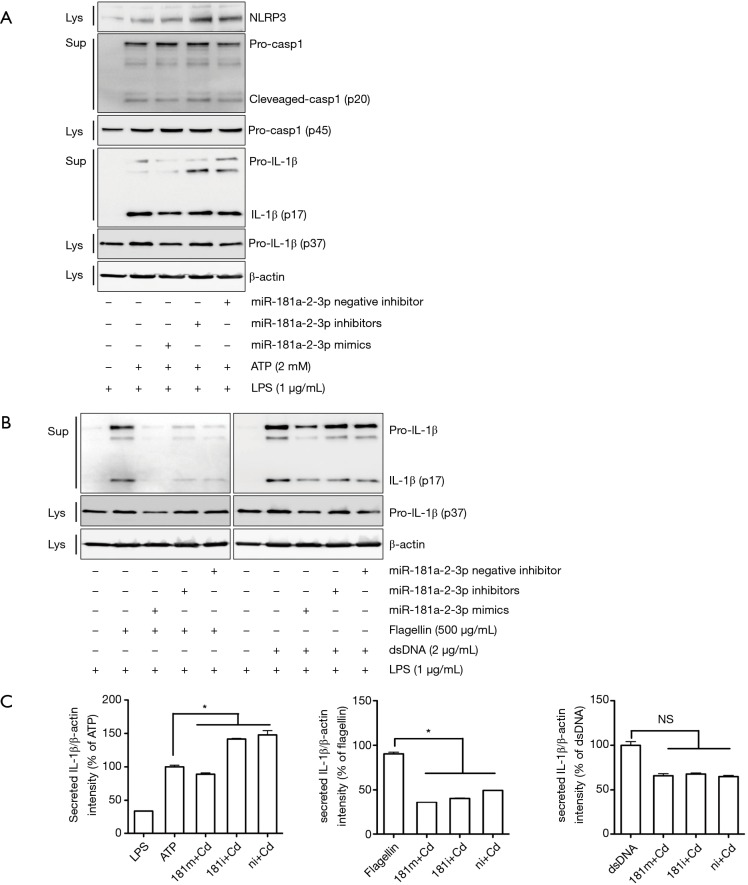
To determine if downregulation of miR-181a-2-3p is involved in regulating inflammatory responses, we measured transcriptional levels of pro-inflammatory cytokines in Cd-treated BEAS-2B cells in the presence/absence of miR-181a-2-3p inhibitor. As shown in Figure 3A, transfection of cells with miR-181a-2-3p inhibitor or negative inhibitor (control miRNA inhibitor) alone did not affect the expression of genes associated with inflammatory responses in BEAS-2B cells. However, knockdown of miR-181a-2-3p followed by Cd-exposure significantly elevated the expression levels of inflammatory cytokines and mediators, such as IL-1α, IL-1β, IL-6, IL-8, TNFα, and COX2 (Figure 3A). To reinforce the biological relevance of this finding in Cd-treated BEAS-2B cells, we investigated whether knockdown of miR-181a-2-3p promotes inflammatory responses in translational level. As seen in mRNA levels, silencing of miR-181a-2-3p resulted in an increase in the production of IL-1β (Figure 3B) in Cd-exposed NHBE cells. In contrast, miR-181a-2-3p mimics ameliorated inflammatory responses (Figure 3B). Inflammasome not only regulates the inflammatory response but also has a central role in the development of COPD (30). To further investigate the mechanism involved in the inflammatory response of miR-181a-2-3p, we evaluated whether overexpression or knockdown of miR-181a-2-3p is able to activate the inflammasome. Although knockdown of miR-181a-2-3p significantly induced NLRP3 and caspase 1 (Casp1) expressions and promoted IL-1β secretion in ATP-treated LPS-primed macrophage-like cells, its overexpression attenuates the secretion of IL-1β through three inflammatory response mechanisms, namely activation of NLRP3, NLRC4, or AIM2 inflammasomes (Figure 4). Our findings indicated that miR-181a-2-3p increased sensitivity to inflammatory responses induced by Cd exposure in human bronchial epithelial cells.
Figure 3.
Effect of miRNA 181a-2-3p on pro-inflammatory cytokines expression. Airway epithelial cells (BEAS-2B and primary NHBE cells) were transfected with miRNA 181a-2-3p mimics or inhibitors (50 nM) when the cells were stimulated with 10 µM Cd for 24 h. mRNA levels of pro-inflammatory cytokines (IL-1α, IL-1β, IL-6, IL-8, TNFα, and COX2) were analyzed using SYBR green-based quantitative real-time PCR. (A) BEAS-2B cells; (B) secreted IL-1β levels were measured in NHBE cells culture supernatants by enzyme-linked immunosorbent assay (ELISA). All data shown are representative of at least three independent experiments. Statistically significant differences were analyzed with the t-test. *, P<0.05; **, P<0.01, and ***, P<0.001 were considered significant. NS, not significant; NHBE, normal human bronchial epithelial; Cd, cadmium; 181m, 181 mimics; 181i, 181 inhibitor; ni, miRNA inhibitor negative control.
Figure 4.
Effect of miR-181a-2-3p on inflammasome activation. miR-181a-2-3p promotes inflammasome activation in human THP-1-derived macrophages. Human THP-1-derived macrophages were pretreated with miR-181a-2-3p mimic/inhibitor (50 nM) and then stimulated with 2 mM ATP, 500 µg/mL flagellin, or 2 µg/mL dsDNA to activate NLRP3, NLRC4 and AIM2 inflammasomes, respectively. (A,B) Western blot analysis of proteins associated with the activation of NLRP3, NLRC4 and AIM2 inflammasomes, such as IL-1β and caspase 1. (C) Quantitation of secreted IL-1β band intensities and their normalization with internal control β-actin by Image J software. All data shown are representative of at least three independent experiments. *, P<0.05 was considered significant. NS, not significant; Lys, lysate; Sup, supernatant; LPS, lipopolysaccharide; 181m, 181 mimics; 181i, 181 inhibitor; ni, miRNA inhibitor negative control; Cd, cadmium.
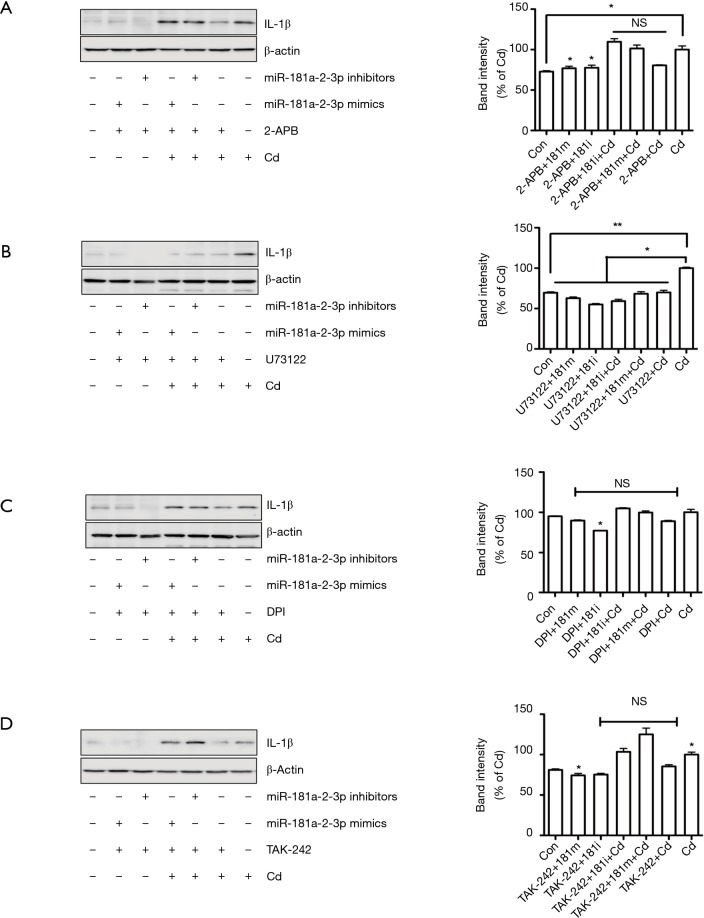
Blockade of intracellular calcium signaling partly attenuates Cd-induced IL-1β expression in human bronchial epithelial cells
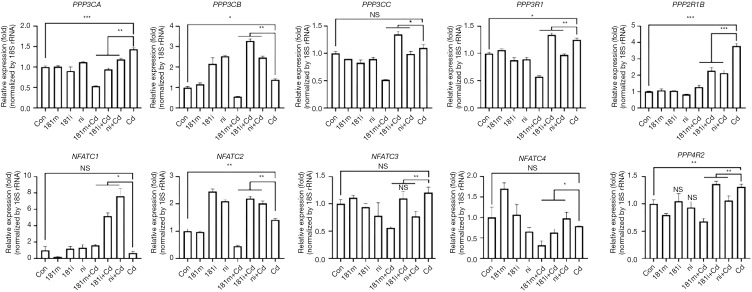
Elevation of intracellular calcium, superoxide generation by the NOXs, and activation of TLR4 signaling are involved in inducing inflammatory responses and inflammation activation in the lung tissues (31,32). Thus, we asked if the above signaling pathways are involved in reducing the negative regulatory effect of miR-181a-2-3p on inflammation in human bronchial epithelial cells treated with Cd. We found that blockage of PLC by U73122 attenuated Cd-induced IL-1β expression in human bronchial epithelial cells, whereas 2-APB (inhibitor of inositol triphosphate receptor and transient receptor potential channels), DPI (NOX inhibitor), and TAK242 (Resatorvid, TRL4 inhibitor) did not inhibit IL-1β production enhanced by silencing of miR-181a-2-3p (Figure 5). We further investigated miR-181a-2-3p target genes by seven well known miRNA target prediction programs, namely TargetScan, miRWalk, MicroCosm, PITA, miRanda, DIANA LAB, or PicTar. After selecting overlapping genes, a total of 52 genes were predicted as targets of miR-181a-2-3p (Tables S2,S3). Once predicted genes were analyzed, we further verified that calcium signaling is associated with calcium/calcineurin (PPP3CA, PPP3CB, PPP3CC, PPP3R1, PPP3R2) and NFATC (NFATC1 to NFATC4) family of genes. As a result, knockdown of miR-181a-2-3p significantly enhances the expression levels of PPPCB, PPPCC, PPP3R1, NFATC2, or PPP4R2 in Cd-exposed cells (Figure S1).This result indicated that the negative regulatory effect of miR-181a-2-3p on Cd-induced inflammation in human bronchial epithelial cells is partly mediated via intracellular calcium release-related signaling pathway.
Figure 5.
Effect of miR-181a-2-3p on multiple intracellular signaling pathways. BEAS-2B cells were pretreated with or without inflammatory signal downstream inhibitors (10 µM) for 1 h. Then, cells were transfected with/without miRNA 181a-2-3p inhibitor and then stimulated with 10 µM Cd for 24 h. 2-APB (A), U73122 (B), DPI (C), and TAK202 (D) were analyzed by immunoblotting, and intensities of bands were quantified and normalized with internal control β-actin by Image J software. All data shown are representative of at least three independent experiments. *, P<0.05; **, P<0.01 were considered significant. NS, not significant; Cd, cadmium; DPI, diphenyleneiodonium; 181m, 181 mimics; 181i, 181 inhibitor.
Table S2. The genes predicted to be down-regulated by miR-181a-2-3p in seven miR-databases.
| Gene | FC | P value | Relative of expression levels |
|---|---|---|---|
| SFTPC | −0.655767365 | 3.61E−09 | 12199.90149 |
| BANF1 | −0.460164268 | 3.36E−14 | 63.73298496 |
| DYNLRB1 | −0.432892943 | 2.60E−15 | 146.1455458 |
| TMED3 | −0.388070092 | 9.23E−08 | 23.74731223 |
| TMEM150A | −0.357928322 | 2.15E−08 | 9.946943966 |
| MTX2 | −0.351826053 | 3.71E−10 | 17.39302484w |
| POLE4 | −0.350029087 | 2.81E−08 | 36.00007791 |
| C11orf73 | −0.33690663 | 4.64E−10 | 14.19046089 |
| CLK4 | −0.317810059 | 1.68E−07 | 16.32098212 |
| MAP2K5 | −0.299291122 | 1.40E−08 | 6.535417212 |
| MRPL1 | −0.289296308 | 4.84E−08 | 13.29145089 |
| RNF220 | −0.287288146 | 4.17E−09 | 10.75798231 |
| BCCIP | −0.226971026 | 1.05E−07 | 25.99758884 |
FC, fold change.
Table S3. The genes predicted to be up-regulated by miR-181a-2-3p in seven miR-databases.
| Gene | FC | P value | Relative of expression levels |
|---|---|---|---|
| BCCIP | 0.141359679 | 2.13E−08 | 30.87231165 |
| YME1L1 | 0.14681086 | 1.24E−07 | 31.3662725 |
| SYNCRIP | 0.152373877 | 1.99E−09 | 13.2009573 |
| TBC1D14 | 0.175317547 | 8.04E−08 | 6.08602146 |
| ZFYVE26 | 0.177817115 | 3.91E−08 | 7.613957172 |
| KDM5B | 0.180621772 | 1.56E−15 | 24.675775 |
| MBTPS1 | 0.190016724 | 4.42E−07 | 10.00839376 |
| NFAT5 | 0.198041404 | 4.68E−10 | 13.62896533 |
| TAOK3 | 0.210266949 | 1.17E−07 | 4.626780497 |
| KIAA2026 | 0.210603089 | 3.45E−10 | 3.837854399 |
| SYNJ1 | 0.218088367 | 2.58E−07 | 21.38963896 |
| ZDHHC20 | 0.218521887 | 1.81E−09 | 9.928368397 |
| SNX18 | 0.220512031 | 5.61E−11 | 5.534562161 |
| CAMSAP1 | 0.223960612 | 8.10E−11 | 9.109183396 |
| SETD2 | 0.231841659 | 4.28E−09 | 44.55153945 |
| TACC1 | 0.232335673 | 6.99E−08 | 14.41540211 |
| GOLGA4 | 0.234400668 | 2.03E−07 | 7.844815734 |
| SMG1 | 0.246868909 | 3.20E−07 | 5.789424095 |
| JHDM1D | 0.250957948 | 6.56E−08 | 7.371499332 |
| STAG1 | 0.258121637 | 4.35E−09 | 10.32410087 |
| PIK3C2A | 0.274096929 | 1.11E−07 | 7.167154236 |
| TAOK1 | 0.280821312 | 1.30E−11 | 7.478640816 |
| MED1 | 0.286434652 | 4.12E−10 | 8.941804783 |
| MAPK8 | 0.30084885 | 9.84E−10 | 5.967073227 |
| RLIM | 0.301335706 | 1.20E−10 | 13.72146289 |
| SETD7 | 0.141359679 | 2.13E−08 | 17.96669999 |
| PPP4R2 | 0.317638925 | 1.15E−07 | 30.87231165 |
| CRK | 0.323066169 | 9.06E−16 | 27.2747069 |
| ADRBK2 | 0.33185984 | 5.10E−13 | 7.138990696 |
| KIAA1549 | 0.345497662 | 3.85E−08 | 0.769423816 |
| KDM5A | 0.362528523 | 1.49E−12 | 5.729807498 |
| TNRC6B | 0.3728883 | 8.88E−19 | 4.348090137 |
| VPS13B | 0.382865278 | 2.72E−12 | 2.740252005 |
| NCOR1 | 0.384445394 | 5.15E−10 | 11.26056239 |
| FBXO48 | 0.405376008 | 2.24E−10 | 0.866248945 |
| C9orf41 | 0.472268719 | 1.75E−11 | 3.66602759 |
| ANTXR2 | 0.501711831 | 6.45E−12 | 12.54870428 |
| TCP11L2 | 0.508754542 | 1.32E−10 | 9.555533445 |
| DOCK5 | 0.744290207 | 3.32E−17 | 4.198296448 |
FC, fold change.
Figure S1.
Expression profiles of miR-181a-2-3p target genes. Quantitative RT-PCR analysis of the expression of different genes in Cd-treated cells, transfected with miR-181a-2-3p mimics/inhibitors. *, P<0.05; **, P<0.01, and ***, P<0.001 were considered significant. NS, not significant; Cd, cadmium; 181m, 181 mimics; 181i, 181 inhibitor; ni, miRNA inhibitor negative control.
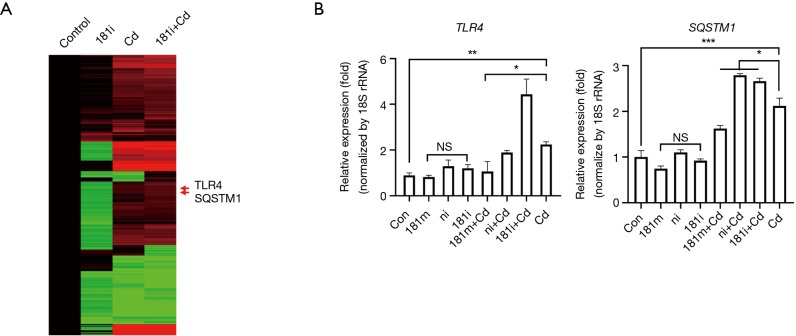
SQSTM1 and TLR4 are potential targets of miR-181α-2-3p
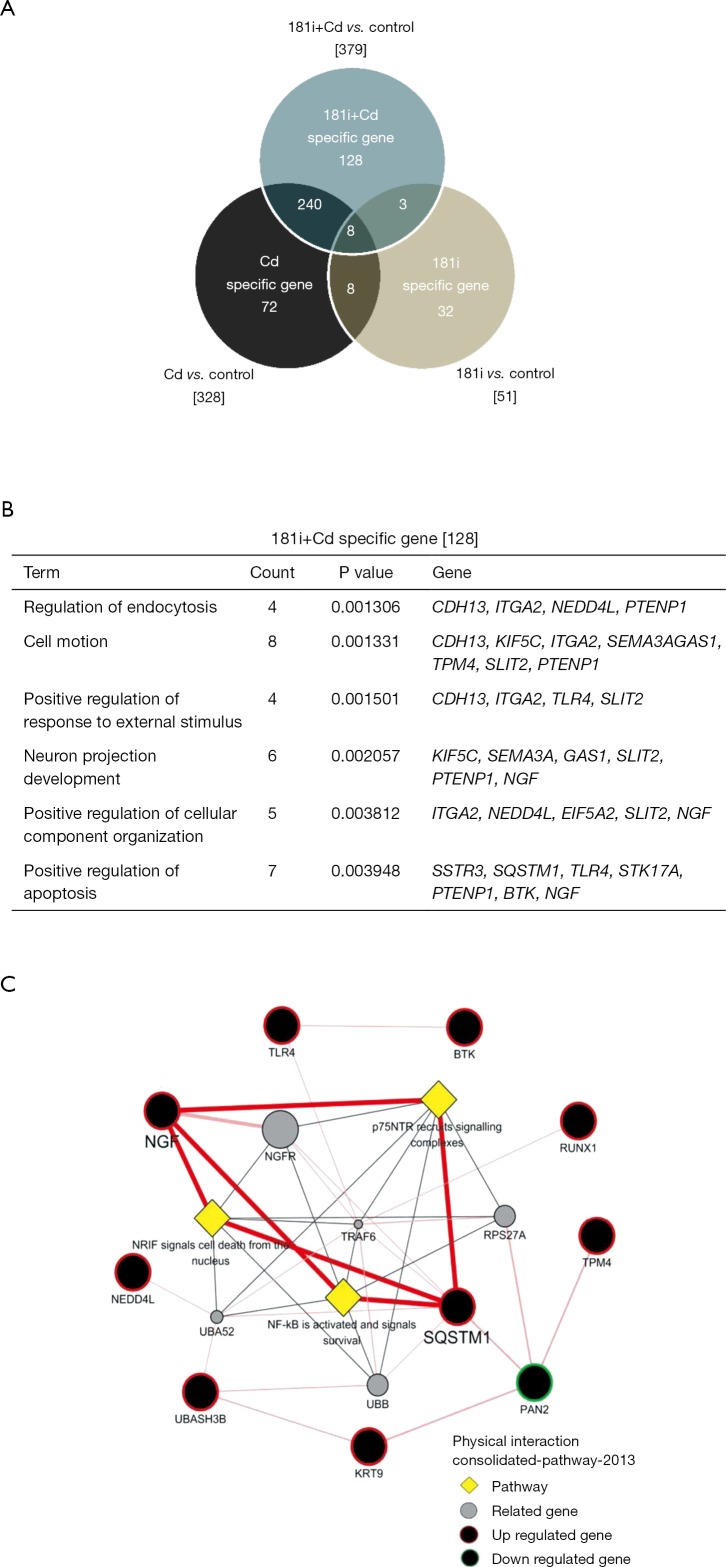
To further identify the potential targets of miR-181a-2-3p contributing to enhancement of the inflammatory response in Cd-treated bronchial epithelial cells, the expression profile of Cd-treated BEAS-2B cells followed by knockdown of miR-181a-2-3p was compared with that of Cd-treated BEAS-2B cells using cDNA microarray. Figure 6A shows a heat map of differentially expressed mRNAs between groups. A total of 128 genes (39 upregulated and 89 downregulated) were altered specifically by miR-181a-2-3p knockdown in Cd-treated cells (−2≤ fc ≤2; Figure S2A). To investigate the biological relevance of these genes, gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes analyses were performed. The main GO categories that included 128 genes were regulation of endocytosis, cell motion, neuron projection development, regulation of response to external stimuli, and apoptosis (Figure S2B). Interactions network analysis revealed that a set of genes, including sequestosome 1 (SQSTM1) and TRL4, are involved in the neurotrophin receptor-interacting factor, p75 neurotrophin receptor, and NF-κB signaling pathway (Figure S2C). We further verified that the expression levels of SQSTM1 and TRL4 genes were elevated by miR-181a-2-3p knockdown in Cd-treated cells compared with nontreated ones (Figure 6B). These data suggested that SQSTM1 and TRL4 could be potential targets of miR-181a-2-3p in Cd-treated bronchial epithelial cells.
Figure 6.
Profiling of global gene expression. The expression patterns for DEGs selected from mRNA profiling analysis on the four groups: control, Cd, miR-181a-2-3p knockdown, and miR-181a-2-3p knockdown treated with Cd. (A) Heat map of differentially expressed mRNAs on the four groups. Green indicates downregulated expression, and red indicates upregulated expression compared with a reference expression level. (B) Validation of miR-181a-2-3p mimics/inhibitors with Cd-treated cells using quantitative RT-PCR. *, P<0.05; **, P<0.01, and ***, P<0.001 were considered significant. NS, not significant; Cd, cadmium; 181m, 181 mimics; 181i, 181 inhibitor; ni, miRNA inhibitor negative control.
Discussion
Numerous studies have recognized miRNAs as major drivers or modifiers in the development and progression of various pulmonary diseases. Thus, elucidating alterations in expression of specific miRNAs and their circulation in blood can be developed to therapeutic targets and biomarkers for pulmonary disease. Although there are limitations on serum miRNA profiling methods and platforms, profiling of serum miRNAs has several advantages. It not only offers new biomarkers for prediction and early detection of COPD but also provides insights into mechanisms of pathogenesis or airway epithelium damage. In our study, we newly identified downregulation of miR-181a-2-3p in lung tissues and serum of COPD patients and demonstrated its negative regulatory role in promoting inflammatory responses and inflammasome activation in human bronchial epithelial cells. These results suggested that miR-181a-2-3p may serve as a new marker in individuals with inflammatory responses and could be a potential target for the development of anti-inflammatory drugs.
miR-181a-2-3p belongs to the miR-181 family, including six members: miR-181a-1/2, miR-181b-1/2, miR-181c, and miR-181d, which are encoded by three independent sequences located on three separate chromosomes (33). The miR-181a-1 and miR-181a-2 clusters locating at chromosomes 1p32.1 and 9q33.3 have a high sequence similarity, which leads to an identical mature miRNA. The miR-181a-2-3p cluster has been reported to be involved in various biological processes, such as development, differentiation, cellular metabolism, and homeostasis (34-36). Detectable miR-181a generally was downregulated in tissues of COPD patients compared with controls and identified as a cytokine-responsive miRNA regulating cellular responses to inflammation (37-39). However, the precise molecular mechanisms by which it exerts this effect have not been fully elucidated.
Aberrant activation of inflammatory responses can lead to cell death and tissue damage caused by activation of multiprotein complexes called inflammasomes, which is constant secretion of cytokines, such as IL-1β. Previous results showed that cigarette smoking induces IL-1β release in the human lung (40). In addition, elevated levels of IL-1α and IL-1β are found in the lungs of COPD patients (30,41), and their secretion is amplified in lungs during disease exacerbations. Several indirect lines of evidence link inflammasome-dependent cytokines to disease pathology of COPD (42,43), but a direct role for the inflammasomes has yet to be clearly shown. Although the effect of miR-181a on inflammatory responses has been reported (37,39), their molecular mechanisms underlying the inflammasome activation have not yet been studied. To our knowledge, ours is the first investigation of the regulatory effect of a miR-181a-2-3p on inflammasome activation. Interestingly, miR-181a-2-3p overexpression attenuated NLRP3, NLRC4, and AIM2 inflammasome-mediated IL-1β production. Unlike miR-181a-2-3p overexpression, its knockdown enhanced NLRP3 inflammasome activation, but did not affect NLRC4, and AIM2 inflammasome-mediated IL-1β production. Thus, understanding the function of miR-181a-2-3p in regulating inflammatory response and inflammasome activation will prompt the development of miRNA-based therapy for COPD.
Under Cd stress, multiple inflammatory signal transduction pathways may get activated. Cd has been shown to be involved in a number of calcium-dependent pathways (44). Recent studies have implicated elevations in cytosolic Ca2+ concentration in NLRP3 inflammasome activation in response to several stimuli (45,46). Although it remains unclear whether the miR-181a-2-3p regulates inflammation via calcium signaling in COPD, there is an accumulating evidence for a significant association of calcium flux with miR-181 family members. In mice, miR-181a has been shown to enhance calcium flux and Erk phosphorylation in T cell (47). Moreover, it has been reported that thapsigargin-induced intracellular Ca2+ release suppressed pre-miR-181a levels through acetylcholinesterase (AChE), protein kinase C (PKC) and protein kinase A (PKA) cascade(s) (48). In another study, it has been demonstrated that miR-181c can directly target the mitochondrial encoded-gene, mt-COX1, which leads to increased ΔΨm and could be associated to increased [Ca2+]m influx and ROS production (49). Although biological studies for its direct role in calcium flux is limited, miR-181a-2-3p has been shown to regulates calcium/calcineurin signaling.
Transcriptomic analysis revealed that TRL4 and SQSTM1 are potential targets of miR-181a-2-3p for elevating inflammatory response in Cd-treated bronchial epithelial cells. TLRs are the first-line sensors that trigger inflammatory cascades (50,51), which leads to upregulation of pro-inflammatory molecules that further amplify the inflammatory response and cell apoptosis. Preclinical studies illustrated that genetic TLR4 knockout was associated with improved outcomes after COPD (36,52). Evidence is now emerging indicating that TLR activation probably affects the expression of miR-181a-2-3p in airway inflammation. However, our results showed that miR-181a-2-3p knockdown did not have any effect on IL-1β secretion after TLR4 inhibitor treatment. In addition, we found no change in the degradation or phosphorylation of IκB αβ, or in the phosphorylation of p65 (data not shown), which indicates that miR-181a-2-3p expression dose not regulate their upstream signaling mediators. These results suggested that miR-181a-2-3p regulation is likely to be complex and redundant or may not be involved in this process. SQSTM1 is known as an ubiquitin-binding adapter protein and promotes tumorigenesis due to autophagy dysfunction and NF-κB activation in Cd-treated BEAS-2B cells (53). Moreover, transcript level of SQSTM1 is significantly increased in COPD-emphysema lungs compared with normal nonsmokers as well as cigarette smoke extract-treated BEAS-2B cells (54). These results suggested that upregulation of SQSTM1 by knockdown of miR-181a-2-3p in Cd-treated BEAS-2B cells may be implicated with the development and progress of COPD.
In conclusion, our study demonstrated that miR-181a-2-3p is downregulated in the lung tissues and serum of COPD patients compared with normal subjects. Downregulation of miR-181a-2-3p in airway cells enhanced inflammatory responses in Cd-treated airway cells. These findings provided an unprecedented biological function of miR-181a-2-3p in human bronchial epithelial cells and their potential target genes (TRL4 and SQSTM1), highlighting its pathological implications related with airway inflammation and inflammasome activation in the COPD.
Figure S2.
Interaction map of miR-181a-2-3p and its target mRNAs using cDNA microarray. Target genes were differentially expressed in normal and miR-181a-2-3p knockdown BEAS-2B cells treated with Cd. (A) Venn diagram of the overlap of mRNA profiles in comparative analyses between different groups. (B) GO term enrichment analysis for biological processes of miR-181a-2-3p knockdown treated with Cd. (C) Interaction network of specific target genes in miR181a-2-3p knockdown cells treated with Cd. Cd, cadmium; 181i, 181 inhibitor.
Acknowledgments
Funding: This study was supported by grants from the National Research Foundation of Korea (NRF) (2017R1A2B4003790) and by a grant from the Global PhD Fellowship Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (2018H1A2A1063441).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was approved by the Institutional Review Board of Kangwon National University Hospital (2012-06-007). All participants provided written informed consent.
Footnotes
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- 1.Kim WJ, Lee CY. Environmental exposures and chronic obstructive pulmonary disease. Mol Cell Toxicol 2017;13:251-5. 10.1007/s13273-017-0027-4 [DOI] [Google Scholar]
- 2.Parker D, Prince A. Innate immunity in the respiratory epithelium. Am J Respir Cell Mol Biol 2011;45:189-201. 10.1165/rcmb.2011-0011RT [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fabbri LM. Smoking, Not COPD, as the Disease. N Engl J Med 2016;374:1885-6. 10.1056/NEJMe1515508 [DOI] [PubMed] [Google Scholar]
- 4.Vogelmeier CF, Criner GJ, Martinez FJ, et al. Global Strategy for the Diagnosis, Management, and Prevention of Chronic Obstructive Lung Disease 2017 Report. GOLD Executive Summary. Am J Respir Crit Care Med 2017;195:557-82. 10.1164/rccm.201701-0218PP [DOI] [PubMed] [Google Scholar]
- 5.Hu G, Zhou Y, Tian J, et al. Risk of COPD from exposure to biomass smoke: a metaanalysis. Chest 2010;138:20-31. 10.1378/chest.08-2114 [DOI] [PubMed] [Google Scholar]
- 6.Van Pottelberge GR, Mestdagh P, Bracke KR, et al. MicroRNA expression in induced sputum of smokers and patients with chronic obstructive pulmonary disease. Am J Respir Crit Care Med 2011;183:898-906. 10.1164/rccm.201002-0304OC [DOI] [PubMed] [Google Scholar]
- 7.Adcock IM, Tsaprouni L, Bhavsar P, et al. Epigenetic regulation of airway inflammation. Curr Opin Immunol 2007;19:694-700. 10.1016/j.coi.2007.07.016 [DOI] [PubMed] [Google Scholar]
- 8.Hassan F, Xu X, Nuovo G, et al. Accumulation of metals in GOLD4 COPD lungs is associated with decreased CFTR levels. Respir Res 2014;15:69. 10.1186/1465-9921-15-69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hutchinson D. Cadmium lung adsorption, citrullination and an enhanced risk of COPD. Eur Respir Rev 2018. doi: . 10.1183/16000617.0054-2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Asker S, Asker M, Yeltekin AC, et al. Serum levels of trace minerals and heavy metals in severe COPD patients with and without pulmonary hypertension. Int J Chron Obstruct Pulmon Dis 2018;13:1803-8. 10.2147/COPD.S164431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou ZH, Lei YX, Wang CX. Analysis of aberrant methylation in DNA repair genes during malignant transformation of human bronchial epithelial cells induced by cadmium. Toxicol Sci 2012;125:412-7. 10.1093/toxsci/kfr320 [DOI] [PubMed] [Google Scholar]
- 12.Liu Q, Zheng C, Shen H, et al. MicroRNAs-mRNAs Expression Profile and Their Potential Role in Malignant Transformation of Human Bronchial Epithelial Cells Induced by Cadmium. Biomed Res Int 2015;2015:902025. 10.1155/2015/902025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liang ZL, Wu DD, Yao Y, et al. Epiproteome profiling of cadmium-transformed human bronchial epithelial cells by quantitative histone post-translational modification-enzyme-linked immunosorbent assay. J Appl Toxicol 2018;38:888-95. 10.1002/jat.3597 [DOI] [PubMed] [Google Scholar]
- 14.Osei ET, Florez-Sampedro L, Timens W, et al. Unravelling the complexity of COPD by microRNAs: it's a small world after all. Eur Respir J 2015;46:807-18. 10.1183/13993003.02139-2014 [DOI] [PubMed] [Google Scholar]
- 15.Perry MM, Moschos SA, Williams AE, et al. Rapid changes in microRNA-146a expression negatively regulate the IL-1beta-induced inflammatory response in human lung alveolar epithelial cells. J Immunol 2008;180:5689-98. 10.4049/jimmunol.180.8.5689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu Q, Han L, Yan W, et al. miR-489 inhibits silica-induced pulmonary fibrosis by targeting MyD88 and Smad3 and is negatively regulated by lncRNA CHRF. Sci Rep 2016;6:30921. 10.1038/srep30921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu TX, Munitz A, Rothenberg ME. MicroRNA-21 is up-regulated in allergic airway inflammation and regulates IL-12p35 expression(). J Immunol 2009;182:4994-5002. 10.4049/jimmunol.0803560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang PX, Cheng J, Zou S, et al. Pharmacological modulation of the AKT/microRNA-199a-5p/CAV1 pathway ameliorates cystic fibrosis lung hyper-inflammation. Nat Commun 2015;6:6221. 10.1038/ncomms7221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hsu ACY, Dua K, Starkey MR, et al. MicroRNA-125a and -b inhibit A20 and MAVS to promote inflammation and impair antiviral response in COPD. JCI Insight 2017;2:e90443. 10.1172/jci.insight.90443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang L, Huang C, Guo Y, et al. MicroRNA-26b modulates the NF-κB pathway in alveolar macrophages by regulating PTEN. J Immunol 2015;195:5404-14. 10.4049/jimmunol.1402933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leidinger P, Keller A, Borries A, et al. Specific peripheral miRNA profiles for distinguishing lung cancer from COPD. Lung Cancer 2011;74:41-7. 10.1016/j.lungcan.2011.02.003 [DOI] [PubMed] [Google Scholar]
- 22.Sato T, Liu X, Nelson A, et al. Reduced miR-146a increases prostaglandin E(2)in chronic obstructive pulmonary disease fibroblasts. Am J Respir Crit Care Med 2010;182:1020-9. 10.1164/rccm.201001-0055OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Conickx G, Mestdagh P, Avila Cobos F, et al. MicroRNA Profiling Reveals a Role for MicroRNA-218-5p in the Pathogenesis of Chronic Obstructive Pulmonary Disease. Am J Respir Crit Care Med 2017;195:43-56. 10.1164/rccm.201506-1182OC [DOI] [PubMed] [Google Scholar]
- 24.Wang K, Yuan Y, Cho JH, et al. Comparing the MicroRNA Spectrum between Serum and Plasma. PLoS One 2012;7:e41561. 10.1371/journal.pone.0041561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee MK, Hong Y, Kim SY, et al. DNA methylation and smoking in Korean adults: epigenome-wide association study. Clin Epigenetics 2016;8:103. 10.1186/s13148-016-0266-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koo HK, Hong Y, Lim MN, et al. Relationship between plasma matrix metalloproteinase levels, pulmonary function, bronchodilator response, and emphysema severity. Int J Chron Obstruct Pulmon Dis 2016;11:1129-37. 10.2147/COPD.S103281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heo HR, Kim J, Kim WJ, et al. Human pluripotent stem cell-derived alveolar epithelial cells are alternatives for in vitro pulmotoxicity assessment. Sci Rep 2019;9:505. 10.1038/s41598-018-37193-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim J, Song H, Heo HR, et al. Cadmium-induced ER stress and inflammation are mediated through C/EBP-DDIT3 signaling in human bronchial epithelial cells. Exp Mol Med 2017;49:e372. 10.1038/emm.2017.125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim WJ, Lim JH, Hong Y, et al. Altered miRNA expression in lung tissues of patients with chronic obstructive pulmonary disease. Mol Cell Toxicol 2017;13:207-12. 10.1007/s13273-017-0022-9 [DOI] [Google Scholar]
- 30.Faner R, Sobradillo P, Noguera A, et al. The inflammasome pathway in stable COPD and acute exacerbations. ERJ Open Res 2016. doi: . 10.1183/23120541.00002-2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu B, Chen S, Luo Y, et al. Calcium Signaling Is Involved in Cadmium-Induced Neuronal Apoptosis via Induction of Reactive Oxygen Species and Activation of MAPK/mTOR Network. PLoS One 2011;6:e19052. 10.1371/journal.pone.0019052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hammad H, Chieppa M, Perros F, et al. House dust mite allergen induces asthma via Toll-like receptor 4 triggering of airway structural cells. Nat Med 2009;15:410-6. 10.1038/nm.1946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ji J, Yamashita T, Budhu A, et al. Identification of microRNA-181 by genome-wide screening as a critical player in EpCAM-positive hepatic cancer stem cells. Hepatology 2009;50:472-80. 10.1002/hep.22989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Henao-Mejia J, Williams A, Goff LA, et al. The microRNA miR-181 is a critical cellular metabolic rheostat essential for NKT cell ontogenesis and lymphocyte development and homeostasis. Immunity 2013;38:984-97. 10.1016/j.immuni.2013.02.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shi Q, Zhou Z, Ye N, et al. MiR-181a inhibits non-small cell lung cancer cell proliferation by targeting CDK1. Cancer Biomark 2017;20:539-46. 10.3233/CBM-170350 [DOI] [PubMed] [Google Scholar]
- 36.Speletas M, Merentiti V, Kostikas K, et al. Association of TLR4-T399I polymorphism with chronic obstructive pulmonary disease in smokers. Clin Dev Immunol 2009;2009:260286. 10.1155/2009/260286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xie W, Li M, Xu N, et al. MiR-181a regulates inflammation responses in monocytes and macrophages. PLoS One 2013;8:e58639. 10.1371/journal.pone.0058639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xie W, Li Z, Li M, et al. miR-181a and inflammation: miRNA homeostasis response to inflammatory stimuli in vivo. Biochem Biophys Res Commun 2013;430:647-52. 10.1016/j.bbrc.2012.11.097 [DOI] [PubMed] [Google Scholar]
- 39.Du Y, Ding Y, Chen X, et al. MicroRNA-181c inhibits cigarette smoke-induced chronic obstructive pulmonary disease by regulating CCN1 expression. Respir Res 2017;18:155. 10.1186/s12931-017-0639-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Doz E, Noulin N, Boichot E, et al. Cigarette smoke-induced pulmonary inflammation is TLR4/MyD88 and IL-1R1/MyD88 signaling dependent. J Immunol 2008;180:1169-78. 10.4049/jimmunol.180.2.1169 [DOI] [PubMed] [Google Scholar]
- 41.Hsu AC, Dua K, Starkey MR, et al. MicroRNA-125a and -b inhibit A20 and MAVS to promote inflammation and impair antiviral response in COPD. JCI Insight 2017;2:e90443. 10.1172/jci.insight.90443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim RY, Pinkerton JW, Gibson PG, et al. Inflammasomes in COPD and neutrophilic asthma. Thorax 2015;70:1199-201. 10.1136/thoraxjnl-2014-206736 [DOI] [PubMed] [Google Scholar]
- 43.Imaoka H, Hoshino T, Takei S, et al. Interleukin-18 production and pulmonary function in COPD. Eur Respir J 2008;31:287-97. 10.1183/09031936.00019207 [DOI] [PubMed] [Google Scholar]
- 44.Choong G, Liu Y, Templeton DM. Interplay of calcium and cadmium in mediating cadmium toxicity. Chem Biol Interact 2014;211:54-65. 10.1016/j.cbi.2014.01.007 [DOI] [PubMed] [Google Scholar]
- 45.Rimessi A, Bezzerri V, Patergnani S, et al. Mitochondrial Ca2+-dependent NLRP3 activation exacerbates the Pseudomonas aeruginosa-driven inflammatory response in cystic fibrosis. Nat Commun 2015;6:6201. 10.1038/ncomms7201 [DOI] [PubMed] [Google Scholar]
- 46.Katsnelson MA, Lozada-Soto KM, Russo HM, et al. NLRP3 inflammasome signaling is activated by low-level lysosome disruption but inhibited by extensive lysosome disruption: roles for K+ efflux and Ca2+ influx. Am J Physiol Cell Physiol 2016;311:C83-100. 10.1152/ajpcell.00298.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li QJ, Chau J, Ebert PJ, et al. miR-181a is an intrinsic modulator of T cell sensitivity and selection. Cell 2007;129:147-61. 10.1016/j.cell.2007.03.008 [DOI] [PubMed] [Google Scholar]
- 48.Guimaraes-Sternberg C, Meerson A, Shaked I, et al. MicroRNA modulation of megakaryoblast fate involves cholinergic signaling. Leuk Res 2006;30:583-95. 10.1016/j.leukres.2005.09.005 [DOI] [PubMed] [Google Scholar]
- 49.Das S, Ferlito M, Kent OA, et al. Nuclear miRNA regulates the mitochondrial genome in the heart. Circ Res 2012;110:1596-603. 10.1161/CIRCRESAHA.112.267732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Janssens S, Beyaert R. Role of Toll-like receptors in pathogen recognition. Clin Microbiol Rev 2003;16:637-46. 10.1128/CMR.16.4.637-646.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Molteni M, Gemma S, Rossetti C. The Role of Toll-Like Receptor 4 in Infectious and Noninfectious Inflammation. Mediators Inflamm 2016;2016:6978936. 10.1155/2016/6978936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Christenson SA, Brandsma CA, Campbell JD, et al. miR-638 regulates gene expression networks associated with emphysematous lung destruction. Genome Med 2013;5:114. 10.1186/gm519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Son YO, Pratheeshkumar P, Roy RV, et al. Nrf2/p62 signaling in apoptosis resistance and its role in cadmium-induced carcinogenesis. J Biol Chem 2014;289:28660-75. 10.1074/jbc.M114.595496 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 54.Tran I, Ji C, Ni I, et al. Role of Cigarette Smoke-Induced Aggresome Formation in Chronic Obstructive Pulmonary Disease-Emphysema Pathogenesis. Am J Respir Cell Mol Biol 2015;53:159-73. 10.1165/rcmb.2014-0107OC [DOI] [PMC free article] [PubMed] [Google Scholar]