Abstract
Background
Several studies have assessed the diagnostic accuracy of serum human epididymis secretory protein 4 (HE4) for lung cancer, but their results were heterogeneous. The aim of this study was to systematically review the available studies and pool their results using meta-analysis.
Methods
PubMed, EMBASE and Web of Science databases were searched up to January 1, 2019 to identify studies investigating the diagnostic accuracy of HE4 for lung cancer. We assessed the quality of eligible studies with the revised Quality Assessment of Diagnostic Accuracy Studies (QUADAS-2) tool. The overall diagnostic sensitivity, specificity, positive and negative likelihood ratios were pooled using a bivariate model. Deeks’s test was applied to detect the degree of publication bias.
Results
A total of 16 studies with 18 cohorts (1,756 lung cancers and 1,446 controls) were included. HE4 had a pooled sensitivity of 0.65 (95% CI: 0.54–0.75), specificity of 0.88 (95% CI: 0.82–0.92), positive likelihood ration of 5.3 (95% CI: 3.7–7.6) and negative likelihood ratio of 0.40 (95% CI: 0.30–0.52). Patient selection bias and partial verification bias were the major design weaknesses of available studies. No publication bias was observed.
Conclusions
HE4 has moderate diagnostic accuracy for lung cancer. Its result should be interpreted in parallel with clinical findings and the results of other conventional tests. Further studies are still needed to rigorously evaluate the diagnostic accuracy of HE4 for lung cancer.
Keywords: Human epididymis secretory protein 4 (HE4), lung cancer, sensitivity, specificity, meta-analysis
Introduction
To improve the prognosis of lung cancer, timely and accurate diagnosis is crucial. Currently, the gold standard for lung cancer diagnosis is biopsy guided by thoracoscopy, bronchoscopy or CT. The major disadvantages of these tools are invasiveness and high cost. In addition, the accuracy of these diagnostic tools is greatly affected by the experience of operators and observers (1). Therefore, it is of great value to develop non-invasive and low-cost tools to detect lung cancer, such as blood tumor markers (2).
During the past decades, several blood tumor markers have been identified for lung cancer diagnosis, such as progastrin-releasing peptide (ProGRP) (3), cytokeratin 19-fragments (CYFRA 21.1) (4) and carcinoma embryonic antigen (CEA) (5). However, the sensitivity and specificity of these tumor markers are far from satisfactory. It seems that multiple tumor markers strategy represents an effective tool for lung cancer diagnosis (6-8). Therefore, developing and evaluating novel tumor markers is promptly needed.
Human epididymis secretory protein 4 (HE4) has been regarded as a tumor marker for ovarian cancer for a long time (9,10). Interestingly, several studies have revealed that it is also a useful diagnostic marker for lung cancer (11-13), but the results of these studies are heterogeneous. Therefore, we performed a systematic review and meta-analysis to assess the diagnostic accuracy of HE4 for lung cancer.
Methods
Databases used for literature searching
This systematic review and meta-analysis was conducted following the PRISMA-DTA (Preferred Reporting Items for a Systematic Review and Meta-analysis of Diagnostic Test Accuracy Studies) guidelines (14) (Tables S1,S2). Three databases, including the PubMed, EMBASE and Web of Science, were searched up to January 1, 2019 to identify eligible studies. The search algorithm in PubMed was: (HE4 OR "Human Epididymis Protein 4" OR "WFDC2 protein, human"[nm]) and ("Lung Neoplasms"[mesh] OR "lung cancer" OR "lung carcinoma*" OR "lung tumor" OR "lung neoplasm*" OR "malignant lung disease*"). Similar search strategy was used for EMBASE and Web of Science. In addition, all references listed in eligible studies were also manually searched.
Table S1. PRISMA-DTA checklist for full-text.
| Section/topic | # | PRISMA-DTA checklist item | Reported on page # |
|---|---|---|---|
| Title/abstract | |||
| Title | 1 | Identify the report as a systematic review (+/− meta-analysis) of diagnostic test accuracy (DTA) studies | 1 |
| Abstract | 2 | Abstract: See PRISMA-DTA for abstracts | Table S2 |
| Introduction | |||
| Rationale | 3 | Describe the rationale for the review in the context of what is already known | 1−2 |
| Clinical role of index test | D1 | State the scientific and clinical background, including the intended use and clinical role of the index test, and if applicable, the rationale for minimally acceptable test accuracy (or minimum difference in accuracy for comparative design) | 1−2 |
| Objectives | 4 | Provide an explicit statement of question(s) being addressed in terms of participants, index test(s), and target condition(s) | 2 |
| Methods | |||
| Protocol and registration | 5 | Indicate if a review protocol exists, if and where it can be accessed (e.g., Web address), and, if available, provide registration information including registration number. | 2 |
| Eligibility criteria | 6 | Specify study characteristics (participants, setting, index test(s), reference standard(s), target condition(s), and study design) and report characteristics (e.g., years considered, language, publication status) used as criteria for eligibility, giving rationale | 2 |
| Information sources | 7 | Describe all information sources (e.g., databases with dates of coverage, contact with study authors to identify additional studies) in the search and date last searched | 2 |
| Search | 8 | Present full search strategies for all electronic databases and other sources searched, including any limits used, such that they could be repeated | 2 |
| Study selection | 9 | State the process for selecting studies (i.e., screening, eligibility, included in systematic review, and, if applicable, included in the meta-analysis) | 2 |
| Data collection process | 10 | Describe method of data extraction from reports (e.g., piloted forms, independently, in duplicate) and any processes for obtaining and confirming data from investigators | 2 |
| Definitions for data extraction | 11 | Provide definitions used in data extraction and classifications of target condition(s), index test(s), reference standard(s) and other characteristics (e.g., study design, clinical setting) | 2 |
| Risk of bias and applicability | 12 | Describe methods used for assessing risk of bias in individual studies and concerns regarding the applicability to the review question | 2 |
| Diagnostic accuracy measures | 13 | State the principal diagnostic accuracy measure(s) reported (e.g., sensitivity, specificity) and state the unit of assessment (e.g., per-patient, per-lesion) | 2 |
| Synthesis of results | 14 | Describe methods of handling data, combining results of studies and describing variability between studies. This could include, but is not limited to: (I) handling of multiple definitions of target condition; (II) handling of multiple thresholds of test positivity; (III) handling multiple index test readers; (IV) handling of indeterminate test results; (V) grouping and comparing tests; (VI) handling of different reference standards | 2 |
| Meta-analysis | D2 | Report the statistical methods used for meta-analyses, if performed | 2 |
| Additional analyses | 16 | Describe methods of additional analyses (e.g., sensitivity or subgroup analyses, meta-regression), if done, indicating which were pre-specified | 2 |
| Results | |||
| Study selection | 17 | Provide numbers of studies screened, assessed for eligibility, included in the review (and included in meta-analysis, if applicable) with reasons for exclusions at each stage, ideally with a flow diagram | 2 |
| Study characteristics | 18 | For each included study provide citations and present key characteristics including: (I) participant characteristics (presentation, prior testing); (II) clinical setting; (III) study design; (IV) target condition definition; (V) index test; (VI) reference standard; (VII) sample size; (VIII) funding sources | 2–3 |
| Risk of bias and applicability | 19 | Present evaluation of risk of bias and concerns regarding applicability for each study | 3 |
| Results of individual studies | 20 | For each analysis in each study (e.g., unique combination of index test, reference standard, and positivity threshold) report 2×2 data (TP, FP, FN, TN) with estimates of diagnostic accuracy and confidence intervals, ideally with a forest or receiver operator characteristic (ROC) plot | 3–4 |
| Synthesis of results | 21 | Describe test accuracy, including variability; if meta-analysis was done, include results and confidence intervals | 4 |
| Additional analysis | 23 | Give results of additional analyses, if done (e.g., sensitivity or subgroup analyses, meta-regression; analysis of index test: failure rates, proportion of inconclusive results, adverse events) | 4–5 |
| Discussion | |||
| Summary of evidence | 24 | Summarize the main findings including the strength of evidence | 6 |
| Limitations | 25 | Discuss limitations from included studies (e.g., risk of bias and concerns regarding applicability) and from the review process (e.g., incomplete retrieval of identified research) | 7 |
| Conclusions | 26 | Provide a general interpretation of the results in the context of other evidence. Discuss implications for future research and clinical practice (e.g., the intended use and clinical role of the index test) | 8 |
| Funding | |||
| Funding | 27 | For the systematic review, describe the sources of funding and other support and the role of the funders | 8 |
For more information, visit: www.prisma-statement.org. TP, true positive; TN, true negative; FP, false positive; FN, false negative.
Table S2. PRISMA-DTA checklist for abstract.
| Section/topic | # | PRISMA-DTA for abstracts checklist item | Reported on page # |
|---|---|---|---|
| Title and purpose | |||
| Title | 1 | Identify the report as a systematic review (+/− meta-analysis) of diagnostic test accuracy (DTA) studies | 1 |
| Objectives | 2 | Indicate the research question, including components such as participants, index test, and target conditions | 1 |
| Methods | |||
| Eligibility criteria | 3 | Include study characteristics used as criteria for eligibility | 1 |
| Information sources | 4 | List the key databases searched and the search dates | 1 |
| Risk of bias & applicability | 5 | Indicate the methods of assessing risk of bias and applicability | 1 |
| Synthesis of results | A1 | Indicate the methods for the data synthesis | 1 |
| Results | |||
| Included studies | 6 | Indicate the number and type of included studies and the participants and relevant characteristics of the studies (including the reference standard) | 1 |
| Synthesis of results | 7 | Include the results for the analysis of diagnostic accuracy, preferably indicating the number of studies and participants. Describe test accuracy including variability; if meta-analysis was done, include summary results and confidence intervals | 1 |
| Discussion | |||
| Strengths and limitations | 9 | Provide a brief summary of the strengths and limitations of the evidence | 1 |
| Interpretation | 10 | Provide a general interpretation of the results and the important implications | 1 |
| Other | |||
| Funding | 11 | Indicate the primary source of funding for the review | NA |
| Registration | 12 | Provide the registration number and the registry name | NA |
For more information, visit: www.prisma-statement.org. NA, not applicable.
Study selection
All retrieved studies were imported into Endnote, a widely-used literature management software, to remove duplicate publications. Two investigators independently reviewed the titles and abstracts of the retrieved studies to verify their eligibility. The inclusion criteria were: (I) studies investigating the diagnostic accuracy of blood HE4 for lung cancer; (II) both sensitivity and specificity were available to construct a two-by-two table. The exclusion criteria were: (I) animal studies; (II) non-English published studies; (III) studies with sample sizes less than 10; (IV) case reports, conference abstracts and letter to the editors. For duplicate studies, only the study with sufficient information or larger sample size was included. All retrieved studies were independently screened by two reviewers and any discrepancies were resolved by consensus and full-text reviewing.
Quality assessment and data extraction
We extracted following data from the included studies: name of the first author; publication year, sources of the subjects, HE4 assays, reference standard for lung cancer diagnosis, sample sizes of lung cancer and control, threshold and its corresponding sensitivity and specificity, area under receiver operating characteristics (ROC) curve (AUC) and characteristics of the control. Two-by-two tables were constructed with sensitivity, specificity, sample sizes of lung cancer and control in each eligible study. The formulas used to construct the two-by-two table were: true positive (TP) = number of lung cancer patients × sensitivity; true negative (TN) = number of control × specificity; false negative (FN) = number of lung cancer patients × (1− sensitivity); false positive (FP) = number of control × (1−specificity). In studies with healthy individuals and benign lung diseases (BLDs) as the control, if the healthy individuals could be removed from final analysis, we constructed the two-by-two tables with BLDs only.
The quality of eligible studies was assessed by the revised Quality Assessment for Studies of Diagnostic Accuracy tool (QUADAS-2) (15). Any discrepancies in quality assessment and data extraction were resolved by consensus.
Statistical analysis
The pooled sensitivity and specificity of HE4 were calculated using a bivariate model (16). A summary ROC (sROC) curve was used to estimate the overall diagnostic accuracy of HE4 (17). A funnel plots and the Deeks’s test were applied to assess the potential publication bias (18). Subgroup analysis was performed to explore the sources of variability. We used the Stata 13.0 (Stata Corp LP, College Station, TX, USA) with the midas command to perform all statistical analyses. Review Manager 5.3 was used to synthesize forest plots.
Results
Summary of eligible studies
Figure S1 is a flowchart depicting the study selecting process. Finally, 16 studies with 3,202 subjects (1,756 lung cancers and 1,446 controls) were identified (8,12,13,19-31). The studies performed by Yoon et al. (29) and Hertlein et al. (23) enrolled two cohorts; therefore, a total of 18 cohorts were included in this systematic review. The characteristics of these studies were summarized in Table 1. Five of the included studies were performed in China (20,21,25,27,30), four were in Turkey (8,12,19,26), two were in Korea (28,29), two were in Japan (22,24). The remaining studies were performed in Hungary (13), Poland (31) and Germany (23). Chemiluminescent immunoassay (CMIA) developed by Architect was used in eight studies (8,12,13,23,26-28,31), and enzyme immunoassay (EIA) developed by Fujirebio was used in six studies (19-22,24,29). Two studies used electrochemiluminescence immunoassay (ECLIA) developed by Roche (25,30). The controls in included studies were various, including healthy individuals (13,20,24,29-31), BLDs (12,23,28), healthy individuals and BLDs (8,19,22,25-27) and tuberculosis (21). Only one study was industry funded (28).
Figure S1.
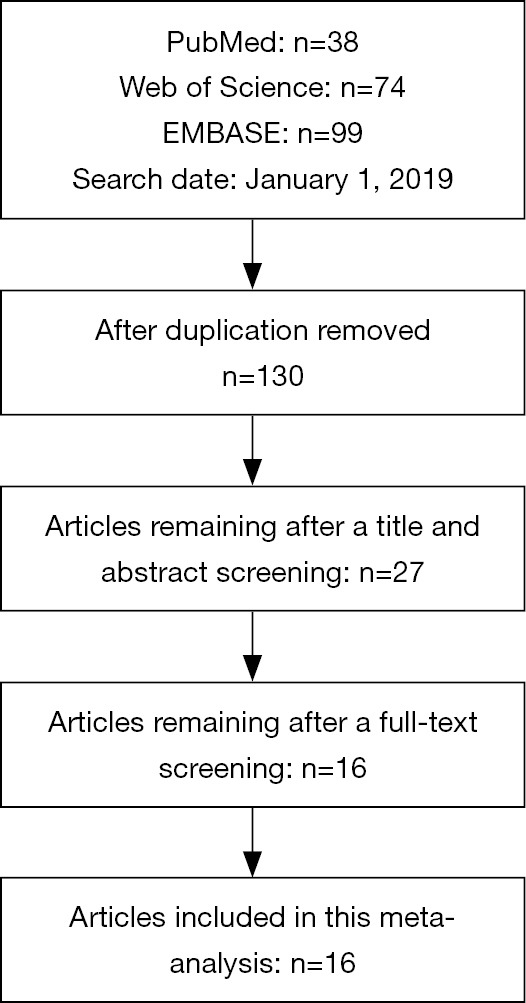
Flow chart illustrating the literature search and study selection process.
Table 1. Summary of eligible studies.
| Author | Year | Country | Disease/control | NSCLC/SCLC | Controls | HE4 assay | Reference | Funding sources |
|---|---|---|---|---|---|---|---|---|
| Korkmaz (8) | 2018 | Turkey | 99/30 | 77/22 | HCs, BLDs | CMIA (Architect) | Clinical course and histology | Non-industry |
| Mo (25) | 2018 | China | 217/80 | 217/0 | HCs, BLDs | ECLIA (Roche) | Unknown | None |
| Kumbasar (26) | 2017 | Turkey | 31/31 | 31/0 | HCs, BLDs | CMIA (Architect) | Unknown | None |
| Huang (27) | 2017 | China | 82/63 | 82/0 | HCs, BLDs | CMIA (Architect) | Histology | Non-industry |
| Choi (28) | 2017 | Korea | 100/57 | 87/7 | BLDs | CMIA (Architect) | Histology | Industry |
| Yoon (29), cohort 1 | 2016 | Korea | 280/515 | 280/0 | HCs | EIA (Fujirebio) | Unknown | None |
| Yoon (29), cohort 2 | 2016 | Korea | 75/75 | 75/0 | HCs | EIA (Fujirebio) | Unknown | None |
| Zeng (30) | 2016 | China | 112/50 | 81/31 | HCs | ECLIA (Roche) | Histology | Non-industry |
| Wojcik (31) | 2016 | Poland | 63/66 | 0/63 | HCs | CMIA (Architect) | Unknown | None |
| Dikmen (12) | 2015 | Turkey | 53/27 | 53/0 | BLDs | CMIA (Architect) | Clinical course and histology | None |
| Ucar (19) | 2014 | Turkey | 64/57 | 40/24 | HCs, BLDs | EIA (Fujirebio) | Histology | Non-industry |
| Wang (20) | 2014 | China | 49/30 | 0/49 | HCs | EIA (Fujirebio) | Histology | Non-industry |
| Nagy (13) | 2014 | Hungary | 90/90 | 69/15 | HCs | CMIA (Architect) | Histology and imaging | Non-industry |
| Liu (21) | 2013 | China | 190/114 | 169/21 | TB | EIA (Fujirebio) | Unknown | Non-industry |
| Yamashita (22) | 2012 | Japan | 102/74 | 102/0 | HCs, BLDs | EIA (Fujirebio) | Histology | None |
| Hertlein (23), female | 2012 | Germany | 23/19 | Unknown | BLDs | CMIA (Architect) | Histology | None |
| Hertlein (23), male | 2012 | Germany | 77/31 | Unknown | BLDs | CMIA (Architect) | Histology | None |
| Iwahori (24) | 2012 | Japan | 49/37 | 40/9 | HCs | EIA (self-made) | Unknown | Non-industry |
HCs, healthy controls; BLDs, benign lung diseases; TB, tuberculosis; NSCLC, non-small cell lung cancer; SCLC, small cell lung cancer; CMIA, chemiluminescent immunoassay; EIA, enzyme immunoassay; ECLIA, electrochemiluminescence immunoassay.
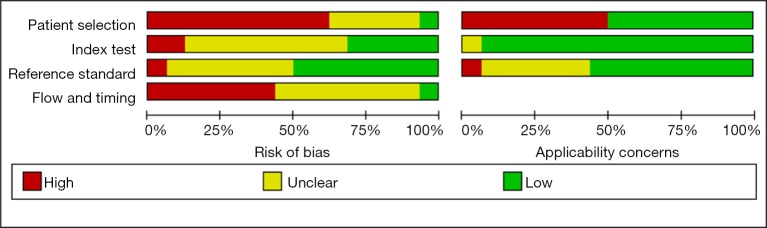
Figure S2 depicts the quality of included studies. Generally, the quality of the included studies was poor. Patient selection and flow and timing domains of some included studies were labeled as high bias because they used healthy individuals as control. Flow and timing domain of some studies were labeled as unclear because the partial verification bias was not reported. Reference domain of some studies was labeled as unclear because criteria used for lung cancer diagnosis were not reported.
Figure S2.
Quality assessment of included studies.
Main findings of included studies and meta-analysis
Table 2 summarizes the main findings of the eligible studies. The AUCs of HE4 in the eligible studies ranged from 0.61 to 0.99. The thresholds used in majority of the eligible studies was around 60 to 100 pmol/L. The sensitivities ranged from 0.12 to 0.90, and specificities ranged from 0.57 to 1.00.
Table 2. Diagnostic accuracy of HE4 in the eligible studies.
| Author | AUC (95% CI) | Cut-off | Sensitivity | Specificity | TP | FP | FN | TN |
|---|---|---|---|---|---|---|---|---|
| Nagy (13) | 0.85 (0.79–0.90) | 97.6 pmol/L | 0.64 | 0.96 | 58 | 4 | 32 | 86 |
| Yoon (29), cohort 1 | 0.82 (unknown) | Unknown | 0.51 | 0.94 | 144 | 31 | 136 | 484 |
| Yoon (29), cohort 2 | 0.84 (unknown) | Unknown | 0.58 | 0.89 | 43 | 8 | 32 | 67 |
| Zeng (30) | 0.82 (0.75–0.89) | 66.8 pmol/L | 0.44 | 0.95 | 49 | 5 | 63 | 45 |
| Liu (21) | 0.75 (0.70–0.80) | 94.01 pmol/L | 0.62 | 0.93 | 98 | 8 | 92 | 106 |
| Korkmaz (8) | 0.61 (0.48–0.73) | 122.5 pmol/L | 0.70 | 0.57 | 69 | 13 | 30 | 17 |
| Wang (20) | 0.85 (0.76–0.94) | 84.19 pmol/L | 0.69 | 0.93 | 34 | 2 | 15 | 28 |
| Yamashita (22) | 0.83 (0.76–0.89) | 50.3 pmol/L | 0.75 | 0.81 | 76 | 14 | 26 | 60 |
| Ucar (19) | 0.78 (0.70–0.87) | 67.5 pmol/L | 0.87 | 0.60 | 56 | 23 | 8 | 34 |
| Mo (25) | 0.81 (0.73–0.88) | 81.26 pmol/L | 0.83 | 0.73 | 180 | 22 | 37 | 58 |
| Huang (27) | 0.76 (0.66–0.82) | 75.0 pmol/L | 0.62 | 0.82 | 51 | 11 | 31 | 52 |
| Wojcik (31) | 0.88 (0.82–0.94) | 77.3 pmol/L | 0.78 | 0.85 | 49 | 10 | 14 | 56 |
| Kumbasar (26) | 0.92 (0.84–1.00) | 70.0 pmol/L | 0.87 | 0.87 | 27 | 4 | 4 | 27 |
| Choi (28) | 0.71 (0.62–0.79) | 70.0 pmol/L | 0.66 | 0.68 | 66 | 18 | 34 | 39 |
| Hertlein (23), female | 0.85 (0.73–0.97) | 77.0 pmol/L | 0.26 | 0.95 | 6 | 1 | 17 | 18 |
| Hertlein (23), male | 0.69 (0.57–0.81) | 89.0 pmol/L | 0.12 | 0.95 | 9 | 1 | 68 | 30 |
| Iwahori (24) | 0.99 (unknown) | 6.56 ng/mL | 0.90 | 1.00 | 44 | 0 | 5 | 37 |
| Dikmen (12) | 0.82 (0.73–0.92) | 70.0 pmol/L | 0.74 | 0.85 | 39 | 4 | 14 | 23 |
AUC, area under receiver operating characteristics curve; TP, true positive; FP, false positive; TN, true negative; FN, false negative.
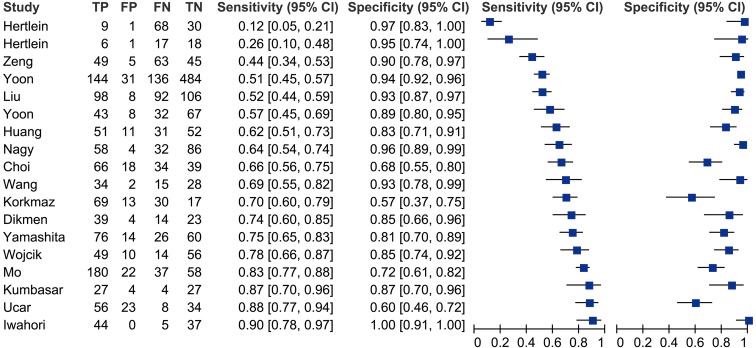
Figure 1 is a forest plot depicting the diagnostic accuracy of HE4 for lung cancer. The pooled sensitivity, specificity, positive likelihood ratio (PLR), negative likelihood ratio (NLR) and diagnostic odds ratio (DOR) of HE4 were 0.65 (95% CI: 0.54–0.75), 0.88 (95% CI: 0.82–0.92), 5.3 (95% CI: 3.7–7.6), 0.40 (95% CI: 0.30–0.52) and 13 (95% CI: 8–21), respectively. Great variability (0.99, 95% CI: 0.98–0.99) was observed among eligible studies.
Figure 1.
Sensitivity and specificity of HE4 in diagnosis of lung cancer assessed by forest plots. HE4, human epididymis secretory protein 4. TP, true positive; FP, false positive; TN, true negative; FN, false negative.
Figure 2 is a sROC plot for HE4, with an AUC of 0.86 (95% CI: 0.82–0.88).
Figure 2.

The summary receiver operating characteristic (sROC) curve of HE4 in lung cancer diagnosis. HE4, human epididymis secretory protein 4.
Subgroup analysis
Considering that great variability was identified among eligible studies and only 37% of them was likely due to threshold effect, we performed a subgroup analysis. The results of subgroup analysis are listed in Table 3. The sensitivity and specificity were not greatly affected by the HE4 test assay and participant sources; however, they were greatly affected by the characteristics of controls. The studies with healthy control had obviously higher AUC than those with BLDs. In the subgroup with EIA assay (Fujirebio), all of the variability could be explained by threshold effect. In addition, in the subgroup with BLD as control, a large portion (83%) of variability could be explained by threshold effect. Taken together, these results indicate that HE4 test assay and control’s characteristics are the potential source of variability.
Table 3. Subgroups analysis.
| Variables | Number of cohorts | AUC (95% CI) | Variability (95% CI) | Proportion of variability likely due to threshold effect | Sensitivity (95% CI) | Specificity (95% CI) |
|---|---|---|---|---|---|---|
| Assays | ||||||
| EIA (Fujirebio) | 6 | 0.84 (0.81–0.87) | 0.97 (0.96–0.99) | 1.00 | 0.66 (0.53–0.77) | 0.87 (0.78–0.93) |
| CMIA (Architect) | 8 | 0.86 (0.83–0.89) | 0.97 (0.96–0.99) | 0.59 | 0.62 (0.43–0.78) | 0.88 (0.79–0.93) |
| Participants | ||||||
| Asian | 10 | 0.85 (0.82–0.88) | 0.98 (0.96–0.99) | 0.08 | 0.66 (0.56–0.74) | 0.88 (0.82–0.93) |
| Europe | 8 | 0.85 (0.82–0.88) | 0.98 (0.97–0.99) | 0.60 | 0.64 (0.41–0.81) | 0.88 (0.74–0.95) |
| Controls | ||||||
| HC only | 7 | 0.92 (0.90–0.94) | 0.72 (0.39–1.00) | 0.13 | 0.66 (0.53–0.77) | 0.93 (0.89–0.96) |
| HC and BLDS | 6 | 0.83 (0.79–0.86) | 0.82 (0.62–1.00) | 0.17 | 0.78 (0.69–0.84) | 0.74 (0.65–0.82) |
| BLDs only | 5 | 0.81 (0.77–0.84) | 0.96 (0.93–0.99) | 0.83 | 0.44 (0.23–0.68) | 0.91 (0.77–0.97) |
AUC, area under curve; CI, confidence interval; HC, healthy control; BLDs,, benign lung diseases; CMIA, chemiluminescent immunoassay; EIA, enzyme immunoassay.
Publication bias
Funnel plot indicated that publication bias was not statistically significant (P=0.97, Figure 3).
Figure 3.

The funnel plot assessment of potential publication bias. ESS, effective sample size.
Discussion
The major findings of present systematic review and meta-analysis are: (I) HE4 had a moderate diagnostic accuracy for lung cancer, with a sensitivity of 0.65 (95% CI: 0.54–0.75), a specificity of 0.88 (95% CI: 0.82–0.92) and an AUC of 0.86 (95% CI: 0.82–0.88) at the threshold between 60 and 100 pmol/L; (II) the quality of available studies were poor because of patient selection bias and partial verification bias; (III) there was no significant publication bias among available studies.
To date, only one study has investigated the diagnostic accuracy of HE4 for lung cancer using meta-analysis (11). Compared with that study, our study has strengths. First, the number of included studies and the overall sample size in our meta-analysis are larger. Therefore, the statistical power of our study is higher. Second, we used a bivariate model to pool the diagnostic accuracy of HE4 while the previous study used a random-effects model with the Meta-Disc software (version 1.4). In the random-effects model, sensitivity and specificity are pooled separately and the trade-off between them is ignored (32). While the bivariate model uses the combination of specificity and sensitivity as the starting point of the analysis (16,33). Therefore, it represents a more reliable method to estimate the diagnostic accuracy of HE4. Third, we explored the sources of variability and found that test assay and characteristics of controls were the potential sources. Fourth, we performed a subgroup analysis and found that using healthy individuals as a control can bias the diagnostic accuracy of HE4.
Sensitivity and specificity are two important characteristics of an index test; however, they have two limitations. The first limitation is that they are greatly affected by the threshold used to define positive and negative results (34,35). By contrast, AUC of sROC is not affected by threshold and thus represents a globe measure of the diagnostic accuracy (17,36). In this meta-analysis, the AUC of HE4 was 0.86 (95% CI: 0.82–0.88), indicating that HE4 has moderate diagnostic accuracy for lung cancer. Another limitation of sensitivity and specificity are that they are not easy to interpret. By contrast, PLR and NLR are considered more clinically meaningful because both pre-test and post-test probabilities are considered (34,37-39). PLR >10 or NLR<0.1 are considered to provide strong evidence to rule in or rule out diagnosis respectively (38). In this meta-analysis, we found the PLR and NLR were 5.3 (95% CI: 3.7–7.6) and 0.40 (95% CI: 0.30–0.52), respectively. These results indicate that HE4, when used alone, is insufficient to rule in or rule out lung cancer, and the serum HE4 concentration should be interpreted in parallel with other clinical findings.
Currently, the diagnosis and classification of lung cancer are based on biopsy guided by thoracoscopy, bronchoscopy or CT. The major limitation of biopsy is that can cause some complications such as infection and bleeding. Therefore, the potential benefit and harm of biopsy should be fully considered before performing biopsy. Previous studies have indicated that HE4 has moderate diagnostic accuracy for lung cancer. However, it should be noted that previous studies only reported the diagnostic characteristics (e.g., sensitivity, specificity, PLR and NLR) at a special threshold. These characteristics, although have been widely used to measure the diagnostic accuracy of an index test, do not incorporate information on consequences. During the past years, decision curve analysis (DCA) (40,41) has been widely used to estimate the net benefit of test for a target disease. To present, none of the studies has used the DCA to estimate the net benefit of HE4 detection for lung cancer. Therefore, further studies with DCA are needed to assess the net benefit of HE4 detection.
The major limitation of this work was that a large portion of included studies has design weaknesses, which might negatively affect the reliability of this meta-analysis. The major design weakness of eligible studies was patient selection bias. All of the included studies did not report the pre-designed inclusion and exclusion criteria, and whether the subjects were enrolled consecutively or randomly was not reported. In other words, all of the included studies were “two-gate” design studies (42). This type of study design may overestimate the diagnostic accuracy of the index test because the studied subjects only represent those who are easy to diagnosis (43-45). Therefore, the conclusions of these studies should be cautiously generalized to other clinical settings. Some diagnostic metrics, such as positive predictive value (PPV) and negative predictive value (NPV), are greatly affected the prevalence of the target disease in the studied cohort (46). These metrics may not be generalized to clinical practice unless the inclusion and exclusion criteria are clearly defined.
In conclusion, our meta-analysis reveals that HE4 seems to be a useful diagnostic marker for lung cancer. Because the currently available studies have study design weakness, especially the patient selection bias, further studies with rigorous design are needed to evaluate the diagnostic accuracy of HE4 for lung cancer.
Acknowledgments
Funding: This work was supported by a grant from the National Natural Science Foundation of China (Grant Number 81860501). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Footnotes
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- 1.Farago AF, Keane FK. Current standards for clinical management of small cell lung cancer. Transl Lung Cancer Res 2018;7:69-79. 10.21037/tlcr.2018.01.16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Velcheti V, Pennell NA. Non-invasive diagnostic platforms in management of non-small cell lung cancer: opportunities and challenges. Ann Transl Med 2017;5:378. 10.21037/atm.2017.08.24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang H, Gu Y, Chen C, et al. Diagnostic value of pro-gastrin-releasing peptide for small cell lung cancer: a meta-analysis. Clin Chem Lab Med 2011;49:1039-46. 10.1515/CCLM.2011.161 [DOI] [PubMed] [Google Scholar]
- 4.Cui C, Sun X, Zhang J, et al. The value of serum Cyfra21-1 as a biomarker in the diagnosis of patients with non-small cell lung cancer: a meta-analysis. J Cancer Res Ther 2014;10 Suppl:C131-4. 10.4103/0973-1482.145835 [DOI] [PubMed] [Google Scholar]
- 5.Okamura K, Takayama K, Izumi M, et al. Diagnostic value of CEA and CYFRA 21-1 tumor markers in primary lung cancer. Lung Cancer 2013;80:45-9. 10.1016/j.lungcan.2013.01.002 [DOI] [PubMed] [Google Scholar]
- 6.Qi W, Li X, Kang J. Advances in the study of serum tumor markers of lung cancer. J Cancer Res Ther 2014;10 Suppl:C95-101. 10.4103/0973-1482.145801 [DOI] [PubMed] [Google Scholar]
- 7.Du Q, Yan C, Wu SG, et al. Development and validation of a novel diagnostic nomogram model based on tumor markers for assessing cancer risk of pulmonary lesions: A multicenter study in Chinese population. Cancer Lett 2018;420:236-41. 10.1016/j.canlet.2018.01.079 [DOI] [PubMed] [Google Scholar]
- 8.Korkmaz ET, Koksal D, Aksu F, et al. Triple test with tumor markers CYFRA 21.1, HE4, and ProGRP might contribute to diagnosis and subtyping of lung cancer. Clin Biochem 2018;58:15-9. 10.1016/j.clinbiochem.2018.05.001 [DOI] [PubMed] [Google Scholar]
- 9.Li F, Tie R, Chang K, et al. Does risk for ovarian malignancy algorithm excel human epididymis protein 4 and ca125 in predicting epithelial ovarian cancer: A meta-analysis. BMC Cancer 2012;12:258. 10.1186/1471-2407-12-258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ferraro S, Panteghini M. Making new biomarkers a reality: The case of serum human epididymis protein 4. Clin Chem Lab Med 2018. [Epub ahead of print] 10.1515/cclm-2018-1111 [DOI] [PubMed] [Google Scholar]
- 11.Cheng D, Sun Y, He H. The diagnostic accuracy of HE4 in lung cancer: a meta-analysis. Dis Markers 2015;2015:352670. 10.1155/2015/352670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dikmen E, Gungor A, Dikmen ZG, et al. Diagnostic efficiency of he4 and cyfra 21-1 in patients with lung cancer. Int J Hematol Oncol 2015;25:44-50. 10.4999/uhod.15739 [DOI] [Google Scholar]
- 13.Nagy B, Bhattoa HP, Steiber Z, et al. Serum human epididymis protein 4 (HE4) as a tumor marker in men with lung cancer. Clin Chem Lab Med 2014;52:1639-48. 10.1515/cclm-2014-0041 [DOI] [PubMed] [Google Scholar]
- 14.McInnes MDF, Moher D, Thombs BD, et al. Preferred Reporting Items for a Systematic Review and Meta-analysis of Diagnostic Test Accuracy Studies: The PRISMA-DTA Statement. JAMA 2018;319:388-96. 10.1001/jama.2017.19163 [DOI] [PubMed] [Google Scholar]
- 15.Whiting PF, Rutjes AW, Westwood ME, et al. QUADAS-2: a revised tool for the quality assessment of diagnostic accuracy studies. Ann Intern Med 2011;155:529-36. 10.7326/0003-4819-155-8-201110180-00009 [DOI] [PubMed] [Google Scholar]
- 16.Reitsma JB, Glas AS, Rutjes AW, et al. Bivariate analysis of sensitivity and specificity produces informative summary measures in diagnostic reviews. J Clin Epidemiol 2005;58:982-90. 10.1016/j.jclinepi.2005.02.022 [DOI] [PubMed] [Google Scholar]
- 17.Walter SD. Properties of the summary receiver operating characteristic (SROC) curve for diagnostic test data. Stat Med 2002;21:1237-56. 10.1002/sim.1099 [DOI] [PubMed] [Google Scholar]
- 18.Deeks JJ, Macaskill P, Irwig L. The performance of tests of publication bias and other sample size effects in systematic reviews of diagnostic test accuracy was assessed. J Clin Epidemiol 2005;58:882-93. 10.1016/j.jclinepi.2005.01.016 [DOI] [PubMed] [Google Scholar]
- 19.Ucar EY, Ozkaya AL, Araz O, et al. Serum and bronchial aspiration fluid HE-4 levels in lung cancer. Tumour Biol 2014;35:8795-9. 10.1007/s13277-014-2134-3 [DOI] [PubMed] [Google Scholar]
- 20.Wang X, Fan Y, Wang J, et al. Evaluating the expression and diagnostic value of human epididymis protein 4 (HE4) in small cell lung cancer. Tumour Biol 2014;35:6847-53. 10.1007/s13277-014-1943-8 [DOI] [PubMed] [Google Scholar]
- 21.Liu W, Yang J, Chi PD, et al. Evaluating the clinical significance of serum HE4 levels in lung cancer and pulmonary tuberculosis. Int J Tuberc lung Dis 2013;17:1346-53. 10.5588/ijtld.13.0058 [DOI] [PubMed] [Google Scholar]
- 22.Yamashita S, Tokuishi K, Moroga T, et al. Serum level of HE4 is closely associated with pulmonary adenocarcinoma progression. Tumour Biol 2012;33:2365-70. 10.1007/s13277-012-0499-8 [DOI] [PubMed] [Google Scholar]
- 23.Hertlein L, Stieber P, Kirschenhofer A, et al. Human epididymis protein 4 (HE4) in benign and malignant diseases. Clin Chem Lab Med 2012;50:2181-8. 10.1515/cclm-2012-0097 [DOI] [PubMed] [Google Scholar]
- 24.Iwahori K, Suzuki H, Kishi Y, et al. Serum HE4 as a diagnostic and prognostic marker for lung cancer. Tumour Biol 2012;33:1141-9. 10.1007/s13277-012-0356-9 [DOI] [PubMed] [Google Scholar]
- 25.Mo D, He F. Serum Human Epididymis Secretory Protein 4 (HE4) is a Potential Prognostic Biomarker in Non-Small Cell Lung Cancer. Clin Lab 2018;64:1421-8. 10.7754/Clin.Lab.2018.180222 [DOI] [PubMed] [Google Scholar]
- 26.Kumbasar U, Dikmen ZG, Yilmaz Y, et al. Serum Human Epididymis Protein 4 (HE4) As A Diagnostic and Follow-Up Biomarker in Patients With Non-Small Cell Lung Cancer. Int J Hematol Oncol 2017;27:137-42. 10.4999/uhod.171830 [DOI] [Google Scholar]
- 27.Huang W, Wu S, Lin Z, et al. Evaluation of HE4 in the Diagnosis and Follow Up of Non-Small Cell Lung Cancers. Clin Lab 2017;63:461-7. 10.7754/Clin.Lab.2016.160818 [DOI] [PubMed] [Google Scholar]
- 28.Choi SI, Jang MA, Jeon BR, et al. Clinical Usefulness of Human Epididymis Protein 4 in Lung Cancer. Ann Lab Med 2017;37:526-30. 10.3343/alm.2017.37.6.526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yoon HI, Kwon OR, Kang KN, et al. Diagnostic Value of Combining Tumor and Inflammatory Markers in Lung Cancer. J Cancer Prev 2016;21:187-93. Erratum in: Erratum: Diagnostic Value of Combining Tumor and Inflammatory Markers in Lung Cancer. [J Cancer Prev 2016]. 10.15430/JCP.2016.21.3.187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zeng Q, Liu M, Zhou N, et al. Serum human epididymis protein 4 (HE4) may be a better tumor marker in early lung cancer. Clin Chim acta 2016;455:102-6. 10.1016/j.cca.2016.02.002 [DOI] [PubMed] [Google Scholar]
- 31.Wojcik E, Tarapacz J, Rychlik U, et al. Human Epididymis Protein 4 (HE4) in Patients with Small-Cell Lung Cancer. Clin Lab 2016;62:1625-32. 10.7754/Clin.Lab.2016.151212 [DOI] [PubMed] [Google Scholar]
- 32.Zamora J, Abraira V, Muriel A, et al. Meta-DiSc: a software for meta-analysis of test accuracy data. BMC Med Res Methodol 2006;6:31. 10.1186/1471-2288-6-31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hu ZD, Wei TT, Yang M, et al. Diagnostic value of osteopontin in ovarian cancer: Meta-analysis and systematic review. PLoS One 2015;10:e0126444. 10.1371/journal.pone.0126444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Linnet K, Bossuyt PM, Moons KG, et al. Quantifying the Accuracy of a Diagnostic Test or Marker. Clin Chem 2012;58:1292-301. 10.1373/clinchem.2012.182543 [DOI] [PubMed] [Google Scholar]
- 35.Dickie GL. Statistical notes. Defining sensitivity and specificity. BMJ 1994;309:539. 10.1136/bmj.309.6953.539a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reitsma JB, Moons KG, Bossuyt PM, et al. Systematic Reviews of Studies Quantifying the Accuracy of Diagnostic Tests and Markers. Clin Chem 2012;58:1534-45. 10.1373/clinchem.2012.182568 [DOI] [PubMed] [Google Scholar]
- 37.Zhou Q, Ye ZJ, Su Y, et al. Diagnostic value of N-terminal pro-brain natriuretic peptide for pleural effusion due to heart failure: a meta-analysis. Heart 2010;96:1207-11. 10.1136/hrt.2009.188474 [DOI] [PubMed] [Google Scholar]
- 38.Deeks JJ, Altman DG. Diagnostic tests 4: likelihood ratios. BMJ 2004;329:168-9. 10.1136/bmj.329.7458.168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Altman DG, Bland JM. Diagnostic tests. 1: Sensitivity and specificity. BMJ 1994;308:1552. 10.1136/bmj.308.6943.1552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vickers AJ, Cronin AM, Elkin EB, et al. Extensions to decision curve analysis, a novel method for evaluating diagnostic tests, prediction models and molecular markers. BMC Med Inform Decis Mak 2008;8:53. 10.1186/1472-6947-8-53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang Z, Rousson V, Lee WC, et al. Decision curve analysis: a technical note. Ann Transl Med 2018;6:308. 10.21037/atm.2018.07.02 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rutjes AW, Reitsma JB, Vandenbroucke JP, et al. Case-control and two-gate designs in diagnostic accuracy studies. Clin Chem 2005;51:1335-41. 10.1373/clinchem.2005.048595 [DOI] [PubMed] [Google Scholar]
- 43.Lijmer JG, Mol BW, Heisterkamp S, et al. Empirical evidence of design-related bias in studies of diagnostic tests. JAMA 1999;282:1061-6. 10.1001/jama.282.11.1061 [DOI] [PubMed] [Google Scholar]
- 44.Schmidt RL, Factor RE. Understanding sources of bias in diagnostic accuracy studies. Arch Pathol Lab Med 2013;137:558-65. 10.5858/arpa.2012-0198-RA [DOI] [PubMed] [Google Scholar]
- 45.Whiting P, Rutjes AW, Reitsma JB, et al. Sources of variation and bias in studies of diagnostic accuracy: a systematic review. Ann Intern Med 2004;140:189-202. 10.7326/0003-4819-140-3-200402030-00010 [DOI] [PubMed] [Google Scholar]
- 46.Hu ZD. Circulating biomarker for malignant pleural mesothelioma diagnosis: pay attention to study design. J Thorac Dis 2016;8:2674-6. 10.21037/jtd.2016.10.94 [DOI] [PMC free article] [PubMed] [Google Scholar]