Abstract
Variations in the HIV genome influence HIV/AIDS epidemiology. We report here a novel HIV-1 unique recombinant form (URF) isolated from an HIV-infected female (NACMR092) in Cameroon, based on the analyses of near-full-length viral genome (partial gag, full-length pol, env, tat, rev, vif, vpr, vpu, and nef genes, and partial 3′-long terminal repeat). Phylogeny, recombination breakpoints, and recombination map analyses showed that NACMR092 was infected with a mosaic URF that had eight breakpoints (two in gag, one in pol, one in vpr, two in env, and two in the nef regions), nine subgenomic regions, and included fragments that had important similarities with HIV-1 subtypes A1, CRF02_AG, and CRF01_AE. This novel mosaic URF underscores complex recombination events occurring between HIV-1 subtypes circulating in Cameroon. Continued monitoring and detection of such recombinants and accurate classification of HIV genotype is important for tracking viral molecular epidemiology and antigenic diversity.
Keywords: HIV-1, near-full-length genome sequence, NACMR092, Cameroon, phylogenetic trees, recombination breakpoints
The human immunodeficiency virus (HIV) is characterized by a very high genetic variability, due to the lack of DNA proofreading activity of the reverse transcriptase (RT) enzyme and pharmacological selective pressure.1,2 This has resulted in mutations, high rates of intra- and intermolecular recombination within infected hosts, and very high clade diversity. HIV-1 accounts for >95% of all infections and includes four groups: M (major), O (outlier), N (non-M non-O), and P.3,4 HIV-1 group M accounts for the vast majority of infections globally and includes nine pure subtypes (A–D, F–H, J, and K), sub-subtypes (A1 and A2, F1 and F2), about 96 circulating recombinant forms (CRFs), and several unique (unclassified) recombinant forms (URFs).4,5 CRFs include CRF01_AE that is prevalent in Southeast Asia, and CRF02_AG that is prevalent in West and Central Africa.5 Some CRFs have originated from a recombination of three or more parental viral strains and are termed complex CRFs or CRF_cpx.5
The epidemiology of HIV/AIDS in Cameroon is characterized by a high genetic diversity, with the predominance of CRF02_AG but also the presence of individuals infected with pure subtypes (A, D, F2, G, H, and K) and other CRFs, including CRF01_AE, CRF22_01A1, CRF09_cpx, CRF11_cpx, and CRF13_cpx.6 Recombination between these subtypes following superinfections can result in the formation of new epidemiologically important recombinant founder strains. Our previous analysis of Tat sequences from HIV-infected persons in Cameroon showed an individual (ID# NACMR092) infected with a novel URF Tat HIV-1 CRF22_01A1/CRF01_AE.7 However, sequencing of other genomic regions of this viral isolate was required for its full characterization and classification. Therefore, our goal in the present study was to sequence the full-length or near-full-length genome of that novel recombinant isolate and fully characterize its probable parental origin. Near-full-length genome analyses demonstrated that this person was infected with a novel HIV-1 URF that had genomic fragments with similarities to subtype A1, CRF01_AE and CRFO2_AG viruses.
The sample analyzed in this study was collected in 2008 as part of a cross-sectional study aimed at analyzing the influence of HIV genetic diversity on viral neuropathogenesis. This study was performed in accordance with the guidelines of Helsinki Declaration and was approved by the Cameroon National Ethics Committee, as well as the institutional review board of the University of Nebraska Medical Center (UNMC). Written informed consent was obtained from the study participants, and data were processed using unique identifiers to ensure confidentiality. Blood sample was collected in 2008 from a 39-year-old female in Yaoundé, Cameroon. At the time of specimen collection, she was antiretroviral therapy-naïve, her plasma viral load was 1,780,208 (6.25 log) copies/mL, and her CD4+ T cell count was 134 cells/μL.
Specimen collection, HIV serology, CD4 cell count, and viral load quantification were performed as previously described.7 HIV-1 RNA was extracted from plasma using QIAamp Viral RNA mini kit (Qiagen, Inc., Germantown, MD) per manufacturer's protocol. Purified RNA (500 ng) was reverse-transcribed in a total volume of 20 μL containing 500 mM dNTP, 2.5 mM primer, 1 × RT buffer, 5 mM MgCl2, 10 mM Dithiothreitol, 40 U RnaseOUT, and 400 U SuperScriptTM III RT (Life Technologies, Grand Island, NY). RNA was mixed with primers, dNTPs, incubated at 65°C for 5 min, snap-chilled (2 min on ice), and the remaining reaction components added. Reverse transcription was carried out at 50°C for 1 h followed by incubation at 70°C for 10 min. The resulting cDNA was cryopreserved at −80°C until further use.
The near-full-length genomic sequence was generated using a modified version of a previously reported protocol.8 The HIV-1 genome was amplified using three overlapping amplicons and a nested PCR strategy. The first amplicon covered nucleotides 769–3338; the second amplicon spanned nucleotides 2483–6231; and the third amplicon spanned nucleotides 5861–9500 of HXB2 reference sequence.9 Each PCR was carried out in a total volume of 50 μL containing 5 μL cDNA, 25 μL 2 × KAPA HiFi HotStart ready mix (KAPA Biosystems, Wilmington, MA), and 10 pmol of forward and reverse primers. For the second-round nested PCR, cDNA was replaced with 5 μL of the first-round PCR amplicon. The thermal cycling condition for each PCR round was 95°C, 5 min, followed by 10 cycles of 98°C, 20 s; 60°C, 30 s; 72°C, 3 min; and then 25 cycles of 94°C, 10 s, 55°C, 30 s; 68°C, 3 min; and an extension step at 72°C, 10 min. Amplicons were detected by agarose (1%) gel electrophoresis.
PCR products were purified using PureLink Quick PCR Purification Kit (Invitrogen, Carlsbad, CA) and subjected to double-strand DNA sequencing to cover the entire amplicon using a set of sequencing primers (Table 1). Sequencing reactions were carried out using the BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA) per manufacturer's instructions, followed by capillary electrophoresis performed on an Applied Biosystems PRISM 3730 Genetic Analyzer at the UNMC DNA Sequencing Core Facility. The primers used for PCR amplifications and DNA sequencing are described in Table 1.
Table 1.
Primers used for the Amplification and Sequencing of Near-Full-Length HIV-1 Genome8
| Primer | Positiona | Sequence (5′ to 3′) |
|---|---|---|
| Fragment 1, first PCR | ||
| msf12b (+/+) | 623–649 | AAATCTCTAGCAGTGGCGCCCGAACAG |
| RT3474R (+/−) | 3477–3453 | GAATCTCTCTGTTTTCTGCCAGTTC |
| Fragment 1, second PCR | ||
| f2nst (+/+) | 769–793 | GCGGAGGCTAGAAGGAGAGAGATGG |
| proRT (+/−) | 3338–3308 | TTTCCCCACTAACTTCTGTATGTCATTGACA |
| Fragment 2, first PCR | ||
| POLoutF1 (+/+) | 2253–2277 | CCTCAAATCACTCTTTGGCARCGAC |
| VIF-VPUR1 (+/−) | 6352–6324 | GGTACCCCATAATAGACTGTRACCCACAA |
| Fragment 2, second PCR | ||
| POLinF1 (+/+) | 2483–2509 | AGGACCTACRCCTGTCAACATAATTGG |
| VIF-VPUinR1 (+/−) | 6231–6207 | CTCTCATTGCCACTGTCTTCTGCTC |
| Fragment 3, first PCR | ||
| ENVoutF1 (+/+) | 5550–5574 | AGARGAYAGATGGAACAAGCCCCAG |
| UNINEF7 (+/ −) | 9632–9605 | GCACTCAAGGCAAGCTTTATTGAGGCTT |
| Fragment 3, second PCR | ||
| ENVinF1 (+/+) | 5861–5884 | TGGAAGCATCCRGGAAGTCAGCCT |
| NefinR2 (+/−) | 9555–9533 | TCTACCTAGAGAGACCCAGTACA |
| Sequencing primers | ||
| F2NST (+/+) | 769–793 | GCGGAGGCTAGAAGGAGAGAGATGG |
| JL19 (+/−) | 1270–1249 | CTTCTATTACTTTTACCCATGC |
| HH (+/+) | 1406–1426 | ATGAGGAAGCTGCAGAATGGG |
| POLCLO1 (+/+) | 2071–2096 | GAGAGACAGGCTAATTTTTTAGGGAA |
| SP2AS (+/−) | 2165–2147 | GGTGGGGCTGTTGGCTCTG |
| SP4AS (+/−) | 2723–2700 | AGTATTGTATGGATTTTCAGGCCC |
| AZT9 (+/+) | 2875–2893 | TGGATGTGGGTGATGCATA |
| AZT4 (+/−) | 3281–3261 | TATAGGCTGTACTGTCCATTT |
| POLinF1 (+/+) | 2483–2509 | AGGACCTACRCCTGTCAACATAATTGG |
| DGPOLF7 (+/+) | 3495–3521 | GGAATATATTATGACCCATCAAAAGAC |
| POLSEQ3 (+/−) | 3567–3546 | GATATGWCCACTGGTCTTGCCC |
| POLI (+/+) | 4116–4136 | GAGCAGTTAATAAAAAAGGAA |
| POLI (+/−) | 4169–4146 | TTTGTGTGCTGGTACCCATGCCAG |
| POLK (+/+) | 4602–4625 | ACGGTTAAGGCCGCCTGTTGGTGG |
| SP1AS (+/−) | 4776–4752 | GGATGAATACTGCCATTTGTACTGC |
| ACC1 (+/−) | 5219–5196 | TTCAGAAGTATACATCCCACTAGG |
| VIFB (+/+) | 5319–5340 | ATATAGCACACAAGTAGACCCT |
| ACC6 (+/−) | 5569–5545 | GCTTGTTCCATCTRTCYTCTGTYAG |
| ED3 (+/+) | 5957–5986 | TTAGGCATCTCCTATGGCAGGAAGAAGCGG |
| ACC8R (+/−) | 5989–5968 | TCTCCGCTTCTTCCTGCCATAG |
| ZFF (+/+) | 6559–6585 | GGGATCAACGCCTAAAGCCATGTGTGA |
| AENVSEQ4 (+/−) | 6838–6817 | CAAGCTTGTGTAATGGCTGAGG |
| E15 (+/+) | 7098–7121 | GTAGAAATTAATTGTACAAGACCC |
| JL98 (+/−) | 7374–7351 | AGAAAAATTCCCCTCCACAATTAA |
| JL109 (+/+) | 7666–7689 | GTGAATTATATAAATATAAAGTAG |
| TUH (+/−) | 7940–7914 | GCCCCAGACTGTGAGTTGCAACAGATG |
| JL104 (+/+) | 8292–8315 | GGAGGCTTGATAGGTTTAAGAATA |
| E8 (+/−) | 8445–8424 | CTCTCTCTCCACCTTCTTCTTC |
| NEF7 (+/+) | 8793–8818 | TAAGATGGGTGGCAAGTGGTCCAAAA |
| JL89 (+/−) | 9089–9064 | TCCAGTCCCCCCTTTTCTTTTAAAAA |
Positions according to HXB2 numbering system.
+/+, sense primer; +/−, antisense primer.
Raw sequence data were manually edited, spliced, and assembled by Sequencher v4.9 to generate the final contig. Multiple sequence alignment of the near-full-length HIV-1 sequence was performed with all known HIV-1 group M reference sequences, using Clustal W.10 The phylogenetic tree of the near-full-length sequence and subregion trees were constructed using the neighbor joining method, as well as the maximum likelihood method and general time reversible model, by testing 1,000 bootstrap replications, using MEGA.v.6.0 software.11 All reference sequences were obtained from the Los Alamos HIV Sequence Database and included HIV isolates from 17 different countries (Cameroon, China, Thailand, France, Democratic Republic of Congo, Sweden, Republic of Korea, Central African Republic, Saudi Arabia, Australia, Rwanda, Uganda, Brazil, Belgium, Afghanistan, Liberia, and Nigeria). Recombination breakpoints relative to HXB2 numbering were determined by boot scanning analysis using Simplot v3.5.1.12 The reliability of plot topologies was assessed by bootstrapping with 100 replicates, 800 bp window, a step size of 50 bp.12 The recombination map was created using the Los Alamos Recombinant HIV-1 Drawing Tool (https://hiv.lanl.gov/content/sequence/DRAW_CRF/recom_mapper.html).
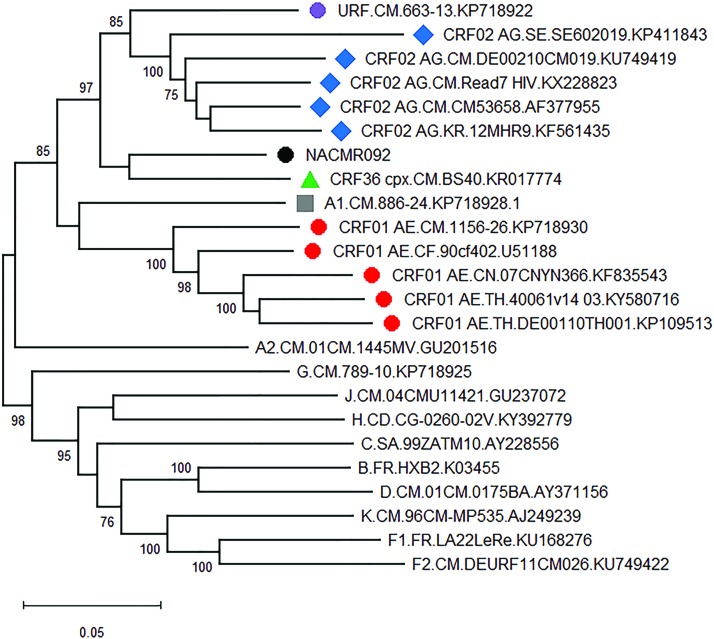
The HIV-1 near-full-length sequence generated from the sample (NACMR092) was 8,524 bp in length, which correspond to nucleotides 874–9403 of the HXB2 genome.9,13 The sequence covered partial gag gene and full-length pol, env, tat, rev, vif, vpr, vpu, and nef genes, as well as a partial 3′-long terminal repeat (LTR). The viral genome from this new isolate had retained intact reading frames for all of its genes, and no frameshift, insertion, or deletion mutation was observed. A nucleoside/nucleotide similarity search in the NCBI sequence database using BLAST14 returned three best matches with >90% similarities with three HIV-1 sequences belonging to different subtypes that had the following accession numbers: AF377955, KR017774, and KP718922—all HIV-1 sequences originating from Cameroon.15–17 Phylogenetic analysis of the near-full-length HIV sequence from NACMR092 performed with group M subtypes and CRFs, using the maximum likelihood or the neighbor joining method of the MEGA 6.0 software,11 gave similar results. Figure 1 are the data obtained using the maximum likelihood method. These data also showed that NACMR092 was distantly related to CRF02_AG, CRF36_cpx, subtype A1, and CRF01_AE.
FIG. 1.
Phylogenetic analysis of the near-full-length genome sequence of the HIV-1 NACMR092 isolate. The maximum likelihood phylogenetic tree was constructed using MEGA 6.0, as described. The black solid circle represents the test sample. NACMR092 clustered together with HIV-1 CRF02_AG (marked by blue solid diamond), URF KP718922 (purple circle), CRF36_cpx (green solid triangle), subtype A1 (grey square), and CRF01_AE (red solid circles). Bootstrap values of 1,000 replicates (>70%) are shown at the corresponding nodes. The scale bar represents 5% genetic distance. Color images are available online.
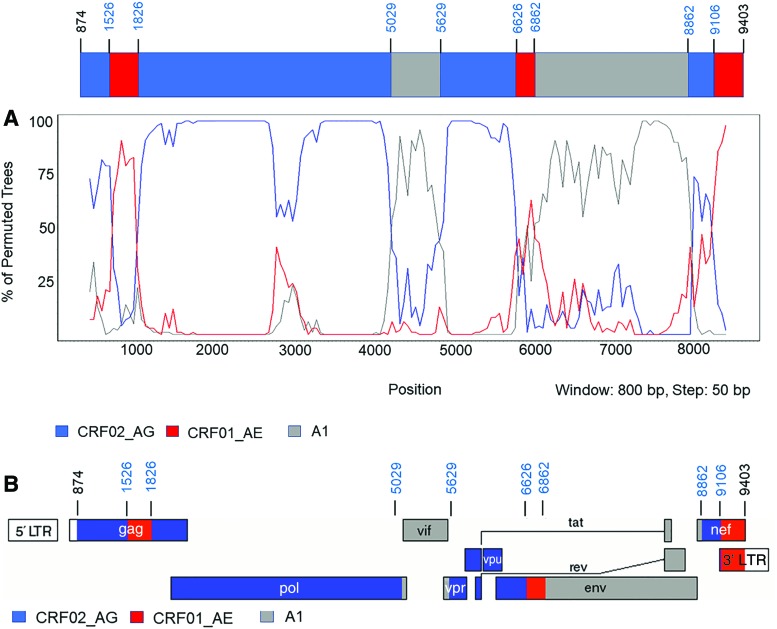
We further used boot scanning analysis with Simplot v3.5.1 to predict recombinant breakpoint positions and intervals. The results showed that NACMR092 is an unknown recombinant form that had eight breakpoints dividing the near-full-length HIV-1 genome into nine fragments (Fig. 2). Fragments I, II, III, IV, V, VI, VII, VIII, and IX of the mosaic genome correspond, respectively, to nucleotides 874–1526, 1527–1826, 1827–5029, 5030–5629, 5630–6629, 6630–6862, 6863–8862, 8863–9106, and nucleotides 9107–9403 of the HXB2 genome.
FIG. 2.
Recombination events and breakpoints identified in NACMR092. (A) Recombinant breakpoint positions were calculated by boot scanning analysis using Simplot v3.5.1, as described. The percentages of permuted trees (y-axis) of the subtypes at each sequence position (x-axis) are shown. (B) Genomic map of the near-full-length sequence of NACMR092 (based on HXB2 numbering). The mosaic map was generated using the Los Alamos Recombinant HIV-1 Drawing Tool (https://hiv.lanl.gov/content/sequence/DRAW_CRF/recom_mapper.html). The start (nucleotide 874) and end (nucleotide 9403) of the near-full-length sequence, as well as the breakpoints are shown. Genomic sequences originating from HIV-1 subtype A1 (grey color), CRF01_AE (red color), and CRF02_AG (blue color) are shown. LTR, long-terminal repeat; gag, group-specific antigen; pol, polymerase; vif, viral infectivity factor; vpr, viral protein R; vpu, viral protein U; env, envelope; tat, trans-activator of transcription; nef, negative factor. Color images are available online.
Nucleotides 874–1526 code for Gag amino acids (aa) 29–246, and nucleotides 1527–1826 code for Gag aa 247–346. Nucleotides 1827–5029 code for Gag aa 347–501, the complete protease (PR, aa 1–99), RT (aa 1–440), RNase H (aa 1–120), and the integrase (aa 1–267). Nucleotides 5030–5629 code for integrase C-terminal (aa 268–288), full-length viral infectivity factor (Vif, aa 1–193), and viral protein R (Vpr, aa 1–24). Nucleotide 5630–6626 code for Vpr aa 25–98, the 5′-end of Tat (aa 1–72), Rev (aa 1–26), Vpu (aa 1–83), and gp120 (aa 1–134). Nucleotides 6627–6862 code for gp120 aa 135–213. Nucleotides 6863–8862 code for gp120 aa 214–511, gp41 aa 1–346, the 3′-end of Tat (aa 73–102), Rev (aa 26–117), and Nef (aa 1–22). Nucleotides 8863–9106 code for Nef aa 23–104 and 5′-end of the 3′-LTR. Nucleotides 9107–9403 code for partial 3′-LTR (based on Los Alamos HIV Sequence Locator; https://hiv.lanl.gov/content/sequence/LOCATE/locate.html).
Recombinant breakpoint analyses also showed that fragments I, III, V, and VIII had similarities with HIV-1 CRF02_AG; that fragments IV and VII had similarities with subtype A1, while fragments II, VI, and IX had similarities with CRF01_AE (Fig. 2A). The mosaic genomic map (Fig. 2B) of the NACMR092 near-full-length HIV sequence confirmed that this URF included parental fragments of HIV-1 subtypes A1 (grey color), CRF02_AG (blue color), and CRF01_AE (red color). The breakpoint positions/intervals of the viral genomic segments (relative to HXB2; Fig. 2, blue letters) are summarized in Table 2. This recombinant structure does not match with any known CRFs or URFs previously reported in public databases.
Table 2.
Breakpoint Nucleotide Positions/Intervals in the NACMR092 Sequence and Genomic Segments (Relative to HXB2)
| Genomic fragment | HXB2 | Fragment subtype | Percentage of the near-full-length genome | |
|---|---|---|---|---|
| Start | End | |||
| I | 874 | 1526 | CRF02_AG | 7.63 |
| II | 1527 | 1826 | CRF01-AE | 3.52 |
| III | 827 | 5029 | CRF02_AG | 37.54 |
| IV | 5030 | 5629 | A1 | 7.04 |
| V | 5630 | 6626 | CRF02_AG | 11.73 |
| VI | 6627 | 6862 | CRF01-AE | 2.35 |
| VII | 6863 | 8862 | A1 | 23.46 |
| VIII | 8863 | 9106 | CRF02_AG | 2.93 |
| IX | 9107 | 9403 | CRF01-AE | 3.80 |
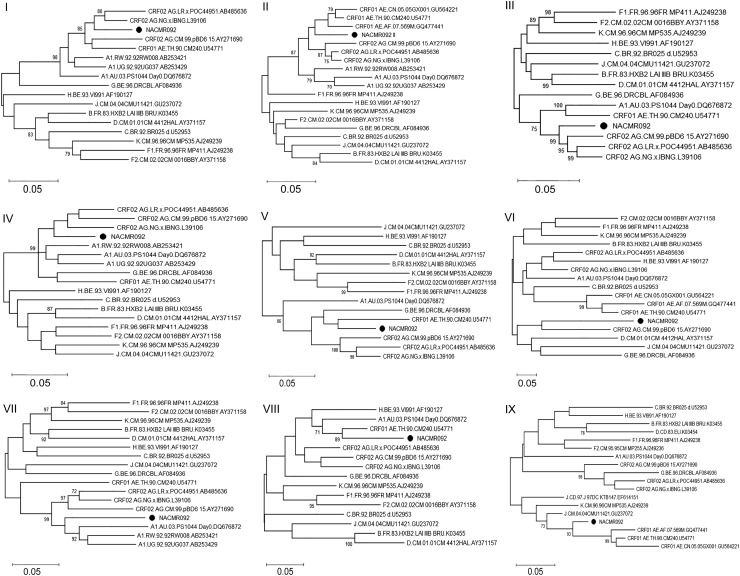
Following phylogenetic analysis of the genomic region spanning fragments I and III, NACMR092 sequences clustered together with CRF02_AG (Fig. 3I, III). Analysis of the genomic region spanning fragments II and IX (Fig. 3II, IX) showed that NACMR092 sequence clustered together with CRF01_AE. For the genomic region spanning fragment IV (Fig. 3IV), NACMR092 clustered together with subtype A1 and CRF02_AG. For genomic regions spanning fragments V and VI (Fig. 3V, VI), NACMR092 clustered together with CRF01_AE and CRF02_AG. For the genomic region spanning fragment VII, NACMR092 clustered together with subtype A1 (Fig. 3VII). For the genomic region spanning fragment VIII, NACMR092 clustered together with CRF01_AE, A1, and CRF02_AG (Fig. 3VIII).
FIG. 3.
Subregion phylogenetic analyses of fragments I–IX of the NACMR092 isolate. The phylogenetic tree for each fragment/subregion (I–IX) of the near-full-length NACMR092 sequence was obtained using MEGA 6.0, as described. Panels (I–IX) correspond respectively to nucleotides 874–1526, 1527–1826, 1827–5029, 5030–5629, 5630–6626, 6627–6862, 6863–8862, 8863–9106, and nucleotides 9107–9403 of the HXB2 genome. For each panel, the black solid circle represents the test sample. Bootstrap values of 1,000 replicates (>70%) are shown at the corresponding nodes. The scale bar represents 5% genetic distance. Figure 3 can be viewed in greater detail online.
Regarding the HIV replication cycle, the error-prone nature of the RT enzyme during reverse transcription,18 pseudodiploidy,19 and point mutations20 increase the risk of variation in the viral genome, which can generate new URFs, CRFs, and subtypes. We previously reported an individual (NACMR092) in Cameroon infected with a novel URF HIV-1 CRF22_01A1/CRF01_AE, based on the analysis of Tat sequences.7 We now report a near-full-length sequence of viral isolates from NACMR092, showing that this person was infected with a mosaic URF that had eight breakpoints (two in the gag, one in the pol, one in vpr, two in the env, and two in nef regions). Phylogeny and bootscan-based recombinant breakpoint analyses of this novel mosaic URF showed that it is a complex inter-subtype recombinant that included fragments of HIV-1 with similarities to subtypes A1 (30.5%), CRF02_AG (59.83%), and CRF01_AE (9.67%).
Phylogeny using the maximum likelihood or neighbor joining methods gave almost similar results. However, whereas breakpoint analyses showed fragments II and VI to be CRF01_AE, these two fragments clustered between CRF01_AE and CRF02_AG in phylogenetic analyses. Similarly, whereas breakpoint analyses showed fragments IV and VII to be A1, phylogeny showed that these fragments clustered between A1 and CRF02_AG. Whereas breakpoint analyses showed that fragments V and VIII where CRF02_AG, these two fragments clustered between CRF02_AG and CRF01_AE in phylogeny analyses. These divergences could be due to limitations in the current phylogeny tools. In fact, there is evidence that current bioinformatics software used to generate phylogenetic trees, including maximum likelihood or neighbor joining methods, are built on the assumption that recombination occurs between loci, which ignores within-loci recombinations.21,22 It has been shown that within-loci recombinations do occur and can introduce heterogeneities that interfere with branch length estimates generated and impact trees obtained with current phylogeny tools.21,22 Mutation rates also vary across different sites and different regions of the genome, but most of the current phylogeny software don't take these variations into account.23,24 It has been shown that the presence of heterogeneity in mutation rates across the genome can influence the estimation of branch lengths, as well as impact phylogenies and evolutionary time scales.23,24 The accuracy of branch length estimation varies with the length of that branch, and the use of longer datasets with maximum likelihood improved estimates.25 Thus, our analysis of all combined fragments (the entire near-full-length genome) of NACMR092 likely represent an accurate estimate of this URF; it showed that NACMR092 clustered with A1, CRF01_AE, CRF02_AG, and a CRF36_cpx isolate.
From a GenBank database search, NACMR092 sequences closely matched (>90% similarity) with sequences associated with accession numbers AF377955, KR017774, and KP718922. AF377955 corresponds to an HIV-1 CRF02_AG that was isolated in 1997 from the peripheral blood mononuclear cells of a 41-year-old male (ID# CM53658) in Douala, Cameroon.15 KR017774 corresponds to an HIV-1 CRF36_cpx that was isolated in 2007 from the plasma of a 26-year-old male (ID# BS40) in Yaoundé, Cameroon.16 KP718922 corresponds to a URF isolated in 2007 from the plasma of a 30-year-old male (ID# 663–13) in Cameroon.17
Proviral DNA from cells or molecular cloning can contain integrated viral sequences that are defective.26 The near-full-length HIV sequences reported in the current study were generated by direct PCR amplification and sequencing of virion RNA from the plasma of patient NACMR092. Thus, the mosaic URF identified reflects a currently replicating viral strain in this person, highlighting its pathogenic and epidemiological significance. Our current data further confirm that Cameroon is a hotspot for HIV genetic recombination and emergence of new URFs and CRFs that contribute to expanding global HIV diversity and driving the HIV/AIDS pandemic. In fact, it is well established that new variants of HIV resulting from genetic recombination are more likely to evade immune control, have increased fitness and higher transmission efficiency, and better adapt to a new host/environment.27 Recombination of viral strains carrying drug resistance mutations can also generate HIV-1 strains that are resistant to different classes of antiretroviral drugs.28,29
An effective strategy for the development of anti-HIV vaccine would require the ability to generate broad neutralizing antibodies that can be effective against many HIV strains and subtypes, including CRFs and URFs. The novel mosaic recombinant reported in this study shows complex recombination events occurring between HIV-1 subtypes A1, CRF02_AG, and CRF01_AE. Continued monitoring and detection of such recombinants as well as accurate classification of HIV genotypes and recombinant forms are important for tracking viral molecular epidemiology and antigenic diversity. Our subsequent studies will characterize the phenotypic properties of this HIV-1 isolate, including its replication fitness, co-receptor usage, and susceptibility to antiretroviral drugs.
Sequence Data
The near-full-length HIV-1 genome sequence generated from this study is available in the NCBI database, GenBank accession number MH667256.
Acknowledgments
This work was supported by grants from the National Institutes of Health, National Institute of Mental Health MH094160, and the Fogarty International Center. We would like to thank the Cameroonian volunteer who donated specimens to this study. We thank Ms. Emilienne Nchindap for technical assistance, and the University of Nebraska Medical Center High-Throughput DNA Sequencing and Genotyping Core Facility for assistance with gene sequencing.
Authors' Contributions
A.A. carried out gene amplification, sequencing, and sequence analyses, made figures and tables, participated in data analysis and writing of methods and results. J.Y.F. carried out subject recruitment, obtained written consent and demographic data from participating human subjects, and helped coordinate the clinical studies in Cameroon. D.M. oversaw serological analyses to determine subject's HIV status, FACS CD4 count, and viral load tests. A.K.N. obtained ethical approval and coordinated subject recruitment along with seeking consent and collection of clinical data. G.D.K. conceived and designed the study, obtained IRB approval, collected and assembled the data, analyzed and interpreted data, and wrote the article.
Author Disclosure Statement
No competing financial interests exist.
References
- 1. Eberle J, Gurtler L: HIV types, groups, subtypes and recombinant forms: Errors in replication, selection pressure and quasispecies. Intervirology 2012;55:79–83 [DOI] [PubMed] [Google Scholar]
- 2. Palaniappan C, Wisniewski M, Wu W, Fay PJ, Bambara RA: Misincorporation by HIV-1 reverse transcriptase promotes recombination via strand transfer synthesis. J Biol Chem 1996;271:22331–22338 [DOI] [PubMed] [Google Scholar]
- 3. Robertson DLAJ, Bradac JA, Carr JK, Foley B, et al. : HIV-1 nomenclature proposal. Science 2000;288:55–56 [DOI] [PubMed] [Google Scholar]
- 4. LANL. HIV and SIV nomenclature. HIV sequence database. Available at https://hiv.lanl.gov/content/sequence/HelpDocs/subtypes-more.html (2017)
- 5. LANL: HIV circulating recombinant forms (CRFs). HIV sequence database. Available at http://hiv.lanl.gov/content/sequence/HIV/CRFs/CRFs.html (2018), accessed April8, 2019
- 6. Lihana RW, Ssemwanga D, Abimiku A, Ndembi N: Update on HIV-1 diversity in Africa: A decade in review. AIDS Rev 2012;14:83–100 [PubMed] [Google Scholar]
- 7. Teto G, Fonsah JY, Tagny CT, et al. : Molecular and genetic characterization of HIV-1 Tat exon-1 gene from cameroon shows conserved Tat HLA-binding epitopes: Functional implications. Viruses 2016;8:pii: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Nadai Y, Eyzaguirre LM, Constantine NT, et al. : Protocol for nearly full-length sequencing of HIV-1 RNA from plasma. PLoS One 2008;3:e1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ratner L, Fisher A, Jagodzinski LL, et al. : Complete nucleotide sequences of functional clones of the AIDS virus. AIDS Res Hum Retroviruses 1987;3:57–69 [DOI] [PubMed] [Google Scholar]
- 10. Aiyar A: The use of CLUSTAL W and CLUSTAL X for multiple sequence alignment. Methods Mol Biol 2000;132:221–241 [DOI] [PubMed] [Google Scholar]
- 11. Tamura K, Stecher G, Peterson D, Filipski A, Kumar S: MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 2013;30:2725–2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lole KS, Bollinger RC, Paranjape RS, et al. : Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J Virol 1999;73:152–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ratner L, Haseltine W, Patarca R, et al. : Complete nucleotide sequence of the AIDS virus, HTLV-III. Nature 1985;313:277–284 [DOI] [PubMed] [Google Scholar]
- 14. Johnson M, Zaretskaya I, Raytselis Y, Merezhuk Y, McGinnis S, Madden TL: NCBI BLAST: A better web interface. Nucleic Acids Res 2008;36(Web Server issue):W5–W9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Carr JK, Torimiro JN, Wolfe ND, et al. : The AG recombinant IbNG and novel strains of group M HIV-1 are common in Cameroon. Virology 2001;286:168–181 [DOI] [PubMed] [Google Scholar]
- 16. Tongo M, Dorfman JR, Abrahams MR, Mpoudi-Ngole E, Burgers WA, Martin DP: Near full-length HIV type 1M genomic sequences from Cameroon: Evidence of early diverging under-sampled lineages in the country. Evol Med Public Health 2015;2015:254–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Luk KC, Berg MG, Naccache SN, et al. : Utility of metagenomic next-generation sequencing for characterization of HIV and human pegivirus diversity. PLoS One 2015;10:e0141723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Abram ME, Ferris AL, Das K, et al. : Mutations in HIV-1 reverse transcriptase affect the errors made in a single cycle of viral replication. J Virol 2014;88:7589–7601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chen J, Nikolaitchik O, Singh J, et al. : High efficiency of HIV-1 genomic RNA packaging and heterozygote formation revealed by single virion analysis. Proc Natl Acad Sci U S A 2009;106:13535–13540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cuevas JM, Geller R, Garijo R, López-Aldeguer J, Sanjuán R: Extremely high mutation rate of HIV-1 in vivo. PLoS Biol 2015;13:e1002251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lyons-Weiler J, Takahashi K: Branch length heterogeneity leads to nonindependent branch length estimates and can decrease the efficiency of methods of phylogenetic inference. J Mol Evol 1999;49:392–405 [DOI] [PubMed] [Google Scholar]
- 22. Lanier HC, Knowles LL: Is recombination a problem for species-tree analyses? Syst Biol 2012;61:691–701 [DOI] [PubMed] [Google Scholar]
- 23. Tateno Y, Takezaki N, Nei M: Relative efficiencies of the maximum-likelihood, neighbor-joining, and maximum-parsimony methods when substitution rate varies with site. Mol Biol Evol 1994;11:261–277 [DOI] [PubMed] [Google Scholar]
- 24. Buckley TR, Simon C, Chambers GK: Exploring among-site rate variation models in a maximum likelihood framework using empirical data: Effects of model assumptions on estimates of topology, branch lengths, and bootstrap support. Syst Biol 2001;50:67–86 [PubMed] [Google Scholar]
- 25. Schwartz RS, Mueller RL: Branch length estimation and divergence dating: Estimates of error in Bayesian and maximum likelihood frameworks. BMC Evol Biol 2010;10:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wei X, Ghosh SK, Taylor ME, et al. : Viral dynamics in human immunodeficiency virus type 1 infection. Nature 1995;373:117–122 [DOI] [PubMed] [Google Scholar]
- 27. Ritchie AJ, Cai F, Smith NM, et al. : Recombination-mediated escape from primary CD8+ T cells in acute HIV-1 infection. Retrovirology 2014;11:69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Moutouh L, Corbeil J, Richman DD: Recombination leads to the rapid emergence of HIV-1 dually resistant mutants under selective drug pressure. Proc Natl Acad Sci U S A 1996;93:6106–6111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nora T, Charpentier C, Tenaillon O, Hoede C, Clavel F, Hance AJ: Contribution of recombination to the evolution of human immunodeficiency viruses expressing resistance to antiretroviral treatment. J Virol 2007;81:7620–7628 [DOI] [PMC free article] [PubMed] [Google Scholar]