Abstract
While the roles in HIV transcription of many cyclin-dependent kinases (CDKs) have been well defined, little is known about the impact of mediator kinases (MDKs), CDK8 and CDK19, in this process. Mediator complexes containing CDK8 or CDK19 repress or activate the expression of selected genes. The aim of this study was to investigate the role of MDKs in HIV transcription. siRNA knockdown of both MDKs had no effect on HIV transcription. This result was confirmed using two MDK inhibitors, Cortistatin A (CA) and Senexin A (SnxA). Furthermore, neither CA nor SnxA inhibited viral reactivation in Jurkat cell models of HIV latency. Taken together, these results indicate that MDKs are not required for HIV transcription.
Keywords: HIV transcription, mediator kinases, CDK8, CDK19, Cortistatin A, Senexin A
Introduction
HIV transcription requires initiation, elongation, and termination machineries, which are also necessary for the expression of host cellular genes. Transcription factors (TFs) such as nuclear factor-kappaB (NF-κB), cyclin-dependent kinase-7 (CDK7), positive transcription elongation factor-b (P-TEFb), CDK11, and RNA polymerase II (RNAPII) are all indispensable for efficient HIV transcription.1 The expression of these TFs is abundant in activated CD4+ T cells, but remarkably low when cells become quiescent, as is the case in resting memory T cells.2,3 The absence of these important TFs promotes and maintains HIV latency.1 In spite of the efficacy of current antiretroviral regimens in suppressing HIV replication, latently infected cells persist and prevent the eradication of infected cells or HIV cure.
The role of CDKs in transcription elongation in HIV transcription has been well established.1 RNAPII has been found at the HIV long-terminal repeat (LTR) in the absence of Tat. It exists in a paused transcription complex with negative elongation (NELF) and DRB sensitivity-inducing factors (DSIF). Following the expression of Tat, HIV transcription elongation ensues.4 Tat promotes the recruitment of P-TEFb to RNAPII, leading to the phosphorylation of NELF, DSIF, and RNAPII by the CDK9 subunit of P-TEFb, resulting in productive elongation. In addition to CDK9, CDK2,5 7,4,6 11,7 and 138 are important to HIV initiation, elongation, and termination.9 However, little is known about the role of CDK8 and its paralogue CDK19, the mediator kinases (MDKs), in HIV transcription.
Mediator (MD) complexes are large multiprotein transcription complexes that form at promoters.10,11 MD complexes are required for pre-initiation complex (PIC) assembly, enhancer to promoter gene looping, and coordination of signals between TFs and RNAPII. Three MD complex proteins were identified in a meta-analysis of nine genome-wide screens that identified host factors that interact with HIV.12 In fact, these MD proteins were among only a few host factors that were identified in three of the nine independent screens. While some MD complexes contain CDKs, their role in transcription elongation is unknown. The binding of MD containing CDK8 (CDK8-module) to RNAPII is independent of and precludes the binding of other MD complexes to RNAPII.13–15 While the CDK8-module was first thought to be a repressive complex, emerging evidence indicates that it can act to repress and/or activate gene expression. Recent evidence implicates the CDK8-module in the function of tumor necrosis factor-alpha-activated NF-κB and hypoxia-induced RNAPII elongation of HIF1A-responsive genes.16,17 CDK19 is a paralogue of CDK8, which can form its own MD complex. It is unclear if the CDK8- and CDK19-modules are functionally redundant; however, only CDK8 knockout is embryonically lethal.18 The inhibition of both MDKs results in global downregulation of genes associated with TFs, transcription, and chromatin regulators.
The inhibition of MDKs, through the use of highly specific chemical inhibitors, has been suggested for use in cancer therapies.19 Cortistatin A (CA) is a steroid alkaloid derived from the marine sponge Corticum simplex. CA is a highly specific inhibitor of Rho-associated protein kinase (ROCK), CDK8, and CDK19, which induces the expression of super enhancer associated genes, including several TFs.15,20,21 The antiproliferative effect of CA was first described in human umbilical vein endothelial cell (HUVEC) lines.20 Differential sensitivity to the antiproliferative effects of CA is observed between long-lived cell lines. Notably, erythroleukemic cell lines are insensitive to the antiproliferative effects of CA.21 Unfortunately, the synthesis of CA is labor-intensive and cost-prohibitive for large-scale therapeutic use. An analog of CA, didehydro-CA (dCA), was synthesized to overcome the production limits of CA.22 dCA blocks HIV through the binding of Tat,23 and this suppression is independent of its ability to inhibit MDKs.24 A second, commercially available MDK inhibitor is Senexin A (SnxA).16,19 SnxA was discovered in a drug screen of small-molecule compounds that inhibit cytomegalovirus (CMV)-green fluorescent protein (GFP) through the induction of the CDK inhibitor p21.19 In contrast to CA, SnxA has no effect on ROCK.19,20 Both CA and SnxA inhibit the ATP binding site and block CDK8 and CDK19 activities.
The aim of this study was to investigate the role of MDKs in HIV transcription. We made use of siRNA knockdowns and specific chemical inhibitors, CA and SnxA, to inhibit MDK expression and activity. We utilized several approaches to assess the requirement of MDKs in HIV transcription: transient co-transfection of a minimal HIV LTR luciferase (LTR-Luc) reporter, transient transfection of the full-length HIV provirus that makes its own Tat, NH1 cells that stably express HIV LTR, and Jurkat cell models that can be activated to re-express latent HIV. In all the systems tested, MDKs were dispensable for HIV transcription.
Results
Knockdowns of CDK8 and CDK19 do not inhibit HIV transcription
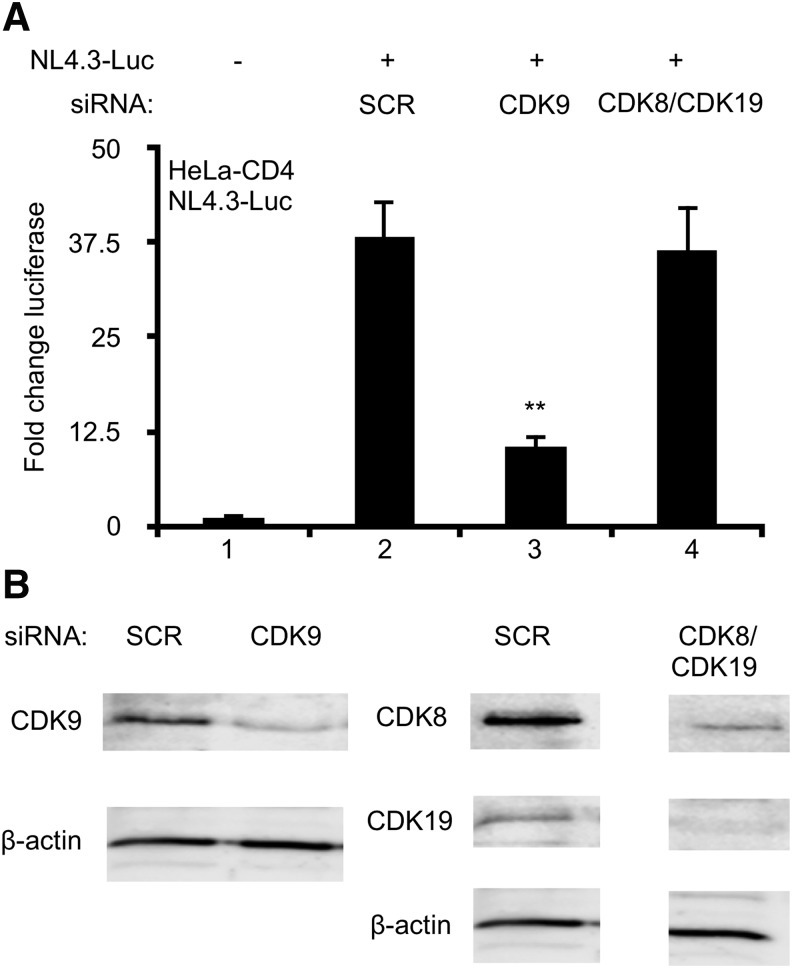
CDK8 and 19 are paralogues and found in separate MDK complexes; therefore, siRNAs against CDK8 and CDK19 were used together for the knockdown of MDKs. CDK9 is essential for HIV transcription, and was used as a positive control. HeLa cells expressing the CD4 T cell receptor (HeLa-CD4) were transfected with siCDK8 and siCDK19, siCDK9, or a scrambled negative control (siSCR). Cells were then transfected with either empty vector negative control or NL4.3-luciferase (NL4.3-Luc), which produces its own Tat to activate efficient HIV transcription. Lysates were harvested 24 h post NL4.3-Luc transfection (48 h post knockdown) and luciferase activity was measured. We observed a 37-fold induction of luciferase activity in cells transfected with NL4.3-Luc (lane 2 in Fig. 1A). Knockdown of CDK9 resulted in a 73% decrease in luciferase activity (lanes 2 and 3 in Fig. 1A), consistent with its requirement for efficient elongation. Luciferase activity was not affected by knockdowns of CDK8 and CDK19 (lanes 2 and 4 in Fig. 1A), indicating that MDKs are not critical for HIV transcription. Efficiency of these knockdowns was confirmed by western blotting (Fig. 1B).
FIG. 1.
Knockdowns of MDKs do not inhibit HIV transcription. HeLa-CD4 cells were transfected with siRNAs against CDK9, CDK8, and CDK19, or scrambled non-specific control (SCR). Twenty-four hours post knockdown, cells were transfected with NL4.3 Luc. At 24 h post NL4.3-Luc transfection (48 h post knockdown), lysates were harvested and examined for luciferase activity or Western blotting. (A) Luciferase activity is presented as a fold change in luciferase activity over untreated control cells (set to 1). (B) Whole-cell lysates were run on 10% SDS-PAGE and blotted with anti-CDK9, anti-CDK8, anti-CDK19 antibodies, with β-actin serving as loading control. Samples from cells treated with scrambled and CDK8/19 siRNAs were run on the same gel. Triplicate stimulations were performed. Error bars represent standard errors of the mean (**p < .01). CDK, cyclin-dependent kinase; MDK, mediator kinase.
Chemical inhibition of MDKs does not inhibit HIV transcription
Two chemical inhibitors, CA and SnxA, were used to observe the effects of chemical inhibition of MDK activity on HIV transcription. Both inhibitors bind the ATP pocket and block CDK8 and CDK19 activation.
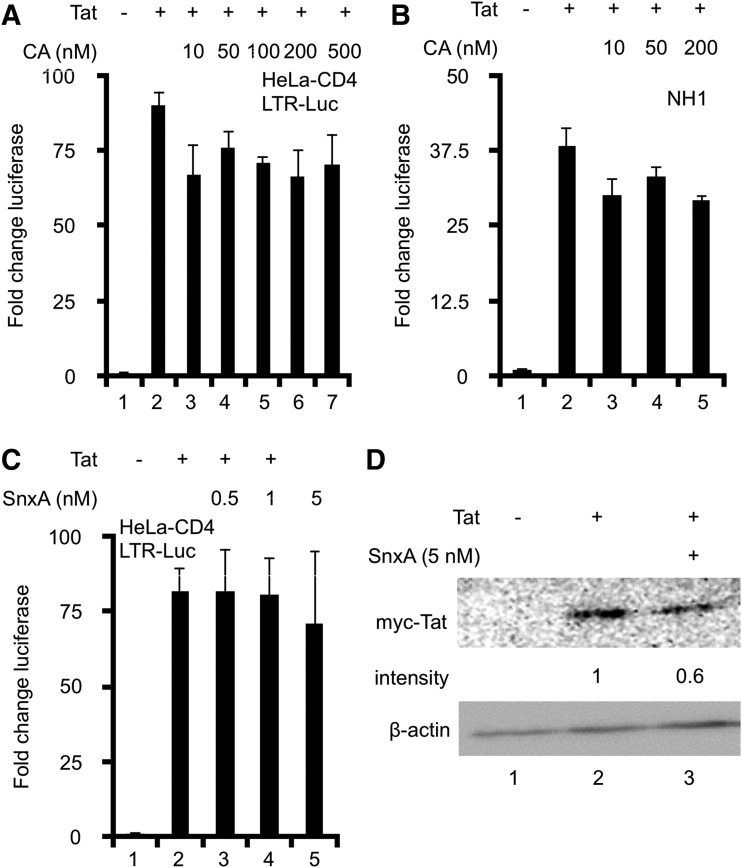
HeLa-CD4 were transiently transfected with LTR-Luc, which expresses the HIV promoter driving luciferase gene expression. Unlike NL4.3-Luc, LTR-Luc does not make its own Tat and requires co-transfection with a plasmid expressing Tat to activate HIV LTR. Cells were given increasing concentrations of CA at the same time as the transfection, and luciferase activity was measured 48 h later. In the absence of Tat, luciferase activity was not detected (lane 1 in Fig. 2A). Tat induced an 88-fold increase in luciferase activity (lane 2 in Fig. 2A). CA did not significantly inhibit Tat-induced LTR activation at any concentration tested (lanes 3–7 in Fig. 2A). Even very high concentrations of CA failed to significantly inhibit Tat transactivation (lanes 5–7 in Fig. 2A). Additionally, we tested whether CA was able to inhibit HIV integrated into chromatin using the HeLa-based NH1 cells, which stably express the luciferase gene driven by HIV LTR. In the absence of Tat, luciferase activity was not detected (lane 1 in Fig. 2B). Tat induced a 37-fold increase in luciferase activity (lane 2 in Fig. 2B), and increasing concentrations of CA did not significantly inhibit this induction (lanes 3–5 in Fig. 2B). We did not observe a greater inhibition with increasing concentrations of CA (lanes 3–5 in Fig. 2B). Taken together, we conclude that Tat-induced activation of HIV LTR is not inhibited by CA in either transient transfection or stable expression systems.
FIG. 2.
Chemical inhibition of MDKs does not inhibit HIV transcription. (A) HeLa-CD4 cells were co-transfected with LTR-Luc and Tat for 48 h. CA was added at the indicated concentrations. Luciferase activity was measured 48 h post transfection, and is presented as a fold change in luciferase activity over untreated control cells (set to 1). (B) NH1 cells, stably expressing LTR-Luc, were transfected with Tat ± CA for 48 h. Luciferase activity was measured 48 h post transfection. (C) HeLa-CD4 cells were co-transfected with LTR-Luc and Tat for 48 h. SnxA was added at indicated concentrations. Luciferase activity was measured 48 h post transfection. (D) Whole-cell lysates of SnxA-treated HeLa-CD4 cells were separated on 15% SDS PAGE. The membrane was probed with anti-myc and anti-β-actin antibodies. Densitometry was calculated and normalized to β-actin. Results are representative of two independent experiments. Triplicate stimulations were performed for all experiments. Error bars represent standard errors of the mean. CA, Cortistatin A; LTR-Luc, long-terminal repeat luciferase; SnxA, Senexin A.
To confirm these results, we repeated the above experiment using a second inhibitor of MDKs, SnxA. In cells co-transfected with LTR-Luc and Tat, Tat induced a 16-fold increase in luciferase activity (lane 2 in Fig. 2C). Increasing concentrations of SnxA had no effect on this luciferase activity induced by Tat (lanes 3–5 in Fig. 2C).
While the inhibition of MDKs with chemical inhibitors did not ablate HIV transcription, we did observe a consistent minor decrease in luciferase activity in all three systems tested. HIV transcription is highly dependent on Tat, and its decreased levels may account for a minor reduction of luciferase activity in cells treated with chemical inhibitors. Indeed, SnxA-treated cells had 40% less Tat expression (lane 3 in Fig. 2D). This decrease was observed with chemical inhibitor treatment (Fig. 2) but not following siRNA knockdown of MDKs (Fig. 1), indicating that a decrease in Tat expression and a subsequent minor decrease in luciferase expression is a side effect of the inhibitors.
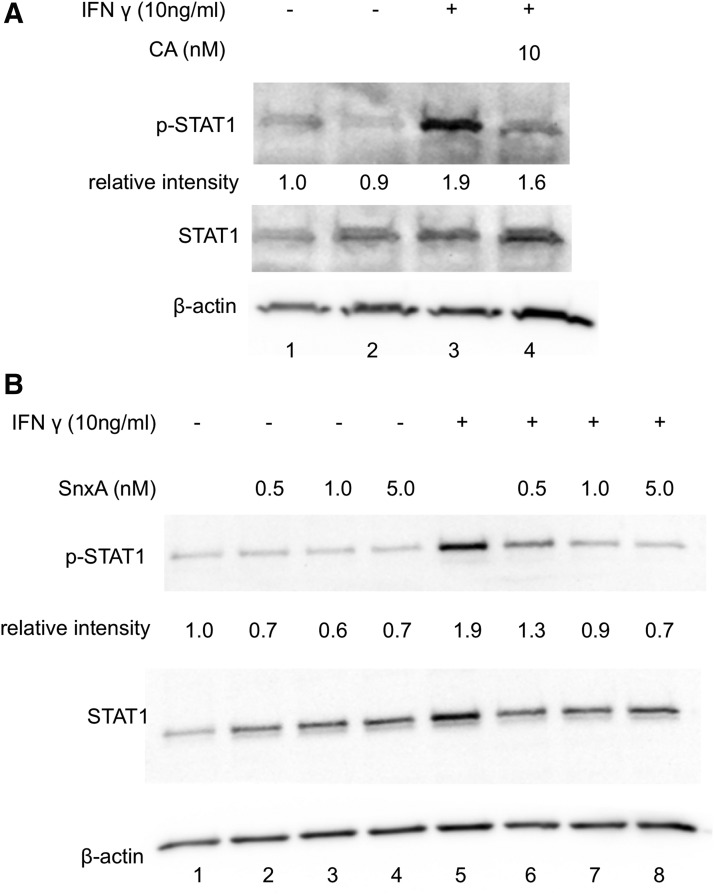
To ensure that these chemical inhibitors inhibited the kinase activity of MDKs, a known target of MDK, interferon-gamma (IFNγ)-induced STAT1 phosphorylation was examined.15 A single concentration of CA was tested due to limiting supplies of the inhibitor. Treatment with inhibitors alone did not affect STAT1 phosphorylation (lane 2 in Fig. 3A and lanes 2–4 in 3B). The addition of IFNγ potently induced phosphorylation of STAT1 (lane 3 in Fig. 3A and lane 5 in Fig. 3B). Importantly, treatment with CA decreased this induced STAT1 phosphorylation (lane 4 in Fig. 3A). Treatment with SnxA also inhibited IFNγ-induced STAT1 phosphorylation in a dose-dependent manner (lanes 6–8 in Fig. 3B). The addition of 5 nM SnxA induced a 64% decrease in this phosphorylation (lane 8 in Fig. 3B). Since CA and SnxA efficiently blocked STAT1 phosphorylation, this finding confirms that these inhibitors potently inhibited MDK function in spite of having no effect on HIV transcription.
FIG. 3.
Chemical inhibition of MDKs decreases STAT1 phosphorylation. HeLa-CD4 cells were pretreated with (A) 10 nM CA for 1 h or (B) indicated concentrations of SnxA, then stimulated with IFNγ (10 ng/mL) for 1 h. Whole-cell lysates were run on 10% SDS-PAGE and blotted with anti-phospho-STAT1 and anti-STAT1 antibodies with β-actin serving as loading control. Representative blots of triplicate stimulations are presented. IFNγ, interferon-gamma.
Chemical inhibition of MDKs does not inhibit HIV reactivation
Although MDKs are not required for HIV transcription, the inhibition of MDKs could prevent reactivation of latent HIV. This would make MDK inhibitors an attractive candidate for “block and lock” therapies, in which HIV is theoretically suppressed to a point where daily antiretroviral therapy is no longer necessary.
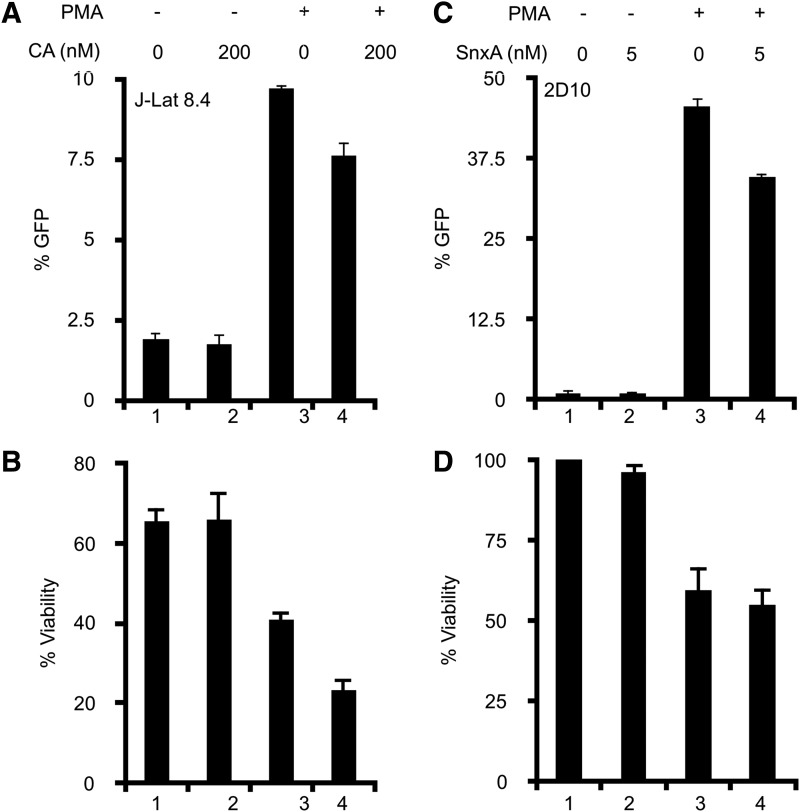
2D10 and J-Lat 8.4 cells are Jurkat T cells that stably express GFP from HIV LTR. Both cell lines are used as models for HIV latency and viruses can be reactivated by a potent protein kinase C agonist, such as phorbol myristate acetate (PMA). In J-Lat 8.4 cells treated with CA, it alone did not induce GFP expression (lane 2 in Fig. 4A). Stimulation with PMA resulted in a fourfold increase in GFP (lane 3 in Fig. 4A). Treatment with CA did not decrease this activation by PMA (lane 4 in Fig. 4A). However, PMA treatment alone did reduce the viability of these cells by 40% (lane 3 in Fig. 4B). CA alone did not affect the viability (lane 2 in Fig. 3B), nor did it increase the toxicity of PMA treatment of J-Lat 8.4 cells (lane 4 in Fig. 4B).
FIG. 4.
Chemical inhibition of MDKs does not inhibit HIV reactivation. (A) J-Lat 8.4 cells were stimulated with PMA (12 nM) ± CA (200 nM) for 24 h. GFP expression of viable cells was measured by flow cytometry. (B) Percent viability was estimated using the percentage of live lymphocytes in 10,000 total cells analyzed. Untreated control cell viability was set at 100% viability. (C) 2D10 cells were stimulated with PMA (12 nM) ± SnxA (5 nM) for 24 h. GFP expression of viable cells was measured by flow cytometry. (D) Percent viability was estimated using the percentage of live lymphocytes in 10,000 total cells analyzed. Untreated control cell viability was set at 100% viability. Triplicate stimulations were performed. Error bars represent standard errors of the mean. GFP, green fluorescent protein; PMA, phorbol myristate acetate.
As observed in previous experiments, CA and SnxA treatments yielded similar results. In 2D10 cells treated with SnxA, it alone did not induce the expression of GFP (lane 2 in Fig. 4AC). Stimulation with PMA resulted in a 44-fold increase in GFP expression (lane 3 in Fig. 4C). Because J-Lat 8.4 cells are transcriptionally interfered25 and less reactivate in response to PMA, GFP expression in 2D10 cells was 10-fold higher than in J-Lat 8.4 cells (lane 3 in Fig. 4A). The addition of SnxA did not significantly inhibit reactivation by PMA (lane 4 in Fig. 4C). PMA treatment alone reduced the viability of these cells by 40% (lane 3 in Fig. 4D). SnxA alone did not affect the viability of 2D10 cells (lane 2 in Fig. 4D), nor did it increase the toxicity of PMA treatment (lane 4 in Fig. 4D). Taken together, we conclude that in addition to having no effect on HIV transcription, the inhibition of MDKs is also unable to inhibit the reactivation of latent HIV.
Discussion
In this study we determined that MDKs are not necessary for HIV transcription. Since transformed cells express adequate levels of TFs necessary for HIV, they were found to be optimal systems to study the effects of MDK deletion and inhibition. Indeed, knockdowns of CDK8 and CDK19 did not affect HIV gene expression. This result was confirmed using two chemical inhibitors of MDKs. Furthermore, chemical inhibition of MDKs did not prevent the reactivation of latent HIV. Taken together, these results indicate that MDKs are not required for HIV transcription.
The role of MDKs in HIV transcription has, in part, been explored previously. While Ruiz et al. determined that the knockdown of MD proteins associated with the CDK8-module did not affect HIV transcription,26 this study did not address CDK8 expression or activity directly. Our study fully assessed the role of MDKs through a direct knockdown of CK8 and CK19, as well as treatment with two highly specific MDK inhibitors. During the preparation of this article, Mediouni et al. published similar observations that the knockdown of CDK8 and CDK19 did not affect viral mRNA production nor the viability of HeLa-CD4.24 Our findings confirm and expand on these results through the use of two chemical inhibitors of MDKs. Both chemical inhibitors of MDKs, CA and SnxA, potently inhibited MDK function, but had no effect on HIV transcription. The efficacy of these inhibitors was confirmed by a known target of CDK8,15 IFNγ-induced STAT3 phosphorylation (Fig. 3). Furthermore, we determined that the inhibition of MDKs also had no effect on the reactivation of latent HIV. Taken together, our results using siRNA knockdown, chemical inhibitors, and cell line models of HIV latency provide definitive evidence that MDKs are not required for HIV transcription.
A slight inhibition of HIV transcription was observed in cell line models treated with CA and SnxA, but not in siRNA knockdown cells (Fig. 2). Concurrently, we also observed reduced levels of Tat following treatment with these inhibitors (Fig. 2D). In this system, Tat was expressed from a separate plasmid and a different promoter. When full-length HIV, which produces its own Tat, was used, we observed no decrease in HIV transcription (Fig. 1). Since Tat is required to recruit P-TEFb to HIV LTR, leading to the release of paused RNAPII and productive elongation,4 and direct knockdowns of MDKs had no effect on HIV gene expression, we conclude that HIV transcription is independent of MDKs. Therefore, the inhibitors may reduce HIV Tat expression from a different promoter.
It was previously reported that an analog of CA, dCA, potently inhibited HIV transcription and reactivation of latent HIV, resulting in prolonged suppression of viral gene expression.23,27,28 dCA binds the basic domain of Tat, which in turn prevents the recruitment of P-TEFb to HIV LTR.23 Although CA and dCA are chemical analogs, the loss of two hydrogens and the addition of a double bond on dCA could contribute to this specific effect of dCA on HIV Tat.23 Furthermore, it has been reported that dCA does not inhibit CDK8 or CDK19,23 suggesting that this chemical may behave very differently in regard to binding and inhibiting MDKs and Tat. While we were unable to obtain dCA, our results using CA were confirmed with a second MDK inhibitor and siRNA knockdowns of CDK8 and CDK19. Together, these data led us to the conclusion that the inhibition of MDKs does not affect HIV transcription.
There is great interest in the role CDKs in HIV transcription and latency, which may provide potential therapeutic targets.29 While CDK8 and 19 do not play a significant role, our study completes the picture of known CDKs in the regulation of HIV transcription. The contributions of CDKs 2, 7, 9, 11, and 13 to HIV transcription have been well described.1,9 A subset of CDKs is required for transcription initiation, including phosphorylation of Tat by CDK25 and phosphorylation of the serine 5 residue on RNAPII by CDK7.4,6 CDK9 is necessary for transcription elongation through phosphorylation of NELF, DSIF, and the serine 2 residue on RNAPII.30,31 CDK11 mediates 3′-end processing of HIV RNA and proper termination of HIV transcription.7 CDK13 is important for HIV RNA splicing.8 Therapeutic agents that target CDK9 have been of interest as HIV latency reversing agents.29 It is feasible that the expression or activity of other CDKs could be manipulated to activate HIV transcription in latently infected cells or inhibited to induce a deeper suppressive state, such as the proposed block-and-lock strategies. As new mechanistic studies emerge that elucidate the function of MDKs,16,17 it was important to determine if they play any role in HIV transcription.1 In this case, however, MDKs that are important for the regulation of some host genes are dispensable for HIV gene expression. Thus, compounds that target MDKs can be excluded as potential HIV therapeutic targets.
Materials and Methods
Cell lines and reagents
HeLa-CD4 cells (NIH AIDS Reagents) are HeLa expressing surface human CD4 protein. NH1 (obtained from Dr. Qiang Zhou at the University of California, Berkley) are HeLa stably expressing HIV LTR luciferase reporter that does not produce its own Tat.32 2D10 cells (obtained from Dr. Jonathan Karn at Case Western Reserve University) are a Jurkat-based HIV latency cell line model that contains attenuated Tat and d2EGFP in the place of Nef.33 J-Lat 8.4 (obtained from Dr. Eric Verdin at the Buck Institute for Research on Aging) are a Jurkat-based HIV latency cell line model that contains full-length HIV expressing GFP in the place of Nef and a frameshift mutation in Env.34 HeLa-CD4, NH1, and 293T cells (American Tissue Culture Collection) were cultured in Dulbecco's modified Eagle's medium (DMEM) (Corning Cellgro). 2D10 were cultured in Roswell Park Memorial Institute (RPMI) (Corning Cellgro). Media was supplemented with 10% fetal calf serum (FCS) (Gibco), 1% Pen/Strep, and cells were grown at 37°C and 5% CO2.
MDK inhibitors
MDK inhibitors, CA (obtained from Dr. Matthew Shair at Harvard University) and SnxA (Tocris Bioscience), were added at the time of transfection.
MDK inhibitor functional assay
To assess the ability of CA and SnxA to inhibit MDK function, HeLa-CD4 cells were pretreated for 1 h with CA (10 nM) or SnXA (0.5, 1, and 5 nM). Cells were then stimulated for 1 h with 10 ng/mL recombinant human IFNγ (PHC4031; Thermo-Fisher). Cells were harvested and whole-cell lysates isolated in Lamelli buffer (Bio-Rad) in the presence of proteinase inhibitor cocktail (Thermo-Fisher).
siRNA knockdowns
One day before transfection, 1 × 105 HeLa-CD4 cells were seeded in 24-well plates to ensure a confluency of 70%–80% on the day of transfection. On the day of transfection, siRNA mix containing 10 μM siRNA and Lipofectamine RNAiMAX reagent (Thermo-Fisher) was prepared in Opti-MEM® I Medium (Gibco). After incubation for 24 h at 37°C, plasmid transfection of NL4.3-Luc was performed as described above. siRNAs used in this study were: control siRNA-A (sc-37007; SCBT), human CDK9 siRNA (sc-29268; SCBT), human cdk19 siRNA (sc-72844; SCBT), and human cdk8 siRNA (6438S; Cell Signaling).
Transfection of cell lines and luciferase assay
The day before transfection, 1 × 105 HeLa-CD4 or NH1 cells were seeded in 24-well plates to ensure a confluency of 70%–80% on the day of transfection. Transfections were performed with Lipofectamine 2000 mix (Thermo-Fisher) prepared in Opti-MEM I Medium (Gibco) using a plasmid mix containing 25 ng LTR-Luc/NL4.3-Luc and 25 ng myc-Tat or a pLINK empty vector. Forty-eight hours post transfection, the medium was removed and cells were washed with PBS. Cells were harvested in 1 × Passive Lysis buffer (Promega), and luciferase activity was measured with the luciferase assay system (Promega) on an EG&G Berthold Microplate luminometer. Experiments were performed in triplicates.
For measuring Tat protein expression, cell number was scaled up to 107 HeLa-CD4 in 10 cm2 plates.
Western blot analysis of protein expression
Whole-cell lysates were generated using Lamelli buffer (Bio-Rad) in the presence of proteinase inhibitor cocktail (Thermo-Fisher). Lysates were run on 10% (Figs. 1 and 3) or 15% (Fig. 2) SDS-PAGE and transferred onto a nitrocellulose membrane. Membranes were blocked in 5% non-fat milk (NFM) for at least 1 h and blotted overnight with rabbit anti-human CDK9 (ab6544; abcam), rabbit anti-human CDK8 (ab15155; abcam), rabbit anti-human CDK19 (HPA007053; Sigma Aldrich), mouse anti-myc (ab32; abcam), rabbit anti-human p-STAT1 (8826S; Cell Signaling), rabbit anti-human STAT1 (14994S; Cell Signaling), rabbit anti-human β-actin (ab8227; abcam) in 5% NFM. Membranes were washed 3 × in TBS with 0.05% Tween-20, and then blotted for 1 h with horseradish peroxidase (HRP) anti-rabbit/mouse IgG in 5% NFM. After washing 3 × with TBS with 0.05% Tween-20, the membranes were treated with ECL Plus chemiluminescence reagent (GE Healthcare) for 5 min and imaged using Odyssey Fc imaging system and Image Studio software (LI-COR). Re-probed membranes were stripped with NewBlot Stripping Buffer (LI-COR) and then washed 3 × with PBS.
Densitometry was obtained using Image Studio software (LI-COR). Relative expression was calculated first normalizing total STAT1 to β-actin, then normalizing p-STAT1 to the normalized STAT1 expression. Untreated conditions were set to 1.
Flow cytometry analysis
Cells were harvested 24 h post reactivation and washed in cold PBS, and 0.5 × 106 cells were allotted to each tube. Cells were fixed in 2% paraformaldehyde and analyzed using the BD Biosciences FACScaliber and CellQuest Pro software at the UCSF Parnassus Flow Cytometry Core. Cells were gated on the live lymphocyte gate using the forward and side scatter plot, and the percentage of live lymphocytes in 10,000 collected total cells was used as an estimate for cell viability.
Statistical analysis
Statistical analysis was performed using Student's t test, two-tailed distribution, and assuming equal variances.
Acknowledgments
We thank the Peterlin Lab, in particular Zeping Luo, Koh Fujinaga, Fang Huang, Rachad Nasr, and Marie Leoz for technical expertise, discussions, and careful reading of the article.
Matthew Shair in the Department of Chemistry and Chemical Biology at Harvard University kindly provided CA for our studies.
This work was supported by the National Institutes of Health grants: R01 AI049104 (Peterlin, PI) and P50 GM082250 (HARC grant, Krogan, PI). DCC was supported by a grant from the National Institutes of Health, University of California, San Francisco-Gladstone Institute of Virology & Immunology Center for AIDS Research, P30-AI027763. MR was supported by the Department of Molecular Biotechnology at the Institute for Pharmacy and Molecular Biotechnology, Ruprecht-Karls-University Heidelberg. AR was supported by the American Slovenian Education Foundation.
Author Disclosure Statement
No competing financial interests exist.
References
- 1. Cary DC, Fujinaga K, Peterlin BM: Molecular mechanisms of HIV latency. J Clin Invest 2016;126:448–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cary DC, Fujinaga K, Peterlin BM: Euphorbia Kansui Reactivates Latent HIV. PLoS One 2016;11:e0168027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bartholomeeusen K, Fujinaga K, Xiang Y, Peterlin BM: Histone deacetylase inhibitors (HDACis) that release the positive transcription elongation factor b (P-TEFb) from its inhibitory complex also activate HIV transcription. J Biol Chem 2013;288:14400–14407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cujec TP, Okamoto H, Fujinaga K, et al. : The HIV transactivator TAT binds to the CDK-activating kinase and activates the phosphorylation of the carboxy-terminal domain of RNA polymerase II. Genes Dev 1997;11:2645–2657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ivanov A, Lin X, Ammosova T, et al. : HIV-1 Tat phosphorylation on Ser-16 residue modulates HIV-1 transcription. Retrovirology 2018;15:39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kim YK, Bourgeois CF, Pearson R, et al. : Recruitment of TFIIH to the HIV LTR is a rate-limiting step in the emergence of HIV from latency. EMBO J 2006;25:3596–3604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pak V, Eifler TT, Jager S, Krogan NJ, Fujinaga K, Peterlin BM: CDK11 in TREX/THOC regulates HIV mRNA 3′ end processing. Cell Host Microbe 2015;18:560–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Berro R, Pedati C, Kehn-Hall K, et al. : CDK13, a new potential human immunodeficiency virus type 1 inhibitory factor regulating viral mRNA splicing. J Virol 2008;82:7155–7166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rice AP: Roles of CDKs in RNA polymerase II transcription of the HIV-1 genome. Transcription 2019;10:111–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Allen BL, Taatjes DJ: The Mediator complex: A central integrator of transcription. Nat Rev Mol Cell Biol 2015;16:155–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yin JW, Wang G: The Mediator complex: A master coordinator of transcription and cell lineage development. Development 2014;141:977–987 [DOI] [PubMed] [Google Scholar]
- 12. Bushman FD, Malani N, Fernandes J, et al. : Host cell factors in HIV replication: Meta-analysis of genome-wide studies. PLoS Pathog 2009;5:e1000437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gold MO, Rice AP: Targeting of CDK8 to a promoter-proximal RNA element demonstrates catalysis-dependent activation of gene expression. Nucleic Acids Res 1998;26:3784–3788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dale T, Clarke PA, Esdar C, et al. : A selective chemical probe for exploring the role of CDK8 and CDK19 in human disease. Nat Chem Biol 2015;11:973–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Poss ZC, Ebmeier CC, Odell AT, et al. : Identification of mediator kinase substrates in human cells using Cortistatin A and quantitative phosphoproteomics. Cell Rep 2016;15:436–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chen M, Liang J, Ji H, et al. : CDK8/19 Mediator kinases potentiate induction of transcription by NFkappaB. Proc Natl Acad Sci U S A 2017;114:10208–10213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Galbraith MD, Allen MA, Bensard CL, et al. : HIF1A employs CDK8-mediator to stimulate RNAPII elongation in response to hypoxia. Cell 2013;153:1327–1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Westerling T, Kuuluvainen E, Makela TP: Cdk8 is essential for preimplantation mouse development. Mol Cell Biol 2007;27:6177–6182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Porter DC, Farmaki E, Altilia S, et al. : Cyclin-dependent kinase 8 mediates chemotherapy-induced tumor-promoting paracrine activities. Proc Natl Acad Sci U S A 2012;109:13799–13804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cee VJ, Chen DY, Lee MR, Nicolaou KC: Cortistatin A is a high-affinity ligand of protein kinases ROCK, CDK8, and CDK11. Angew Chem Int Ed Engl 2009;48:8952–8957 [DOI] [PubMed] [Google Scholar]
- 21. Pelish HE, Liau BB, Nitulescu II, et al. : Mediator kinase inhibition further activates super-enhancer-associated genes in AML. Nature 2015;526:273–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Shi J, Manolikakes G, Yeh CH, et al. : Scalable synthesis of cortistatin A and related structures. J Am Chem Soc 2011;133:8014–8027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mousseau G, Clementz MA, Bakeman WN, et al. : An analog of the natural steroidal alkaloid cortistatin A potently suppresses Tat-dependent HIV transcription. Cell Host Microbe 2012;12:97–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mediouni S, Chinthalapudi K, Ekka MK, et al. : Didehydro-Cortistatin A inhibits HIV-1 by specifically binding to the unstructured basic region of Tat. MBio 2019;10:e02662–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lenasi T, Contreras X, Peterlin BM: Transcriptional interference antagonizes proviral gene expression to promote HIV latency. Cell Host Microbe 2008;4:123–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ruiz A, Pauls E, Badia R, et al. : Characterization of the influence of mediator complex in HIV-1 transcription. J Biol Chem 2014;289:27665–27676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mediouni S, Jablonski J, Paris JJ, et al. : Didehydro-cortistatin A inhibits HIV-1 Tat mediated neuroinflammation and prevents potentiation of cocaine reward in Tat transgenic mice. Curr HIV Res 2015;13:64–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kessing CF, Nixon CC, Li C, et al. : In vivo suppression of HIV rebound by didehydro-Cortistatin A, a “Block-and-Lock” strategy for HIV-1 treatment. Cell Rep 2017;21:600–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rice AP: Cyclin-dependent kinases as therapeutic targets for HIV-1 infection. Expert Opin Ther Targets 2016;20:1453–1461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wei P, Garber ME, Fang SM, Fischer WH, Jones KA: A novel CDK9-associated C-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity, loop-specific binding to TAR RNA. Cell 1998;92:451–462 [DOI] [PubMed] [Google Scholar]
- 31. Garber ME, Wei P, KewalRamani VN, et al. : The interaction between HIV-1 Tat and human cyclin T1 requires zinc and a critical cysteine residue that is not conserved in the murine CycT1 protein. Genes Dev 1998;12:3512–3527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yang Z, Zhu Q, Luo K, Zhou Q: The 7SK small nuclear RNA inhibits the CDK9/cyclin T1 kinase to control transcription. Nature 2001;414:317–322 [DOI] [PubMed] [Google Scholar]
- 33. Kim YK, Mbonye U, Hokello J, Karn J: T-cell receptor signaling enhances transcriptional elongation from latent HIV proviruses by activating P-TEFb through an ERK-dependent pathway. J Mol Biol 2011;410:896–916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Jordan A, Bisgrove D, Verdin E: HIV reproducibly establishes a latent infection after acute infection of T cells in vitro. EMBO J 2003;22:1868–1877 [DOI] [PMC free article] [PubMed] [Google Scholar]