Abstract
Biofilms provide cells favorable growth conditions, which have been exploited in industrial biotechnological processes. However, industrial application of the biofilm has not yet been reported in Escherichia coli, one of the most important platform strains, though the biofilm has been extensively studied for pathogenic reasons. Here, we engineered E. coli by overexpressing the fimH gene, which successfully enhanced its biofilm formation under industrial aerobic cultivation conditions. Subsequently, a biofilm-based immobilized fermentation strategy was developed. L-threonine production was increased from 10.5 to 14.1 g/L during batch fermentations and further to 17.5 g/L during continuous (repeated-batch) fermentations with enhanced productivities. Molecular basis for the enhanced biofilm formation and L-threonine biosynthesis was also studied by transcriptome analysis. This study goes beyond the conventional research focusing on pathogenic aspects of E. coli biofilm and represents a successful application case of engineered E. coli biofilm to industrial processes.
Keywords: Escherichia coli, L-threonine, fimH gene, biofilm, transcriptome analysis
Introduction
L-threonine is one of the most essential amino acids in human body, and its demand is sharply increasing due to its wide application in food, chemical, and pharmaceutical industries (Leuchtenberger et al., 2005). Currently, microbial fermentation is widely employed for industrial L-threonine production with Escherichia coli as the best candidate strain (Lee et al., 2006; Dong et al., 2011). However, L-threonine fermentation has been operated in a free-cell batch fermentation mode, wherein cells cannot be reused after fermentation (Rajkumar et al., 2013; Boelee et al., 2014). This batch fermentation and single-use of cells would increase the cost of operation and reduce productivities. Meanwhile, the free cells dispersed in fermentation media are often challenged by stress conditions such as shear forces during aerobic fermentation, resulting in decreased cell viability over the fermentation process. These problems need to be solved urgently to improve the fermentation efficiency. Alternatively, biofilm-based immobilized fermentation has been proposed as an alternative to free-cell fermentation owing to its advantages such as protection by biofilm matrix, enhanced metabolic activities, and repeated use of cells compared with free-cell fermentation processes (Zhao et al., 2015). The biofilms of some microorganisms such as Clostridium acetobutylicum, Corynebacterium glutamicum, Aspergillus niger, and Saccharomyces cerevisiae have been applied to immobilized batch or continuous (repeated-batch) fermentation effectively (Liu et al., 2013; Shi et al., 2014; Yang et al., 2018; Yu et al., 2018). However, for E. coli, one of the most important platform strains, industrial application of the biofilm has not yet been reported, though the biofilm has been extensively studied for pathogenic reasons.
Biofilms are complex cell communities living in close association with biological or abiotic surfaces (Sauer, 2003). For pathogenic bacteria, formation of biofilms is one of the most important factors leading to medical infection which is difficult to be removed (Stoodley et al., 2002). Type I fimbriae is one of the most important factors for biofilm formation in Gram-negative bacteria such as E. coli (Tripathi et al., 2013). In E. coli, a fimH-encoded protein that is secreted and located at the top of type I fimbriae plays a key function to generate biofilm structures by serving as an adhesin (Nishiyama et al., 2008; Le Trong et al., 2010). Cells could use these structures to obtain nutrients and withstand shear forces. It was found that E. coli cells covered by biofilms could tolerate stricter conditions such as high osmotic pressure, oxygen limitation, and high cell density, which is a desired characteristic during fermentation (Prigent-Combaret et al., 1999; Weissman et al., 2006).
In this study, E. coli was first metabolically engineered with overexpression of fimH gene to enhance biofilm formation (Figure 1). A biofilm-based fermentation system was constructed using a carrier to support the biofilm. Cells adhered to the surface of the carrier and formed a large amount of biofilm so that it could withstand high-speed shaking. Moreover, the biofilm cells that attached to the carrier surface could be renewed when the fermentation broth was replaced with fresh medium (Huang et al., 2002; Kim et al., 2014). Due to high cell activities and repeated use of cells in the biofilm-immobilization fermentation, no seed culture was needed and cellular lag phase and fermentation period were reduced substantially. Overall, this study represents a successful case of development of biofilm-based immobilized fermentation under aerobic industrial conditions for efficient biochemical production.
FIGURE 1.
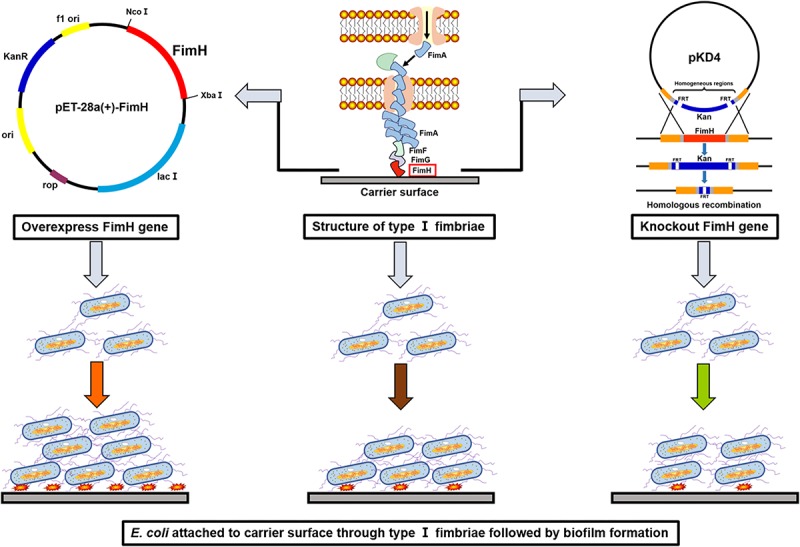
A schematic illustration of constructions for the overexpression or knockout of fimH in Escherichia coli W1688 and the effects on the biofilm formation.
Materials and Methods
Strains and Plasmids
Escherichia coli W1688 (CCTCC M2015233) was an L-threonine producer obtained from E. coli MG1655 (ATCC47076) by mutation and molecular modification. It could not produce biofilms apparently. All strains and plasmids used in this work are listed in Tables 1, 2, respectively. The fimH gene was amplified from the genomic DNA of E. coli W1688. The fimH gene and plasmid pET28a (with restriction enzyme XbaI and NcoI) were ligated by using the ClonExpress II One Step Cloning Kit C112-01 (Vazyme, Nanjing, China), resulting in a plasmid pET28a-fimH. The final engineered strain was named E. coli W1688-fimH* with Kanamycin resistance for screening. On the other hand, fimH from E. coli W1688 was deleted by Red homologous recombination, resulting in an E. coli W1688-ΔfimH (Madyagol et al., 2011). Briefly, a PCR-generated Kanamycin resistance marker was used as knock-in DNA fragment. The Kanamycin resistance marker consisted of a Kanamycin resistance sequence in plasmid pKD4 and homologous regions (50–100 bp) flanking the target locus. The knock-in component was transformed into strain E. coli W1688-pKD46 using Bio-Rad electroporation system set at 2.0 kV, 25 mF with a 200 Ohm pulse controller.
TABLE 1.
Strains and plasmids used in this study.
| Strains or plasmids | Relevant characteristics | Sources |
| Strains | ||
| E. coli W1688a | L-threonine producing strain | Prof. Sheng Yang |
| E. coli W1688-fimH* | E. coli W1688 harboring plasmid pET28a-fimH | This study |
| E. coli W1688-ΔfimH | E. coli W1688 with the deletion of fimH | This study |
| E. coli W1688-pKD46 | E. coli W1688 harboring plasmid pKD46 | This study |
| Plasmids | ||
| pET28a | Kan resistance | Stored in our lab |
| pET28a-fimH | pET28a containing fimH | This study |
| pKD46 | Amp resistance | Stored in our lab |
| pKD4 | Kan resistance | Stored in our lab |
aA gift from Prof. Sheng Yang (Institute of plant physiology and ecology, CAS, Shanghai, China).
TABLE 2.
Primers used in this study.
| Primer | Sequence |
| fimH-F | ATGAAACGAGTTATTACCCTG |
| fimH-R | GTGACTTTTGTTTATCAATAA |
| fimH-Kan-F | GATTAGCATCACCTATACCTACAGCTGAACCCGAAGAGAT GATTGTAATGGTGTAGGCTGGAGCTGCTTC |
| fimH-Kan-R | TACCAGCATTAGCAATGTCCTGTGATTTCTTTATTGATAAA CAAAAGTCAGCCATGGTCCATATGAATATCCTCC |
| fimH-Kan-cx-F | GTGTAGGCTGGAGCTGCTTC |
| fimH-Kan-cx-R | GCCATGGTCCATATGAATATCCTCC |
| fimH-mq-F | GGATAACAATTCCCCTCTAGAATGAAACGAGTTAT TACCCTGTTTG |
| fimH-mq-R | GATGATGGCTGCTGCCCATGGTTATTGATAAACAAA AGTCACGCCA |
| fimH-cx-F | TATAGGCGCCAGCAACCGCACC |
| fimH-cx-R | CCGCGACCCATTTGCTGTCCA |
Media and Growth Conditions
Escherichia coli W1688, E. coli W688-fimH* and E. coli W1688-ΔfimH were cultured in LB medium, which contained 5 g/L yeast extract, 10 g/L tryptone and 10 g/L NaCl. Solid media was prepared in all cases by adding 1.5% (w/v) agar. Fermentation medium contained 30 g/L glucose, 2 g/L yeast extract, 1 g/L KH2PO4, 20 g/L (NH4)2SO4, 0.8 g/L MgSO4⋅7H2O, 0.2 g/L FeSO4⋅37H2O, 0.2 g/L MnSO4⋅5H2O, and 15 g/L CaCO3. Acetic acid was added to the medium to adjust its initial pH to 7.2. All media were sterilized at 115°C for 20 min. Kanamycin (50 mg/mL), ampicillin (100 mg/mL), isopropyl-β-D-thiogalactopyranoside (IPTG) 0.5 mM or L-arabinose (100 mM) were added as necessary. The fermentation culture was grown at 37°C with a shaking speed of 200–220 rpm.
Carrier Preparation
A new type of polymer porous foam named Y-11 which was made of polyurethane, was prepared in the laboratory. The carrier Y-11 had a density of 0.32 g/cm3 with a pore diameter of 0.2 to 0.6 mm sheared to a size of 10 mm × 10 mm × 10 mm. This carrier was pretreated with the previously reported method. The carriers were rinsed in 1 M NaOH and then1 M HCl before being washed by ddH2O (sterile water) until the pH-value reached 7.0. All carriers were sterilized at 115 C for 20 min (Zhao et al., 2015).
Free-Cell Fermentation and Immobilized Fermentation
For free-cell fermentation, the flasks were inoculated with 5% seed culture and then run at 37°C with shaking at 220 rpm. Samples were centrifugated at 8000 rpm, 4°C for 5 min, then the supernatants were used for the quantification of L-threonine and residual sugar.
For immobilized continuous (repeated-batch) fermentation, the same conditions for free-cell fermentation were employed and 30 g/L of the carrier was added into the fermentation medium. After the first batch, 80% of the fermented broth was removed from the flask and 20% of the broth with the carrier that was covered by biofilm was left for the second batch. After adding fresh culture medium, the second batch was initiated under the same conditions described above until the L-threonine titer was stable. The subsequent batches were operated in the same way as above.
Analytical Methods
L-threonine concentrations were measured by high-performance liquid chromatography (Agilent 1260 series; Hewlett-Packard, Palo Alto, CA, United States) with a UV detector, using a Sepax AAA ion exclusion column (250 × 4.6 mm; Bio-Rad Laboratories, Hercules, CA, United States), with 0.1 M sodium acetate and 80% acetonitrile as the mobile phase (1 mL/min) at 36°C. Glucose concentrations were measured by a refractive index detector, using an Aminex HPX-87H column (300 × 7.8 mm), with 5 mM H2SO4 as the mobile phase (1 mL/min) at 55°C.
The basic dye crystal violet binds to negatively charged surface molecules and polysaccharides in the biofilm matrix which can be applied for total biomass staining (Li et al., 2003). Strains were grown in LB medium or fermentation medium at 37°C for 12 h. Then they were diluted to the ratio of 1:200 in 96-well plates (200 μL in each well) and kept at 37°C without shaking. PBS (1%) was prepared for washing the biofilm-containing wells twice to remove free cells. Methanol was used for fixing biofilm for 15 min at 4°C. Biofilms were then stained with 200 μL of 1% crystal violet for 15 min at 37°C, after which wells were washed by PBS 3–4 times repeatedly. For quantification, 200 μL of 33% acetic acid was added to release the crystal violet and the plates were incubated by shaking slowly for 30 min. The absorbance of crystal violet was an indicator of biofilm amount and was measured at 570 nm by using a multiscan spectrum (SpectraMax® iD5; Molecular Devices; United States).
In some cases, coverslips were used as carriers to observe biofilms by microscopy. The coverslips were removed gently from the fermentation broth after culture of 30 h. PBS (1%) was used to wash off the free cells on the coverslips. The biofilm was fixed by 4% paraformaldehyde for 12 h at 4°C. The coverslips with the biofilm were dehydrated by vacuum freeze-drying device (Labconco Corporation, Fort Scott, KS, United States), then coated with gold-palladium before scanning electron microscopy (SEM) analysis (TM3000, Hitachi, Japan). Fresh carriers or carriers after the continuous (repeated-batch) fermentation were prepared in the same way as above for SEM analysis.
Fluorescence microscope was used to visualize the adhesion property of bacteria and distribution of cells in biofilms. The carriers were removed gently from the fermentation broth after culture for 30 h. PBS (1%) was used to wash off the free cells. The biofilm was fixed by 4% paraformaldehyde for 30 min at 4°C. Then 0.2 μg/mL of 4′,6-Diamidino-2-phenylindole (DAPI) (Sigma, St. Louis, MO, United States) was prepared to stain the cells in the biofilm for 30 min at room temperature.
Transcription Analysis
Samples of the three different strains were centrifuged at 8000 rpm for 5 min at 4°C after 30 h of immobilized batch fermentation. Three biological replicates were prepared. Sediments were collected and washed twice with PBS, and then frozen in liquid nitrogen immediately and stored at −80°C. The level of false discovery rate (FDR) ≤ 0.05 and absolute value of Log2Ratio ≥ 1 were selected as criteria for assessing expression levels of different genes.
Quantitative Reverse Transcription PCR (qRT-PCR) Analysis
The E. coli wild-type strain and two recombinant strains were harvested at exponential phase. Total RNA was isolated using RNAprep Pure Cell/bacteria Kit (TianGen Biotech, China) and the residual DNA was digested by using TianGen RNase-Free DNase. After reverse transcription, cDNAs were prepared for qRT-PCR. Primer Express software was used for primer design. The analyzed genes and primers used in the analysis are listed in Table 3. qRT-PCR assays were performed by using SYBR Green PCR Master Mix (Applied Biosystems, United States) in a StepOnePlus Real-Time PCR System (Applied Biosystems, United States). Gene transcript levels were determined according to the 2–ΔΔCt method, using 16s RNA as a reference gene for normalizing the gene expression levels. Reactions were performed according to the manufacturer’s instructions, and three technical replicates with one negative control were performed for each sample. Values and error bars represent the mean and the s.d. ∗∗∗p < 0.001, ∗∗p < 0.01, *p < 0.05 as determined by two-tailed t-test.
TABLE 3.
Genes and primers used for quantitative real-time PCR.
| Gene | Forward primer sequence | Reverse primer sequence |
| fimH | GATGCGGGCAACTCGATT | CGCCCTGTGCAGGTGAA |
| flu | CAGCGTGGAAAAATCAGGAAGT | ACGGCTTTCTGGGTGAGTGT |
| cyaA | TGCCTGGTAGGTAGCGTTGAC | GCAGCGTACGCACTTCGTT |
| csgD | CGGAATCAGCCCTCCTTACTC | GCGCCGATACGCAGCTTAT |
| luxS | GTGTTCGATCTGCGCTTCTG | GGATCCCTCTTTCTGGCATCA |
| lsrR | CGGTGGCGTCGGTTCTT | CTGCACGCCGCGTTAAG |
| ackA | GTATTTGACACCGCGTTCCA | GGCAGGGCGTAGAGGTAAGA |
| aspA | TGCAGGCGGGCTCTTC | TCCGGAACAACCGGGTTT |
| lysA | TCTCACCGCCGAAAATCTG | ACACCGGGCAGCCAAAT |
| tdcB | ATCCAAAGTAGCGGCAACGT | CGTTGAAGTTATCACCATGCAGAA |
| thrA | CTTCACCCCCGCACCAT | ATCAGGCAAGGGATCTGGAA |
| thrB | CGAGCTGGAAGGCCGTATC | AACACGGTGCCACGTTGTC |
| thrC | AAGCGACTCAGGCGACGTTA | CACACGCGGCCAGTTGT |
| rhtA | TCGTCGCCCGGTAGATTTC | GCAGGAACCACAGACCAAGAA |
| 16s RNA | TCGGGAACCGTGAGACAGG | CCGCTGGCAACAAAGGATAAG |
Results and Discussion
Characterization of Biofilm Formation in Engineered Strains
PCR and sequencing results confirmed that recombinant strains, in which fimH gene was overexpressed (E. coli W1688-fimH*) or knocked out (E. coli W1688-ΔfimH) were constructed successfully. The 96-well plates experiment showed that the biofilm formation abilities of these strains were different. The optical density from crystal violet staining (which was an indicator of biofilm amount) for E. coli W1688-fimH* in LB medium increased greatly by 75.9% compared with that of the original strain (1.34 vs. 2.35), which could be attributed to the overexpression of fimH gene (Figure 2A). On the contrary, the optical density of E. coli W1688-ΔfimH was decreased by 38.8% due to the deletion of fimH gene (1.34 vs. 0.82). Similar results were also observed in fermentation medium. Furthermore, SEM and fluorescence microscope images showed that biofilm formation and cell adhesion were more obvious in E. coli W1688-fimH* compared with the original strain (Figures 2B,C). In E. coli W1688-ΔfimH, biofilm formation was apparently reduced and a sparse bacterial distribution was observed. Taken together, these results indicated that overexpression of fimH gene facilitated cell adhesion to abiotic surfaces and contributed to the clustering effects of E. coli and resulted in the biofilm formation. Whereas, deletion of fimH gene had a negative effect on the biofilm formation. So, the fimH gene had a significant regulatory effect on E. coli biofilm formation.
FIGURE 2.
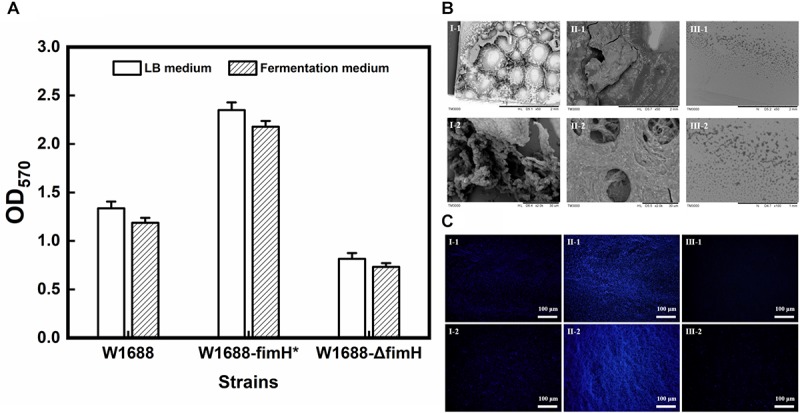
Quantitative analysis for biofilm formation in the three strains with different characterization methods. (A) Crystal violet staining for LB medium and fermentation medium; (B) Scanning electron microscope (SEM) images of biofilm formation and cell adhesion under different magnification levels; and (C) Fluorescence microscope images of biofilm and cell adhesion under different magnification levels. I: E. coli W1688, II: E. coli W1688-fimH*, and III: E. coli W1688-ΔfimH.
Biofilm-Based Fermentation for Enhanced L-Threonine Production
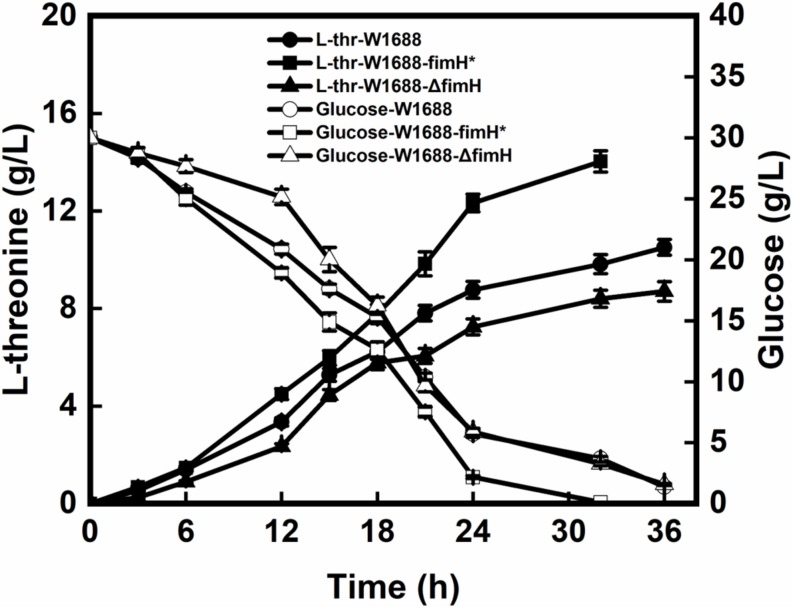
The recombinant and original strains with different capabilities for the biofilm formation were investigated in batch fermentations. As seen in Figure 3, L-threonine production was increased by 42.9% in E. coli W1688-fimH* compared with that in the original strain (14.1 g/L vs. 10.5 g/L). Besides, the fermentation period was shortened from 36 h to 32 h. In contrast, L-threonine production in E. coli W1688-ΔfimH showed a decrease compared with the original strain (8.7 g/L vs. 10.5 g/L) as well as a delay in glucose consumption at the initial phase of fermentation. Also, the final cell density showed a reduction of 21% compared with the fimH overexpression strain. Since some enzymes involved in L-threonine biosynthesis, the different level of expression in three strains might affect cell growth. All these observations suggested that L-threonine production and productivity were enhanced in strain E. coli W1688-fimH*.
FIGURE 3.
L-threonine production and glucose consumption in batch fermentation by E. coli W1688, E. coli W1688-fimH*, and E. coli W1688-ΔfimH.
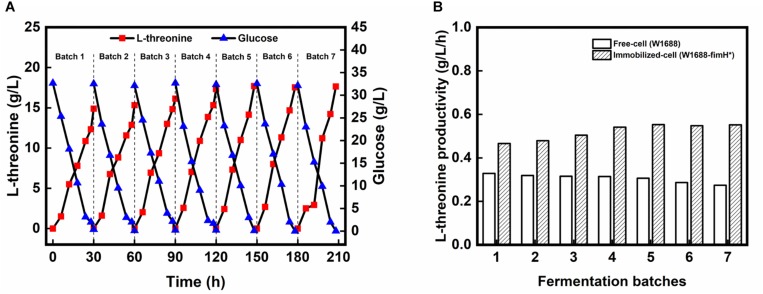
To further improve the fermentation efficiency, a biofilm-based immobilized fermentation strategy was developed. The polyurethane carrier, which could be beneficial to cell aggregation owing to its high strength and toughness was used to support the biofilm (Zhao et al., 2015). The pore size of the carrier was also important for biofilm immobilization (Yu et al., 2018). Here, immobilized fermentations by above-mentioned three strains were carried out with 10 mm × 10 mm × 10 mm polyurethane sponge pieces. In the immobilized continuous (repeated-batch) fermentation, L-threonine production in the first four batches was improved gradually in strain E. coli W1688-fimH* (Figure 4A), while L-threonine production did not show obvious improvement in strain E. coli W1688 (around 10.4 g/L) and E. coli W1688-ΔfimH (around 9.5 g/L; Data not shown). After the 4th batch, L-threonine production was maintained at an average of 17.5 g/L during a fermentation period decreased from 30 to 28 h. L-threonine productivity was kept at about 0.63 g/L/h from fourth batch, which was much higher compared with that from free-cell fermentation by the original strain (0.63 g/L/h vs. 0.35 g/L/h) (Figure 4B). Near 1-fold improved productivity contributed to a less time in a batch fermentation and we could achieve more products in the same fermentation time. These indicated that the continuous (repeated-batch) immobilized fermentation strategy taking advantage of biofilm formation in fimH overexpression strain could enhance L-threonine titer and productivity.
FIGURE 4.
(A) L-threonine production by E. coli W1688-fimH* in continuous immobilized fermentation. (B) Comparison of L-threonine productivities in continuous (repeated batch) immobilized fermentation with those in free-cell fermentation.
To further confirm biofilm formation by strain E. coli W1688-fimH* under industrial fermentation conditions, scanning electron microscope experiments were performed. The images of carriers during the immobilized fermentation with different strains are shown in Figure 5. Biofilm formation could be observed obviously when using E. coli W1688-fimH*. It could be concluded that the carrier could fix bacterial cells on the surface and provided good conditions for oxygen- and mass-transfer during the cell growth process (Lan et al., 2013). Furthermore, it was shown that the carrier could provide surfaces for cell adhesion and facilitated biofilm formation during the fermentation process. Hence, seed culture was avoided before each batch of fermentation owing to the existence of cells in the biofilm. In contrast, strains E. coli W1688 and E. coli W1688-ΔfimH did not show noticeable adhesion and biofilm formation on the surface (Figures 5II,IV). As a result, the immobilized fermentation by E. coli W1688-fimH* biofilm could be continually operated to produce L-threonine. In such a fermentation mode, cell degeneration and cell growth were supposed to be in an equilibrium, suggesting that an ideal state of balance was achieved (De Ory et al., 2004). This combination of biofilm and immobilized fermentation generates a new idea, which is also applicable to other industrial fermentation processes.
FIGURE 5.
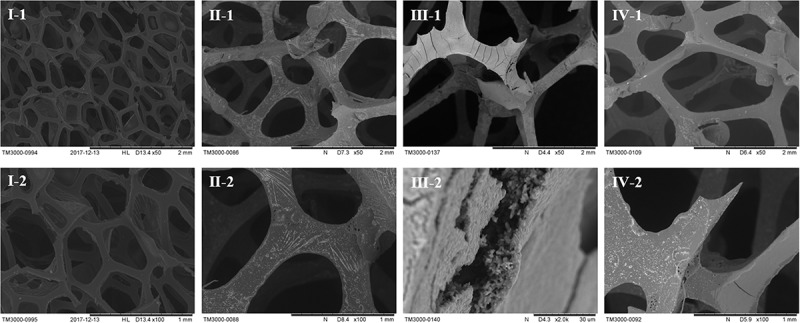
Scanning electron microscopy images of carrier in the immobilized fermentation by three different strains. I: fresh carrier, II: E. coli W1688, III: E. coli W1688-fimH*, IV: E. coli W1688-ΔfimH.
Transcriptome Analysis for Enhanced Biofilm Formation
To investigate the mechanism of enhanced biofilm formation, transcriptome analysis was performed for wild-type, E. coli W1688-fimH* and E. coli W1688-ΔfimH. A total of over 22.7, 23.0, and 21.3 million raw reads were obtained, respectively. The expression ratios of genes involved in biofilm biosynthesis which showed significant differences among these strains were calculated in Figure 6A, where the regulated genes could be classified into six distinct clusters. The fimH (encoding type I fimbriae adhesin) and flu genes guide the secretion of adhesins responsible for cell adhesion to surfaces (Reisner et al., 2003; Schembri et al., 2003). FlhD and FlhC are transcriptional activators involved in flagellar assembly and regulon, which is related to cell motility and biofilm formation (Beloin et al., 2004). CsgD is in charge of curli assembly, transport and structural components biosynthesis for biofilm formation together with csgA, csgB, and csgC (Reisner et al., 2003). The glgA, glgC, and glgP control glycogen biosynthesis (Beloin et al., 2004). The luxS, metK, speD, and lsrR are involved in biosynthesis of quorum sensing (QS) signal molecule AI-2 (autoinducer-2), which can activate transcription factors to promote formation of biofilm when the cell density reaches to a threshold (Vendeville et al., 2005). Actually, gene luxS concerning catalyzing the reaction to AI-2 was widely spread in Gram-negative and -positive bacterium, which shows a high homologous conservation. All these genes were up-regulated in varying degrees in the fimH-overexpressed strain E. coli W1688-fimH*, while they were down-regulated in the fimH-deleted strain compared with the wild-type. Concentration of extracellular AI-2 could also be decreased rapidly by lsr’s ABC (ATP-binding cassette) transporter, which can transport AI-2 into the cell. The transcriptome data showed that some genes in lsr operon were down-regulated, which might further lead to accumulation of extracellular AI-2 and promote the expression of biofilm-related genes (Schauder et al., 2001; Barrios et al., 2006). The genes showed various degrees of upregulation in fimH overexpression strain, which was validated by qRT-PCR analysis (Figure 6B). Notably, the transcription level by fimH was 6.2-fold higher than that in E. coli W1688-fimH* compared to the wild type, which was close to 0-fold in fimH deletion strain, indicating that the deletion of fimH was successful in E. coli W1688-ΔfimH. Besides, flu gene expression showed more than 2-fold in E. coli W1688-fimH* by qRT-PCR analysis. CyaA, csgD, luxS, and lsrR involved in flagellar assembly and signal secretion and reception of quorum sensing were up-regulated with more than 1-fold with original strain, while these genes showed down-regulated with varying degrees in E. coli W1688-ΔfimH. Taken together, the overexpression of fimH gene triggered modulation of related genes to enhance biofilm formation in E. coli W1688-fimH*.
FIGURE 6.
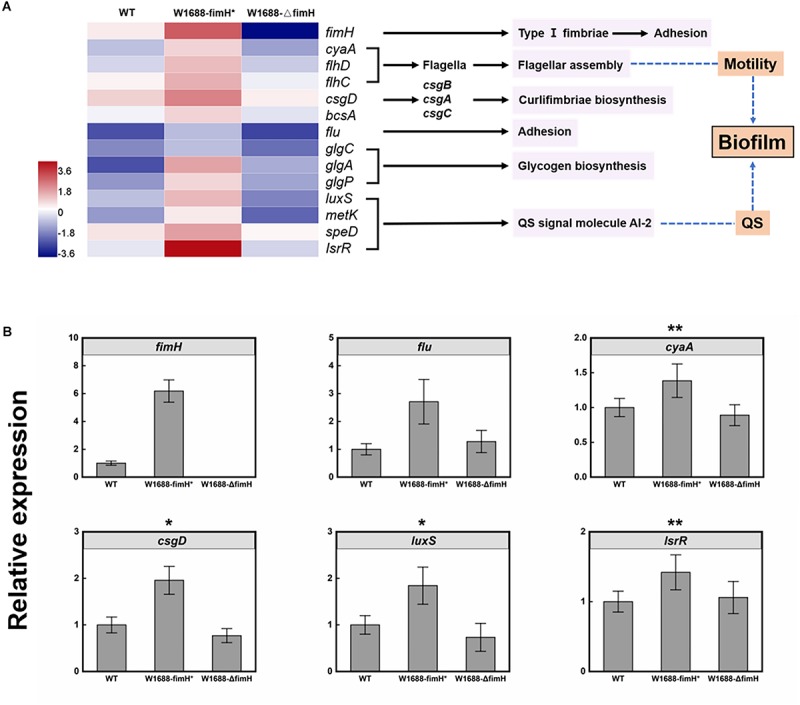
(A) Transcriptome analysis of genes involved in biofilm biosynthesis pathway in E. coli W1688, E. coli W1688-fimH*, and E. coli W1688-ΔfimH. Red and blue indicate up- and down-regulated genes, respectively. (B) qRT-PCR verification of the genes related to biofilm biosynthesis. Values and error bars represent the mean and the s.d. (n = 3). ∗∗∗p < 0.001, ∗∗p < 0.01, *p < 0.05 as determined by two-tailed t-test.
Transcriptome Analysis for Overexpressing FimH Gene to Increase L-Threonine Production in E. coli
The overexpression of fimH resulted in oversecretion of adhesion protein, which was beneficial for the gather of cells to biofilm production. Moreover, the immobilized fermentation system based on biofilm formation for L-threonine production was applied in increasing L-threonine production. To figure out whether the increase in L-threonine production was also associated with regulation of biosynthetic pathway genes and elaborate the molecular mechanism linking biofilm formation to L-threonine biosynthesis, further transcriptome analysis was performed. Fortunately, it was found that overexpression of fimH gene resulted in not only the accumulation of biofilm, but also regulation of genes in the L-threonine biosynthetic pathway (Figure 7A). The key genes such as thrA, lysC, metL, and asd that dominate the pathway from L-aspartate to L-homoserine, and thrB and thrC that dominate the pathway from L-homoserine to L-threonine (Lee et al., 2003; Livshits et al., 2003) were all up-regulated by an average of 11-fold in E. coli W1688-fimH* compared with those in wild-type strain and E. coli W1688-ΔfimH. The L-threonine transporter-encoding genes rhtA, rhtB, and rhtC were also up-regulated, which would facilitate the extracellular accumulation of L-threonine (Kruse et al., 2002). These results indicated that the enhanced biofilm formation affected the enzymes expression of L-threonine pathway and facilitated the central carbon flux. On the other hand, the tdcB gene encoding threonine dehydratase was down-regulated, which would benefit the accumulation of target products. In addition, down-regulated genes were lysA and metA, which catalyzed the last step in L-lysine biosynthesis and generated the L-methionine, respectively. More precursor substances involved in central carbon metabolism diverted to L-threonine formation. Obviously, down-regulation of these genes could beneficially facilitate the accumulation of L-threonine. Furthermore, expression of genes in competing branch pathways such as aspA, mediating the pathway from L-aspartate to fumarate were all decreased apparently in E. coli W1688-fimH*. Indeed, aspA encoding aspartase to synthesize target chemicals in TCA cycle always brings a competitive effect on carbon flux. Therefore, the overexpression of FimH adhesin protein could facilitate the redistribution of carbon flux via down-regulation of aspA. The gene expression levels of acs, pta, and ackA (Lee et al., 2006; Nahku et al., 2010) were down-regulated notably by 3.7, 5.9, and 6.1-fold, respectively. Since acetate accumulation has a detrimental effect on biofilm formation, cell growth and production, the transcriptional level in whole module of acetate pathway was down-regulated which could be beneficial for L-threonine production. As a result, it could be conducive to keeping the pH of broth relatively stable due to decreased acetate flux decreased. This would create a favorable condition for L-threonine biosynthesis and glucose consumption (De Mey et al., 2007). The genes for branched metabolic pathway showed various degrees of downregulation, while the genes related to central carbon metabolism and L-threonine transportation were significantly upregulated in E. coli W1688-fimH*, which were quantified by qRT-PCR (Figure 7B). The relative expressions of ackA, aspA, lysA, and tdcB were decreased at least 23% in E. coli W1688-fimH*, which these genes related to biosynthesis of acetate, the degradation of L-threonine and branch by-products like fumarate, L-lysine and L-methionine. The genes involved in L-threonine direct synthesis (thrA/B/C) showed more than 2.3-fold expressions than that in original strain. Significantly, the relative expression of rhtA was 10.7-fold higher in E. coli W1688-fimH* than that in wild-type strain. It meant that more carbon flux contributed to L-threonine biosynthesis and the target product could be transported out of cell membrane easier. Meanwhile, the four genes didn’t show any obvious differences in E. coli W1688-ΔfimH compared with original strain. Overall, these results showed that the overexpression of fimH gene also enhanced the metabolic flux toward L-threonine by up- or down-regulating related pathway genes in E. coli W1688-fimH*.
FIGURE 7.
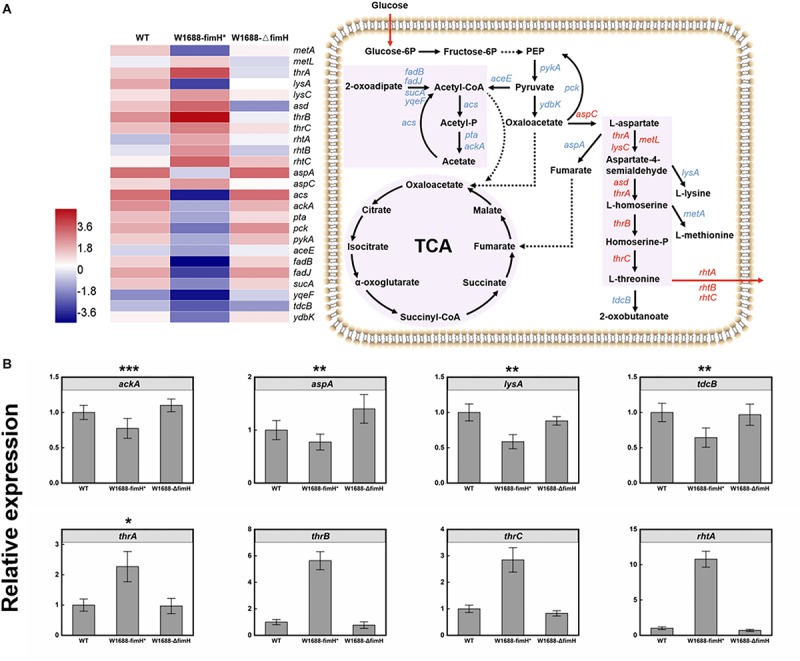
(A) Transcriptome analysis of L-threonine biosynthesis pathway genes in three different strains and the expression levels of these genes in E. coli W1688-fimH*. Red and blue indicate up- and down-regulated genes, respectively. (B) qRT-PCR verification of the genes related to L-threonine biosynthesis and transportation. Values and error bars represent the mean and the s.d. (n = 3). ∗∗∗p < 0.001, ∗∗p < 0.01, *p < 0.05 as determined by two-tailed t-test.
Comparison of L-Threonine Production by E. coli W1688-fimH* With Other Studies
Currently, production of L-threonine in E. coli can be enhanced by metabolic engineering such as inactivation in TCA cycle, increasing glycolysis pathway flux and facilitating L-threonine central carbon metabolism. Besides these established metabolic pathway approaches and strategies, there are still several limiting factors hampering further improvement of L-threonine productivity such as fermentation strategy of optimization. ThrB and thrC are clustered with thrA in the thrABC operon, which is mainly responsible for central carbon metabolism to L-threonine (Lee et al., 2003). Increased rhtA, rhtB, and rhtC expression will hence L-threonine exported from intracellular to extracellular through transmembrane protein (Yuzbashev et al., 2013). Recently, fermentation optimization has been proved in improving L-threonine production effectively such as two-stage feeding strategy and fed-batch fermentation mode (Table 4). Compared to the original strain E. coli W1688, the cell growth and biofilm formation of E. coli W1688-fimH* in the medium containing 30 g/L glucose was improving. Moreover, glucose was completely consumed in 28 h after 4th batch in biofilm-based immobilized repeated-fed batch fermentation. The highest yield (0.59 g/g) of L-threonine was achieved owing to the high expression of genes in the central carbon metabolism and decrease of by-products, which was a 60% increase compared to the original strain E. coli W1688. Since the biofilm formation performance can be varied depending on many genes, we further performed to screen and excavate potential genes to facilitate biofilm formation and increase the L-threonine production.
TABLE 4.
Comparison of L-threonine production in engineered E. coli strains.
| Strains | Carbon source | Time (h) | L-threonine (g/L) | Productivity (g/L/h) | Yield (g/g) | Fermentation mode | References |
| E. coli βIM4 (pBR322-thrAr) | Glucose | 72 | 13.4 | 0.186 | 0.45 | Batch | Miwa et al., 1983 |
| E. coli TWF006/pFW01-thrA*BC-asd | Glucose | 36 | 15.9 | 0.44 | 0.53 | Batch | Zhao et al., 2018 |
| E. coli TH28C (pBRThrABCR3) | Glucose | 50 | 82.4 | 1.648 | 0.46 | Fed-batch | Lee et al., 2007 |
| E. coli MT201 | Glucose | 28 | 102 | 3.643 | 0.38 | Fed-batch | Lee et al., 2018 |
| E. coli THPE5 | Glucose | 40 | 70.8 | 1.77 | 0.404 | Fed-batch | Liu et al., 2019 |
| E. coli W1688 | Glucose | 36 | 10.5 | 0.292 | 0.367 | Batch | This study |
| E. coli W1688-fimH* | Glucose | 28 | 17.5 | 0.63 | 0.59 | Repeated batch | This study |
Conclusion
An immobilized fermentation system for L-threonine production by E. coli was developed by taking advantages of biofilm formation. The engineered strain overexpressing fimH successfully enhanced biofilm formation under industrial cultivation conditions, which could also apply to continuous (repeated-batch) immobilized fermentation. L-threonine production was increased from 10.5 to 14.1 g/L using E. coli W1688-fimH* during batch fermentations and was further improved to 17.5 g/L during continuous (repeated-batch) fermentations, with a productivity of 0.63 g/L/h. Transcriptome profiles indicated that the biofilm formation was enhanced by regulation of biofilm-related genes. Meanwhile, L-threonine biosynthesis was also enhanced by up- or down-regulating related genes in L-threonine metabolic pathway. The engineered E. coli W1688-fimH* would be of great value for immobilized fermentation of L-threonine. This study will also provide a reference for developing more biochemical-producing processes based on E. coli biofilm.
Data Availability
The reads and the HiSeq transcriptomic reads generated for E. coli W1688, E. coli W1688-fimH*, and E. coli W1688-ΔfimH, respectively, have been submitted to the BioProject database of National Center for Biotechnology Information (NCBI) under the accession numbers SRR8335002, SRR8335001, and SRR8334999, respectively.
Author Contributions
TC and NL conceived and designed the experiments, performed the laboratory work, analyzed and interpreted the data, and drafted the manuscript. PR constructed the plasmids and strains, participated in the fermentation experiments, performed the shooting of electron microscope, analyzed the metabolic products, and performed the statistical analysis. XX, LY, and WS performed the transcriptome analysis of different strains and revised the manuscript. BY supplied the carriers. HY and PO critically revised the manuscript. YC and DL contributed to the experimental design, data interpretation, and critically revised the manuscript. All authors read and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Many thanks to Frasergen Inc. (http://www.frasergen.com/) for providing sequencing platform and technical assistance in data analyses. We thank Zhongxue Dai, Ph.D., for the help in formatting and language editing in this manuscript. We also thank Prof. Sheng Yang for providing strain E. coli W1688.
Footnotes
Funding. This work was supported by the Major Research Plan of the National Natural Science Foundation of China (21390204), the Key Program of the National Natural Science Foundation of China (21636003), the Program for Changjiang Scholars and Innovative Research Team in University (IRT_14R28), the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD), the Technology Support Program of Jiangsu (BE2014715), the Outstanding Youth Foundation of Jiangsu (SBK2017010373), the Nantong Applied Research Program, the Research Start-up Cost Project for Introduced Talent of Nantong University (03081134), and the China Postdoctoral Innovative Talents Support Program (BX20180140).
References
- Barrios A. F. G., Zuo R., Hashimoto Y., Yang L., Bentley W. E., Wood T. K. (2006). Autoinducer 2 controls biofilm formation in Escherichia coli through a novel motility quorum-sensing regulator (MqsR, B3022). J. Bacteriol. 188 305–316. 10.1128/JB.188.1.305-316.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beloin C., Valle J., Latour-Lambert P., Faure P., Kzreminski M., Balestrino D., et al. (2004). Global impact of mature biofilm lifestyle on Escherichia coli K-12 gene expression. Mol. Microbiol. 51 659–674. 10.1046/j.1365-2958.2003.03865.x [DOI] [PubMed] [Google Scholar]
- Boelee N., Temmink H., Janssen M., Buisman C., Wijffels R. (2014). Balancing the organic load and light supply in symbiotic microalgal-bacterial biofilm reactors treating synthetic municipal wastewater. Ecol. Eng. 64 213–221. 10.1016/j.ecoleng.2013.12.035 [DOI] [Google Scholar]
- De Mey M., Lequeux G. J., Beauprez J. J., Maertens J., Van Horen E., Soetaert W. K., et al. (2007). Comparison of different strategies to reduce acetate formation in Escherichia coli. Biotechnol. Progr. 23 1053–1063. 10.1021/bp070170g [DOI] [PubMed] [Google Scholar]
- De Ory I., Romero L. E., Cantero D. (2004). Optimization of immobilization conditions for vinegar production. Siran, wood chips and polyurethane foam as carriers for Acetobacter aceti. Process. Biochem. 39 547–555. 10.1016/S0032-9592(03)00136-5 28401461 [DOI] [Google Scholar]
- Dong X., Quinn P. J., Wang X. (2011). Metabolic engineering of Escherichia coli and Corynebacterium glutamicum for the production of L–threonine. Biotechnol. Adv. 29 11–23. 10.1016/j.biotechadv.2010.07.009 [DOI] [PubMed] [Google Scholar]
- Huang Y. L., Wu Z., Zhang L., Cheung C. M., Yang S.-T. (2002). Production of carboxylic acids from hydrolyzed corn meal by immobilized cell fermentation in a fibrous-bed bioreactor. Bioresour. Technol. 82 51–59. 10.1016/S0960-8524(01)00151-1 [DOI] [PubMed] [Google Scholar]
- Kim K. H., Choi I. S., Kim H. M., Wi S. G., Bae H.-J. (2014). Bioethanol production from the nutrient stress-induced microalga Chlorella vulgaris by enzymatic hydrolysis and immobilized yeast fermentation. Bioresour. Technol. 153 47–54. 10.1016/j.biortech.2013.11.059 [DOI] [PubMed] [Google Scholar]
- Kruse D., Krämer R., Eggeling L., Rieping M., Pfefferle W., Tchieu J., et al. (2002). Influence of threonine exporters on threonine production in Escherichia coli. Appl. Microbiol. Biotechnol. 59 205–210. 10.1007/s00253-002-0987-7 [DOI] [PubMed] [Google Scholar]
- Lan T.-Q., Wei D., Yang S.-T., Liu X. (2013). Enhanced cellulase production by Trichoderma viride in a rotating fibrous bed bioreactor. Bioresour. Technol. 133 175–182. 10.1016/j.biortech.2013.01.088 [DOI] [PubMed] [Google Scholar]
- Le Trong I., Aprikian P., Kidd B. A., Forero-Shelton M., Tchesnokova V., Rajagopal P., et al. (2010). Structural basis for mechanical force regulation of the adhesin FimH via finger trap-like β sheet twisting. Cell 141 645–655. 10.1016/j.cell.2010.03.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H.-W., Lee H.-S., Kim C.-S., Lee J.-G., Kim W.-K., Lee E.-G., et al. (2018). Enhancement of L–threonine production by controlling sequential carbon-nitrogen ratios during fermentation. J. Microbiol. Biotechnol. 28 293–297. 10.4014/jmb.1708.08072 [DOI] [PubMed] [Google Scholar]
- Lee J.-H., Lee D.-E., Lee B.-U., Kim H.-S. (2003). Global analyses of transcriptomes and proteomes of a parent strain and an L–threonine-overproducing mutant strain. J. Bacteriol. 185 5442–5451. 10.1128/JB.185.18.5442-5451.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K. H., Park J. H., Kim T. Y., Kim H. U., Lee S. Y. (2007). Systems metabolic engineering of Escherichia coli for L–threonine production. Mol. Syst. Biol. 3:149. 10.1038/msb4100196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M.-H., Lee H.-W., Park J.-H., Ahn J.-O., Jung J.-K., Hwang Y.-I. (2006). Improved L–threonine production of Escherichia coli mutant by optimization of culture conditions. J. Biosci. Bioeng. 101 127–130. 10.1263/jbb.101.127 [DOI] [PubMed] [Google Scholar]
- Leuchtenberger W., Huthmacher K., Drauz K. (2005). Biotechnological production of amino acids and derivatives: current status and prospects. Appl. Microbiol. Biotechnol. 69 1–8. 10.1007/s00253-005-0155-y [DOI] [PubMed] [Google Scholar]
- Li X., Yan Z., Xu J. (2003). Quantitative variation of biofilms among strains in natural populations of Candida albicans. Microbiology 149 353–362. 10.1099/mic.0.25932-0 [DOI] [PubMed] [Google Scholar]
- Liu D., Chen Y., Li A., Ding F., Zhou T., He Y., et al. (2013). Enhanced butanol production by modulation of electron flow in Clostridium acetobutylicum B3 immobilized by surface adsorption. Bioresour. Technol. 129 321–328. 10.1016/j.biortech.2012.11.090 [DOI] [PubMed] [Google Scholar]
- Liu J., Li H., Xiong H., Xie X., Chen N., Zhao G., et al. (2019). Two-stage carbon distribution and cofactor generation for improving l–threonine production of Escherichia coli. Biotechnol. Bioeng. 116 110–120. 10.1002/bit.26844 [DOI] [PubMed] [Google Scholar]
- Livshits V. A., Zakataeva N. P., Aleshin V. V., Vitushkina M. V. (2003). Identification and characterization of the new gene rhtA involved in threonine and homoserine efflux in Escherichia coli. Res. Microbiol. 154 123–135. 10.1016/S0923-2508(03)00036-6 [DOI] [PubMed] [Google Scholar]
- Madyagol M., Al-Alami H., Levarski Z., Drahovská H., Turňa J., Stuchlík S. (2011). Gene replacement techniques for Escherichia coli genome modification. Folia Microbiol. 56 253–263. 10.1007/s12223-011-0035-z [DOI] [PubMed] [Google Scholar]
- Miwa K., Tsuchida T., Kurahashi O., Nakamori S., Sano K., Momose H. (1983). Construction of L-threonine overproducing strains of Escherichia coli K–12 using recombinant DNA techniques. Agric. Biol. Chem. 47 2329–2334. 10.1080/00021369.1983.10865958 [DOI] [Google Scholar]
- Nahku R., Valgepea K., Lahtvee P.-J., Erm S., Abner K., Adamberg K., et al. (2010). Specific growth rate dependent transcriptome profiling of Escherichia coli K12 MG1655 in accelerostat cultures. J. Biotechnol. 145 60–65. 10.1016/j.jbiotec.2009.10.007 [DOI] [PubMed] [Google Scholar]
- Nishiyama M., Ishikawa T., Rechsteiner H., Glockshuber R. (2008). Reconstitution of pilus assembly reveals a bacterial outer membrane catalyst. Science 320 376–379. 10.1126/science.1154994 [DOI] [PubMed] [Google Scholar]
- Prigent-Combaret C., Vidal O., Dorel C., Lejeune P. (1999). Abiotic surface sensing and biofilm-dependent regulation of gene expression in Escherichia coli. J. Bacteriol. 181 5993–6002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajkumar R., Yaakob Z., Takriff M. S. (2013). Potential of micro and macro algae for biofuel production: a brief review. Bioresources 9 1606–1633. [Google Scholar]
- Reisner A., Haagensen J. A., Schembri M. A., Zechner E. L., Molin S. (2003). Development and maturation of Escherichia coli K-12 biofilms. Mol. Microbiol. 48 933–946. 10.1046/j.1365-2958.2003.03490.x [DOI] [PubMed] [Google Scholar]
- Sauer K. (2003). The genomics and proteomics of biofilm formation. Genome Biol. 4:219. 10.1186/gb-2003-4-6-219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauder S., Shokat K., Surette M. G., Bassler B. L. (2001). The LuxS family of bacterial autoinducers: biosynthesis of a novel quorum–sensing signal molecule. Mol. Microbiol. 41 463–476. 10.1046/j.1365-2958.2001.02532.x [DOI] [PubMed] [Google Scholar]
- Schembri M. A., Kjaergaard K., Klemm P. (2003). Global gene expression in Escherichia coli biofilms. Mol. Microbiol. 48 253–267. 10.1046/j.1365-2958.2003.03432.x [DOI] [PubMed] [Google Scholar]
- Shi X., Chen Y., Ren H., Liu D., Zhao T., Zhao N., et al. (2014). Economically enhanced succinic acid fermentation from cassava bagasse hydrolysate using Corynebacterium glutamicum immobilized in porous polyurethane filler. Bioresour. Technol. 174 190–197. 10.1016/j.biortech.2014.09.137 [DOI] [PubMed] [Google Scholar]
- Stoodley P., Sauer K., Davies D. G., Costerton J. W. (2002). Biofilms as complex differentiated communities. Annu. Rev. Microbiol. 56 187–209. 10.1146/annurev.micro.56.012302.160705 [DOI] [PubMed] [Google Scholar]
- Tripathi P., Beaussart A., Alsteens D., Dupres V., Claes I., Von Ossowski I., et al. (2013). Adhesion and nanomechanics of pili from the probiotic Lactobacillus rhamnosus GG. ACS Nano 7 3685–3697. 10.1021/nn400705u [DOI] [PubMed] [Google Scholar]
- Vendeville A., Winzer K., Heurlier K., Tang C. M., Hardie K. R. (2005). Making’sense’of metabolism: autoinducer-2, LuxS and pathogenic bacteria. Nat. Rev. Microbiol. 3 383–396. 10.1038/nrmicro1146 [DOI] [PubMed] [Google Scholar]
- Weissman S. J., Chattopadhyay S., Aprikian P., Obata-Yasuoka M., Yarova-Yarovaya Y., Stapleton A., et al. (2006). Clonal analysis reveals high rate of structural mutations in fimbrial adhesins of extraintestinal pathogenic Escherichia coli. Mol. Microbiol. 59 975–988. 10.1111/j.1365-2958.2005.04985.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Zheng C., Chen Y., Ying H. (2018). FLO genes family and transcription factor MIG1 regulate Saccharomyces cerevisiae biofilm formation during immobilized fermentation. Front. Microbiol. 9:1860. 10.3389/fmicb.2018.01860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu B., Zhang X., Sun W., Xi X., Zhao N., Huang Z., et al. (2018). Continuous citric acid production in repeated-fed batch fermentation by Aspergillus niger immobilized on a new porous foam. J. Biotechnol. 276 1–9. 10.1016/j.jbiotec.2018.03.015 [DOI] [PubMed] [Google Scholar]
- Yuzbashev T., Vybornaya T., Larina A., Gvilava I., Voyushina N., Mokrova S., et al. (2013). Directed modification of Escherichia coli metabolism for the design of threonine-producing strains. Appl. Biochem. Microbiol. 49 723–742. 10.1134/S0003683813090056 [DOI] [Google Scholar]
- Zhao H., Fang Y., Wang X., Zhao L., Wang J., Li Y. (2018). Increasing L-threonine production in Escherichia coli by engineering the glyoxylate shunt and the L-threonine biosynthesis pathway. Appl. Microbiol. Biotechnol. 102 5505–5518. 10.1007/s00253-018-9024-3 [DOI] [PubMed] [Google Scholar]
- Zhao N., Ren H., Li Z., Zhao T., Shi X., Cheng H., et al. (2015). Enhancement of nuclease P1 production by Penicillium citrinum YL104 immobilized on activated carbon filter sponge. Appl. Microbiol. Biotechnol. 99 1145–1153. 10.1111/j.1471-4159.1969.tb10388.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The reads and the HiSeq transcriptomic reads generated for E. coli W1688, E. coli W1688-fimH*, and E. coli W1688-ΔfimH, respectively, have been submitted to the BioProject database of National Center for Biotechnology Information (NCBI) under the accession numbers SRR8335002, SRR8335001, and SRR8334999, respectively.