Abstract
Transfer RNAs (tRNAs) are key components of the translation machinery. They read codons on messenger RNAs (mRNAs) and deliver the appropriate amino acid to the ribosome for protein synthesis. The human genome encodes more than 500 tRNA genes but their individual contribution to the cellular tRNA pool is unclear. In recent years, novel methods were developed to improve the quantification of tRNA gene expression, most of which rely on next-generation sequencing such as small RNA-Seq applied to tRNAs (tRNA-Seq). In a previous study, we presented a bioinformatics strategy to analyse tRNA-Seq datasets that we named ‘isodecoder-specific tRNA gene contribution profiling’ (Iso-tRNA-CP). Using Iso-tRNA-CP, we showed that tRNA gene expression is cell type- and tissue-specific and that this process can regulate tRNA-derived fragments abundance. An additional observation that stems from that work is that approximately half of human tRNA genes appeared silent or poorly expressed. In this commentary, I discuss this finding in light of the current literature and speculate on potential functions that transcriptionally silent tRNA genes may play. Studying silent tRNA genes may offer a unique opportunity to unravel novel mechanisms of cell regulation associated to tRNA biology.
Keywords: Transfer RNA, tRNA-Seq, tRNA-derived fragments, tRNA function, gene expression
Comment on: Torres AG, Reina O, Stephan-Otto Attolini C, Ribas de Pouplana L. Differential expression of human tRNA genes drives the abundance of tRNA-derived fragments [published online ahead of print April 8, 2019]. Proc Natl Acad Sci U S A. 2019;116(17):8451-8456. doi:10.1073/pnas.1821120116. PubMed PMID: 30962382. PubMed Central PMCID: PMC6486751. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6486751
Transfer RNAs (tRNAs) are small non-coding RNAs that deliver amino acids to the ribosome for protein synthesis. They are transcribed by RNA Polymerase III (Pol III) and go through a series of maturation steps and posttranscriptional modifications to become fully active.1 Once mature tRNAs are charged with their cognate amino acid, they read nucleotide triplets on messenger RNAs (codons) via base-pairing with nucleotide triplets present on the tRNA molecule (anticodon). Transfer RNAs carrying the same amino acid are termed isoacceptors (e.g. tRNAArg), while those isoacceptors that share the same anticodon sequence are called isodecoders (e.g. tRNAArgACG).
The human genome contains more than 500 tRNA genes to decode 61 codons.2 The reason for such a high genetic redundancy is to date unclear. As tRNA genes contain internal promoter regions that are in principle sufficient for Pol III-mediated transcription,1 it was once believed that all tRNA genes would be similarly expressed. In this sense, tRNA gene copy number can correlate with codon usage (i.e. codons recognised by high gene copy tRNAs are more frequently used than those recognised by low gene copy tRNAs).3 However, clear evidence pointing at differential expression of tRNA genes now exist.4-9 Moreover, natural genomic loss of tRNA genes have been described in the human population,10 resulting in no obvious phenotypes.11 This suggests that some tRNA genes may be constitutively silent transcriptionally. Whether these genes may have extra-transcriptional functions remain to be explored (see below).
Transfer RNA gene expression can be quantified by next-generation sequencing (tRNA-Seq). However, the tRNA structure, the presence of posttranscriptionally modified residues, the sequence similarity between different tRNA species, and the difficulty in discriminating between precursor tRNA transcripts and mature tRNAs, can result in quantification biases.9,12-15 We have recently developed a bioinformatics pipeline to help overcome some of these biases that we termed ‘isodecoder-specific tRNA gene contribution profiling’ (Iso-tRNA-CP).9 Isodecoder-specific tRNA-CP is based on the assumption that the sequences of isodecoder tRNA genes, that share similar structures and modifications patterns, are subjected to similar tRNA-Seq-dependent quantification biases. Hence, the method compares the expression of individual tRNA genes within their specific isodecoder set, and results are expressed as the proportional (%) transcript contribution of each gene to their corresponding isodecoder pool.
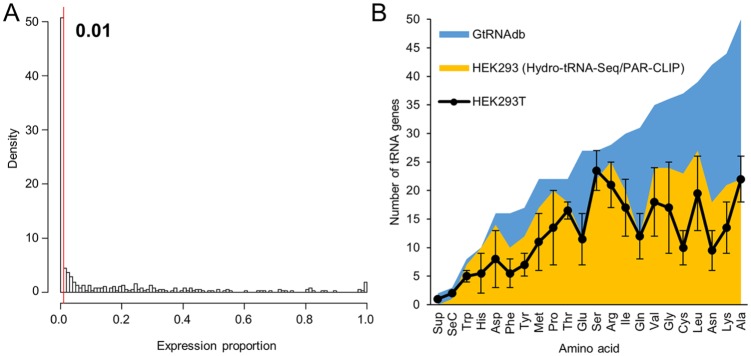
Analyses of the HEK293T tRNAome by Iso-tRNA-CP9 revealed that approximately half of human tRNA genes contribute less than 1% to their respective isodecoder pools (Figure 1A). This minor contribution could be interpreted as those genes being silent or poorly expressed in this cell line. A recent study performed in the closely related HEK293 cell line using alternative methods to estimate tRNA abundance (i.e. Hydro-tRNA-Seq and PAR-CLIP) reported that only 359 tRNA genes were transcriptionally active.15 Results obtained by Iso-tRNA-CP in HEK293T cells are in good agreement with this report when defining ‘active’ tRNA genes as those contributing more than 1% to their isodecoder pools (Figure 1B).
Figure 1.
Number of tRNA genes significantly expressed in HEK293T cells: (A) histogram analysis of number of tRNA genes (Y-axis; density) distributed across ranges of proportional contribution to their isodecoder sets (X-axis; expression proportion); based on Iso-tRNA-CP analyses obtained for HEK293T cells.9 A defined threshold (expression proportion = 0.01 = 1%) is indicated. Note that ‘tRNA genes’ are defined as single tRNA genes or tRNA families comprising variable number of tRNA genes with identical mature tRNA sequences as defined by Torres et al.9 (B) Number of tRNA genes per isoacceptor set (tRNAs charged with the same amino acid) sorted by their gene copy number according to the GtRNAdb v2.0,2 (dark-area graph). Light-area graph represents the number of tRNA genes for each isoacceptor set expressed in HEK293 cells as evidenced by Hydro-tRNA-Seq and PAR-CLIP.15 Black points represent the number of tRNA genes expressed in HEK293T cells based on a proportional contribution of at least 1% to their isodecoder set as calculated by Iso-tRNA-CP.9 As, in this analysis, all isoacceptor sets contain tRNA gene families, the error bars represent the minimum and maximum number of expressed tRNA genes considering that at least one, and potentially all, members of the tRNA family are expressed. The line connecting each isoacceptor set point is shown for profile comparisons against the dark- and light-area graphs.
Abbreviation: Iso-tRNA-CP, isodecoder-specific tRNA gene contribution profiling.
Expression of tRNA genes is cell type and tissue specific.4-9 Therefore, silent tRNA genes found in HEK293/HEK293T cells could potentially be transcriptionally active in other cell types or under specific physiological scenarios. Analyses using genome-wide chromatin state data from the Roadmap Epigenomics Consortium in 127 human tissues and cell lines, revealed that out of the 596 human tRNA genes analysed, 254 were found consistently inactive (i.e. 342 active human tRNA genes).16 This is also in agreement with reported Pol III occupancy patterns in human cells and tissues.7,17-19 While the evaluation of chromatin states or patterns of Pol III occupancy are indirect measurements of tRNA gene expression (e.g. Pol III binding to a tRNA gene may not necessarily result in tRNA gene transcription), these studies also support that nearly half of human tRNA genes are constitutively silent or poorly expressed.
The observation that so many of the predicted human tRNA genes are in a silent state is intriguing. Some of these genes may be tRNA pseudogenes.15 However, mounting evidence suggest that tRNA genes may have extra-transcriptional functions.20,21 For example, human tRNA genes were shown to play a role as insulators: they can block heterochromatin, repression mediated by the polycomb group proteins, and enhancer-mediated transcription activation.22 Notably, while recruitment of transcription factors are critical for the insulator function of tRNA genes, transcription of such tRNA genes is not a requirement.23 Transfer RNA genes are also sites for replication fork pauses and have been suggested to participate in preventing genetic instability in tumour formation.24 A role in directing transposon integration for tRNA genes has also been described.25 In addition, in yeast, binding of the transcription factor TFIIIC (without Pol III) to tRNA genes was shown to participate in the three-dimensional organisation of the genome, by tethering distant loci to the nuclear periphery.26 Finally, comparative genomic studies revealed that tRNA genes tend to be associated to sites of gene gain and evolutionary breakpoints,27 suggesting a role for tRNA genes in recombination events in evolving yeast genomes.20
Altogether, these observations suggest that nuclear-encoded tRNA genes (both transcriptionally active and silent ones) may be playing an important role in genome structure. In this sense, it is worth drawing a parallelism with mitochondrial-encoded tRNA (mt-tRNA) genes. In the mitochondrial genome, mt-tRNA genes are located specifically between mt-messenger RNA (mt-mRNA) and mt-ribosomal RNA (mt-rRNA) genes. The mitochondrial genome is transcribed as long polycistronic transcripts and upon 5′- and 3′-end processing of the tRNA, the corresponding mt-mRNA and mt-rRNA transcripts are released.28 Although mt-tRNA genes can structurally differ from nuclear-encoded tRNA genes,28 it is possible that, in general, tRNA genes can serve as optimal intervening DNA sequences to regulate cellular functions (via diverse mechanisms) at genomic level.
Concluding Remarks
Mapping of tRNA-Seq reads to the genome is not perfect.29 This makes the definition of an ‘active’ versus ‘inactive’ tRNA gene based on the absolute number of sequencing reads mapping to a given tRNA gene very challenging. Isodecoder-specific tRNA-CP may serve as a method to address this issue, as the number of significantly expressed tRNA genes can be estimated by defining a threshold of transcript contribution that tRNA genes generate towards their respective isodecoder pool (e.g. 1% contribution threshold as used in this work).
Direct tRNA transcript measurements,9,12,13,15 as well as indirect tRNA gene expression analyses,7,16-19 in several cell lines and tissues now support the observation that approximately half of human tRNA genes are silent or poorly expressed. While evidence for extra-transcriptional function for tRNA genes is emerging, a number of outstanding issues remain to be addressed: is there a correlation between the transcriptional state of tRNA genes and their chromosomal positioning?30 Given that tRNA genes are highly conserved, are genes homologous to human silent tRNAs also conserved and silent in other species? Do silent tRNA genes present specific structural features? What other extra-transcriptional functions for tRNA genes are awaiting to be described? Addressing these questions will be key to expand our understanding on the multiple roles tRNA genes can play and will pave the way towards consolidating the study of silent tRNA genes as a developing playground waiting to be enjoyed.
Acknowledgments
The author thanks Dr Lluís Ribas de Pouplana (IRB Barcelona) for critical reading of this manuscript and helpful discussions; and Dr Oscar Reina and the Bioinformatics and Biostatistics Unit at IRB Barcelona for support in data analyses.
Footnotes
Funding:The author(s) received no financial support for the research, authorship, and/or publication of this article.
Declaration of Conflicting Interests:The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Author Contributions: AGT conceived the project, analyzed data and wrote the article.
ORCID iD: Adrian Gabriel Torres  https://orcid.org/0000-0002-5479-1325
https://orcid.org/0000-0002-5479-1325
References
- 1. Piñeyro D, Torres AG, Ribas de Pouplana L. Biogenesis and evolution of functional tRNAs. In: Sesma A, Von der Haar T, eds. Fungal RNA Biology. Cham, Switzerland: Springer; 2014:233-267. [Google Scholar]
- 2. Chan PP, Lowe TM. GtRNAdb 2.0: an expanded database of transfer RNA genes identified in complete and draft genomes. Nucleic Acids Res. 2016;44:D184-D189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Novoa EM, Pavon-Eternod M, Pan T, Ribas de, Pouplana L. A role for tRNA modifications in genome structure and codon usage. Cell. 2012;149:202-213. [DOI] [PubMed] [Google Scholar]
- 4. Dittmar KA, Goodenbour JM, Pan T. Tissue-specific differences in human transfer RNA expression. PLoS Genet. 2006;2:e221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Goodarzi H, Nguyen HCB, Zhang S, Dill BD, Molina H, Tavazoie SF. Modulated expression of specific tRNAs drives gene expression and cancer progression. Cell. 2016;165:1416-1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ishimura R, Nagy G, Dotu I, et al. RNA function. Ribosome stalling induced by mutation of a CNS-specific tRNA causes neurodegeneration. Science. 2014;345:455-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kutter C, Brown GD, Goncalves A, et al. Pol III binding in six mammals shows conservation among amino acid isotypes despite divergence among tRNA genes. Nat Genet. 2011;43:948-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sagi D, Rak R, Gingold H, et al. Tissue- and time-specific expression of otherwise identical tRNA genes. PLoS Genet. 2016;12:e1006264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Torres AG, Reina O, Stephan-Otto Attolini C, Ribas de, Pouplana L. Differential expression of human tRNA genes drives the abundance of tRNA-derived fragments. Proc Natl Acad Sci U S A. 2019;116:8451-8456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Iben JR, Maraia RJ. tRNA gene copy number variation in humans. Gene. 2014;536:376-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yang P, Beltramo DM, de Pouplana LR, Soria NW, Torres AG. Loss of the tRNA(Lys) CUU encoding gene, Chr-11 tRNA-Lys-CUU, is not associated with Type 2 diabetes mellitus. Biomark Med. 2019;13:259-266. [DOI] [PubMed] [Google Scholar]
- 12. Cozen AE, Quartley E, Holmes AD, Hrabeta-Robinson E, Phizicky EM, Lowe TM. ARM-seq: AlkB-facilitated RNA methylation sequencing reveals a complex landscape of modified tRNA fragments. Nat Methods. 2015;12:879-884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zheng G, Qin Y, Clark WC, et al. Efficient and quantitative high-throughput tRNA sequencing. Nat Methods. 2015;12:835-837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Torres AG, Pineyro D, Rodriguez-Escriba M, et al. Inosine modifications in human tRNAs are incorporated at the precursor tRNA level. Nucleic Acids Res. 2015;43:5145-5157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gogakos T, Brown M, Garzia A, Meyer C, Hafner M, Tuschl T. Characterizing expression and processing of precursor and mature human tRNAs by hydro-tRNAseq and PAR-CLIP. Cell Rep. 2017;20:1463-1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Thornlow BP, Hough J, Roger JM, Gong H, Lowe TM, Corbett-Detig RB. Transfer RNA genes experience exceptionally elevated mutation rates. Proc Natl Acad Sci U S A. 2018;115:8996-9001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Canella D, Praz V, Reina JH, Cousin P, Hernandez N. Defining the RNA polymerase III transcriptome: genome-wide localization of the RNA polymerase III transcription machinery in human cells. Genome Res. 2010;20:710-721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Moqtaderi Z, Wang J, Raha D, et al. Genomic binding profiles of functionally distinct RNA polymerase III transcription complexes in human cells. Nat Struct Mol Biol. 2010;17:635-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Oler AJ, Alla RK, Roberts DN, et al. Human RNA polymerase III transcriptomes and relationships to Pol II promoter chromatin and enhancer-binding factors. Nat Struct Mol Biol. 2010;17:620-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. McFarlane RJ, Whitehall SK. tRNA genes in eukaryotic genome organization and reorganization. Cell Cycle. 2009;8:3102-3106. [DOI] [PubMed] [Google Scholar]
- 21. Donze D. Extra-transcriptional functions of RNA Polymerase III complexes: TFIIIC as a potential global chromatin bookmark. Gene. 2012;493:169-175. [DOI] [PubMed] [Google Scholar]
- 22. Raab JR, Chiu J, Zhu J, et al. Human tRNA genes function as chromatin insulators. EMBO J. 2012;31:330-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Valenzuela L, Dhillon N, Kamakaka RT. Transcription independent insulation at TFIIIC-dependent insulators. Genetics. 2009;183:131-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Clelland BW, Schultz MC. Genome stability control by checkpoint regulation of tRNA gene transcription. Transcription. 2010;1:115-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chalker DL, Sandmeyer SB. Transfer RNA genes are genomic targets for de Novo transposition of the yeast retrotransposon Ty3. Genetics. 1990;126:837-850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Noma K, Cam HP, Maraia RJ, Grewal SI. A role for TFIIIC transcription factor complex in genome organization. Cell. 2006;125:859-872. [DOI] [PubMed] [Google Scholar]
- 27. Gordon JL, Byrne KP, Wolfe KH. Additions, losses, and rearrangements on the evolutionary route from a reconstructed ancestor to the modern Saccharomyces cerevisiae genome. PLoS Genet. 2009;5:e1000485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Suzuki T, Nagao A. Human mitochondrial tRNAs: biogenesis, function, structural aspects, and diseases. Annu Rev Genet. 2011;45:299-329. [DOI] [PubMed] [Google Scholar]
- 29. Hoffmann A, Fallmann J, Vilardo E, Morl M, Stadler PF, Amman F. Accurate mapping of tRNA reads. Bioinformatics. 2018;34:1116-1124. [DOI] [PubMed] [Google Scholar]
- 30. Van Bortle K, Phanstiel DH, Snyder MP. Topological organization and dynamic regulation of human tRNA genes during macrophage differentiation. Genome Biol. 2017;18:180. [DOI] [PMC free article] [PubMed] [Google Scholar]