Abstract
Purpose of Review:
Allergen-antibody complexes are extremely valuable in describing the detailed molecular features of epitopes. This review summarizes insights gained from recently published co-structures and what obstacles impede the acquisition of further data.
Recent Findings:
Structural epitope data helped define the epitopes of two anti-Fel d 1 antibodies undergoing phase I clinical trials, providing a greater level of detail than was possible through hydrogen exchange protection studies. Separately, a human camelid-like antibody structure with lysozyme described several unique features in a long variable loop that interacted with the active site cleft of Gal d 4. Finally, a co-structure conclusively demonstrated that Phl p 7 could function as a superantigen, and that an antibody could simultaneously recognize two epitopes. These remarkable assertions would not have been possible without visualization of the complex. Only 3 new complexes have appeared in the last few years, suggesting there are major impediments to traditional production and crystallization.
Summary:
The structural data was extremely valuable in describing epitopes. New techniques like cryo-EM may provide an alternative to crystallography.
Keywords: Allergens, antibodies, structure, complex, epitope, cystallography
Introduction
An allergen, by definition, is a protein that binds to the IgE antibody isotype. The cross-linking of IgE on effector cells leads to the symptoms associated with allergy. There are more than 100 different allergen structures in the protein data base [1], and the information provided has been utilized in many different ways to better understand allergic disease [2]. Structural data on allergens has helped predict biological function, which in turn suggested hypotheses about how allergens promote sensitization either through proteolytic activity or immune mimicry [3–5]. These structures have also proven useful in understanding patient cross-reactivity, [6–8] and have assisted in the mapping of patient epitopes and the development of hypoallergens [9]. A hypoallergen is an idealized allergen with reduced IgE or symptomatic epitopes, but still promotes tolerance to the original allergen when used in immunotherapy. [9]
A recent review of allergen structures found very few co-structures of allergens with antibodies [1]. This comes with a slight caveat in that there were many structures of lysozyme (also known as the egg allergen Gal d 4) with various antibodies. Lysozyme was crucial to early crystallography development as it was readily available and an x-ray structure was solved in 1965 [10]. These features allowed immunologists to utilize lysozyme as model antigen, with the structure of the HY/HEL-10 FAB in complex with Gal d 4 being the first allergen-antibody complex to be solved via X-ray crystallography in 1989 [11]. The next 26 years saw the release of 11 structures of antibodies in complex with only 7 more allergens. These included Api m 2 [12], Bos d 5 [13], Bet v 1 [14], Phl p 2 [15], Der p 1 [16], Der f 1 [17] and Bla g 2 [18, 19]. Most of the structural studies discuss how the epitope information can be used for future development of immunotherapy by designing hypo-allergens. Some studies also provide a structural understanding of antibody specificity and cross-reactivity, or the basis for designing mutants able to modulate T-cell responses. [20, 21] In one case the structure demonstrated carbohydrate recognition by the anti-Bla g 2 antibodies [19, 22].
The major utility of these co-structures is in the unambiguous epitope information obtained [23]. Currently, there are many different empirical and computational methods available to study antigen-antibody interactions, and each comes with their unique set of advantages and disadvantages [24]. The co-structures and the corresponding studies discussed herein however, are unique in their ability to provide structural information on both the allergen epitope and mode of antibody binding at the atomic level. In this review, we will to focus on three papers with antibody-allergen co-structures that have appeared since 2015 which, highlight the value of such information. These new complexes were used to understand epitopes in a phase I trial of two anti-Fel d 1 antibodies [25], for describing the unique epitope properties of human camelid-like antibodies with lysozyme as the model antigen [26], and finally for demonstrating that Phl p 7 could function as a superantigen with a novel mode of binding [27]. Despite the wealth of information provided from these co-structures, only a few similar studies have been reported in the literature. In recognition of this scarcity, we will also discuss some of the technical limitations that make these co-structures difficult, and suggest new avenues that might be explored.
Therapeutic Epitope Information
A recently described innovation in allergen immunotherapy utilized two anti-Fel d 1 antibodies in conjunction with the typical cat allergen therapy. To understand the significance, successful allergen immunotherapy and patient desensitization is usually associated with increased allergen-specific IgG4 levels [28, 29]. While the exact mechanism for this is still not entirely clear, it has been suggested that IgG4 blocks the allergen at the mast cell level and/or at the level of the antigen-presenting cell preventing IgE-facilitated activation of T-cells [30–32]. IgG4 has the additional benefit that it does not induce effector functions like IgG1 promoting inflammation. There have been many attempts to design immunotherapies that promote IgG4 through the direct stimulation of the patient’s immune system with allergen or allergen-like compounds. The private company Regeneron attempted a different approach wherein IgG4 levels were artificially enhanced with recombinant allergen-specific antibodies. The hypothesis was that augmenting the IgG4/IgE ratio would enhance the efficacy of traditional immunotherapies. This was tested in cat allergic patients utilizing two anti-Fel d 1 IgG4’s: REGN1908 and REGN1909. Based on the efficacy observed in a mouse model, patients were given 600 mg of IgG4 (300 mg of each of the two antibodies). While this amount might seem excessive even by the standards of structural biologists who typically employ milligram quantities of protein in their assays, this dosage produced anti-Fel d 1 serum levels of 75 mg/L at day 8 post-injection and >11 mg/L throughout the trial. For comparison, a study of patients after birch pollen immunotherapy reported increases in IgG4 against Bet v 1 averaging 2 mg/L [33]. In a more extreme example, bee keepers display anti-Api m 1 IgG4 in the 100 mg/L range during the summer season [34]. To summarize the results from the Regeneron therapy trial, administration of the recombinant IgG4 antibodies provided significant clinical benefit [25].
While these findings have significant implications in the field of immunology and immunotherapy, this review will focus on the role of the antibody-allergen structure in the development of the IgG4-based treatment itself. The allergen Fel d 1 is composed of peptide chains from 2 non-identical gene products that come together to form a uteroglobulin-like fold, [35] in contrast to most uteroglobulins, which are dimers of 2 identical chains. As an aside, it is interesting to note that Fel d 1 is one of the few allergens derived from more than 1 gene. A major innovation in determining the original structure of Fel d 1 was to create a construct which placed chain 2 at the N-terminus, connected to chain 1 via a short peptide linker (PDB code: 1PUO [35]). This strategy was also followed by Regeneron in their structure 5VYF [25].
The authors first attempted to map epitopes of REGN1908 and REGN1909 with hydrogen-deuterium exchange protection experiments (HDX) using mass spectrometry (MS) to compare 1H and 2H isotope levels, and thus the degree of solvent exposure in the bound versus free allergen [25], similar to what has been previously reported for other allergens like Pru du 6 [36], Ano o 1 [37], and Bet v 1 [38]. The level of 1H/2H exchange can also be monitored using NMR, which takes advantage of the property that 1H produces a strong NMR signal while 2H does not [39, 40]. While this technique has been used to successfully map epitopes on Der p 2 and Gal d 4, it does not enjoy the same popularity as MS, likely due to the much larger sample quantities required. Using MS HDX, the antibody REGN1908 was found to protect residues on alpha helices 1, 2, and 3 of chain 1 of Fel d 1 which suggested a large discontinuous epitope. In contrast, only a few residues spanning helices 1 and 2 of chain 2 were protected by REGN1909. Since most antibody epitopes occupy an area of 600–900 Å2, it was likely that additional residues were involved in REGN1909 binding.
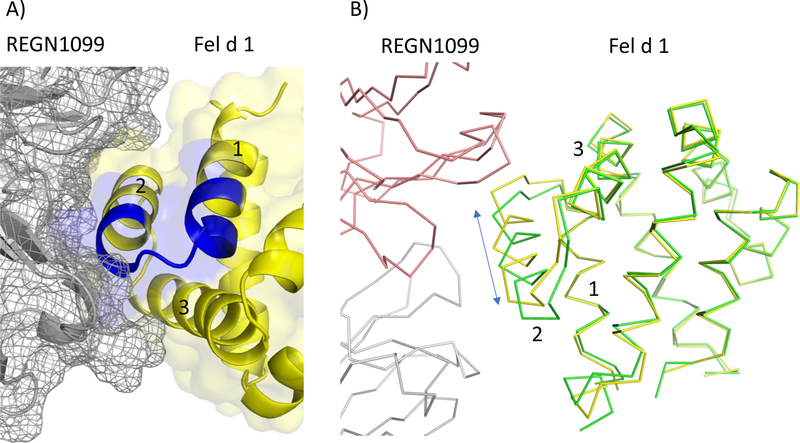
To obtain a more definitive understanding of the epitopes, crystal structures of both REGN1908 and REGN1909 in complex with the single chain Fel d 1 construct were attempted. Only the structure of REGN1909 in complex with Fel d 1 could be obtained, highlighting further the technical difficulties in obtaining antibody-allergen complexes. The antibody REGN1909 was found to interact with all of helix 2 and part of helix 3 of Chain 2 of Fel d 1, and did not interact with helix 1 at all in contrast to the HDX results that reported residues in helix 1 were protected from exchange (Figure 1A). This highlights an important consideration when interpreting HDX mapping experiments, as the results can be influenced by conformational changes in remote parts of the protein. This was previously noted in the NMR HDX results of Gal d 4, where residues distal from the epitope can vary in protection levels and thermodynamic properties [39]. While the structure of Fel d 1 is generally well conserved upon REGN1909 binding, a closer examination reveals that helix 2 is shifted almost 2 Å along its helical axis in the antibody complex compared to the unbound structure (Figure 1B). This translocation could potentially enhance the stability of this adjacent region of the protein, thus leading to greater HDX protection extending into helix 1. Summarizing this section, the complex structure provided new epitope details that were ambiguous or unavailable from other techniques.
Figure 1.
Fel d 1 bound to REGN1099. A- Fab REGN1099 is rendered with a mesh surface and cartoon secondary structure in grey, and Fel d 1 is shown with a cartoon secondary structure and semitransparent surface in yellow (5OTJ). Residues in blue were protected from exchange in MS HDX. Helices of Fel d 1 are labeled. B- Panel B is rotated relative to A to highlight changes in helix 2 of Fel d 1 upon binding REGN1099. The structure of Fel d 1 alone 1PUO (green) and in complex (yellow) with REGN1099 (peach: heavy chain, white: light chain) are rendered in as Cα traces.
In this study, it was reported that REGN1908 or REGN1909 alone could block up to 51% of some patients’ IgE binding to Fel d 1, while tandem administration reduced IgE binding by 83%, sufficient to produce a protective effect in most patients. This level of protection is impressive when we consider that the total surface area of the Fel d 1 structure is approximately 7,100 Å2, and typical epitopes are 600–900 Å2. As such, a single Fel d 1 could theoretically support the binding of 8–12 antibodies. While it is likely that the number that could simultaneously bind without sterically hindering each other is smaller, the ability of two antibodies (REGN1908 and REGN1909) to block 83% of the IgE binding suggests that the majority of IgE binding occurs over a limited fraction of the accessible Fel d 1 surface area. There is precedent for this in previous studies of the grass pollen allergen Phl p 2, where one antibody could block up to 80% of the patient IgE [41]. Similarly, high levels of IgE antibody binding (up to ~40–45%) were obtained with each of two IgG antibodies that bind to opposite lobes of the cockroach allergen Bla g 2 [22, 19]. Analogous evidence might be found in studies of patients undergoing oral immunotherapy for peanut allergy. Antibodies cloned from several patients appeared to show convergent evolution in high affinity Ara h 2-binding antibodies, suggesting that a similar epitope was driving the response [42]. Further studies of allergen-antibody complexes could provide insight into which epitopes are important for the immune response, potentially enabling us to augment the above therapy with hypoallergens that would further reduce the risk in immunotherapy.
Camelid-like Human VH Domains
Humans produce paired antibodies from two gene products, wherein a light chain pairs with a heavy chain to form the antibody combining site. Camels and llamas forgo the light chain, and utilize only the heavy chain for the combining site and effector functions; these are termed camelid antibodies [43]. The simplicity of this design and the ease of single gene manipulation has attracted the development of camelid antibodies for various therapies [44]. In the paper by Rouet et al., a similar design is suggested using only the heavy chain of human antibodies [26]. Other groups have attempted to ‘camelize’ human heavy chains by modifying the framework residues to improve the biophysical properties of the isolated domains [45]. In contrast, Rouet et al. developed a phage display system that focused exclusively on the CDRs of the heavy chain variable domain (VH) for optimization of stability [26]. In this way, VH could still theoretically pair with a human light chain if desired, increasing the range of therapeutic techniques available.
This phage display process also included a step with incubation of the proteins at high temperatures, allowing the authors to select for VH products that were exceptionally stable [26]. They found that residues in the variable loops had a significant effect on stability. This was rather unexpected in that it seems evolution would favor creating a highly stable scaffold (the immunoglobulin fold) that, even in isolation, would be tolerant to hypervariability in the loops as in naturally-produced antibodies. However, this may explain a number of anecdotal experiences, whereby grafting new variable loops onto a previously successful single chain fraction variable (ScFv) constructs yielded poor expression or insoluble proteins (personal observations).
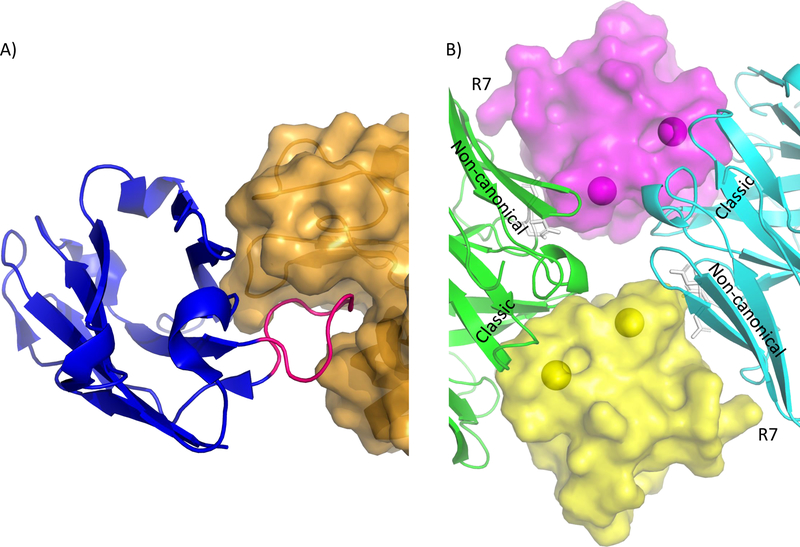
The phage display system used by Rouet et al. allowed for an artificially longer than usual CDR3, Figure 2A [26]. In the crystal structure of one such VH complexed with lysozyme (4U3X) the CDR3 contributes 535 Å2 of the buried surface area out of a total of 783 Å2. The length of the loop allowed it to extend into the active site cleft of the antigen in a manner similar to some other camelid and shark single chain antibodies [46–48]. The CDR3 of the VH was only 9 residues long compared to 12–18 in the other structures but was in the typical range of buried surface area, 561–906 Å2. These comparisons are only possible with the aid of a co-structure, and highlights the importance of this type of analysis in understanding the structural basis of antibody-epitope interactions, particularly those involving specific antigen features such as binding in clefts or hydrophobic pockets.
Figure 2.
Antibody complexes with allergens. A) VH (blue) in complex with lysozyme (Gal d 4, orange) rendered as cartoon diagrams with the surface of lysozyme shown semi-transparent. CDR3 of the VH is highlighted in pink in the lysozyme active site cleft. B) Phl p 7 bound to 101.1F10. Two Phl p 7 molecules are rendered as semi-transparent surfaces and bound Ca2+ ions are shown as spheres. Arg 7 of Phl p 7 is labeled. Two 101.1F10 molecules are rendered as cartoon secondary structures in green and cyan, while CDR L2 is shown with white stick renderings. The Classical (canonical) and Noncanonical antibody interacting regions are labeled.
In conclusion, the authors demonstrated that CDR composition can have a significant effect on the stability of an isolated VH, and that the affinity and functionality of camelids can be engineered into a fully human VH [26]. This opens up a number of opportunities for future therapeutic design. For example, these may prove useful in the design of anti-allergen IgG4 therapies like that described by Regeneron.
Superantigens
As a rule, antigens interact with the hypervariable loops of antibodies; superantigens interact instead with the constant domains or framework residues of the antibody [49]. Recently, Mitropoulou et al. reported a co-structure which utilized both modes of antigen-antibody binding. The antibody in question was an IgG antibody called 101.1F10 cloned from a grass pollen allergic patient. The antibody was specific for the grass allergen Phl p 7, and the resulting co-structure was revealed two 101.1F10 antibodies interacting simultaneously with Phl p 7 (Figure 2B) [27]. Concurrently, two Phl p 7 molecules could interact with two 101.1F10, running counter to the generally accepted idea that only dimeric antigens could interact with two identical antibodies at a given time. The authors also demonstrated that an IgE version of 101.1F10 alone could crosslink and signal effector functions with monomeric Phl p 7, further verifying this unique mode of binding. Adding interest to the paper, the authors noticed that Phl p 7 interacts with two epitopes, one of which is canonical in that it interacts with 5 of the hypervariable loops. The second noncanonical epitope interacts with the complimentary determining region loop 2 (CDR-L2, white sticks Figure 2B) and framework residues thus making it technically a superantigen.
Closer examination of the 101.1F10 -Phl p 7 co-structure reveals a possible structural basis for this unique interaction. The CDR-L2 in the noncanonical epitope was mutated from the germline gene at residue 54, suggesting that it was optimized for the interaction observed in the crystal. Supporting this, the NH2 group of N54 appears to make 2 hydrogen-bonds with Phl p 7, which the germline serine would be incapable of doing. The authors interpret this as optimization of the noncanonical site for a second epitope, allowing one antibody to recognize and bind Phl p 7 at two separate regions simultaneously. However, since the antigen:antibody stoichiometry could not be determined in the cell-based experiment, the available data cannot confirm that only one Phl p 7 could functionally crosslink two IgE. To elaborate, it may be that one Phl p 7 can crosslink two IgE, but it may also be that two Phl p 7 are required to operate in tandem as in the crystal. The noncanonical epitope is only 330 Å2, which is relatively small and may not bind tightly enough to crosslink 2 IgE with only a single Phl p 7. Indeed, the crosslinking was abrogated by mutating only R7 of the noncanonical epitope. Two noncanonical epitopes operating cooperatively appears more plausible, as together they would represent a binding surface of similar size to typical high affinity epitopes. The development of assays to test the stoichiometry required for crosslinking Phl p 7 with 101.1F10 represents a promising area for future work, as it could provide conclusive evidence that only one Phl p 7 is needed for crosslinking.
Clearly the claim that Phl p 7 functions as a superantigen with 101.1F10 is merited based on the literal definition of its binding site outside the hypervariable region of the antibody. Closer examination of the co-structure reveals that this interaction is mediated by only 3 residues in the framework region of 101.1F10. The resulting modest binding area of 300 Å2 stands in stark contrast with other superantigens like protein A binding to the constant region of an antibody (1DEE, [49]), where the entire protein interacts solely with framework residues to produce a binding area of ~600 Å2 [50]. In another superantigen protein L, there is more than 700 Å2 of interactions with framework residues (1HEZ [51]). Thus, while 101.1F10 can be classified as a superantigen, its mode of binding coupled with its dual reactivity against Phl p 7 is significantly different from previous precedent, and opens up new avenues of research with regards to other non-standard antibody interactions.
The study demonstrated that a non-dimeric antigen can crosslink with a single IgE molecule, although the stoichometry is not absolutely certain. This challenges a number of existing paradigms about antigens in general and suggests this type of interaction could be possible in IgG crosslinking as well. The value of allergen-antibody complex structures is apparent in that it is unlikely that other epitope mapping techniques would have noticed so few residues functioning as a superantigen, or even suggested two epitopes could be recognized by one antibody. Visualization of the allergen-antibody complex at atomic resolution was required to make these assertions.
Challenges and Future Directions
The detailed information content of these complexes makes them highly desirable, but obtaining crystal structures of antibody-allergen complexes is fraught with challenges. A primary problem is producing enough antibody. Many efforts turn to recombinant expression after sequencing of the specific desired clones from patients [52, 53]. Crystallography often requires milligram quantities of both the allergen and antibody to form complexes. Additionally, the design of the construct is crucial, as full-length IgG or IgE have a high degree of flexibility between the Fc and Fab region, which increases the structural heterogeneity and represents a significant detriment for sample crystallization. For this reason, the majority of structural biologist have opted for crystallization of antibody fragments using Fab or Fv regions [54] obtained from either bacterial or mammalian expression systems.
To this end, we have explored and researched a number of prokaryotic expression options. Escherichia coli expression systems can be used for production of recombinant ScFv and Fab [55]. ScFv constructs contain the VH and VL chains linked to together by an extended linker (usually about 18 residues) that allow for proper pairing of the immunoglobulin domains [56]. Shorter linkers result in formation of diabodies that form dimers with the VH of one monomer interacting with the VL of the other, generating a bivalent antibody [57, 58]. In addition, Fab constructs can be formed by co-expression of the VHCH1 and VLCL1 chains either using separate vectors or dual expression vectors [59, 55]. Due to the reducing environment, expression in the cytoplasmic space in E. coli often results in formation of insoluble inclusion bodies, requiring inefficient refolding procedures. Engineered E. coli strains with mutations in the glutathione and thioredoxin reductase (trxB gor mutants) in combination with chaperone co-expression can improve the yield of Fab to the mg/L range [60]. Stability and yield can also be improved by introducing a disulfide bond-stabilized Fv (ds-Fv), in which the unstable VL and VH heterodimers are stabilized by forming inter-domain disulfide bonds [61]. This method has been combined with grafting, where the CDRs of one antibody are spliced into the structured framework of another antibody, to successfully crystalize and solve several antibody-antigen complexes, including one against vascular endothelial growth factor (VEGF) [62]. Expression in the periplasmic space in E. coli can also facilitate formation of the correct disulfide bond pattern and production of soluble protein [59]. Finally, use of a Gram-positive bacterial expression organism in place of the traditional gram-negative E. coli-based systems can facilitate the production of recombinant antibodies [59, 55]. Here, the lack of an outer membrane allows the direct secretion of recombinant proteins into the growth medium, which can reduce purification steps. In summary, there exists a vast literature on improving prokaryotic expression of antibodies; this is a testament to the importance of this process as an obstacle for structural studies, and the recent progress that has been made towards overcoming the challenges discussed above.
Despite the advancement in bacterial antibody fragment expression, it is interesting to note that all but one (Bos d 5) of the crystal structures of allergen-antibody complexes have used antibody Fab or Fab′ fragments generated from mammalian expression systems (Table 1). This is consistent with our experience that obtaining soluble functional antibody constructs from patient-derived sequences expressed in bacteria remains difficult. To elaborate, many phage display technologies and animal systems can select for proteins with optimized characteristics. But expressing a particular cloned sequence against a specific epitope has been problematic. Even when we have successfully obtained a protein with favorable biophysical properties through grafting a desired CDR region onto a known soluble ScFv, the resulting product did not display strong affinity for its corresponding antigen (unpublished results). This may be a product of protein instability, consistent with the camelid paper above showing that the loops can influence stability [26].
Table 1.
Crystal structures of allergen-antibody complexes
| Antigen | Allergen expression system | Antibody | Antibody expression system | Co-purified by SEC | Crystallization Conditions | PDB code |
|---|---|---|---|---|---|---|
| Api m 2 | Insect cells (High Five)d | Fab; IgG1 | Mus musculus hybridoma cells | Yes | 0.1M CHES pH 9.5, 0.2M NaCl, 10% PEG 8000 | 2j88 [12] |
| Bet v 1 | E. coli | Fab′; IgG1 | Mus musculus hybridoma cells | Yes | 0.1M sodium citrate pH 4.0, 12% PEG 6000, 0.01% sodium azide | 1FSK [14] |
| Bla g 2 | Pichia pastoris | Fab′, 7C11, IgG1 | Mus musculus hybridoma cells | Yes | 0.1M sodium citrate pH 5.6, 16% PEG 10,000 | 2NR6 [18] |
| Bla g 2 | Pichia pastoris | Fab′, 4C3, IgG1 | Mus musculus hybridoma cells | Yes | Buffer unknown, 20% PEG 8000, 8% ethylene glycol, 0.2mM CdCl, 5mM DTT | 3LIZ [22] |
| Bos d 5 | E. coli | Fab | E. coli RV308 | No | 0.1M BTP pH 5.5, 14% PEG 3350 | 2R56 [13] |
| Der f 1 | D. farinae mite culture | Fab; IgG1 | Mus musculus hybridoma cells | Yes | 0.1M HEPES pH 7.0,18% PEG 12,000 | 3RVV [17] |
| Der p 1 | D. pteronyssinus mite culture | Fab; IgG1 | Mus musculus hybridoma cells | Yes | 1)0.1M sodium cacodylate pH 6.0, 15% PEG 4000 2)0.1M HEPES pH 7.0, 10% PEG4000, 0.05M GlyGlyGly |
1)3RVW 2)3RVX [17] |
| Der p 1 | D. pteronyssinus mite culture | Fab; IgG1 | Mus musculus hybridoma cells | yes | 0.1M MES pH 7.0, 10% PEG 6000, 5% MBP | 4PP1 [16] |
| Der p 1 | D. pteronyssinus mite culture | Fab; IgG1 | Mus musculus hybridoma cells | Yes | 0.1M sodium citrate pH 5.5, 15% PEG 6000 | 4PP2 [16] |
| Fel d 1 | CHO | Fab; IgG4f | CHOc | Yes | 0.1M sodium cacodylate pH 6.5, 0.2M calcium acetate, 40% PEG 300 | 5VYF [25] |
| Gal d 4 (lysozyme) | unknown | VH domain; Phage displayed | E. coli BL21 Goldb | Yes | 1) 0.1M sodium citrate pH 5.4, 28% PEG 1500 2) 0.3M sodium citrate pH 5.5, 16% PEG 3350 |
1)4PGJ 2)4U3X [26] |
| Phl p 2 | E. coli BL21e | Fab; IgE variable, IgG constant | CHO-K1 | Yes | 0.2M NaFormate, 20% PEG 3350 | 2VXQ [15] |
| Phl p 7 | Ecoli BL21 star DE3a | Fab; IgG1 | FreeStyle 293Fa | unknown | 18 C, 0.1 M MES pH 6.5, 15% PEG 6000, 10% MPD | 5OTJ [27] |
Life Technologies
Stratagene
Regeneron
ThermoFisher
Purchased from BIOMAY
S228P, to promote disulfide bond stabilization
The expression systems that have been most successful for Fabs used in allergen-antibody complexes have been mouse hybridoma and Chinese hamster ovarian (CHO) cell lines, Table 1. (This analysis disregards the numerous lysozyme antibody structures.) Immortalized B lymphocytes generated by hybridoma technology has been used to produce monoclonal antibodies since the 1970s [63]. These hybridoma cells can continuously generate pure and functional monoclonal antibodies in culture medium, and were used in the production of 9 of the allergen complex crystal structures in Table 1. Currently, the most common production system for monoclonal antibodies is recombinant expression in mammalian cells including CHO and Human Embryonic Kidney (HEK) 293 cells. These systems were used to solve antibody complex structures with Phl p 7, Fel d 1, and Phl p 2 (Table 1). HEK 293 are commonly used due to high transient transfection efficiency and human-like glycosylation. Antibody production in these cells can reach 0.1~0.3 g/L in batch and 0.1~0.6 g/L in fed-batch processes [64]. That said, CHO cells remain the predominate host cell line used for monoclonal antibody production, with yields of ~1 g/L in batch and 1~13 g/L in fed-batch processes [64]. This is enhanced by commercially available ExpiCHO-S expression systems containing improved media to help reach higher cell densities and potentially increased protein yields [65, 66]. Once antibodies have been obtained using these systems, bead immobilized papain can be used to remove the Fc domain, generating the Fab fragment for crystallization.
The end product of these steps is the generation of purified antibody/antigen complexes with biochemical and biophysical properties amenable for structural studies. However, even at this stage there is no guarantee that complex crystal structures will be obtained. Crystallization often requires screening of thousands of conditions as well as many modifications of the protein construct to obtain diffraction quality crystals. Despite the typically tight nM affinity of antibodies to antigens and co-purification of the complex using gel filtration, on more than one occasion, crystals we obtained from a purified complex contained only antibody fragment (unpublished and Mueller et al. 2014) [58]. Another approach to obtaining complex structures may be cryo-electron microscopy (cryo-EM) which requires significantly less material than crystallography [67]. However, difficulties remain in terms of sample preparation, particularly the identification of appropriate freezing conditions, and data analysis. These difficulties are counterbalanced by recent advancements in cryo-EM that suggest the feasibility of solving molecular structures to 3.2 Å resolution on proteins as small as 64 kDa [68]. While this resolution is still unable to match those obtainable from crystal structures, they can provide an understanding of specific antibody-allergen interactions while avoiding some of the difficulties associated with crystallization. Given these advancements, obtaining near-atomic resolution structures of complexes between Fab fragments (50kDa) to larger allergens is now feasible. Additionally, structures of complete antibody (150–190 kDa) complexes may also be possible, though the flexibility between the Fc and Fab regions present additional challenges for data processing and image analysis. Nonetheless, the rapid pace and development of cryo-EM technologies yields optimism that this technique may be useful for gleaning more antibody epitope information than is currently possible with crystallography.
Conclusions
Recent complex structures of allergens with antibodies are providing detailed epitope information. The data have been useful in describing the epitope of an antibody used in a phase I clinical trial, understanding antibody recognition of complex molecular surfaces, and redefining the number of epitopes that can be recognized by one antibody. Clearly more of this epitope data could be useful in a number of future applications and new technologies may promote this kind of research.
Acknowledgements
Research reported in this publication was supported in part by the Intramural Research Program of the National Institute of Environmental Health Sciences, National Institutes of Health (Research Project nos. Z01- ES102906–01, G.A.M. and ZIA- ES102645, L.C.P.), and in part by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under Award Number R01AI077653 (to A.P., contact PI). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
References
- 1.Pomés A, Chruszcz M, Gustchina A, Minor W, Mueller GA, Pedersen LC et al. 100 Years later: Celebrating the contributions of x-ray crystallography to allergy and clinical immunology. J Allergy Clin Immun 2015;136(1):29–U87. doi: 10.1016/j.jaci.2015.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Comprehensive review of many allergen structures
- 2.Mueller GA. Contributions and Future Directions for Structural Biology in the Study of Allergens. Int Arch Allergy Imm 2017;174(2):57–66. doi: 10.1159/000481078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chapman MD, Wunschmann S, Pomés A. Proteases as Th2 adjuvants. Curr Allergy Asthm R 2007;7(5):363–7. [DOI] [PubMed] [Google Scholar]
- 4.Karp CL. Guilt by intimate association: what makes an allergen an allergen? J Allergy Clin Immunol 2010;125(5):955–60; quiz 61–2. doi: 10.1016/j.jaci.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thomas WR. Innate affairs of allergens. Clinical and Experimental Allergy 2013;43(2):152–63. doi: 10.1111/j.1365-2222.2012.04059.x. [DOI] [PubMed] [Google Scholar]
- 6.Alessandri C, Ferrara R, Bernardi ML, Zennaro D, Tuppo L, Giangrieco I et al. Diagnosing allergic sensitizations in the third millennium: why clinicians should know allergen molecule structures. Clin Transl Allergy 2017;7:21. doi: 10.1186/s13601-017-0158-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mueller GA, Pedersen LC, Glesner J, Edwards LL, Zakzuk J, London RE et al. Analysis of glutathione S-transferase allergen cross-reactivity in a North American population: Relevance for molecular diagnosis. J Allergy Clin Immunol 2015. doi: 10.1016/j.jaci.2015.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ghosh D, Mueller GA, Schramm G, Edwards LL, Petersen A, London RE et al. Primary Identification, Biochemical Characterization, and Immunologic Properties of the Allergenic Pollen Cyclophilin Cat r 1. Journal of Biological Chemistry 2014;289(31):21374–85. doi:Doi 10.1074/Jbc.M114.559971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tscheppe A, Breiteneder H. Recombinant Allergens in Structural Biology, Diagnosis, and Immunotherapy. Int Arch Allergy Imm 2017;172(4):187–202. doi: 10.1159/000464104. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Comprehensive review of the utitily of recombinant allergens.
- 10.Blake CC, Koenig DF, Mair GA, North A, Phillips DC, Sarma VR. Sturcture of hen egg-white lysozyme. A three-dimensional Fourier synthesis at 2 Angstrom resolution. Nature 1965;206(4986):757–61. [DOI] [PubMed] [Google Scholar]
- 11.Padlan EA, Silverton EW, Sheriff S, Cohen GH, Smith-Gill SJ, Davies DR. Structure of an antibody-antigen complex: crystal structure of the HyHEL-10 Fab-lysozyme complex. Proc Natl Acad Sci U S A 1989;86(15):5938–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Padavattan S, Schirmer T, Schmidt M, Akdis C, Valenta R, Mittermann I et al. Identification of a B-cell epitope of hyaluronidase, a major bee venom allergen, from its crystal structure in complex with a specific Fab. J Mol Biol 2007;368(3):742–52. doi: 10.1016/j.jmb.2007.02.036. [DOI] [PubMed] [Google Scholar]
- 13.Niemi M, Jylha S, Laukkanen ML, Soderlund H, Makinen-Kiljunen S, Kallio JM et al. Molecular interactions between a recombinant IgE antibody and the beta-lactoglobulin allergen. Structure 2007;15(11):1413–21. doi: 10.1016/j.str.2007.09.012. [DOI] [PubMed] [Google Scholar]
- 14.Mirza O, Henriksen A, Ipsen H, Larsen JN, Wissenbach M, Spangfort MD et al. Dominant epitopes and allergic cross-reactivity: Complex formation between a Fab fragment of a monoclonal murine IgG antibody and the major allergen from birch pollen Bet v 1. J Immunol 2000;165(1):331–8. [DOI] [PubMed] [Google Scholar]
- 15.Padavattan S, Flicker S, Schirmer T, Madritsch C, Randow S, Reese G et al. High-Affinity IgE Recognition of a Conformational Epitope of the Major Respiratory Allergen Phl p 2 As Revealed by X-Ray Crystallography. J Immunol 2009;182(4):2141–51. doi: 10.4049/jimmunol.0803018. [DOI] [PubMed] [Google Scholar]
- 16.Osinski T, Pomés A, Majorek KA, Glesner J, Offermann LR, Vailes LD et al. Structural Analysis of Der p 1-Antibody Complexes and Comparison with Complexes of Proteins or Peptides with Monoclonal Antibodies. J Immunol 2015;195(1):307–16. doi: 10.4049/jimmunol.1402199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chruszcz M, Pomés A, Glesner J, Vailes LD, Osinski T, Porebski PJ et al. Molecular determinants for antibody binding on group 1 house dust mite allergens. J Biol Chem 2012;287(10):7388–98. doi: 10.1074/jbc.M111.311159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li M, Gustchina A, Alexandratos J, Wlodawer A, Wunschmann S, Kepley CL et al. Crystal structure of a dimerized cockroach allergen Bla g 2 complexed with a monoclonal antibody. Journal of Biological Chemistry 2008;283(33):22806–14. doi:Doi 10.1074/Jbc.M800937200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li M, Gustchina A, Glesner J, Wunschmann S, Vailes LD, Chapman MD et al. Carbohydrates contribute to the interactions between cockroach allergen Bla g 2 and a monoclonal antibody. J Immunol 2011;186(1):333–40. doi: 10.4049/jimmunol.1002318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Glesner J, Vailes LD, Schlachter C, Mank N, Minor W, Osinski T et al. Antigenic Determinants of Der p 1: Specificity and Cross-Reactivity Associated with IgE Antibody Recognition. J Immunol 2017;198(3):1334–44. doi: 10.4049/jimmunol.1600072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Woodfolk JA, Glesner J, Wright PW, Kepley CL, Li M, Himly M et al. Antigenic Determinants of the Bilobal Cockroach Allergen Bla g 2. J Biol Chem 2016;291(5):2288–301. doi: 10.1074/jbc.M115.702324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Glesner J, Wunschmann S, Li M, Gustchina A, Wlodawer A, Himly M et al. Mechanisms of Allergen-Antibody Interaction of Cockroach Allergen Bla g 2 with Monoclonal Antibodies That Inhibit IgE Antibody Binding. Plos One 2011;6(7). doi:ARTN e22223 DOI 10.1371/journal.pone.0022223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pomés A, Chruszcz M, Gustchina A, Wlodawer A. Interfaces between allergen structure and diagnosis: know your epitopes. Curr Allergy Asthma Rep 2015;15(8):506. doi: 10.1007/s11882-014-0506-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dall’Antonia F, Pavkov-Keller T, Zangger K, Keller W. Structure of allergens and structure based epitope predictions. Methods 2014;66(1):3–21. doi: 10.1016/j.ymeth.2013.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Excellent review of empirical and computational epitope mapping strategies
- 25.Orengo JM, Radin AR, Kamat V, Badithe A, Ben LH, Bennett BL et al. Treating cat allergy with monoclonal IgG antibodies that bind allergen and prevent IgE engagement. Nat Commun 2018;9(1):1421. doi: 10.1038/s41467-018-03636-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rouet R, Dudgeon K, Christie M, Langley D, Christ D. Fully Human VH Single Domains That Rival the Stability and Cleft Recognition of Camelid Antibodies. J Biol Chem 2015;290(19):11905–17. doi: 10.1074/jbc.M114.614842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mitropoulou AN, Bowen H, Dodev TS, Davies AM, Bax HJ, Beavil RL et al. Structure of a patient-derived antibody in complex with allergen reveals simultaneous conventional and superantigen-like recognition. Proc Natl Acad Sci U S A 2018;115(37):E8707–E16. doi: 10.1073/pnas.1806840115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wachholz PA, Durham SR. Mechanisms of immunotherapy: IgG revisited. Curr Opin Allergy Clin Immunol 2004;4(4):313–8. [DOI] [PubMed] [Google Scholar]
- 29.Shamji MH, Ljorring C, Francis JN, Calderon MA, Larche M, Kimber I et al. Functional rather than immunoreactive levels of IgG4 correlate closely with clinical response to grass pollen immunotherapy. Allergy 2012;67(2):217–26. doi: 10.1111/j.1398-9995.2011.02745.x. [DOI] [PubMed] [Google Scholar]
- 30.Aalberse RC, Stapel SO, Schuurman J, Rispens T. Immunoglobulin G4: an odd antibody. Clinical and Experimental Allergy 2009;39(4):469–77. doi: 10.1111/j.1365-2222.2009.03207.x. [DOI] [PubMed] [Google Scholar]
- 31.James LK, Shamji MH, Walker SM, Wilson DR, Wachholz PA, Francis JN et al. Long-term tolerance after allergen immunotherapy is accompanied by selective persistence of blocking antibodies. J Allergy Clin Immunol 2011;127(2):509–16 e1–5. doi: 10.1016/j.jaci.2010.12.1080. [DOI] [PubMed] [Google Scholar]
- 32.Bachmann MF, Kundig TM. Allergen-specific immunotherapy: is it vaccination against toxins after all? Allergy 2017;72(1):13–23. doi: 10.1111/all.12890. [DOI] [PubMed] [Google Scholar]
- 33.Subbarayal B, Schiller D, Mobs C, de Jong NW, Ebner C, Reider N et al. Kinetics, cross-reactivity, and specificity of Bet v 1-specific IgG4 antibodies induced by immunotherapy with birch pollen. Allergy 2013;68(11):1377–86. doi: 10.1111/all.12236. [DOI] [PubMed] [Google Scholar]
- 34.Varga EM, Kausar F, Aberer W, Zach M, Eber E, Durham SR et al. Tolerant beekeepers display venom-specific functional IgG4 antibodies in the absence of specific IgE. J Allergy Clin Immunol 2013;131(5):1419–21. doi: 10.1016/j.jaci.2012.08.037. [DOI] [PubMed] [Google Scholar]
- 35.Kaiser L, Gronlund H, Sandalova T, Ljunggren HG, Achour A, Schneider G et al. Three-dimensional structure of Fel d 1, the major allergen in cat. Int Arch Allergy Imm 2003;132(1):25–6. doi:Doi 10.1159/000073261. [DOI] [PubMed] [Google Scholar]
- 36.Willison LN, Zhang Q, Su MN, Teuber SS, Sathe SK, Roux KH. Conformational epitope mapping of Pru du 6, a major allergen from almond nut. Mol Immunol 2013;55(3–4):253–63. doi: 10.1016/j.molimm.2013.02.004. [DOI] [PubMed] [Google Scholar]
- 37.Guan XY, Noble KA, Tao YQ, Roux KH, Sathe SK, Young NL et al. Epitope mapping of 7S cashew antigen in complex with antibody by solution-phase H/D exchange monitored by FT-ICR mass spectrometry. J Mass Spectrom 2015;50(6):812–9. doi: 10.1002/jms.3589. [DOI] [PubMed] [Google Scholar]
- 38.Brier S, Le Mignon M, Jain K, Lebrun C, Peurois F, Kellenberger C et al. Characterization of epitope specificities of reference antibodies used for the quantification of the birch pollen allergen Bet v 1. Allergy 2018;73(5):1032–40. doi: 10.1111/all.13364. [DOI] [PubMed] [Google Scholar]
- 39.Williams DC Jr., Benjamin DC, Poljak RJ, Rule GS. Global changes in amide hydrogen exchange rates for a protein antigen in complex with three different antibodies. J Mol Biol 1996;257(4):866–76. [DOI] [PubMed] [Google Scholar]
- 40.Mueller GA, Smith AM, Chapman MD, Rule GS, Benjamin DC. Hydrogen exchange nuclear magnetic resonance spectroscopy mapping of antibody epitopes on the house dust mite allergen Der p 2. J Biol Chem 2001;276(12):9359–65. doi: 10.1074/jbc.M010812200. [DOI] [PubMed] [Google Scholar]
- 41.Flicker S, Steinberger P, Norderhaug L, Sperr WR, Majlesi Y, Valent P et al. Conversion of grass pollen allergen-specific human IgE into a protective IgG(1) antibody. Eur J Immunol 2002;32(8):2156–62. doi:Doi . [DOI] [PubMed] [Google Scholar]
- 42.Patil SU, Ogunniyi AO, Calatroni A, Tadigotla VR, Ruiter B, Ma A et al. Peanut oral immunotherapy transiently expands circulating Ara h 2-specific B cells with a homologous repertoire in unrelated subjects. J Allergy Clin Immun 2015;136(1):125–U253. doi: 10.1016/j.jaci.2015.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hamers-Casterman C, Atarhouch T, Muyldermans S, Robinson G, Hamers C, Songa EB et al. Naturally occurring antibodies devoid of light chains. Nature 1993;363(6428):446–8. doi: 10.1038/363446a0. [DOI] [PubMed] [Google Scholar]
- 44.Muyldermans S Nanobodies: natural single-domain antibodies. Annu Rev Biochem 2013;82:775–97. doi: 10.1146/annurev-biochem-063011-092449. [DOI] [PubMed] [Google Scholar]
- 45.Riechmann L, Muyldermans S. Single domain antibodies: comparison of camel VH and camelised human VH domains. J Immunol Methods 1999;231(1–2):25–38. doi:Doi 10.1016/S0022-1759(99)00138-6. [DOI] [PubMed] [Google Scholar]
- 46.De Genst E, Silence K, Decanniere K, Conrath K, Loris R, Kinne J et al. Molecular basis for the preferential cleft recognition by dromedary heavy-chain antibodies. Proc Natl Acad Sci U S A 2006;103(12):4586–91. doi: 10.1073/pnas.0505379103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.De Genst E, Silence K, Ghahroudi MA, Decanniere K, Loris R, Kinne J et al. Strong in vivo maturation compensates for structurally restricted H3 loops in antibody repertoires. J Biol Chem 2005;280(14):14114–21. doi: 10.1074/jbc.M413011200. [DOI] [PubMed] [Google Scholar]
- 48.Stanfield RL, Dooley H, Flajnik MF, Wilson IA. Crystal structure of a shark single-domain antibody V region in complex with lysozyme. Science 2004;305(5691):1770–3. doi: 10.1126/science.1101148. [DOI] [PubMed] [Google Scholar]
- 49.Graille M, Stura EA, Corper AL, Sutton BJ, Taussig MJ, Charbonnier JB et al. Crystal structure of a Staphylococcus aureus protein A domain complexed with the Fab fragment of a human IgM antibody: structural basis for recognition of B-cell receptors and superantigen activity. Proc Natl Acad Sci U S A 2000;97(10):5399–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. Journal of molecular biology 2007;372(3):774–97. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 51.Graille M, Stura EA, Housden NG, Beckingham JA, Bottomley SP, Beale D et al. Complex between Peptostreptococcus magnus protein L and a human antibody reveals structural convergence in the interaction modes of Fab binding proteins. Structure 2001;9(8):679–87. [DOI] [PubMed] [Google Scholar]
- 52.James LK. The Cloning and Expression of Human Monoclonal Antibodies: Implications for Allergen Immunotherapy. Curr Allergy Asthma Rep 2016;16(2):15. doi: 10.1007/s11882-015-0588-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Robinson WH. Sequencing the functional antibody repertoire--diagnostic and therapeutic discovery. Nat Rev Rheumatol 2015;11(3):171–82. doi: 10.1038/nrrheum.2014.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kovari LC, Momany C, Rossmann MG. The use of antibody fragments for crystallization and structure determinations. Structure 1995;3(12):1291–3. [DOI] [PubMed] [Google Scholar]
- 55.Gupta SK, Shukla P. Microbial platform technology for recombinant antibody fragment production: A review. Crit Rev Microbiol 2017;43(1):31–42. doi: 10.3109/1040841X.2016.1150959. [DOI] [PubMed] [Google Scholar]
- 56.Long NE, Sullivan BJ, Ding HM, Doll S, Ryan MA, Hitchcock CL et al. Linker engineering in anti-TAG-72 antibody fragments optimizes biophysical properties, serum half-life, and high-specificity tumor imaging. Journal of Biological Chemistry 2018;293(23):9030–40. doi: 10.1074/jbc.RA118.002538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Perisic O, Webb PA, Holliger P, Winter G, Williams RL. Crystal structure of a diabody, a bivalent antibody fragment. Structure 1994;2(12):1217–26. [DOI] [PubMed] [Google Scholar]
- 58.Mueller GA, Ankney JA, Glesner J, Khurana T, Edwards LL, Pedersen LC et al. Characterization of an anti-Bla g 1 scFv: epitope mapping and cross-reactivity. Mol Immunol 2014;59(2):200–7. doi: 10.1016/j.molimm.2014.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Frenzel A, Hust M, Schirrmann T. Expression of recombinant antibodies. Front Immunol 2013;4:217. doi: 10.3389/fimmu.2013.00217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Levy R, Weiss R, Chen G, Iverson BL, Georgiou G. Production of correctly folded Fab antibody fragment in the cytoplasm of Escherichia coli trxB gor mutants via the coexpression of molecular chaperones. Protein Expr Purif 2001;23(2):338–47. doi: 10.1006/prep.2001.1520. [DOI] [PubMed] [Google Scholar]
- 61.Schmiedl A, Breitling F, Winter CH, Queitsch I, Dubel S. Effects of unpaired cysteines on yield, solubility and activity of different recombinant antibody constructs expressed in E. coli. J Immunol Methods 2000;242(1–2):101–14. [DOI] [PubMed] [Google Scholar]
- 62.Yu CM, Peng HP, Chen IC, Lee YC, Chen JB, Tsai KC et al. Rationalization and design of the complementarity determining region sequences in an antibody-antigen recognition interface. PLoS One 2012;7(3):e33340. doi: 10.1371/journal.pone.0033340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kohler G, Milstein C. Continuous cultures of fused cells secreting antibody of predefined specificity. Nature 1975;256(5517):495–7. [DOI] [PubMed] [Google Scholar]
- 64.Kunert R, Reinhart D. Advances in recombinant antibody manufacturing. Appl Microbiol Biotechnol 2016;100(8):3451–61. doi: 10.1007/s00253-016-7388-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jain NK, Barkowski-Clark S, Altman R, Johnson K, Sun F, Zmuda J et al. A high density CHO-S transient transfection system: Comparison of ExpiCHO and Expi293. Protein Expr Purif 2017;134:38–46. doi: 10.1016/j.pep.2017.03.018. [DOI] [PubMed] [Google Scholar]
- 66.Dangi AK, Sinha R, Dwivedi S, Gupta SK, Shukla P. Cell Line Techniques and Gene Editing Tools for Antibody Production: A Review. Front Pharmacol 2018;9:630. doi: 10.3389/fphar.2018.00630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Merk A, Bartesaghi A, Banerjee S, Falconieri V, Rao P, Davis MI et al. Breaking Cryo-EM Resolution Barriers to Facilitate Drug Discovery. Cell 2016;165(7):1698–707. doi: 10.1016/j.cell.2016.05.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Khoshouei M, Radjainia M, Baumeister W, Danev R. Cryo-EM structure of haemoglobin at 3.2 A determined with the Volta phase plate. Nat Commun 2017;8:16099. doi: 10.1038/ncomms16099. [DOI] [PMC free article] [PubMed] [Google Scholar]