Abstract
Human innate immune lectins that recognize microbial glycans can conduct microbial surveillance and thereby help prevent infection. Structural analysis of soluble lectins has provided invaluable insight into how these proteins recognize their cognate carbohydrate ligands and how this recognition gives rise to biological function. In this opinion, we cover the structural features of lectins that allow them to mediate microbial recognition, highlighting examples from the collectin, Reg protein, galectin, pentraxin, ficolin and intelectin families. These analyses reveal how some lectins (e.g., human intelectin-1) can recognize glycan epitopes that are remarkably diverse, yet still differentiate between mammalian and microbial glycans. We additionally discuss strategies to identify lectins that recognize microbial glycans and highlight tools that facilitate these discovery efforts.
INTRODUCTION
The health of humans and other mammals is influenced by their microbial residents. A host must therefore distinguish between microorganisms that are benign (commensal), beneficial (mutualist/symbiont), or pathogenic to tolerate or resist their colonization. The mechanisms involved in surveillance of bacteria, fungi, viruses, and parasites have not been fully delineated. On the host side of the dialog, mounting evidence supports important roles for lectins [1,2]. Some host lectins recognize distinct features within the carbohydrate coats of select populations of microbes. As a consequence, lectins have been implicated in the regulation of microbial colonization and in protection against infection. Seminal research on the acute response to bacterial infection led to the identification of secreted factors that include C-reactive protein (CRP) and mannose-binding lectin (MBL) [1,3]. Both CRP and MBL can recognize carbohydrate antigens on the surface of pathogens, including Streptococcus pneumoniae and Staphylococcus aureus and then promote complement-mediated opsonization and cell killing [4]. Since these initial observations, other lectins have been implicated in microbial recognition. Like MBL some of these proteins are C-type lectins, while others are members of the ficolin, pentraxin, galectin, or intelectin families. Many of the lectins that function in microbial surveillance are present on the surface of immune cells such as dendritic cells and macrophages, but many are also soluble proteins. While the former has been reviewed extensively elsewhere [5,6], this review focuses on recent advances in our understanding of soluble lectins that recognize microbial glycans. Here, we highlight the structural features of soluble lectins in the context of biological function. Additionally, we delineate strategies to facilitate the identification of novel microbe-binding lectins and outline the experimental challenges.
Tools for the discovery of lectins that bind microbial glycans
The cell surface glycans of microorganisms perform multiple functions. They help maintain cellular osmolarity, protect the organism from dessication, transduce signals, and mediate adhesion of the organism to cells or surfaces in its environment [7]. For a host, this carbohydrate coat could represent a cellular ID, an identification system used to discriminate between organisms. Even within a single species of bacteria, individual strains/serotypes can be distinguished by differences in the composition and structure of surface-exposed carbohydrates [7]. Thus, host lectins that recognize surface glycans may function as sentinels for detecting and identifying organisms seeking to establish residency.
The structure, ligand specificity, recognition mechanism, and biological function of a lectin are not readily determined from its primary sequence alone. Tools such as glycan microarrays, which display hundreds of structurally defined glycans have thus been indispensable in characterizing the carbohydrate binding profiles of lectins [8,9••]. Individual protein–carbohydrate interactions can be weak, thus surface arraying of glycans is valuable as lectins often rely on multivalent binding in physiological settings [10]. Indeed, glycan arrays can mimic the cell’s dense coat of glycans [8,11]. Various array platforms have been developed that display glycans at different densities [8], but the most widely used arrays present synthetic glycans derived from known mammalian glycan sequences. In 2014, a glycan microarray was disclosed whose composition differed dramatically from those described previously [9••]. Specifically, this array was composed of more than 300 purified microbial glycans. The microbial glycan array spans a structural space that differs markedly from that of the mammalian arrays because microbial glycans are often composed of building blocks absent from mammals [12,13]. Results from glycan-binding studies also can inform lectin-ligand cocrystallization or modeling experiments to elucidate the molecular basis of their ligand selectivity.
Common structural elements of soluble lectins
Soluble lectins that can recognize microbial glycans or whole microbes share several features that facilitate their function (highlighted in Box 1). Lectins are typically classified by their carbohydrate recognition domain, or CRD [14]. Many CRDs possess protein-bound cations, especially calcium ions, which stabilize the CRD structure. Moreover, calcium ions present in the binding sites of C-type lectins, pentraxins, and intelectins facilitate carbohydrate recognition [5,15•,16••]. Many lectins interact with their carbohydrate ligands using a “lock-and-key” binding mode, that is, there is little change in lectin conformation upon carbohydrate binding. Divalent calcium ions or other cations may therefore organize and rigidify the CRD and its binding site. This pre-organization may minimize the entropic penalty paid upon carbohydrate complexation.
Box 1. Common Attributes of Soluble Lectins that Bind Microbial Glycans.
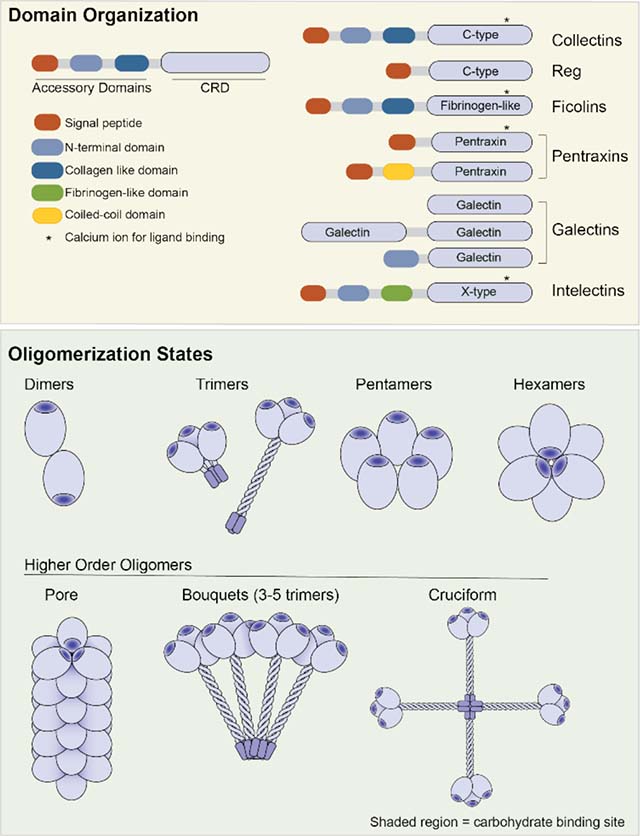
The type of carbohydrate recognition domain (CRD) of a lectin can be inferred from its primary sequence. Structural analysis of soluble innate immune lectins has revealed that in addition to a CRD many lectins possess accessory domains that facilitate lectin asseembly and/or protein-protein interactions between lectin monomers and other host immune proteins. Lectin oligomerization state can modulate the functional affinity (observed affinity) of the lectin for cells displaying glycan ligands. These features are outlined for the lectin classes whose constituents participate in microbial glycan recognition.
-
Carbohydrate Recognition Domain (CRD)
The CRD mediates recognition and binding to carbohydrate ligands, and lectins are classified by their CRD sequence. The sequence and structure of the CRD dictates the carbohydrate specificity of the lectin and the mechanism by which it engages its ligands. The collectin and Reg lectins belong to the large C-type lectin class. The collectins use binding site calcium ions to coordinate to the hydroxyl groups of their carbohdrate ligands but the Reg proteins do not. The ficolin CRD is a fibrinogen-like domain (FBG domain) that binds calcium, but the ion does not participate directly in saccharide binding. The jelly-roll fold of the pentraxin CRD requires calcium ions for ligand binding, while the galectin CRD, which also has a jelly-roll fold, does not. An alternative CRD fold that also uses calcium ions for glycan coordination is found in the X-type lectins or intelectins
-
Accessory protein-protein interaction domains
Domains outside the lectin CRD can facilitate oligomerization and/or mediate interactions with host immune response proteins. The collagen-like domain found in the collectin and ficolin classes of lectins is an example The collagen-like domain mediates homotrimerization and facilitates interactions with with mannose-binding lectin-associated serine proteases (MASPs) to initiate complement mediated cell-killing in response to microbial binding Cysteine-containing N-terminal oligomerization domains are involved in the stabilization of higher order oligomeric structures that are commonly observed among the lectin classes. Reg and pentraxin proteins possess a CRD that can mediate oligomerization in the absence of other domains.
-
Lectin Oligomerization
The innate immune lectins reviewed herein are oligomeric. As their carbohydrate binding sites are solvent exposed and shallow, the affinity of a single CRD for a monosaccharide ligand is often low (Kd ~ 10−3 M). Lectin oligomerization allows for multivalent binding therefore, the functional affinity, or avidity, of a lectin for its substrate is orders of magnitude tighter than the corresponding monovalent interactions. The oligomerization states of various lectins can differ greatly. The collectins. intelectins and ficolins are most commonly observed as trimers, while the galectins often form dimers and the pentraxins assemble into pentamers. Lectins often oligomerize further when binding to a cell. For example, Reg proteins can associate into hexameric cyclic structures that function as membrane pene-trating pores. Collectins and ficolins can form bouquets of trimers, which result in recruitment of complement. The collectin SP-D has been observed to form a cruciform-like structure, rather than the bouquet assembly generated from the other collectins.
A second attribute of many lectins that bind microbes is this presence of domains that mediate oligomerization. Soluble lectins often exist as multimers, and many undergo further oligomerization upon binding to surface-exposed glycans. For example, many collectins are multimers of trimers that undergo further oligomerization once they interact with microbial surfaces. Oligomerization facilitates multivalent binding and increases the functional affinity (apparent affinity or avidity) of a lectin for the microbe [17,18]. This oligomerization can have another consequence. Specifically, many lectins also have accessory structural domains often employed to effect a biological function. This domain structure is exemplified by the collectin and ficolin lectin classes; they possess collagen-like domains that facilitate oligomerization and can recruit mannose-binding lectin-associated serine proteases (MASPs) to initiate the lectin pathway of complement [1,19]. The following sections outline the relevant features of each lectin class and delineate what is known about lectin recognition of microbial glycans and the biological consequences.
Collectins
A number of soluble human collectins bind microbial glycans including the paradigmatic innate immune lectin, MBL, but also the surfactant proteins A and D (SP-A and SP-D), collectin liver 1 (CL-L1; collectin-10), and collectin kidney 1 (CL-K1; collectin-11) [1]. The best studied collectin, MBL, can interact with clinically important microbes including viruses, the bacterial pathogens Escherichia coli, group A Streptococci, and Staphylococcus aureus, fungal pathogens belonging to the Aspergillus and Candida genus, and trypanosomes belonging to Leishmania [20]. MBL deficiencies in humans are associated with increased susceptibility to chronic and recurring infections, implicating the lectin in protecting against pathogen invasion [1].
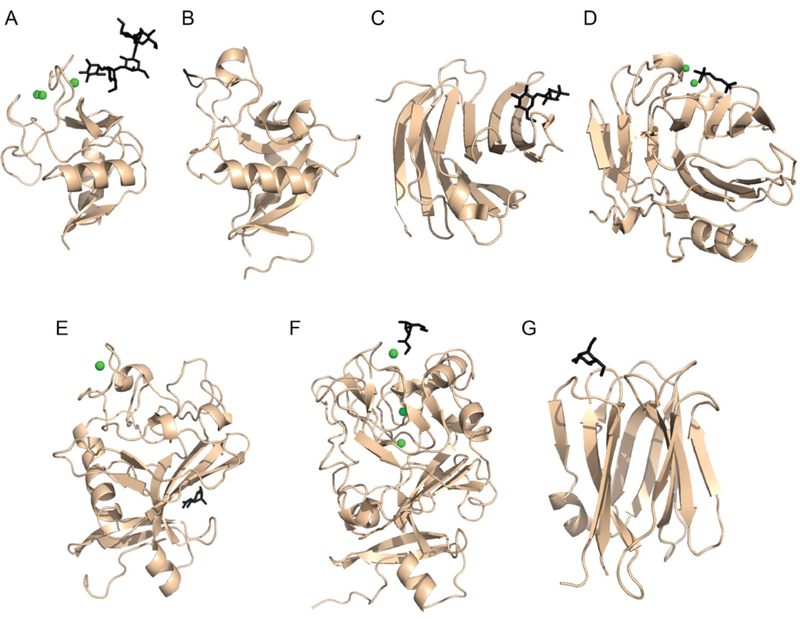
Collectins have a C-type lectin superfamily CRD, and C-type lectins typically bind their glycan ligands via direct coordination of carbohydrate ring vicinal hydroxyl groups to a protein-bound calcium ion [5,21]. The C-type lectin domain is a compact, globular CRD that can accommodate significant sequence variation; this variation can dictate the saccharide specificity of each lectin (Figure 1A) [5,22]. All human collectins contain a conserved glutamic acid-proline-asparagine (EPN) tripeptide motif that positions side chain carbonyl groups in the binding site to engage in calcium ion coordination and to interact with diequatorial vicinal ring hydroxyl groups of the glycan [22,23]. Saccharide residues with the requisite stereochemical arrangement of hydroxyl groups include D-mannose, L-fucose (L-Fuc), D-glucose (Glc), D-N-acetyl-mannosamine (ManNAc), and D-N-acetyl-glucosamine (GlcNAc) [21]. Most collectins recognize the terminal glycan residue. Still, some collectins can recognize more complex epitopes, as was observed in the structure of CL-K1 complexed with a Man(α1–2)Man disaccharide [24]. In this case, the canonical collectin binding site was occupied by the penultimate rather than terminal mannose residue while the terminal mannose residue engaged in hydrogen-bonding to the surface in the protein. The ability of CL-K1 to specifically interact with Man(α1–2)Man disaccharides expands the binding modes for collectin recognition [24].
Figure 1.
Structural representation of CRDs present in human soluble lectins. A) C-type lectin domain of rat MBL (PDB:2MSB) bound to oligosaccharide ligand [23]. B) C-type lectin domain of human RegIIIα (PDB:4MTH) with residues proposed to function in peptidoglycan binding highlighted in black [28••]. C) Beta-sandwich or jelly-roll fold from human galectin-3 (PDB:3ZSJ) bound to lactose [29]. D) Jelly-roll fold of human C-reactive protein bound to phosphorylcholine (PDB: 1B09) [30] E) Fibrinogen-like domain from human L-Ficolin bound to N-acetyl-glucosamine (PDB: 2J3U) [31]. F) Intelectin domain from human intelectin-1 bound to β-Galactofuranose (PDB: 4WMY) [16••]. G) Beta-prism fold from human ZG16p bound to mannose (PDB: 3VZF) [32]. Bound calcium ions are shown as green spheres and carbohydrate ligands are depicted in black.
Collectins are produced as single polypeptides that can undergo trimerization and eventually self-associate to form higher-order oligomers [21]. Moreover, collectins can form heterooligomers with other lectins [4,25], which could provide a means of targeting cells with higher specificity. Specifically, higher order mixed lectin oligomers would form only on cells displaying glycan ligands for multiple lectins. In this way, lectin oligomerization could influence both cell-binding avidity and specificity. In addition to influencing lectin cell binding, oligomerization is important for the recruitment of MASPs and activation of the lectin pathway of complement [4]. Collectins assemble through a cysteine-containing N-terminal oligomerization domain, a central collagen-like domain, and a coiled-coil neck region [1,4]. Functional collectins exist in macromolecular complexes that can vary in size, with the most commonly observed species of MBL containing three to five trimers (9–15 CRDs) [19] (Figure 1). Post-translational modifications stabilize the collagen-like domain, facilitate collectin trimerization, and modulate interactions with both host and microbial proteins.
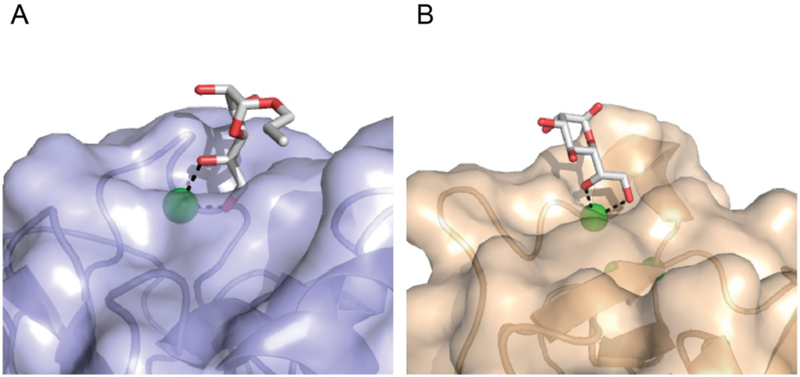
Amongst collectins, surfactant protein-D (SP-D) forms a unique macromolecular assembly and has multiple ligand binding modes [26]. Specifically, the oligomerization of most collectins results in a bouquet-like structure, while the assembly of SP-D resembles a cruciform (Figure 1). In the SP-D complex, the N-terminal oligomerization domains of four trimeric units are captured through disulfide bonds. The lectin has been reported to interact with multiple glycan ligands—including mannosides and lipopolysaccharides. A recent structural study of SP-D revealed the CRD could interact with L-glycero-D-manno-heptose (L-D-Hep) through two distinct binding modes: the binding site calcium ion can coordinate either to adjacent equatorial hydroxyl groups on the carbohydrate ring, the traditional recognition mechanism, or alternatively to the exocyclic 1,2-vicinal hydroxyl groups [27]. Though there is little data on the relative preferences of SP-D for different microbial glycans, these structural studies suggest that SP-D can recognize a broad suite of carbohydrates.
Reg Proteins
Reg proteins are secreted lectins that play roles in the inflammatory/immune response. Humans encode five Reg homologs; RegIα, RegIβ, RegIIIα or hepatocarcinoma intestine-pancreas/pancreatic associated protein (HIP/PAP), RegIIIγ, and RegIV [33]. Most of what is known about the Reg protein function comes from studies of murine RegIIIγ, the ortholog of human RegIIIα [34]. Colonization of gnotobiotic mice induces RegIIIγ expression in a myeloid differentiation primary-response protein 88 (MyD88)- and toll-like receptor 4 (TLR4)-dependent manner [34]. RegIIIγ expression can protect mice from Listeria monocytogenes infection and vancomycin-resistant enterococci, but the lectin also supports mucus layer integrity within the mammalian GI tract [34–36]. These functional roles have been rationalized by the finding that mouse RegIIIγ and human RegIIIα and RegIV are bactericidal.
Reg proteins, like the collectins, contain a C-type CRD. Unlike the collectins, however, Reg proteins do not have an obvious oligomerization or complement-recruiting domain. Other than a trypsin-sensitive propeptide immediately N-terminal to the CRD, Reg proteins consist almost entirely of a C-terminal C-type lectin domain [37]. A carbohydrate-binding function for Reg proteins was proposed initially because of the C-type lectin domain motif within their sequence and their ability to agglutinate some bacterial species. In contrast to canonical C-type lectins, Reg proteins lack the conserved side chains that typically mediate calcium ion and ligand binding (Figure 2B) [38]. Nuclear magnetic resonance (NMR) spectroscopy analysis of human RegIIIα and RegIV bound to glycoconjugate ligands, including purified soluble peptidoglycan (sPG), chitooligosaccharides (β1-4-GlcNAc polymers), and mannose, indicates that Reg proteins do not use a traditional C-type binding site nor do they require calcium ions for interaction with glycans [38,39].
Figure 2.
Lectin recognition of exocyclic 1,2-diols. A) Surface rendering of human intelectin-1 binding the exocyclic diol of allyl-β-D-Galf (PDB: 4WMY) [16]. The aromatic box in the ligand binding site, which is constrained by tryptophan 288 and tyrosine 297, is highlighted. B) Surface rendering of human SP-D binding the exocyclic diol of L-D-heptose (PDB: 2RIB) [27]. Calcium ions (green), oxygen atoms (red), and nitrogen atoms (blue) are highlighted. The protein-bound carbohydrate ligands are depicted in the stick representation.
As mentioned, Reg proteins can be bactericidal, and this function requires proteolysis of the N-terminal propeptide. This cleavage unleashes Reg toxicity toward Gram-positive bacteria [34]. Data from a combination of synthetic liposomes, cryo-electron microscopy, and protein X-ray crystallography suggest that mature RegIIIα can interact with acidic lipid assemblies, oligomerize into a hexameric ring, and thereby generate a pore [28••]. Pore formation depolarizes Gram-positive cell membranes and thereby results in cell killing. The hexameric arrangement situates all six putative peptidoglycan binding sites toward the center of the ring with the propeptide cleavage site near the edge. Given that Reg proteins bind carbohydrates, a model has emerged in which peptidoglycan binding precedes pore formation.
Despite advances in understanding Reg protein function and recognition, questions remain. Other peptidoglycan-binding proteins can cause cell killing by activation of two-component signaling pathways [40•] and whether Reg activates these pathways is not known. Alternatively, RegIIIα can form pores in the absence of carbohydrate ligands. Thus, whether specific or multiple ligands are relevant for Reg function is not yet clear. The ability of RegIIIα to bind multiple different ligands (i.e., carbohydrate and lipid) provides further impetus to understand Reg recognition of bacterial glycans and bacterial cells, as such knowledge could lead to new antimicrobial strategies for selective cell killing.
Galectins
Galectins are soluble lectins, whose microbe-binding abilities can have different consequences that depend on the identity of the galectin and the microbe [2,41,42]. Although galectins can recognize mammalian glycans and mediate cell signaling processes, this review focuses on extracellular galectins that interact with microbial glycoconjugates. In some cases, galectins can bridge microbe and host cells, thereby promoting microbial cell uptake and infection. Alternatively, galectins also can function as antimicrobial agents. Their interactions with microbes can promote immune cell signaling or even microbial cell death [43–45]. The complexity of galectin roles identified to date provides incentive to better understand this intriguing class of proteins.
Galectins share a beta-sandwich or jelly-roll fold containing CRD and a conserved affinity for β-galactose containing glycans (Figure 1C). They do not contain a signal peptide, though some are secreted as soluble, aglycosylated proteins [46]. Galectins are categorized into three groups according to their primary sequence and domain organization [46]. Prototype galectins (human Gal-1, −2, −7, −10, −13, and −14) are expressed as a single CRD and assemble into functional homodimers whose carbohydrate binding surfaces appear on opposite faces of the dimer. Chimera galectins (human Gal-3) possess a single C-terminal CRD connected to an N-terminal oligomerization domain, which facilitates trimer and pentamer formation. Tandem-repeat galectins (human Gal-4, −8, −9, and −12) are composed of two distinct CRDs within the same polypeptide chain. Ligand binding by galectins is mediated by a preorganized hydrogen bonding network and a conserved tryptophan residue that contributes a key CH-pi interaction with the α-face of β-D-Gal-containing ligands [29,47•]. NMR spectroscopy has revealed a second mechanism for galectin binding to larger polysaccharides, such as galactomannan and mannan, Human Gal-3 interacts with these ligands using the galectin face opposite that employed in traditional binding modes [48]. This alternative binding site can be occupied simultaneously with the canonical binding site, which provides a rational for binding of Gal-3 to fungal pathogens such as Candida species [43].
The roles for galectins in innate microbial immunity have been accumulating. After a report noting Gal-3 could bind lipopolysaccharide [49], galectin interactions with many microbial species including bacteria, viruses, fungi, protozoans and helminths has been described [2]. For example, multiple galectins were found to interact with ABO(H) blood group antigens when assayed at low protein concentrations (less than 0.5 μM) [44]. Galectins-3, −4, and −8 bind blood group B antigen, while galectins-4 and −8 interact with blood group A antigen. Furthermore, Gal-4 and −8 were shown to be directly bactericidal to E. coli expressing blood group B carbohydrate antigens. The C-terminal carbohydrate binding activity of Gal-4 and −8 was necessary and sufficient for this activity. The scope of galectin mediated antimicrobial activity was expanded when Gal-4 and −8 were shown to also kill bacteria that express the Gal-α(1–3)-Gal epitope, and galectin-8 was shown to inhibit viability of Gram-positive Streptococcus expressing host-like antigens [9••]. Similarly, Gal-3 can bind Heliobacter pylori LPS, and the lectin is bactericidal to this species [45]. Galectin-mediated cell killing is proposed to proceed via membrane disruption and permeabilization, although the molecular mechanisms remain unknown [50]. In the case of those galectins that bind blood group antigens, how the lectins refrain from collaterally damaging mammalian cells is unclear [50,51].
Pentraxins
Pentraxins are among the earliest identified human lectins to function in innate immunity [3,25]. This familyMembers includes the short pentraxins, such as C-reactive protein (CRP) and serum amyloid P component (SAP), and the long pentraxin PTX3. Pentraxin structure is characterized by a globular flattened jelly-roll fold that contains the eight amino acid pentraxin motif (HxCxS/TWxS, where x represents any amino acid) (Figure 1D). This domain mediates carbohydrate recognition, homo-pentamerization into a cyclic complex, and engagement with host immune proteins, including Fc receptors and the complement component C1q [3]. The long pentraxin PTX3 additionally possesses an N-terminal coiled-coil domain that facilitates further oligomerization [52]. Although the structure of PTX3 has not yet been determined, biochemical analysis suggests that PTX3 forms a disulfide-linked octamer (dimer of tetramers) [52].
While pentraxins are often classified as lectins, they can bind other types of ligands, including lipids. Both CRP and SAP bind varied ligands, including the phosphorylated epitopes present in lipoteichoic acid and LPS. CRP binds phosphocholine, a major constituent of the C-polysaccharide of S. pneumoniae, and repeating units of phosphate-6Gal(β1–4)Manα1, a disaccharide prevalent in the lipophosphoglycan of intracellular parasitic Leishmania species [53]. An X-ray crystal structure of CRP bound to phosphocholine revealed that each CRP monomer binds two calcium ions that directly coordinate the phosphate group of phosphocholine. Additionally, the trimethylammonium group of the ligand fit snuggly into a hydrophobic region in the pentraxin ligand binding site [54]. SAP binds the ligands, phosphoethanolamine and methyl 4,6-O-(1-carboxyethylidene)-β-D-galactopyranoside, using a similar calcium-dependent mechanism [55,56]. SAP also can bind LPS from Gram-negative bacteria and the lectin adheres to the fungus Candida albicans [3]. The long pentraxin PTX3 can interact with outer membrane protein A from K. pneumoniae, and multiple bacterial, fungal, and viral species; however, the molecular mechanism by which it recognizes these microbes remains unclear [57]. The specificity of pentraxin function and/or recognition may be altered by interaction with other microbe-binding lectins on the cell surface. Taken together, these data indicate that pentraxins may provide protection against a broad range of microorganisms.
Most studies of pentraxins have focused on their biological roles. Pentraxins use protein-protein interactions with host immune proteins to effect function. They can interact with C1q to enhance the classical pathway of complement-mediated cell killing and interact directly with Fcγ receptors on immune cells to enhance phagocytosis of microbes [57]. An understanding of the molecular mechanisms underlying their function is beginning to emerge. Specifically, a structure of the SAP-FcγR complex revealed that SAP binds to the IgG binding site on FcγR using the face opposite of the pentraxin carbohydrate binding site [58]. These data suggest that pentraxins share functional overlap with antibodies in the human innate immune system. Pentraxins also are found in complexes with other mediators of the lectin pathway of complement, including MBL and ficolins [25,59,60]. Pentraxins may therefore facilitate crosstalk between complement pathways. Understanding how pentraxins collaborate to modulate complement pathways may help elucidate their roles in regulating microbial colonization.
Ficolins
Humans express three ficolins, M-ficolin (ficolin-1), L-ficolin (ficolin-2), and H-ficolin (ficolin-3) that can bind a range of microbes including Gram-positive and Gram-negative bacteria, viruses, and eukaryotic protozoa [1]. Like collectins, ficolins can activate the lectin pathway of complement. As might be expected from these shared functional roles, the ficolins and collectins are similar in structure. Both classes possess a cysteine-containing N-terminal domain and a central collagen-like domain, components that stabilize ficolin oligomers and engage in protein-protein interactions with MASPs [1].
A key difference between collectins and ficolins is the nature of their C-terminal CRD. Whereas collectins use a C-type CRD, ficolins use a fibrinogen-like domain (FBG domain) to bind N-acetylated glycans such as GlcNAc and GalNAc. The FBG domain is structurally related to the C-terminal halves of the beta and gamma chains of fibrinogen and contains a series of conserved cysteines and aromatic residues that assemble the canonical ficolin carbohydrate binding pocket (S1 binding site). Proximal to this binding site is a protein-ligated calcium ion that appears to be critical for carbohydrate binding but, in contrast to the collectins, is not directly involved in ligand coordination [31]. Structural analysis of M-ficolin and L-ficolin demonstrates that even highly related members of this lectin family may use distinct ligand binding surfaces. Whereas M-ficolin uses the S1 binding site, L-ficolin appears to use additional surfaces (S2–S4) adjacent to the S1 site for ligand binding (Figure 1E)[31]. The extended binding surface of L-ficolin is believed to contribute to the lectin’s ability to broadly recognize acetylated, phosphorylated, and sulfated molecules, in addition to its ability to bind extended polysaccharide ligands including peptidoglycan (PG), beta-glucan of yeast, and lipotechoic acid (LTA) of Gram positive bacteria [61,62]. Glycan array data indicate the ficolins differ not only in their binding modes but also in their saccharide selectivity [63,64]. Specifically, L-ficolin recognizes sulfated saccharides and those with N-acetyl groups, while M-ficolin preferentially binds sialylated glycans. In contrast, effective ligands for H-ficolin were not apparent from screening a glycan array composed mostly of mammalian glycans [63,65], but other studies suggest that this lectin can bind LPS from Hafnia alvei, a commensal found in the human gastrointestinal tract. Thus, the ficolins also bind a broad range of glycans.
Although ficolins and collectins share structural and functional features, the disparate carbohydrate recognition profiles of the two classes and their divergent tissue expression profiles, suggest that they have distinct functions. A recent phylogenetic analysis of proteins that function in complement pathways suggests that the lectin pathway of complement pre-dates the emergence of adaptive immune responses and the classical, antibody-initiated pathway of complement activation [66]. Thus, the recognition of microbes by ficolins and collectins appears to be an ancestral mechanism to regulate microbial colonization. The evolutionary conservation of these proteins highlights their importance in maintaining host health. Elucidating the biological significance of the observed crosstalk between ficolins and other lectins that function in complement pathways and host immunity will be important in understanding how soluble lectins regulate microbial colonization at host-microbe interfaces.
Intelectins
Intelectins, or X-type lectins, were classified relatively recently as a discrete group of animal lectins. They were first identified in Xenopus laevis oocytes (hence the label of “X-type” lectins) and subsequently found to occur in most chordates [67]. As with most mammals, humans encode two intelectins, intelectin-1 (hIntL-1) and intelectin-2 (hIntL-2), which share greater than 80% sequence identity [68]. The function of human intelectins is not known, but the available evidence implicates a role in innate immunity. The lectins are produced at mucosal barriers, and human variants are linked to asthma and Crohn’s disease [69,70].
Although intelectins possess a small FBG domain, they lack sequence homology with the ficolins, which also have an FBG domain. Indeed, a 1.6 Å resolution protein X-ray crystal structure revealed that intelectins differ from other lectin classes in their secondary and tertiary structure and ligand binding mechanism [15•,16••]. These findings led to the proposal that intelectins possess a distinct CRD, the intelectin domain (Figure 1F). Intelectins are calcium ion-dependent, non-C-type lectins. They bind three divalent calcium ions: two of which are buried and proposed to serve structural roles and a third that is involved in glycan binding [16••]. Human IntL-1 was originally described as a furanose-binding lectin [71]. Subsequently, hIntL-1 was shown not to be a generic furanose binder, but rather a lectin that recognizes a vicinal exocyclic 1,2-diol motif [16••].
The ligand specificity of hIntL-1 was elucidated through the use of glycan microarrays. When a mammalian glycan microarray [65] was screened no carbohydrate ligands were detected, but a microbial glycan microarray [9••] revealed multiple, disparate microbial glycoconjugates could function as ligands. Each bound microbial glycan contained an exocyclic 1,2-diol. This epitope is found in multiple monosaccharide ligands specific to microbes including D-galactofuranose (D-Galf) residues, D-phospho glycerol-modified glycans, heptoses, D-glycero-D-talo-oct-2-ulosonic acid (KO) and 3-deoxy-D-manno-oct-2-ulosonic acid (KDO).
Information about the ligand specificity of hIntL-1 led to determination of an X-ray structure of the complex of hIntL-1 and allyl-β-D-Galf [16••]. This structure revealed the exocyclic 1,2-diol sits inside an aromatic box generated by tryptophan and tyrosine residues, and the diol hydroxyl groups coordinate directly to a surface accessible calcium ion. The residues comprising the aromatic box are conserved between human and mouse intelectin-1, and both orthologs interact with Galf residues. Moreover, like hIntL-1, a Xenopus intelectin (XEEL) can engage Galf residues and phosphoglycerol moieties [15•]. A structure of XEEL complexed to phosphoglycerol revealed the hIntL-1 and XEEL binding sites are remarkably similar. Even the most apparent difference is subtle: XEEL forms its aromatic box using two tryptophan residues rather than the hIntL-1 tyrosine and tryptophan. Thus, the aromatic box is a conserved feature of both of these lectins. The successful identification of hIntL-1 ligands highlights how glycan microarrays can be used to inform protein structural biology studies of lectin—carbohydrate recognition.
The hIntL-1 residues that coordinate calcium ions are highly conserved amongst intelectins from diverse species; in contrast, the aromatic box of hIntL-1 and XEEL is not conserved in all other intelectins [15•]. We postulate that the residues corresponding to tryptophan and tyrosine of hIntL-1 are critical determinants of intelectin ligand specificity. Intriguingly, hIntL-2 contains a tryptophan and serine combination, suggesting the glycan specificity of hIntl-2 differs.
Most intelectin proteins do not contain well-defined or annotated domains outside of their CRD. In the case of XEEL, which is upregulated in frog eggs prior to hatching [72], a coiled-coil region could oligomerize the lectin to form a dumbbell-like structure and agglutinate bacteria displaying XEEL binding motifs [15•]. In contrast, hIntL-1 has almost 20 residues between the signal peptide cleavage site and the beginning of the FBG domain, yet it lacks GXY repeats like the collagen-like domains present in collectins and ficolins. Still, hIntL-1 forms disulfide-linked homotrimers. The cysteines that engage in intermolecular disulfide bonds in hIntL-1 are not strictly conserved among other intelectins. Additionally, these residues are not strictly required for oligomerization, as a Xenopus intelectin (XEEL) construct lacking intermolecular cysteines residues could form high affinity trimers in solution [15•]. Similarly, we anticipate the murine IntL-1, which lacks analogous cysteines and has been considered monomeric [73], is a trimer in solution.
Human IntL-1 selectively recognizes carbohydrate ligands synthesized by microbes, and therefore binds microbial cells [16••,71]. This selectivity provides additional support for a role for hIntL-1 in innate immunity. Soluble or cell-bound proteins might function with the intelectins in innate immunity. While a hIntL-1 trimer is involved in carbohydrate binding both the N-termini and the FBG domain are available to participate in protein–protein interactions.
The ability to recognize an exocyclic 1,2-diol motif is not restricted to the intelectins, as the collectin SP-D can bind a similar epitope [27,74•]. Specifically, X-ray crystallography revealed L-D-Hep can interact with SP-D through coordination of the C7 and C8 exocyclic sugar hydroxyl groups (Figure 2). Whether SP-D can recognize other exocyclic diol containing hIntL-1 ligands is unknown. Still, this example raises an interesting attribute of the soluble lectins — ligand redundancy. While the lectin sequences can be tuned to engage select ligands, there is redundancy in the ligands lectins bind, especially within a structural class. Whether further studies that examine a wider array of glycans will reveal finer differences in lectin binding properties is unclear. The ability of different lectins to recognize similar microbial glycan epitopes may be beneficial, as many pathogens have related carbohydrate antigens [12]. In this way, different microbes can engage multiple types of lectins to give rise to different downstream immune responses [75]. Moreover, the cell-type or tissue-selective expression of different lectins elicit responses to relevant microbes. For example, in the human gut, hIntL-1 may play the predominant role in recognizing exocyclic 1,2-diols of commensals while SP-D is induced during inflammation.
Challenges and Advances in Characterizing Lectin Recognition
The properties of lectins raise challenges for identifying their ligands. Lectins have a propensity to oligomerize and aggregate. When coupled with the low affinity of many protein–carbohydrate interactions, these properties together can complicate the analysis of lectin specificity. In this section, we outline some of the issues we have encountered and some of the strategies we have found effective in overcoming difficulties in determining lectin selectivity.
The identification of the important roles of mucosal lectins (e.g., Reg proteins) in host defense provides impetus to re-evaluate the structure and function of known lectins. Ligand binding to lectins was historically assessed using columns displaying immobilized polysaccharides or monosaccharides and/or elution with competitive carbohydrates or chelating agents that complex calcium or other cations. With access to mammalian and microbial glycan arrays [9••], it now is possible to use unbiased routes to identify lectins that bind self and non-self glycan epitopes. The advantages of continuing to probe lectin function are illustrated by recent studies of the lectin zymogen granule protein 16 (ZG16p) [32,76•,77], which revealed that it has a jacalin-related beta-prism fold. This lectin architecture is well documented in plant-derived lectins but no mammalian lectin adopting this fold had been described (Figure 1G). Recognition of peptidoglycan on Gram-positive bacteria by ZG16 was recently shown to be critical in colonic mucosal barrier function and spatial microbiota regulation within the murine gut [78••]. These findings provide impetus to search for and explore other microbe-binding lectins.
Many of the lectins discussed in this review are glycosylated or function as disulfide-linked oligomers. In generating soluble recombinant lectins, successful lectin production is typically judged by protein binding to immobilized carbohydrates. We have observed a curious phenomenon that can give rise to irreproducible results: misfolded and aggregated lectins can retain carbohydrate-binding activity and this activity can depend on the presence of calcium ions. Still, the ligand specificity for such non-native lectins differs dramatically from that observed for properly folded lectins. Methods to verify that recombinant lectins are folded and adopt a relevant oligomerization state are therefore critical. Multiple techniques can be employed, including native polyacrylamide gels, chemical crosslinking, size exclusion chromatography, small angle X-ray scattering, and analytical ultracentrifugation [15•].
With assays designed to assess lectin binding to the glycans of intact bacteria, fungi, or parasites, we have found irrelevant lectin—cell binding can occur through interaction with glycans of other proteins present under assay conditions (i.e., antibodies, cell surface glycoproteins) or through non-specific binding of aggregated lectins. Reproducible binding can be monitored (i.e., using flow cytometry or microscopy) by visualizing lectin distribution and localization and by quantifying the percentage of cells bound. Keeping the lectin concentration as low as possible also avoids aggregation.
Many lectins depend on divalent ions for their structure and/or for glycan binding. Experimental controls such as addition of the chelator ethylenediaminetetraacetic acid (for calcium-dependent lectins) or exposure to competitive ligands can separate interactions mediated by the lectin CRD from those that occur through other binding modes. In testing carbohydrate competitors, we have found it beneficial to employ glycosides with defined linkages rather than free monosaccharide competitors. The latter can undergo ring opening and interconversion; and in the open form, they can react with surface lysine residues and thereby inhibit binding through non-specific mechanisms. These strategies can facilitate reproducible lectin binding to cells or glycans.
Conclusion
The development of new tools to probe lectin specificity and facilitate lectin discovery suggest exciting new avenues for understanding and exploiting lectin function. A number of soluble lectins can recognize microbial glycans, but many of these have been tested for binding to a small number of possible glycan epitopes. The advent of glycan microarrays displaying glycans from microbes offers new opportunities to explore lectin specificity [9••]. Expanding the arrays to include commensal and mutualistic bacteria as well as fungal and parasite-derived polysaccharides would provide the means to further elucidate the specificity of the lectins that function in microbial surveillance. We anticipate that the advances described above can be used to augment understanding of the host factors that regulate microbial colonization and prevent infection.
Highlights.
Elucidating lectin selectivity for microbial glycans could yield new antibiotic strategies
Microbe-binding lectins can bind an unexpectedly wide range of glycan epitopes
Mammalian microbe-binding lectins are oligomeric or form oligomers upon binding
Expanding glycan microarray composition is critical for defining lectin selectivity
Strategies are presented for identifying reproducible glycan–lectin interactions
Acknowledgments
LLK acknowledges support from the University of Wisconsin and National Institutes of Health (NIH) (R01GM55984 and R01AI063596). DAW is a recipient of a National Science Foundation Graduate Research Fellowship and a trainee of the NIH Chemistry-Biology Interface Training Program (T32 GM008505), which is supported the University of Wisconsin–Madison, Office of the Vice Chancellor for Research and Graduate Education with funding from the Wisconsin Alumni Research Foundation.
Footnotes
Conflict of Interest
The authors declare no conflict of interest.
References and recommended reading
Papers of particular interest, published within the period of review have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Holmskov U, Thiel S, Jensenius JC: Collectins and ficolins: humoral lectins of the innate immune defense. Annu Rev Immunol 2003, 21:547–578. [DOI] [PubMed] [Google Scholar]
- 2.Vasta GR: Roles of galectins in infection. Nat Rev Microbiol 2009, 7:424–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bottazzi B, Doni A, Garlanda C, Mantovani A: An Integrated View of Humoral Innate Immunity: Pentraxins as a Paradigm. Annu Rev Immunol 2010, 28:157–183. [DOI] [PubMed] [Google Scholar]
- 4.Bajic G, Degn SE, Thiel S, Andersen GR: Complement activation, regulation, and molecular basis for complement-related diseases. EMBO J 2015, 34:2735–2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Drickamer K, Taylor ME: Recent insights into structures and functions of C-type lectins in the immune system. Curr Opin Struct Biol 2015, 34:26–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iwasaki A, Medzhitov R: Control of adaptive immunity by the innate immune system. Nat Immunol 2015, 16:343–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tytgat HLP, Lebeer S: The Sweet Tooth of Bacteria: Common Themes in Bacterial Glycoconjugates. Microbiol Mol Biol Rev 2014, 78:372–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rillahan CD, Paulson JC: Glycan microarrays for decoding the glycome. Annu Rev Biochem 2011, 80:797–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stowell SR, Arthur CM, McBride R, Berger O, Razi N, Heimburg-Molinaro J, Rodrigues LC, Gourdine JP, Noll AJ, von Gunten S, et al. : Microbial glycan microarrays define key features of host-microbial interactions. Nat Chem Biol 2014, 10:470–476. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Describes the generation and utility of a glycan microarray composed of bacterial cell surface glycans. The utility of the array was illustrated by probing galectin binding to microbial mimics of mammalian glycans.
- 10.Kiessling LL, Cairo CW: Hitting the sweet spot. Nat Biotechnol 2002, 20:234–235. [DOI] [PubMed] [Google Scholar]
- 11.Wang D, Liu S, Trummer BJ, Deng C, Wang A: Carbohydrate microarrays for the recognition of cross-reactive molecular markers of microbes and host cells. Nat Biotechnol 2002, 20:275–281. [DOI] [PubMed] [Google Scholar]
- 12.Herget S, Toukach PV, Ranzinger R, Hull WE, Knirel YA, von der Lieth CW: Statistical analysis of the Bacterial Carbohydrate Structure Data Base (BCSDB): characteristics and diversity of bacterial carbohydrates in comparison with mammalian glycans. BMC Struct Biol 2008, 8:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Adibekian A, Stallforth P, Hecht ML, Werz DB, Gagneux P, Seeberger PH: Comparative bioinformatics analysis of the mammalian and bacterial glycomes. Chem Sci 2011, 2:337–344. [Google Scholar]
- 14.Drickamer K, Taylor ME: Biology of Animal Lectins. Annu Rev Cell Biol 1993, 9:237–264. [DOI] [PubMed] [Google Scholar]
- 15.Wangkanont K, Wesener DA, Vidani JA, Kiessling LL, Forest KT: Structures of Xenopus Embryonic Epidermal Lectin Reveal a Conserved Mechanism of Microbial Glycan Recognition. J Biol Chem 2016, 291:5596–5610. [DOI] [PMC free article] [PubMed] [Google Scholar]; • The structure of a frog intelectin XEEL bound to phosphoglycerol reveals that terminal 1,2-diol recognition is conserved between XEEL and human intelectin-1.
- 16.Wesener DA, Wangkanont K, McBride R, Song X, Kraft MB, Hodges HL, Zarling LC, Splain RA, Smith DF, Cummings RD, et al. : Recognition of microbial glycans by human intelectin-1. Nat Struct Mol Biol 2015, 22:603–610. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Uses a multidisciplinary approach to define the structure and carbohydrate binding specificity of human intelectin-1, revealing the first human lectin specific for microbial monosaccharides. This work also suggests that intelectin proteins comprise a structurally distinct family of lectins.
- 17.Mann DA, Kanai M, Maly DJ, Kiessling LL: Probing low affinity and multivalent interactions with surface plasmon resonance: Ligands for concanavalin A. J Am Chem Soc 1998, 120:10575–10582. [Google Scholar]
- 18.Mammen M, Choi SK, Whitesides GM: Polyvalent interactions in biological systems: Implications for design and use of multivalent ligands and inhibitors. Angew Chem Int Edit 1998, 37:2755–2794. [DOI] [PubMed] [Google Scholar]
- 19.Degn SE, Kjaer TR, Kidmose RT, Jensen L, Hansen AG, Tekin M, Jensenius JC, Andersen GR, Thiel S: Complement activation by ligand-driven juxtaposition of discrete pattern recognition complexes. Proc Natl Acad Sci U S A 2014, 111:13445–13450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Neth O, Jack DL, Dodds AW, Holzel H, Klein NJ, Turner MW: Mannose-binding lectin binds to a range of clinically relevant microorganisms and promotes complement deposition. Infect Immun 2000, 68:688–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weis WI, Taylor ME, Drickamer K: The C-type lectin superfamily in the immune system. Immunol Rev 1998, 163:19–34. [DOI] [PubMed] [Google Scholar]
- 22.Drickamer K: Engineering Galactose-Binding Activity into a C-Type Mannose-Binding Protein. Nature 1992, 360:183–186. [DOI] [PubMed] [Google Scholar]
- 23.Weis WI, Drickamer K, Hendrickson WA: Structure of a C-type mannose-binding protein complexed with an oligosaccharide. Nature 1992, 360:127–134. [DOI] [PubMed] [Google Scholar]
- 24.Venkatraman Girija U, Furze CM, Gingras AR, Yoshizaki T, Ohtani K, Marshall JE, Wallis AK, Schwaeble WJ, El-Mezgueldi M, Mitchell DA, et al. : Molecular basis of sugar recognition by collectin-K1 and the effects of mutations associated with 3MC syndrome. BMC Biol 2015, 13:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Foo SS, Reading PC, Jaillon S, Mantovani A, Mahalingam S: Pentraxins and Collectins: Friend or Foe during Pathogen Invasion? Trends Microbiol 2015, 23:799–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kishore U, Greenhough TJ, Waters P, Shrive AK, Ghai R, Kamran MF, Bernal AL, Reid KBM, Madan T, Chakraborty T: Surfactant proteins SP-A and SP-D: Structure, function and receptors. Mol Immunol 2006, 43:1293–1315. [DOI] [PubMed] [Google Scholar]
- 27.Wang H, Head J, Kosma P, Brade H, Muller-Loennies S, Sheikh S, McDonald B, Smith K, Cafarella T, Seaton B, et al. : Recognition of heptoses and the inner core of bacterial lipopolysaccharides by surfactant protein D. Biochemistry 2008, 47:710–720. [DOI] [PubMed] [Google Scholar]
- 28.Mukherjee S, Zheng H, Derebe MG, Callenberg KM, Partch CL, Rollins D, Propheter DC, Rizo J, Grabe M, Jiang QX, et al. : Antibacterial membrane attack by a pore-forming intestinal C-type lectin. Nature 2014, 505:103–107. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Defines the mechanism of bacterial cell killing by the antimicrobial C-type lectin RegIIIα.
- 29.Saraboji K, Hakansson M, Genheden S, Diehl C, Qvist J, Weininger U, Nilsson UJ, Leffler H, Ryde U, Akke M, et al. : The Carbohydrate-Binding Site in Galectin-3 Is Preorganized To Recognize a Sugarlike Framework of Oxygens: Ultra-High-Resolution Structures and Water Dynamics. Biochemistry 2012, 51:296–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thompson D, Pepys MB, Wood SP: The physiological structure of human C-reactive protein and its complex with phosphocholine. Structure 1999, 7:169–177. [DOI] [PubMed] [Google Scholar]
- 31.Garlatti V, Belloy N, Martin L, Lacroix M, Matsushita M, Endo Y, Fujita T, Fontecilla-Camps JC, Arlaud GJ, Thielens NM, et al. : Structural insights into the innate immune recognition specificities of L- and H-ficolins. EMBO J 2007, 26:623–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kanagawa M, Satoh T, Ikeda A, Nakano Y, Yagi H, Kato K, Kojima-Aikawa K, Yamaguchi Y: Crystal structures of human secretory proteins ZG16p and ZG16b reveal a Jacalin-related beta-prism fold. Biochem Biophys Res Commun 2011, 404:201–205. [DOI] [PubMed] [Google Scholar]
- 33.Zhang YW, Ding LS, Lai MD: Reg gene family and human diseases. World J Gastroenterol 2003, 9:2635–2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mukherjee S, Hooper LV: Antimicrobial Defense of the Intestine. Immunity 2015, 42:28–39. [DOI] [PubMed] [Google Scholar]
- 35.Vaishnava S, Yamamoto M, Severson KM, Ruhn KA, Yu XF, Koren O, Ley R, Wakeland EK, Hooper LV: The Antibacterial Lectin RegIII gamma Promotes the Spatial Segregation of Microbiota and Host in the Intestine. Science 2011, 334:255–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Caballero S, Pamer EG: Microbiota-mediated inflammation and antimicrobial defense in the intestine. Annu Rev Immunol 2015, 33:227–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mukherjee S, Partch CL, Lehotzky RE, Whitham CV, Chu H, Bevins CL, Gardner KH, Hooper LV: Regulation of C-type lectin antimicrobial activity by a flexible N-terminal prosegment. J Biol Chem 2009, 284:4881–4888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lehotzky RE, Partch CL, Mukherjee S, Cash HL, Goldman WE, Gardner KH, Hooper LV: Molecular basis for peptidoglycan recognition by a bactericidal lectin. Proc Natl Acad Sci U S A 2010, 107:7722–7727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ho MR, Lou YC, Wei SY, Luo SC, Lin WC, Lyu PC, Chen C: Human RegIV protein adopts a typical C-type lectin fold but binds mannan with two calcium-independent sites. J Mol Biol 2010, 402:682–695. [DOI] [PubMed] [Google Scholar]
- 40.Kashyap DR, Rompca A, Gaballa A, Helmann JD, Chan J, Chang CJ, Hozo I, Gupta D, Dziarski R: Peptidoglycan Recognition Proteins Kill Bacteria by Inducing Oxidative, Thiol, and Metal Stress. PLoS Path 2014, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]; • A mechanism of microbial cell killing by peptidoglycan recognition proteins (PGRPs) sis described. Instead of mechanical or enzymatic membrane destabilization, PGRPs activate two-component signaling systems involved in stress sensing.
- 41.Baum LG, Garner OB, Schaefer K, Lee B: Microbe-host interacions are positively and negatively regulated by galectin glycan interactions. Front Immunol 2014, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen HY, Weng IC, Hong MH, Liu FT: Galectins as bacterial sensors in the host innate response. Curr Opin Microbiol 2014, 17:75–81. [DOI] [PubMed] [Google Scholar]
- 43.Kohatsu L, Hsu DK, Jegalian AG, Liu FT, Baum LG: Galectin-3 induces death of Candida species expressing specific beta-1,2-linked Mannans. J Immunol 2006, 177:4718–4726. [DOI] [PubMed] [Google Scholar]
- 44.Stowell SR, Arthur CM, Dias-Baruffi M, Rodrigues LC, Gourdine JP, Heimburg-Molinaro J, Ju T, Molinaro RJ, Rivera-Marrero C, Xia B, et al. : Innate immune lectins kill bacteria expressing blood group antigen. Nat Med 2010, 16:295–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Park AM, Hagiwara S, Hsu DK, Liu FT, Yoshie O: Galectin-3 Plays an Important Role in Innate Immunity to Gastric Infection by Helicobacter pylori. Infect Immun 2016, 84:1184–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thiemann S, Baum LG: Galectins and Immune Responses-Just How Do They Do Those Things They Do? Annu Rev Immunol 2016, 34:243–264. [DOI] [PubMed] [Google Scholar]
- 47.Hudson KL, Bartlett GJ, Diehl RC, Agirre J, Gallagher T, Kiessling LL, Woolfson DN: Carbohydrate-Aromatic Interactions in Proteins. J Am Chem Soc 2015, 137:15152–15160. [DOI] [PMC free article] [PubMed] [Google Scholar]; • A bioinformatic and structural analysis of carbohydrate interactions with protein aromatic amino acid side chains. Highlights the importance and preferred location of aromatic resides in protein – carbohydrate recognition.
- 48.Miller MC, Ippel H, Suylen D, Klyosov AA, Traber PG, Hackeng T, Mayo KH: Binding of polysaccharides to human galectin-3 at a noncanonical site in its carbohydrate recognition domain. Glycobiology 2016, 26:88–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mey A, Leffler H, Hmama Z, Normier G, Revillard JP: The animal lectin galectin-3 interacts with bacterial lipopolysaccharides via two independent sites. J Immunol 1996, 156:1572–1577. [PubMed] [Google Scholar]
- 50.Stowell SR, Arthur CM, Slanina KA, Horton JR, Smith DF, Cummings RD: Dimeric Galectin-8 induces phosphatidylserine exposure in leukocytes through polylactosamine recognition by the C-terminal domain. J Biol Chem 2008, 283:20547–20559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Knirel YA, Gabius HJ, Blixt O, Rapoport EM, Khasbiullina NR, Shilova NV, Bovin NV: Human tandem-repeat-type galectins bind bacterial non-beta Gal polysaccharides. Glycoconjugate J 2014, 31:7–12. [DOI] [PubMed] [Google Scholar]
- 52.Inforzato A, Rivieccio V, Morreale A, Bastone A, Salustri A, Scarchilli L, Verdoliva A, Vincenti S, Gallo G, Chiapparino C, et al. : Structural characterization of PTX3 disulfide bond network and its multimeric status in cumulus matrix organization. J Biol Chem 2008, 283:10147–10161. [DOI] [PubMed] [Google Scholar]
- 53.Culley FJ, Bodman-Smith KB, Ferguson MAJ, Nikolaev AV, Shantilal N, Raynes JG: C-reactive protein binds to phosphorylated carbohydrates. Glycobiology 2000, 10:59–65. [DOI] [PubMed] [Google Scholar]
- 54.Thompson D, Pepys MB, Wood SP: The physiological structure of human C-reactive protein and its complex with phosphocholine. Structure 1999, 7:169–177. [DOI] [PubMed] [Google Scholar]
- 55.Emsley J, White HE, O’Hara BP, Oliva G, Srinivasan N, Tickle IJ, Blundell TL, Pepys MB, Wood SP: Structure of pentameric human serum amyloid P component. Nature 1994, 367:338–345. [DOI] [PubMed] [Google Scholar]
- 56.Hind CRK, Collins PM, Renn D, Cook RB, Caspi D, Baltz ML, Pepys MB: Binding Specificity of Serum Anyloid P Component for the Pyruvate Acetal of Galactose. J Exp Med 1984, 159:1058–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ma YJ, Doni A, Romani L, Jurgensen HJ, Behrendt N, Mantovani A, Garred P: Ficolin-1–PTX3 Complex Formation Promotes Clearance of Altered Self-Cells and Modulates IL-8 Production. J Immunol 2013, 191:1324–1333. [DOI] [PubMed] [Google Scholar]
- 58.Lu J, Marnell LL, Marjon KD, Mold C, Du Clos TW, Sun PD: Structural recognition and functional activation of FcgammaR by innate pentraxins. Nature 2008, 456:989–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ma YJ, Doni A, Skjoedt MO, Honore C, Arendrup M, Mantovani A, Garred P: Heterocomplexes of Mannose-binding Lectin and the Pentraxins PTX3 or Serum Amyloid P Component Trigger Cross-activation of the Complement System. J Biol Chem 2011, 286:3405–3417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gout E, Moriscot C, Doni A, Dumestre-Perard C, Lacroix M, Perard J, Schoehn G, Mantovani A, Arlaud GJ, Thielens NM: M-Ficolin Interacts with the Long Pentraxin PTX3: A Novel Case of Cross-Talk between Soluble Pattern-Recognition Molecules. J Immunol 2011, 186:5815–5822. [DOI] [PubMed] [Google Scholar]
- 61.Laffly E, Lacroix M, Martin L, Vassal-Stermann E, Thielens NM, Gaboriaud C: Human ficolin-2 recognition versatility extended: an update on the binding of ficolin-2 to sulfated/phosphated carbohydrates. FEBS Lett 2014, 588:4694–4700. [DOI] [PubMed] [Google Scholar]
- 62.Ma YG, Cho MY, Zhao M, Park JW, Matsushita M, Fujita T, Lee BL: Human mannose-binding lectin and L-ficolin function as specific pattern recognition proteins in the lectin activation pathway of complement. J Biol Chem 2004, 279:25307–25312. [DOI] [PubMed] [Google Scholar]
- 63.Gout E, Garlatti V, Smith DF, Lacroix M, Dumestre-Perard C, Lunardi T, Martin L, Cesbron JY, Arlaud GJ, Gaboriaud C, et al. : Carbohydrate Recognition Properties of Human Ficolins: glycan array screening reveals the sialic acid binding specificity of M-ficolin. J Biol Chem 2010, 285:6612–6622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Krarup A, Mitchell DA, Sim RB: Recognition of acetylated oligosaccharides by human L-ficolin. Immunol Lett 2008, 118:152–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Blixt O, Head S, Mondala T, Scanlan C, Huflejt ME, Alvarez R, Bryan MC, Fazio F, Calarese D, Stevens J, et al. : Printed covalent glycan array for ligand profiling of diverse glycan binding proteins. Proc Natl Acad Sci U S A 2004, 101:17033–17038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Doni A, Musso T, Morone D, Bastone A, Zambelli V, Sironi M, Castagnoli C, Cambieri I, Stravalaci M, Pasqualini F, et al. : An acidic microenvironment sets the humoral pattern recognition molecule PTX3 in a tissue repair mode. J Exp Med 2015, 212:905–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lee JK, Baum LG, Moremen K, Pierce M: The X-lectins: a new family with homology to the Xenopus laevis oocyte lectin XL-35. Glycoconj J 2004, 21:443–450. [DOI] [PubMed] [Google Scholar]
- 68.Lee JK, Schnee J, Pang M, Wolfert M, Baum LG, Moremen KW, Pierce M: Human homologs of the Xenopus oocyte cortical granule lectin XL35. Glycobiology 2001, 11:65–73. [DOI] [PubMed] [Google Scholar]
- 69.Barrett JC, Hansoul S, Nicolae DL, Cho JH, Duerr RH, Rioux JD, Brant SR, Silverberg MS, Taylor KD, Barmada MM, et al. : Genome-wide association defines more than 30 distinct susceptibility loci for Crohn’s disease. Nat Genet 2008, 40:955–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pemberton AD, Rose-Zerilli MJ, Holloway JW, Gray RD, Holgate ST: A single-nucleotide polymorphism in intelectin 1 is associated with increased asthma risk. J Allergy Clin Immunol 2008, 122:1033–1034. [DOI] [PubMed] [Google Scholar]
- 71.Tsuji S, Uehori J, Matsumoto M, Suzuki Y, Matsuhisa A, Toyoshima K, Seya T: Human intelectin is a novel soluble lectin that recognizes galactofuranose in carbohydrate chains of bacterial cell wall. J Biol Chem 2001, 276:23456–23463. [DOI] [PubMed] [Google Scholar]
- 72.Dubaissi E, Rousseau K, Lea R, Soto X, Nardeosingh S, Schweickert A, Amaya E, Thornton DJ, Papalopulu N: A secretory cell type develops alongside multiciliated cells, ionocytes and goblet cells, and provides a protective, anti-infective function in the frog embryonic mucociliary epidermis. Development 2014, 141:1514–1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tsuji S, Yamashita M, Nishiyama A, Shinohara T, Zhongwei U, Myrvik QN, Hoffman DR, Henriksen RA, Shibata Y: Differential structure and activity between human and mouse intelectin-1: Human intelectin-1 is a disulfide-linked trimer, whereas mouse homologue is a monomer. Glycobiology 2007, 17:1045–1051. [DOI] [PubMed] [Google Scholar]
- 74.Reinhardt A, Wehle M, Geissner A, Crouch EC, Kang Y, Yang Y, Anish C, Santer M, Seeberger PH: Structure binding relationship of human surfactant protein D and various lipopolysaccharide inner core structures. J Struct Biol 2016, 195:387–395. [DOI] [PubMed] [Google Scholar]; • The mechanism and specificity of SP-D binding to the exocyclic diol of heptoses was probed using a custom glycan microarray and surface plasmon resonance.
- 75.Kuperman DA, Lewis CC, Woodruff PG, Rodriguez MW, Yang YH, Dolganov GM, Fahy JV, Erle DJ: Dissecting asthma using focused transgenic modeling and functional genomics. J Allergy Clin Immunol 2005, 116:305–311. [DOI] [PubMed] [Google Scholar]
- 76.Kanagawa M, Liu Y, Hanashima S, Ikeda A, Chai W, Nakano Y, Kojima-Aikawa K, Feizi T, Yamaguchi Y: Structural basis for multiple sugar recognition of Jacalin-related human ZG16p lectin. J Biol Chem 2014, 289:16954–16965. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Biochemical and structural analysis of ZG16p to define its carbohydrate specificity. Compares ZG16p to other Jacalin-related lectins.
- 77.Hanashima S, Gotze S, Liu Y, Ikeda A, Kojima-Aikawa K, Taniguchi N, Silva DV, Feizi T, Seeberger PH, Yamaguchi Y: Defining the Interaction of Human Soluble Lectin ZG16p and Mycobacterial Phosphatidylinositol Mannosides. ChemBioChem 2015, 16:1502–1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bergstrom JH, Birchenough GM, Katona G, Schroeder BO, Schutte A, Ermund A, Johansson ME, Hansson GC: Gram-positive bacteria are held at a distance in the colon mucus by the lectin-like protein ZG16. Proc Natl Acad Sci U S A 2016, 113:13833–13838. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• ZG16 binding to peptidoglycan on Gram-positive bacteria is revealed. Transgenic mouse experiments suggest a role in integrity of the colonic mucus barrier and spatial regulation of the gut microbiota.