Abstract
In 2016, the Frank R. Lautenberg Chemical Safety for the 21st Century Act became the first US legislation to advance chemical safety evaluations by utilizing novel testing approaches that reduce the testing of vertebrate animals. Central to this mission is the advancement of computational toxicology and artificial intelligence approaches to implementing innovative testing methods. In the current big data era, the terms volume (amount of data), velocity (growth of data), and variety (the diversity of sources) have been used to characterize the currently available chemical, in vitro, and in vivo data for toxicity modeling purposes. Furthermore, as suggested by various scientists, the variability (internal consistency or lack thereof) of publicly available data pools, such as PubChem, also presents significant computational challenges. The development of novel artificial intelligence approaches based on public massive toxicity data is urgently needed to generate new predictive models for chemical toxicity evaluations and make the developed models applicable as alternatives for evaluating untested compounds. In this procedure, traditional approaches (e.g., QSAR) purely based on chemical structures have been replaced by newly designed data-driven and mechanism-driven modeling. The resulting models realize the concept of adverse outcome pathway (AOP), which can not only directly evaluate toxicity potentials of new compounds, but also illustrate relevant toxicity mechanisms. The recent advancement of computational toxicology in the big data era has paved the road to future toxicity testing, which will significantly impact on the public health.
INTRODUCTION
Traditional experimental testing procedures, both in vitro and in vivo, to identify compounds that can induce chemical toxicity are generally expensive and time-consuming.1,2 Computational modeling is a promising alternative method for chemical toxicity evaluations. Existing computational models for risk assessment, such as quantitative structure–activity relationship (QSAR) models for various toxicity end points, can be used to quickly predict large numbers of new compounds in the risk assessment process and prioritize potential toxic compounds for experimental testing. However, critical issues of previous computational toxicology modeling studies, such as the small size of the data sets often being used in model development inducing coverage of a limited chemical space,3 activity cliffs,4 and overfitting,5 limit the applicability of existing models (e.g., QSAR models). The primary hypothesis of QSAR modeling (e.g., similar compounds will have similar activities) sometimes proves to be flawed and is the primary reason for activity cliffs.6,7
Despite these limitations, regulatory acceptance of computational models remains an urgent demand in modern toxicology.8,9 In 2016, the Frank R. Lautenberg Chemical Safety for the 21st Century Act (LCSA) became the first legislation since the Toxic Substances Control Act of 1976 (TSCA) to progress chemical risk assessment.10,11 An essential component of the LCSA is a call for applicable computational approaches and associated predictive models for safety evaluation purposes.10 In the past decade, the development of new experimental protocols, especially high-throughput screening (HTS) assays, and the progress of combinatorial chemistry generated various biological data for millions of compounds.12 Data sharing projects, such as PubChem,13,14 have made chemical ”big data” publically available, which advanced modern toxicology studies into a big data era.1,2,15,16 The available massive public data bring urgent requests for the development of innovative modeling approaches, driven by the recent progress of artificial intelligence, which can fulfill the current needs of chemical risk assessment.
On the basis of the Organization of Economic Co-operation and Development (OECD) guidance of QSAR model development for chemical toxicity, the predictions of computational models for new compounds need to be mechanistically explainable.17 However, the recently popular neural network approach to deal with big data typically performs as a “black box” algorithm,18,19 which brings an uncertain future to computational toxicology.12 Many HTS assays utilize human cells and tissues and have quantitative results that allow for mechanistic interpretation.12 This data landscape enables researchers to create in silico models that incorporate the concept of the adverse outcome pathway (AOP)20 with publically available big data, resulting in mechanism-driven modeling studies.1,15,21 The resulting models of these studies can not only predict the toxicity of new compounds, but also illustrate toxicity mechanisms of importance in humans and animals, thereby filling the gap created by speculation about a possible lack of concordance between animal and human test data.22 The urgent need for advanced computational methods, availability of abundant HTS big data, and opportunity for incorporation of mechanistic analysis introduce new challenges and prospects to the modern computational toxicology area.
BIG DATA IN CHEMICAL TOXICOLOGY
The term “big data” refers to data sets, structured or unstructured, that multiply quickly and are so large and multifaceted that they are impossible to treat using personal computers and traditional computational approaches.23 Data sets with big data require advanced tools such as heterogeneous and cloud computing24 that have capabilities beyond those of conventional data processing and handling techniques as well as dynamic data curation and sharing using algorithms such as those used to handle data streams.25,26 These advanced techniques allow for rapid identification of target entities in these massive data sets in ways that manual data compilation and curation could never efficiently match, which has radical implications for the improvement of traditional computational toxicology modeling techniques like read-across.15,16
Recent HTS programs and their associated data sharing efforts have revolutionized the landscape in many health fields, highlighted by the Big Data to Knowledge (BD2K) initiative by the National Institutes of Health (NIH), which emphasizes the usefulness of big data in biomedical research and critical need to capitalize on the amount of data available in the health field.27,28 A significant HTS effort in toxicology is the United States Environmental Protection Agency (US EPA) research program called Toxicity Forecaster (ToxCast), which employed in vitro HTS tests and toxicogenomics techniques to quickly evaluate the toxicity of compounds and prioritize compounds for experimental testing.29–31 Phase I of this project evaluated 300 unique compounds, mostly of agricultural interest (i.e., pesticides), using about 500 HTS assays.30 Phase II evaluated an additional 767 compounds, including some failed pharmaceutical compounds, using about 700 HTS assays.31 Recently, the ToxCast initiative advanced to the Tox21 collaboration between the US EPA Office of Research and Development/National Center for Computational Toxicology (NCCT), NIH/National Institute of Environmental Health Sciences (NIEHS)/National Toxicology Program (NTP), and the NIH/National Chemical Genomics Center (NCGC), now part of the National Center for Advancing Translational Sciences (NCATS).32–35 Phase I of Tox21 used 75 HTS assays, which were selected and refined from ToxCast assays, to screen an initial set of about 2800 compounds.32 Phase II began in 2010 to screen a more extensive set of approximately 10 000 environmental compounds.32,34,35 As of 2018, the Tox21 program generated over 120 million data points for approximately 8500 chemicals.33
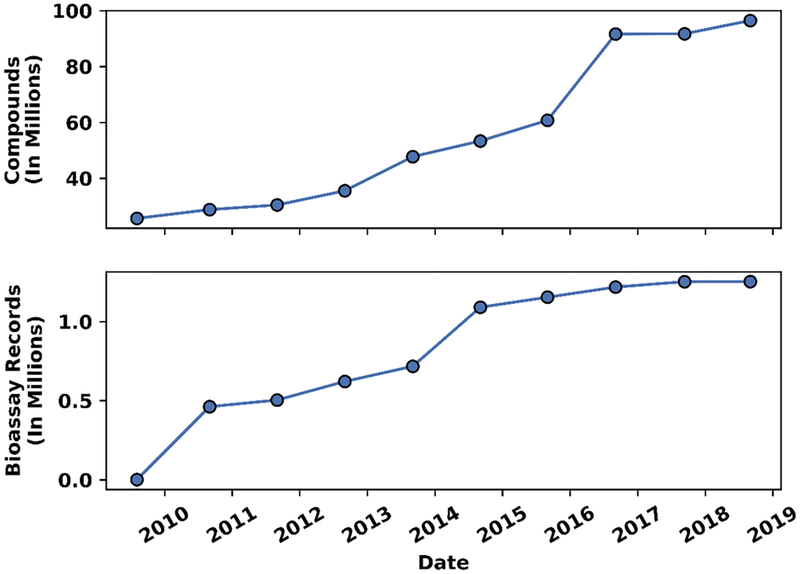
Publicly available databases store much of the data obtained from the toxicology community, including data from HTS programs such as the ToxCast and Tox21 programs.29–31,36 Table 1 describes a selection of significant sources representing publically available big data in the toxicology field. Among them, Aggregated Computational Toxicology Resource (ACToR),37,38 Registration, Evaluation, Authorization, and Restriction of Chemicals (REACH),16,39–42 RepDose,43 Safety Evaluation Ultimately Replacing Animal Testing (SEURAT),44 and Toxicology Data Network (ToxNET)45 were specifically developed to share toxicity data. Chemical Effects in Biological Systems (CEBS),46 ChEMBL,47 Connectivity Map,48,49 Comparative Toxicogenomics Database (CTD),50 DrugMatrix,51 Gene Expression Omnibus (GEO),52,53 and PubChem13,14 share general biological data, including toxicity data. Most of these data portals are being updated frequently, and the total number of available data is increasing quickly with the above-mentioned HTS programs. In 2013, the total toxicity data pool contained over 70 million compounds and around 1 million assays.54 Figure 1 shows the increase of the numbers of compound and bioassay records in PubChem since 2008.54–64 From 2008 to 2018, the number of compounds in PubChem increased over three-fold from 25.6 million54 to 96.5 million.56 Similarly, the number of bioassay records increased from approximately 150054 to over 1 million.56
Table 1.
Selected Publically Available Big Data Repositories
| database | sizea | data type | web access |
|---|---|---|---|
| ACToR37,38 | over 800 000 compounds and 500 000 assays | toxicity data (in vitro and in vivo) | https://actor.epa.gov/ |
| CEBS46 | over 11 000 compounds and 8000 studies | gene expression data | https://www.niehs.nih.gov/research/resources/databases/index.cfm |
| ChEMBL47 | 1.1 million bioassays, 1.8 million compounds, over 15 million activities | literature data for binding, function, and toxicity of drugs and drug-like compounds | https://www.ebi.ac.uk/chembl/ |
| Connectivity Map48,49 | about 1300 compounds and 7000 genes | gene expression data | https://portals.broadinstitute.org/cmap/ |
| CTD50 | over 14 000 compounds, 42 000 genes, 6000 diseases | relationships among compounds, genes, and diseases | https://ctdbase.org/ |
| DrugMatrix51 | about 600 drug molecules and 10 000 genes | gene expression data | https://ntp.niehs.nih.gov/results/toxfx/index.html |
| GEO52,53 | over 4300 subdata sets | microarray, next-generation sequencing and other forms of high-throughput functional genomics data | https://www.ncbi.nlm.nih.gov/geo/ |
| PubChem13,14,76 | over 96 million compounds, 1 million bioassays, and 13 billion data points | toxicology, genomics, pharmacology, and literature data | https://pubchem.ncbi.nlm.nih.gov/ |
| REACH16,39–42 | 21 405 unique substances with information from 89 905 dossiers | data submitted in european union chemical legislation | https://echa.europa.eu/information-on-chemicals/registered-substances/ |
| RepDose43 | 364 chemicals investigated in 1017 studies, which resulted in 6002 specific effects | repeat-dose study data for dog, mouse, and rat | https://repdose.item.fraunhofer.de/ |
| SEURAT44 | over 5500 cosmetic-type compounds in the current COSMOS database web portal | animal toxicity data | http://www.seurat-1.eu/ |
| ToxNET45 | over 50 000 environmental compounds from 16 different resources | toxicity data (in vitro and in vivo) | https://toxnet.nlm.nih.gov/ |
On the basis of live web counts or most recent literature articles as of October 2018; ACToR, Aggregated Computational Toxicology Resource; CTD, Comparative Toxicogenomics Database; CEBS, Chemical Effects in Biological Systems; GEO, Gene Expression Omnibus; REACH, Registration, Evaluation, Authorization, and Restriction of Chemicals; SEURAT, Safety Evaluation Ultimately Replacing Animal Testing; ToxNET, Toxicology Data Network.
Figure 1.
Increase of the number of (a) compound and (b) bioassay records in PubChem in the recent ten year period (from September 2008 to September 2018).
MODELING CHALLENGES CREATED BY BIG DATA: THE FOUR “V”S
The available big data for chemical toxicity brings new challenges to the future computational toxicology studies.36,65 As the “big data” concept suggests, the volume of data is a critical characteristic. The nature of data that is relevant to various toxicity end points creates a large data volume.1,15,65 These data stem from information obtained from compounds and original testing protocols including chemical information,13,14 physicochemical properties, in vitro data,16,36–42 in vivo data,16,37–45,47 and various–omics data46–53 (Table 1). For example, the current PubChem bioassay database has around 240 million bioactivities as 30 gigabytes of Extensive Markup Language (XML) files. It is not feasible to apply traditional computational approaches or even Personal Computers (PCs) to deal with data with this kind of volume for modeling purposes. The recent progress of computer hardware, especially the application of Graphics Processing Unit (GPU),67 makes it possible to deal with toxicity data with significant volume.
The progress of testing technology determines the velocity of big data. In the 1990s, combinatorial chemistry began to progress rapidly, creating large chemical libraries for screening in drug discovery.68 The advancement of HTS protocols in the past decades makes the screening of these large chemical libraries (i.e., over one million compounds) feasible.69,70 Automatic data analysis and the application of robots to replace humans in the testing procedures considerably lower the cost of testing a compound and rapidly grow the current big data sources.32 As a result, a substantial number of compounds have been tested against many assays. Table 2 shows the 20 compounds obtained from the Tox21 program with the most active responses in the PubChem bioassay database14 (accessed December 2018). For example, doxorubicin (CAS 25316–40–9), a drug that is used to treat cancer by killing cancer cells, showed 4452 active responses (Table 2). Vorinostat (CAS 149647–78–9), which is used to treat T-cell lymphoma that persists after treatment with other drugs, showed 4278 active responses (Table 2). Other well-characterized compounds, such as drugs and well-known pesticides, have similarly prolific response information available.
Table 2.
Twenty Tox21 Compounds with the Most Active Responses in PubChem Bioassays
| chemical | CAS | number of active responses | number of inactive responses |
|---|---|---|---|
| Doxorubicin | 25316-40-9 | 4452 | 119 |
| Vorinostat | 149647-78-9 | 4278 | 760 |
| Paclitaxel | 33069-62-4 | 3043 | 801 |
| Colchicine | 64-86-8 | 2043 | 1581 |
| Etoposide | 33419-42-0 | 1907 | 525 |
| Fluorouracil | 51-21-8 | 1804 | 1887 |
| Acetazolamide | 59-66-5 | 1794 | 2091 |
| Sunitinib | 341031-54-7 | 1702 | 138 |
| Methotrexate | 59-05-2 | 1687 | 1120 |
| Lestaurtinib | 111358-88-4 | 1414 | 35 |
| Gefitinib | 184475-35-2 | 1379 | 661 |
| Diazepam | 439-14-5 | 1320 | 628 |
| Haloperidol | 52-86-8 | 1309 | 1820 |
| Bortezomib | 179324-69-7 | 1276 | 212 |
| Zidovudine | 30516-87-1 | 1251 | 1964 |
| Clozapine | 5786-21-0 | 1204 | 1687 |
| Efavirenz | 154598-52-4 | 1184 | 537 |
| Dasatinib | 302962-49-8 | 1078 | 380 |
| Mitomycin | 50-07-7 | 1048 | 664 |
| Nicotine | 54-11-5 | 983 | 1283 |
Traditional modeling studies, usually using small in-house data sets for modeling purposes, often had a risk of overfitting and made poor predictions to new compounds.2 The modeling community expected models to improve with more available data used for modeling purposes, thereby increasing knowledge about activity cliffs71 and decreasing the chance of overfitting.72 However, when using large data sets for modeling purposes, traditional machine learning approaches usually have flaws such as extended computational time and memory requirements that require adaptation of commonly used algorithms.73 As a potential solution, deep learning with neural networks using GPUs might be more suitable for big data processing.74
Additionally, the variety of big data brings new challenges to modeling procedures. Traditional modeling studies only deal with one object (i.e., a toxicity end point) using one type of attributes (i.e., chemical descriptors). However, existing big data repositories (e.g., PubChem) contain a diverse variety of information for compounds of interest, such as quantitative data obtained directly from assays and qualitative data as the screening read-out, which requires different data processing techniques. Integrating and curating data with high variety requires advanced artificial intelligent approaches.75
Each source of big data contains a certain degree of data variability. PubChem, for example, contains data deposited by different sources including academia, pharmaceutical companies, government agencies, chemical vendors, screening centers, and journal publishers.13–15,70,76 The information obtained from each of these data sources is not consistent across assays or compounds, which creates an inherent variability that creates challenges in the following modeling procedure. For example, data generated from a Tox21 quantitative HTS (qHTS) assay to measure genotoxicity induced by small molecules in human embryonic kidney cells77 exists in PubChem twice as (1) original data (AID 651632) and (2) conclusions by counting cytotoxicity (AID 720516). Under this condition, automatic data mining tools need to be able to distinguish this difference.
Furthermore, inconsistencies may also arise due to inaccurate chemical structures16,78–80 and inherent experimental errors1,80,81 resulting from data quality control (QC) issues of various experimental across sources. When aggregating data from multiple sources, it is common to encounter different representations to represent the same compounds (e.g., implicit versus explicit hydrogens and tautomeric forms). The quality of experimental data is also likely to vary across sources due to differences in protocols, compound purities, and other experimental errors. For example, Luechtefeld et al. reported that animal toxicity data obtained from various sources for the same compounds have consistency ranging from 70% to 90%, depending on the nature of testing protocols.39,42 Therefore, the data curation of both chemical structures and experimental data is critical before using big data for the computational modeling procedure.79,80,82,83 Because of the size of public data sets, automated curation workflows, such as those described in previous publications,79,80,82,84 are necessary prior to modeling.
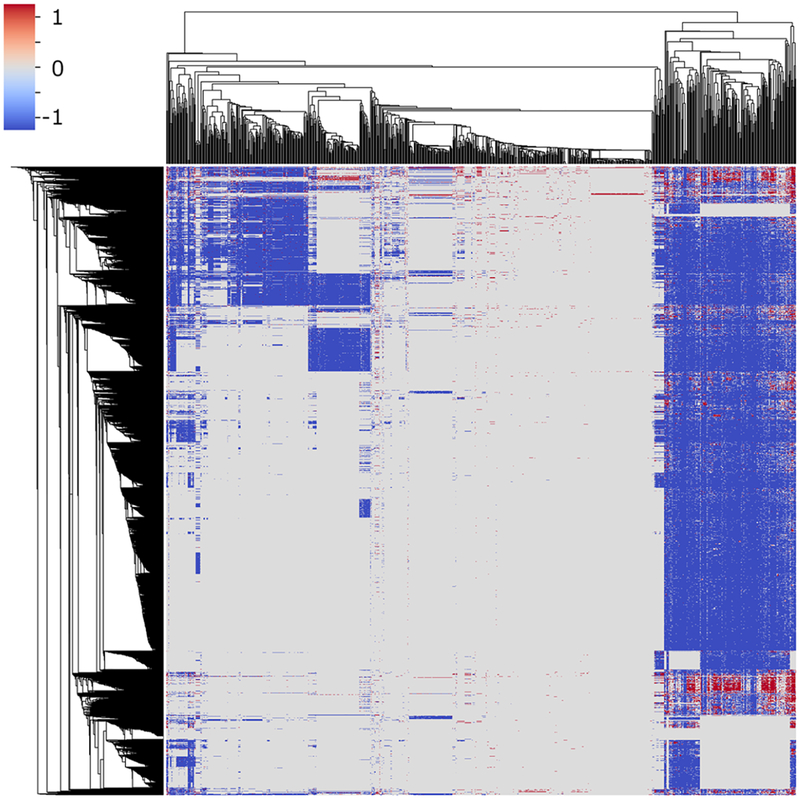
Another data variability issue is due to the complex, disorganized nature of public data and unbalanced distribution of HTS testing results (i.e., many more inactive results than actives). Although a wealth of data exists, there are many data gaps for compounds of interest since no compound has been tested against all assays, and many tests returned inconclusive results. For most existing data, a bias exists toward inactive responses due to the nature of HTS assays. For example, searching the recent PubChem database for 8367 Tox21 compounds yielded 812 assays that have at least 25 active results within these compounds including assays carried out by the Tox21 program and other sources (accessed in November 2018) (Figure 2). There are approximately 6.8 million data points in this bioprofile. However, the ratio of active versus inactive results is 1:11 (2% vs 22%) and the remaining data (76%) represent results from which no conclusion can be made(i.e., either “inconclusive” or “untested”). It is understandable that inactive results are much less informative than active results to determine chemical toxicity. In this situation, novel modeling approaches are needed to deal with missing data such as the method described by Zhang et al.6 and biased data,including cost-sensitive learning,85 under-sampling,85 and oversampling86,87 algorithms.
Figure 2.
Bioprofile of 8367 Tox21 compounds represented by data from 812 PubChem assays. Active results (1) were represented by red; inactive results (−1) were represented by blue; and inconclusives or untested results (0) were represented by gray.
DATA-DRIVEN COMPUTATIONAL TOXICOLOGY MODELING
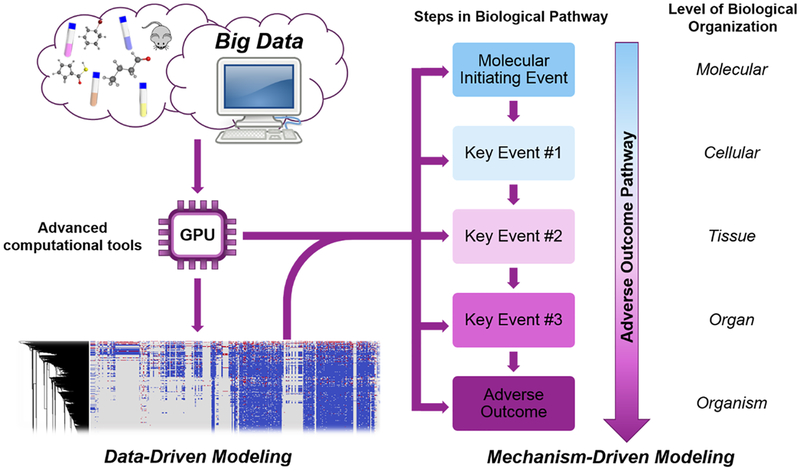
Despite the challenges posed by big data in computational toxicological modeling, the advancement of data-driven technology and development of computational tools to overcome the challenges of the four “V”s create new opportunities for existing model improvement and novel model development (Figure 3). For example, some early developed data mining tools, such as Chem2BioRDF88 and HTS Navigator,89 can link various public data sources to target compounds. Additionally, some data sharing portals have relevant data mining tools, such as rpubchem90 and ToxCast pipeline (“tcpl”),91 which exist to simplify and optimize the parsing and collection of data from PubChem90 and the ToxCast database,91 respectively. The recently developed online tools REACHacross2,16 and the Chemical In Vitro–In Vivo Profiling portal (CIIPro)92 provide new methods to extract data from the REACH and PubChem databases automatically. Additionally, online tools such as Chembench93 and the Chemistry at Harvard Macromolecular Mechanics web-user interface (CHARM-Ming)94 are available to streamline the development and distribution of curated toxicity data and QSAR models.
Figure 3.
General workflow for construction of data-driven and mechanism-driven models for chemical toxicity.
In 2014, the National Center for Advancing Translational Sciences (NCATS) launched a challenge for the development of computational models of nuclear receptor and stress response pathways using HTS data generated by the Tox21 program.95 This challenge inspired the creation of around 400 models using a variety of different modeling techniques based on 8043 compounds, which have been tested against 12 HTS assays.95 All the models were used to predict an external test set of 296 new compounds, which were experimentally tested using the same protocols but kept aside until all models were submitted to NCATS. The top model, which has the best prediction accuracy of these new compounds, employed a neural network approach.96 The high performance of the resulting ensemble model demonstrates the potential of neural networks for the improvement of big data models. In 2017, another similar initiative of big toxicity data modeling was organized by the National Toxicology Program Interagency Center for the Evaluation of Alternative Toxicological Methods (NICEATM) and the Interagency Coordinating Committee on the Validation of Alternative Methods (ICCVAM).97 Thirty-two international research groups submitted models using deep learning, classic QSAR, and cluster-based methods. These models were developed based on 8994 compound training set tested in rodent studies for acute toxicity. Consensus predictions were made for a large set of 40 000 compounds of environmental interest as a resource for the toxicology community.
In recent studies, neural networks, as a popular artificial intelligent approach, showed advantages to deal with large data sets. In 2017, Xu et al. reported three neural network models developed to predict acute oral toxicity end points based on a training set of 8080 compounds.98 All three models (i.e., a regression model for LD50 values, a multiclassification model for US EPA hazard categories, and a multitask model to simultaneously predict both of these end points) simultaneously outperformed previously reported models for these end points. Wen et al. also reported a deep learning model developed to predict interactions between drugs and their biological targets based on 15 524 drug–target pairs obtained from the DrugBank database.99 This model employed a pretraining feature extraction step to predict whether specific drug-target pairs will interact and overall outperformed classic QSAR approaches. The high performance of these models demonstrates not only the advantages of using neural networks to model large data sets, but also to advance feature selections. On the other hand, there is a study that showed that neural network models are not better than traditional models using machine learning approaches.100 Currently, there is still no universal criterion to select modeling approaches for big data sets.
Read-across was initially introduced as a technique to fill toxicity data gaps by making predictions based on similar compounds.101 Traditionally, read-across relied only on chemical similarity calculations. In some recent studies, extra parameters, such as physicochemical properties, biological interactions (e.g., metabolism potential), including QSAR predictions, were also used to identify similar compounds.102,103 However, in the big data era, the inclusion of comprehensive toxicity data in the read-across study may improve model quality.15,16,104,105 Luechtefeld et al. recently described a method for automating the read-across process using read-across structure–activity relationship (RASAR) models.105 The RASAR models developed in this study were based on the REACH database consisting of over 10 000 unique compounds with various toxicity end points, making use of hazard classifications rather than the raw data from in vivo tests. According to this study, due to the integration of large collections of toxicity data and the application associated with new modeling approaches, such as automatic feature selection, the resulted RASAR models showed better average sensitivity than the reproducibility of experimental animal tests, which indicated potential abilities to overcome experimental errors and data inconsistencies by computational modeling approaches.
PREDICTIONS AND INTERPRETATIONS OF NEW TOXICANTS BY MECHANISM-DRIVEN MODELING
In 2007, the National Research Council (NRC) laid out the framework for risk assessment using modern techniques, urgently calling for mechanistic rather than empirical interpretation.12 Traditional QSAR models often perform as “black boxes”, which provide toxicity predictions without clear mechanistic interpretations.19 Although the neural network modeling showed certain advantages when dealing with big toxicity data, the resulted neural network models still cannot resolve the above challenge. In vitro assays often investigate mechanistically relevant information on toxicants. However, one or few assays cannot represent the complexity of whole organisms, and the results obtained from in vitro and in vivo tests always have obscure relationships, making in vitro–in vivo extrapolation (IVIVE) challenging.106
Mechanism-driven modeling, initiated and advanced by the concept of the adverse outcome pathway (AOP), allows for mechanistic extrapolation of toxicity evaluations of new compounds, filling a critical need of applying alternatives for regulatory toxicology studies.20 An AOP starts with a molecular feature (e.g., a chemical fragment), which indicates potential interactions with biomolecules such as receptors. This molecular initiating event (MIE) that triggers a cascade of measurable key events at the cellular level, and lead to tissue, organ, and eventually in vivo organism level adverse outcomes20 (Figure 3). The identification and organization of MIEs and key events in a pathway that leads to an adverse outcome define the associated toxicity mechanisms of interest for risk assessments. Mechanism-based assays outcomes can be used within this pathway to systematically assess whether a compound is likely to induce the target adverse outcome.107 Currently, AOPs are being developed for various types of toxicities such as acute inhalation toxicity,108 neurotoxicity,109–111 skin sensitization,112–114 estrogen receptor bindings,115,116 forestomach tumors not induced by genotoxic events,117 and drug-induced cholestatic liver injury.118
One of the major research goals of toxicology HTS programs, such as ToxCast and Tox21, is to perform mechanism-driven computational toxicology modeling, which presents a practical way to increase the quality of IVIVE by employing comprehensive testing batteries consisting of associated in vitro assays related to animal toxicity end points.1 These programs generate a large amount of mechanistic data that paves the way for AOP modeling that are more interpretable than traditional computational toxicology studies, which are always questioned as “black boxes.” Carefully considering the biological relevance of the experimental data selected, including associated biological pathway information, for modeling (i.e., HTS assay measurements and readouts) the target animal toxicity end point of interest is to integrate the AOP concept into the development of computational models with both high prediction accuracy and a meaningful biological interpretations. Therefore, the current critical features of resulting AOP models are (1) biological relevance of data to target toxicity; (2) computational approaches to identify/organize mechanistic assays; and (3) both predictive and interpretable pathway models.
The current literature also documents the results from profiling of ToxCast and Tox21 assay data using computational clustering techniques to elucidate previously unknown compound–receptor interactions, pathway perturbations, and toxicity mechanisms.119–123 One such profiling effort was described by Sipes et al. in 2013.119 This study clustered 976 compounds from 330 ToxCast Phase I and II bioassays based on chemical structure and bioassay responses, which led to the identification of possible modes of action of compounds. For example, a pharmaceutical compound Anthralin (CAS 1143–38–0) that has a therapeutic use for the treatment of psoriasis with unknown mechanism of action was identified to show active responses in the same assays as a known inhibitor of inflammation, tannic acid. This connection gives insight into the possible mode of action of Anthralin and demonstrates the value of data generated from the ToxCast program in identifying previously unknown mechanistic information for target compounds.
The ToxCast initiative and Tox21 program have also inspired the creation of mechanistic models for developmental toxicity,124,125 estrogen receptor activity,126–128 and acute oral toxicity129 that incorporate mechanistically relevant HTS assay data by computational models into pathways leading to adverse outcomes. For example, Browne et al. developed a computational model that incorporates HTS data from 18 ToxCast assays that comprise an adverse outcome pathway leading to endocrine disruption.127 The authors of this study also demonstrated a generalizable performance-based validation procedure to evaluate the robustness of a computational AOP model for regulatory use. To be considered as a viable alternative for regulatory evaluation purposes, computational models must perform equivalently or better than the existing approved protocols. This computational endocrine disruption AOP model was validated by predicting a set of in vitro reference chemicals identified by ICCVAM and in vivo reference chemicals curated through literature review.75 The computational model pre-dictions of the 42 in vivo reference chemicals with at least two independent concordant results (active or inactive) from guideline-like uterotrophic studies showed 84% accuracy. A comparison of the computational predictions with the results from in vivo protocols identified false negative, which showed activity in multiple independent in vivo tests but inactivity in the 18 ToxCast assays, potentially due to its volatility.
The strong potential for computational approaches to support risk assessment grounded in mechanistic interpretation has inspired the creation of other mechanism-based studies. These studies, similar to AOP models developed by using mechanistic ToxCast and Tox21 assays, can predict chemical toxicity by interpretable results. However, instead of manually selected assays to be integrated into pathways, these studies relied on computational approaches to prioritize useful biological data, which are suitable for big data modeling. For example, Virtual AOP (vAOP) models were developed by using the currently available big data for hepatotoxicity.130 In this study, an automatic profiling tool that can extract bioassay data from PubChem was used to identify assays relevant to hepatotoxicity and oxidative stress.92,130 Data from several PubChem assays can be combined to predict hepatotoxicity for compounds with specific structural alerts. The resulting vAOP models provided insight into possible new mechanisms leading to hepatotoxicity.130 The identified vAOP contained two chemical fragments as MIEs and four PubChem assays (AIDs 686978, 743067, 743140, and 743202), which are all relevant to oxidative stress, such that if a new compound contains one of these MIEs and has an active response in at least one of the four assays, it is predicted to be hepatotoxic by inducing oxidative stress.
Luechtefeld et al. reported a procedure for the recursive importance-based elimination of chemical and biological features that are irrelevant to target toxicity.131 The application of this technique involves ranking features based on relative importance to identify the assays and chemical fragments that contribute the most critical information to a resulting model of an in vivo toxicity end point. In this study, they identified in vitro assays and chemical fragments of mechanistic significance to skin sensitization and then used this information to train models that incorporate dose–response data, which showed an advantage when compared to models trained without these data. The success of this modeling process was validated by better predictivity and mechanism interpretations.
Computational techniques and models based on chemical structures, such as structural alerts, also could advance traditional QSAR studies by predicting toxicity mechanisms for large data sets.107,132 For example, recently, there have been reports of modeling studies to predict MIEs relevant to hepatic steatosis.133,134 Mellor et al. evaluated binding interactions of 12 713 compounds in the ChEMBL database with nuclear receptor structure files in Protein Data Bank to develop a basic alert-based workflow to identify compounds that may bind to nuclear receptors and induce hepatic steatosis.133 Another study by Gadaleta et al. involved the development of QSAR models to predict compound activity in ToxCast assays that are relevant to MIEs that lead to hepatic steatosis.134
OTHER AREAS OF COMPUTATIONAL TOXICOLOGY IN THE BIG DATA ERA
A critical aspect of mechanism-based toxicity evaluation is the incorporation of toxicokinetic information on compounds of interest to predict dose-dependent Effects of compounds. For example, Bhhatarai et al. recently reported a modeling study of acute toxicity, which incorporated simulations of absorption and metabolism into the modeling process.129 Strope et al. also reported a modeling study that resulted in an ionization constant (pKa) model for a set of 32 413 chemicals.135 The applicability of this model was evaluated by using the pKa predictions to estimate distribution ratio into tissues for 22 compounds with steady-state volume of distribution data. Modeling studies that incorporate toxicokinetic information are becoming applicable with tools such as the High-Throughput Toxicokinetics (“httk”) package in R that was designed to make use of the data from programs such as ToxCast and Tox21.136
Toxicogenomics uses techniques such as proteomics, metabolomics, and genetic sequencing to study the toxicity of compounds and provides insight into how cells exposed to a toxic chemical express genes, proteins, and metabolites, which yields critical information for elucidating and understanding toxicity pathways.17,137 The toxicogenomics data complement the results of in vitro assays that focus on interactions with and activation of nuclear receptors and specific cellular stress responses.138 In the current big data era, the toxicogenomics data landscape continues to grow. For example, in 2008, the CTD contained 116 067 compound–gene interactions.66 By 2016, this number increased more than 10-fold to 1 379 105 compound–gene interactions.50 A collaboration among Agilent, Inc., Brown University, Georgetown University, the Hamner Institute, the Johns Hopkins Center for Alternatives to Animal Testing (CAAT), and the US EPA led the Human Toxome project that also aims to generate omics data, with an end goal of developing a process to map and evaluate the specific molecular mechanisms that underlie AOPs.139 As toxicogenomics technologies continue to progress and molecular mechanisms become well-understood, it will become feasible to assess differences in chemical toxicity pathways that may arise due to the genetic variation that is inherent among individuals.
CONCLUSIONS
Computational modeling is a promising alternative method to replace, reduce, and refine traditional animal models for chemical toxicity evaluations, especially in the current big data era. As big data repositories continue to grow at rapid velocity and new techniques to deal with big data sets are being developed, computational models become applicable to large chemical space analysis, diverse biological data optimization, and complex mechanism studies. This innovative movement will allow not only for the predictions of new compounds, but also for toxicity mechanism illustrations of potential toxicants. This currently growing big toxicity data landscape and the advancements in modeling technique developments to handle the wealth of toxicity information available together create a new direction that is optimal for the integration of computational models into the mechanism-based chemical risk assessments, which are urgently required by regulatory agencies.
Funding
This work was partially supported by the National Institute of Environmental Health Sciences [Grant No. R15ES023148], the Colgate-Palmolive Grant for Alternative Research, and the Johns Hopkins Center for Alternatives to Animal Testing (CAAT) grant.
ABBREVIATIONS
- ACToR
Aggregated Computational Toxicology Resource
- AID
BioAssay Identifier
- AOP
Adverse Outcome Pathway
- BD2K
Big Data to Knowledge
- CAAT
Center for Alternatives to Animal Testing
- CAS
Chemical Abstracts Service
- CEBS
Chemical Effects in Biological Systems
- CHARMMing
Chemistry at Harvard Macromolecular Mechanics
- CIIPro
Chemical In Vitro–In Vivo Profiling
- CTD
Comparative Toxicogenomics Database
- GPU
Graphics Processing Unit
- GPU
Graphics Processing Unit
- HTS
High-Throughput Screening
- ICCVAM
Interagency Coordinating Committee on the Validation of Alternative Methods
- IVIVE
In Vitro–In Vivo Extrapolation
- LCSA
Frank R. Lautenberg Chemical Safety for the 21st Century Act
- LD50
Median Lethal Dose
- MIE
Molecular Initiating Event
- NCATS
National Center for Advancing Translational Sciences
- NCCT
National Center for Computational Toxicology
- NCGC
NIH Chemical Genomics Center
- NICEATM
National Toxicology Program Interagency Center for the Evaluation of Alternative Toxicological Methods
- NIEHS
National Institute of Environmental Health Sciences
- NIH
National Institutes of Health
- NRC
National Research Council
- NTP
National Toxicology Program
- OECD
Organization for Economic Cooperation and Development
- PC
Personal Computer
- pKa
Ionization Constant
- QC
Quality Control
- QSAR
Quantitative Structure–Activity Relationship
- RASAR
Read-Across Structure–Activity Relationship
- REACH
Registration, Evaluation, Authorization and Restriction of Chemicals
- SEURAT
Safety Evaluation Ultimately Replacing Animal Testing
- TGx
Toxicogenomics
- Tox21
Toxicity Testing in the 21st Century
- ToxCast
Toxicity Forecaster
- ToxNET
Toxicology Data Network
- TSCA
Toxic Substances Control Act of 1976
- US EPA
United States Environmental Protection Agency
- vAOP
Virtual Adverse Outcome Pathway
- XML
Extensive Markup Language
Biographies
Heather L. Ciallella currently is a Ph.D. student in CCIB under the mentorship of Dr. Hao Zhu, where her research focuses on the applications of deep learning algorithms to chemical toxicity predictions.
Hao Zhu is an Associate Professor in the Chemistry Department and Center for Computational and Integrative Biology (CCIB) at Rutgers, The State University of New Jersey in Camden. He received his Ph.D. in Computational Chemistry from Case Western Reserve University in 2002. Dr. Zhu has authored or coauthored over 60 peer-reviewed publications and book chapters in the applications of cheminformatics to chemical toxicity assessments, computer-aided drug discovery, and rational nanomaterial design.
Footnotes
The authors declare no competing financial interest.
REFERENCES
- (1).Zhu H, Zhang J, Kim MT, Boison A, Sedykh A, and Moran K (2014) Big Data in Chemical Toxicity Research: The Use of High-Throughput Screening Assays to Identify Potential Toxicants. Chem. Res. Toxicol 27 (10), 1643–1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Luechtefeld T, Rowlands C, and Hartung T (2018) Big-Data and Machine Learning to Revamp Computational Toxicology and Its Use in Risk Assessment. Toxicol. Res. (Cambridge, U. K.) 7, 732–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Stouch TR, Kenyon JR, Johnson SR, Chen XQ, Doweyko A, and Li Y (2003) In Silico ADME/Tox: Why Models Fail. J. Comput.-Aided Mol. Des 17 (2–4), 83–92. [DOI] [PubMed] [Google Scholar]
- (4).Maggiora GM (2006) On Outliers and Activity Cliffs - Why QSAR Often Disappoints. J. Chem. Inf. Model 46 (4), 1535. [DOI] [PubMed] [Google Scholar]
- (5).Dearden JC, Cronin MTD, and Kaiser KLE (2009) How Not to Develop a Quantitative Structure-Activity or Structure-Property Relationship (QSAR/QSPR). SAR QSAR Environ. Res 20 (3–4), 241–266. [DOI] [PubMed] [Google Scholar]
- (6).Zhang J, Hsieh JH, and Zhu H (2014) Profiling Animal Toxicants by Automatically Mining Public Bioassay Data: A Big Data Approach for Computational Toxicology. PLoS One 9 (6), e99863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Wang W, Kim MT, Sedykh A, and Zhu H (2015) Developing Enhanced Blood-Brain Barrier Permeability Models: Integrating External Bio-Assay Data in QSAR Modeling. Pharm. Res 32 (9), 3055–3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Piersma AH, Burgdorf T, Louekari K, Desprez B, Taalman R, Landsiedel R, Barroso J, Rogiers V, Eskes C, Oelgeschläger M,et al. (2018) Workshop on Acceleration of the Validation and Regulatory Acceptance of Alternative Methods and Implementation of Testing Strategies. Toxicol. In Vitro 50, 62–74. [DOI] [PubMed] [Google Scholar]
- (9).Schiffelers MJWA, Blaauboer BJ, Bakker WE, Beken S, Hendriksen CFM, Koëter HBWM, and Krul C (2014) Regulatory Acceptance and Use of 3R Models for Pharmaceuticals and Chemicals: Expert Opinions on the State of Affairs and the Way Forward. Regul. Toxicol. Pharmacol 69 (1), 41–48. [DOI] [PubMed] [Google Scholar]
- (10).Frank R Lautenberg Chemical Safety for the 21st Century Act. Public Law 114–182, 2016. [Google Scholar]
- (11).Toxic Substances Control Act. U.S. Code, Section 2601, Title 15, 1976. [Google Scholar]
- (12).National Research Council. Toxicity Testing in the 21st Century: A Vision and a Strategy; National Academies Press: Washington, D.C., 2007. [Google Scholar]
- (13).Wang Y, Xiao J, Suzek TO, Zhang J, Wang J, and Bryant SH (2009) PubChem: A Public Information System for Analyzing Bioactivities of Small Molecules. Nucleic Acids Res 37, W623–W633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Wang Y, Bolton E, Dracheva S, Karapetyan K, Shoemaker BA, Suzek TO, Wang J, Xiao J, Zhang J, and Bryant SH (2010) An Overview of the PubChem BioAssay Resource. Nucleic Acids Res 38, D255–D266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Zhu H, Bouhifd M, Donley E, Egnash L, Kleinstreuer N, Kroese ED, Liu Z, Luechtefeld T, Palmer J, Pamies D, et al. (2016) Supporting Read-across Using Biological Data. ALTEX 33 (2), 167–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Hartung T (2016) Making Big Sense from Big Data in Toxicology by Read-Across. ALTEX 33 (2), 83–93. [DOI] [PubMed] [Google Scholar]
- (17).Organization for Economic Co-operation and Development. Guidance Document for the Use of Adverse Outcome Pathways in Developing Integrated Approaches to Testing and Assessment (IATA) Ser. Test. Assess 2016, 260. [Google Scholar]
- (18).Sjöberg J, Zhang Q, Ljung L, Benveniste A, Deylon B, Glorennee P-Y, Hjalmarsson H, and Juditsky A (1995) Nonlinear Black-Box Modeling in System Identification: A Unified Overview. Automatica 31 (12), 1691–1724. [Google Scholar]
- (19).Guha R (2008) On the Interpretation and Interpretability of Quantitative Structure-Activity Relationship Models. J. Comput.-Aided Mol. Des 22, 857–871. [DOI] [PubMed] [Google Scholar]
- (20).Ankley GT, Bennett RS, Erickson RJ, Hoff DJ, Hornung MW, Johnson RD, Mount DR, Nichols JW, Russom CL, Schmieder PK, et al. (2010) Adverse Outcome Pathways: A Conceptual Framework to Support Ecotoxicology Research and Risk Assessment. Environ. Toxicol. Chem 29 (3), 730–741. [DOI] [PubMed] [Google Scholar]
- (21).Wittwehr C, Aladjov H, Ankley G, Byrne HJ, de Knecht J, Heinzle E, Klambauer G, Landesmann B, Luijten M, MacKay C, et al. (2017) How Adverse Outcome Pathways Can Aid the Development and Use of Computational Prediction Models for Regulatory Toxicology. Toxicol. Sci 155 (2), 326–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Clark M, and Steger-Hartmann T (2018) A Big Data Approach to the Concordance of the Toxicity of Pharmaceuticals in Animals and Humans. Regul. Toxicol. Pharmacol 96, 94–105. [DOI] [PubMed] [Google Scholar]
- (23).Gandomi A, and Haider M (2015) Beyond the Hype: Big Data Concepts, Methods, and Analytics. Int. J. Inf. Manage 35 (2), 137–144. [Google Scholar]
- (24).Schadt EE, Linderman MD, Sorenson J, Lee L, and Nolan GP (2011) Cloud and Heterogeneous Computing Solutions Exist Today for the Emerging Big Data Problems in Biology. Nat. Rev. Genet 12, 224. [DOI] [PubMed] [Google Scholar]
- (25).Liu W, Schmidt B, Voss G, and Müller-Wittig W (2007) Streaming Algorithms for Biological Sequence Alignment on GPUs. IEEE Trans. Parallel Distrib. Syst 18 (9), 1270–1281. [Google Scholar]
- (26).Charikar M, O’Callaghan L, and Panigrahy R (2003) Better Streaming Algorithms for Clustering Problems. Proc. thirty-fifth ACM Symp. Theory Comput. - STOC’ 03, 30–39. [Google Scholar]
- (27).National Institutes of Health. Big Data to Knowledge; National Institutes of Health, 2018. https://commonfund.nih.gov/bd2k (accessed Nov 10, 2018).
- (28).Margolis R, Derr L, Dunn M, Huerta M, Larkin J, Sheehan J, Guyer M, and Green ED (2014) The National Institutes of Health’s Big Data to Knowledge (BD2K) Initiative: Capitalizing on Biomedical Big Data. J. Am. Med. Informatics Assoc 21 (6), 957–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Dix DJ, Houck KA, Martin MT, Richard AM, Setzer RW, and Kavlock RJ (2007) The ToxCast Program for Prioritizing Toxicity Testing of Environmental Chemicals. Toxicol. Sci 95 (1), 5–12. [DOI] [PubMed] [Google Scholar]
- (30).Judson RS, Houck KA, Kavlock RJ, Knudsen TB, Martin MT, Mortensen HM, Reif DM, Rotroff DM, Shah I, Richard AM, et al. (2010) In Vitro Screening of Environmental Chemicals for Targeted Testing Prioritization: The Toxcast Project. Environ. Health Perspect 118 (4), 485–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Kavlock R, Chandler K, Houck K, Hunter S, Judson R, Kleinstreuer N, Knudsen T, Martin M, Padilla S, Reif D, et al. (2012) Update on EPA’s ToxCast Program: Providing High Throughput Decision Support Tools for Chemical Risk Management. Chem. Res. Toxicol 25 (7), 1287–1302. [DOI] [PubMed] [Google Scholar]
- (32).Attene-Ramos MS, Miller N, Huang R, Michael S, Itkin M, Kavlock RJ, Austin CP, Shinn P, Simeonov A, Tice RR, et al. (2013) The Tox21 Robotic Platform for the Assessment of Environ-mental Chemicals – from Vision to Reality. Drug Discovery Today 18 (15–16), 716–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Thomas RS, Paules RS, Simeonov A, Fitzpatrick SC, Crofton KM, Casey WM, and Mendrick DL (2018) The US Federal Tox21 Program: A Strategic and Operational Plan for Continued Leadership. ALTEX 35 (2), 163–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Shukla SJ, Huang R, Austin CP, and Xia M (2010) The Future of Toxicity Testing: A Focus on in Vitro Methods Using a Quantitative High-Throughput Screening Platform. Drug Discovery Today 15 (23–24), 997–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Hsu C-W, Huang R, Attene-Ramos MS, Austin CP, Simeonov A, and Xia M (2017) Advances in high-throughput screening technology for toxicology. Int. J. Risk Assess. Manage 20, 109. [Google Scholar]
- (36).Zhao L, and Zhu H Big Data in Computational Toxicology: Challenges and Opportunities. In Computational Toxicology; Ekins S, Ed.; John Wiley & Sons, Inc.: Hoboken, NJ, 2018; pp 291–312. [Google Scholar]
- (37).Judson RS, Martin MT, Egeghy P, Gangwal S, Reif DM, Kothiya P, Wolf M, Cathey T, Transue T, Smith D, et al. (2012) Aggregating Data for Computational Toxicology Applications: The U.S. Environmental Protection Agency (EPA) Aggregated Computational Toxicology Resource (ACToR) System. Int. J. Mol. Sci 13 (2), 1805–1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Judson R, Richard A, Dix D, Houck K, Elloumi F, Martin M, Cathey T, Transue TR, Spencer R, and Wolf M (2008) ACToR - Aggregated Computational Toxicology Resource. Toxicol. Appl. Pharmacol 233 (1), 7–13. [DOI] [PubMed] [Google Scholar]
- (39).Luechtefeld T, Maertens A, Russo DP, Rovida C, Zhu H, and Hartung T (2016) Analysis of Publically Available Skin Sensitization Data from REACH Registrations 2008–2014. ALTEX 33 (2), 135–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Luechtefeld T, Maertens A, Russo DP, Rovida C, Zhu H, and Hartung T (2016) Analysis of Public Oral Toxicity Data from REACH Registrations 2008–2014. ALTEX 33 (2), 111–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Luechtefeld T, Maertens A, Russo DP, Rovida C, Zhu H, and Hartung T (2016) Global Analysis of Publicly Available Safety Data for 9,801 Substances Registered under REACH from 2008–2014. ALTEX 33 (2), 95–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Luechtefeld T, Maertens A, Russo DP, Rovida C, Zhu H, and Hartung T (2016) Analysis of Draize Eye Irritation Testing and Its Prediction by Mining Publicly Available 2008–2014 REACH Data. ALTEX 33 (2), 123–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Bitsch A, Jacobi S, Melber C, Wahnschaffe U, Simetska N, and Mangelsdorf I (2006) REPDOSE: A Database on Repeated Dose Toxicity Studies of Commercial Chemicals-A Multifunctional Tool. Regul. Toxicol. Pharmacol 46 (3), 202–210. [DOI] [PubMed] [Google Scholar]
- (44).Vinken M, Pauwels M, Ates G, Vivier M, Vanhaecke T, and Rogiers V (2012) Screening of Repeated Dose Toxicity Data Present in SCC(NF)P/SCCS Safety Evaluations of Cosmetic Ingredients. Arch. Toxicol 86 (3), 405–412. [DOI] [PubMed] [Google Scholar]
- (45).Fonger GC, Stroup D, Thomas PL, and Wexler P (2000) TOXNET: A Computerized Collection of Toxicological and Environ-mental Health Information. Toxicol. Ind. Health 16 (1), 4–6. [DOI] [PubMed] [Google Scholar]
- (46).Lea IA, Gong H, Paleja A, Rashid A, and Fostel J (2017) CEBS: A Comprehensive Annotated Database of Toxicological Data. Nucleic Acids Res 45 (D1), D964–D971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Gaulton A, Bellis LJ, Bento AP, Chambers J, Davies M, Hersey A, Light Y, McGlinchey S, Michalovich D, Al-Lazikani B, et al. (2012) ChEMBL: A Large-Scale Bioactivity Database for Drug Discovery. Nucleic Acids Res 40 (D1), 1100–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Subramanian A, Narayan R, Corsello SM, Peck DD, Natoli TE, Lu X, Gould J, Davis JF, Tubelli AA, Asiedu JK, et al. (2017) A Next Generation Connectivity Map: L1000 Platform and the First 1,000,000 Profiles. Cell 171 (6), 1437–1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Lamb J, Crawford ED, Peck D, Modell JW, Blat IC, Wrobel MJ, Lerner J, Brunet J, Subramanian A, Kenneth N, et al. (2006) The Connectivity Map: Using Gene-Expression Signatures to Connect Small Molecules, Genes, and Disease. Science (Washington, DC, U. S.) 313 (5795), 1929–1935. [DOI] [PubMed] [Google Scholar]
- (50).Davis AP, Grondin CJ, Johnson RJ, Sciaky D, King BL, McMorran R, Wiegers J, Wiegers TC, and Mattingly CJ (2017) The Comparative Toxicogenomics Database: Update 2017. Nucleic Acids Res 45 (D1), D972–D978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Ganter B, Tugendreich S, Pearson CI, Ayanoglu E, Baumhueter S, Bostian KA, Brady L, Browne LJ, Calvin JT, Day GJ, et al. (2005) Development of a Large-Scale Chemogenomics Database to Improve Drug Candidate Selection and to Understand Mechanisms of Chemical Toxicity and Action. J. Biotechnol 119 (3), 219–244. [DOI] [PubMed] [Google Scholar]
- (52).Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, Holko M, et al. (2012) NCBI GEO: Archive for Functional Genomics Data Sets - Update. Nucleic Acids Res 41 (D1), D991–D995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Edgar R, Domrachev M, and Lash AE (2002) Gene Expression Omnibus: NCBI Gene Expression and Hybridization Array Data Repository. Nucleic Acids Res 30 (1), 207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Sayers EW, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V, Church DM, DiCuccio M, Edgar R, Federhen S, et al. (2009) Database Resources of the National Center for Biotechnology Information. Nucleic Acids Res 37 (D1), D5–D15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Sayers EW, Barrett T, Benson DA, Bolton E, Bryant SH, Canese K, Chetvernin V, Church DM, DiCuccio M, Federhen S, et al. (2010) Database Resources of the National Center for Biotechnology Information. Nucleic Acids Res 38 (D1), D5–D16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Sayers EW, Agarwala R, Bolton EE, Brister JR, Canese K, Clark K, Connor R, Fiorini N, Funk K, Hefferon T, et al. (2019) Database Resources of the National Center for Biotechnology Information. Nucleic Acids Res 47 (D1), D23–D28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Sayers EW, Barrett T, Benson DA, Bolton E, Bryant SH, Canese K, Chetvernin V, Church DM, DiCuccio M, Federhen S, et al. (2011) Database Resources of the National Center for Biotechnology Information. Nucleic Acids Res 39 (D1), D38–D51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Sayers EW, Barrett T, Benson DA, Bolton E, Bryant SH, Canese K, Chetvernin V, Church DM, DiCuccio M, Federhen S, et al. (2012) Database Resources of the National Center for Biotechnology Information. Nucleic Acids Res 40 (D1), D13–D25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).NCBI Resource Coordinators (2012) Database Resources of the National Center for Biotechnology Information. Nucleic Acids Res 41 (D1), D8–D20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).NCBI Resource Coordinators (2014) Database Resources of the National Center for Biotechnology Information. Nucleic Acids Res 42 (D1), D7–D17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).NCBI Resource Coordinators (2015) Database Resources of the National Center for Biotechnology Information. Nucleic Acids Res 43 (D1), D6–D17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).NCBI Resource Coordinators (2016) Database Resources of the National Center for Biotechnology Information. Nucleic Acids Res 44 (D1), D7–D19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).NCBI Resource Coordinators (2017) Database Resources of the National Center for Biotechnology Information. Nucleic Acids Res 45 (D1), D12–D17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (64).NCBI Resource Coordinators, et al. (2018) Database Resources of the National Center for Biotechnology Information. Nucleic Acids Res 46 (D1), D8–D13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).Andreu-Perez J, Poon CCY, Merrifield RD, Wong STC, and Yang G-Z (2015) Big Data for Health. IEEE J. Biomed. Heal. Informatics 19 (4), 1193–1208. [DOI] [PubMed] [Google Scholar]
- (66).Davis AP, King BL, Mockus S, Murphy CG, Saraceni-Richards C, Rosenstein M, Wiegers T, and Mattingly CJ (2011) The Comparative Toxicogenomics Database: Update 2011. Nucleic Acids Res 39, D1067–D1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (67).Nickolls J, and Dally WJ (2010) The GPU Computing Era. IEEE Micro 30 (2), 56–69. [Google Scholar]
- (68).Lehn J-M (1999) Dynamic Combinatorial Chemistry and Virtual Combinatorial Libraries. Chem. - Eur. J 5 (9), 2455–2463. [Google Scholar]
- (69).Malo N, Hanley JA, Cerquozzi S, Pelletier J, and Nadon R (2006) Statistical Practice in High-Throughput Screening Data Analysis. Nat. Biotechnol 24 (2), 167–175. [DOI] [PubMed] [Google Scholar]
- (70).Inglese J, Auld DS, Jadhav A, Johnson RL, Simeonov A, Yasgar A, Zheng W, and Austin CP (2006) Quantitative High-Throughput Screening: A Titration-Based Approach That Efficiently Identifies Biological Activities in Large Chemical Libraries. Proc. Natl. Acad. Sci. U. S. A 103 (31), 11473–11478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (71).Hu Y, Maggiora GM, and Bajorath J (2013) Activity Cliffs in PubChem Confirmatory Bioassays Taking Inactive Compounds into Account. J. Comput.-Aided Mol. Des 27 (2), 115–124. [DOI] [PubMed] [Google Scholar]
- (72).Gudivada VN, Baeza-Yates R, and Raghavan VV (2015) Big Data: Promises and Problems. Computer 48 (3), 20–23. [Google Scholar]
- (73).Wang C, Chen M-H, Schifano E, Wu J, and Yan J (2016) Statistical Methods and Computing for Big Data. Stat. Interface 6 (4), 399–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (74).Najafabadi MM, Villanustre F, Khoshgoftaar TM, Seliya N, Wald R, and Muharemagic E (2015) Deep Learning Applications and Challenges in Big Data Analytics. J. Big Data 2, 1. [Google Scholar]
- (75).Kleinstreuer NC, Ceger PC, Allen DG, Strickland J, Chang X, Hamm JT, and Casey WM (2016) A Curated Database of Rodent Uterotrophic Bioactivity. Environ. Health Perspect 124 (5), 556–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (76).About PubChem; PubChem, 2018. https://pubchemdocs.ncbi.nlm.nih.gov/about (accessed Oct 11, 2018).
- (77).Fox JT, Sakamuru S, Huang R, Teneva N, Simmons SO, Xia M, Tice RR, Austin CP, and Myung K (2012) High-Throughput Genotoxicity Assay Identifies Antioxidants as Inducers of DNA Damage Response and Cell Death. Proc. Natl. Acad. Sci. U. S. A 109 (14), 5423–5428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (78).Young D, Martin T, Venkatapathy R, and Harten P (2008) Are the Chemical Structures in Your QSAR Correct? QSAR Comb. Sci 27 (11–12), 1337–1345. [Google Scholar]
- (79).Fourches D, Muratov E, and Tropsha a. (2010) Trust but Verify: On the Importance of Chemical Structure Curation in Chemoinformatics and QSAR Modeling Research. J. Chem. Inf. Model 50 (7), 1189–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (80).Zhao L, Wang W, Sedykh A, and Zhu H (2017) Experimental Errors in QSAR Modeling Sets: What We Can Do and What We Cannot Do. ACS Omega 2 (6), 2805–2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (81).Zhu H, Kim M, Zhang L, and Sedykh A Computers Instead of Cells: Computational Modeling of Chemical Toxicity In Reducing, Refining and Replacing the Use of Animals in Toxicity Testing; Allen DG, and Waters MD, Eds.; Royal Society of Chemistry: Cambridge, UK, 2013; pp 163–182. [Google Scholar]
- (82).Fourches D, Muratov E, and Tropsha A (2016) Trust, but Verify II: A Practical Guide to Chemogenomics Data Curation. J. Chem. Inf. Model 56 (7), 1243–1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (83).Papadatos G, Gaulton A, Hersey A, and Overington JP (2015) Activity, Assay and Target Data Curation and Quality in the ChEMBL Database. J. Comput.-Aided Mol. Des 29 (9), 885–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (84).Kim MT, Wang W, Sedykh A, and Zhu H Curating and Preparing High Throughput Screening Data for Quantitative Structure Activity Relationship Modeling In Methods Mol. Biol; Zhu H, and Xia M, Eds.; Humana Press: New York, 2016; Vol. 1473, pp 161–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (85).Zakharov AV, Peach ML, Sitzmann M, and Nicklaus MC (2014) QSAR Modeling of Imbalanced High-Throughput Screening Data in PubChem. J. Chem. Inf. Model 54 (3), 705–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (86).Chang CY, Hsu MT, Esposito EX, and Tseng YJ (2013) Oversampling to Overcome Overfitting: Exploring the Relationship between Data Set Composition, Molecular Descriptors, and Predictive Modeling Methods. J. Chem. Inf. Model 53 (4), 958–971. [DOI] [PubMed] [Google Scholar]
- (87).Hao M, Wang Y, and Bryant SH (2014) An Efficient Algorithm Coupled with Synthetic Minority Over-Sampling Technique to Classify Imbalanced PubChem BioAssay Data. Anal. Chim. Acta 806, 117–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (88).Chen B, Dong X, Jiao D, Wang H, Zhu Q, Ding Y, and Wild D (2010) Chem2Bio2RDF: A Sematic Framework for Linking and Data Mining Chemogenomic and Systems Chemical Biology Data. BMC Bioinf 11, 255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (89).Fourches D, Sassano MF, Roth BL, and Tropsha A (2014) HTS Navigator: Freely Accessible Cheminformatics Software for Analyzing High-Throughput Screening Data. Bioinformatics 30 (4), 588–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (90).Guha R (2007) Chemical Informatics Functionality in R. J. Stat. Softw 18 (5), 1–16. [Google Scholar]
- (91).Filer DL, Kothiya P, Woodrow Setzer R, Judson RS, and Martin MT (2016) Tcpl: The ToxCast Pipeline for High-Throughput Screening Data. Bioinformatics 33 (4), 618–620. [DOI] [PubMed] [Google Scholar]
- (92).Russo DP, Kim MT, Wang W, Pinolini D, Shende S, Strickland J, Hartung T, and Zhu H (2016) CIIPro: A New Read-across Portal to Fill Data Gaps Using Public Large-Scale Chemical and Biological Data. Bioinformatics 33 (3), 464–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (93).Capuzzi SJ, Kim ISJ, Lam WI, Thornton TE, Muratov EN, Pozefsky D, and Tropsha A (2017) Chembench: A Publicly Accessible, Integrated Cheminformatics Portal. J. Chem. Inf. Model 57(2), 105–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (94).Weidlich IE, Pevzner Y, Miller BT, Filippov IV, Woodcock HL, and Brooks BR (2015) Development and Implementation of (Q)SAR Modeling within the CHARMMing Web-User Interface. J. Comput. Chem 36 (1), 62–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (95).Huang R, Xia M, Nguyen D-T, Zhao T, Sakamuru S, Zhao J, Shahane SA, Rossoshek A, and Simeonov A (2016) Tox21 Challenge to Build Predictive Models of Nuclear Receptor and Stress Response Pathways as Mediated by Exposure to Environmental Chemicals and Drugs. Front. Environ. Sci 3, 1–9. [Google Scholar]
- (96).Mayr A, Klambauer G, Unterthiner T, and Hochreiter S DeepTox: Toxicity Prediction Using Deep Learning. Front. Environ. Sci 2016, 3 DOI: 10.3389/fenvs.2015.00080 [DOI] [Google Scholar]
- (97).Kleinstreuer NC, Karmaus AL, Mansouri K, Allen DG, Fitzpatrick JM, and Patlewicz G (2018) Predictive Models for Acute Oral Systemic Toxicity: A Workshop to Bridge the Gap from Research to Regulation. Comput. Toxicol 8 (11), 21–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (98).Xu Y, Pei J, and Lai L (2017) Deep Learning Based Regression and Multiclass Models for Acute Oral Toxicity Prediction with Automatic Chemical Feature Extraction. J. Chem. Inf. Model 57(11), 2672–2685. [DOI] [PubMed] [Google Scholar]
- (99).Wen M, Zhang Z, Niu S, Sha H, Yang R, Yun Y, and Lu H (2017) Deep-Learning-Based Drug-Target Interaction Prediction. J. Proteome Res 16 (4), 1401–1409. [DOI] [PubMed] [Google Scholar]
- (100).Russo DP, Zorn KM, Clark AM, Zhu H, and Ekins S (2018) Comparing Multiple Machine Learning Algorithms and Metrics for Estrogen Receptor Binding Prediction. Mol. Pharmaceutics 15, 4361–4370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (101).Ball N, Cronin MT, Shen J, Blackburn K, Booth ED, Bouhifd M, Donley E, Egnash L, Hastings C, Juberg DR, et al. (2016) Toward Good Read-Across Practice (GRAP) Guidance. ALTEX - Altern. to Anim. Exp 33 (2), 149–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (102).Cronin MTD, Madden JC, Enoch SJ, and Roberts DW Chemical Toxicity Prediction: Category Formation and Read-Across; The Royal Society of Chemistry: Cambridge, UK, 2013. [Google Scholar]
- (103).Berggren E, Amcoff P, Benigni R, Blackburn K, Carney E, Cronin M, et al. (2015) Chemical Safety Assessment Using Read-Across: Assessing the Use of Novel Testing Methods to Strengthen the Evidence Base for Decision Making. Environ. Health Perspect 123 (12), 1232–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (104).Low Y, Sedykh A, Fourches D, Golbraikh A, Whelan M, Rusyn I, and Tropsha A (2013) Integrative Chemical – Biological Read-Across Approach for Chemical Hazard Classi Fi Cation. Chem. Res. Toxicol 26, 1199–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (105).Luechtefeld T, Marsh D, Rowlands C, and Hartung T (2018) Machine Learning of Toxicological Big Data Enables Read-Across Structure Activity Relationships (RASAR) Outperforming Animal Test Reproducibility. Toxicol. Sci 165 (1), 198–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (106).Yoon M, Campbell JL, Andersen ME, and Clewell HJ (2012) Quantitative in Vitro to in Vivo Extrapolation of Cell-Based Toxicity Assay Results. Crit. Rev. Toxicol 42 (8), 633–652. [DOI] [PubMed] [Google Scholar]
- (107).Patlewicz G, Simon TW, Rowlands JC, Budinsky RA, and Becker RA (2015) Proposing a Scientific Confidence Framework to Help Support the Application of Adverse Outcome Pathways for Regulatory Purposes. Regul. Toxicol. Pharmacol 71 (3), 463–477. [DOI] [PubMed] [Google Scholar]
- (108).Clippinger AJ, Allen D, Behrsing H, BéruBé KA, Bolger MB, Casey W, DeLorme M, Gaça M, Gehen SC, Glover K, et al. (2018) Pathway-Based Predictive Approaches for Non-Animal Assessment of Acute Inhalation Toxicity. Toxicol. In Vitro 52, 131–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (109).Bal-Price A, Lein PJ, Keil KP, Sethi S, Shafer T, Barenys M, Fritsche E, Sachana M, and Meek ME (2017) Bette. Developing and Applying the Adverse Outcome Pathway Concept for Understanding and Predicting Neurotoxicity. NeuroToxicology 59, 240–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (110).Bal-Price A, and Meek ME (2017) Bette. Adverse Outcome Pathways: Application to Enhance Mechanistic Understanding of Neurotoxicity. Pharmacol. Ther 179, 84–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (111).Sachana M, Rolaki A, and Bal-Price A (2018) Development of the Adverse Outcome Pathway (AOP): Chronic Binding of Antagonist to N-Methyl-D-Aspartate Receptors (NMDARs) during Brain Development Induces Impairment of Learning and Memory Abilities of Children. Toxicol. Appl. Pharmacol 354, 153–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (112).Maxwell G, MacKay C, Cubberley R, Davies M, Gellatly N, Glavin S, Gouin T, Jacquoilleot S, Moore C, Pendlington R, et al. (2014) Applying the Skin Sensitization Adverse Outcome Pathway (AOP) to Quantitative Risk Assessment. Toxicol. In Vitro 28(1), 8–12. [DOI] [PubMed] [Google Scholar]
- (113).Patlewicz G, Kuseva C, Kesova A, Popova I, Zhechev T, Pavlov T, Roberts DW, and Mekenyan O (2014) Towards AOP Application - Implementation of an Integrated Approach to Testing and Assessment (IATA) into a Pipeline Tool for Skin Sensitization. Regul. Toxicol. Pharmacol 69 (3), 529–545. [DOI] [PubMed] [Google Scholar]
- (114).Organization for Economic Co-operation and Development. The Adverse Outcome Pathway for Skin Sensitisation Initiated by Covalent Binding to Proteins Part 1: Scientific Evidence. Ser. Test. Assess 2012, 168. [Google Scholar]
- (115).Browne P, Noyes PD, Casey WM, and Dix DJ (2017) Application of Adverse Outcome Pathways to U.S. EPA’s Endocrine Disruptor Screening Program. Environ. Health Perspect 125 (9), 096001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (116).Benigni R, Battistelli CL, Bossa C, Giuliani A, and Tcheremenskaia O (2017) Endocrine Disruptors: Data-Based Survey of in Vivo Tests, Predictive Models and the Adverse Outcome Pathway. Regul. Toxicol. Pharmacol 86, 18–24. [DOI] [PubMed] [Google Scholar]
- (117).Proctor DM, Suh M, Chappell G, Borghoff SJ, Thompson CM, Wiench K, Finch L, and Ellis-Hutchings R (2018) An Adverse Outcome Pathway (AOP) for Forestomach Tumors Induced by Non-Genotoxic Initiating Events. Regul. Toxicol. Pharmacol 96 (April), 30–40. [DOI] [PubMed] [Google Scholar]
- (118).Vinken M, Landesmann B, Goumenou M, Vinken S, Shah I, Jaeschke H, Willett C, Whelan M, and Rogiers V (2013) C from Drug-Mediated Bile Salt Export Pump Inhibition to Cholestatic Liver Injury. Toxicol. Sci 136 (1), 97–106. [DOI] [PubMed] [Google Scholar]
- (119).Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, and Knudsen TB (2013) Profiling 976 ToxCast Chemicals across 331 Enzymatic and Receptor Signaling Assays. Chem. Res. Toxicol 26 (6), 878–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (120).Kleinstreuer NC, Yang J, Berg EL, Knudsen TB, Richard AM, Martin MT, Reif DM, Judson RS, Polokoff M, Dix DJ, et al. (2014) Phenotypic Screening of the ToxCast Chemical Library to Classify Toxic and Therapeutic Mechanisms. Nat. Biotechnol 32 (6), 583–591. [DOI] [PubMed] [Google Scholar]
- (121).Huang R, Xia M, Sakamuru S, Zhao J, Shahane SA, Attene-Ramos M, Zhao T, Austin CP, and Simeonov A (2016) Modelling the Tox21 10 K Chemical Profiles for in Vivo Toxicity Prediction and Mechanism Characterization. Nat. Commun 7, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (122).Attene-Ramos MS, Huang R, Sakamuru S, Witt KL, Beeson GC, Shou L, Schnellmann RG, Beeson CC, Tice RR, Austin CP, et al. (2013) Systematic Study of Mitochondrial Toxicity of Environmental Chemicals Using Quantitative High Throughput Screening. Chem. Res. Toxicol 26 (9), 1323–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (123).Attene-Ramos MS, Huang R, Michael S, Witt KL, Richard A, Tice RR, Simeonov A, Austin CP, and Xia M (2015) Profiling of the Tox21 Chemical Collection for Mitochondrial Function to Identify Compounds That Acutely Decrease Mitochondrial Membrane Potential. Environ. Health Perspect 123 (1), 49–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (124).Sipes NS, Martin MT, Reif DM, Kleinstreuer NC, Judson RS, Singh AV, Chandler KJ, Dix DJ, Kavlock RJ, and Knudsen TB (2011) Predictive Models of Prenatal Developmental Toxicity from ToxCast High-Throughput Screening Data. Toxicol. Sci 124 (1), 109–127. [DOI] [PubMed] [Google Scholar]
- (125).Kleinstreuer N, Dix D, Rountree M, Baker N, Sipes N, Reif D, Spencer R, and Knudsen T (2013) A Computational Model Predicting Disruption of Blood Vessel Development. PloS Comp. Biol 9(4), 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (126).Judson RS, Magpantay FM, Chickarmane V, Haskell C, Tania N, Taylor J, Xia M, Huang R, Rotroff DM, Filer DL, et al. (2015) Integrated Model of Chemical Perturbations of a Biological Pathway Using 18 in Vitro High-Throughput Screening Assays for the Estrogen Receptor. Toxicol. Sci 148 (1), 137–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (127).Browne P, Judson RS, Casey WM, Kleinstreuer NC, and Thomas RS (2015) Screening Chemicals for Estrogen Receptor Bioactivity Using a Computational Model. Environ. Sci. Technol 49(14), 8804–8814. [DOI] [PubMed] [Google Scholar]
- (128).Ruiz P, Sack A, Wampole M, Bobst S, and Vracko M (2017) Integration of in Silico Methods and Computational Systems Biology to Explore Endocrine-Disrupting Chemical Binding with Nuclear Hormone Receptors. Chemosphere 178, 99–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (129).Bhhatarai B, Wilson DM, Bartels MJ, Chaudhuri S, Price PS, and Carney EW (2015) Acute Toxicity Prediction in Multiple Species by Leveraging Mechanistic ToxCast Mitochondrial Inhibition Data and Simulation of Oral Bioavailability. Toxicol. Sci 147 (2), 386–396. [DOI] [PubMed] [Google Scholar]
- (130).Kim MT, Huang R, Sedykh A, Wang W, Xia M, and Zhu H (2016) C. Environ. Health Perspect 124 (5), 634–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (131).Luechtefeld T, Maertens A, Mckim JM, Hartung T, Kleensang A, and Sá-Rocha V (2015) Probabilistic Hazard Assessment for Skin Sensitiza Tion Potency by Dose-Response Modeling Using Feature Elimination Instead of Quantitative Structure-Activity Relationships. J. Appl. Toxicol 35 (11), 1361–1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (132).Xia M, Huang R, Shi Q, Boyd WA, Zhao J, Sun N, Rice JR, Dunlap PE, Hackstadt AJ, Bridge MF, et al. (2018) Comprehensive Analyses and Prioritization of Tox21 10K Chemicals Affecting Mitochondrial Function by In-Depth Mechanistic Studies. Environ. Health Perspect 126 (7), No. 077010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (133).Mellor CL, Steinmetz FP, and Cronin MTD (2016) Using Molecular Initiating Events to Develop a Structural Alert Based Screening Workflow for Nuclear Receptor Ligands Associated with Hepatic Steatosis. Chem. Res. Toxicol 29 (2), 203–212. [DOI] [PubMed] [Google Scholar]
- (134).Gadaleta D, Manganelli S, Roncaglioni A, Toma C, Benfenati E, and Mombelli E (2018) QSAR Modeling of ToxCast Assays Relevant to the Molecular Initiating Events of AOPs Leading to Hepatic Steatosis. J. Chem. Inf. Model 58 (8), 1501–1517. [DOI] [PubMed] [Google Scholar]
- (135).Strope CL, Mansouri K, Clewell HJ, Rabinowitz JR, Stevens C, and Wambaugh JF (2018) High-Throughput in-Silico Prediction of Ionization Equilibria for Pharmacokinetic Modeling. Sci. Total Environ 615, 150–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (136).Pearce RG, Setzer RW, Strope CL, Sipes NS, and Wambaugh JF (2017) Httk: R Package for High-Throughput Toxicokinetics. J. Stat. Softw 79 (4), 1–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (137).Applications of Toxicogenomic Technologies to Predictive Toxicology and Risk Assessment; National Research Council (US) Committee on Applications of Toxicogenomic Technologies to Predictive Toxicology, National Academies Press: Washington, D.C., 2007. [PubMed] [Google Scholar]
- (138).Merrick BA, Paules RS, and Tice RR (2015) Intersection of Toxicogenomics and High Throughput Screening in the Tox21 Program: An NIEHS Perspective. Int. J. Biotechnol 14 (1), 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (139).Bouhifd M, Andersen ME, Baghdikian C, Boekelheide K, Kevin MAJF Jr., Kleensang A, Li H, Livi C, Mcmullen PD, et al. (2015) The Human Toxome Project. ALTEX 32 (2), 112–124. [DOI] [PMC free article] [PubMed] [Google Scholar]