Abstract
Lactobacillus rhamnosus is a lactic acid bacterium (LAB) that survives diverse ecological niches, including the human oral cavity and gastrointestinal tract. L. rhamnosus is an acidogenic bacterium that produces copious amounts of lactic acid. The organism is also considered as an aciduric, since it can survive prolonged exposure to an acidic environment. For a probiotic bacterium such as L. rhamnous, it is necessary to understand how this organism survives acid stress. In this study we used L. rhamnous LRB to isolate one spontaneous mutant that was sensitive to acid stress. The mutant, which we named RBM1, also displayed sensitivity to a wide range of stresses including osmotic, thermal, and other stresses. Using whole genome sequencing, we mapped the putative mutations in the mutant strain. It appears that three single nucleotide substitutions occurred in the mutant as compared to the wild type LRB strain. Among those, the most relevant mutation occurred in the ftsH gene that created a single amino acid change in the protein. We performed a comparative proteomic study to understand the molecular basis for stress sensitivity and found that ~ 15% of the proteome is altered in the mutant strain. Our study suggests that generation of spontaneous mutants during L. rhamnosus colonization could drastically affect bacterial physiology and survival under stress conditions.
INTRODUCTION
Lactobacilli are a highly diverse group of lactic acid producing bacteria that dwell in nutrient rich niches, such as animals, food, humans, and plants because of the fastidious growth requirement (Duar et. al. 2017; Walter 2008). The genus Lactobacillus is composed of 200 or more species. In terms of both phylogenetic and metabolic diversity, the genus Lactobacillus exceeds an archetypal bacteria family (Sun et. al. 2015). Because of their importance in fermentation, earlier classification was based on carbohydrate utilization. These bacteria employ the classical glycolytic pathway (homofermentive) and/or phosphoketolase pathway (heterofermentative) for the conversion of hexoses (Duar et. al. 2017). These two groups also differ in the presence or absence of other major metabolic pathways for utilization of carbohydrates and amino acids, and represent two separate phylogenetic clades (Zheng et. al. 2015). Lactobacillus spp. are also highly diverse at the genomic level as well. The overall genome sizes could vary from as small as 1.23 Mb (e.g. L. sanfranciscensis) to as large as 4.91 Mb (e.g. L. parakefiri). In addition to varied genome sizes, the GC content also differs drastically, ranging from ~32% to 57.0%. Surprisingly, the core-genome for Lactobacillus is very small (less than 100 genes), whereas it comprises a large open pan-genome with at least 44,668 gene families (Sun et. al. 2015). It seems that lactobacilli drastically reduced their genome sizes during evolution from a common ancestor by expunging about 3000 genes, of which more than 1000 genes are in individual groups or species (Makarova et. al. 2006; Sun et. al. 2015; Zheng et. al. 2015). This reduction in genome sizes has some severe consequences, including loss of critical metabolic functions such as carbohydrate utilization, biosynthesis of amino acids and cofactors; thereby, turning the species fastidious in nature (Makarova et. al. 2006). The reduction of genome size also strongly correlated with the host adaptation. Lactobacillus spp. that are nomadic or free-living tend to encode larger genome sizes, whereas host adapted species have significantly reduced genome sizes. Furthermore, the genomes of host adapted are generally with lower GC content. Some host adapted species show more “promiscuous” lifestyles, including utilization of various hosts and body sites. Many lactobacilli that are often found in the oral cavity are also found in gastrointestinal tracts and feces (Caufield et. al. 2015; Dal Bello and Hertel 2006).
L. rhamnosus is one such lactobacilli that commonly colonizes in the oral cavity, as well as gastrointestinal tracts and the vagina. The organism displays a strong nomadic life-style and can be found in a wide range of ecological niches, including fermented dairy products, plant associated materials, and vertebrate animals. Due to its cheese-ripening properties and lactate production, L. rhamnosus is frequently used in the food industry (Caggia et. al. 2015; Succi et. al. 2005). Some strains of L. rhamnosus are used as probiotic organisms since they possess immunomodulatory activity and have been extensively used for treatment of various human diseases (for reviews see, (Banna et. al. 2017; Segers and Lebeer 2014). Among the probiotic strains that are currently used, L. rhamnosus GG (LGG) and ATCC 53103 are perhaps the best characterized strains, which were originally isolated from human feces (Lebeer et. al. 2007; Silva et. al. 1987). In general L. rhamnosus is considered as a commensal and/or a beneficial organism, but occasionally the bacterium can cause clinical manifestations such as bacteremia and meningitis (Gouriet et. al. 2012; Martinez et. al. 2014))(Robin et. al. 2010). Recent comparative genome studies have identified nearly 20 highly variable regions on the chromosome that encode functions related to the commensal lifestyle. These functions are typically related to carbohydrate transport and utilization, capsule production, pili production, altered transcriptional regulations, and bile salt resistance (Douillard et. al. 2013; Nadkarni et. al. 2014; Petrova et. al. 2018; Westerik et. al. 2018). Though comparative genome studies are somewhat helpful to understand why L. rhamnosus has adapted to different environmental niches, these studies do not provide information regarding how the organism tolerates and survives under various stressful environmental conditions, such as the acidic pH encountered during fermentation, the antimicrobial effect of bile during gastrointestinal colonization, and the oxidative stresses during clinical manifestations, among others. While general stress tolerance response has been studied in various lactobacilli (for recent reviews see (De Angelis and Gobbetti 2004; Papadimitriou et. al. 2016; van de Guchte et. al. 2002), very little is known regarding how L. rhamnosus alleviates various stresses encountered during growth in such drastically different ecological niches.
We have recently isolated an oral commensal L. rhamnosus strain LRB, which produces antimicrobial compounds that inhibit wide range bacteria belonging to both Gram-negative and Gram-positive families (Biswas and Biswas 2016; Biswas et. al. 2018). The antimicrobial compounds secreted by LRB are small peptides in nature (bacteriocins); however, the exact sequences of the peptides are yet to be known. Since LRB is also closely related to the probiotic strain LGG, which mainly colonizes the gastrointestinal (GI) tract, we wanted to explore whether LRB could be used as a functional probiotic strain. One advantage of using LRB is that it colonizes oral cavity that is ecologically much different than the GI tract. In this study we wanted to generate and characterize a low acid-producing variant of LRB that retains its antimicrobial activity; thus, well suited for oral health. Toward this end, we have isolated a spontaneous mutant that is highly sensitive to low pH and therefore low acid producer. In addition, the mutant also displayed enhanced sensitivity to various stresses and increased susceptibility to numerous antibiotics. Here we discuss the molecular basis for the observed stress sensitive phenotypes based on the genomic and proteomic data.
MATERIALS AND METHODS
Bacterial strains and growth media:
Lactobacillus rhamnosus LRB and its derivatives were routinely grown in Mann Rogosa Sharpe (MRS; Difco). For some stress related studies, MRS broth was supplemented with 2% glucose and 1% casein hydrolysate (designated as MRS+), where indicated. L. rhamnosus strains were also grown in Todd-Hewitt medium supplemented by 0.2% yeast extract (THY). Strains were incubated at 37°C grown under microaerophilic conditions (no oxygen) using candle jar. For some stress studies, strains were incubated at different temperatures where indicated.
Isolation of mutants:
L. rhamnosus LRB was grown at 37°C in MRS+ broth in the presence of 2% glycine. Next day, cultures were diluted (1:20 fold) in fresh MRS+ broth supplemented with 2% glycine and incubated at 37°C until the OD600 reached 0.5. Cells were harvested and washed twice in ice cold buffer containing 0.5M sucrose, 7mM KHPO4 (pH 7.5) and 1mM MgCl2. Cells were resuspended in the same buffer to a final volume of 1:100 of the original cultures to prepare electro-competent cells. Cells were then electroporated (2mm gap cuvette and 1.7kV) using ~500ng of pGhost9::ISS1 plasmid DNA. Electroporated cells were recovered in MRS+ broth containing 2mM CaCl2 and 20mM MgCl2 for an hour at 30°C, plated onto MRS+ agar containing 5 μg/ml erythromycin (Em) and incubated at 30°C under anaerobic condition. An Em resistant colony was selected, grown at 30°C in the presence of Em and proceeded toward transposon library preparation as described previously (Biswas and Biswas 2011). About 10000 mutants were replica patched on MRS+ agar and MRS+ agar adjusted to pH 4.5 with citrate-phosphate buffer as described previously (Biswas et. al. 2008). Briefly, the initial pH of the MRS+ agar medium was adjusted prior to sterilization to pH 5.5 with HCl. After sterilization, citrate-phosphate buffer (50mM) of the desired pH was added to media. Sensitive colonies were recovered from the MRS+ agar plate and verified again using fresh media.
Acid and thermal stress tolerance response:
For growth on MRS+ agar plates containing stress-inducing chemicals, cultures were grown to exponential phase in MRS+ broth, pelleted via centrifugation, washed twice with 0.85% NaCl, and resuspended in 0.85% NaCl. The cultures were adjusted to an optical density (A600) of 5.0 and 10-fold serially diluted, and 10 μl of each dilution was spotted onto MRS+ agar containing the stress-inducing chemicals or pH adjusted to pH 4.5 with citrate-phosphate buffer. Plates were incubated overnight at 37°C, under microaerophilic conditions, and bacterial growth was evaluated as previously described (Biswas and Biswas 2011).
Sensitivity to chemical reagents (stressors).
Sensitivity of the L. rhamonosus mutant strains to various chemicals was evaluated primarily by using the disk diffusion method. MRS agar plates inoculated with the wild-type or the mutant cultures (as described above) were overlaid with disks containing various toxic or stress-inducing chemicals. The chemicals (all from Sigma) tested included acriflavine (10mg/ml), benzidine (4mM), benzalkonium chloride (10mg/ml), chlorhexidine gluconate (2%), 2,2′-dipyridyl (50mM), ethidium bromide (1.5%), ethyl viologen (1M), hydrogen peroxide (10%), menadione (1%), methyl viologen (1M), mitomycin C (12.5μg/ml), 1,10′-phenanthroline (40mg/ml), potassium telurite (1.0%), puromycin (5mg/ml), pyronin B (0.5%), and tert-butyl hydroperoxide (70%). The chemicals are listed in Table 1.
Table 1:
Sensitivity of LRB and RBM1 strains against various chemicals
| Stressors | Zone of inhibition diameter (mm) | |
|---|---|---|
| LRB | RBM1 | |
| Acryflavin (10 mg/ml) * | 14.0 ± 1.0 | 22.0 ± 1.0 |
| Benzidine (4 mM) | 0.0 | 0.0 |
| Benzalkonium chloride (10 mg/ml) | 14.0 ± 1.0 | 16.0 ± 1.0 |
| Chlorhexidine gluconate (2%) * | 15.0 ± 1.0 | 20.0 ± 1.0 |
| Dipyridyl (50 mM) | 0.0 | 0.0 |
| Diquat dibromide (500 mg/ml) * | 28.0 ± 2.0 | 35.0 ± 2.0 |
| Ethidium bromide (10 mg/ml) * | 11.0 ± 0.5 | 17.0 ± 1.0 |
| Ethyl viologen (1 M) * | 20.0 ± 1.0 | 34.0 ± 2.0 |
| Menadione (1%) * | 25.0 ± 1.0 | 43.0 ± 1.0 |
| Methyl viologen (1.0 M) * | 25.0 ± 2.0 | 37.0 ± 2.0 |
| Mitomycin C1 (1.25 mg/ml) * | 33.0 ± 1.0 | 39.0 ± 0.5 |
| Puromycin (5 mg/ml) * | 16.0 ± 1.0 | 21.0 ± 1.0 |
| Hydrogen peroxide (10%) | 40.0 ± 1.0 | 39.0 ± 1.0 |
| 1,10’-phenanthroline (40 mg/ml) * | 22.0 ± 1.0 | 25.0 ± 1.0 |
| Potassium tellurite | 8.0 ± 1.0 | 8.0 ± 1.0 |
| Pyronin B (0.5%) * | 12.0 ± 1.0 | 16.0 ± 1.0 |
| tert-butyl hydroperoxide (70%) | 0.0 | 0.0 |
, significant difference
Antibiotic susceptibility stress.
Disk diffusion assays were performed to evaluate antibiotic susceptibility of the L. rhamonosus mutants. Three or four fresh bacterial colonies grown on MRS agar plate were resuspended in 0.85% NaCl and the initial optical density (A600) was adjusted to 0.2. Cultures were spread onto MRS plates with a cotton swab. Antibiotic disks (6 mm in diameter; Becton and Dickinson Laboratories) were then placed on the inoculated plates. The zones of inhibition were measured after overnight incubation under anaerobic conditions. The antibiotics selected for this study targeted all five major biosynthesis pathways and they are listed in Table 2.
Table 2:
Inhibitory zones for various antimicrobials
| Antibiotics $ | Zone of inhibition diameter (mm) | |
|---|---|---|
| LRB | RBM1 | |
| Bacitracin (10 mcg) | 18.0 ± 1.0 | 24.0 ± 1.0 |
| Carbenicillin (5 mg/ml) # | 17.0 ± 1.0 | 17.0 ± 1.0 # |
| Cefixime (5 mcg) | 0.0 | 0.0 |
| Cefoxitin (30 mcg) | 0.0 | 0.0 |
| Ceftazidime (30 mcg) * | 20.0 ± 1.0 | 22.0 ± 1.0 |
| Cefuroxime (30 mcg) | 21.0 ± 1.0 | 22.0 ± 1.0 |
| Cephalothin (30 mcg) * | 23.0 ± 2.0 | 26.0 ± 2.0 |
| Cephazolin (30 mcg) * | 21.0 ± 1.0 | 29.0 ± 1.0 |
| Chlomphenicol (5 mcg) * | 19.0 ± 1.0 | 22.0 ± 2.0 |
| Ciprofloxaxin (5 mcg) * | 18.0 ± 2.0 | 26.0 ± 2.0 |
| Clindamycin (2 mcg) * | 25.0 ± 1.0 | 27.0 ± 1.0 |
| Erythromycin (5mg/ml) * | 40.0 ± 1.0 | 45.0 ± 2.0 |
| Flavomycin (5 mg/ml) * | 12.0 ± 2.0 | 15.0 ± 1.0 |
| Fosfomycin (200 mcg) | 0.0 | 0.0 |
| Imipenem (10 mcg) * | 26.0 ± 2.0 | 30.0 ± 3.0 |
| Meropenem (10 mcg) * | 21.0 ± 2.0 | 24.0 ± 1.0 |
| Mezlocillin (75 mcg) * | 40.0 ± 2.0 | 46.0 ± 2.0 |
| Nalidixic acid (30 mcg) | 0.0 | 0.0 |
| Nisin (40 mg/ml) * | 13.0 ± 1.0 | 18.0 ± 1.0 |
| Oxacillin (10 mcg) | 0.0 | 0.0 |
| Penicillin (10 mcg) * | 25.0 ± 1.0 | 30.0 ± 1.0 |
| Rifampin (5 mcg) * | 31.0 ± 2.0 | 37.0 ± 2.0 |
| Streptomycin (10mg/ml) * | 11.0 ± 1.0 | 21.0 ± 1.0 |
| Tetracycline (10 mg/ml) * | 34.0 ± 1.0 | 40 ± 2.0 |
| Trimethoprime (5 mcg) | 0.0 | 0.0 |
| Tunicamycin (5 mg/ml) | 0.0 | 0.0 |
| Vancomycin (5 mcg) | 0.0 | 0.0 |
, per disc (mcg);
, significant difference;
, a secondary halo
UV- sensitivity assay.
UV sensitivity was determined by swabbing two lines of each test culture across an MRS agar plate, after which half of the petri dish was covered with a glass plate to block UV radiation and the agar plate was exposed to UV short-wave radiation (254 nm) for 15 s. Plates were then wrapped with aluminum foil and incubated at 37°C under microaerophilic conditions.
Whole genome sequencing to map mutations.
Genomic DNA from L. rhamnosus strains was purified using the MasterPure DNA purification kit (Epibio) from 10 mL of overnight cultures grown in THY broth. DNA concentration and purity were assessed with a NanoDrop ND-1000 spectrophotometer. DNA libraries were constructed for paired-end 151 bases (PE150) following manufacturer’s protocol (Illumina) and sequencing was performed at the Institute for Genome Sciences (Univ. Maryland) using the Illumina HiSeq2000 platform. The average genome coverage provided by each library was nearly 60X. To identify single nucleotide polymorphism (SNP) or short insertion/deletions (indels), we used Geneious Prime software. For each mutant, Illumina reads were directly mapped on to the reference genome sequence of L. rhamnosus LRB (CP016823) with the default alignment parameters. Short indels and SNPs were called based on the read mapping of the SNP calling procedures following the default setting.
Preparation of cell lysate.
L. rhamnosus strains were grown until exponential phase (OD600nm = 0.7) in THY broth. Cells were harvested by centrifugation, washed once in sterile PBS, and then lysed in PBS containing protease inhibitors (Roche) using a bead beater. The lysate was collected by centrifugation at 12,000 ×g, 4°C for 10 min. The total protein concentration was measured by Bradford assay using BSA as standard.
Comparative proteomic studies using tandem mass tag (TMT) and level free quantitation (LFQ).
Whole cell protein extracts were reduced, alkylated, and subjected to proteolytic digestion using Filter-Aided Sample Preparation (FASP) essentially as described previously (McDowell et. al. 2013). About 100μg protein was reduced (10mM TCEP) in a reaction volume of 105μL, then diluted with 600μL 8M urea, 100mM Tris-Cl pH 7.5, and transferred to a microcon filter (10k MWCO); all centrifugation steps were at 12,000 rcf. The retained protein was alkylated with 50mM chloracetamide for 30 minutes and then rinsed with 8M urea. The buffer was exchanged to 100mM Tri-Ethyl-Ammonium Bicarbonate (TEAB) and 0.5μg Trypsin/LysC (Promega V5073) was added in the same buffer (100μL) and incubated at 370C overnight. Peptidomes were harvested by centrifugation, dried down, and resuspended in pure water.
For TMT quantitation, peptidomes were labeled with a TMT reagent (Thermo Fisher, San Jose, CA), then purified by solid phase extraction (SPE) using Isolute C18 (EC) spin columns (BioTage, Charlotte, NC). An equal quantity of each was mixed for mudpit offline fractionation (1st dimension) on a C18 column under basic conditions (basic reversed phase, or BRP). Peptides were fractionated and five-minute fractions were taken. The resulting 26 fractions were pooled into 13 samples as described earlier (Wang et. al. 2011). The pooled fractions were resolved by standard acidic reversed phase nanoLC for data dependent acquisition with MS survey scans (QExactive Plus, MS system), and 18 dependent scans per cycle, both at 35k resolution. Data files from mudpit were merged and searched with Mascot version 2.6 (Matrix Science), against the Lactobacillus rhamnosus (ATCC 53103) database, with a decoy search included, applying a significance threshold of p< 0.03 (97% confidence), resulting in a 0.81% False Discovery Rate. TMT ratios were normalized using the average ratio of all peptides, with at least two unique identified peptides required for quantitation. Peptide mass tolerance was 8 ppm, and MS/MS peak tolerance was 0.02, allowing up to one missed cleavage for identification.
For label free quantitation (LFQ), pure peptidomes were analyzed in a single long gradient for standard acidic (0.2% formic acid) reversed phase on a C18 column. Each sample was run in triplicate, generating three data files each for the pairwise comparison (LRB vs RBM1). Data was acquired in data dependent mode essentially as described above on a QExactive Plus mass spectrometer, but with 17k resolution in the MS/MS scans. For database search and LFQ analysis, MaxQuant (version 1.6.3.3, Max Plank Institute for Biochemistry) was used, enabling match between runs and searched against the complete proteome Lactobacillus rhamnosus (ATCC 53103) with yeast proteome included for complexity (8,874 protein sequences). In this case, FDR was set at 1% for identification purposes within Maxquant. Main search tolerances were 4.5 ppm for MS and 8 ppm MS/MS. Match between runs was enabled with a three min time window. Two missed cleavages were allowed with carbamidomethyl Cysteine (fixed) and variable deamidation of Asn and Gln. Cys carbamidomethyl modification was allowed for quantitation. LFQ was enabled with no changes to default settings. Data was further analyzed using the Perseus software package 62.
RESULTS
Isolation of a stress sensitive mutant:
L. rhamnosus LGG has long been used as a probiotic strain because it is highly acid tolerant and survives in the gastrointestinal tract. LRB is very closely related to LGG, and to understand the molecular mechanisms of acid tolerance, we wanted to isolate acid sensitive mutants that do not survive low pH but are still able to produce antimicrobial activity. We generated a potential transposon library by electroporating plasmid pGh9::ISS1, which is thermosensitive and carries ISS1 sequence capable of inserting randomly in the genome of gram-positive bacteria including streptococci (Biswas and Biswas 2011; 2014; Thibessard et. al. 2002). We then screened nearly 10,000 erythromycin-resistant transformants by replica patching onto both MRS+ and on MRS+ with pH 4.5. We initially obtained only three mutants that were unable to grow MRS+ with pH 4.5. Out of the three, only one mutant, which we named RBM1-R, passed the secondary screening that was performed using cell density adjusted dilution spotting (Fig. 1). We selected RBM1-R for further characterization.
Figure 1.
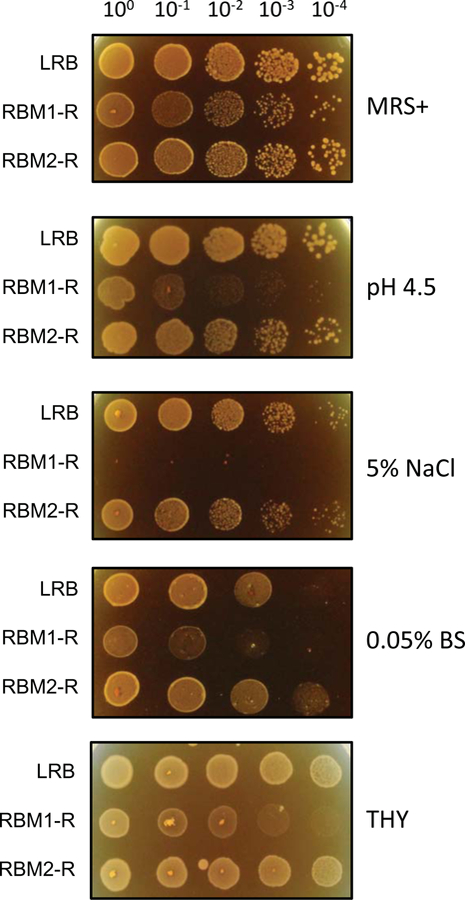
Characterization of an acid sensitive LRB mutant. Fresh overnight cultures were adjusted to a starting optical density (A600) of 5.0 made in 0.85% NaCl, and a 10-fold dilution series were made. 10 μl from the dilution series were spotted onto MRS+ agar plates containing 5% NaCl, containing 0.05% bile salt (BS) or MRS+ agar adjusted to pH 4.5 with sodium citrate buffer. As a control, cultures were also spotted on MRS+ agar plates. Strains used for the assays are wild type L. rhamnosus LRB; RBM1, an isogenic mutant derived from LRB displaying the acid sensitive phenotype; and RBM2, another isogenic mutant derived from LRB. Experiments were repeated at least three times, and relevant areas of representative plates are shown.
We found that RBM1-R also shows growth defect when grown in THY medium instead of MRS or MRS+ (Fig. 1). We also found that RBM1-R showed sensitivity towards osmotic stress since it was unable to grow on MRS containing 5% sodium chloride (Fig. 1). Unlike LGG, the LRB strain was more sensitive when exposed to 1% bile salt containing MRS plates (Biswas et. al. 2018). However, LRB could grow well on MRS agar containing bile salt if the final concentration was less than 0.2% (data not shown). We then tested RBM1-R and whether it can tolerate exposure to bile salt (0.05%). As shown in Fig. 1, we found that RBM1-R was highly sensitive to bile salt. Since LRB is known to produce antimicrobial activity, we then tested antimicrobial activity against some selected streptococci isolates and ESKAPE pathogens. We found that RBM1 was as capable of inhibiting all the tested organisms with equal efficiency as the wild type LRB, indicating that antimicrobial production and stress sensitivity phenotypes were not intimately connected (data not shown).
To identify the location of putative ISS1 insertion on RBM1-R, we followed the standard identification methods (Biswas and Biswas 2011). After several attempts, we were unable to identify the insertion site on RBM1-R. We checked the presence of ISS1 in RBM1-R cells by PCR amplification and found a positive amplification (data not shown). Therefore, we decided to perform whole genome sequencing to map the putative insertion site. For this, we first cured the resident pGhost9:: ISS1 plasmid (either free or integrated) by serially passaging the strain without antibiotic selection and under non permissive temperature (37°C). We selected one isolate that had lost the erythromycin-resistance cassette, and hence the plasmid. This isolate, which we called RBM1 (without R) was used for further verification. We first confirmed the acid sensitive phenotype and found that RBM1 is sensitive to pH 4.5 like the original mutant RBM1-R (data not shown). Genomic DNA was isolated from RBM1 and subjected to whole genome sequencing by the Illumina platform. We achieved a genome coverage of nearly 60X. We then mapped the Illumina sequencing reads on the parent LRB genome and looked for insertion/deletion or single nucleotide changes as described in the method section. We found three single nucleotide changes in the RBM1 genome. The first polymorphism was mapped on the ftsH gene at position 791 of the open reading frame. A nucleotide alteration from C to T generated a change of amino acid from Gly to Asp at codon position 263 in the FtsH protein. The second polymorphism, which resulted in a C to A change, was mapped on a putative ABC transporter encoding gene at position 175 that caused a Pro to Thr change at the amino acid level. The third change, which was from G to T, was mapped on a gene encoding a putative bacitracin ABC transporter; however, this nucleotide substitution did not alter the amino acid residue.
RBM1 is sensitive to various environmental stresses.
To characterize RBM1 further, we first tested whether RBM1 is sensitive to thermal stress. For this, we spotted different dilutions of the cultures on MRS agar plates and incubated the plates at 42°C. As shown in Fig. 2, RBM1 indeed displayed a strong growth defect as compared to the wild type LRB when grown on high temperature. Based on the dilution spotting, it seemed RBM1 was nearly ten-fold more sensitive to exposure to 42°C compared to LRB. To confirm the thermal stress phenotype, we used a disc diffusion assay with puromycin. Puromycin causes premature chain termination during protein synthesis and thus mimics thermal stress. As expected, the zone of inhibition (ZOI) produced by puromycin was 21±1 mm for RBM1 whereas the ZOI was 16±1mm for the wild type strain (Fig. 2B and Table 1). Taken together, our results indicated that RBM1 was sensitive to thermal stress.
Figure 2.
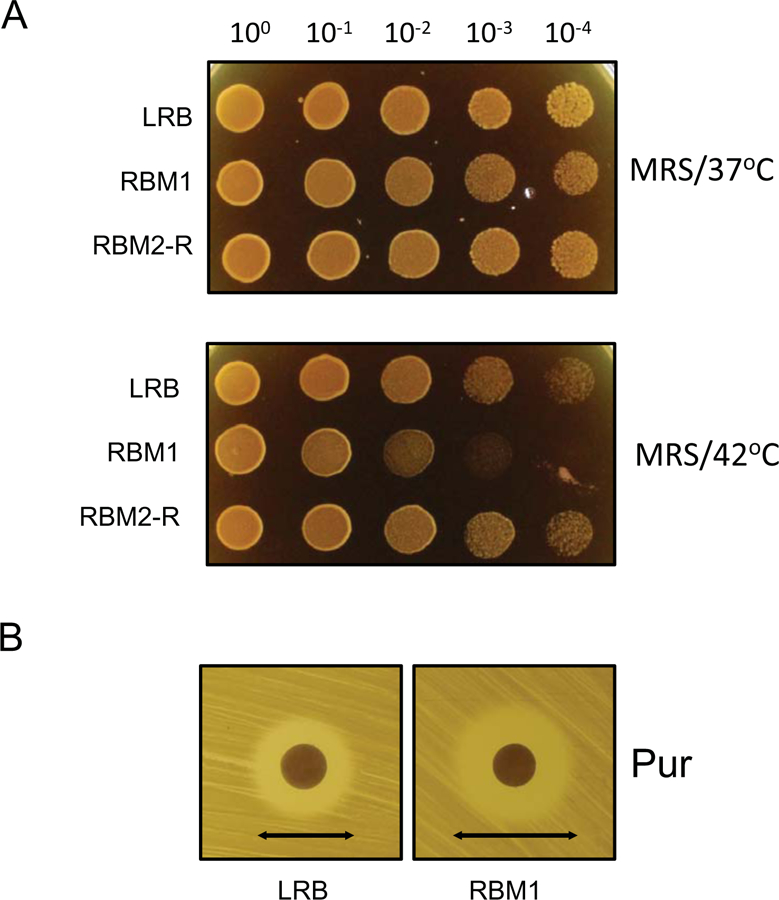
Thermo-sensitive phenotypes of RBM1. A) Dilutions of fresh overnight cultures were prepared as described in Fig. 1 and spotted on MRS or THY agar plates. Plates were incubated at 37°C or 42°C under microaerophilc conditions. B) Disk diffusion assay to measure the susceptibility against puromycin. Plates were prepared by spreading bacterial cultures as described in the text. A filter paper disk (6 mm) was placed on the plates and puromycin was added to the discs. The plates were then incubated under microaerophilic conditions at 37°C for 16 h. The inhibitory-zone diameters (indicated by arrow) for LRB and RMB1 were measured and compared. Experiments were repeated no fewer than three times, and relevant areas of representative plates are shown.
Since we found that the RBM1 mutant was sensitive to acid, osmotic, and thermal stresses, we wondered whether the mutant was sensitive to DNA damage. For this assay, we first exposed the cells to UV-induced DNA damage. As shown in Fig. 3, RBM1 cells were more susceptible to UV-induced DNA damage than the parental LRB cells. To confirm the DNA damage repair defect, we also used mitomycin C, which alkylates double-stranded DNA and blocks DNA replication to generate double stranded DNA breaks. We found that RBM1 was more sensitive to mitomycin C as compared to LRB (ZOI 33±1 mm and 39±0.5 mm for LRB and RBM1, respectively; Table 1). We also found that RBM1 was more sensitive to ciprofloxacin, which targets DNA gyrase. The mutant strain RBM1 was much more sensitive to ciprofloxacin as compared to LRB (ZOI 18±2 mm and 26±2 mm for LRB and RBM1, respectively; Table 2). Taken together our data indicated that RBM1 had reduced ability to withstand DNA damage insults.
Figure 3.
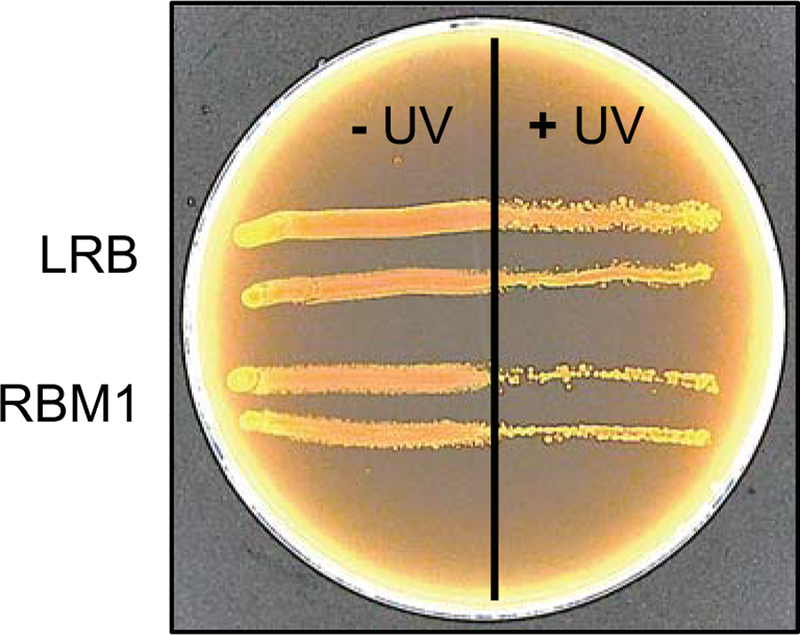
RBM1 is sensitive to DNA damage-inducing stress. Freshly swabbed bacterial cultures on the MRS agar surface were exposed to UV short-wave radiation for 15 s. Half of the plate was covered using a glass plate to block UV irradiation. The irradiated plate was incubated at 37°C under microaerophilic conditions, and growth was monitored for up to 48 h. UV sensitivity was assessed by observing the growth defect on the UV- irradiated portion of the plate. Experiments repeated at least three times and a representative plate is shown.
Since L. rhamnosus is a facultative anaerobic organism, we wanted to verify whether RBM1 has any growth defect when grown under ambient air. We found both RBM1 and LRB grew equally well on MRS agar plates under ambient air, albeit the growth was more robust when grown under microaerophilic condition (data not shown). This observation suggested that RBM1 is not more sensitive to the oxidative stress response compared to LRB. Along the same lines, we then tested the strains’ ability to withstand exposure to hydrogen peroxide. We found that both RBM1 and LRB were equally sensitive to hydrogen peroxide (~40 mm; Table 1). We next tested for the ability to withstand superoxide stress generated by ethyl viologen (EV) and methyl viologen (MV). We found that the RBM1 mutant was highly sensitive to both EV and MV. The ZOIs for EV and MV were 34±1 mm and 37±2 mm, respectively for the RBM1 strain; whereas ZOIs for the LRB strain for EV and MV were 20±1 mm and 25±1 mm, respectively. Interestingly, there was no halo formation when RBM1 and LRB were subjected to superoxide stress generated by tert-butyl hydroperoxide (Table 1). However, when we used menadione, which is structurally and chemically different from viologens, we found that RBM1 is highly sensitive to menadione (Fig. 4 and Table 1). We noticed that sensitivity to menadione is highly variable and depends on the age of the culture. The difference between RBM1 and LRB is reduced when the cultures are from the stationary phase. In addition to menadione, we also tested the sensitivity of RBM1 to diquat, since it is structurally similar to viologen and generates superoxide. We found that the ZOI was larger for RBM1 compared to LRB (Fig. 4 and Table 1). However, we also noticed that the ZOI for RMB1 was more diffused than the wild type LRB strain. Taken together our results suggested that RBM1 was highly sensitive to superoxide stress generated by viologens, diquat and menadione.
Figure 4.
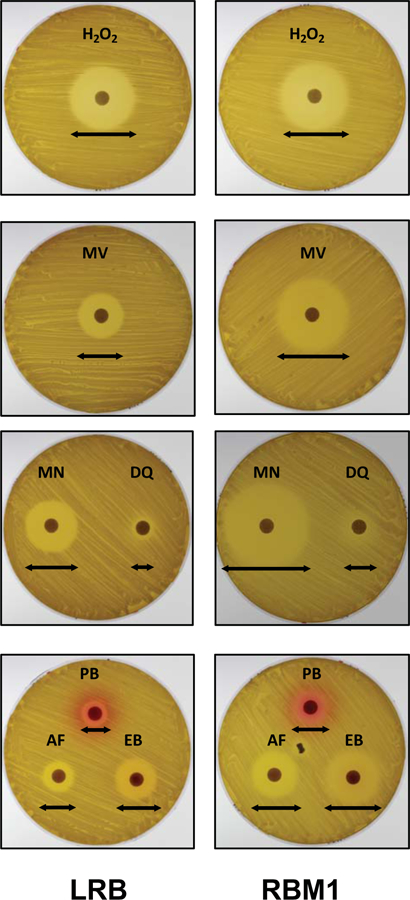
Susceptibility of RBM1 to various stressors as determined by disk diffusion assay. Plates with overlaid cultures were prepared as described in the text. Filter paper disks (6 mm) containing indicated stressors were placed on the agar plates. The plates were then incubated under microaerophilic conditions at 37°C for 16 h and inhibitory-zone diameters (indicated by arrows) were measured and compared. The stressors used are acriflavin (AF; 10 mg/ml), diquat dibromide (DQ; 500 mg/ml), ethidium bromide (EB; 10 mg/ml), hydrogen peroxide (H2O2; 10%), menadione (MN; 1%) methyl viologen (MV; 1.0 M), or pyronin B (PB; 0.5%). Assays were repeated twice and representative plates are shown.
Both viologen and diquat are charged molecules, structurally similar and contain the dipyridyl group. We wanted to examine whether RBM1 was more sensitive to compounds that are similar to the dipyridyl group but do not generate superoxide radicals. Thus, we tested the sensitivity of the RBM1 strain against 2,2′-dipyridyl, benzidine and 1,10′-phenanthroline. Both dipyridyl and benzadine did not produce any ZOI on either of the strains. On the other hand, 1,10′-phenanthroline produced a larger ZOI on RBM1 lawns (25±1 mm) as compared to LRB lawns (22±1) [Table 1].
Since the reagents that we used are all quaternary ammonium compounds (QAC), we wanted to know whether RBM1 was sensitive to other QACs. Therefore, we tested RBM1 for sensitivity against commonly used QACs, such as acriflavine (used as disinfectant), benzalkonium chloride (used in mouthwash), ethidium bromide, and pyronin B (used as biological dye). As shown in Fig. 4, RBM1 consistently showed enhanced sensitivity to all the tested QACs, with ZOIs between 115–155% larger than those of the wild type LRB strain (Table 1). Thus, our results showed that RBM1 was more sensitive to QAC compounds.
RBM1 is susceptible to certain antibiotics.
We next examined the sensitivity of the RBM1 strain towards antibiotics that target all five known biosynthesis pathways. For this we used a disc diffusion assay with freshly grown bacterial lawns and commercially available antibiotic discs as well as lab-made discs. The list of antibiotics used for the testing is shown in Table 2. We found that RBM1 was more sensitive than LRB to antibiotics that target the cell membrane (such as bacitracin and nisin) and cell wall biosynthesis (such as penicillin, imipenem, meropenem, and other cephalosporins). We also found that RBM1 was more sensitive to rifampicin, which targets transcription. In addition, we also found that RMB1 was more sensitive to the antibiotics that selectively target protein biosynthesis (Table 2). We noticed that for some of the antibiotics (such as cefixime, trimethoprime, vancomycin et. al.; Table 2) there was no ZOI on both the LRB and RBM1 strains. It could be due to the insufficient amounts of drugs present on the discs or L. rhamnosus could be intrinsically resistant to these antibiotics. We also tested chlorohexidine gluconate, which disrupts the cell membrane and potassium tellurite, which inhibits the cell growth by producing excess amounts reactive oxygen. We found that chlorohexidine produced larger ZOI on RBM1 lawns (20±1 mm) compared to LRB lawns (15±1 mm) [see Table 1]. On the other hand the ZOIs for potassium tellurite were similar on both the strains (8±1 mm, Table 1). Taken together, our data suggested that RBM1 was more sensitive to certain antibiotics as compared to the wild type LRB strain.
RBM1 displays an altered proteome.
To gain an overview at the molecular level for such stress sensitive phenotypes, we performed proteomic studies using tandem-mass tag (TMT) with multidimensional protein identification technology (mudpit) as described in the method section. For this assay, we used THY medium instead of MRS medium to avoid intensive alcohol and acetaldehyde fermentation during growth (Laakso et. al. 2011). Furthermore, RBM1 displays a growth defect when grown in THY (Fig. 1). We grew both LRB and RBM1 strains under microaerophilic condition at 37°C until mid-logarithmic phase (OD600 = 0.7), at which point samples were collected for TMT analysis. LRB and other L. rhamnosus strains encode approximately 2000 proteins (Koskenniemi et. al. 2011; Laakso et. al. 2011; Savijoki et. al. 2011). In this study, we could only identify ~950 proteins with TMT. Approximately 160 proteins were upregulated at least 2-fold or more in the RBM1 mutant strain as compared to the wild type LRB strain. The highest differentially expressed protein was the F0/F1 ATPase subunit C, which was nearly 10-fold less abundant in the mutant (Table 3A). Interestingly many ribosomal associated proteins were found to be downregulated in the mutant. At least seven 50S ribosomal proteins (L2, L5, L13, L19, L21, L22, and L35) were less abundant in RBM1 (Table 3A and Table S1). Similarly, five 30S ribosomal proteins (S7, S11, S12, S13, and S21) were less abundant in the RBM1 strain. Among the other notable proteins that were found in lower amounts in RBM1 (> 2-fold) are D-alanyl-D-alanine carboxypeptidase, D-alanine--D-alanyl carrier protein ligase (DltA) flavodoxin, a putative oxidoreductase, putative dehydrogenase, several ABC transporters, RecA, and several conserved hypothetical proteins (Table 3A and Table S1).
Table 3A:
Differentially expressed proteins identified by Tandem mass tag (TMT) [Downregulated in RBM1]
| Protein ID | Description | Fold change (RBM1/LRB) | # Unique peptides | # MS/MS scans scored |
|---|---|---|---|---|
| BAI41653.1 | F0F1- ATPase subunit C | 0.102 | 2 | 5 |
| BAI41046.1 | Putative hydrolase | 0.236 | 3 | 6 |
| BAI41533.1 | Flavodoxin | 0.245 | 4 | 7 |
| BAI43237.1 | Argininosuccinate synthase | 0.286 | 5 | 14 |
| BAI42025.1 | Aspartyl-tRNA synthetase | 0.305 | 11 | 27 |
| BAI41740.1 | MreB | 0.306 | 10 | 26 |
| BAI42395.1 | Oligopeptide ABC transporter | 0.311 | 15 | 93 |
| BAI42125.1 | Hypothetical protein | 0.312 | 2 | 6 |
| BAI40637.1 | Amidohydrolase | 0.331 | 2 | 2 |
| BAI43157.1 | Dipeptidase | 0.333 | 2 | 3 |
| BAI42020.1 | 30S ribosomal protein S21 | 0.337 | 5 | 53 |
| BAI42898.1 | 30S ribosomal protein S11 | 0.342 | 5 | 30 |
| BAI42063.1 | Hypothetical protein | 0.342 | 9 | 55 |
On the other hand, we found nearly 100 proteins that were found in 2-fold or more in the RBM1 mutant strain (Table 3B and Table S1). We noticed that many tRNA synthetases were more abundant in the RBM1 mutant strain as compared to the wild type LRB strain. Surprisingly, we found that two F0/F1 ATPase subunits (α and γ) and a different flavodoxin were more abundant in RBM1. We also found that a glutathione reductase was upregulated in the RBM1 mutant. Taken together, our proteomic data suggested that ~15% of the proteins were differentially expressed in the RBM1 mutant strain.
Table 3B:
Differentially expressed proteins identified by Tandem mass tag (TMT) [Upregulated in RBM1]
| Protein ID | Description | Fold change (RBM1 / LRB) | # Unique peptides | #MS/MS scans Sscored |
|---|---|---|---|---|
| BAI41433.1 | Putative metal dependent hydrolase | 2.897 | 5 | 17 |
| BAI43325.1 | Ribose-phosphate pyrophosphokinase | 2.948 | 6 | 14 |
| BAI40835.1 | Deoxyribose-phosphate aldolase | 3.032 | 9 | 31 |
| BAI41170.1 | Tagatose-1,6-bisphosphate aldolase | 3.087 | 11 | 68 |
| BAI41122.1 | Glutamate dehydrogenase | 3.117 | 10 | 17 |
| BAI41362.1 | Putative rRNA methyltransferase | 3.141 | 3 | 6 |
| BAI41418.1 | Phosphoglycerate kinase | 3.158 | 22 | 152 |
| BAI41650.1 | Uracil phosphoribosyltransferase | 3.183 | 4 | 25 |
| BAI43319.1 | Phage major capsid protein | 3.188 | 10 | 16 |
| BAI40747.1 | Flavodoxin | 3.182 | 2 | 5 |
| BAI42971.1 | CTP synthase | 3.193 | 7 | 30 |
| CAR87156.1 | Valyl-tRNA synthetase | 3.211 | 27 | 100 |
| BAI42423.1 | Dipeptidase | 3.261 | 10 | 48 |
| BAI40696.1 | Hypothetical protein | 3.313 | 2 | 3 |
| BAI41958.1 | Asparaginyl-tRNA synthetase | 3.317 | 9 | 32 |
| BAI41401.1 | HPr kinase/phosphorylase | 3.325 | 7 | 23 |
| BAI41732.1 | Cysteine desulfurase | 3.376 | 9 | 34 |
| BAI42985.1 | Ribose-phosphate pyrophosphokinase | 3.411 | 8 | 28 |
| BAI40837.1 | Purine-nucleoside phosphorylase | 3.476 | 7 | 26 |
| CAR88403.1 | Lysyl-tRNA synthetase | 3.489 | 16 | 62 |
| BAI43305.1 | Hypothetical protein | 3.479 | 3 | 6 |
| BAI42948.1 | L-lactate dehydrogenase | 3.522 | 11 | 100 |
| BAI41327.1 | S-adenosylmethionine synthetase | 3.599 | 10 | 32 |
| BAI42999.1 | Tagatose 1,6-diphosphate aldolase | 3.652 | 17 | 200 |
| BAI41815.1 | Elongation factor Tu | 3.673 | 16 | 200 |
| BAI41160.1 | UDP-glucose 4-epimerase | 3.726 | 6 | 27 |
| BAI43010.1 | Proline iminopeptidase | 3.804 | 11 | 45 |
| BAI41656.1 | F0F1-type ATP synthase subunit α | 3.83 | 20 | 120 |
| BAI42026.1 | Histidyl-tRNA synthetase | 3.9 | 14 | 65 |
| BAI42447.1 | dTDP-glucose-4,6-dehydratase | 3.942 | 10 | 56 |
| BAI40874.1 | Hypothetical protein | 3.945 | 5 | 15 |
| BAI41657.1 | F0F1-type ATP synthase subunit γ | 3.979 | 8 | 37 |
| BAI43119.1 | Ribose-5-phosphate isomerase | 4.009 | 10 | 45 |
| BAI43038.1 | Glutathione reductase | 4.03 | 6 | 26 |
| BAI41034.1 | Fructose-bisphosphate aldolase | 4.057 | 7 | 81 |
| BAI40660.1 | Adenylosuccinate synthase | 4.109 | 14 | 74 |
| CAR87079.1 | ATP synthase B chain | 4.151 | 15 | 91 |
| BAI41558.1 | Adenylosuccinate lyase | 4.19 | 18 | 76 |
| BAI42159.1 | Glutamine synthetase | 4.223 | 17 | 90 |
| BAI41346.1 | Hypothetical protein | 4.312 | 7 | 25 |
| BAI42241.1 | Putative hydrolase | 4.367 | 5 | 35 |
| BAI41405.1 | Phosphoglucomutase | 4.412 | 21 | 122 |
| BAI41238.1 | Nucleoside diphosphate kinase | 4.773 | 3 | 10 |
| BAI41359.1 | GMP oxidoreductase | 4.851 | 11 | 56 |
| BAI40798.1 | Gluconate kinase | 5.287 | 8 | 19 |
| BAI41002.1 | NAD-dependent dehydrogenase | 5.358 | 8 | 62 |
| BAI41443.1 | Phosphotransacetylase | 5.719 | 6 | 42 |
| CAR86129.1 | Putative protein without homology | 6.069 | 2 | 5 |
| BAI42940.1 | Hypoxanthine p-ribosyltransferase | 8.063 | 4 | 15 |
| BAI40758.1 | Hypothetical protein | 9.791 | 2 | 7 |
While TMT coupled with mudpit offers certain advantages, some of the proteins are often missed due to limited dynamic range and contaminating peptides (Singh et. al. 2016). It is sometimes beneficial to apply more than one technique for quantification, as the complementarity of various approaches might generate a greater proteome coverage (O’Connell et. al. 2018)). Towards this end, we used level-free quantitation (LFQ) that was supposed to have a higher dynamic range and offer a higher proteome coverage depending on the nature of the samples. For LFQ studies, we grew the cells (RBM1 and LRB) in THY broth at 37°C till the culture reached the mid-logarithmic growth phase. At this growth point, samples were taken for LFQ analysis as described in the method section. With LFQ, we could identify approximately 740 proteins. We found over 200 proteins were differentially regulated in the RBM1 mutant strain as compared with the wild type LRB strain. The differentially regulated proteins are shown in Fig. 6 as a volcano plot. We did not detect any signals for at least eight proteins in RMB1 strain: BAI41051.1, BAI42059.1, BAI42276.1, BAI42439.1, BAI42568.1, BAI42730.1, BAI42735.1, and BAI43280.1 (Table 4).
Figure 6.
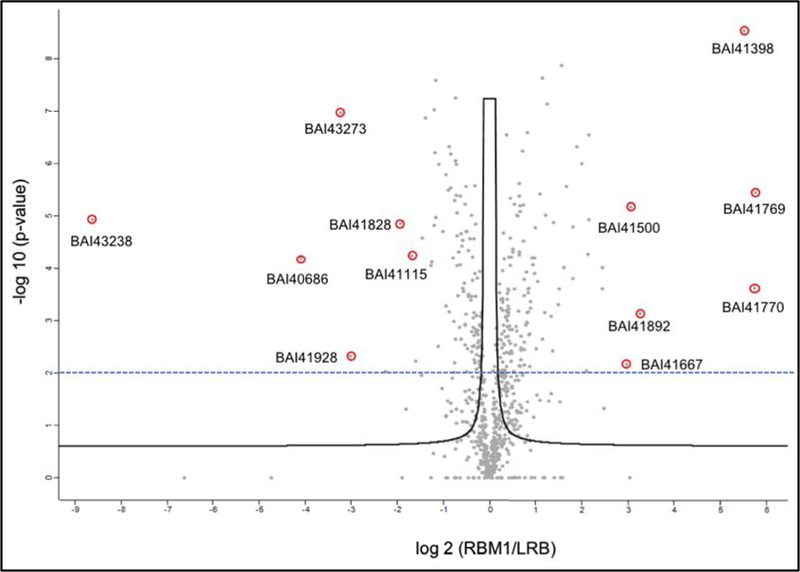
Label-free quantitative analysis differentially expressed proteins. Volcano plot representing the logarithmic ratio of protein LFQ intensities in the RBM1/LRB experiments plotted against negative logarithmic p-values of the t-test performed from triplicates (FDR threshold = 0.1, S0 = 0.5). A hyperbolic threshold curve separates significant differentially expressed proteins from background. Some of the most differentially expressed proteins are indicated. Dashed line indicates a more stringent p-value cutoff (2.0). Proteins that are completely absent in one of the strains are not indicated in the plot.
Table 4:
Differentially expressed proteins identified by Label Free Quantitation (LFQ)
| Protein ID | Description | Score | Sequence coverage (%) | Peptides |
|---|---|---|---|---|
| Absent or Downregulated in LRB | ||||
| BAI40759.1 | Putative protein without homology (DUF979) | 3.16 | 8.7 | 2 |
| BAI40813.1 | Transcriptional regulator, LytR family | 3.96 | 4.9 | 2 |
| BAI40849.1 | Beta-galactosidase (DU507_09460) | 10.94 | 8.6 | 5 |
| BAI41212.1 | Conserved protein (DU507_11915) | 134.28 | 16.4 | 4 |
| BAI41385.1 | Cell division protein FtsX | 2.98 | 7.5 | 2 |
| BAI41399.1 | Conserved protein | 6.13 | 18.6 | 1 |
| BAI41659.1 | F0F1-type ATP synthase subunit epsilon | 62.87 | 23.1 | 1 |
| BAI41759.1 | Cell division protein, FtsA | 9.43 | 13.5 | 4 |
| BAI41771.1 | NUDIX hydrolase | 10.74 | 13.7 | 2 |
| BAI41853.1 | Ribosomal large subunit pseudouridine synthase B | 6.22 | 17.7 | 4 |
| BAI42028.1 | ABC transporter | 61.49 | 25.4 | 4 |
| BAI42242.1 | Conserved protein | 6.2 | 21.8 | 1 |
| BAI42505.1 | Conserved membrane protein | 2.69 | 12.0 | 2 |
| BAI42666.1 | Putative protein without homology | 8.22 | 7.2 | 2 |
| BAI42949.1 | ErfK/YbiS/YcfS/YnhG family protein | 7.27 | 10.3 | 3 |
| BAI43089.1 | Putative 2-dehydro-3-deoxyphosphogluconate aldolase | 33.87 | 30.8 | 5 |
| Absent or Downregulated in RBM1 | ||||
| BAI41051.1 | Transcriptional regulator, HxlR family (DU507_01520) | 4.59 | 14.9 | 1 |
| BAI42059.1 | GTP cyclohydrolase (DUF960) | 2.85 | 8.6 | 1 |
| BAI42276.1 | Phosphoribosylaminoimidazole carboxylase | 26.88 | 15.4 | 2 |
| BAI42439.1 | GTP-binding protein, HflX subfamily (DU507_04765) | 71.01 | 27.6 | 10 |
| BAI42568.1 | (3R)-hydroxymyristoyl-ACP dehydratase (DU507_07220) | 4.60 | 12.0 | 2 |
| BAI42730.1 | Amidase | 19.17 | 14.9 | 5 |
| BAI42735.1 | Nucleoside deoxyribosyltransferase (DU507_10485) | 4.38 | 8.2 | 1 |
| BAI43280.1 | Amino acid ABC transporter permease component (DU507_14610) | 7.73 | 10.1 | 5 |
On the other hand, we found about 16 proteins that were present in the mutant but were not detected in the wild type LRB strain. These are: BAI40759.1, BAI40813, BAI40849.1, BAI41212.1, BAI41385.1, BAI41399.1, BAI41659.1, BAI41759.1, BAI41771.1, BAI41853.1, BAI42028.1, BAI42242.1, BAI42505.1, BAI42666.1, BAI42949.1, and BAI43089.1 (Table 4). Among these, six are uncharacterized conserved proteins with no known function. We also found that the F0F1 ATP synthase subunit epsilon (BAI41659.1) was not detected in LRB while it was highly expressed in RMB1. Surprisingly, we found that FtsH was overexpressed in the RBM1 mutant as compared to LRB (detected in both).
Differential expression analysis by these two different methods (LFQ, and TMT) resulted in about 10% overlap. Mudpit was applied only to the TMT dataset, whereas a single long gradient was used in the LFQ analysis. In addition, the TMT analysis in Mascot utilizes reporter ions in the MS2 scans of matches that meet significance threshold, whereas MaxQuant uses the MS2 only for identification, but tracks MS peak intensity in the data for quantitation. Although it is possible that more overlap would have been seen if mudpit were applied to the LFQ analysis, the results of the proteomic studies provide evidence for a significantly altered proteome in the RBM1 mutant strain compared to the wild type LRB strain.
DISCUSSION:
L. rhamnosus is known to be a highly acidogenic and aciduric organism which predominates in various low pH ecological niches, including the oral cavity and gastrointestinal tract of humans. In an environment where sugar is plentiful, L. rhamnosus readily brings down the surrounding pH below 4.0 by producing copious amounts of lactic acid (Piwat et. al. 2012). Furthermore, a comparative study indicates that L. rhamnosus, along with a few other lactobacilli, can grow well even when the external pH is less than 5.0 (Piwat et. al. 2012). It is suggested that survival of L. rhamnosus in acid stress conditions is regulated by multiple cellular processes.
However, a low acid producing strain is more suitable for dental health as long as it retains its antimicrobial properties. To generate such strain and gain insight into the acid stress tolerance response, we first performed transposon mutagenesis in L. rhamnosus LRB using the pGhost9:ISS1 plasmid. After screening nearly 10,000 mutants we only obtained one mutant that had passed the secondary screening. The number of transposon mutants that showed the desirable phenotype seemed to be very low as compared to other cases. We believe this low frequency is due to the fact that ISS1 failed to insert into the LRB genome. Since ISS1 is a replicative transposon, after excision from the chromosome, it leaves a copy of ISS1 at the original insertion site. We were unable to identify the integrated ISS1 sequence in the mutant genome suggesting that ISS1 was never inserted into the LRB genome. This was surprising since ISS1 is highly efficient in many Gram-positive bacteria including lactobacilli (Biswas and Scott 2003; Gury et. al. 2004; Hossain and Biswas 2012; Liu et. al. 2011; Maguin et. al. 1996). One reason could be that the L. rhamnosus genome, specifically LGG and LRB, contains numerous IS and IS-like elements (Biswas and Biswas 2016; Morita et. al. 2009). These resident IS elements could interfere with the ISS1 transposition. Alternatively, ISS1 insertion could have been prevented by host encoded factors such as nucleoid associated proteins (Vandecraen et. al. 2017). Whatever the reason, it appears that ISS1 is not a suitable tool for mutagenesis in L. rhamnosus.
The mutant (RBM1) that we obtained was not only sensitive to acid stress but also to other stresses including thermal and osmotic stresses (Fig 1–3). But fortunately, the mutant did not display any growth defect under ambient oxygen, which is critical for an oral commensal.
The whole genome sequencing of RBM1 indicated that the strain harbors three mutations in three different genes. One of these mutations, which we mapped in a gene encoding a putative transporter protein, was silent and therefore it should not have any effect on the observed phenotype. The second mutation was mapped in a gene that encodes a putative ABC-transporter protein (WP_005685245.1). In this gene, a single base pair substitution resulted in a Pro to Thr change at the amino acid level. While this putative ABC transporter is highly conserved among all the L. rhamnosus strains as well as present in many lactobacilli. We believe this ABC-transporter is not responsible for the observed stress sensitive phenotypes; although we cannot rule out the possibility that this ABC transporter might have roles in some of the observed antibiotic sensitivities. The third mutation that we found was mapped on the ftsH gene (BFC96_10340). The mutation resulted in change of a Gly residue to an Asp residue at codon position 263 (G263D) in the FtsH protein.
FtsH is a membrane associated ATP-dependent zinc metalloprotease for both cytoplasmic and membrane proteins. This protein has been extensively studied in E. coli where it is involved in multiple biological processes including degradation of membrane proteins such as F0 ATPase subunit a and SecY; role in heat shock response, LPS biosynthesis, and cell division [for reviews see, (Bittner et. al. 2017; Ito and Akiyama 2005; Janska et. al. 2013; Langklotz et. al. 2012)]. FtsH resides in the inner membrane as a homohexamer that uses its ATPase domain to unfold and translocate substrates that are subsequently degraded in the proteolytic chamber of the protease domain. Thus, FtsH eliminates misfolded proteins in the context of general quality control and properly folded proteins for regulatory reasons. Because of its involvement in protein quality control and other biological processes, FtsH is an essential protein in E. coli.
E. coli FtsH protein, which is 647 residues long, encodes two N-terminal transmembrane segments that are followed by a central cytoplasmic ATPase domain and a C-terminal cytoplasmic protease domain (Ito and Akiyama 2005). The cytoplasmic domains are highly conserved across several bacterial species. Numerous mutational studies with the FtsH protein in E. coli established several key residues in both the ATPase and the protease domains. One such residue is a Gly at position 230 (G230). This residue along with conserved Phe228 (F228) was predicted to lie in the central pore region of the FtsH hexamer. Ogura and colleagues have shown that substituting a Gly with an Ala reside at position 230 (G230A) markedly reduced ATPase activity and was unable to degrade a natural substrate heat shock sigma factor σ32 (Yamada-Inagawa et. al. 2003). However, G230A was able to degrade an unfolded substrate. Thus, a single amino change has a profound effect on FtsH function.
While FtsH is an essential protein in E. coli, the essentiality of FtsH varies in Gram-positive bacteria. It appears that FtsH is not essential in Bacillus subtilis, Lactobacillus plantarum and Lactococcus lactis, (Fiocco et. al. 2009; Lysenko et. al. 1997; Nilsson et. al. 1994). On the other hand, FtsH is essential in Streptococcus mutans and a few other streptococci, but not all (Shields et. al. 2018). At present we do not know whether FtsH is essential for survival in L. rhamnosus and other lactobacilli. L. rhamnosus FtsH protein is 717 residues long. We found that the G263 residue of FtsH of L. rhamnosus, which corresponds to the G230 residue of E. coli FtsH, is also very important for its proper function. In fact, a G263D mutation displayed a wide range of stress sensitive phenotypes in L. rhamnosus suggesting that FtsH indeed plays a key role in stress response.
Since we have generated a stress sensitive strain, our next step was to create a molecular map of the stress tolerance processes. Our proteomic studies indicated that nearly one fifth of the L. rhamnosus proteome is differentially regulated in the RBM1 mutant strains. Considering that the RBM1 strain contains only G263D mutation in the FtsH protein, it seems that the number of proteins that are regulated by FtsH is very large. We speculate that not all these proteins are directly regulated by FtsH. It is possible that some of the altered proteome could also be due to the ABC transporter mutation. Since we found that several subunits of the F0F1 ATPase complex were expressed more in the mutant as compared with the wild type strain (Table 3–4), we speculate that these subunits could be the direct target for FtsH, similar to what was seen in E. coli (Akiyama et. al. 1996). We also found several proteins were down-regulated in the mutant strain. We speculate that these proteins are indirectly regulated by FtsH via some unknown factors. Additional studies are needed to identify the substrates that are directly regulated by FtsH in L. rhamnosus.
In general, lactobacilli seem to have slightly higher levels of spontaneous mutation frequency (Curragh 1992; Machielsen et. al. 2010). The exact reason for this enhanced frequency is not clearly understood. To understand the evolutionary bias, an experiment has been conducted by de Vos and colleague using L. rhamnosus GG strain (Douillard et. al. 2016). These authors found accumulation of several single nucleotide polymorphisms (SNPs) in the GG genome after approximately 1,000 generations of growth. Surprising, they also observed genome rearrangements when the cells were exposed to osmotic or shear stresses. These authors concluded that the frequency at which SNP accumulates is not higher than expected and genome rearrangements were mediated by the IS elements.
It was surprising for us to isolate a mutant strain with a mutation in one of the critical stress response proteins. Since we obtained this non-targeted mutation in this ftsH gene, it suggests that LRB and perhaps other L. rhamsnosus strains have inclination of accumulating mutations in ftsH, which could have a deleterious effect. Indeed, de Vos and colleagues have found a mutation in one of the DNA polymerase III subunits (Douillard et. al. 2016). Genome stability is an important issue that needs to be carefully assessed. We are currently performing experiments employing LRB and GG strains to evaluate the genome stability in L. rhamnosus.
Supplementary Material
Figure 5.
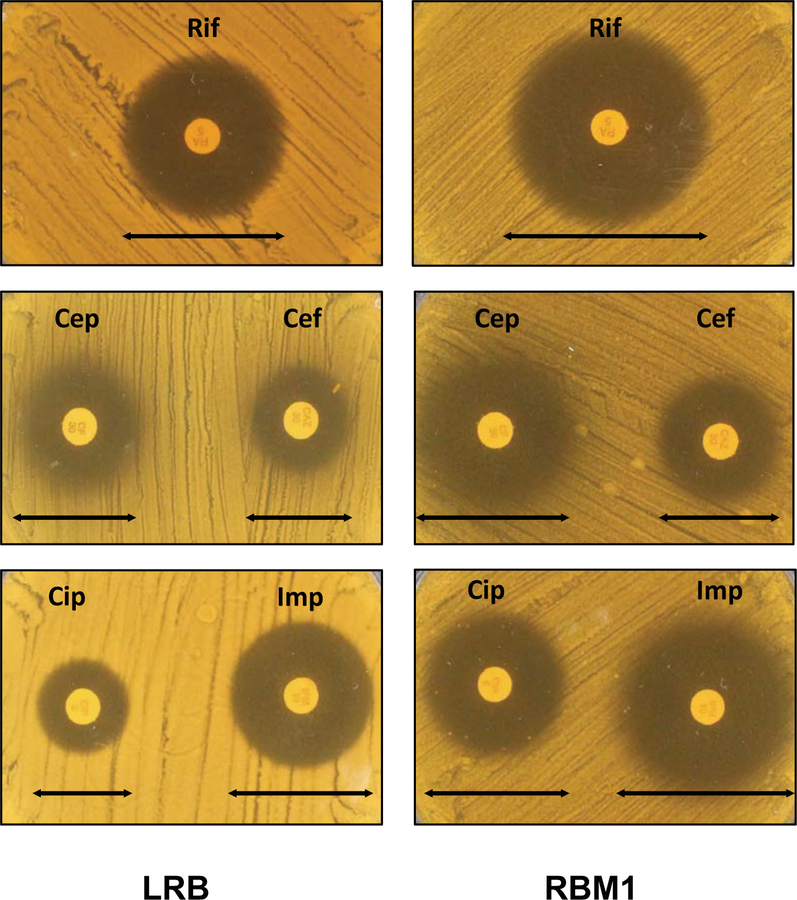
Antibiotic sensitivity assay. Disk diffusion assay was used to measure the susceptibility of LRB and RBM1 against different antibiotic discs. MRS+ agar containing bacterial lawns of LRB and RBM1 strains were prepared as described in the text. Antibiotics discs were placed and the plates were incubated further. Symbols are: Cef, ceftazidime; Cep, cephalothin; Cip, ciprofloxacin; Imp, imipenem; and Rif, rifampicin. Experiments were repeated no fewer than three times, and relevant areas of representative plates are shown.
ACKNOWLEDGEMENT:
This research is sponsored by NIDCR (NIH) under the award number DE026995.
Footnotes
CONFLICTS OF INTEREST
The authors declare no potential conflicts of interest with respect to the authorship and/or publication of this article.
REFERENES:
- Akiyama Y, Kihara A, and Ito K (1996) Subunit a of proton ATPase F0 sector is a substrate of the FtsH protease in Escherichia coli. FEBS Lett 399: 26–8. [DOI] [PubMed] [Google Scholar]
- Banna GL, Torino F, Marletta F, Santagati M, Salemi R, Cannarozzo E, Falzone L, Ferrau F, and Libra M (2017) Lactobacillus rhamnosus GG: An Overview to Explore the Rationale of Its Use in Cancer. Front Pharmacol 8: 603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas I, Drake L, Erkina D, and Biswas S (2008) Involvement of sensor kinases in the stress tolerance response of Streptococcus mutans. J Bacteriol 190: 68–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas I and Scott JR (2003) Identification of rocA, a Positive Regulator of covR Expression in the Group A Streptococcus. J Bacteriol 185: 3081–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas S and Biswas I (2011) Role of VltAB, an ABC transporter complex, in viologen tolerance in Streptococcus mutans. Antimicrob Agents Chemother 55: 1460–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas S and Biswas I (2014) A conserved streptococcal membrane protein, LsrS, exhibits a receptor-like function for lantibiotics. J Bacteriol 196: 1578–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas S and Biswas I (2016) Complete Genome Sequence of Lactobacillus rhamnosus Strain LRB. Genome Announc 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas S, Turner L, and Biswas I (2018) Lactobacillus rhamnosus LRB mediated inhibition of oral streptococci. Mol Oral Microbiol 33: 396–405. [DOI] [PubMed] [Google Scholar]
- Bittner LM, Arends J, and Narberhaus F (2017) When, how and why? Regulated proteolysis by the essential FtsH protease in Escherichia coli. Biol Chem 398: 625–635. [DOI] [PubMed] [Google Scholar]
- Caggia C, De Angelis M, Pitino I, Pino A, and Randazzo CL (2015) Probiotic features of Lactobacillus strains isolated from Ragusano and Pecorino Siciliano cheeses. Food Microbiol 50: 109–17. [DOI] [PubMed] [Google Scholar]
- Caufield PW, Schon CN, Saraithong P, Li Y, and Argimon S (2015) Oral Lactobacilli and Dental Caries: A Model for Niche Adaptation in Humans. J Dent Res 94: 110S–8S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curragh HJ, and Collins MA (1992) High levels of spontaneous drug resistance in Lactobacillus. Journal of Applied Microbiology 73: 31–36. [Google Scholar]
- Dal Bello F and Hertel C (2006) Oral cavity as natural reservoir for intestinal lactobacilli. Syst Appl Microbiol 29: 69–76. [DOI] [PubMed] [Google Scholar]
- De Angelis M and Gobbetti M (2004) Environmental stress responses in Lactobacillus: a review. Proteomics 4: 106–22. [DOI] [PubMed] [Google Scholar]
- Douillard FP, Ribbera A, Kant R, Pietila TE, Jarvinen HM, Messing M, Randazzo CL, Paulin L, Laine P, Ritari J, Caggia C, Lahteinen T, Brouns SJ, Satokari R, von Ossowski I, Reunanen J, Palva A, and de Vos WM (2013) Comparative genomic and functional analysis of 100 Lactobacillus rhamnosus strains and their comparison with strain GG. PLoS Genet 9: e1003683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douillard FP, Ribbera A, Xiao K, Ritari J, Rasinkangas P, Paulin L, Palva A, Hao YL, and de Vos WM (2016) Polymorphisms, Chromosomal Rearrangements, and Mutator Phenotype Development during Experimental Evolution of Lactobacillus rhamnosus GG. Applied and Environmental Microbiology 82: 3783–3792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duar RM, Lin XB, Zheng J, Martino ME, Grenier T, Perez-Munoz ME, Leulier F, Ganzle M, and Walter J (2017) Lifestyles in transition: evolution and natural history of the genus Lactobacillus. FEMS Microbiol Rev 41: S27–S48. [DOI] [PubMed] [Google Scholar]
- Fiocco D, Collins M, Muscariello L, Hols P, Kleerebezem M, Msadek T, and Spano G (2009) The Lactobacillus plantarum ftsH gene is a novel member of the CtsR stress response regulon. J Bacteriol 191: 1688–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouriet F, Million M, Henri M, Fournier PE, and Raoult D (2012) Lactobacillus rhamnosus bacteremia: an emerging clinical entity. Eur J Clin Microbiol Infect Dis 31: 2469–80. [DOI] [PubMed] [Google Scholar]
- Gury J, Barthelmebs L, and Cavin JF (2004) Random transposon mutagenesis of Lactobacillus plantarum by using the pGh9:IS S1 vector to clone genes involved in the regulation of phenolic acid metabolism. Arch Microbiol 182: 337–45. [DOI] [PubMed] [Google Scholar]
- Hossain MS and Biswas I (2012) An Extracelluar Protease, SepM, Generates Functional Competence-Stimulating Peptide in Streptococcus mutans UA159. Journal of Bacteriology 194: 5886–5896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito K and Akiyama Y (2005) Cellular functions, mechanism of action, and regulation of FtsH protease. Annu Rev Microbiol 59: 211–31. [DOI] [PubMed] [Google Scholar]
- Janska H, Kwasniak M, and Szczepanowska J (2013) Protein quality control in organelles - AAA/FtsH story. Biochim Biophys Acta 1833: 381–7. [DOI] [PubMed] [Google Scholar]
- Koskenniemi K, Laakso K, Koponen J, Kankainen M, Greco D, Auvinen P, Savijoki K, Nyman TA, Surakka A, Salusjarvi T, de Vos WM, Tynkkynen S, Kalkkinen N, and Varmanen P (2011) Proteomics and transcriptomics characterization of bile stress response in probiotic Lactobacillus rhamnosus GG. Mol Cell Proteomics 10: M110 002741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laakso K, Koskenniemi K, Koponen J, Kankainen M, Surakka A, Salusjarvi T, Auvinen P, Savijoki K, Nyman TA, Kalkkinen N, Tynkkynen S, and Varmanen P (2011) Growth phase-associated changes in the proteome and transcriptome of Lactobacillus rhamnosus GG in industrial-type whey medium. Microb Biotechnol 4: 746–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langklotz S, Baumann U, and Narberhaus F (2012) Structure and function of the bacterial AAA protease FtsH. Biochim Biophys Acta 1823: 40–8. [DOI] [PubMed] [Google Scholar]
- Lebeer S, Verhoeven TL, Perea Velez M, Vanderleyden J, and De Keersmaecker SC (2007) Impact of environmental and genetic factors on biofilm formation by the probiotic strain Lactobacillus rhamnosus GG. Appl Environ Microbiol 73: 6768–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Gao Y, Yu LR, Jones RC, Elkins CA, and Hart ME (2011) Inhibition of Staphylococcus aureus by lysostaphin-expressing Lactobacillus plantarum WCFS1 in a modified genital tract secretion medium. Appl Environ Microbiol 77: 8500–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lysenko E, Ogura T, and Cutting SM (1997) Characterization of the ftsH gene of Bacillus subtilis. Microbiology 143 ( Pt 3): 971–8. [DOI] [PubMed] [Google Scholar]
- Machielsen R, van Alen-Boerrigter IJ, Koole LA, Bongers RS, Kleerebezem M, and Van Hylckama Vlieg JE (2010) Indigenous and environmental modulation of frequencies of mutation in Lactobacillus plantarum. Appl Environ Microbiol 76: 1587–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maguin E, Prevost H, Ehrlich SD, and Gruss A (1996) Efficient insertional mutagenesis in lactococci and other gram-positive bacteria. Journal of Bacteriology 178: 931–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova K, Slesarev A, Wolf Y, Sorokin A, Mirkin B, Koonin E, Pavlov A, Pavlova N, Karamychev V, Polouchine N, Shakhova V, Grigoriev I, Lou Y, Rohksar D, Lucas S, Huang K, Goodstein DM, Hawkins T, Plengvidhya V, Welker D, Hughes J, Goh Y, Benson A, Baldwin K, Lee JH, Diaz-Muniz I, Dosti B, Smeianov V, Wechter W, Barabote R, Lorca G, Altermann E, Barrangou R, Ganesan B, Xie Y, Rawsthorne H, Tamir D, Parker C, Breidt F, Broadbent J, Hutkins R, O’Sullivan D, Steele J, Unlu G, Saier M, Klaenhammer T, Richardson P, Kozyavkin S, Weimer B, and Mills D (2006) Comparative genomics of the lactic acid bacteria. Proc Natl Acad Sci U S A 103: 15611–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez RM, Hulten KG, Bui U, and Clarridge JE 3rd. (2014) Molecular analysis and clinical significance of Lactobacillus spp. recovered from clinical specimens presumptively associated with disease. J Clin Microbiol 52: 30–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDowell GS, Gaun A, and Steen H (2013) iFASP: combining isobaric mass tagging with filter-aided sample preparation. J Proteome Res 12: 3809–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita H, Toh H, Oshima K, Murakami M, Taylor TD, Igimi S, and Hattori M (2009) Complete genome sequence of the probiotic Lactobacillus rhamnosus ATCC 53103. J Bacteriol 191: 7630–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadkarni MA, Chen Z, Wilkins MR, and Hunter N (2014) Comparative genome analysis of Lactobacillus rhamnosus clinical isolates from initial stages of dental pulp infection: identification of a new exopolysaccharide cluster. PLoS One 9: e90643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson D, Lauridsen AA, Tomoyasu T, and Ogura T (1994) A Lactococcus lactis gene encodes a membrane protein with putative ATPase activity that is homologous to the essential Escherichia coli ftsH gene product. Microbiology 140 ( Pt 10): 2601–10. [DOI] [PubMed] [Google Scholar]
- O’Connell JD, Paulo JA, O’Brien JJ, and Gygi SP (2018) Proteome-Wide Evaluation of Two Common Protein Quantification Methods. J Proteome Res 17: 1934–1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadimitriou K, Alegria A, Bron PA, de Angelis M, Gobbetti M, Kleerebezem M, Lemos JA, Linares DM, Ross P, Stanton C, Turroni F, van Sinderen D, Varmanen P, Ventura M, Zuniga M, Tsakalidou E, and Kok J (2016) Stress Physiology of Lactic Acid Bacteria. Microbiol Mol Biol Rev 80: 837–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrova MI, Macklaim JM, Wuyts S, Verhoeven T, Vanderleyden J, Gloor GB, Lebeer S, and Reid G (2018) Comparative Genomic and Phenotypic Analysis of the Vaginal Probiotic Lactobacillus rhamnosus GR-1. Front Microbiol 9: 1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piwat S, Teanpaisan R, Dahlen G, Thitasomakul S, and Douglas CW (2012) Acid production and growth by oral Lactobacillus species in vitro. J Investig Clin Dent 3: 56–61. [DOI] [PubMed] [Google Scholar]
- Robin F, Paillard C, Marchandin H, Demeocq F, Bonnet R, and Hennequin C (2010) Lactobacillus rhamnosus meningitis following recurrent episodes of bacteremia in a child undergoing allogeneic hematopoietic stem cell transplantation. J Clin Microbiol 48: 4317–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savijoki K, Lietzen N, Kankainen M, Alatossava T, Koskenniemi K, Varmanen P, and Nyman TA (2011) Comparative proteome cataloging of Lactobacillus rhamnosus strains GG and Lc705. J Proteome Res 10: 3460–73. [DOI] [PubMed] [Google Scholar]
- Segers ME and Lebeer S (2014) Towards a better understanding of Lactobacillus rhamnosus GG--host interactions. Microb Cell Fact 13 Suppl 1: S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shields RC, Zeng L, Culp DJ, and Burne RA (2018) Genomewide Identification of Essential Genes and Fitness Determinants of Streptococcus mutans UA159. mSphere 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva M, Jacobus NV, Deneke C, and Gorbach SL (1987) Antimicrobial substance from a human Lactobacillus strain. Antimicrob Agents Chemother 31: 1231–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh SA, Aikawa E, and Aikawa M (2016) Current Trends and Future Perspectives of State-of-the-Art Proteomics Technologies Applied to Cardiovascular Disease Research. Circ J 80: 1674–83. [DOI] [PubMed] [Google Scholar]
- Succi M, Tremonte P, Reale A, Sorrentino E, Grazia L, Pacifico S, and Coppola R (2005) Bile salt and acid tolerance of Lactobacillus rhamnosus strains isolated from Parmigiano Reggiano cheese. FEMS Microbiol Lett 244: 129–37. [DOI] [PubMed] [Google Scholar]
- Sun Z, Harris HM, McCann A, Guo C, Argimon S, Zhang W, Yang X, Jeffery IB, Cooney JC, Kagawa TF, Liu W, Song Y, Salvetti E, Wrobel A, Rasinkangas P, Parkhill J, Rea MC, O’Sullivan O, Ritari J, Douillard FP, Paul Ross R, Yang R, Briner AE, Felis GE, de Vos WM, Barrangou R, Klaenhammer TR, Caufield PW, Cui Y, Zhang H, and O’Toole PW (2015) Expanding the biotechnology potential of lactobacilli through comparative genomics of 213 strains and associated genera. Nat Commun 6: 8322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thibessard A, Fernandez A, Gintz B, Decaris B, and Leblond-Bourget N (2002) Transposition of pGh9:ISS1 is random and efficient in Streptococcus thermophilus CNRZ368. Can J Microbiol 48: 473–8. [DOI] [PubMed] [Google Scholar]
- van de Guchte M, Serror P, Chervaux C, Smokvina T, Ehrlich SD, and Maguin E (2002) Stress responses in lactic acid bacteria. Antonie Van Leeuwenhoek 82: 187–216. [PubMed] [Google Scholar]
- Vandecraen J, Chandler M, Aertsen A, and Van Houdt R (2017) The impact of insertion sequences on bacterial genome plasticity and adaptability. Crit Rev Microbiol 43: 709–730. [DOI] [PubMed] [Google Scholar]
- Walter J (2008) Ecological role of lactobacilli in the gastrointestinal tract: implications for fundamental and biomedical research. Appl Environ Microbiol 74: 4985–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Yang F, Gritsenko MA, Wang Y, Clauss T, Liu T, Shen Y, Monroe ME, Lopez-Ferrer D, Reno T, Moore RJ, Klemke RL, Camp DG 2nd, and Smith RD (2011) Reversed-phase chromatography with multiple fraction concatenation strategy for proteome profiling of human MCF10A cells. Proteomics 11: 2019–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerik N, Kort R, Sybesma W, and Reid G (2018) Lactobacillus rhamnosus Probiotic Food as a Tool for Empowerment Across the Value Chain in Africa. Front Microbiol 9: 1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada-Inagawa T, Okuno T, Karata K, Yamanaka K, and Ogura T (2003) Conserved pore residues in the AAA protease FtsH are important for proteolysis and its coupling to ATP hydrolysis. J Biol Chem 278: 50182–7. [DOI] [PubMed] [Google Scholar]
- Zheng J, Ruan L, Sun M, and Ganzle M (2015) A Genomic View of Lactobacilli and Pediococci Demonstrates that Phylogeny Matches Ecology and Physiology. Appl Environ Microbiol 81: 7233–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


