Abstract
Western populations use prescription and nonprescription drugs extensively, but large-scale population usage is rarely assessed objectively in epidemiological studies. Here we apply statistical methods to characterize structural pathway connectivities of metabolites of commonly used drugs detected routinely in 1H NMR spectra of urine in a human population study. 1H NMR spectra were measured for two groups of urine samples obtained from U.S. participants in a known population study. The novel application of a statistical total correlation spectroscopy (STOCSY) approach enabled rapid identification of the major and certain minor drug metabolites in common use in the population, in particular, from acetaminophen and ibuprofen metabolites. This work shows that statistical connectivities between drug metabolites can be established in routine “high-throughput” NMR screening of human samples from participants who have randomly self-administered drugs. This approach should be of value in considering interpopulation patterns of drug metabolism in epidemiological and pharmacogenetic studies.
The application of modern analytical and “omics” technologies to study large-scale population-based biological sample collections offers new opportunities to investigate many aspects of human biology and population phenotyping that were not envisaged at the time such epidemiological studies were initiated.1 Systems biology tools, such as metabonomics, can provide suitable analytical platforms for studying the varied responses of individuals and populations to drug therapy.1 For the majority of these epidemiological studies, intake of nutrients and other xenobiotics is self-reported via questionnaires and almost never validated. Thus, the researcher is ultimately reliant upon the cooperation and recall by study participants to obtain important environmental data. Detecting and monitoring drug intake and metabolism in such epidemiological studies can be particularly problematic since many over-the-counter analgesics contain a mixture of pharmaceutical components, of which the participant may be unaware. Furthermore, not only can these compounds and their major and minor metabolites be detected with modern analytical technologies used for phenotyping, but treatment with these compounds can impact directly on the endogenous metabolic profile via pharmacological interactions. We define the new concept of the xenometabolome as the multivariate description of the xenobiotic (foreign compound) metabolite profile of an individual or sample from an individual that has been exposed through any route (either deliberately or accidentally) to drugs, environmental pollutants, or dietary components that cannot be completely catabolized by endogenous metabolic enzyme systems. Thus, for a population of individuals, the xenometabolome becomes an exogenous part (not under direct mammalian genomic control) of the metabolic phenotype (metabotype) of that group that is characteristic of their general environmental exposures to chemicals or their behavioral choices relating to drug use. The ability to monitor exposure to xenobiotics, which may not be toxic in their own right, but which may carry a risk to certain individuals in populations after long-term exposure, is important in the broad context of epidemiological risk assessment and allows connection between “real-world” analytical data and epidemiological factors. Recently, we have applied NMR spectral tools to the investigation of biochemical features of the urine samples collected as part of the INTERSALT and INTERMAP studies,2,3 which investigate the role of nutrition in adverse population blood pressure levels.4–9 These samples were obtained from many populations and carry unique biochemical information about those groups and individuals.4–9
Ongoing work on animal and human metabolic phenotyping10–12 has revealed characteristic metabolic signatures relating to specific genotypes and strains of animal. These metabotypes are also heavily influenced by diet10 and gut microfloral contributions.13,14 In addition to metabolic phenotyping, we have previously shown that NMR spectroscopy is an effective tool for profiling the broad metabolic composition of biological samples15,16 and can also be applied as a discovery tool in drug metabolism studies.17,18 NMR profiling approaches and techniques that disperse overlapping signals such as two-dimensional NMR spectroscopic experiments and partial purification using chromatographic methods have been extensively applied to aid the isolation and identification of xenobiotics.19,20 Alternative approaches to detecting drug metabolites in biological samples include mass spectrometric methods such as searching for specific drug metabolite fragment patterns21,22 and chromatographic assays.23,24 LC-NMR and LC-MS-NMR approaches have proved particularly effective metabolic probes.25–27
Combining the capacity of NMR spectroscopy to simultaneously profile endogenous metabolites relating to specific phenotypes, and also to provide information on the intake and metabolism of xenobiotics such as drugs, we have used a new metabonomic approach to probe the detection of major urinary metabolites of xenobiotics in population studies. We and others have shown the power of pattern recognition tools for studying the metabolic effects of toxins and drugs28–30 and the use of “statistical spectroscopic” methods for biomarker recovery and molecular structure assignment. In particular, orthogonal projection to latent structure discriminant analysis (OPLS-DA),31 when applied to NMR data, statistical total correlation spectroscopy (STOCSY),32 and statistical heterospectroscopy (SHY)33 have proven useful for improved detection of potential biomarkers and for removing confounding analytical or biological variation from data sets. Since these methods operate using full NMR spectral resolution (digital point space), they allow direct identification of metabolites from the mathematical models via spectral reconstruction.
Here we present a strategy for directly detecting drugs and their metabolites in the biofluids of individuals sampled from free living human populations; this method is based on statistical correlation matrices calculated for individual spectral data points. Acetaminophen34 and ibuprofen35 are used to exemplify this method based on extensive knowledge of their metabolism and their popularity as over-the-counter analgesics.
METHODS
Study Design.
Urine samples from free living humans and corresponding information on analgesic intake were sourced from the INTERMAP study.36 Methods of data collection have been described.4 Each individual attended a specified clinic center on four occasions; the first two and last two visits were on adjacent days with a period of 3–6 weeks between these pairs of visits. Data collection included blood pressure on eight occasions (2/visit), four in-depth 24-h dietary recalls, two 7-day daily alcohol assessments by interview, and extensive questionnaire data on medication (including analgesic use), smoking, and other variables. At the first and third visits, a timed 24-h urinary collection was initiated, which was completed the following day in the clinic, according to a standardized protocol. Boric acid preservative (~5g/L) was incorporated in the urine collections. Specimens were mixed thoroughly and total volumes recorded. Aliquots (9 mL) were taken, frozen at −20 °C within 24 h, and shipped frozen to the central laboratory in Leuven, Belgium. The samples were stored at −40 °C before shipment on dry ice to the metabonomic laboratory at Imperial College. For this study, two subsets of samples were selected from 4290 urine spectra (first and repeat clinic visits for 2145 U.S. participants) based on the presence or absence of acetaminophen. A subset of 70 samples was selected from 413 urine spectra containing acetaminophen. A further subset of spectra was selected from the remaining 3877 urine spectra that did not contain acetaminophen metabolites. A subset of 70 samples (for consistency in comparison with the acetaminophen data set) was selected from 353 urine spectra containing ibuprofen and its metabolites, and a further subset of 70 spectra was selected as control using the remaining 3937 spectra.
Preparation of Samples for 1H NMR Spectroscopy.
Urine samples stored at −40 °C were thawed completely before mixing 500 μL of the sample with 250 μL of phosphate buffer, for stabilization of the urinary pH 7.4 ((0.5), and 75 μL of sodium 3-trimethylsilyl-(2,2,3,3-2H4)-1-propionate (TSP) in D2O (final concentration 0.1 mg/mL) solution for internal calibration at δ 0.0. The D2O provided an NMR lock signal for the NMR spectrometer. Each sample was placed into a 96-well plate for analysis and the remaining sample refrozen. Samples were mixed and the 96-well plate was left to stand for 10 min before centrifuging at 4000 rpm for 10 min to remove any precipitate from the sampling volume.
1H NMR Spectroscopic Analysis of Urine Samples.
Conventional 1H NMR spectra of the urine sample were obtained using a Bruker (Bruker Biospin, Rheinstetten, Germany) Avance 600 spectrometer operating at 600.29 MHz in flow injection mode. Samples were automatically delivered to the spectrometer using a Gilson 215 robot incorporated into the Bruker efficient sample transfer system. The one-dimensional 1H NMR spectra of urine were acquired using a standard one-dimensional pulse sequence NOSEYPR1D (recycle delay−90°−1−90°−tm−9−°-acquisition; XWIN-NMR 3.5) with water presaturation during both the recycle delay (2 s) and mixing time (tm, 150 ms) (See Supporting Information 1 for all NMR acquisition parameters). The 90° pulse length was adjusted to ~10 μs, and t1 was set to 3 μs, providing an acquisition time of 2.73 s and a total pulse repetition time of 4.88 s. For each sample, 64 free induction decays (FIDs) were collected into 32k data points using a spectral width of 20 ppm. The FIDs were multiplied by an exponential weighting function corresponding to a line broadening of 0.3 Hz, and data were zero-filled to 64k data points prior to Fourier transformation (FT).
Two-dimensional NMR analyses were performed on selected samples for identification of drug metabolites. 1H–1H total correlation spectroscopy (TOCSY), 1H–13C heteronuclear multiple bond correlation (HMBC), and 1H–13C heteronuclear single-quantum correlation (HSQC) 2D NMR spectra were acquired on two urine samples containing typical levels of acetaminophen metabolites and another urine sample containing ibuprofen metabolites. For the 2D TOCSY, spectra of urine were acquired using an mlevphpr pulse sequence (XWIN-NMR 3.5) where 56 transients per increment and 120 increments were collected into 2k data points over a spectral width of 10.5 ppm in both dimensions using the MLEV17 spin-lock scheme with a spin-lock power of 6 kHz. A recycle delay of 1.2 s was used, during which water presaturation pulse was switched on. The total measurement time per sample was ~3 h.
1H–13C HMBC (hmbcgplpndqfpr, XWIN-NMR 3.5) and HSQC (hsqcetgppr, XWIN-NMR 3.5) spectra were acquired at 600 MHz for 1H and 150 MHz for 13C with the spectral width of 10.5 ppm for 1H and 220 ppm for 13C. Experimental parameters for HSQC were as follows: 100 increments, 280 transients, and 2k data points in the 1H detection dimension. The experimental parameters for 1H–13C HMBC were as follows: 100 increments, 416 transients, and 2k data points in the 1H detection dimension. For processing, TOCSY and HSQC data were apodized using shifted sine-bell; HMBC spectra were processed using sine-bell weighting in both dimensions. All data were zero-filled to 2k data points in both dimensions prior to FT. The total measurement time was approximately 6 and 18 h per sample for HSQC and HMBC respectively.
1H NMR Spectral Processing and Multivariate Analysis.
Baseline correction and phasing of the spectra were achieved with in-house software (T. Ebbels and H. Keun). All spectra were referenced to TSP automatically using an in-house routine written in MATLAB 7.0.1 (MathWorks, Natick, MA), and each spectrum was collected into 18 001 data points with a data point resolution of 0.0005 ppm prior to data analysis. The regions of the spectra containing the water (δ 4.7−4.9) and urea (δ 5.6−6.0) resonances were removed to eliminate both the variation in water suppression and the variation in the integral of the urea signal due to partial cross-saturation via the solvent exchanging protons. The remaining data point (n = 16 599) intensities were normalized to the sum of the row (total spectrum) prior to further data analysis.
Multivariate Analysis and Experimental Design. Orthogonal Projection to Latent Structure Discriminant Analysis. A principal component analysis (PCA) model was constructed for a subset of INTERMAP participants from the United States in order to identify a subset of urine specimens from participants who had ingested acetaminophen and those who had not. To optimize visualization of drug resonances from spectra exhibiting highdegree biochemical variation, a projection to latent structure algorithm with an inbuilt orthogonal filter (OPLS-DA)31,37 was then applied to spectra that were randomly selected from the two subsets of the data that were identified from the PCA model, comprising a group who had spectral evidence of acetaminophen intake (n) 70) and a control group of nonusers (n = 70). The variation common to the class variable matrix Y (i.e., consumer/non consumer of acetaminophen) can be observed from the R2Y, and the class validity of the OPLS-DA model was tested against overfitting using the cross-validation parameter, Q2Y.
The analysis was repeated for a subset of INTERMAP participants whose urine spectra contained evidence of ibuprofen ingestion and a second group of randomly sampled control samples.
Statistical Total Correlation Spectroscopy. STOCSY was performed on the subset of samples from acetaminophen users. Correlation matrices were calculated for data points selected to coincide with the peak maximums of acetaminophen-related NMR signals with a view to establishing correlations between the separate proton environments for each metabolite and generating molecular structural information. Because the NMR peak intensities of different peaks from the same molecule are directly related to the number of protons contributing to the signal, the relative intensity of signals from the same molecule should in theory be linear (r = 1). However, in reality the correlation between any two data points in a sample set will be somewhat weaker, depending on degree of overlap from other molecules contributing to the spectrum.
Correlations between the selected resonance and all the other data points of the spectra are presented using color coding (corresponding to the r2 = 1 instead of r) for ease of visualization. Upward-oriented data points reflect a positive correlation to the selected resonance of interest, whereas downward-oriented data points signify negative correlations. The signal intensity and shape is used to represent the covariance matrix, which provides a graphical similarity to NMR spectra, thereby facilitating ease of interpretation by the NMR spectroscopist.
The bootstrapped38 correlation coefficients were computed on all samples containing acetaminophen metabolites. The mean and the median of correlation coefficient between each pair of the selected data points, together with the 95% confidence interval for the sampled population, were computed. The bootstrapped process, an iterative resampling with replacement method was applied using a randomly selected sample set containing the same number of samples as the original OPLS-DA model defining the differences between acetaminophen intakes, i.e., 70 from the 413 samples containing acetaminophen metabolites. Random selection of the sample set was repeated a 1000 times in order to assess the probability of obtaining the significance of the OPLS-DA model by chance. The bootstrapping process was repeated for all 353 samples containing ibuprofen and its metabolites.
RESULTS
Acetaminophen Metabolites.
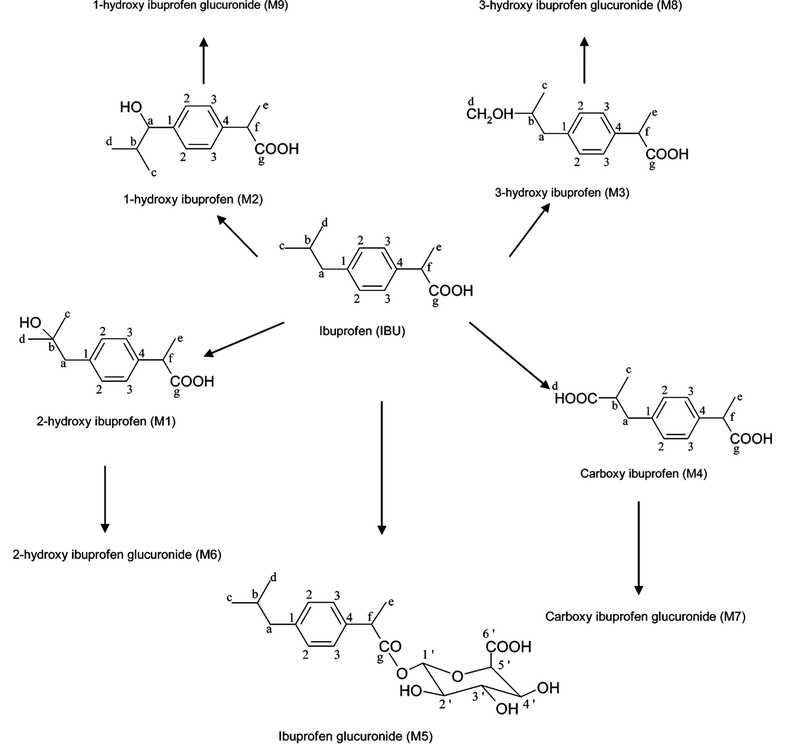
The metabolism of acetaminophen is well-documented;34,39 major metabolites are shown in Figure 1. Resonances arising from the parent drug and many of its metabolites were observed in the 1D NMR urine spectra from participants who had not ingested any drug (Figure 2A) or had ingested either acetaminophen (Figure 2B,C) or ibuprofen (Figure 2D,E; as discussed below). Assignments were made from literature values40,41 and from a series of 2D NMR measurements on selected samples containing acetaminophen. 1H–1H (TOCSY) and 1H–13C (HSQC, HMBC) spectra are also illustrated for acetaminophen and its NMR-detected metabolites (Supporting Information 2). Chemical shifts are provided for each of the reported metabolites (Table 1).
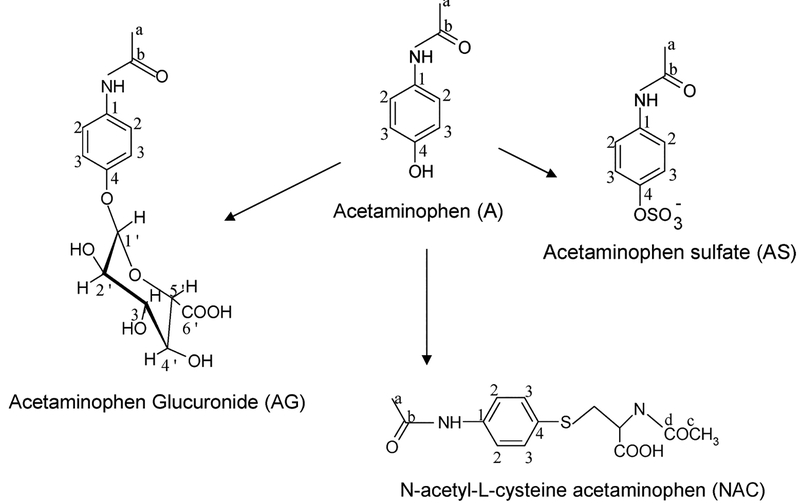
Figure 1.
Schematic showing the chemical structures of acetaminophen (A) and its metabolites. The numbers and letters refer to the position of 13C and 1H in the molecules.
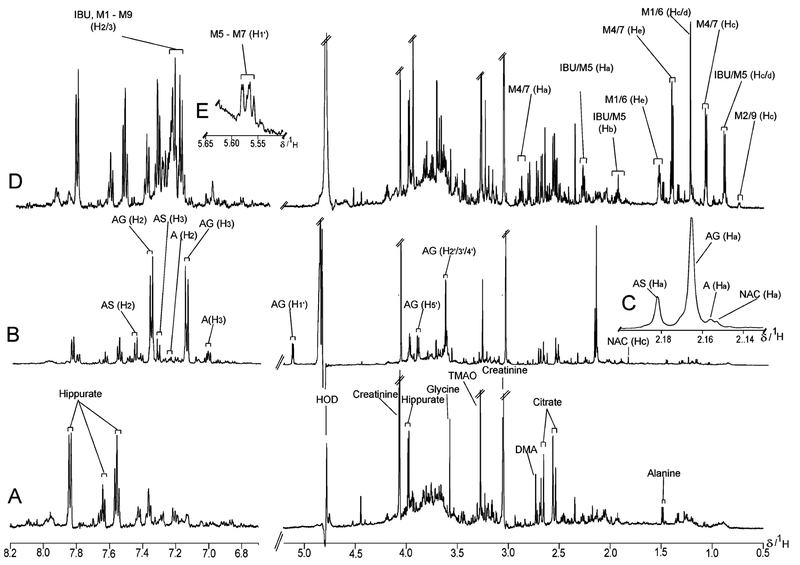
Figure 2.
Typical 1H NMR spectra of urine from participants: (A) no analgesic intake; (B) after ingestion of acetaminophen; (C) expansion on acetaminophen region at δ 2.16; (D) after ingestion of ibuprofen; (E) showing the characteristic of the anomeric proton signal of the ibuprofen glucuronide rings. For a key to the identity of ibuprofen metabolites, refer to Figures 1 and 6.
Table 1.
1H and 13C NMR Chemical Shifts and Coupling Constants (J/Hz) for Acetaminophen and Its Metabolites
| metabolites |
1H shift (δ) |
coupling constant (J/Hz) | appearancea | assignmentb | 13C shift (δ)c |
|---|---|---|---|---|---|
| A | 2.16 | s | Ha | 25.8 (Ca) | |
| 6.91 | 8.8 | d | H3 | 120 (C3) | |
| 7.25 | 8.8 | d | H2 | 128.2d (C2) | |
| AG | 2.17 | s | Ha | 25.8 (Ca) | |
| 3.62 | m | H2′3′4′ | 75.4e (C2′/3′) | ||
| 78.8 (C4′) | |||||
| 3.89 | 8.9 | d | H5′ | 76.5 (C1′) | |
| 5.10 | 6.8 | d | H1′ | 103.4 (C1′) | |
| 7.13 | 8.8 | d | H3 | 120.3 (C3) | |
| 7.36 | 8.8 | d | H2 | 127.3 (C2) | |
| AS | 2.18 | s | Ha | 25.8 (Ca) | |
| 7.31 | 8.8 | d | H3 | 125.2 (C3) | |
| 7.46 | 8.8 | d | H2 | 126.7 (C2) | |
| NAC | 1.86 | s | Hc | 176.2 (Cd) | |
| 2.15 | s | Ha | 25.8 (Ca) |
d = doublet; s = singlet; m = multiplet.
These assignments were based on 1H-1H TOCSY NMR.
Assignments were based on 1H-13C HSQC and HMBC NMR.
Observed in only one of the sample.
Two signals observed.
The PCA model which based on all the 4290 urine spectra was used to identify 413 samples containing acetaminophen and its metabolites with the remaining 3877 urine spectra not showing acetaminophen metabolites. OPLS-DA analysis on a subset of 70 samples from the two groups (acetaminophen users or nonusers) generated a scores plot showing clear differentiation of the spectra based on acetaminophen intake of the participants (Figure 3). The OPLS-DA model calculated for the acetaminophen versus control groups generated one predictive component with two orthogonal components and explained 89% of the variance of on the class variable matrix (Y) with a predictive value of 72% according to the cross-validation parameter (Q2Y = 0.72). The coefficients plot shows the differential contributions of spectral regions in contributing to discrimination between the two groups (acetaminophen ingestion vs noningestion). The resonances exerting the strongest influence on the class discrimination relate to the acetaminophen parent compound, its glucuronide, and sulfate conjugates, with a slightly weaker contribution from the N-acetylcysteine conjugate (Figure 3A–C). The glucuronide conjugate was the dominant urinary excretion product of acetaminophen in all 413 cases with a mean ratio of 1:2.1 for sulfate/glucuronide. In addition, unchanged acetaminophen compound and its N-acetylcysteine conjugate were also observed in the majority of the spectra but were present at much lower concentrations.
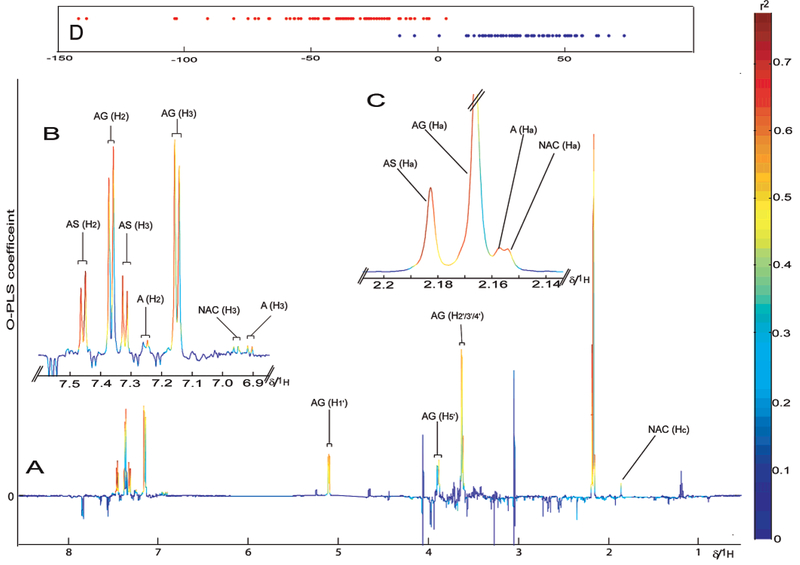
Figure 3.
(A) The OPLS-DA loading coefficient plot showing that the basis of differentiation between these two groups (acetaminophen ingestion versus controls) primarily results from the presence of acetaminophen (A), acetaminophen glucuronide (AG), acetaminophen sulfate (AS), and the N-acetylcysteine conjugate of acetaminophen (NAC) denoted by the red/orange color of the resonances indicating a r2 > 0.5 in discriminating between the two groups. (B) Expansion for the aromatic region showing signals for acetaminophen and its related metabolites (C) Expansion on acetaminophen region at δ 2.16. (D) The O-PLS-DA scores plot showing discrimination between two classes (blue, control; red, participants who had taken acetaminophen).
Having modeled the spectral features characterizing acetaminophen ingestion, statistical correlation was performed for prominent acetaminophen-related resonances to determine correlations between protons within the same molecule and to ascertain the statistical relationship between resonances from the parent compound and resonances from acetaminophen metabolites. Statistical correlations were computed on full-resolution spectra for the 413 samples containing acetaminophen metabolites. The correlation matrix derived from correlations between the data point corresponding to δ 2.17, at the apex of the N-acetyl proton signal from acetaminophen glucuronide and all other data points, showed high-correlation values with most of the other glucuronide metabolite resonances (Figure 4A–C and Supporting Information 3). This includes the pair of doublets in the aromatic region of the spectrum (δ 7.13, with a mean correlation coefficient, r = 0.84 and a 95% confidence interval (CI) of 0.79 − 0.88, and 7.36, r = 0.98; with 95% CI of 0.97−0.99), the characteristic doublet at δ 5.10 (r = 0.95; with 95% CI of 0.93−0.97) for the anomeric proton signal of the glucuronide ring and the signals from the glucuronide ring protons at δ 3.62 (r = 0.89; with 95% CI of 0.85−0.93). Correlation values between the N-acetyl δ 2.17 signal and signal at δ 3.89 derived from the protons H5` on the glucuronide ring were somewhat weaker (r = 0.72; 95% CI of 0.61–0.82). These weaker correlation values were influenced by partial overlap of signals of glucose at δ ~3.89.
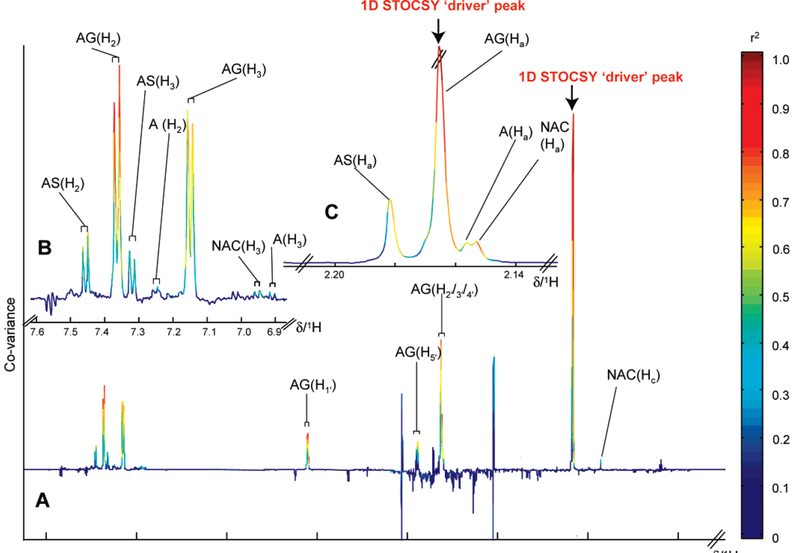
Figure 4.
(A) STOCSY plot derived from the correlation matrix calculated between the data point at the peak maximums of the N-acetyl proton signal of AG resonance (δ 2.17) and all other data points, as indicated by the arrow, showing strong correlation (red/orange data points) with resonances at δ 3.62, 3.89, 5.1, 7.13, and 7.36. Slightly weaker correlations with A and AS. (B) Expansion for the aromatic region showing signals for acetaminophen and its related metabolites. (C) Expansions for the δ 2.17 N-acetyl resonance of acetaminophen metabolites.
In addition, the N-acetyl peak for the glucuronide was highly correlated with the N-acetyl peak for the sulfate at δ 2.18 (r = 0.81; 95% CI of 0.74–0.87), N-acetylcysteine at δ 2.15 (r = 0.82; 95% CI of 0.77–0.87), and parent drug molecule at δ 2.16 (r = 0.87; 95% CI of 0.82−0.91). Positive correlations were also observed between the N-acetyl of the glucuronide conjugate and the aromatic protons of the sulfate conjugate at δ 7.31 and 7.46 (r = 0.76; 95% CI of 0.66−0.83 and 0.90; 95% CI of 0.85−0.93, respectively). The correlation value between the N-acetyl peak for the glucuronide conjugate and the N-acetylcysteine at ä 1.86 (r = 0.61; 95% CI of 0.5−0.72) or the aromatic proton of the N-acetylcysteine (H3) at ä 6.96 (r = 0.63; 95% CI of 0.53−0.73) was somewhat weaker when compared to the correlation between the glucuronide conjugate and the N-acetylcysteine resonance at δ 2.15 (r = 0.82; 95% CI of 0.77−0.87). The difference in correlation values for the two N-acetylcysteine resonances at δ 2.15 and 1.86 or 6.96 with the N-acetyl glucuronide peak was most likely caused by overlap of the N-acetylcysteine resonance at δ 2.15 with the N-acetyl signal from acetaminophen itself.
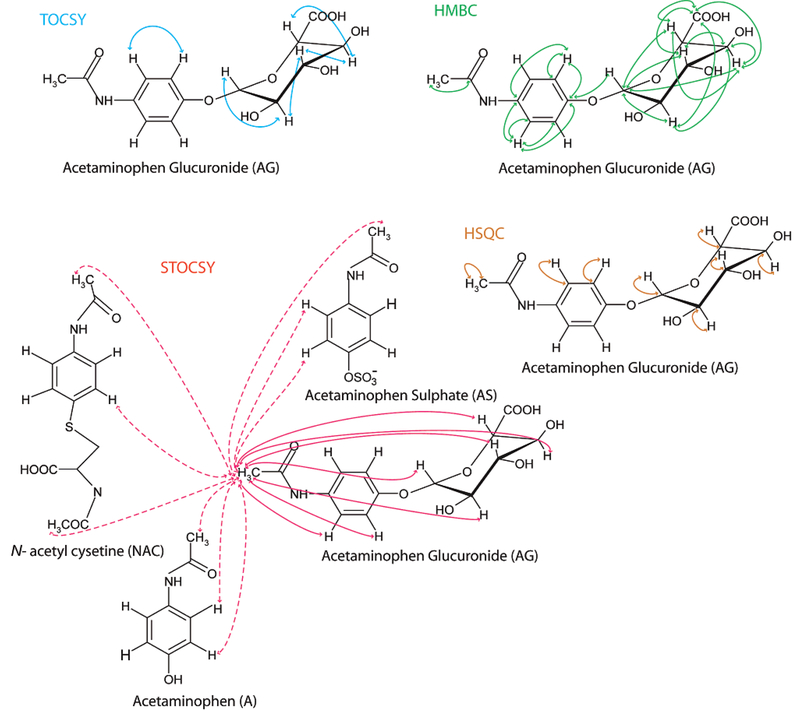
The schematic diagram in Figure 5 compares the molecular structural information obtained by TOCSY, HSQC, HMBC, and STOCSY analyses, respectively, for acetaminophen glucuronide. Many of the correlations observed by the statistical correlation method, for example, between the N-acetyl proton and the aromatic protons, cannot be observed by the NMR pulse sequence experiments. Thus, the STOCSY method in this case was able to provide “constitutional” correlation/connectivity for all nuclei that are present in a given molecule, regardless of the number or the existence of covalent chemical bonds or the magnetization transfer between the nuclei. This statistical correlation procedure is also potentially more efficient than 2D measurements, in some circumstances. For example, the analysis time taken to acquire this data set (n = 413) for the STOCSY was ~56 h (8 min × 413 samples) and encompassed all samples. In contrast the series of 2D NMR spectra took a total of 27 h to acquire for a single sample, which would not necessarily be a representative sample for all metabolites profiled.
Figure 5.
Schematic for acetaminophen glucuronide comparing the intramolecular 1H-1H and 1H-13C correlations detected in the TOCSY (blue), HSQC (brown), HMBC (green), and intra-and intermolecular (dotted line) correlations given by STOCSY (red) analyses. Note that these are the actual observed, and not the theoretical correlations. Although the NMR parameters were optimized for this study by adjusting the acquisition/processing parameters, it may be possible to observe further correlation structures from each of these measurements.
The correlation matrix for the acetaminophen sulfate conjugate was calculated at the apex corresponding to the proton from the aromatic ring at δ 7.46 as this peak was deemed to be the least overlapped of the acetaminophen sulfate resonances. A high correlation was demonstrated between this resonance and all the other resonances of the sulfate conjugate, e.g., with the doublet derived from the aromatic proton at δ 7.31 (r = 0.9; 95% CI of 0.86−0.93) and with the singlet derived from the methyl of the acetyl at δ 2.18 (r = 0.94; 95% CI of 0.91−0.96) (Supporting Information).
Ibuprofen Metabolites.
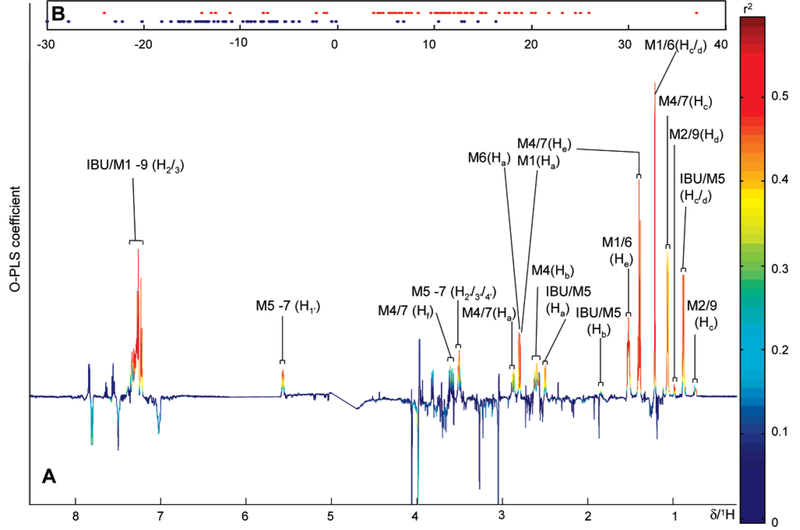
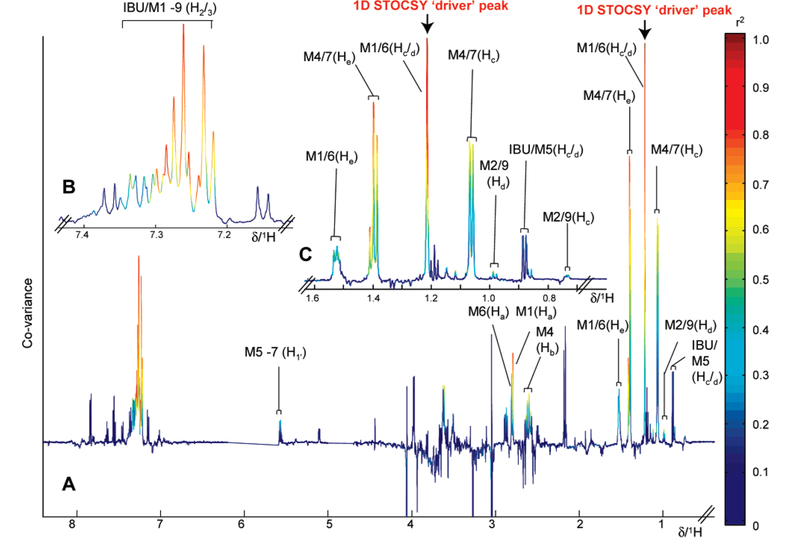
After acetaminophen and diabetic outliers were removed, a significant number of ibuprofen-containing samples (n = 353) were identified by PCA. OPLS-DA was performed on a subset of 70 samples selected randomly from participants who had taken ibuprofen compared with a control group. As with acetaminophen, the metabolism of ibuprofen is well documented35,42 (Figure 6) and produces distinctive resonances in the 1H NMR urine spectrum(Figure 2D). Participants who had ingested ibuprofen were clearly differentiated from controls as visualized in the OPLS-DA scores and coefficients plots (Figure 7). The OPLS-DA model, which used one predictive component and one orthogonal component explained 54% of the variance within Y matrix (R2Y) 0.54) with a predictive value of 49% according to the cross-validation parameter (Q2Y) 0.49). Similarly, correlation matrices calculated for each of the data points at the apex of an ibuprofen-related signal showed strong correlation between signals derived from protons on the same molecule and to a lesser extent from signals from other ibuprofen-related molecules. The correlation coefficients calculated from the data point corresponding to δ 1.21, at the apex of the methyl group signal from the isobutyl side chain of 2-hydroxy ibuprofen (Figure 6, Hc/Hd position for M1) and its glucuronide conjugate (M6) are shown in Figure 8. High correlations values were observed between this data point and those corresponding to the methyl groups (δ 1.52; r = 0.94; 95% CI of 0.92−0.97) and the aromatic protons (around δ 7.24; r = 0.94; 95% CI of 0.92−0.9) of both metabolites. The −CH2 resonance of 2-hydroxy ibuprofen (M1) at δ 2.79 (r = 0.96; 95% CI of 0.94–0.97) and the-CH2 resonance of 2-hydroxy ibuprofen glucuronide conjugate (M6) at δ 2.80 (r = 0.92; 95% CI of 0.9–0.94) were also strongly correlated with the δ 1.21 methyl signal (Figure 8). Additionally, STOCSY driven from the apex of the resonance at δ 1.21 also highlighted correlations between 2-hydroxy ibuprofen and some of the other dominant ibuprofen metabolites, e.g., the carboxy ibuprofen (Figure 6, M4) and its glucuronide conjugate (M7). For example, strong correlation values describing the relationship between the 2-hydroxy ibuprofen methyl at δ 1.21 and resonances of the carboxy ibuprofen were found; CH3 (δ 1.40, r = 0.97; 95% CI of 0.95−0.98), the-CH group (δ 2.61, r = 0.88; 95% CI of 0.85−0.91), and the methyl group from the propionic acid side chain (δ 1.07, r) 0.87; 95% CI of 0.92–0.9). Strong correlation was also observed between the 2-hydroxy ibuprofen methyl and the anomeric proton from the ibuprofen glucuronides resonance at δ 5.56 (, r = 0.92; 95% CI of 0.88−0.95).
Figure 6.
Schematic showing the chemical structures of ibuprofen and its major urinary metabolites. The letters and numbers refer to the position of 13C and 1H of the molecules.
Figure 7.
(A) OPLS-DA loading coefficient plot showing that the basis of differentiation between these two groups (ibuprofen users and nonusers) primarily results from the presence of 2-hydroxy, carboxy, and glucuronide conjugates of ibuprofen. (B) The OPLS-DA scores plot showing discrimination between two population classes (blue, control; red, participants who had consumed ibuprofen).
Figure 8.
(A) STOCSY plot derived from the correlation matrix between all data points and the data point at the peak maxima relating to the combined resonances from 2-hydroxy signal of ibuprofen and its glucuronide resonance (δ 1.21), as indicated by the arrow. Strong correlations (red/orange data points) between this resonance and many related ibuprofen resonances can be observed. (B) Expansion for the aromatic region showing resonances for ibuprofen and its related metabolites. (C) Expansions for the δ 0.6–1.6 region showing resonances of ibuprofen metabolites. (IBU, ibuprofen; M1, 2-hydroxy ibuprofen; M2, 1-hydroxy ibuprofen; M3, 3-hydroxy ibuprofen; M4, carboxy ibuprofen; M5, ibuprofen glucuronide; M6, 2-hydroxy ibuprofen glucuronide conjugate; M7, carboxy ibuprofen glucuronide conjugate; M8, 3-hydroxy iburprofen glucuronide conjugate; and M9, 1-hydroxy ibuprofen glucuronide conjugate).
DISCUSSION
Understanding the patterns of use and differential metabolic profiles of drugs in populations would provide important background genetic (drug-metabolizing enzymes) and social information in epidemiological studies designed to understand the demographics of human disease. The concept of the “xenometabolome” is useful as it provides potential metrics of population drug (and even environmental pollutant) exposure in an epidemiological context. In the majority of such epidemiological studies, this information is difficult or impossible to derivesbut the recent advent of spectroscopy-based population phenotyping methods coupled with statistical information recovery tools provides the possibility for the first time as illustrated here for the widely used drugs acetaminophen and ibuprofen. The combined application of OPLS-DA and statistical correlation (STOCSY) to 1D NMR urine spectra from a human population study enabled discrimination of samples based on analgesic intake and allowed structural characterization of the analgesics and their metabolites. Strong statistical correlations between signals deriving from protons on the same molecule but in different chemical environments were observed in most cases, as demonstrated for the glucuronide and sulfate conjugates of acetaminophen and the 2-hydroxy, carboxy, and glucuronide conjugates of ibuprofen (Figures 3 and 7). Where a drug metabolite was present in low concentrations and prone to overlap with resonances from other endogenous or exogenous molecules, for example the N-acetyl cysteine conjugate of acetaminophen, correlations obtained between discrete proton signals on the same molecule were lower, but nevertheless detectable. In contrast to conventional 2D NMR pulse sequences, which are most efficient at identifying crosspeaks or correlations between proton groups in physical proximity, the statistical correlation method detects covariance of resonances and is, therefore, not influenced by distances in terms of bond length. For example, from the 2D 1H-1H TOCSY spectrum, it was possible to identify coupling between the H2/H3 and H1’/H2’/H3’/H4’/H5’ moieties of acetaminophen glucuronide (Figure 5). However, unlike the STOCSY spectrum, no cross-peak or correlation between the N-acetyl (Ha) and aromatic protons (H2/H3 and H1’/H2’/H3’/H4’/H5’) on the acetaminophen glucuronide molecule proton groups could be observed as these are separated by 5 and 8 bond (lengths), respectively, for H3 and H1’. In addition, an obvious advantage of the mathematical correlation method over 2D NMR spectroscopy is the time required to generate structural information for the metabolites. Each 1D NMR spectrum is generated in 8 min, whereas a typical 2D TOCSY requires ~3 h of acquisition time, and an HMBC up to 18 h, to obtain an adequate signal-to-noise ratio. Therefore, the collation of rapidly acquired 1D spectra can offer a more efficient solution to analyzing drug metabolism in human populations since the use of STOCSY often negates the requirement for performing extensive 2D NMR experiments and is therefore more time efficient.
Although back-scaling the unit variance-scaled correlation values to the covariance matrix restores much of the original spectral structure,31 the weighting afforded to highly correlated signals results in an emphasis on the drug metabolites producing a down weighting or flattening of signal contributions from non-correlated molecules, in this case, the majority of the endogenous molecules. This has the effect of simplifying the interpretation of the correlated structures for the molecule of interest, as is apparent in the correlation spectra where the glucuronide ring resonances at δ 3.62 and 3.89 are clearly visible. These signals are often difficult to assign in a standard 1D 1H NMR spectrum due to overlapping resonances from glucose, CH groups of amino and organic acids, and other sugar moieties.
Most importantly, the STOCSY application provides structural information not just pertaining to protons on the same molecule, but also connects signals for protons deriving from closely related molecules. For example, metabolites derived from the same parent compound, e.g., acetaminophen, acetaminophen glucuronide, acetaminophen sulfate, and N-acetyl cysteine acetaminophen, all show correlations that vary in strength according to the degree of overlap with other endogenous or exogenous signals. This may prove a particularly useful feature in cases where the metabolism of a xenobiotic is not well characterized and may help to differentiate xenobiotic metabolites from drug-induced changes in endogenous molecules. STOCSY can differentiate between endogenous and exogenous signals based on the structure of the pharmacological connections. The negative correlation matrix generated from any drug metabolite resonance will contain only endogenous responses to the drug. The positive correlation matrix will indicate either drug-related or endogenous correlations with drug metabolites. However, one would expect that in most cases the correlations between drug metabolites would be stronger than correlations observed between drug metabolites and endogenous molecules. In the case of the positive correlation matrix, differentiation can be made on the basis of molecular structural information, since endogenous molecular structures will be largely implausible for drug biotransformations. It is also possible to draw inferences regarding the strength of intermolecular correlations. The correlation between acetaminophen and its sulfate conjugate is weaker than the correlation between the acetaminophen parent and the glucuronide. Factors that may influence drug metabolite ratios include saturation of metabolic pathways, interaction with other pharmaceuticals, nutritional status, and time of administration of the drug in relation to sampling time. For example, acetaminophen sulfate is known to have a longer half-life (~3.5 h) than acetaminophen glucuronide (~2 h), and the capacity for sulfation is more readily saturated.43
In addition to applying the STOCSY method in conjunction with projection methods for detection and structural identification of drug metabolites in human biofluid samples, correlation between drugs and endogenous metabolites can also be visualized. Metabolic effects of over-the-counter therapeutics can impact on disease risk or development. For example, heavy or prolonged use of analgesics such as aspirin or phenacetin has been implicated in the development of renal papillary necrosis and associated end stage renal failure.44–46 It is therefore important to understand variation in the metabolism of these xenobiotics both at the individual and at the population level.
In summary, the combination of NMR spectroscopic profiling of biological fluids with the STOCSY method for identification of correlated signals confers molecular structure elucidation potential both for xenobiotics and endogenous molecules. It can be a powerful tool for the identification of candidate biomarkers generated from metabonomic studies. Detection and identification of drug metabolites in urine or plasma can be used to verify questionnaire data in human clinical studies and may also be useful for exploring idiosyncratic responses to drugs. Measurement of major xenometabolome components, which can be performed as part of a general exploratory metabolic profiling procedure, is of value because it carries extra environmental chemical information that might be important in the assessment of overall disease risk profiles of populations and cannot be currently assessed using questionnaire data (performed as part of epidemiological studies) alone. Furthermore, correlations between drug metabolites and molecules of endogenous origin may uncover important information on mechanisms of action and potential population exposure risks. This approach should be extendable to investigate differential metabolism of “self-dosed” individuals in populations of different ethnic/lifestyle backgrounds.
Supplementary Material
ACKNOWLEDGMENT
We are grateful to the U.S. National Heart, Lung, and Blood Institute, Bethesda, Maryland, MD, for their financial support of the project. The INTERMAP study is supported by grant 2-RO1-HL50490, US NHLBI; and national and local agencies in the four countries, and the INTERMAP Metabonomics study was supported by grant 5-RO1-HL71950–2 US NHLBI. It is a pleasure also to express appreciation to the many colleagues who collected and processed the INTERMAP data; for a listing of many of them, see ref 4. H.T. acknowledges financial support from the Chinese Academy of Sciences (100T programme: 2005[35]-T12508–06S138), NSFC (20575074) and National Basic Research Program of China (2006CB503908). E.H. and R.L.L. contributed equally to this work.
Abbreviations:
- CI:
confidence interval
- NMR
nuclear magnetic resonance
- STOCSY
statistical total correlation spectroscopy
- OPLS-DA
orthogonal projection to latent structure discriminant analysis
- PCA
principal component analysis
- SHY
statistical heterospectroscopy
- TOCSY
total correlation spectroscopy
- HMBC
1H-13C heteronuclear multiple bond correlation
- HSQC
1H-13C heteronuclear single-quantum correlation
- A
acetaminophen
- AG
acetaminophen glucuronide
- AS
acetaminophen sulfate
- NAC
N-acetylcysteine acetaminophen conjugate
- IBU
ibuprofen
- M1
2-hydroxyibuprofen
- M2
1-hydroxyibuprofen
- M3
3-hydroxyibuprofen
- M4
carboxyibuprofen
- M5
ibuprofen glucuronide conjugate
- M6
2-hydroxyibuprofen glucuronide conjugate
- M7
carboxyibuprofen glucuronide conjugate
- M8
3-hydroxyibuprofen glucuronide conjugate
- M9
1-hydroxyibuprofen glucuronide conjugate
Footnotes
SUPPORTING INFORMATION AVAILABLE
Supplement 1: Spectrometer acquisition parameters for standard 1D (NOSEYPR1D), TOCSY, HSQC, and HMBC. Supplement 2: Selected 2D NMR spectra of urine from a participant after ingestion of acetaminophen. (A) TOCSY spectrum; (B) HSQC spectrum; (C) HMBC spectrum; (D) 1D spectrum. Supplement 3: Bootstrapped correlation coefficient showing the mean and median correlation coefficient (together with the 95% confidence interval) for 413 1H NMR spectra samples based on 1000 resamplings) with replacement. Shaded columns indicate the intramolecular correlation coefficient. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- (1).Nicholson JK Mol. Syst. Biol 2006, 2, 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Dumas M-E; Maibaum EC; Teague C; Ueshima H; Zhou B; Lindon JC; Nicholson JK; Stamler J; Elliott P; Chan Q; Holmes E Anal. Chem 2006, 78 (7), 2199–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Teague C; et al. Analyst 2004, 129 (3), 259–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Stamler J; et al. J. Hum. Hypertens 2003, 17 (9), 591–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Stamler J Am. J. Clin. Nutr 1997, 65 (2 Suppl), 626S–42S. [DOI] [PubMed] [Google Scholar]
- (6).Zhao L; et al. Hypertension, 2004, 43 (6), 1332–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Stamler J; et al. J. Hum. Hypertens 2003, 17 (9), 655–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Stamler J; et al. Circulation 1996, 94 (7), 1629–34. [DOI] [PubMed] [Google Scholar]
- (9).Elliott P; et al. Arch. Intern. Med 2006, 166 (1), 79–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Dumas ME; et al. Proc. Natl. Acad. Sci. U.S.A. 2006, 103 (33), 12511–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).GavaghanMcKee CL; Wilson ID; Nicholson JK J. Proteome Res. 2006, 5 (2), 378–84. [DOI] [PubMed] [Google Scholar]
- (12).Gavaghan CL; et al. FEBS Lett, 2000, 484 (3), 169–74. [DOI] [PubMed] [Google Scholar]
- (13).Robosky LC; et al. Toxicol. Sci 2005, 87 (1), 277–84. [DOI] [PubMed] [Google Scholar]
- (14).Holmes E; Nicholson J Toxicol. Sci 2005, 87 (1), 1–2. [DOI] [PubMed] [Google Scholar]
- (15).Bollard ME; et al. NMR Biomed. 2005, 18 (3), 143–62. [DOI] [PubMed] [Google Scholar]
- (16).Brindle JT; et al. Analyst 2003, 128 (1), 32–6. [DOI] [PubMed] [Google Scholar]
- (17).Wilson ID; Nicholson JK Anal. Chem 1987, 59 (23), 2830–2. [DOI] [PubMed] [Google Scholar]
- (18).Chen CN; et al. Chem. Pharm. Bull. (Tokyo), 1996, 44 (1), 204–7. [DOI] [PubMed] [Google Scholar]
- (19).Shockcor JP; et al. Xenobiotica 1996, 26 (2), 189–99. [DOI] [PubMed] [Google Scholar]
- (20).Nicholls AW; Holmes E; Lindon JC; Shockcor JP; Farrant RD,; Haselden JN; Damment SJP; Waterfield C,J; Nicholson JK Chem. Res. Toxicol 2001, 14 (8), 975–87. [DOI] [PubMed] [Google Scholar]
- (21).Josefsson M; et al. J. Chromatogr., B 2003, 789 (1), 151–67. [DOI] [PubMed] [Google Scholar]
- (22).Atsumi R; et al. Arzneimittelforschung, 2001, 51 (3), 253–7. [DOI] [PubMed] [Google Scholar]
- (23).Chen M; et al. J. Chromatogr., B 2005, 820 (1), 147–50. [DOI] [PubMed] [Google Scholar]
- (24).Fernandez P; et al. J. Appl. Toxicol 2005, 25 (3), 200–4. [DOI] [PubMed] [Google Scholar]
- (25).Spraul M; Freund AS; Nast RE; Withers RS; Mass WE; Corcoran O Anal. Chem 2003, 75 (6), 1536–41. [DOI] [PubMed] [Google Scholar]
- (26).Clayton E; et al. Chromatographia, 1998, 47 (5/6), 264–70. [Google Scholar]
- (27).Lindon JC; Nicholson JK; Wilson IDJ J.Chromatogr., B 2000, 748(1), 233–58. [DOI] [PubMed] [Google Scholar]
- (28).Zhang X; et al. Food Chem. Toxicol 2006, 44 (7), 1006–14. [DOI] [PubMed] [Google Scholar]
- (29).Antti H; et al. Chemom. Intell. Lab. Syst 2004, 73 (1), 139–49. [Google Scholar]
- (30).Beckwith-Hall BM; Nicholson JK; Nicholls AW; Foxall PJD; Lindon JC; Connor SC; Abdi M; Connelly J Holmes E Chem. Res. Toxicol 1998, 11 (4), 260–72. [DOI] [PubMed] [Google Scholar]
- (31).Cloarec O; Dumas ME; Trygg J; Craig A; Barton RH; Lindon JC; Nicholson JK; Holmes E Anal. Chem 2005, 77 (2), 517–26. [DOI] [PubMed] [Google Scholar]
- (32).Cloarec O; Dumas M-E; Craig A; Barton RH; Trygg J; Hudson J; Blancher C; Gauguier D; Lindon JS Holmes E; Nicholson J Anal. Chem 2005, 77 (5), 1282–9. [DOI] [PubMed] [Google Scholar]
- (33).Crockford DJ; Holmes E; Lindon JC; Plumb RS; Zirah S; Bruce SJ; Rainville P,; Stumpf CL; Nicholson JK Anal. Chem 2006, 78 (2), 363–71. [DOI] [PubMed] [Google Scholar]
- (34).Graham GG; Scott KF Am. J. Ther 2005, 12 (1), 46–55. [DOI] [PubMed] [Google Scholar]
- (35).Davies NM Clin. Pharmacokinet 1998, 34 (2), 101–4. [DOI] [PubMed] [Google Scholar]
- (36).Beevers DG; Stamler JJ Hum. Hypertens 2003, 17 (9), 589–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Johan Trygg SW J. Chemom 2002, 16 (3), 119–28. [Google Scholar]
- (38).Efron B; Tibshirani RJ An Introduction to the Bootstrap; Chapman & Hall: New York, 1993. [Google Scholar]
- (39).Barkin RL Am. J. Ther 2001, 8 (6), 433–42. [DOI] [PubMed] [Google Scholar]
- (40).Bales JR; et al. Clin. Chem 1984, 30 (10), 1631–6. [PubMed] [Google Scholar]
- (41).Bales JR; Nicholson JK; Sadler PJ Clin. Chem 1985, 31 (5), 757–62. [PubMed] [Google Scholar]
- (42).Spraul M; Hofmann M; Dvortsak P; Nicholson JK; Wilson ID Anal. Chem 1993, 65 (4), 327–30. [DOI] [PubMed] [Google Scholar]
- (43).Forrest JA; Clements JA; Prescott LF Clin. Pharmacokinet 1982, 7 (2), 93–107. [DOI] [PubMed] [Google Scholar]
- (44).Prescott LF Br. J. Clin. Pharmacol 1980, 10 (Suppl 2), 291S–298S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).McLaughlin JK; et al. Kidney Int 1998, 54 (3), 679–86. [DOI] [PubMed] [Google Scholar]
- (46).Kincaid-Smith P Drugs 1986, 32 (Suppl 4), 109–28. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.