Abstract
Rhizoma drynariae is the main traditional Chinese medicine used for the treatment of osteoporosis, but its anti-osteoporotic targeting mechanism has not been fully elucidated due to the complexity of its active ingredients. In this study, the pharmacological mechanism of action of Rhizoma drynariae against osteoporosis was studied by integrating pharmacological concepts.
The pharmacokinetic characteristics of selected major active constituents of Rhizoma drynariae and the SMILES structural similarity were used to predict related targets. A literature search was conducted to identify known osteoporosis treatment targets, which were then combined with the predicted targets to construct the direct or indirect target interaction network map of Rhizoma drynariae against osteoporosis. Finally, data on the key targets of the interactions, ranked according to relevant node parameters obtained through pathway enrichment analysis and screening of key targets and active ingredients of Rhizoma drynariae, were used to perform molecular docking simulation.
We screened 16 active ingredients of Rhizoma drynariae, and 7 key targets with direct or indirect effects with a high frequency were obtained. These main pathways were found to play important roles in the PI3k-akt signaling pathway, osteoclast differentiation, Wnt signaling pathway, and estrogen signaling pathway. Molecular docking showed that most active ingredients of Rhizoma drynariae had strong binding efficiency with key targets.
Based on network pharmacology, we conclude that Rhizoma drynariae plays an anti-osteoporotic role by directly or indirectly targeting multiple major signaling pathways and influencing the proliferation and differentiation of multiple types of cells.
MeSH Keywords: Medicine, Chinese Traditional; Molecular Mechanisms of Pharmacological Action; Osteoporosis; Protein Interaction Maps
Background
Osteoporosis (OP) is a systemic bone disease that involves decreased bone density and bone quality. The destruction of the bone microstructure and an increase in bone fragility eventually lead to an increase of fracture risk and pose a serious threat to the health of middle-aged and elderly people [1]. In terms of pathogenesis, osteogenesis-osteoclast coupling is unbalanced, with increased osteoclasts, increased lipogenesis, and decreased osteogenesis, which are closely related to proliferation and differentiation of bone marrow mesenchymal stem cells (BMSCs), hematopoietic stem cells, osteoblasts, osteoclasts, adipocytes and other cells, and involves multiple systems such as osteogenesis, hematopoiesis, and immunity. The pathogenesis of OP is complex, and there is interaction and crossover between OP and a variety of related signaling pathways. At present, conventional anti-osteoporotic drugs have disadvantages such as targeting only a single target, poor efficacy, and serious adverse effects [2]. Therefore, an increasing number of scholars are paying attention to research on the prevention and treatment of osteoporosis using traditional Chinese medicine [3].
Traditional Chinese medicine contains a large number of ingredients and uses extremely complex mechanisms to intervene in the progression of disease. Rhizoma drynariae, from the Drynaria fortunei dry rhizome, is a traditional Chinese medicine commonly used in orthopedics departments and widely used in the treatment of osteoporosis. Although previous studies have confirmed the efficacy of Rhizoma drynariae in treating OP [4,5], since its ingredients are complex and its action targets numerous, it is difficult to fully elucidate its mechanism of action. Therefore, it is necessary to study the biological processes of drug, gene, and protein interactions at a systematic level to reveal the molecular mechanisms related to the efficacy of TCM (Figure 1).
Figure 1.
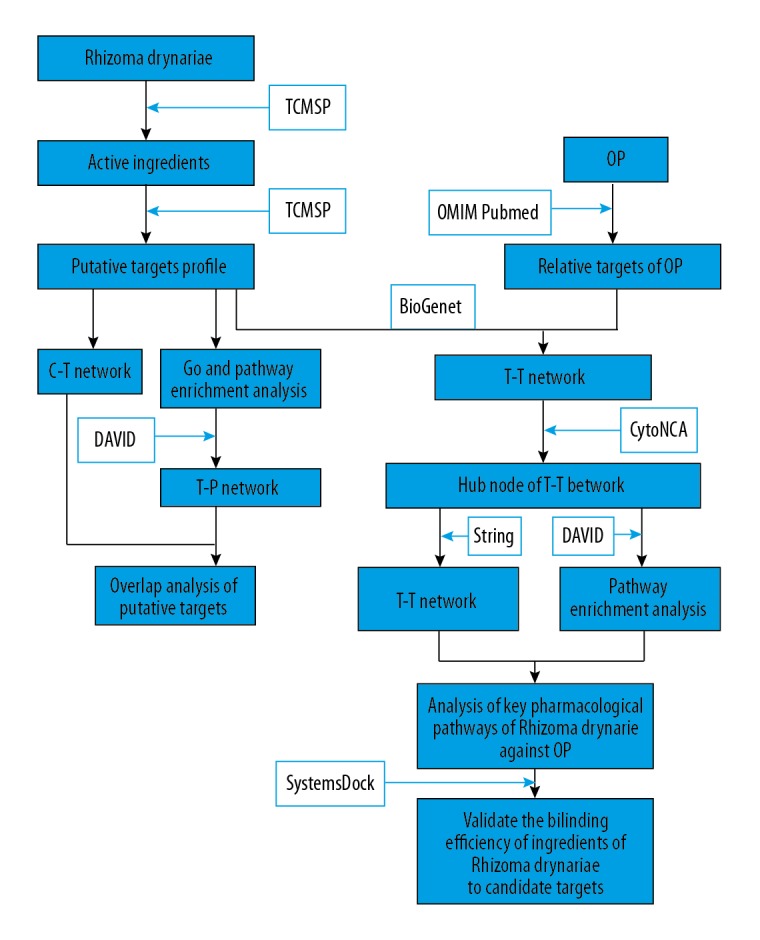
The technical strategy of this research based on network pharmacology for deciphering key pharmacological pathways of Rhizoma drynariae against OP.
In recent years, along with continuous innovation and the development of systems biology, network pharmacology, and computer technology, it can be seen that network pharmacology has a wide applicational value in many fields, such as drug-target identification, the discovery of effective ingredients, research on the mechanism of action, preclinical study of drug efficacy, and safety evaluation [6]. We sought to further assess the anti-osteoporotic mechanism of Rhizoma drynariae, to assist in development of new drugs, and to improve conventional anti-osteoporosis drug treatments for OP defects and bottlenecks. We based our study on the concepts of network pharmacology, data searching, and data mining through the establishment of screening for Rhizoma drynariae active ingredients and the osteoporosis targets of the protein–protein interaction (PPI) network diagram.
Material and Methods
Screening for active ingredients of Rhizoma drynariae
According to the parameters of ADME (absorption, distribution, metabolism, excretion) [7,8], we screened for the active ingredients of Rhizoma drynariae by searching the pharmacological database of the traditional Chinese medicine system and analyzing platforms (TCMSP) (http://ibts.hkbu.edu.hk/LSP/tcmsp.php) [9], last updated on 2014-05-31. The screening conditions were oral bioavailability (OB) greater than 30% and drug similarity (DL) greater than 0.18 [10,11]. A total of 16 active ingredients and their corresponding typical SMILES structures were obtained by searching relevant literature and the PubChem network database (https://pubchem.ncbi.nlm.nih.gov/) [12].
Prediction of targets of the active ingredients in Rhizoma drynariae
The SMILES structures of ingredients obtained from PubChem were imported into the Prediction Target of Swiss Target Prediction network database (http://www.swisstargetprediction.ch/) [13]. Predictions can be carried out using 5 marker organisms, and predictions can be mapped using the principle of homology, within and between different species for close paralogs and orthologs. To improve prediction accuracy, only 118 predicted targets with high probability were selected and duplicates were deleted. After importing the 118 targets into the Therapeutic Target Database (TTD) (http://bidd.nus.edu.sg/group/cjttd/), Comparative Toxicogenomics Database (CTD) (http://ctdbase.org/), and PharmGKB (https://www.pharmgkb.org/), the disease target network database was used to verify whether they were related to osteoporosis. Then, Gene Ontology (GO) enrichment and Kyoto Encyclopedia of Genes and Genomes(KEGG) network pathway analysis were performed on the targets obtained. Cytoscape [14] was used to construct Compound Target and Compound Pathway network maps for this data.
Targets associated with osteoporosis
The Online Mendelian Inheritance in Man (OMIM) [15], PubMed, and other databases were used to search for relevant research reports, and all known targets related to osteoporosis were found. “Osteoporosis” was used as the keyword to search and screen for OP-related targets, and known targets in the pathogenesis of OP were obtained. Since target names in each database are irregular, official symbols were converted into Uniprot IDs for subsequent analysis. Then, the OP related targets were imported into the DAVID database (http://david.abcc.ncifcrf.gov version 6.7) for GO enrichment and KEGG network pathway analysis.
Establishing a T-T network
PPI network mapping was performed on obtained bioactive ingredients and disease targets using BisoGenet software. Then, Cytoscape was used to fuse and extract the intersection of the 2 network graphs drawn, so as to obtain a direct and an indirect target-target regulatory network graph of Rhizoma drynariae treatment of OP. Again, along with the analysis of network topology obtained using the CytoNCA [16] plug-in and literature, indicators such as the degree of centricity (DC), medium degree centrality (BC), close to the central (CC), the eigenvector centrality (EC), the network centricity (NC), and local edge connectivity topology selection (LAC) were used. Where nodes with a DC value greater than 2 times the median DC value of all nodes [17] are important nodes in the network, the nodes whose other indexes are larger than the median of all nodes are termed “Big hubs” [18]. To obtain more node information and higher node information transfer efficiency in the network as much as possible, the direct or indirect targets or mechanisms of the anti-osteoporotic effects of Rhizoma drynariae were further explored. The PPI network was constructed for the hub nodes obtained through the String database (https://string-db.org/) [19]. At the same time, GO enrichment and KEGG pathway analysis were also conducted. After extensive pathways were excluded, the top 20 signaling pathways related to osteoporosis were screened out.
Molecular docking
To further screen for the most active interactions between the active ingredients of Rhizoma drynariae and the key nodes, the reliability of this study was verified through computer simulation docking technology. To obtain the SDF structure of active ingredients from the PubChem database (https://pubchem.ncbi.nlm.nih.gov/), data were imported into the SystemsDock online docking software (http://systemsdock.unit.oist.jp/) [20] for molecular docking simulation.
Results
Screening of active ingredients of Rhizoma drynariae
A total of 71 reported active ingredients of Rhizoma drynariae were retrieved by searching the TCMSP database, and 18 of these bioactive ingredients were preliminarily screened out using ADME parameters such as OB and DL. After finding the relevant structure and target data from PubChem and the Swiss Target Prediction database, 16 active ingredients with corresponding targets were obtained after final screening, as shown in Table 1. Most of the selected active ingredients are flavonoids such as naringin, (2R)-5,7-dihydroxy-2-(4-hydroxyphenyl)chroman-4-one, kaempferol, eriodictyol, luteolin, and a small amount of lignans and steroids such as β-sitosterol. Most of these ingredients possess potent pharmacological activity. For example, β-sitosterol has good anti-oxidation and anti-inflammatory effects [21,22], luteolin has antibacterial, anti-oxidant, and other broad biological and pharmacological properties [23], and kaempferol has an anti-inflammatory effect [24], and naringenin was determined to be the quality markers of Rhizoma drynariae in the Chinese Pharmacopoeia (the State Pharmacopoeia Commission of China, 2015). The OB of these compounds varies greatly, presenting an approximately normal distribution, mostly concentrated at about 40%, and the OB is generally inversely proportional to the molecular weight (MW). The lower the MW, the higher the OB is. The route of administration of most drugs is oral. In addition to OB, the intestinal epithelial permeability (Caco-2) of the drug plays an important role in its efficacy. Overall, the higher the DL, the higher the Caco-2, such as Stigmasterol DL: 1.76, Caco-2: 1.44, luteolin DL: 0.25, and Caco-2: 0.19.
Table 1.
Active Ingredients and ADME parameters of Rhizoma drynariae.
| No | Name | Structure | OB (%) | DL |
|---|---|---|---|---|
| MOL001040 | (2R)-5,7-dihydroxy-2-(4-hydroxyphenyl)chroman-4-one |
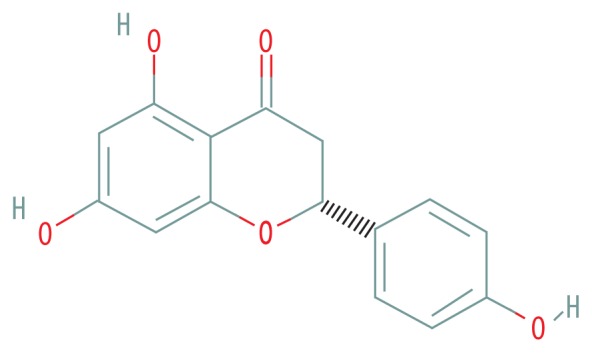
|
42.36 | 0.21 |
| MOL001978 | Aureusidin |
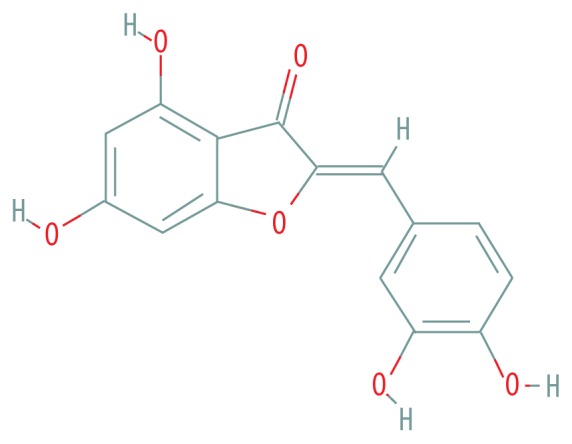
|
53.42 | 0.24 |
| MOL002914 | Eriodyctiol (flavanone) |
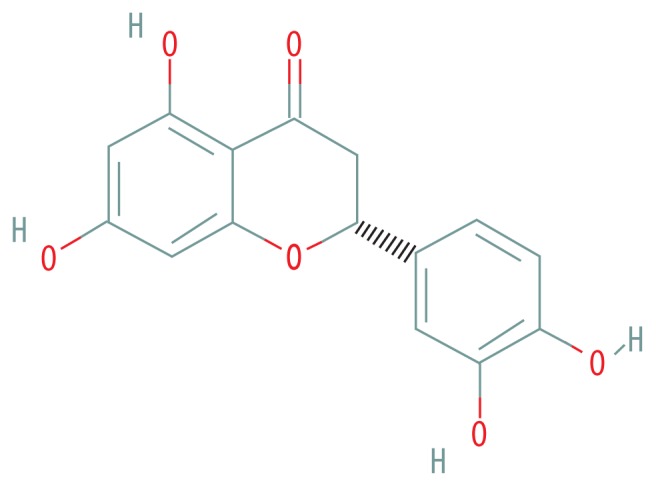
|
41.35 | 0.24 |
| MOL000449 | Stigmasterol |
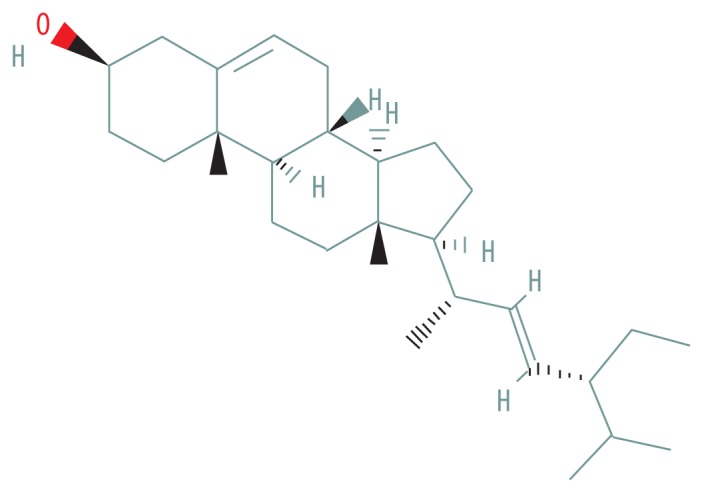
|
43.83 | 0.76 |
| MOL000358 | beta-sitosterol |
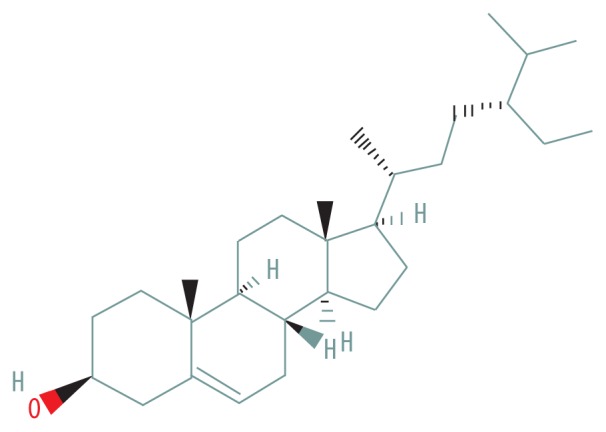
|
36.91 | 0.75 |
| MOL000422 | Kaempferol |
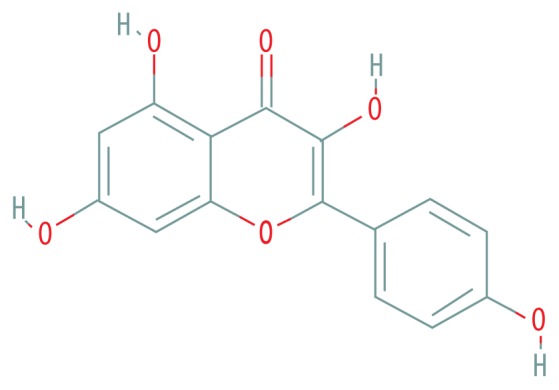
|
41.88 | 0.24 |
| MOL004328 | Naringenin |
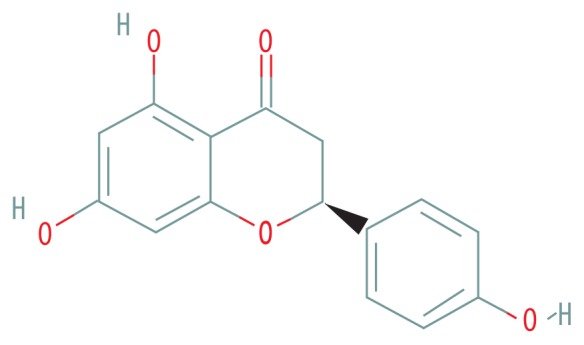
|
59.29 | 0.21 |
| MOL000492 | (+)-Catechin |
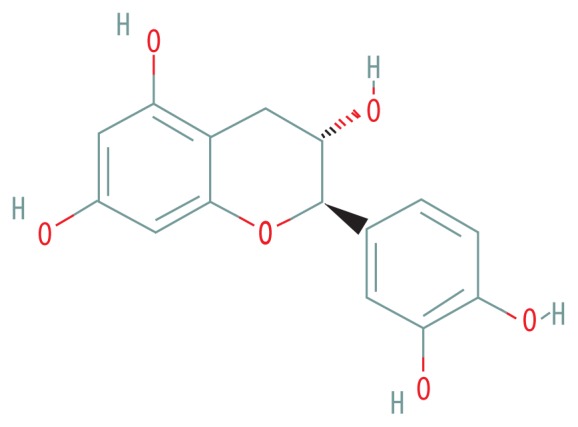
|
54.83 | 0.24 |
| MOL005190 | Eriodictyol |
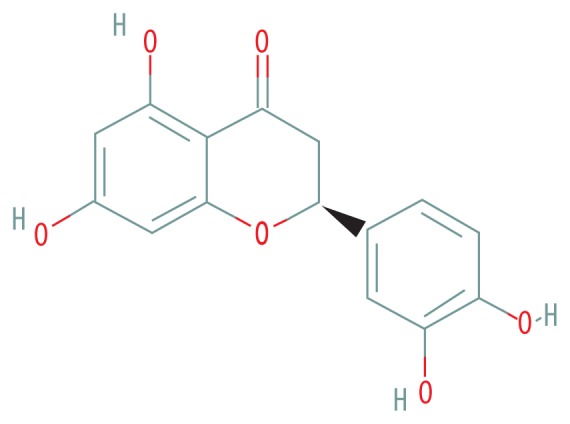
|
71.79 | 0.24 |
| MOL000569 | Digallate |
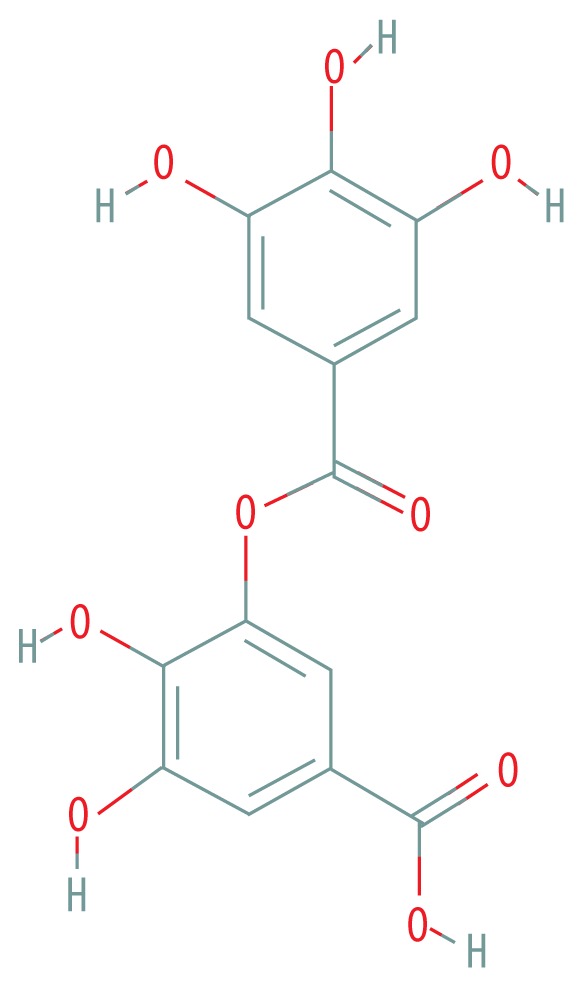
|
61.85 | 0.26 |
| MOL000006 | Luteolin |
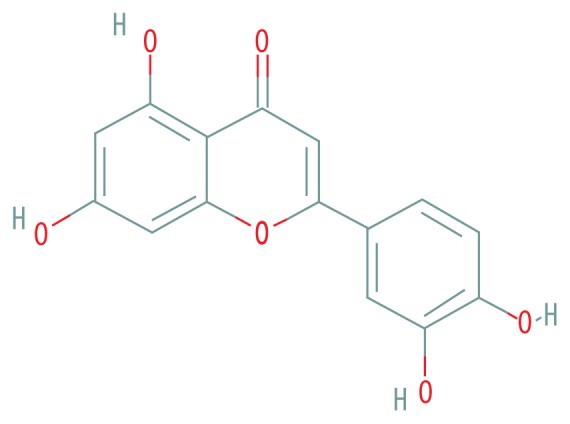
|
36.16 | 0.25 |
| MOL009061 | 22-Stigmasten-3-one |
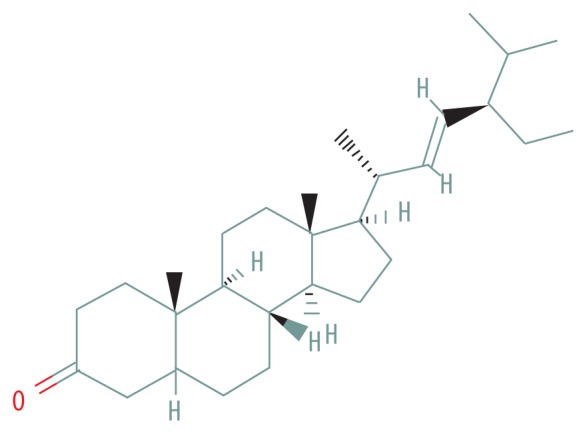
|
39.25 | 0.76 |
| MOL009063 | Cyclolaudenol acetate |
|
41.66 | 0.79 |
| MOL009075 | Cycloartenone |
|
40.57 | 0.79 |
| MOL009076 | Cyclolaudenol |
|
39.05 | 0.79 |
| MOL009091 | Xanthogalenol |
|
41.08 | 0.32 |
The putative target of active ingredients of Rhizoma drynariae
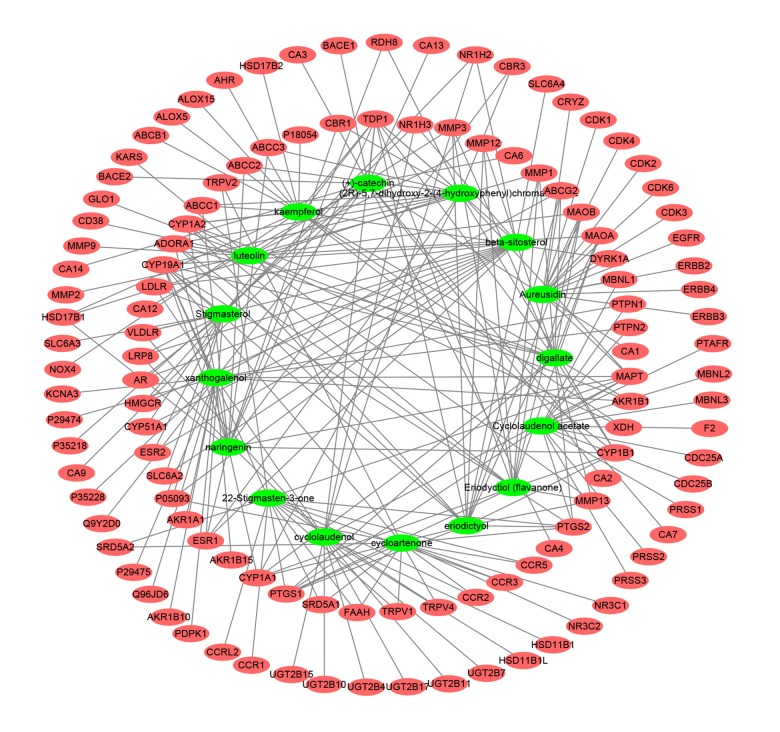
Swiss Target Prediction was used to predict the targets of 16 chemical ingredients of Rhizoma drynariae, and a total of 118 targets were obtained after high-possibility screening and de-duplication. We found that 50 of the 118 putative targets were common to 2 or more of these ingredients, suggesting that these ingredients act on some of the same biological processes or pathways, which reflects the synergistic effect of multiple ingredients on bone fractionation, and the targets also play an important role in the entire pathogenesis of OP. The Compound target network is shown in Figure 2. Figure 3 shows the 32 KEGG enriched signaling pathways and the constructed Target-Pathway network.
Figure 2.
Compound Target network (C-T network). Network of 16 active ingredients of Rhizoma drynariae and 118 putative targets.
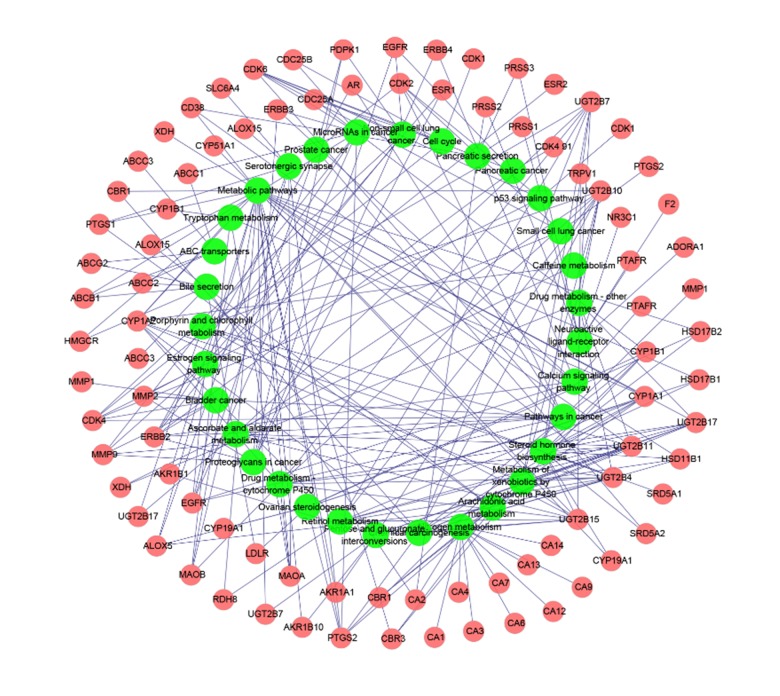
Figure 3.
Target-Pathway network (T-P network). Network of putative targets of Rhizoma drynariae and 32 KEGG pathways.
OP-related targets
By searching the OMIM and PubMed databases for related research reports, we obtained 95 and 256 targets, respectively, which are closely related to the occurrence and development of OP. A total of 316 targets were screened after double checking and removal of false-positive information. Since database names are irregular, the official symbol was converted into Uniprot IDs for use in subsequent analyses. Then, the obtained OP-related targets were imported into the DAVID database for GO enrichment and KEGG network pathway analysis. By excluding a wide range of pathways from KEGG enrichment results, the following top 10 signal transduction pathways were selected: PI3K-Akt signaling pathway, osteoclast differentiation, MAPK signaling pathway, TNF signaling pathway, Rap1 signaling pathway, Ras signaling pathway, FoxO signaling pathway, thyroid hormone signaling pathway, signaling pathways regulating pluripotency of stem cells, and the Wnt signaling pathway (Table 2).
Table 2.
Known related targets of OP.
| LRP5 | MAPK1 | HLA-DQA1 | SOX6 | WNT3A | CRY2 | CER1 | TEC |
|---|---|---|---|---|---|---|---|
| TNFRSF11B | IL17A | CYP1B1 | NOS1 | MST1R | SLC37A4 | GREM2 | CASP3 |
| VDR | SERPINE1 | NLRP3 | NPY | IL37 | ZYX | NMUR2 | STAT5A |
| COL1A1 | CYP19A1 | HSPA5 | CDX2 | MIR214 | MIR503 | BMP2K | IRF8 |
| TGFB1 | GSTT1 | ALDH7A1 | CCNE1 | PTGER4 | GNPTAB | SNX10 | INPP5D |
| TNF | RAP1A | FGF23 | WRN | PTN | FRZB | NPNT | S1PR1 |
| ESR1 | NPPB | TYMS | SLC34A1 | MTNR1B | LGMN | MIR422A | SH3BP2 |
| MTHFR | ICAM1 | CD40 | ALPL | ITGA1 | GALNT3 | THSD4 | ZMPSTE24 |
| TP53 | MIR21 | RUNX2 | ETS1 | UGT2B17 | LRRC4C | RNF185 | TCIRG1 |
| IL6 | HLA-A | VKORC1 | POMC | PER3 | TSC22D3 | DCAF13 | HS6ST1 |
| APOE | DKK1 | TWIST1 | SCG2 | EPHB2 | MIR449A | FBXO33 | CYLD |
| COL1A2 | PIK3CA | CNR1 | PTHLH | MEF2C | KDM5A | MIR2861 | CRTAP |
| VEGFA | HLA-DQB1 | DPP4 | FGF21 | INSL3 | PLOD1 | ZNF410 | ACKR1 |
| IGF1 | CD44 | MMP14 | NOX4 | ATF6 | CA8 | IL4 | FUT7 |
| IL10 | KIT | CNR2 | PTK2B | SLC11A2 | IL18BP | SP7 | XPA |
| ACE | CALCR | THBS1 | SAA1 | PLS3 | IBSP | LMNA | MIR34C |
| ADIPOQ | ADRB2 | LTA | CX3CL1 | MT-TL1 | SPRY1 | TERT | SLC26A2 |
| APP | CYP1A1 | ANXA2 | GSN | SLC1A3 | FTCDNL1 | IL1RN | TPH1 |
| BRCA1 | SIRT1 | GH1 | TGFB2 | SATB2 | SLC7A7 | CTSK | HRAS |
| NOS3 | SPP1 | SHBG | JAG1 | KAT2A | DOK6 | ITGB2 | LMNB2 |
| ABCB1 | SOD2 | IL15 | WNT1 | G6PC | NMU | CBS | ACTG2 |
| AR | BMP2 | AHSG | MIR27A | LEPQTL1 | SMS | IGF1 | PROK2 |
| AKT1 | MAPK3 | GC | CTSK | SPP2 | PUM1 | CALCA | TGIF2 |
| BDNF | SOST | HP | ITLN1 | MIR140 | DANCR | BGN | RPL5 |
| IL1B | IGF1R | TNFRSF1B | DNM1L | COLEC10 | SLC22A11 | EIF2AK3 | B4GALT7 |
| TLR4 | FGF2 | BGLAP | PRKAR1A | FSHB | VPS13B | TERC | MTND5 |
| CTNNB1 | MBL2 | MC4R | ALOX15 | AMFR | RXFP2 | RECQL2 | NOG |
| CXCL8 | IL1A | KL | CCL20 | MIR34B | QPCT | MIR34A | CAVIN1 |
| GSTM1 | FTO | CCR2 | RARRES2 | SFRP4 | GPR68 | POLG | GPR48 |
| SLC6A4 | CYP3A4 | SPARC | SFRP1 | MIR133A1 | LGR4 | ZAP70 | RIN2 |
| TNFSF11 | P2RX7 | PTH | NOTCH2 | MIR23B | GDF11 | MYC | GHR |
| LEP | HGF | TRPV1 | CYP27B1 | CSTA | TBC1D8 | NOTCH1 | SERPINA12 |
| HLA-B | RHOA | BMP4 | LCT | DVL2 | TSPAN12 | ERCC2 | TYROBP |
| SLC9A3R1 | ALDH2 | ITGA2B | IRS2 | TUBA1B | XYLT2 | HDC | CA10 |
| MTOR | IGF2 | LTF | BIRC2 | CARTPT | FGFBP1 | IL1R1 | CCKBR |
| ESR2 | LEPR | SMAD2 | IL16 | NPY2R | LRP4 | MGP | CST3 |
| PON1 | STAT1 | TRAF6 | OSBPL1A | ANKH | ATP6V1H | ITGAV | |
| CCL2 | ALB | HSD11B1 | LRP6 | P2RX4 | THSD7A | PPP3CA | |
| BRCA2 | MMP3 | ADORA2A | CD47 | MCM6 | ARHGEF3 | NFKB1 | |
| MMP2 | CYP17A1 | TNFRSF11A | PDLIM4 | WNK4 | SQOR | BTK |
Hub node screening and PPI network graph construction of Rhizoma drynariae action on OP
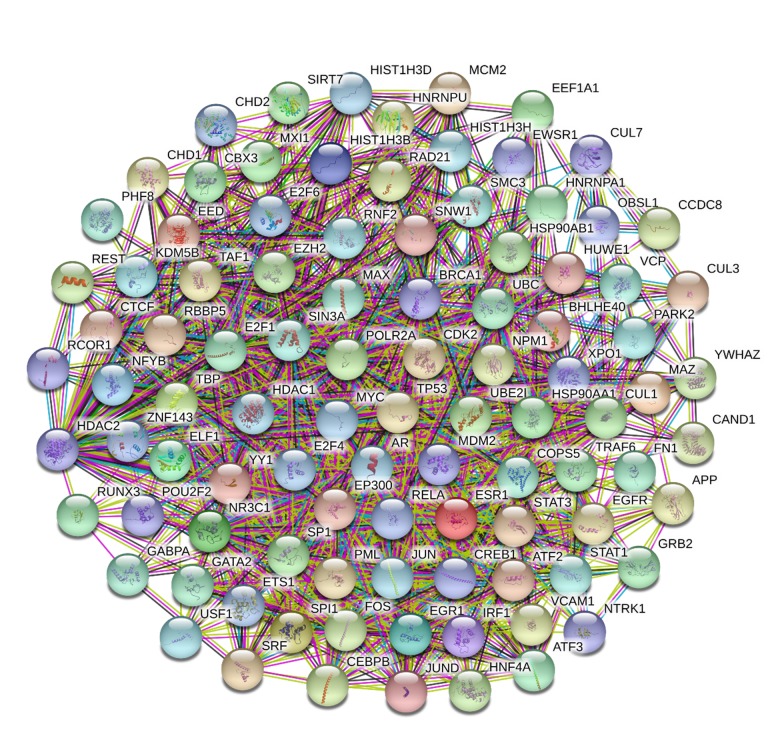
Regulation within the body is usually not dominated by a single signaling pathway but rather through a complex regulatory network. Complex crossover networks usually exist between different signaling pathways and targets. Therefore, the active ingredients of a drug not only directly combine with the target, but also combine with other targets in direct and indirect ways. Therefore, the construction of the PPI network and text mining can further explore the direct or indirect regulation relationship between the active ingredients of Rhizoma drynariae and the regulation of OP. Through the construction of the T-T network on the predicted targets of the biological active ingredients of Rhizoma drynariae using BisoGenet, it can be seen that there are 5019 targets that have direct or indirect effects on them, while there are 213 839 correlations between them. There are 13 500 targets directly or indirectly related to osteoporosis, while 523 812 targets are related to each other. Using Cytoscape, the 2 T-T graphs drawn above were extracted and intersected, resulting in a total of 4421 targets and 199 264 correlations. Then, according to the topology of DC, BC, CC, EC, NC, and LAC, CytoNCA was used to obtain 97 hub nodes and 2200 relationships, including 5 putative targets of Rhizoma drynariae and 9 targets of osteoporosis. Among them, 2 targets were common to both and 85 targets did not belong to either, suggesting that bone fragments mainly regulate osteoporosis through indirect targets. The hub node was imported into the string to build the T-T network, as shown in Figure 4.
Figure 4.
PPI Network graph of 97 Hub Nodes based on their direct interactions.
Hub node enrichment analysis of OP with Rhizoma drynariae
After G0 enrichment analysis of the hub node through the DAVID database (p<0.05), a total of 139 enrichment results were obtained. These include 80 Biological processes (BP), 32 Molecular functions (MF), and 27 Cellular components (CC). The top 10 enrichment results listed in each category are shown in Figure 2. The top 20 related signaling pathways, after removal of the extensive pathway of KEGG pathway enrichment, are shown in Table 3. Topological signaling pathways that coincide with OP target-related pathways or very important pathways include: PI3K-Akt signaling pathway, osteoclast differentiation, Wnt signaling pathway, estrogen signaling pathway, MAPK signaling pathway, FoxO signaling pathway, TNF signaling pathway, Thyroid hormone signaling pathway, and the cell cycle. Of these, the putative targets of 16 active ingredients of Rhizoma drynariae are directly involved in the estrogen signaling pathway, thyroid hormone signaling pathway, PI3K Akt signaling pathway, cell cycle, FoxO signaling pathway, MAPK signaling pathway, and other OP-related signaling pathways, involving all cells related with the occurrence of osteoporosis, such as osteoblasts, osteoclasts, adipocytes, and bone marrow stromal stem cells. These are also the mainstream pathways of clinical research and drug treatment for OP, indicating that the results of this study are reliable. The anti-OP targets of Rhizoma drynariae are abundant and the mechanism reliability is high (Figure 5).
Table 3.
The Top 20 enrichment pathways of 97 hub nodes.
| Term | Count | Value |
|---|---|---|
| Cell cycle | 14 | 8.70E-11 |
| PI3K-Akt signaling pathway | 14 | 1.80E-05 |
| MAPK signaling pathway | 12 | 1.60E-05 |
| Estrogen signaling pathway | 10 | 2.30E-07 |
| Thyroid hormone signaling pathway | 9 | 1.00E-05 |
| Osteoclast differentiation | 8 | 1.50E-04 |
| Prolactin signaling pathway | 7 | 5.10E-05 |
| FoxO signaling pathway | 6 | 8.40E-03 |
| TNF signaling pathway | 6 | 2.80E-03 |
| Neurotrophin signaling pathway | 6 | 5.40E-03 |
| Wnt signaling pathway | 5 | 3.70E-02 |
| TGF-beta signaling pathway | 5 | 6.90E-03 |
| Toll-like receptor signaling pathway | 5 | 1.90E-02 |
| Jak-STAT signaling pathway | 5 | 6.50E-02 |
| Notch signaling pathway | 4 | 1.10E-02 |
| NOD-like receptor signaling pathway | 4 | 1.20E-02 |
| B cell receptor signaling pathway | 4 | 2.90E-02 |
| ErbB signaling pathway | 4 | 5.00E-02 |
| NF-kappa B signaling pathway | 4 | 5.00E-02 |
| HIF-1 signaling pathway | 4 | 6.70E-02 |
| T cell receptor signaling pathway | 4 | 7.20E-02 |
Figure 5.
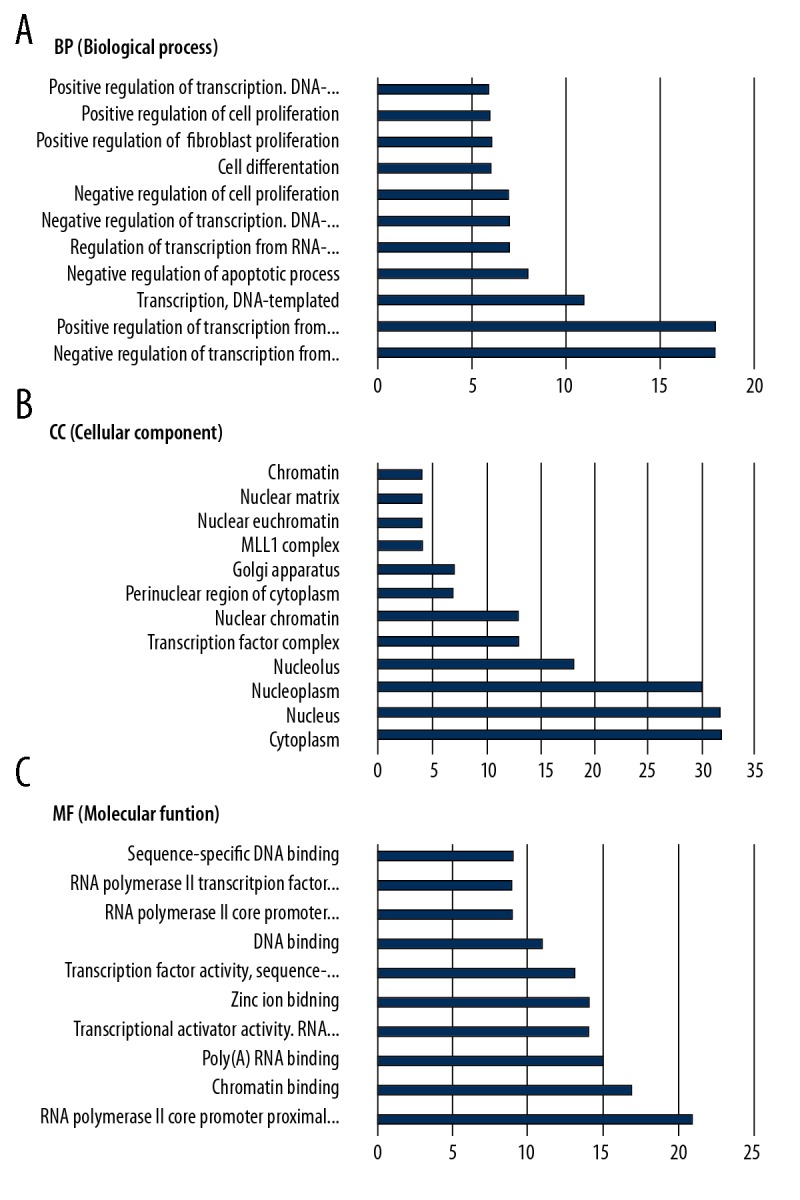
GO enrichment analysis of the putative targets of Rhizoma drynariae. The top 10 significantly enriched terms in CC, BP, and MF categories. Cellular component (A), Biological process (B), Molecular function (C).
The main active ingredients of Rhizoma drynariae are docked with key targets
In these 9 KEGG pathways, a total of 36 targets were enriched, in which TP53 frequency and connectivity were the highest. EP300 and RELA appeared frequently and were abundantly expressed in bone marrow. In recent years, the pathogenesis of oxidative stress in OP has attracted more and more attention. STAT3 not only participates in the oxidative stress FoxO pathway, but also acts as a downstream transcription factor to mediate the stress response caused by multiple pathways. The estrogen pathway and thyroid hormone pathway are the main pathways for clinical use of drugs to inhibit bone resorption and promote bone formation. ESR1 is an important target for the treatment of OP and participates in the enrichment of estrogen and thyroid receptor pathways. Osteoblast proliferation plays an important role in improving osteoporosis. CDK2 is an important kinase that regulates the cell cycle and has a high frequency in the enrichment pathway. TRAF appears in the osteoclast differentiation pathway and is upstream of the signaling pathway, which mediates multiple pathways involved in osteoporosis. Therefore, among the 9 main pathways, key targets with high repetition frequency, high clinical research value, and wide coverage were selected: CDK-2, EP300, ESR1, RELA, STAT3, TP53, and TRAF6. Molecular docking validation was conducted on all active ingredients of Rhizoma drynariae. The binding strength was evaluated according to the score, and the mechanism of action was predicted. Binding efficiency is shown in the table below. However, these results need further experimental verification in order to evaluate the reliability of this study and further reveal the active ingredients and the mechanism of action of Rhizoma drynariae (Table 4).
Table 4.
Molecular docking between the 16 compounds of Drynaria rhizome and the candidate major hub targets.
| Ligand | Receptor | score | Ligand | Receptor | Score |
|---|---|---|---|---|---|
| (+)-catechin | CDK2 | 8.341 | Digallate | CDK2 | 6.062 |
| (+)-catechin | EP300 | 8.257 | Digallate | EP300 | 6.542 |
| (+)-catechin | ESR1 | 7.497 | Digallate | ESR1 | 6.41 |
| (+)-catechin | RELA | 6.337 | Digallate | RELA | 6.288 |
| (+)-catechin | STAT3 | 7.454 | Digallate | STAT3 | 7.556 |
| (+)-catechin | TP53 | 6.284 | Digallate | TP53 | 6.242 |
| (+)-catechin | TRAF6 | 7.137 | Digallate | TRAF6 | 4.934 |
| (2R)-5,7-dihydroxy-2-(4-hydroxyphenyl)chroman-4-one | CDK2 | 6.143 | Eriodictyol | CDK2 | 8.331 |
| (2R)-5,7-dihydroxy-2-(4-hydroxyphenyl)chroman-4-one | EP300 | 6.339 | Eriodictyol | EP300 | 8.278 |
| (2R)-5,7-dihydroxy-2-(4-hydroxyphenyl)chroman-4-one | ESR1 | 6.368 | Eriodictyol | ESR1 | 8.072 |
| (2R)-5,7-dihydroxy-2-(4-hydroxyphenyl)chroman-4-one | RELA | 6.099 | Eriodictyol | RELA | 7.835 |
| (2R)-5,7-dihydroxy-2-(4-hydroxyphenyl)chroman-4-one | STAT3 | 7.749 | Eriodictyol | STAT3 | 4.942 |
| (2R)-5,7-dihydroxy-2-(4-hydroxyphenyl)chroman-4-one | TP53 | 6.328 | Eriodictyol | TP53 | 4.898 |
| (2R)-5,7-dihydroxy-2-(4-hydroxyphenyl)chroman-4-one | TRAF6 | 6.532 | Eriodictyol | TRAF6 | 2.180 |
| 22-Stigmasten-3-one | CDK2 | 6.077 | Eriodyctiol (flavanone) | CDK2 | 5.683 |
| 22-Stigmasten-3-one | EP300 | 6.452 | Eriodyctiol (flavanone) | EP300 | 6.502 |
| 22-Stigmasten-3-one | ESR1 | 6.431 | Eriodyctiol (flavanone) | ESR1 | 6.333 |
| 22-Stigmasten-3-one | RELA | 6.786 | Eriodyctiol (flavanone) | RELA | 7.204 |
| 22-Stigmasten-3-one | STAT3 | 7.531 | Eriodyctiol (flavanone) | STAT3 | 4.985 |
| 22-Stigmasten-3-one | TP53 | 6.404 | Eriodyctiol (flavanone) | TP53 | 4.953 |
| 22-Stigmasten-3-one | TRAF6 | 6.408 | Eriodyctiol (flavanone) | TRAF6 | 7.112 |
| Aureusidin | CDK2 | 6.013 | kaempferol | CDK2 | 5.706 |
| Aureusidin | EP300 | 6.546 | kaempferol | EP300 | 6.602 |
| Aureusidin | ESR1 | 6.351 | kaempferol_ | ESR1 | 6.429 |
| Aureusidin | RELA | 7.651 | kaempferol | RELA | 6.337 |
| Aureusidin | STAT3 | 4.958 | kaempferol | STAT3 | 7.543 |
| Aureusidin | TP53 | 4.927 | kaempferol | TP53 | 6.405 |
| Aureusidin | TRAF6 | 5.015 | kaempferol | TRAF6 | 6.513 |
| beta-sitosterol | CDK2 | 8.373 | luteolin | CDK2 | 6.092 |
| beta-sitosterol | EP300 | 8.258 | luteolin | EP300 | 6.493 |
| beta-sitosterol | ESR1 | 7.602 | luteolin | ESR1 | 6.338 |
| beta-sitosterol | RELA | 6.19 | luteolin | RELA | 6.291 |
| beta-sitosterol | STAT3 | 7.514 | luteolin | STAT3 | 7.723 |
| beta-sitosterol | TP53 | 6.434 | luteolin | TP53 | 6.387 |
| beta-sitosterol | TRAF6 | 4.969 | luteolin | TRAF6 | 6.446 |
| cycloartenone | CDK2 | 8.367 | naringenin | CDK2 | 8.345 |
| cycloartenone | EP300 | 8.351 | naringenin | EP300 | 8.232 |
| cycloartenone | ESR1 | 7.842 | naringenin | ESR1 | 7.602 |
| cycloartenone | RELA | 7.85 | naringenin | RELA | 6.224 |
| cycloartenone | STAT3 | 4.961 | naringenin | STAT3 | 7.339 |
| cycloartenone | TP53 | 4.946 | naringenin | TP53 | 6.316 |
| cycloartenone | TRAF6 | 7.134 | naringenin | TRAF6 | 6.258 |
| Cyclolaudenol acetate | CDK2 | 8.419 | Stigmasterol | CDK2 | 6.045 |
| Cyclolaudenol acetate | EP300 | 8.376 | Stigmasterol | EP300 | 6.555 |
| Cyclolaudenol acetate | ESR1 | 7.681 | Stigmasterol | ESR1 | 6.652 |
| Cyclolaudenol acetate | RELA | 7.853 | Stigmasterol | RELA | 7.385 |
| Cyclolaudenol acetate | STAT3 | 4.818 | Stigmasterol | STAT3 | 4.902 |
| Cyclolaudenol acetate | TP53 | 4.778 | Stigmasterol | TP53 | 4.86 |
| Cyclolaudenol acetate | TRAF6 | 4.974 | Stigmasterol | TRAF6 | 4.99 |
| cyclolaudeno | CDK2 | 3.64 | xanthogalenol | CDK2 | 7.291 |
| cyclolaudeno | EP300 | 4.538 | xanthogalenol | EP300 | 5.738 |
| cyclolaudeno | ESR1 | 3.897 | xanthogalenol | ESR1 | 6.396 |
| cyclolaudeno | RELA | 3.896 | xanthogalenol | RELA | 7.599 |
| cyclolaudeno | STAT3 | 1.914 | xanthogalenol | STAT3 | 8.323 |
| cyclolaudeno | TP53 | 2.917 | xanthogalenol | TP53 | 8.514 |
| cyclolaudeno | TRAF6 | 4.968 | xanthogalenol | TRAF6 | 8.075 |
Discussion
The theory of traditional Chinese medicine is based on systematic medical methodology. It is difficult to fully reveal the mechanisms of the action of traditional Chinese medicine through the study of single ingredients or action targets. It is of great significance to integrate the molecular mechanism of traditional Chinese medicine used in the treatment of diseases through network pharmacology, which is based on big data bioinformatics. Rhizoma drynariae is one of the main drugs used in the treatment of OP with Chinese herbal medicine. The chemical ingredients isolated from Rhizoma drynariae mainly are flavonoids, triterpenoids, phenolic acids, and glycosides. Flavonoids such as naringin can play an anti-osteoporotic role by promoting osteogenesis and inhibiting osteoclast formation [25], while neoeriocitrin can play a stronger anti-osteoporotic role by promoting osteogenesis [26].
After enrichment analysis of the predicted targets of the active ingredients of the bone-crushing tonic, the main signaling pathways related to osteoporosis were found to include: nitrogen metabolism, ovarian steroidogenesis and serotonergic synapses, calcium signaling pathway, cell cycle, estrogen signaling pathway, inflammatory signaling pathways, and other pathways closely related to the occurrence and development of OP. This suggests that Rhizoma drynariae may have a variety of active ingredients that prevent osteoporosis through multiple pathways.
The KEGG enrichment analysis of the hub node further revealed that Rhizoma drynariae can regulate stem cells, osteoblasts, osteoclasts, and immune cells through multiple pathways, including proliferation, differentiation, immunity, and oxidative stress. GO enrichment analysis showed that Rhizoma drynariae can play an anti-osteoporotic role through the bi-directional regulation of upstream transcription processes, promoting an osteogenic differentiation of cells, and regulating cell proliferation and apoptosis, but its bi-directional regulation mechanism still needs further experimental verification. After the KEGG enrichment analysis, it was found that relevant molecular mechanisms were mainly concentrated in 3 categories of pathways related to cell proliferation and differentiation, immune, and inflammation pathways, with the main pathway being the classical Wnt signaling pathway, which plays a dual role in osteoblast and osteoclast differentiation [27]. Increased levels of oxidative stress induced by various factors can promote osteoclast activity, inhibit osteoblastic activity, and promote apoptosis of bone cells [28,29]. Experiments have proven that the FoxO pathway plays an important role in the anti-oxidative stress response of osteoblasts [30]. However, its antagonistic Wnt signal increases with age [31]. On one hand, the decrease in estrogen will lead to an increase of osteoclasts and osteoclastic activity; on the other hand, it can promote the secretion of inflammatory factors, such as that of the interleukin and tumor necrosis factor (TNF) family, further promoting bone resorption, and affecting the balance of the RANKL-RANK-OPG axis, leading to increased osteoclast differentiation and activation of osteoclastic activity [32]. Rap1 is critical for resorptive function, and its selective inhibition in mature osteoclasts retards pathological bone loss [33]. In vivo and in vitro studies indicate that the PI3K/Akt cell signaling pathway is involved in the inhibition of osteoporosis by promoting osteoblast proliferation, differentiation, and bone formation [34]. Activated MAPK is involved in physiological processes such as cell proliferation, differentiation, migration, apoptosis, and stress response. It has been found that the ERK1/2 pathway, JNK pathway, and P38 pathway play important roles in increasing osteoblastic generation, reducing osteoclast differentiation, and anti-osteoporotic activity [35–37]. Studies have shown that thyroid hormones increase bone turnover in the body and promote osteogenesis, but stimulate bone resorption in vitro [38,39]. The renin-angiotensin system (RAS) also plays an important role in the regulation of bone metabolism in osteoporosis [40,41]. These studies all indicate that Rhizoma drynariae can act on OP at multiple levels through multiple mainstream signaling pathway networks that are mediated by direct or indirect targets. Furthermore, the advantages of Rhizoma drynariae in the treatment of osteoporosis and its potential in new drug development are further illustrated by this study.
Molecular docking is a quick method of predicting binding strength between TCM ingredients and targets based on the spatial structure of ligands and receptors. SystemsDock [20] online docking software uses a docking score to evaluate the binding strength of molecules, and a number from 0 to 10 indicates binding strength from weak to strong. In the signaling pathway network, cross-linking or key nodes often play an important role in mediation. Through screening and integration of hub nodes, related signal pathways, and OP experimental research, we screened out CDK2, ESR1, EP300, RELA, TRAF6, TP53, and STAT3 by performing computer simulations of molecular docking in order to verify the enrichment pathway and T-T network. The docking results show that multiple active ingredients of Rhizoma drynariae have a tight binding strength on the hub nodes, which further indicates that the anti-OP mechanism of Rhizoma drynariae is diverse and active. Estrogen deficiency is the main factor leading to PMOP, and existing estrogen-like drugs are important in the treatment of OP. Genome-wide association studies have also revealed that ESR1 may be a key gene that is highly associated with osteoporosis [42]. The docking results show that many ingredients of Rhizoma drynariae have high binding strength with estrogen receptors, suggesting that Rhizoma drynariae may have estrogen-like effects. Existing research has also confirmed that naringin in Rhizoma drynariae extract can promote osteogenesis through estrogen receptor stimulation [43], which also further confirms the reliability of this study. Cyclin-dependent kinases (Cdks) are a family of serine/threonine kinases that are key regulators of transcription processes underling coordinated cell cycle entry and sequential progression in nearly all types of proliferative cells [44], and can be combined with Cyclin. To play a role in regulating the proliferation cycle of osteoblasts, multiple active ingredients of Rhizoma drynariae bind tightly to CDK2, and it is speculated that this promotes osteoblast proliferation by increasing the expression of CDK2 in osteoblasts. EP300 is highly expressed in bone marrow and may be involved in the regulation of cell osteogenic differentiation and osteopenia through chromatin remodeling. It has also been found to be regulated by BMP2 and BMP7 in nucleus pulposus cells, which may be closely related to the occurrence of osteoporosis [45,46]. Studies have shown that Rhizoma drynariae not only has an anti-OP effect, but can also improve the degeneration of nucleus pulposus cells, which can influence the expression of EP300. Increased nuclear transcription factor kappa B (NF-κB) signaling activity can induce osteoclast differentiation gene expression and increase osteoclast activity and bone resorption [47,48]. RELA belongs to the NF-κB family and is abundant in bone marrow. It has been found that plastin 3 (PLS3) mutation can significantly enhance susceptibility to osteoporosis and postmenopausal osteoporosis in the elderly, while the upregulation of RELA (NFκB subunit p65) in PLS3-overexpressing mice is known to stimulate osteoclastogenesis, but strikingly reduces osteoclast resorption [49]. Multiple ingredients of Rhizoma drynariae have strong binding capacity with RELA, suggesting that its mechanism of improving osteoporosis may be related to inhibiting the expression of RELA and inhibiting the activity of osteoclasts. The TRAF (TNF receptor associated factor) family of molecules, as intracellular adaptor proteins, are involved in various biological processes such as bone metabolism, immunity, and stress response. TRAF6 may be involved in inducing the activation of NF-κB, thereby promoting osteoporosis [50]. Studies have found that TP53 plays a role in regulating tissue aging [51], while p53 induces cell cycle arrest during aging [52], and the activation of autophagy can restore degenerative properties of aged BMMSCs via regulating p53 expression [53]. STAT3 is a member of the STAT family and acts as a downstream target of multiple pathways such as MAPK, MTOR, and JAK. It can be stimulated by upstream signaling molecules such as IL5, IL6, and BMP2 to act as a transcription factor to mediate the expression of various genes in cell growth and cell death. It plays a key role in the process of cell death. The activation of STAT3 can promote upregulation of Runx2 and ALP and decrease the expression of DKK1 [54], suggesting that some active ingredients in Rhizoma drynariae may be specific ligands for regulating STAT3 expression.
Due to the complexity of Chinese native medicine ingredients and limitations in experimental research methods, there is at present a lack of relevant research reports on the effect of the ingredients of Rhizoma drynariae on the targets and its activity of treating OP alone. However, this study shows that it is valuable to use network pharmacology and molecular docking technology to screen for the active ingredients of traditional Chinese medicines and predict their relevant mechanisms of action. Multiple targets and multiple pathways that play a role in OP were used to decipher, for the first time, the possible molecular mechanisms of Rhizoma drynariae. This work shows the holistic characteristics of OP treated with Rhizoma drynariae. According to the docking results of SystemsDock, the active chemical ingredients were filtered, which provided the basis for subsequent research. The medicinal ingredients with a high score obtained by screening can provide a scientific and practical basis for the establishment of a multi-index quantitative fingerprint and improvement of the quality evaluation of compound ingredients.
Conclusions
Traditional Chinese medicine itself is a mixture of complex ingredients acting on multiple targets and pathways, which makes it difficult to study the mechanism of action of the active ingredients of traditional Chinese medicines. Network pharmacology provides a new approach and ideas that are of high research significance and value for screening the active ingredients of traditional Chinese medicines. Based on bioinformatics analysis and computer simulation technology, we analyzed the active materials and the possible molecular mechanism of Rhizoma drynariae on OP, providing an important theoretical basis for subsequent experimental studies.
Acknowledgment
The authors would like to acknowledge Cao Yixun, national famous Traditional Chinese Medicine expert inheritance studio (state administration of Traditional Chinese Medicine 2018) and Xu Zhanwang, Shandong famous Traditional Chinese Medicine expert inheritance studio for their strong support to this research; Thanks to Yali Gan of Traditional Chinese Medicine College, Shandong University of Traditional Chinese Medicine for the technical guidance of this study.
Footnotes
Source of support: This work was supported by a grant from the National Natural Science Foundation of China (No. 81473709)
References
- 1.Rachner TD, Khosla S, Hofbauer LC. Osteoporosis: Now and the future. Lancet (London, England) 2011;377(9773):1276–87. doi: 10.1016/S0140-6736(10)62349-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gupta T, Das N, Imran S. The prevention and therapy of osteoporosis: A review on emerging trends from hormonal therapy to synthetic drugs to plant-based bioactives. J Diet Suppl. 2018 doi: 10.1080/19390211.2018.1472715. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 3.Jia M, Nie Y, Cao DP, et al. Potential antiosteoporotic agents from plants: A comprehensive review. Evid Based Complement Alternat Med. 2012;2012 doi: 10.1155/2012/364604. 364604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang Y, Liu X, Zhao L, et al. Kidney tissue targeted metabolic profiling of glucocorticoid-induced osteoporosis and the proposed therapeutic effects of Rhizoma drynariae studied using UHPLC/MS/MS. Biomed Chromatogr. 2014;28(6):878–84. doi: 10.1002/bmc.3194. [DOI] [PubMed] [Google Scholar]
- 5.Liu X, Zhang S, Lu X, et al. Metabonomic study on the anti-osteoporosis effect of Rhizoma drynariae and its action mechanism using ultra-performance liquid chromatography-tandem mass spectrometry. J Ethnopharmacol. 2012;139(1):311–17. doi: 10.1016/j.jep.2011.11.017. [DOI] [PubMed] [Google Scholar]
- 6.Ye H, Liu Q, Wei J. Construction of drug network based on side effects and its application for drug repositioning. PLoS One. 2014;9(2):e87864. doi: 10.1371/journal.pone.0087864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu H, Wang J, Zhou W, et al. Systems approaches and polypharmacology for drug discovery from herbal medicines: An example using licorice. J Ethnopharmacol. 2013;146(3):773–93. doi: 10.1016/j.jep.2013.02.004. [DOI] [PubMed] [Google Scholar]
- 8.Xu X, Zhang W, Huang C, et al. A novel chemometric method for the prediction of human oral bioavailability. Int J Mol Sci. 2012;13(6):6964–82. doi: 10.3390/ijms13066964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ru J, Li P, Wang J, et al. TCMSP: A database of systems pharmacology for drug discovery from herbal medicines. J Cheminform. 2014;6:13. doi: 10.1186/1758-2946-6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li J, Zhao P, Li Y, et al. Systems pharmacology-based dissection of mechanisms of Chinese medicinal formula Bufei Yishen as an effective treatment for chronic obstructive pulmonary disease. Sci Rep. 2015;5:15290. doi: 10.1038/srep15290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang S, Wang H, Lu Y. Tianfoshen oral liquid: A CFDA approved clinical traditional Chinese medicine, normalizes major cellular pathways disordered during colorectal carcinogenesis. Oncotarget. 2017;8(9):14549–69. doi: 10.18632/oncotarget.14675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang Y, Bryant SH, Cheng T, et al. PubChem BioAssay: 2017 update. Nucleic Acids Res. 2017;45(D1):D955–63. doi: 10.1093/nar/gkw1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gfeller D, Grosdidier A, Wirth M, et al. SwissTargetPrediction: Aweb server for target prediction of bioactive small molecules. Nucleic Acids Res. 2014;42(Web Server issue):W32–38. doi: 10.1093/nar/gku293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shannon P, Markiel A, Ozier O, et al. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Amberger JS, Hamosh A. Searching online mendelian inheritance in man (OMIM): A knowledgebase of human genes and genetic phenotypes. Curr Protoc Bioinformatics. 2017;58:1.2.1–2.12. doi: 10.1002/cpbi.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tang Y, Li M, Wang J, et al. CytoNCA: A cytoscape plugin for centrality analysis and evaluation of protein interaction networks. Biosystems. 2015;127:67–72. doi: 10.1016/j.biosystems.2014.11.005. [DOI] [PubMed] [Google Scholar]
- 17.Li S, Zhang ZQ, Wu LJ, et al. Understanding ZHENG in traditional Chinese medicine in the context of neuro-endocrine-immune network. IET Syst Biol. 2007;1(1):51–60. doi: 10.1049/iet-syb:20060032. [DOI] [PubMed] [Google Scholar]
- 18.Yong W, Zhongyang L, Chun L, et al. Drug target prediction based on the herbs components: the study on the multitargets pharmacological mechanism of qishenkeli acting on the coronary heart disease. Evid Based Complement Alternat Med. 2012;2012 doi: 10.1155/2012/698531. 69853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Szklarczyk D, Morris JH, Cook H, et al. The STRING database in 2017: Quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45(D1):D362–68. doi: 10.1093/nar/gkw937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hsin KY, Matsuoka Y, Asai Y, et al. systemsDock: A web server for network pharmacology-based prediction and analysis. Nucleic Acids Res. 2016;44(W1):W507–13. doi: 10.1093/nar/gkw335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ambavade SD, Misar AV Ambavade PDJOP, Medicine E. Pharmacological, nutritional, and analytical aspects of β-sitosterol: A review. 2014;14(3):193–211. [Google Scholar]
- 22.Valerio M, Awad AB. beta-Sitosterol down-regulates some pro-inflammatory signal transduction pathways by increasing the activity of tyrosine phosphatase SHP-1 in J774A.1 murine macrophages. Int Immunopharmacol. 2011;11(8):1012–17. doi: 10.1016/j.intimp.2011.02.018. [DOI] [PubMed] [Google Scholar]
- 23.Lin Y, Shi R, Wang X, Shen HM. Luteolin, a flavonoid with potential for cancer prevention and therapy. Curr Cancer Drug Targets. 2008;8(7):634–46. doi: 10.2174/156800908786241050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu ZK, Xiao HB, Fang J. Anti-inflammatory properties of kaempferol via its inhibition of aldosterone signaling and aldosterone-induced gene expression. Can J Physiol Pharmacol. 2014;92(2):117–23. doi: 10.1139/cjpp-2013-0298. [DOI] [PubMed] [Google Scholar]
- 25.Wei M, Yang Z, Li P, et al. Anti-osteoporosis activity of naringin in the retinoic acid-induced osteoporosis model. Am J Chin Med. 2007;35(4):663–67. doi: 10.1142/S0192415X07005156. [DOI] [PubMed] [Google Scholar]
- 26.Lina L, Zhen Z, Guoping CJP. Comparison of neoeriocitrin and naringin on proliferation and osteogenic differentiation in MC3T3-E1. Phytomedicine. 2011;18(11):985–89. doi: 10.1016/j.phymed.2011.03.002. [DOI] [PubMed] [Google Scholar]
- 27.Albers J, Keller J, Baranowsky A, et al. Canonical Wnt signaling inhibits osteoclastogenesis independent of osteoprotegerin. J Cell Biol. 2013;200(4):537–49. doi: 10.1083/jcb.201207142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu Q, Zhong ZM, Pan Y, et al. Advanced oxidation protein products as a novel marker of oxidative stress in postmenopausal osteoporosis. Med Sci Monit. 2015;21:2428–32. doi: 10.12659/MSM.894347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Manolagas SC. From estrogen-centric to aging and oxidative stress: A revised perspective of the pathogenesis of osteoporosis. Endocr Rev. 2010;31(3):266–300. doi: 10.1210/er.2009-0024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ambrogini E, Almeida M, Martin-Millan M, et al. FoxO-mediated defense against oxidative stress in osteoblasts is indispensable for skeletal homeostasis in mice. Cell Metab. 2010;11(2):136–46. doi: 10.1016/j.cmet.2009.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Manolagas SC, Almeida M. Gone with the Wnts: beta-catenin, T-cell factor, forkhead box O, and oxidative stress in age-dependent diseases of bone, lipid, and glucose metabolism. Mol Endocrinol (Baltimore, Md) 2007;21(11):2605–14. doi: 10.1210/me.2007-0259. [DOI] [PubMed] [Google Scholar]
- 32.Chou CW, Chiang TI, Chang IC, et al. Expression levels of estrogen receptor α mRNA in peripheral blood cells are an independent biomarker for postmenopausal osteoporosis. BBA Clin. 2016;5:124–29. doi: 10.1016/j.bbacli.2016.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zou W, Izawa T, Zhu T, et al. Talin1 and Rap1 are critical for osteoclast function. Mol Cell Biol. 2013;33(4):830–44. doi: 10.1128/MCB.00790-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xi JC, Zang HY, Guo LX, et al. The PI3K/AKT cell signaling pathway is involved in regulation of osteoporosis. J Recept Signal Transduct Res. 2015;35(6):640–45. doi: 10.3109/10799893.2015.1041647. [DOI] [PubMed] [Google Scholar]
- 35.Hye Kyung K, Myung-Gyou K, Kang-Hyun LJM. Osteogenic activity of collagen peptide via ERK/MAPK pathway mediated boosting of collagen synthesis and its therapeutic efficacy in osteoporotic bone by back-scattered electron imaging and microarchitecture analysis. Molecules. 2013;18(12):15474–89. doi: 10.3390/molecules181215474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li G, Wang M, Hao L, et al. Angiotensin II induces mitochondrial dysfunction and promotes apoptosis via JNK signalling pathway in primary mouse calvaria osteoblast. Arch Oral Biol. 2014;59(5):513–23. doi: 10.1016/j.archoralbio.2014.02.015. [DOI] [PubMed] [Google Scholar]
- 37.Choi SW, Son YJ, Yun JM, Kim SH. Fisetin inhibits osteoclast differentiation via downregulation of p38 and c-Fos-NFATc1 signaling pathways. Evid Based Complement Alternat Med. 2012;2012 doi: 10.1155/2012/810563. 810563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Britto JM, Fenton AJ, Holloway WR, Nicholson GC. Osteoblasts mediate thyroid hormone stimulation of osteoclastic bone resorption. Endocrinology. 1994;134(1):169–76. doi: 10.1210/endo.134.1.8275930. [DOI] [PubMed] [Google Scholar]
- 39.Cray JJ, Jr, Khaksarfard K, Weinberg SM, et al. Effects of thyroxine exposure on osteogenesis in mouse calvarial pre-osteoblasts. PLoS One. 2013;8(7):e69067. doi: 10.1371/journal.pone.0069067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kwok T, Leung J, Zhang YF, et al. Does the use of ACE inhibitors or angiotensin receptor blockers affect bone loss in older men? Osteoporos Int. 2012;23(8):2159–67. doi: 10.1007/s00198-011-1831-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Diao TY, Pan H, Gu SS, et al. Effects of angiotensin-converting enzyme inhibitor, captopril, on bone of mice with streptozotocin-induced type 1 diabetes. J Bone Miner Metab. 2014;32(3):261–70. doi: 10.1007/s00774-013-0500-7. [DOI] [PubMed] [Google Scholar]
- 42.Qin L, Liu Y, Wang Y, et al. Computational characterization of osteoporosis associated SNPs and genes identified by genome-wide association studies. PLoS One. 2016;11(3):e0150070. doi: 10.1371/journal.pone.0150070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pang WY, Wang XL, Mok SK, et al. Naringin improves bone properties in ovariectomized mice and exerts oestrogen-like activities in rat osteoblast-like (UMR-106) cells. 2010;159(8):1693–703. doi: 10.1111/j.1476-5381.2010.00664.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.He G, Yang X, Wang G, et al. Cdk7 is required for activity-dependent neuronal gene expression, long-lasting synaptic plasticity and long-term memory. Front Mol Neurosci. 2017;10:365. doi: 10.3389/fnmol.2017.00365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hu Z, Wang Y, Sun Z, et al. miRNA-132-3p inhibits osteoblast differentiation by targeting Ep300 in simulated microgravity. Sci Rep. 2015;5:18655. doi: 10.1038/srep18655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Leung VY, Zhou L, Tam WK, et al. Bone morphogenetic protein -2 and -7 mediate the anabolic function of nucleus pulposus cells with discrete mechanisms. Connect Tissue Res. 2017;58(6):573–85. doi: 10.1080/03008207.2017.1282951. [DOI] [PubMed] [Google Scholar]
- 47.Boyce BF, Yao Z, Xing L. Functions of nuclear factor kappaB in bone. Ann NY Acad Sci. 2010;1192:367–75. doi: 10.1111/j.1749-6632.2009.05315.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bone YA-AJ. NF-κB signaling and bone resorption. Osteoporos Int. 2013;24(9):2377–86. doi: 10.1007/s00198-013-2313-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Neugebauer J, Heilig J, Hosseinibarkooie S, et al. Plastin 3 influences bone homeostasis through regulation of osteoclast activity. Hum Mol Genet. 2018;27(24):4249–62. doi: 10.1093/hmg/ddy318. [DOI] [PubMed] [Google Scholar]
- 50.Zhang W, Zhang X, Wu XL, et al. Competition between TRAF2 and TRAF6 regulates NF-kappaB activation in human B lymphocytes. Chin Med Sci J. 2010;25(1):1–12. doi: 10.1016/s1001-9294(10)60013-2. [DOI] [PubMed] [Google Scholar]
- 51.Tyner SD, Venkatachalam S, Choi J, et al. p53 mutant mice that display early ageing-associated phenotypes. Nature. 2002;415(6867):45–53. doi: 10.1038/415045a. [DOI] [PubMed] [Google Scholar]
- 52.Vigneron A, Vousden KH. p53, ROS and senescence in the control of aging. Aging. 2010;2(8):471–74. doi: 10.18632/aging.100189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ma Y, Qi M, An Y, et al. Autophagy controls mesenchymal stem cell properties and senescence during bone aging. Aging Cell. 2017;17:e12709. doi: 10.1111/acel.12709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nicolaidou V, Wong MM, Redpath AN, et al. Monocytes induce STAT3 activation in human mesenchymal stem cells to promote osteoblast formation. PLoS One. 2012;7(7):e39871. doi: 10.1371/journal.pone.0039871. [DOI] [PMC free article] [PubMed] [Google Scholar]