The crystal structures of Hsp104 N-terminal domains from S. cerevisiae (ScHsp104NTD) and C. albicans (CaHsp104NTD) were determined to high resolution. The structures of ScHsp104NTD and CaHsp104NTD reveal that the yeast Hsp104 N-terminal domain may utilize a conserved putative peptide-binding groove to interact with misfolded polypeptides.
Keywords: Hsp104, crystal structure, peptide-binding groove, prions, yeast, Saccharomyces cerevisiae, Candida albicans, amyloidogenic neurodegenerative diseases
Abstract
Hsp104 is a yeast member of the Hsp100 family which functions as a molecular chaperone to disaggregate misfolded polypeptides. To understand the mechanism by which the Hsp104 N-terminal domain (NTD) interacts with its peptide substrates, crystal structures of the Hsp104 NTDs from Saccharomyces cerevisiae (ScHsp104NTD) and Candida albicans (CaHsp104NTD) have been determined at high resolution. The structures of ScHsp104NTD and CaHsp104NTD reveal that the yeast Hsp104 NTD may utilize a conserved putative peptide-binding groove to interact with misfolded polypeptides. In the crystal structures ScHsp104NTD forms a homodimer, while CaHsp104NTD exists as a monomer. The consecutive residues Gln105, Gln106 and Lys107, and Lys141 around the putative peptide-binding groove mediate the monomer–monomer interactions within the ScHsp104NTD homodimer. Dimer formation by ScHsp104NTD suggests that the Hsp104 NTD may specifically interact with polyQ regions of prion-prone proteins. The data may reveal the mechanism by which Hsp104 NTD functions to suppress and/or dissolve prions.
1. Introduction
AAA+ (ATPases associated with various cellular activities) proteins belong to a family of molecular machines with related structures but diverse functions. Some of these proteins utilize ATP hydrolysis to facilitate the degradation of proteins and nucleic acids, as well as the remodeling of aggregated macromolecules (Hanson & Whiteheart, 2005 ▸; Erzberger & Berger, 2006 ▸). The heat-shock protein 100 (Hsp100) family represents a large group of AAA+ ATPases that perform molecular-chaperone activities, including the degradation of misfolded polypeptides, regulation of transcription, reactivation of aggregated proteins and protection of cells from thermal and chemical stresses (Schirmer et al., 1996 ▸; Wawrzynow et al., 1996 ▸). Bacterial ClpB and yeast Hsp104 are members of the Hsp100 family and are hexameric molecular motors that couple the energy generated from ATP binding and/or hydrolysis to the disaggregation of protein aggregates in vivo (Doyle & Wickner, 2009 ▸; Mogk et al., 2008 ▸; Zolkiewski, 2006 ▸).
Yeast Hsp104 facilitates the disaggregation and protein folding of non-native proteins, usually by collaborating with the Hsp70 and Hsp40 chaperones (Glover & Lindquist, 1998 ▸; Parsell et al., 1994 ▸; Shorter & Lindquist, 2004 ▸; Kummer et al., 2016 ▸; Klosowska et al., 2016 ▸). Hsp104 is essential for yeast survival in stressful conditions such as elevated temperatures (Sanchez & Lindquist, 1990 ▸; Sanchez et al., 1992 ▸). Hsp104 is essential for the propagation of yeast prions [PSI +], [URE3] and [RNQ1+] (Chernoff et al., 1995 ▸; Wickner, 1994 ▸; Cox et al., 2003 ▸). These yeast prions are self-replicating, amyloid-like aggregates of the normal functional proteins. Hsp104 helps yeast prions to replicate mainly by fragmenting the aggregates into smaller pieces, thereby breeding new propagons or seeds (Satpute-Krishnan et al., 2007 ▸; Winkler et al., 2012 ▸). However, overexpression of Hsp104 in yeast dissolves these seeds completely and accordingly cures the [PSI+] prion state (Park et al., 2014 ▸). A recent study has demonstrated that the amyloid-dissolution activity of Hsp104 can be potentiated by mutations in the middle domain (Jackrel et al., 2014 ▸; Torrente et al., 2016 ▸), thus providing the possibility that a modified Hsp104 could serve as a therapeutic approach against amyloidogenic neurodegenerative diseases in humans, such as Alzheimer’s disease (AD), Parkinson’s disease (PD) and Huntington’s disease (HD) (Shorter, 2008 ▸; Jackrel & Shorter, 2015 ▸).
The Hsp104 protomer contains five domains: an N-terminal domain (NTD), nucleotide-binding domain 1 (NBD1), a coiled-coil middle domain (MD), nucleotide-binding domain 2 (NBD2) and a short acidic C-terminal domain (CTD). Cryo-EM studies indicated that Hsp104 may form a dynamic hexameric ring structure and harbors a globular NTD that is connected to NBD1 by an unstructured linker sequence (Wendler et al., 2007 ▸; Lee et al., 2010 ▸). Recent cryo-EM data showed that Hsp104 constitutes a unusual spiral hexamer with a three-tiered domain arrangement, with the NTD forming the top layer (Yokom et al., 2016 ▸). The ClpB NTD has been demonstrated to have a substrate-binding groove that specifically recognizes exposed hydrophobic stretches in unfolded or aggregated client proteins, thus playing a regulatory role in protein disaggregation (Li & Sha, 2003 ▸; Rosenzweig et al., 2015 ▸). The roles of the Hsp104 NTD in protein disaggregation and prion dissolution have yet to be fully understood. Previous studies have shown that the Hsp104 NTD is dispensable for thermotolerance and prion propagation (Hung & Masison, 2006 ▸; Lum et al., 2008 ▸; Tipton et al., 2008 ▸; Reidy et al., 2012 ▸). However, recent data support the idea that the Hsp104 NTD enables cooperative substrate translocation by Hsp104, which is critical for potentiated activity and prion dissolution, but not for prion fragmentation (Sweeny et al., 2015 ▸). The Hsp104 NTD might be involved directly in the binding of substrates, and its action might introduce trimming forces that are critical for the disaggregation activity of Hsp104 (Sweeny et al., 2015 ▸).
Extensive structural studies have been performed for ClpB (Li & Sha, 2002 ▸, 2003 ▸), while limited structural investigations, particularly crystallographic studies, are available for yeast Hsp104 (Wendler et al., 2007 ▸; Lee et al., 2010 ▸; Yokom et al., 2016 ▸). To understand the mechanism by which the Hsp104 NTD interacts with its peptide substrates, we have determined the crystal structures of the Hsp104 NTDs from Saccharomyces cerevisiae (ScHsp104NTD) and Candida albicans (CaHsp104NTD). The structures of ScHsp104NTD and CaHsp104NTD reveal that the yeast Hsp104 NTD may utilize a conserved putative peptide-binding groove to interact with misfolded polypeptides. More importantly, the crystal structure of ScHsp104NTD also suggests that the Hsp104 NTD may specifically interact with polyQ regions of prion-prone proteins. These studies may reveal the mechanisms by which the Hsp104 NTD functions to suppress and/or dissolve prions.
2. Materials and methods
2.1. Protein expression and purification
The coding sequences for the N-terminal domains of Hsp104 from the yeasts C. albicans (residues 1–164; CaHsp104NTD; GenBank accession No. AAK60625) and S. cerevisiae (residues 1–166; ScHsp104NTD; GenBank accession No. CAA97475) were obtained by PCR. The genes were then digested with NheI and XhoI, and subsequently subcloned into the pET-28b expression vector (Novagen). All proteins were expressed in Escherichia coli strain BL21 (DE3) grown in LB medium at 37°C. Protein expression was induced with 0.5 mM IPTG when the cells reached an OD600 of 0.6. The cells were harvested, washed in 100 ml binding buffer (10 mM Tris–HCl pH 7.5, 150 mM NaCl) and then lysed by sonication on ice. The lysates were clarified by centrifugation at 12 000 rev min−1 for 30 min. The supernatant was loaded onto an Ni2+–NTA column pre-equilibrated with binding buffer. The resin was washed with ice-cold washing buffer (20 mM Tris–HCl pH 7.9, 0.5 M NaCl, 60 mM imidazole) and the target proteins were eluted with elution buffer (20 mM Tris–HCl buffer pH 7.9, 0.5 M NaCl, 400 mM EDTA). The typical yield of the purified soluble proteins (>95% purity from SDS–PAGE analysis) from a 1 l culture was about 50 mg. The proteins were then treated with thrombin (one unit per milligram of protein) at 4°C overnight to remove the polyhistidine purification tag. Digestion took place and was stopped by the addition of 0.2 mM PMSF. Following thrombin treatment, the samples were purified again using an Ni2+–NTA column to simultaneously remove unprocessed tagged protein, tag and thrombin. The final eluates were subsequently loaded onto a Superdex 200 column (Amersham Biosciences) in 10 mM Tris–HCl pH 7.5, 100 mM NaCl. The eluted recombinant CaHsp104NTD and ScHsp104NTD were concentrated to 20 mg ml−1 by centrifugal ultrafiltration (Millipore) for crystallization assays.
2.2. The ELISA assay
The binding between the two Hsp104 NTDs (CaHsp104NTD and ScHsp104NTD) and two denatured proteins (rhodanese and citrate synthase) were tested using the ELISA method. CaHsp104NTD or ScHsp104NTD (100 µl of 1 µg ml−1 protein in PBS) was incubated in the wells of 96-well EIA/RIA plates (Corning, New York, USA) for 1 h at room temperature. The protein solutions were then decanted and the wells were washed three times with 200 µl 0.2% BSA in PBST (PBS with 0.05% Tween 20). To block the wells, 200 µl 0.2% BSA in PBS was incubated in the wells for 1 h at room temperature. After blocking, the wells were washed three times with 200 µl 0.2% BSA in PBST. Rhodanese (25 mg ml−1, Sigma) and citrate synthase (15 mg ml−1, Sigma) were diluted 100 times into the denaturing solution [50 mM Tris–HCl pH 8.0, 6 M guanidine, 0.1%(v/v) β-mercaptoethanol] and incubated at room temperature for 2 h. The denatured proteins were then diluted to two concentrations (0.1 and 1 µg ml−1) with PBS, added to wells coated with CaHsp104NTD, coated with ScHsp104NTD or only blocked with BSA (control), and then incubated at room temperature for 1 h. In order to test whether CaHsp104NTD and ScHsp104NTD can bind native proteins, native rhodanese (1 µg ml−1 in PBS) and citrate synthase (1 µg ml−1 in PBS) were also added to the CaHsp104NTD- or ScHsp104NTD-coated wells in this step. After decanting all liquid, all wells were washed three times with 200 µl 0.2% BSA in PBST. Following this, 100 µl anti-rhodanese goat polyclonal IgG (Santa Cruz Biotechnology, Santa Cruz, California, USA) or 100 µl anti-citrate synthase rabbit polyclonal IgG (Thermo Scientific, Madison, Wisconsin, USA) was added to the corresponding wells and incubated for 1 h at room temperature. After washing three times with 200 µl 0.2% BSA in PBST, the wells were then incubated with 100 µl donkey anti-goat IgG conjugated to horseradish peroxidase (IgG-HRP) for rhodanese (Santa Cruz Biotechnology) or 100 µl donkey anti-rabbit IgG-HRP for citrate synthase (Thermo Scientific) for 30 min at room temperature. The wells were then washed six times with 200 µl PBST and the amount of secondary antibody was measured using the TMB peroxidase substrate (Thermal Scientific). The absorbance at 450 nm was measured using a UV microplate reader (Bio-Rad, Hercules, California, USA).
2.3. Crystallization of CaHsp104NTD and ScHsp104NTD
CaHsp104NTD was concentrated to 20 mg ml−1 and crystals of about 0.4 × 0.02 × 0.02 mm in size were obtained at 22°C with a well condition consisting of 25% PEG 1000, 50 mM Tris–HCl pH 8.5, 200 mM ammonium citrate using the hanging-drop vapor-diffusion method. The crystals were cryoprotected in mother liquor supplemented with 25%(v/v) glycerol. All crystals were then mounted in a cryoloop and immediately flash-cooled in liquid nitrogen. The cooled crystals were instantly transported to the SER-CAT beamline at APS for data collection (Table 1 ▸). The native CaHsp104NTD crystals diffracted to 1.66 Å resolution. To solve the crystal structure of CaHsp104NTD using the SAD method, selenomethionyl (SeMet) CaHsp104NTD was produced following previously described protocols (Sha et al., 2000 ▸). The preparation of SeMet CaHsp104NTD crystals was the same as that used to grow the native crystals. ScHsp104NTD crystals were obtained at 22°C using protein at a concentration of 20 mg ml−1 with a well solution consisting of 0.1 M Tris–HCl pH 8.5, 25%(w/v) PEG 3350. Data for the ScHsp104NTD crystals were collected to a resolution of 2.54 Å (Table 1 ▸).
Table 1. Data collection and structure determination of CaHsp104NTD and ScHsp104NTD.
Values in parentheses are for the highest resolution shell.
| Native CaHsp104NTD | SeMet CaHsp104NTD | Native ScHsp104NTD | |
|---|---|---|---|
| Data collection | |||
| Space group | P3221 | P3221 | C2 |
| Unit-cell parameters | |||
| a (Å) | 55.21 | 54.84 | 148.59 |
| b (Å) | 55.21 | 54.84 | 66.26 |
| c (Å) | 109.45 | 109.10 | 74.58 |
| α (°) | 90.00 | 90.00 | 90.00 |
| β (°) | 90.00 | 90.00 | 107.37 |
| γ (°) | 120.00 | 120.00 | 90.00 |
| Wavelength (Å) | 1.000 | 0.9790 | 1.000 |
| Resolution (Å) | 1.66 (1.69–1.66) | 1.78 (1.81–1.78) | 2.54 (2.58–2.54) |
| R merge | 0.072 (0.47) | 0.056 (0.28) | 0.078 (0.17) |
| 〈I/σ(I)〉 | 59.3 (2.1) | 89.1 (9.9) | 30.7 (4.7) |
| Completeness (%) | 99.4 (70.5) | 99.1 (80.5) | 96.2 (89.7) |
| Multiplicity | 16.1 | 21.4 | 3.5 |
| Refinement | |||
| Resolution (Å) | 1.66 | 2.54 | |
| No. of reflections | 23417 | 21839 | |
| R work | 0.170 (0.183) | 0.199 (0.221) | |
| R free | 0.203 (0.237) | 0.256 (0.313) | |
| No. of atoms | |||
| Protein | 1206 | 3807 | |
| Water | 243 | 11 | |
| B factors (Å2) | |||
| Protein | 29.4 | 47.7 | |
| Water | 47.0 | 48.3 | |
| R.m.s. deviations | |||
| Bond lengths (Å) | 0.006 | 0.009 | |
| Bond angles (°) | 0.970 | 1.348 | |
2.4. Structure determination and refinement
The SAD method was utilized to determine the structure of CaHsp104NTD. Se atoms were located and the resultant phases were calculated using PHENIX (Adams et al., 2010 ▸). The resultant electron-density map had continuous electron density and was readily interpreted. Residues 4–160 of CaHsp104NTD were modeled into the electron-density map using Coot (Emsley & Cowtan, 2004 ▸). The native CaHsp104NTD structure was refined with PHENIX (Table 1 ▸). The CaHsp104N model was used to search for molecular-replacement solutions for the crystals of ScHsp104NTD using PHENIX. The initial model was built using PHENIX, followed by manual model building using Coot. The final model was refined using PHENIX (Table 1 ▸).
2.5. Sequence alignment
The amino-acid sequence alignment was conducted with ClustalX (ftp://ftp.ebi.ac.uk/pub/software/clustalw2; Larkin et al., 2007 ▸) and the ESPript 3.0 web tool (http://espript.ibcp.fr/ESPript/ESPript/; Robert & Gouet, 2014 ▸).
3. Results and discussion
3.1. CaHsp104NTD and ScHsp104NTD can directly bind denatured proteins
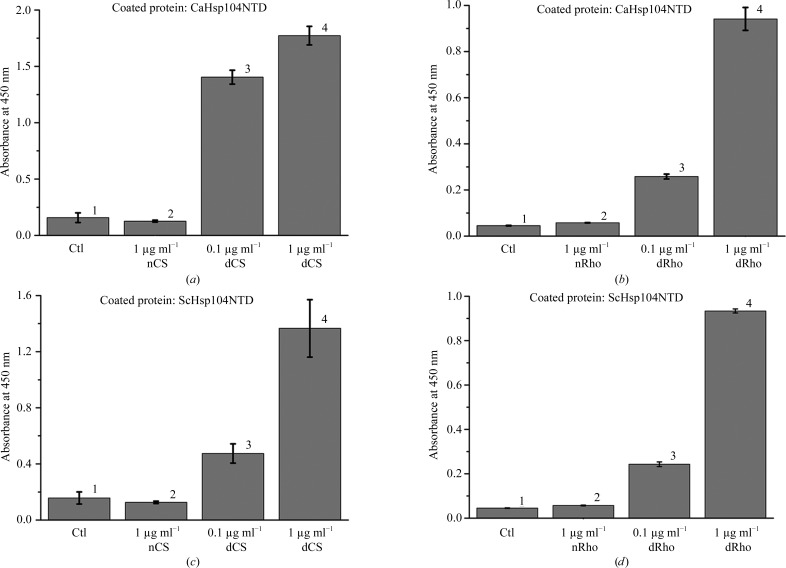
To examine whether CaHsp104NTD and ScHsp104NTD can interact with denatured proteins, we performed a series of ELISA experiments. Two model proteins, rhodanese and citrate synthase, were chemically denatured and utilized as substrates to monitor the binding. The plate was first coated with either CaHsp104NTD or ScHsp104NTD. The denatured rhodanese or citrate synthase was then applied to the corresponding wells at various concentrations. We found that both CaHsp104NTD and ScHsp104NTD can efficiently bind denatured rhodanese and citrate synthase (Fig. 1 ▸). In contrast, the data showed that CaHsp104NTD and ScHsp104NTD do not interact with native rhodanese and citrate synthase (Fig. 1 ▸). The data suggest that Hsp104 NTD can function as a molecular chaperone to specifically recognize and interact with non-native polypeptides over native proteins.
Figure 1.
The yeast Hsp104 N-terminal domain can selectively interact with denatured model proteins but not with native proteins, as shown by ELISA assays. (a) CaHsp104NTD can interact with denatured citrate synthase (CS). CaHsp104NTD was coated onto the ELISA plates and 1 µg ml−1 native CS (nCS) and 0.1 µg ml−1 and 1 µg ml−1 denatured CS (dCS) were added after blocking with 1% BSA. Wells coated with BSA only were utilized as controls (Ctl). After extensive washing, the bound CS can be detected by HRP-conjugated polyclonal CS antibody and the OD450 readings are shown as gray bars. The standard derivations of three independent experiments are shown in the figure. (b) CaHsp104NTD preferably binds denatured rhodanese (dRho) over native rhodanese (nRho). The bound rhodanese can be detected with an HRP-conjugated polyclonal antibody against rhodanese. (c) ScHsp104NTD can interact with denatured citrate synthase but not with native citrate synthase. (d) ScHsp104NTD selectively interacts with denatured rhodanese (dRho) over native rhodanese.
3.2. The crystal structures of CaHsp104NTD and ScHsp104NTD
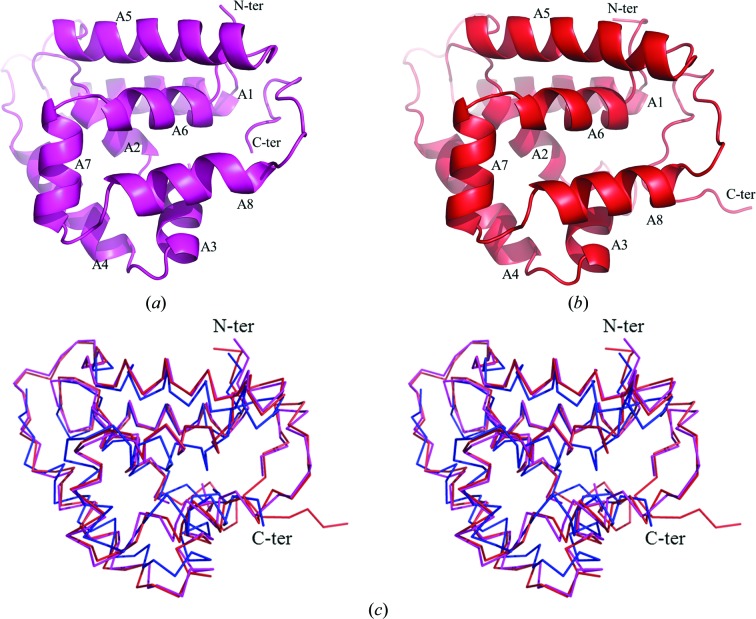
The crystal structure of CaHsp104NTD was determined to 1.66 Å resolution using the SAD method (Table 1 ▸). The crystal structure of ScHsp104NTD was solved to 2.54 Å resolution by the molecular-replacement method using the CaHsp104NTD structure as the search model (Table 1 ▸). In the CaHsp104NTD crystal structure, residues 2–158 are visible in the electron-density map. In the ScHsp104NTD structure, residues 4–166 can be modeled into the electron-density map. Both the CaHsp104NTD and the ScHsp104NTD monomer structures contain one single domain, which is composed of eight α-helices (A1–A8) and no β-strands (Figs. 2 ▸ a and 2 ▸ b). CaHsp104NTD and ScHsp104NTD show high primary-sequence homology, with a sequence identity of 54%. The two structures share very similar protein folds and the crystal structures superimpose well, with a root-mean-square deviation (r.m.s.d.) of 0.799 Å for the main-chain atoms of 156 equivalent residues (Fig. 2 ▸ c; CaHsp104NTD and ScHsp104NTD are shown in magenta and red, respectively).
Figure 2.
The monomer structures of CaHsp104NTD and ScHsp104NTD shown as ribbon drawings. Helices A1–A8 are labeled. The N-termini and C-termini of the molecules are denoted. (a) The monomer structure of CaHsp104NTD. (b) A ribbon drawing of the ScHsp104NTD monomer. (c) Structure alignment of CaHsp104NTD, ScHsp104NTD and the E. coli ClpB NTD shown as a Cα trace in stereo mode. The CaHsp104NTD structure is shown in magenta, the ScHsp104NTD structure is shown in red and the ClpB NTD structure is shown in blue. The orientations of CaHsp104NTD and ScHsp104NTD in (c) are very similar to those in (a) and (b).
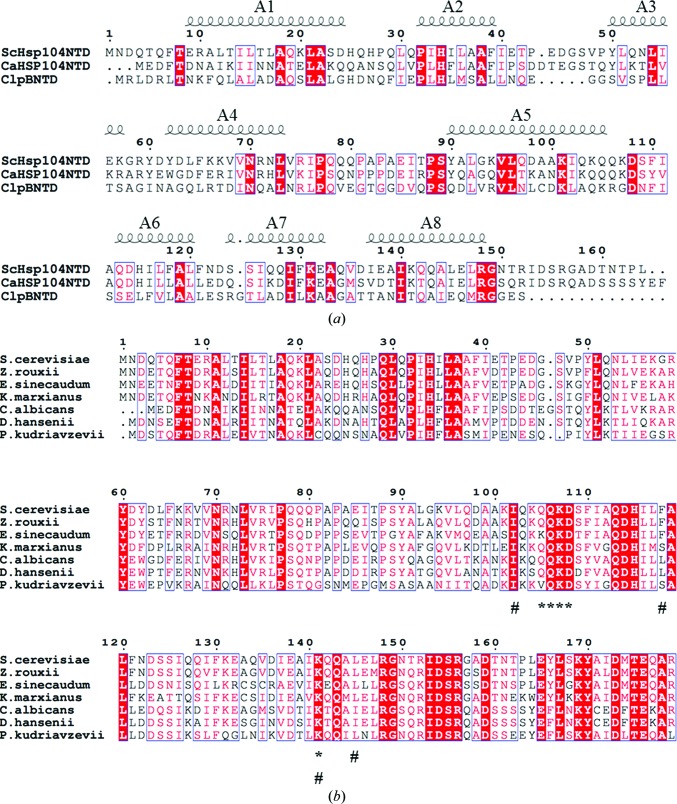
When the CaHsp104NTD and ScHsp104NTD structures are compared with the structure of the N-terminal domain of E. coli ClpB, significant differences can be observed, particularly at the C-terminus (Li & Sha, 2003 ▸). The C-terminal 15 residues of CaHsp104NTD and ScHsp104NTD constitute a stable loop that form extensive hydrogen bonds with the loop between helices A5 and A6 and with the N-terminal loop. In contrast, the ClpB N-terminal domain lacks this C-terminal loop in the structure. The sequence identity between the N-terminal domains of yeast Hsp104 and E. coli ClpB is ∼20% (Fig. 3 ▸ a). When the ClpB N-terminal domain structure was superimposed with that of ScHsp104NTD, the root-mean-square deviation (r.m.s.d.) for the main-chain atoms of 139 equivalent residues was 1.776 Å (Fig. 2 ▸ c; ClpB NTD and ScHsp104NTD are shown in red and blue, respectively).
Figure 3.
(a) Sequence alignment of ScHsp104NTD, CaHsp104NTD and the E. coli ClpB NTD. The numbering is based on the ScHsp104NTD sequence. The positions of helices A1–A8 are marked and labeled. (b) Sequence alignment of Hsp104 N-terminal domains from various yeast species. The numbering is based on the ScHsp104NTD sequence. The residues (Gln105, Gln106, Lys107, Asp108 and Lys141) involved in ScHsp104NTD dimerization are labeled with an asterisk. The residues involved in forming the putative peptide-binding grooves of Hsp104 are indicated by a hash symbol (#).
3.3. The putative peptide-binding groove of the Hsp104 N-terminal domain
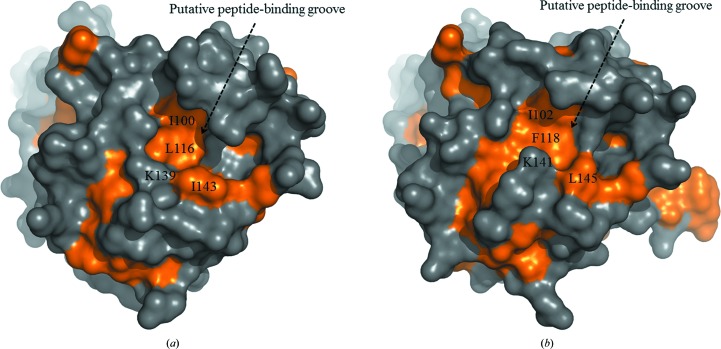
Our data suggested that the yeast Hsp104 N-terminal domain may function as a molecular chaperone that interacts with non-native polypeptides. The structure of the ClpB N-terminal domain revealed a putative peptide-binding groove on the molecular surface (Li & Sha, 2003 ▸). In the CaHsp104NTD and ScHsp104NTD structures, putative peptide-binding grooves can be identified at similar locations (Fig. 4 ▸). The putative peptide-binding groove of the Hsp104 NTD was formed by residues from helices A5, A6 and A8 and the C-terminal loop. In the ClpB NTD structure, the putative peptide-binding groove has a hydrophobic nature. Mutations of the hydrophobic residues that form the putative peptide-binding groove in the ClpB NTD compromised the molecular-chaperone activity of ClpB (Li & Sha, 2003 ▸). The putative peptide-binding grooves in the CaHsp104NTD and ScHsp104NTD structures are partially hydrophobic (Fig. 4 ▸). The side chains of residues Ile100, Leu116, Lys139 and Ile143 constitute the hydrophobic patch in the putative peptide-binding groove of CaHsp104NTD. The side chains of residues Ile102, Phe118, Lys141 and Leu145 form the hydrophobic region in the putative peptide-binding groove of ScHsp104NTD. The hydrophobicity of these residues is well conserved among yeast species, indicating a common feature of yeast Hsp104 N-terminal domains (Fig. 3 ▸ b). It is likely that the yeast Hsp104 NTD utilizes the hydrophobic residues within its putative peptide-binding groove to interact with the exposed hydrophobic stretches of the misfolded proteins. Interestingly, the putative peptide-binding grooves of yeast Hsp104 and E. coli ClpB exhibit quite different hydrophobic patterns, which suggest that they may interact with polypeptide substrates using distinct mechanisms (Fig. 4 ▸).
Figure 4.
The putative peptide-binding grooves of CaHsp104NTD and ScHsp104NTD shown as molecular-surface drawings. The hydrophobic regions of the molecular surface are shown in gold. (a) The putative peptide-binding groove of CaHsp104NTD is indicated. The hydrophobic side chains of residues (Ile100, Leu116, Lys139 and Ile143) which are located within the putative peptide-binding groove are labeled. (b) The putative peptide-binding groove of ScHsp104NTD.
3.4. Dimer formation of ScHsp104NTD
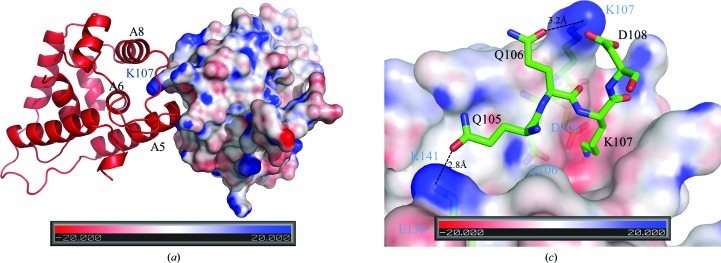
The E. coli ClpB NTD and CaHsp104NTD form a monomer in the crystal structure. In contrast, ScHsp104NTD forms a homodimer in our crystal structure. The two monomers within the homodimer are related by a twofold axis (Fig. 5 ▸ a). The gel-filtration profiles of CaHsp104NTD and ScHsp104NTD showed that CaHsp104NTD exists as a monomer in solution, while ScHsp104NTD may form an equilibrium between homodimers and monomers in solution (Supplementary Fig. S1). Interestingly, ScHsp104NTD utilizes the putative peptide-binding groove as the sole dimerization interface to constitute the homodimer. The C-terminal end of helix A5 of one monomer (named monomer A) is docked into the putative peptide-binding groove of the other monomer (named monomer B) within the homodimer, while the C-terminal end of helix A5 of monomer B is positioned into the putative peptide-binding groove of monomer A. The side chain of Lys107 from monomer A is deeply buried in the peptide-binding groove of monomer B (Fig. 5 ▸ b). Lys107 of monomer A forms a strong salt bridge with Asp108 of monomer B. The side chain of Lys107 of monomer A also forms a hydrogen bond to that of Gln106 from monomer B. Owing to the twofold axis, the side chain of Gln106 of monomer A also forms a hydrogen bond to the side chain of Lys107 of monomer B. The side chain of Gln105 of monomer A establishes a hydrogen bond to the side chain of Lys141 of monomer B. The dimer interface is quite limited within the peptide-binding groove region and has a modest surface area of ∼300 Å2, which accounts for ∼2% of the surface area of one monomer. Based on these observations, we hypothesize that this localized hydrogen-bond and salt-bridge network in ScHsp104NTD may give rise to a monomer–dimer equilibrium in solution. These data further support the hypothesis that the Hsp104 N-terminal domain can utilize its putative peptide-binding groove to interact with non-native polypeptide substrates. Moreover, the residues involved in the association of the two monomers, such as Gln105, Gln106, Lys107, Asp108 and Lys141, are very well conserved among yeast species (Fig. 3 ▸).
Figure 5.
The dimerization interface of ScHsp104NTD. (a) Within the ScHsp104NTD homodimer, monomer A is shown as a red ribbon drawing and monomer B is shown as a surface-potential drawing using PyMOL (v.1.3; Schrödinger). Helices A5, A6 and A8 of monomer A which are involved in forming the putative peptide-binding groove are labeled in black. The C-terminal end of A5 docks into the peptide-binding groove of monomer B. The protruding Lys107 from monomer B is buried in the peptide-binding groove of monomer A and is labeled in blue. In this figure, the twofold axis lies vertically to the paper and points towards the reader. (b) In this figure, Gln105, Gln106, Lys107 and Asp108 of monomer A of the ScHsp104NTD homodimer are shown in stick mode and labeled in black. Monomer B is shown as a surface-potential drawing. Gln106, Lys107, Asp108 and Lys141 of monomer B are shown in stick mode under the semi-transparent surface and are labeled in blue. The hydrogen bonds between Gln105 and Gln106 from monomer A and Lys141 and Lys107 from monomer B are shown as dotted lines.
3.5. Indications that the yeast Hsp104 NTD interacts with polyQ peptides
Recent data have suggested that the Hsp104 NTD might be directly involved in the binding of glutamine-rich substrates (polyQ peptides) and play critical roles in disaggregating prions (Sweeny et al., 2015 ▸). It has been demonstrated that the Hsp104 NTD enables cooperative substrate translocation, which is critical for prion dissolution and potentiated disaggregase activity (Sweeny et al., 2015 ▸). In the crystal structure of the ScHsp104NTD homodimer, the consecutive residues Gln105, Gln106 and Lys107 from monomer A make interactions with the putative peptide-binding groove of monomer B to constitute the homodimer. This structural observation may suggest the mechanism by which Hsp104 NTD binds the polyQ peptide to disaggregate prions. Several lines of evidence may support this hypothesis. (i) The side chain of Lys107 of ScHsp104NTD fully sticks out of the molecular surface of the monomer and forms a hydrogen bond to Gln106 from the other monomer. The conserved Lys107 may function as a sensor for polyQ peptides and provide easy access to aggregated prions. (ii) The side chain of Lys141 of ScHsp104NTD also points outward from the molecular surface of the monomer and is stabilized by forming a salt bridge with the nearby Glu138. The side chain of Lys141 approaches Gln105 of the other monomer from an opposite angle relative to Lys107 (Fig. 5 ▸). The positions of Lys107 and Lys141 may allow them to attack the polyQ peptide in the β-sheet formation simultaneously. (iii) The side chains of Gln105 and Gln106 are not deeply buried in the putative peptide-binding groove of the other monomer. It is likely that the polyQ peptide in the β-sheet does not have much flexibility and mobility. The Hsp104 NTD may be able to gain access to the side chains of the polyQ peptide in the β-sheet from the surface. This association of Hsp104 with polyQ peptide may facilitate the dissolution of prion-prone amyloid formations.
The crystal structure of ScHsp104NTD suggests that the Hsp104 NTD may specifically interact with polyQ regions of prion-prone proteins. The data may shed light on the mechanism by which the Hsp104 NTD functions to suppress and/or dissolve prions. Gln105, Gln106, Lys107 and Lys141 of ScHsp104NTD play critical roles in associating the two monomers into a homodimer. These residues may facilitate direct binding between the Hsp104 NTD and the polyQ peptide within the β-sheet formation for subsequent disaggregation. Interestingly, none of these residues are conserved in the primary sequence of E. coli ClpB, which may provide an explanation of why ClpB lacks the amyloid-dissolution activity for polyQ peptides (DeSantis et al., 2012 ▸). Moreover, Hsp104 may function as an oligomer in solution (Yokom et al., 2016 ▸). It is possible that the NTDs within the Hsp104 oligomer may exhibit more potent remodeling abilities for polyQ peptides in a cooperative fashion compared with the Hsp104 NTD monomer.
Supplementary Material
PDB reference: CaHsp104NTD, 5u2l
PDB reference: ScHsp104NTD, 5u2u
Supporting Information: Supplementary Figure S1. DOI: 10.1107/S2059798317002662/qh5044sup1.pdf
Acknowledgments
We are grateful to the staff scientists at the SER-CAT beamline at APS for their help in data collection.
Funding Statement
This work was funded by National Institutes of Health grant R01 GM080261.
References
- Adams, P. D. et al. (2010). Acta Cryst. D66, 213–221.
- Chernoff, Y. O., Lindquist, S. L., Ono, B., Inge-Vechtomov, S. G. & Liebman, S. W. (1995). Science, 268, 880–884. [DOI] [PubMed]
- Cox, B., Ness, F. & Tuite, M. (2003). Genetics, 165, 23–33. [DOI] [PMC free article] [PubMed]
- DeSantis, M. E., Leung, E. H., Sweeny, E. A., Jackrel, M. E., Cushman-Nick, M., Neuhaus-Follini, A., Vashist, S., Sochor, M. A., Knight, M. N. & Shorter, J. (2012). Cell, 151, 778–793. [DOI] [PMC free article] [PubMed]
- Doyle, S. M. & Wickner, S. (2009). Trends Biochem. Sci. 34, 40–48. [DOI] [PubMed]
- Emsley, P. & Cowtan, K. (2004). Acta Cryst. D60, 2126–2132. [DOI] [PubMed]
- Erzberger, J. P. & Berger, J. M. (2006). Annu. Rev. Biophys. Biomol. Struct. 35, 93–114. [DOI] [PubMed]
- Glover, J. R. & Lindquist, S. (1998). Cell, 94, 73–82. [DOI] [PubMed]
- Hanson, P. I. & Whiteheart, S. W. (2005). Nature Rev. Mol. Cell Biol. 6, 519–529. [DOI] [PubMed]
- Hung, G.-C. & Masison, D. C. (2006). Genetics, 173, 611–620. [DOI] [PMC free article] [PubMed]
- Jackrel, M. E., DeSantis, M. E., Martinez, B. A., Castellano, L. M., Stewart, R. M., Caldwell, K. A., Caldwell, G. A. & Shorter, J. (2014). Cell, 156, 170–182. [DOI] [PMC free article] [PubMed]
- Jackrel, M. E. & Shorter, J. (2015). Prion, 9, 90–109. [DOI] [PMC free article] [PubMed]
- Klosowska, A., Chamera, T. & Liberek, K. (2016). eLife, 5, e15159. [DOI] [PMC free article] [PubMed]
- Kummer, E., Szlachcic, A., Franke, K. B., Ungelenk, S., Bukau, B. & Mogk, A. (2016). J. Mol. Biol. 428, 4378–4391. [DOI] [PubMed]
- Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., McGettigan, P. A., McWilliam, H., Valentin, F., Wallace, I. M., Wilm, A., Lopez, R., Thompson, J. D., Gibson, T. J. & Higgins, D. G. (2007). Bioinformatics, 23, 2947–2948. [DOI] [PubMed]
- Lee, S., Sielaff, B., Lee, J. & Tsai, F. T. (2010). Proc. Natl Acad. Sci. USA, 107, 8135–8140. [DOI] [PMC free article] [PubMed]
- Li, J. & Sha, B. (2002). J. Mol. Biol. 318, 1127–1137. [DOI] [PubMed]
- Li, J. & Sha, B. (2003). Structure, 11, 323–328. [DOI] [PubMed]
- Lum, R., Niggemann, M. & Glover, J. R. (2008). J. Biol. Chem. 283, 30139–30150. [DOI] [PMC free article] [PubMed]
- Mogk, A., Haslberger, T., Tessarz, P. & Bukau, B. (2008). Biochem. Soc. Trans. 36, 120–125. [DOI] [PubMed]
- Park, Y.-N., Zhao, X., Yim, Y.-I., Todor, H., Ellerbrock, R., Reidy, M., Eisenberg, E., Masison, D. C. & Greene, L. E. (2014). Eukaryot. Cell, 13, 635–647. [DOI] [PMC free article] [PubMed]
- Parsell, D. A., Kowal, A. S., Singer, M. A. & Lindquist, S. (1994). Nature (London), 372, 475–478. [DOI] [PubMed]
- Reidy, M., Miot, M. & Masison, D. C. (2012). Genetics, 192, 185–193. [DOI] [PMC free article] [PubMed]
- Robert, X. & Gouet, P. (2014). Nucleic Acids Res. 42, W320–W324. [DOI] [PMC free article] [PubMed]
- Rosenzweig, R., Farber, P., Velyvis, A., Rennella, E., Latham, M. P. & Kay, L. E. (2015). Proc. Natl Acad. Sci. USA, 112, E6872–E6881. [DOI] [PMC free article] [PubMed]
- Sanchez, Y. & Lindquist, S. L. (1990). Science, 248, 1112–1115. [DOI] [PubMed]
- Sanchez, Y., Taulien, J., Borkovich, K. A. & Lindquist, S. (1992). EMBO J. 11, 2357–2364. [DOI] [PMC free article] [PubMed]
- Satpute-Krishnan, P., Langseth, S. X. & Serio, T. R. (2007). PLoS Biol. 5, e24. [DOI] [PMC free article] [PubMed]
- Schirmer, E. C., Glover, J. R., Singer, M. A. & Lindquist, S. (1996). Trends Biochem. Sci. 21, 289–296. [PubMed]
- Sha, B., Lee, S. & Cyr, D. M. (2000). Structure, 8, 799–807. [DOI] [PubMed]
- Shorter, J. (2008). Neurosignals, 16, 63–74.
- Shorter, J. & Lindquist, S. (2004). Science, 304, 1793–1797. [DOI] [PubMed]
- Sweeny, E. A., Jackrel, M. E., Go, M. S., Sochor, M. A., Razzo, B. M., DeSantis, M. E., Gupta, K. & Shorter, J. (2015). Mol. Cell, 57, 836–849. [DOI] [PMC free article] [PubMed]
- Tipton, K. A., Verges, K. J. & Weissman, J. S. (2008). Mol. Cell, 32, 584–591. [DOI] [PMC free article] [PubMed]
- Torrente, M. P., Chuang, E., Noll, M. M., Jackrel, M. E., Go, M. S. & Shorter, J. (2016). J. Biol. Chem. 291, 5101–5115. [DOI] [PMC free article] [PubMed]
- Wawrzynow, A., Banecki, B. & Zylicz, M. (1996). Mol. Microbiol. 21, 895–899. [DOI] [PubMed]
- Wendler, P., Shorter, J., Plisson, C., Cashikar, A. G., Lindquist, S. & Saibil, H. R. (2007). Cell, 131, 1366–1377. [DOI] [PMC free article] [PubMed]
- Wickner, R. B. (1994). Science, 264, 566–569. [DOI] [PubMed]
- Winkler, J., Tyedmers, J., Bukau, B. & Mogk, A. (2012). J. Struct. Biol. 179, 152–160. [DOI] [PubMed]
- Yokom, A. L., Gates, S. N., Jackrel, M. E., Mack, K. L., Su, M., Shorter, J. & Southworth, D. R. (2016). Nature Struct. Mol. Biol. 23, 830–837. [DOI] [PMC free article] [PubMed]
- Zolkiewski, M. (2006). Mol. Microbiol. 61, 1094–1100. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: CaHsp104NTD, 5u2l
PDB reference: ScHsp104NTD, 5u2u
Supporting Information: Supplementary Figure S1. DOI: 10.1107/S2059798317002662/qh5044sup1.pdf