Abstract
Adipocyte differentiation is controlled by multiple signaling pathways. To identify new adipogenic factors, C3H10T1/2 adipocytes were treated with previously known antiadipogenic phytochemicals (resveratrol, butein, sulfuretin, and fisetin) for 24 hours. Commonly regulated genes were then identified by transcriptional profiling analysis. Three genes (chemokine (C-X-C motif) ligand 1 [Cxcl1], heme oxygenase 1 [Hmox1], and PHD (plant homeo domain) finger protein 16 [Phf16]) were upregulated while two genes (G0/G1 switch gene 2 [G0s2] and patatin-like phospholipase domain containing 3 [Pnpla3]) were downregulated by these four antiadipogenic compounds. Tissue expression profiles showed that the G0s2 and Pnpla3 expressions were highly specific to adipose depots while the other three induced genes were ubiquitously expressed with significantly higher expression in adipose tissues. While Cxcl1 expression was decreased, expressions of the other four genes were significantly increased during adipogenic differentiation of C3H10T1/2 cells. Small interfering RNA–mediated knockdown including Phf16 and Pnpla3 indicated that these genes might play regulatory roles in lipid accumulation and adipocyte differentiation. Specifically, the silencing of two newly identified adipogenic genes, Phf16 or Pnpla3, suppressed lipid accumulation and expression of adipocyte markers in both 3T3-L1 and C3H10T1/2 cells. Taken together, these data showed previously uncovered roles of Phf16 and Pnpla3 in adipogenesis, highlighting the potential of using phytochemicals for further investigation of adipocyte biology.
Keywords: adipocyte, adipocyte differentiation, PHD (plant homeo domain) finger protein 16, Phytochemical, patatin-like phospholipase domain containing 3
1 |. INTRODUCTION
Adipocyte serves as an energy reservoir by storing excess energy in the form of triglycerides.1 It also acts as an endocrine organ to regulate whole-body energy homeostasis by secreting fatty acids and adipokines including leptin, adiponectin, and tumor necrosis factor α (TNF-α).2–4 The generation of adipocytes can be dictated by transcriptional cascades including peroxisome proliferator-activated receptor γ (Pparγ) and CCAAT/enhancer-binding proteins (C/ebpα).5 Pparγ also regulates whole-body glucose metabolism. It is a direct target of thiazolidinediones (TZD), a class of drugs used to treat type 2 diabetes.5 The extracellular signal-regulated kinases (ERK)/cyclin-dependent kinase 5 (Cdk5) axis also controls diabetogenic actions of Pparγ by affecting its phosphorylation status,6–8 suggesting that selective modulation on Pparγ activity can provide therapeutic potentials against insulin resistance. Therefore, there is a need to identify new factors that act on adipogenesis to further understand adipocyte biology. Such new factors might show potential to treat type 2 diabetes.
Phytochemicals identified from natural products have been widely studied for their effects on obesity and molecular mechanisms involved in their effects.9,10 For example, epigallocatechin gallate, a phenolic antioxidant found in green and black tea, can inhibit adipocyte proliferation and increase fatty acid oxidation.11 Resveratrol from red wine exhibits inhibitory effects on lipid accumulation with stimulatory actions on lipid oxidation.12 Fisetin, sulfuretin, and butein from Rhus verniciflua stokes have numerous biological activities including antiadipogenic effects.13,14 They have antiadipogenic actions on adipocyte differentiation by inhibiting lipid accumulation and reducing expression of adipogenic markers in multiple adipogenic cell lines.15 It has been demonstrated that resveratrol can mediate antiadipogenic effects by activating Pparγ through sirtuin 1 (SIRT1) and Pparγ coactivator 1α (PGC-1α).16 Similarly, mammalian target of rapamycin complex 1 or SIRT1 targeted by fisetin and Prdm4 targeted by butein have been identified as the axis for their antiadipogenic effects. We have previously identified complement factor D as an adipogenic modulator in a transcriptional profiling assay using proadipogenic small molecules.19 These studies indicate that antiadipogenic phytochemicals are useful tools to uncover genes involved in adipogenesis.
The aim of this study was to identify genes regulated by four antiadipogenic phytochemicals (resveratrol, butein, sulfuretin, and fisetin). Transcriptional profiling assays of these antiadipogenic phytochemicals revealed five commonly regulated genes including chemokine (C-X-C motif) ligand 1 (Cxcl1), heme oxygenase 1 (Hmox1), PHD (plant homeo domain) finger protein 16 (Phf16), G0/G1 switch gene 2 (G0s2), and patatin-like phospholipase domain containing 3 (Pnpla3). Of these five genes, the roles of Phf16 and Pnpla3 in adipocyte differentiation are currently unknown. Thus, we focused on Phf16 and Pnpla3 to investigate their roles in adipogenesis. Our results showed that Phf16 and Pnpla3 regulated by antiadipogenic phytochemicals could modulate adipocyte differentiation. We found that Pnpla3 was a downstream target of Pparγ whereas Phf16 could facilitate Pparγ activity on its target promoters. These data provide evidence for unknown roles of phytochemically regulated Phf16 and Pnpla3 in adipogenesis, further suggesting the potential of using phytochemicals to understand adipocyte biology.
2 |. MATERIALS AND METHODS
2.1 |. Cell culture
Mouse C3H10T1/2 (CCL-226) and 3T3-L1 (CL-173) cells were purchased from the American Type Culture Collection (ATCC, Rockville, MD) and cultured in Dulbecco modified Eagle medium (DMEM) (SH30243.01; Hyclone, Logan, UT) supplemented with 10% fetal bovine serum (FBS) (SH30397.03; Hyclone) and penicillin/streptomycin (SV30010; Hyclone) as described previously.20 3T3-L1 preadipocytes were maintained in DMEM supplemented with 10% fetal calf serum (SH30401.01 l; Hyclone). C3H10T1/2 cells were maintained in DMEM supplemented with 10% FBS. To induce adipocyte differentiation of 3T3-L1 preadipocytes, cells were seeded in six-well tissue culture plates and induced to adipocytes by differentiation medium (1 µM dexamethasone [D-2915; Sigma, St Louis, MO], 0.5 mM 3-isobutyl-1-methylxanthine [I-7018; Sigma], and 5 µg/mL insulin [I-0516; Sigma] in DMEM containing 10% FBS) for 2 days after confluence. DMEM containing 10% FBS and 5 µg/mL insulin was refreshed every 2 days up to 6 to 8 days. For C3H101/2 adipocytes, 20 nM GW1929 (G5668; Sigma) was further supplemented during the differentiation period.
Cell viability was determined using 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazoliumbromide (MTT) (Affymetrix, Inc, Cleveland, OH) assay. Briefly, 3T3-L1 cells were seeded into 96-well plates and cultured. After reaching confluence, cells were treated with 0.1, 1, 5, 10, 20, and 40 μM of butein, sulfuretin, resveratrol, and fisetin in triplicates. After 24 hours, MTT (5 mg/mL in phosphate-buffered saline [PBS]) was added and formazan crystals were measured at 520 nm using a microplate reader.
2.2 |. Expression analysis
Total RNA was isolated using TRIzol reagent (Invitrogen, Carlsbad, CA). RNA samples were separated on agarose gels and relatively sharp 28S and 18S ribosomal RNA bands were verified. The purity and integrity of RNA samples were further assessed using a spectrophotometer. RNA samples with A260/A280 ratios close to 2.0 (range, 1.8 to 1.9) were used in the expression analysis. Total RNA (500 ng) and random primers were reverse transcribed using ReverTra Ace qPCR RT Master Mix (Toyobo, Osaka, Japan) at 37°C for 50 minutes. Complementary DNA (cDNA) was heat inactivated at 70°C for 10 minutes and amplified with a Thermal Cycler Dicer (Takara, Shiga, Japan) using THUNDERBIRD SYBR qPCR Mix (Toyobo). The Δ cycle threshold (Ct) was used to calculate differences between target gene Ct value and control gene (Rplp0) Ct value for each sample, where ΔCt = Ct (target gene) − Ct (control gene). Relative expression level was calculated using . Gene-specific primers were described previously.15,19
For microarray analysis, total RNAs from C3H10T1/2 adipocytes treated with 10 μM of butein, sulfuretin, resveratrol, or fisetin for 24 hours were prepared using RNeasy Mini Kit (Qiagen, Venlo, The Netherlands). cDNA preparation and hybridization to Affymetrix Mouse Genome Arrays (430 version 2.0) were performed by GenoChek (Ansan, Korea). Data were analyzed using GeneSpring GX 7.3 software (Agilent Technologies, Santa Clara, CA).
2.3 |. Oil Red O staining
Differentiated adipocytes were fixed with 4% paraformaldehyde in PBS at room temperature for 4 hours and then stained with 0.5% Oil Red O (Sigma) in a mixture of isopropanol and distilled water (at 3:2 ratio) for 45 minutes. To quantify intracellular triglyceride content, stained cells from at least three independent experiments were resolved with isopropanol and measured with a spectrophotometer at 520 nm.
2.4 |. Knockdown experiments
Small interfering RNAs (siRNA) against candidate genes were purchased from Genolution Pharmaceutical Inc (Seoul, Korea). Sequences included the following: Pnpla3 si#1: 5′-CCGUUCUUCAACAUUAACAUU-3′, Pnpla3 si#2: 5′-CAAAUUAUAGUUACAAAUAU U-3′; Phf16 si#1: 5′-GCUAUUACCUCUUUACUGAUU-3′, Phf16 si#2: 5′-CUAGAAGAAGAAUUCUAUAUU-3′; Cxcl1 si#1: 5′-CCUA UUUAUUUAUGUAUUUUU-3′, Cxcl1 si#2: 5′- GAGAUA GAGUUUAGUAUUAUU-3′; G0s2 si#1: 5′-GCUAUCAC UUUGCAUUAGAUU-3′, GOs2 si#2: 5′- GGCUUUUUAU ACAGUU AUUUU-3′; Hmox1 si#1: 5′-CUAACUU CUGU GUGAAAUAUU-3′, and Hmox1 si#2: 5′-CUCUGUAAGG GAGAAUCUUUU-3′. The sense sequence of nonspecific scramble RNA as control was 5′-CCUCGUGCCGUUCC AUCAGGUAGUU-3′. C3H10T1/2 or 3T3-L1 cells in serum-free media were transfected with two independent siRNAs using Lipofectamine RNAiMAX (13778-075; Invitrogen). siRNA at a concentration of 30 nM was transfected into 50% confluent C3H10T1/2 or 3T3-L1 cells. At 16 hours after transfection, medium was replaced with fresh medium containing 10% FBS. At 48 hours posttransfection, cells were differentiated into adipocytes for 6 to 8 days followed by Oil Red O staining and gene expression analysis. To repress expression of Pparγ messenger RNA, short hairpin RNAs (shRNAs) were introduced into 3T3-L1 cells using retroviral transduction as described previously.21 Target sequences for Pparγ were 5′-GTTTGAGTTTGCTGT-GAAGTT-3′. Phoenix cells were transfected with pBabeshGFP or shPparγ retroviral vector. Viruses were collected at 48 hours posttransfection. Target cells were infected with viruses for 24 hours and then selected with puromycin for 2 weeks.
2.5 |. Luciferase assay
Ppar responsive element (3XPPRE) containing luciferase vector, Pparγ, and RXR vector were cotransfected with Phf16 expressing vector or an empty vector into 293A cells using Lipofectamine 2000 (Invitrogen). For luciferase reporter assay, luciferase vector (100 ng), Pparγ vector (100 ng), RXR vector (10 ng), Phf16 expressing vector (100 ng), or empty vector, 1 μL Lipofectamine 2000, and 50 μL Opti-MEM (Gibco, Grand Island, NY) were used for transfection in each well of a 24-well plate. At 48 hours posttransfection, cells were harvested and reporter gene activity was measured using Dual-luciferase Reporter Assay System (Promega, Madison, WI). Pparγ ligands (Rosiglitazone 0.5 μM; GW1929 100 nM) were added and incubated for 24 hours before cell harvest. Luciferase activity was normalized by Renilla luciferase activity.
2.6 |. Statistical analysis
All statistical analyses were conducted using PASW Statistics 17 (SPSS Inc, Chicago, IL). Data are presented as mean ± SD or SEM. Statistical differences of gene expression and lipid accumulation were analyzed using one-way analysis of variance or two-tailed unpaired Student t test (PASW Statistics 17, SPSS Inc., Chicago, IL). Statistical significance was defined at P < 0.05.
3 |. RESULTS
3.1 |. Identification of adipogenic genes regulated by phytochemicals
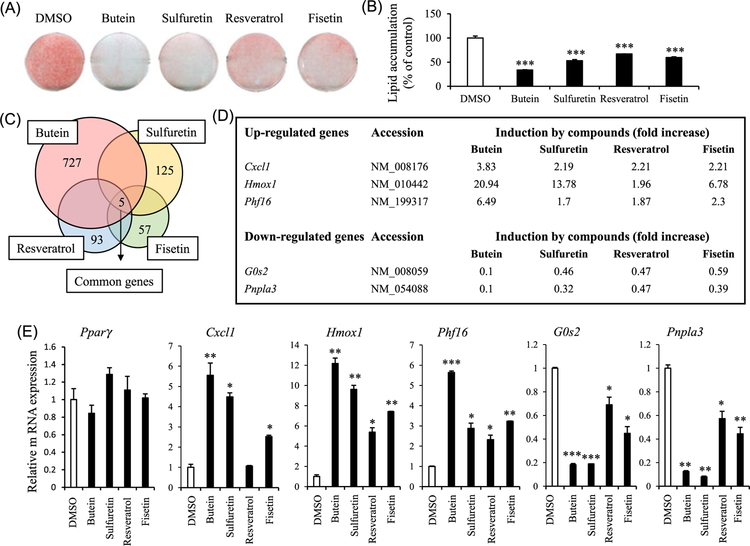
We performed MTT assays to select optimal doses of four antiadipogenic phytochemicals (butein, sulfuretin, resveratrol, and fisetin) in C3H10T1/2 cells (Figures 1A and 1B). Treatments with these phytochemicals up to 20 μM for 24 hours did not significantly affect cell viabilities. All four phytochemicals at 40 μM exhibited slight but significant cellular toxicity (Supporting Information Figure S1). Based on these data, concentration of 20 μM or lower was chosen to analyze actions of these phytochemicals in adipogenesis.
FIGURE 1.
Identification of genes controlled by antiadipogenic phytochemicals. A, Antiadipogenic effects of butein, sulfuretin, resveratrol, and fisetin at 20 μM in C3H10T1/2 cells. B, Lipid accumulation was quantified by measuring the extracted Oil Red O dye at 520 nm. Data shown represent the mean ± SEM from three independent experiments. C, Diagram showing genes commonly induced by antiadipogenic compounds. C3H10T1/2 adipocytes were treated with previously known antiadipogenic phytochemicals (butein, sulfuretin, resveratrol, and fisetin at 10 μM) for 24 hours and commonly regulated genes were identified by transcriptional profiling analysis. D, Cxcl1, Hmox1, and Phf16 were induced at least 1.7 folds and G0s2 and Pnpla3 were down regulated at least 0.6 folds by the four antiadipogenic compounds. E, Changes in expression of Pparγ, Cxcl1, Hmox1, Phf16, G0s2, and Pnpla3 by phytochemicals were verified in C3H10T1/2 cells. C3H10T1/2 adipocytes were treated with compounds for 24 hours and then mRNA expression level was assessed by real-time PCR. Data are expressed as means ± SEM and statistical analysis was performed using one-way ANOVA and Student t test (*P < 0.05; **P < 0.005; ***P < 0.0005). ANOVA, analysis of variance; Cxcl1, chemokine (C-X-C motif) ligand 1; DMSO, dimethyl sulfoxide; G0s2, G0/G1 switch gene 2; Hmox1, heme oxygenase 1; mRNA, messenger RNA; PCR, polymerase chain reaction; Phf16, PHD (plant homeo domain) finger protein 16; Pnpla3, patatin-like phospholipase domain containing 3; Pparγ, peroxisome proliferator-activated receptor γ
To identify new adipogenic genes, we performed transcriptional profiling analysis for C3H10T1/2 adipocytes treated with 10 µM of four known antiadipogenic phytochemicals (resveratrol, butein, sulfuretin, and fisetin) for 24 hours (Figure 1C). Numerous genes controlled by a single compound were identified but Pparγ expression is not affected by any of the phytochemicals in this analysis (Figures 1D and 1E). Multiple signaling pathways are known to be involved in the regulation of adipocyte differentiation. Thus, we hypothesized that genes commonly controlled by these four phytochemicals might have direct effects on adipocyte differentiation. Three genes, Cxcl1, Hmox1, and Phf16, were induced by at least 1.7 folds and two genes, G0s2 and Pnpla3, were downregulated by at least 0.6 folds by any of the four antiadipogenic compounds (Figure 1D). We then verified the differential expression of these five genes by real-time quantitative polymerase chain reaction (qPCR) in C3H10T1/2 adipocytes (Figure 1E).
3.2 |. Expression profiles of these phytochemically regulated genes
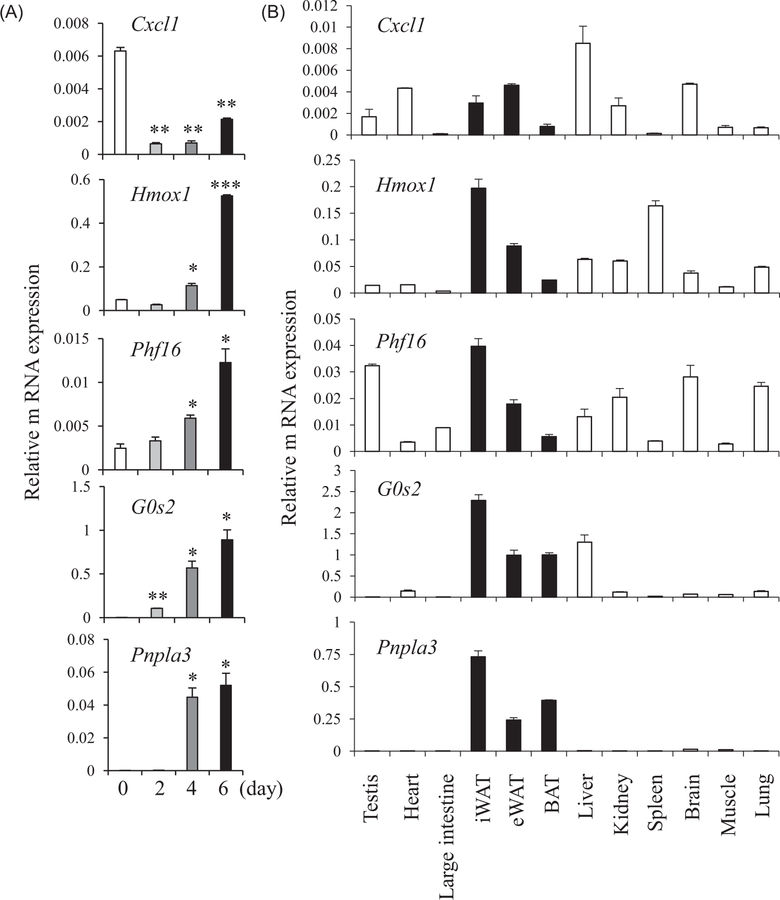
We considered that if these genes were involved in the regulation of adipocyte differentiation, expression of these genes would change over the period of differentiation. Thus, we investigated the expression of these five genes during adipocyte differentiation of C3H10T1/2 adipocytes. We found that expression levels of Hmox1, Phf16, G0s2, and Pnpla3 were increased during adipocyte differentiation whereas the expression level of Cxcl1 was decreased at the beginning of differentiation but moderately increased during the remaining differentiation period of C3H10T1/2 adipocytes (Figure 2A). We also examined tissue distribution of these genes. Hmox1, Phf16, G0s2, and Pnpla3 were expressed in adipose tissues, showing the highest expression in inguinal white adipose tissue (iWAT). Expression of G0s2 and Pnpla3 were found to be highly specific to adipose depots (iWAT, epididymal WAT [eWAT], and brown adipose tissue [BAT]) while the other three induced genes were ubiquitously expressed, yet with significant expression in adipose tissues. Expression of Cxcl1 in adipose tissue showed relatively lower expression levels than the other four genes (Figure 2B).
FIGURE 2.
Expression profiles in tissues and during adipocyte differentiation. A, Induction of expression during differentiation of C3H10T1/2 cells treated with differentiation cocktail including dexamethasone, IBMX, and insulin. The mRNA levels were assessed by real-time PCR at various time points for 6 days after treatment. B, Tissue distributions of Cxcl1, Hmox1, Phf16, G0s2, and Pnpla3. Relative mRNA expression was quantified by real-time PCR (n = 3). Data are expressed as means ± SEM and statistical analysis was performed using one-way ANOVA and Student t test (*P < 0.05; **P < 0.005; ***P < 0.0005). ANOVA, analysis of variance; BAT, brown adipose tissue; Cxcl1, chemokine (C-X-C motif) ligand 1; G0s2, G0/G1 switch gene 2; Hmox1, heme oxygenase 1; iWAT, inguinal white adipose tissue; eWAT, epididymal white adipose tissue; mRNA, messenger RNA; PCR, polymerase chain reaction; Phf16, PHD (plant homeo domain) finger protein 16; Pnpla3, patatin-like phospholipase domain containing 3
3.3 |. Roles of Cxcl1, Hmox1, Phf16, G0s2, and Pnpla3 in adipocyte differentiation
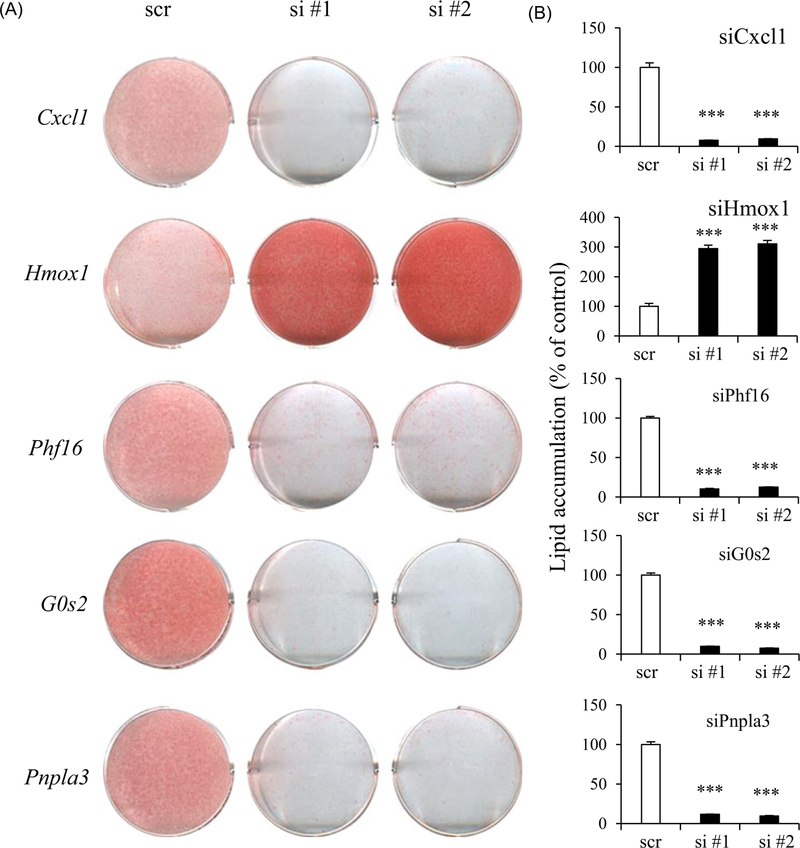
To investigate the roles of Cxcl1, Hmox1, Phf16, G0s2, and Pnpla3 in adipocytes, we silenced their expression in 3T3-L1 using siRNA and assessed lipid accumulation based on Oil Red O staining (Figure 3A). Silencing of Phf16, G0s2, Pnpla3, and Cxcl1 knockdown reduced lipid accumulation but Hmox1 knockdown induced lipid accumulation, validating our approach for identifying genes involved in adipogenesis by using phytochemicals as tools (Figure 3A and 3B)
FIGURE 3.
Adipogenic potential of Cxcl1, Hmox1, Phf16, G0s2, and Pnpla3. 3T3-L1 cells were transfected with control siRNA or two independent siRNAs against each gene. A, Differentiation levels were assessed by Oil Red O staining. B, Oil Red O staining of cells on day 6 was quantified by measuring extracted dye at 520 nm. Data are expressed as means ± SEM and statistical analysis was performed using Student t test (***P < 0.0005). Cxcl1, chemokine (C-X-C motif) ligand 1; G0s2, G0/G1 switch gene 2; Hmox1, heme oxygenase 1; Phf16, PHD (plant homeo domain) finger protein 16; Pnpla3, patatin-like phospholipase domain containing 3; siRNA, small interfering RNA
3.4 |. Proadipogenic functions of Phf16 and Pnpla3 during adipocyte differentiation
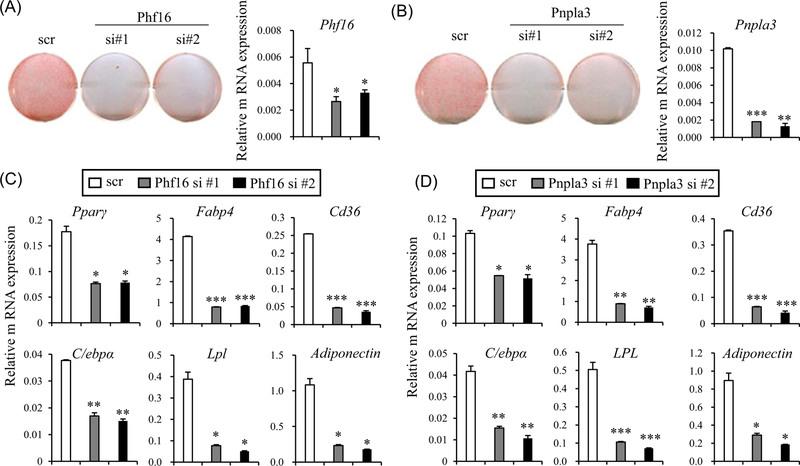
Roles of Phf16 and Pnpla3 in adipogenesis are currently unknown. Thus, we further investigated the function of Phf16 and Pnpla3 in adipocyte differentiation. Our data showed that silencing of Phf16 and Pnpla3 in 3T3-L1 prevented adipocyte differentiation (Figure 3). To further address whether Phf16 and Pnpla3 could contribute to adipocyte differentiation, we used siRNAs to knockdown Phf16 and Pnpla3 expression in C3H10T1/2 cells. Consistent with their effects in 3T3-L1 cells, both Phf16 and Pnpla3 knockdown in C3H10T1/2 cells reduced differentiation compared with control (nonspecific control siRNA transfection) (Figures 4A and 4B). Furthermore, expression levels of Pparγ, the master regulator of adipogenesis, and its target genes cluster of differentiation 36 (Cd36), fatty acid binding protein 4 (Fabp4), C/ebpα, lipoprotein lipase (Lpl), and adiponectin were also reduced in Phf16 and Pnpla3 siRNA–transfected cells (Figures 4C and 4 D). These data demonstrate that Phf16 and Pnpla3 are new adipogenic factors.
FIGURE 4.
siRNA-mediated silencing of Phf16 and Pnpla3 inhibits adipocyte differentiation. A, C3H10T1/2 cells were transfected with control siRNA or two independent siRNAs against Phf16. Differentiation levels were assessed by Oil Red O staining (left) and Oil Red O staining of cells on day 6 was quantified (right). The cells were differentiated into adipocytes for 6 days. B, C3H10T1/2 cells were transfected with control siRNA or two independent siRNAs against Pnpla3. Differentiation levels were assessed by Oil Red O staining (left) and Oil Red O staining of cells on day 6 was quantified (right). The cells were differentiated into adipocytes for 6 days. C, Gene expression in C3H10T1/2 cells expressing two independent siRNAs against Phf16 was measured. Cells were differentiated into adipocytes for 6 days. D, Expression of adipogenic markers in Pnpla3 siRNA expressing cells was quantified by real-time PCR. Data are representative of the two independent experiments. Data shown are means ± SEM and statistical analysis was performed using one-way ANOVA and Student t test (*P < 0.05; **P < 0.005; ***P < 0.0005). ANOVA, analysis of variance; Cd36, cluster of differentiation 36; C/ebpα, CCAAT/enhancer-binding protein α; Fabp4, fatty acid binding protein 4; Lpl, lipoprotein lipase; PCR, polymerase chain reaction; Phf16, PHD (plant homeo domain) finger protein 16; Pnpla3, patatin-like phospholipase domain containing 3; Pparγ, peroxisome proliferator-activated receptor γ; siRNA, small interfering RNA
3.5 |. Pnpla3 is a Pparγ downstream gene
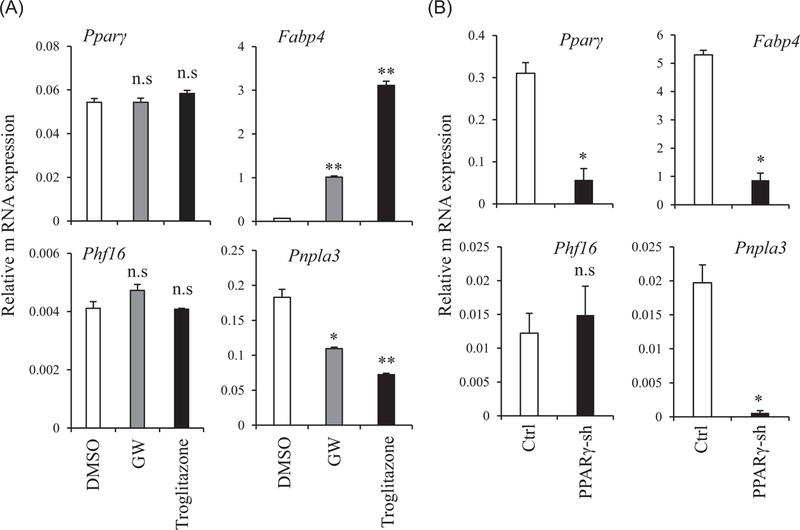
Since Pparγ is a coordinator of adipogenesis, it is reasonable to suspect that Phf16 and Pnpla3 might be transcriptional target genes of Pparγ. To this end, we stimulated C3H10T1/2 adipocytes with Pparγ agonists and measured expression levels of Phf16 and Pnpla3 along with Fabp4, a direct target of Ppaγ. As expected, Fabp4 was significantly induced either by GW1929 or TZD treatment. Interestingly, TZD treatment also resulted in significantly reduced expression of Pnpla3, suggesting that Pnpla3 might be another Pparγ downstream gene. However, the expression of Phf16 was not affected by GW1929 or TZD treatment (Figure 5A). To complement this, we compared gene expression in Pparγ shRNA–expressing cells and control shRNA–expressing (ctrl) cells. Similar to adipose selective expression, the expression of Pnpla3 and Fabp4 was reduced in Pparγ-deficient cells. However, Phf16 expression was not affected in the absence of Pparγ (Figure 5B). These results suggest that Pnpla3, but not Phf16, is an adipogenic gene acting downstream of Pparγ cascades.
FIGURE 5.
Pnpla3 is a Pparγ downstream gene. A, Real-time PCR analysis of Phf16 and Pnpla3 in C3H10T1/2 preadipocytes treated with 30 nM GW1929 and 10 μM troglitazone for 48 hours. B, Expression of Phf16 and Pnpla3 in 3T3-L1 cells stably expressing Pparγ-shRNA and control shRNA (ctrl) was determined by real-time PCR. Data are present as means ± SEM and statistical analysis was performed using Student t test (*P < 0.05; **P < 0.005). 3XPPRE, Ppar responsive element; DMSO, dimethyl sulfoxide; Fabp4, fatty acid binding protein 4; PCR, polymerase chain reaction; Phf16, PHD (plant homeo domain) finger protein 16; Pnpla3, patatin-like phospholipase domain containing 3; Pparγ, peroxisome proliferator-activated receptor γ; shRNA, short hairpin RNa
3.6 |. Phf16 acts as a coactivator of Pparγ
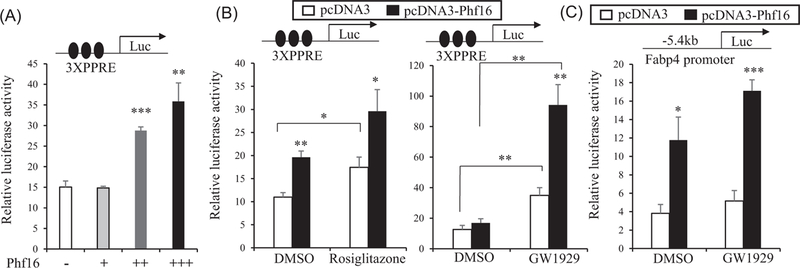
Phf16 contains a zinc finger motif often found in transcriptional regulators. This observation suggests that Phf16 might regulate adipocyte differentiation by acting as a cofactor for Pparγ. To investigate transcriptional coactivation by Phf16, 293A cells were transfected with 3XPPRE containing luciferase reporter, Pparγ, and RXR. Cells were then exposed to Pparγ ligands (GW1929 and rosiglitazone) in the presence or absence of a Phf16 expressing vector. Cells transfected with Pparγ and RXR expression vectors showed increased luciferase activity in the presence of Phf16 (Figure 6A) or Pparγ ligands (Figure 6B). Reporter activity was more robustly enhanced when both Phf16 and Pparγ ligands are present (Figure 6B). Similarly, Phf16 showed enhanced transcription when a 5.4 kb Fabp4-luciferase reporter was used in the assay, indicating that Phf16 could stimulate Pparγ in the synthetic promoter and upstream promoter region of Fabp4 (Figure 6C). These results strongly suggest that Phf16 may coactivate Pparγ activity to induce target gene expression.
FIGURE 6.
Phf16 is a positive transcriptional cofactor of Pparγ. A, Phf16 promotes Pparγ/RXR activation of luciferase driven by minimal 3XPPRE. Cells were transfected with Pparγ/RXR with various amounts of Phf16. B, Analysis of 3XPPRE-luciferase reporter activity by coexpressing Pparγ/RXR and Phf16 in 293A cells treated with DMSO, 0.5 μM rosiglitazone, or 100 nM GW1929. C, Fabp4 promoter/enhancer (−5.4 kb) driven luciferase reporter activity was stimulated in Phf16 and Pparγ/RXR coexpressing cells treated with 100 nM GW1929. Data shown are means ± SEM and statistical analysis was performed using one-way ANOVA and Student t test (*P < 0.05; **P < 0.005; ***P < 0.0005). 3XPPRE, PPAR responsive elements; ANOVA, analysis of variance; DMSO, dimethyl sulfoxide; Fabp4, fatty acid binding protein 4; Phf16, PHD (plant homeo domain) finger protein 16; Pparγ, peroxisome proliferator-activated receptor γ
4 |. DISCUSSION
Bioactive compounds from natural products including resveratrol, sulfuretin, fisetin, fustin, kaempferol, gallic acid, quercetin, protocatechuic acid, and butein have been widely investigated for their beneficial effects on obesity.9 In this study, Cxcl1, Hmox1, Phf16, G0s2, and Pnpla3 were found to be genes commonly regulated by four antiadipogenic phytochemicals sulfuretin, fisetin, resveratrol, and butein. In agreement with our findings, previous studies have shown that enhanced Hmox1 expression has a beneficiary effect on adiposity by increasing preadipocytes and reducing enlarged adipocytes.22,23 G0s2 expression is upregulated during adipocyte differentiation in 3T3-L1 cells while suppression of endogenous G0s2 can reduce adiposity.24–26 Other reports indicate that Cxcl1 is a dominant chemokine in preadipocytes and downregulated during adipogenesis.27,28 Cxcl1 can be induced by epidermal growth factor, a secreted peptide closely related to obesity.29 Pnpla3, a member of patatin-like phospholipases, is upregulated during adipocyte differentiation.30–32 Phf16 is a member of a family of large proteins containing plant homeo domain (PHD)-type zinc fingers.33 However, functions of Pnpla3 and Phf16 in adipocyte differentiation remain unclear. Therefore, further analyses may provide insights into the roles of Pnpla3 and Phf16 genes in adipocyte differentiation.
It has been demonstrated that Pnpla3 can function as both lipase and transacylase and that its expression is tightly regulated by nutritious status.30,34 Although Pnpla3 is involved in lipid metabolism and expressed in the adipose tissue at the highest level,35 functions of Pnpla3 in adipocyte differentiation have not been reported yet. Here, we observed that Pnpla3 was upregulated during adipocyte differentiation and that silencing Pnpla3 reduced lipid accumulation. Moreover, the expression of Pnpla3 was responsive to Pparγ agonizts, consistent with previous publication.36,37 However, direct regulation of Pparγ on Pnpla3 transcription should be carefully performed in promoter analysis.
Further, our data suggest that Pnpla3 regulated by Pparγ can also control adipogenesis. There are precedents that a set of Pparγ target genes can also behave as suppressors or inducers of Pparγ and adipocyte differentiation. For example, C/EBPα is a direct target of Pparγ. It can also increase Pparγ expression and adipogenesis. In addition, other Pparγ target genes such as Hrasl3, Dlk (dual leucine zipper-bearing kinase), adiponectin, G0s2, and chemerin can similarly increase adipogenesis and Pparγ expression.5,38 Alternatively, since Pnpla2 and Pnpla3 share high homology, they may exhibit similar effects on Pparγ activation. G0s2, an inhibitor of Pnpla2, is a regulator of lipid droplet formation. It can also increase adipocyte differentiation and Pparγ expression.39 CGI-58, an activator of Pnpla2, promotes adipogenesis. Pnpla3 also induces a reduction in LD size upon coexpression with ABDH5/CGI-58.39 However, Pnpla2 and Pnpla3 homologous genes are usually regulated in the conflicting direction from each other, suggesting nonredundant or divergent actions of these related genes. Our results showed that Pnpla3 was suppressed by Pparγ activation while its deficiency in cells decreased adipocyte differentiation. By contrast, Pnpla2 is transcriptionally induced by Pparγ.40 It has been shown that lipolytic products of Pnpla2 can activate Ppar family members in response to cyclic adenosine monophosphate stimulation in brown adipocytes.41 Furthermore, knockdown of G0s2 or Pnpla3 in this study exhibited similar antiadipogenic effects, implying that G0s2 may not inhibit Pnpla3 activity, unlike its regulatory effects on Pnpla2.42 Thus, Pnpla3 and Pnpla2 may possess different regulatory effects on Pparγ and adipocyte differentiation. Together, it is obvious that Pparγ regulation by various growth, transcription factors, and lipid metabolic enzymes including Pnpla3 is complicated. This should be further elucidated in the future.
It was expected that antiadipogenic chemicals would reduce the expression of proadipogenic genes. However, we observed that Phf16 induced by antiadipogenic chemicals acted as an adipogenic gene. Of note, all four antiadipogenic compounds utilized in the current study have been shown to be able to affect Wnt or transforming growth factor β (TGF-β) signaling pathways. It is thus possible that the four antiadipogenic compounds might exert effects on TGF or Wnt signaling pathways followed by induction of Wnt or Tgf-target genes. Large numbers of known Wnt-induced target genes including Axin, Sfrp, chemokine receptor 1, and Dickkopf-1 can also act as Wnt suppressors to inactivate Wnt signaling and increase adipogenesis.43 Similarly, TGF-induced Smad7 negatively controls TGF receptor TβRI activity.44 Interestingly, Phf16 has been recently shown as a Wnt-induced gene,45 suggesting that it may either directly or indirectly affect Wnt pathways. Therefore, Phf16 induction by antiadipogenic chemicals may negatively act on Wnt or TGF-signaling pathways, which may in turn affect adipocyte differentiation and Pparγ. In addition to this possibility, Phf16 is a general transcriptional regulator. As such, chemically induced Phf16 may affect a wide range of transcriptional activators or repressors that direct transcriptions toward stimulation of adipogenesis. Additional studies are needed to provide mechanistic explanations for action of Phf16 in adipocyte differentiation.
We also demonstrated that Phf16 controlled adipocyte differentiation. Phf16 (JADE family PHD zinc finger 3) encoding 823-amino acids belongs to a family of small JADE proteins. Roles of Phf14 (JADE1) in cytokinesis and regulation of histone acetylation have been reported.46–48 JADE1 functions through interaction with other transcriptional partners. Phf16 has PHD zinc finger domains found in transcription regulating proteins.49,50 Thus, we speculated that Phf16 might act as a coactivator of Pparγ in adipocytes. We found that coexpression of Phf16 with Pparγ and RXR increased Pparγ driven transcription activity, which could be further enhanced in the presence of Pparγ ligand. These data demonstrate that Phf16 could be a component of the protein complex that works cooperatively with Ppaγ to stimulate adipocyte differentiation. A probable direct physical interaction and specificity with Pparγ need to be determined in the future to provide mechanistic explanations for action of Phf16 in adipocyte differentiation.
In summary, we identified previously uncovered roles of Phf16 and Pnpla3 in adipogenesis. Results of this study further highlight the potential use of phytochemicals as tools to further dissect adipocyte biology.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF), funded by the Ministry of Education, Science, and Technology (NRF-2016M3A9B6903451 and NRF-2017R1A2B4002005 to KWP and NRF-2017R1A6 A3A11029584 to UJY).
Funding information
National Research Foundation of Korea, Grant/Award Numbers: NRF-2016M3A9B6903451, NRF-2017R1A6A3A11029584, NRF-2017R1A2B4002005
Abbreviations:
- BAT
brown adipose tissue
- Cd36
cluster of differentiation 36
- Cdk5
cyclin-dependent kinase 5
- C/ebpα
CCAAT/enhancer-binding protein α
- Cxcl1
chemokine (C-X-C motif) ligand 1
- ERK
extracellular signal-regulated kinases
- eWAT
epididymal white adipose tissue
- Fabp4
fatty acid binding protein 4
- G0s2
G0/G1 switch gene 2
- Hmox1
heme oxygenase 1
- iWAT
inguinal white adipose tissue
- Lpl
lipoprotein lipase
- Phf16
PHD (plant homeo domain) finger protein 16
- Pnpla3
patatin-like phospholipase domain containing 3
- Pparγ
peroxisome proliferator-activated receptor γ
- siRNA
small interfering RNA
- TNF-α
tumor necrosis factor α
Footnotes
CONFLICTS OF INTEREST
The authors declare that there are no conflicts of interest.
SUPPORTING INFORMATION
Additional supporting information may be found online in the Supporting Information section at the end of the article.
REFERENCES
- 1.Rosen CJ, Bouxsein ML. Mechanisms of disease: is osteoporosis the obesity of bone? Nat Clin Pract Rheumatol 2006;2:35–43. 10.1038/ncprheum0070 [DOI] [PubMed] [Google Scholar]
- 2.Halaas J, Gajiwala K, Maffei M, et al. Weight-reducing effects of the plasma protein encoded by the obese gene. Science 1995;269:543–546. 10.1126/science.7624777 [DOI] [PubMed] [Google Scholar]
- 3.Steppan CM, Bailey ST, Bhat S, et al. The hormone resistin links obesity to diabetes. Nature 2001;409:307–312. 10.1038/35053000 [DOI] [PubMed] [Google Scholar]
- 4.Yamauchi T, Kamon J, Waki H, et al. The fat-derived hormone adiponectin reverses insulin resistance associated with both lipoatrophy and obesity. Nature Med 2001;7:941–946. 10.1038/90984 [DOI] [PubMed] [Google Scholar]
- 5.Tontonoz P, Spiegelman BM. Fat and beyond: the diverse biology of PPARgamma. Annu Rev Biochem 2008;77:289–312. 10.1146/annurev.biochem.77.061307.091829 [DOI] [PubMed] [Google Scholar]
- 6.Banks AS, McAllister FE, Camporez JPG, et al. An ERK/Cdk5 axis controls the diabetogenic actions of PPARgamma. Nature 2015;517:391–395. 10.1038/nature13887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Prusty D, Park BH, Davis KE, Farmer SR. Activation of MEK/ERK signaling promotes adipogenesis by enhancing peroxisome proliferator-activated receptor gamma (PPARgamma) and C/EBPalpha gene expression during the differentiation of 3T3-L1 preadipocytes. J Biol Chem 2002;277:46226–46232. 10.1074/jbc.M207776200 [DOI] [PubMed] [Google Scholar]
- 8.Choi JH, Banks AS, Estall JL, et al. Anti-diabetic drugs inhibit obesity-linked phosphorylation of PPARgamma by Cdk5. Nature 2010;466:451–456. 10.1038/nature09291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hasani-Ranjbar S, Nayebi N, Larijani B, Abdollahi M. A systematic review of the efficacy and safety of herbal medicines used in the treatment of obesity. World J Gastroenterol 2009;15:3073–3085 10.3748/wjg.15.3073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jung SR, Kim YJ, Gwon AR, et al. Genistein mediates the antiadipogenic actions of Sophora japonica L. extracts. J Med Food 2011;14:360–368. 10.1089/jmf.2010.1324 [DOI] [PubMed] [Google Scholar]
- 11.Hwang JT, Park IJ, Shin JI, et al. Genistein, EGCG, and capsaicin inhibit adipocyte differentiation process via activating AMP-activated protein kinase. Biochem Biophys Res Commun 2005;338:694–699. 10.1016/j.bbrc.2005.09.195 [DOI] [PubMed] [Google Scholar]
- 12.Baur JA, Pearson KJ, Price NL, et al. Resveratrol improves health and survival of mice on a high-calorie diet. Nature 2006;444:337–342. 10.1038/nature05354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jeon WK, Lee JH, Kim HK, et al. Anti-platelet effects of bioactive compounds isolated from the bark of Rhus verniciflua Stokes. J Ethnopharmacol 2006;106:62–69. 10.1016/j.jep.2005.12.015 [DOI] [PubMed] [Google Scholar]
- 14.Lee JC, Lee KY, Kim J, et al. Extract from Rhus verniciflua Stokes is capable of inhibiting the growth of human lymphoma cells. Food Chem Toxicol 2004;42:1383–1388. 10.1016/j.fct.2004.03.012 [DOI] [PubMed] [Google Scholar]
- 15.Song NJ, Yoon HJ, Kim KH, et al. Butein is a novel antiadipogenic compound. J Lipid Res 2013;54:1385–1396 10.1194/jlr.M035576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feige JN, Lagouge M, Canto C, et al. Specific SIRT1 activation mimics low energy levels and protects against diet-induced metabolic disorders by enhancing fat oxidation. Cell Metab 2008;8:347–358. 10.1016/j.cmet.2008.08.017 [DOI] [PubMed] [Google Scholar]
- 17.Jung CH, Kim H, Ahn J, Jeon TI, Lee DH, Ha TY. Fisetin regulates obesity by targeting mTORC1 signaling. J Nutr Biochem 2013;24:1547–1554. 10.1016/j.jnutbio.2013.01.003 [DOI] [PubMed] [Google Scholar]
- 18.Kim JH, Song J, Park KW. The multifaceted factor peroxisome proliferator-activated receptor gamma (PPARgamma) in metabolism, immunity, and cancer. Arch Pharmacal Res 2015;38:302–312. 10.1007/s12272-015-0559-x [DOI] [PubMed] [Google Scholar]
- 19.Song NJ, Kim S, Jang BH, et al. Small molecule-induced complement factor D (adipsin) promotes lipid accumulation and adipocyte differentiation. PLOS One 2016;11:e0162228 10.1371/journal.pone.0162228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Won park K, Waki H, Choi SP, Park KM, Tontonoz P. The small molecule phenamil is a modulator of adipocyte differentiation and PPARgamma expression. The Journal of Lipid Research 2010;51:2775–2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kang S, Bennett CN, Gerin I, Rapp LA, Hankenson KD, Macdougald OA. Wnt signaling stimulates osteoblastogenesis of mesenchymal precursors by suppressing CCAAT/enhancer-binding protein alpha and peroxisome proliferator-activated receptor gamma. J Biol Chem 2007;282:14515–14524. 10.1074/jbc.M700030200 [DOI] [PubMed] [Google Scholar]
- 22.Wagner G, Lindroos-Christensen J, Einwallner E, et al. HO-1 inhibits preadipocyte proliferation and differentiation at the onset of obesity via ROS dependent activation of Akt2. Sci Rep 2017;7:40881 10.1038/srep40881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cao J, Peterson SJ, Sodhi K, et al. Heme oxygenase gene targeting to adipocytes attenuates adiposity and vascular dysfunction in mice fed a high-fat diet. Hypertension 2012;60:467–475. 10.1161/HYPERTENSIONAHA.112.193805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang X, Lu X, Lombès M, et al. The G(0)/G(1) switch gene 2 regulates adipose lipolysis through association with adipose triglyceride lipase. Cell Metab 2010;11:194–205. 10.1016/j.cmet.2010.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zandbergen F, Mandard S, Escher P, et al. The G0/G1 switch gene 2 is a novel PPAR target gene. Biochem J 2005;392:313–324. 10.1042/BJ20050636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang X, Xie X, Heckmann BL, Saarinen AM, Czyzyk TA, Liu J. Targeted disruption of G0/G1 switch gene 2 enhances adipose lipolysis, alters hepatic energy balance, and alleviates high-fat diet-induced liver steatosis. Diabetes 2014;63:934–946. 10.2337/db13-1422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ignacio RMC, Gibbs CR, Lee ES, Son DS. Differential chemokine signature between human preadipocytes and adipocytes. Immune Netw 2016;16:189–194. 10.4110/in.2016.16.3.189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kabir SM, Lee ES, Son DS. Chemokine network during adipogenesis in 3T3-L1 cells: Differential response between growth and proinflammatory factor in preadipocytes vs. adipocytes. Adipocyte 2014;3:97–106. 10.4161/adip.28110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bolitho C, Hahn MA, Baxter RC, Marsh DJ. The chemokine CXCL1 induces proliferation in epithelial ovarian cancer cells by transactivation of the epidermal growth factor receptor. Endocr Relat Cancer 2010;17:929–940. 10.1677/ERC-10-0107 [DOI] [PubMed] [Google Scholar]
- 30.Baulande S, Lasnier F, Lucas M, Pairault J. Adiponutrin, a transmembrane protein corresponding to a novel dietary- and obesity-linked mRNA specifically expressed in the adipose lineage. J Biol Chem 2001;276:33336–33344. 10.1074/jbc.M105193200 [DOI] [PubMed] [Google Scholar]
- 31.Wilson PA, Gardner SD, Lambie NM, Commans SA, Crowther DJ. Characterization of the human patatin-like phospholipase famil. J Lipid Res 2006;47:1940–1949. 10.1194/jlr.M600185-JLR200 [DOI] [PubMed] [Google Scholar]
- 32.Chamoun Z, Vacca F, Parton RG, Gruenberg J. PNPLA3/adiponutrin functions in lipid droplet formation. Biol Cell 2013;105:219–233. 10.1111/boc.201200036 [DOI] [PubMed] [Google Scholar]
- 33.Panchenko MV. Structure, function and regulation of jade family PHD finger 1 (JADE1). Gene 2016;589:1–11. 10.1016/j.gene.2016.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lake AC, Sun Y, Li JL, et al. Expression, regulation, and triglyceride hydrolase activity of Adiponutrin family members. J Lipid Res 2005;46:2477–2487 10.1194/jlr.M500290-JLR200 [DOI] [PubMed] [Google Scholar]
- 35.Hoekstra M, Li Z, Kruijt JK, Eck MV, Berkel TJCV, Kuiper J. The expression level of non-alcoholic fatty liver disease-related gene PNPLA3 in hepatocytes is highly influenced by hepatic lipid status. J Hepatol 2010;52:244–251. 10.1016/j.jhep.2009.11.004 [DOI] [PubMed] [Google Scholar]
- 36.Kim JY, Tillison K, Lee JH, Rearick DA, Smas CM. The adipose tissue triglyceride lipase ATGL/PNPLA2 is downregulated by insulin and TNF-alpha in 3T3-L1 adipocytes and is a target for transactivation by PPARgamma. Am J Physiol-Endocrinol Metab 2006;291:E115–E127. 10.1152/ajpendo.00317.2005 [DOI] [PubMed] [Google Scholar]
- 37.Tardelli M, Bruschi FV, Claudel T, et al. AQP3 is regulated by PPARgamma and JNK in hepatic stellate cells carrying PNPLA3 I148M. Sci Rep 2017;7:14661 10.1038/s41598-017-14557-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim SC, Kim YH, Son SW, Moon EY, Pyo S, Um SH. Fisetin induces Sirt1 expression while inhibiting early adipogenesis in 3T3-L1 cells. Biochem Biophys Res Commun 2015;467:638–644. 10.1016/j.bbrc.2015.10.094 [DOI] [PubMed] [Google Scholar]
- 39.Zhang X, Heckmann BL, Campbell LE, Liu J. G0S2: a small giant controller of lipolysis and adipose-liver fatty acid flux. Biochim Biophys Acta 2017;1862:1146–1154. 10.1016/j.bbalip.2017.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roy D, Farabaugh KT, Wu J, et al. Coordinated transcriptional control of adipocyte triglyceride lipase (Atgl) by transcription factors Sp1 and peroxisome proliferator-activated receptor gamma (PPARgamma) during adipocyte differentiation. J Biol Chem 2017;292:14827–14835. 10.1074/jbc.M117.783043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mottillo EP, Bloch AE, Leff T, Granneman JG. Lipolytic products activate peroxisome proliferator-activated receptor (PPAR) alpha and delta in brown adipocytes to match fatty acid oxidation with supply. J Biol Chem 2012;287:25038–25048. 10.1074/jbc.M112.374041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Choi H, Lee H, Kim TH, et al. G0/G1 switch gene 2 has a critical role in adipocyte differentiation. Cell Death Differ 2014;21:1071–1080. 10.1038/cdd.2014.26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ross SE, Hemati N, Longo KA, et al. Inhibition of adipogenesis by Wnt signaling. Science 2000;289:950–953. 10.1126/science.289.5481.950 [DOI] [PubMed] [Google Scholar]
- 44.Lu H, Li X, Mu P, Qian B, Jiang W, Zeng L. Dickkopf-1 promotes the differentiation and adipocytokines secretion via canonical Wnt signaling pathway in primary cultured human preadipocytes. Obesity Res Clin Pract 2016;10:454–464. 10.1016/j.orcp.2015.08.016 [DOI] [PubMed] [Google Scholar]
- 45.Jian Y, Wang M, Zhang Y, et al. Jade family PHD finger 3 (JADE3) increases cancer stem cell-like properties and tumorigenicity in colon cancer. Cancer Lett 2018;428:1–11. 10.1016/j.canlet.2018.04.012 [DOI] [PubMed] [Google Scholar]
- 46.Siriwardana NS, Meyer R, Havasi A, Dominguez I, Panchenko MV. Cell cycle-dependent chromatin shuttling of HBO1-JADE1 histone acetyl transferase (HAT) complex. Cell Cycle 2014;13:1885–1901. 10.4161/cc.28759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Siriwardana NS, Meyer RD, Panchenko MV. The novel function of JADE1S in cytokinesis of epithelial cells. Cell Cycle 2015;14:2821–2834. 10.1080/15384101.2015.1068476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tzouanacou E, Tweedie S, Wilson V. Identification of Jade1, a gene encoding a PHD zinc finger protein, in a gene trap mutagenesis screen for genes involved in anteroposterior axis development. Mol Cell Biol 2003;23:8553–8562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lalonde ME, Avvakumov N, Glass KC, et al. Exchange of associated factors directs a switch in HBO1 acetyltransferase histone tail specificity. Genes Dev 2013;27:2009–2024. 10.1101/gad.223396.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Saksouk N, Avvakumov N, Champagne KS, et al. HBO1 HAT complexes target chromatin throughout gene coding regions via multiple PHD finger interactions with histone H3 tail. Mol Cell 2009;33:257–265. 10.1016/j.molcel.2009.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.