SUMMARY
There is an unmet need for new antimitotic drug combinations that target cancer-specific vulnerabilities. Based on our finding of elevated biomolecule oxidation in mitotically arrested cancer cells, we combined Plk1 inhibitors with TH588, an MTH1 inhibitor that prevents detoxification of oxidized nucleotide triphosphates. This combination showed robust synergistic killing of cancer, but not normal, cells that, surprisingly, was MTH1-independent. To dissect the underlying synergistic mechanism, we developed VISAGE, a strategy integrating experimental synergy quantification with computational pathway-based gene expression analysis. VISAGE predicted, and we experimentally confirmed, that this synergistic combination treatment targeted the mitotic spindle. Specifically, TH588 binding to β-tubulin impaired microtubule assembly, which when combined with Plk1 blockade, synergistically disrupted mitotic chromosome positioning to the spindle midzone. These findings identify a cancer-specific mitotic vulnerability that is targetable using Plk1 inhibitors with microtubule-destabilizing agents, and highlight the general utility of the VISAGE approach to elucidate molecular mechanisms of drug synergy.
Graphical Abstract
eTOC Blurb
We identified the combination of TH588 and Plk1 inhibition as synergistic for the killing of tumor cells, but not normal cells, and used a combined experimental/computational method to identify defective spindle assembly and chromosome segregation in co-treated tumor cells as the synergistic mechanism.
INTRODUCTION
The vast majority of tumors are treated with some type of combination chemotherapy (DeVita and Chu, 2008). Synergistic combination therapies, in particular, are of substantial clinical interest due to their potential for increasing efficacy and cancer cell selectivity, reducing the development of resistance, and allowing for decreases in individual drug dosage, possibly avoiding toxicity (Keith et al., 2005; Lehar et al., 2009). The individual drugs in these combinations are generally selected based on either their ability to target pathways required for unrestrained cell proliferation, or their involvement in the acquisition and maintenance of cancer-cell specific traits, exemplified by the hallmarks of cancer (Hanahan and Weinberg, 2011), and can be combined to target orthogonal cancer vulnerabilities.
A particularly useful class of anticancer therapeutics is antimitotic drugs, which includes microtubule targeting agents and inhibitors of mitotic kinases (Dominguez-Brauer et al., 2015). Microtubule-directed agents have shown impressive clinical activity against a wide variety of epithelial cancers, and are currently used as a standard of care in the treatment of breast, lung, ovarian, and prostate cancer, among others. However, toxicity and side effects remain major problems with these agents, since they show little discrimination between cancer cells and normal cells, and target a wide variety of mitotic and non-mitotic microtubule-based cell processes. In contrast, inhibitors of mitotic kinases including Plk1, the Aurora kinases, and mitotic cyclin-dependent kinases, which target molecules required mainly for unrestrained cell proliferation, have fared poorly in clinical trials, despite repeatedly demonstrating impressive efficacy in pre-clinical studies (Dominguez-Brauer et al., 2015). Given these limitations of current antimitotic drugs, “the challenge is to identify cell cycle regulators that are essential for mitosis of cancer cells rather than normal cells,” as stated in a seminal and comprehensive review by Tak Mak and colleagues, (Dominguez-Brauer et al., 2015).
We were particularly interested in anti-cancer drug combinations that included inhibitors of Plk1, a kinase well known for its pleiotropic role in all stages of mitosis. Structurally, Plk1 consists of an N-terminal kinase domain and a C-terminal phosphopeptide-binding Polo-box domain (PBD) phosphopeptide-binding domain that targets Plk1 to substrates primed via phosphorylation by CDKs and other kinases (Elia et al., 2003a; Elia et al., 2003b) allowing it to function, in part, as a molecular integrator of mitotic and DNA damage kinase signaling pathways (Alexander et al., 2011; Archambault and Glover, 2009; Bruinsma et al., 2012; Macurek et al., 2008; Reinhardt and Yaffe, 2013; van Vugt et al., 2010). Based on positive data from a Phase II trial, the Plk1 inhibitor volasertib was designated by the FDA as a “breakthrough therapy” for acute myeloid leukemia, fulfilling an unmet medical need (Dohner et al., 2014). However, the development of neutropenia-associated infections in a subset of patients treated with this drug counterbalanced its utility in a subsequent Phase III trial (NCT: ) (Döhner et al., 2016). Another Plk1 inhibitor, onvansertib, is currently being tested in multiple clinical trials (NCT: and NCT: ). These data demonstrate that inhibition of Plk1 has clear clinical utility, and suggest that high-dose toxicity might be overcome by inclusion of a Plk1 inhibitor as part of a synergistic drug combination.
For the second component of a Plk1-containing anti-cancer combination treatment, we considered the consequences that sustained cell proliferation has on metabolic demands and cellular energetics (Hanahan and Weinberg, 2011; Vander Heiden and DeBerardinis, 2017). It has long been known that cancer cells produce excessive amounts of reactive oxygen species (ROS) relative to non-cancer cells (Liou and Storz, 2010; Szatrowski and Nathan, 1991), and we and others have observed that mitotic cancer cells, in particular, have higher levels of ROS and oxidative stress than interphase cells (Havens et al., 2006; Kawamura, 1960; Mazia, 1958; Patterson et al., 2019). We recently showed that prolonged mitotic arrest further increases oxidative damage to proteins and nucleotides (Patterson et al., 2019). Therefore, we investigated whether combining Plk1 kinase inhibitors with drugs that increase the lethality of ROS-dependent DNA damage could be used to synergistically enhance tumor cell killing. We were particularly interested in the nudix hydrolase MTH1 (NUDT1), an enzyme that hydrolyzes the oxidatively-damaged nucleotide triphosphates 8-oxo-deoxyguanosine triphosphate (8-oxodGTP) and 2-hydroxy-deoxyadenosine triphosphate (2-OH-dATP) to their monophosphate forms (Speina et al., 2005), since we and others had observed elevated levels of 8-oxoguanine after mitotic arrest (Crea et al., 2011; Patterson et al., 2019).
Several inhibitors of MTH1 have recently been highlighted for their potential broad utility as monotherapies in cancer (Gad et al., 2014; Huber et al., 2014) based on the proposal that cancer cells in general have a markedly increased dependence on MTH1 activity to mitigate oxidative DNA damage (Rai et al., 2009; Tsuzuki et al., 2001), and a phase I clinical trial testing the safety of a MTH1 inhibitor is currently underway (NCT: ). Paradoxically, however, Mth1−/− mice are fully viable, and display increased rather than decreased tumor incidence (Tsuzuki et al., 2001). Tumors in these mice entirely lack MTH1 activity, indicating that MTH1 is not universally essential for tumor viability (Tsuzuki et al., 2001). Moreover, recent reports have, in a seeming contradiction, both cast doubt on the utility of MTH1 as a cancer therapeutic target (Ellermann et al., 2017; Kettle et al., 2016) but also reinforced the observations of cancer-cell specific essentiality of this enzyme (Pudelko et al., 2017; Samaranayake et al., 2017; Warpman Berglund et al., 2016), adding to the confusion in this area. We therefore set out to examine whether biomolecule oxidation induced by Plk1 inhibitors would enhance cancer cells’ dependence on MTH1 and, in turn, increase their sensitivity to a well-described MTH1 inhibitor.
RESULTS
Inhibition of the mitotic kinase Plk1 synergizes with the MTH1 inhibitor TH588 to induce apoptotic tumor cell death in cell culture and in human tumor xenografts.
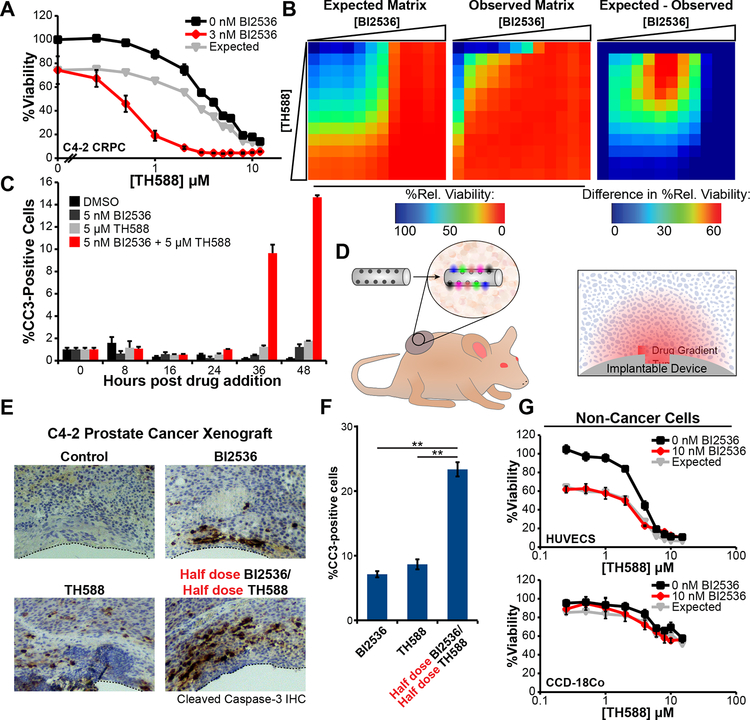
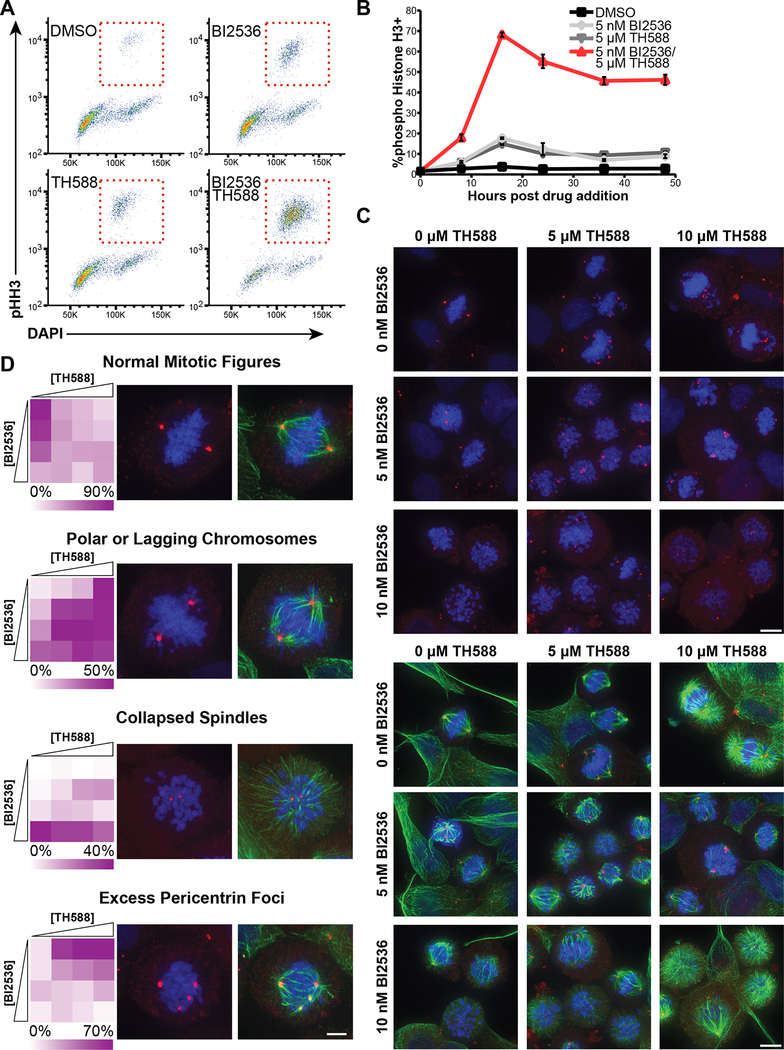
Based on our recent finding of elevated oxidative stress in mitotically-arrested cancer cells (Patterson et al., 2019), we investigated the combined effects of the MTH1 inhibitor TH588 with BI2536, an inhibitor of the mitotic kinase Plk1. The castration-resistant prostate cancer (CRPC) cell line C4–2 was chosen as a starting point for examination of TH588 synergy because: (1) Plk1 expression is elevated in prostate cancer and shown to correlate with Gleason grade (Weichert et al., 2004); (2) antimitotic drugs are widely used for treatment of advanced prostate cancer (Tannock et al., 2004); and (3) we had previously demonstrated elevated levels of cysteine and guanine oxidation following mitotic arrest in these cells (Patterson et al., 2019). As shown in Figure 1A, the Plk1 inhibitor BI2536 displayed substantial synergy with TH588 in this cell type. To assess synergy, the expected cell viability was calculated according to the Bliss independence model of drug additivity (Bliss, 1939), and decreases in viability beyond these expected values were used to indicate a greater-than-additive effect of the drugs in combination. The synergy observed is not specific to the Bliss model of drug independence, but is also observed under the Loewe additivity model (CI 0.58 at 0.8 fraction affected for 1 μM TH588 per 1 nM BI2536). Synergy between BI2536 and TH588 was observed over a broad area of a dose-response matrix centered around the EC50 of each drug (Figure 1B). This loss of cell viability was further demonstrated by showing a synergistic induction of apoptosis by the combination of BI2536 and TH588, as measured by cleaved-caspase-3 positive cells in a flow cytometric assay (Figure 1C), and a synergistic decrease in cell number using SYTO 60 staining (Figure S1A).
Figure 1. The Plk1 inhibitor BI2536 and MTH1 inhibitor TH588 synergistically kill cancer cells in cell culture and in tumor xenografts.
(A) C4–2 CRPC cells were subjected to a dose-response matrix of the MTH1 inhibitor, TH588, and BI2536. Viability relative to vehicle control was measured 5 days after treatment. TH588 dose-response curves in the absence (black line) or presence (red line) of BI2536 are shown. Expected viability according to the Bliss independence model of drug additivity is shown in grey. Mean ± SEM for n = 3 experiments is shown. The dose of the Plk1 inhibitor shown was chosen based on a 10–20% decrease in relative viability when that drug was used as a single agent.
(B) Heat map representation of the observed and expected BI2536-TH588 dose-response matrices and calculated difference between observed and expected viabilities after treatment of C4–2 cells for 5 days. Drug doses included all combinations of 1.25 fold serial dilutions starting from 14 nM and 12 μM for BI2536 and TH588, respectively, along with undrugged and single-drug controls.
(C) C4–2 cells were treated with the indicated drugs, fixed at various times, and analyzed for the apoptotic marker cleaved-caspase-3 by flow cytometry. Mean ± SEM for n = 3 experiments is shown.
(D) Illustration of the tumor implantable device for multiplexed drug sensitivity testing in vivo. Colored ovals (inset) indicate diffusion of distinct drug or drug combinations into the surrounding tissue. Right panel shows orientation of the device and drug gradients with respect to the fixed and stained tissue sections in panel (E).
(E) C4–2 CRPC tumor xenografts on the hind flanks of castrated male NCR nude mice were implanted with devices. Fixed sections of the tissue adjacent to microwells containing the indicated drug were stained with antibodies directed against cleaved caspase-3 as a readout for apoptosis. Clear zones at the bottom of each micrograph (dotted lines) indicate where the device was located.
(F) Percentage of cells positive for cleaved-caspase-3 within a 400 μm radius of the given well in the device was measured. Bars indicate the mean of measurements in three tumors ± SEM, ** p < 0.01 using a two-tailed Student’s t-test.
(G) Non-cancer cell lines, HUVECs and CCD-18Co colon fibroblasts, were treated and analyzed as in panel (A). No synergy was observed. Mean ± SEM (n = 3) is shown.
To validate this combination drug treatment for greater-than-additive effects in vivo, a tumor-implantable device that can achieve multiplexed drug sensitivity testing within single human tumor xenografts in a murine model through spatially isolated drug delivery was used (Jonas et al., 2015). This tumor-implantable device contains multiple microwells, each of which is loaded with a drug or combination of drugs in PEG hydrogel. Following implantation into a single tumor, the drugs are solvated and diffuse, creating a concentration gradient in the immediately surrounding tissue (Figure 1D). Subsequently, the tumor tissue surrounding the device is harvested, fixed, sectioned, and stained for markers of cell death in the region surrounding each of the drugs. Microwells in these tumor-implantable devices were loaded with PEG containing either no drug, 25% BI2536, 25% TH588 or 12.5% BI2536 and 12.5% TH588 by weight, and implanted into C4–2 human CRPC xenografts grown on the flanks of castrated NCR nude male mice. As shown in Figures 1E and 1F, we consistently observed significantly more apoptotic cell death in the tissue immediately surrounding the wells containing both drugs, despite the fact that these drug-combination wells contained only one-half of the amount of each drug relative to the wells containing the single drugs alone, consistent with greater-than-additive tumor cell killing.
In contrast to the substantial synergy that we observed in C4–2 CRPC cells, we did not observe synergy in two non-cancer cell types. Human umbilical vein endothelial cells (HUVECs), while sensitive to each of the individual drugs alone, did not display any synergy (Figure 1G). Similarly, human fibroblasts (CCD-18Co) isolated from normal colon were largely resistant to either of the drugs in isolation and also did not display any synergy (Figure 1G). The lack of synergy in either of these non-cancer cell lines suggests that this combination of drugs is not universally cytotoxic but may synergistically target a cancer-specific trait present in a subset of cancer cell lines (see below).
The cytotoxicity of TH588 and the synergy between Plk1 inhibitors and TH588 are independent of MTH1 activity.
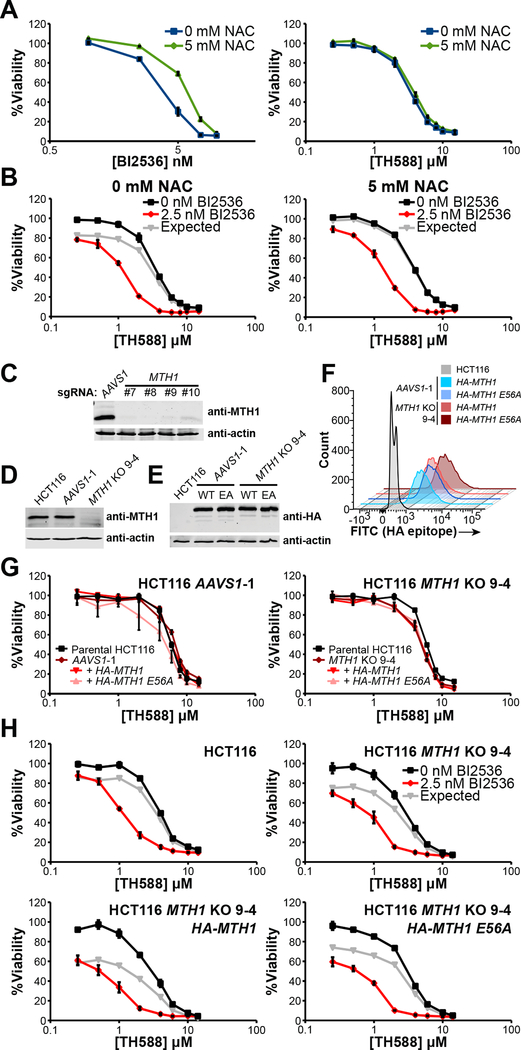
This combination of a Plk1 inhibitor with the MTH1 inhibitor TH588 was chosen based on a model of enhanced ROS and oxidative damage induced by mitotic arrest, resulting in a subsequent increased reliance on MTH1. To confirm this causal relationship between ROS and sensitivity to BI2536, TH588, or the combination, we examined the effect of the antioxidant N-acetyl cysteine (NAC) on tumor cell viability after treatment with these agents. Dose-response curves for the drugs were measured for C4–2 CRPC cells in the presence or absence of NAC. The addition of NAC resulted in increased resistance to the Plk1 inhibitor (Figure 2A, left panel), consistent with a model where antimitotic drugs induce cell death in a manner that involves accumulation of ROS (Alexandre et al., 2006). In contrast, however, the addition of NAC had no effect on the sensitivity of tumor cells to TH588 (Figure 2A, right panel), a finding that appears to conflict with this drug’s purported mechanism of action. Moreover, NAC did not prevent nor decrease the synergy between these drugs (Figure 2B). These data suggest that the MTH1 inhibitor TH588, and the synergy between this drug and Plk1 inhibitors, do not necessarily derive their cytotoxic effects from ROS-mediated accumulation of toxic metabolites.
Figure 2. MTH1 is dispensable for cancer cell viability, sensitivity to the MTH1 inhibitor TH588, and Plk1 inhibitor - TH588 synergy.
(A) C4–2 prostate cancer cells were treated with the indicated doses of BI2536 (left) or TH588 (right) in the presence (blue) or absence (green) of 5 mM NAC, added at the time of drug dosing. Relative viability was determined 5 days after drug addition. NAC decreased the sensitivity to the Plk1 inhibitor BI2536 but not to the MTH1 inhibitor TH588. Mean ± SEM (n = 3) is shown.
(B) Synergy between TH588 and BI2536 was determined by culturing C4–2 cells with various doses of TH588 in the absence (black) or presence (red) of BI2536, with (right) or without (left) 5 mM NAC. Relative viability was measured at 5 days. Expected viability (gray lines) was calculated using the Bliss independence model of drug additivity. Mean ± SEM (n = 3) is shown.
(C) Immunoblot analysis of lysates from polyclonal populations of HCT116 transductants co-expressing Cas9 and the indicated MTH1-targeting sgRNA. An sgRNA directed against AAVS1 served as a control.
(D) Immunoblot analysis of MTH1 expression in HCT116 parental cells and single-cell clones derived from the populations shown in panel (C), denoted AAVS1-1 and MTH1 KO 9–4, respectively.
(E, F) AAVS1-1 and MTH1 KO 9–4 single-cell clones were transduced with constructs containing HA-tagged wildtype (WT) MTH1 or a catalytically inactive mutant, MTH1 E56A (EA). Immunoblot analysis of HA-tagged MTH1 is shown in (E). Distribution of HA-tagged MTH1 expression measured by flow cytometry is shown in (F).
(G) TH588 dose-response curves for the parental HCT116 cells and AAVS1-1 (left) or MTH1 KO 9–4 (right) clones without or with overexpression of HA-MTH1 or HA-MTH1 E56A. Cells were treated with TH588 for 5 days before measurement of relative viability. Mean ± SEM (n = 3) is shown.
(H) The indicated cell lines from panels (D) and (E) were treated with increasing doses of TH588 in the presence (red lines) or absence (black lines) of 2.5 nM BI2536 for 5 days and assessed for relative viability. Gray lines depict expected viability according to the Bliss independence model of drug additivity. Synergy was observed regardless of either the total absence of MTH1 or its robust overexpression. Mean ± SEM (n = 3) is shown.
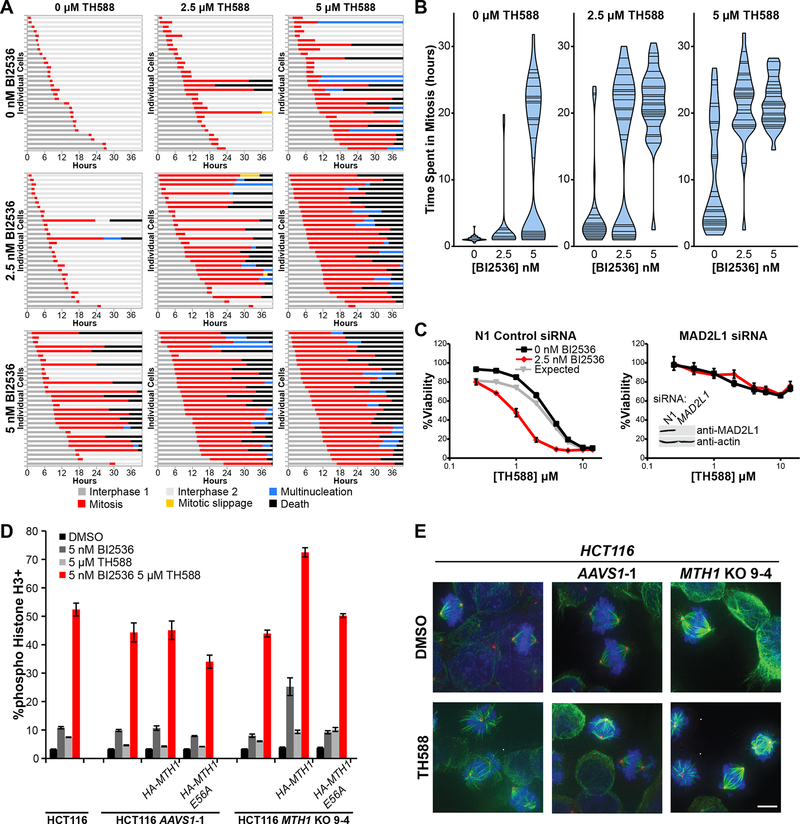
The observation that TH588 cytotoxicity was independent of antioxidant treatment cast doubt that this drug kills cells through MTH1 inhibition. We therefore tested for synergy between Plk1 inhibitors and a second reported MTH1 inhibitor, (S)-crizotinib (Huber et al., 2014). Not only was no synergy observed between these drugs, but they were highly antagonistic at intermediate doses (Figure S1B). We confirmed that both TH588 and (S)-crizotinib physically engage and stabilize MTH1 in whole cells using a cellular thermal shift assay (Huber et al., 2015) (Figure S1C). Since both TH588 and (S)-crizotinib inhibit MTH1 in vitro (Gad et al., 2014; Huber et al., 2014) and both bind to MTH1 in cells, their differential effects in combination with BI2536 raised doubt that MTH1 inhibition underlies the mechanism of the observed TH588/Plk1 inhibitor synergy. To further explore this, we sought to create viable MTH1 knockouts using the CRISPR-Cas9 system (Hsu et al., 2013). We transduced Cas9-expressing C4–2 CRPC cells, HCT116 colorectal cancer cells, and MDA-MB-231 breast cancer cells with a construct containing the MTH1 targeting sgRNA. Polyclonal populations of transductants in all these cell lines displayed reduced or absent expression of MTH1 (Figure S1D), with no overall change in proliferation, sensitivity to TH588, or synergy between TH588 and Plk1 inhibition (Figure S1E). To further explore this in detail in HCT116 cells, the line that displayed the lowest MTH1 expression as a polyclonal population, we used four different sgRNAs against the first constitutive coding exon of MTH1 (Figure S2A) or a control guide that targets the adeno-associated virus integration site 1 (AAVS1) locus. All four MTH1 guides led to substantial loss of MTH1 expression without loss of viability (Figure 2C), as further validated by analysis of single cell clones that were confirmed to contain null alleles of MTH1 by DNA sequencing (Figure S2B–E). These findings, together with recent reports from others (Kettle et al., 2016; Wang et al., 2016), provide strong evidence that MTH1 activity is not essential for cancer cell viability.
To separate the effects of MTH1 loss from those of interclonal heterogeneity, we rescued MTH1 expression in single HCT116 cell clones lacking functional MTH1 by transduction with constructs that express either HA-tagged wild-type (WT) or catalytically inactive (E56A, (Fujii et al., 1999)) MTH1 (Figures 2D–2F and S2F–S2G). We observed no difference in sensitivity to TH588 among parental HCT116 cells, MTH1 KO cells, or MTH1 KO cells rescued with wild-type or catalytically inactive HA-MTH1 (Figure 2G). Additionally, there was an equivalent sensitivity to BI2536 (Figure S2H) and equivalent synergy between TH588 and BI2536 in MTH1 KO 9–4 cells compared to parental HCT116 cells (Figure 2H, upper panels). Furthermore, overexpression of catalytically inactive MTH1 in MTH1 knockout clones had no effect on either BI2536 or on synergistic cell killing by the combination with TH588, while overexpression of wild-type MTH1 appeared to actually slightly increase sensitivity to Plk1 inhibition while preserving the synergy of the combination treatment (Figures 2H, S2I). These observations were confirmed using a second MTH1 knockout clone (10–2) generated using a different sgRNA targeting the first constitutive exon of MTH1 (Figures S2B–F and S2I). Taken together, these findings indicate that MTH1 is dispensable both for cancer cell viability and for the substantial synergy in cell killing observed between TH588 and Plk1 inhibitors.
A combined experimental and computational approach implicates the mitotic spindle as the molecular target of drug synergy.
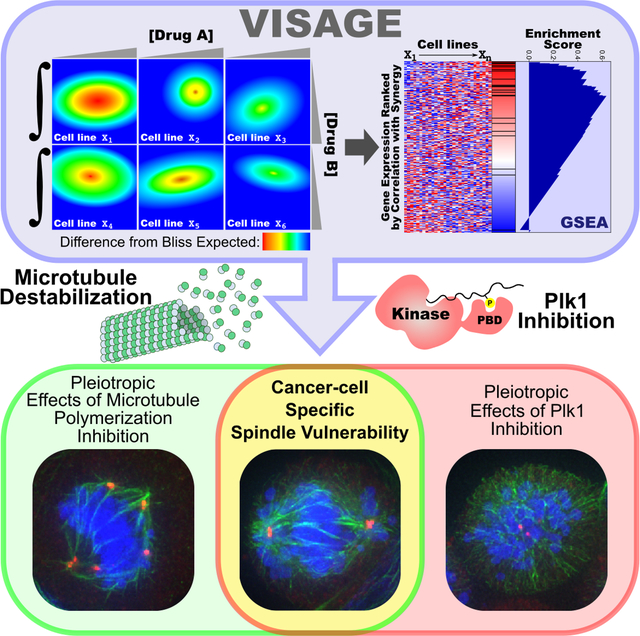
Plk1 inhibitors and TH588 display marked synergy in an MTH1-independent, but apparently cancer-specific, manner. Understanding the mechanisms behind this synergy and TH588’s cytotoxicity could uncover a potentially useful novel cancer-specific vulnerability. To achieve this goal, we devised and implemented a novel approach for mechanistically linking quantification of synergy to specific pathways and biological functions using existing computational methods, termed ‘VISAGE’ (Volumetric Interrogation of Synergy and Gene set Enrichment, R script available at https://github.com/yaffelab/visage). The aim of the VISAGE strategy is to quantify synergy in a robust manner that facilitates experimental scaling to large numbers of cell lines exhibiting varying degrees of synergy, and then leverage basal transcriptional heterogeneity among those lines in order to identify potentially causal correlates of synergy at the pathway level (Figures 3A–B, S3 and STAR Methods).
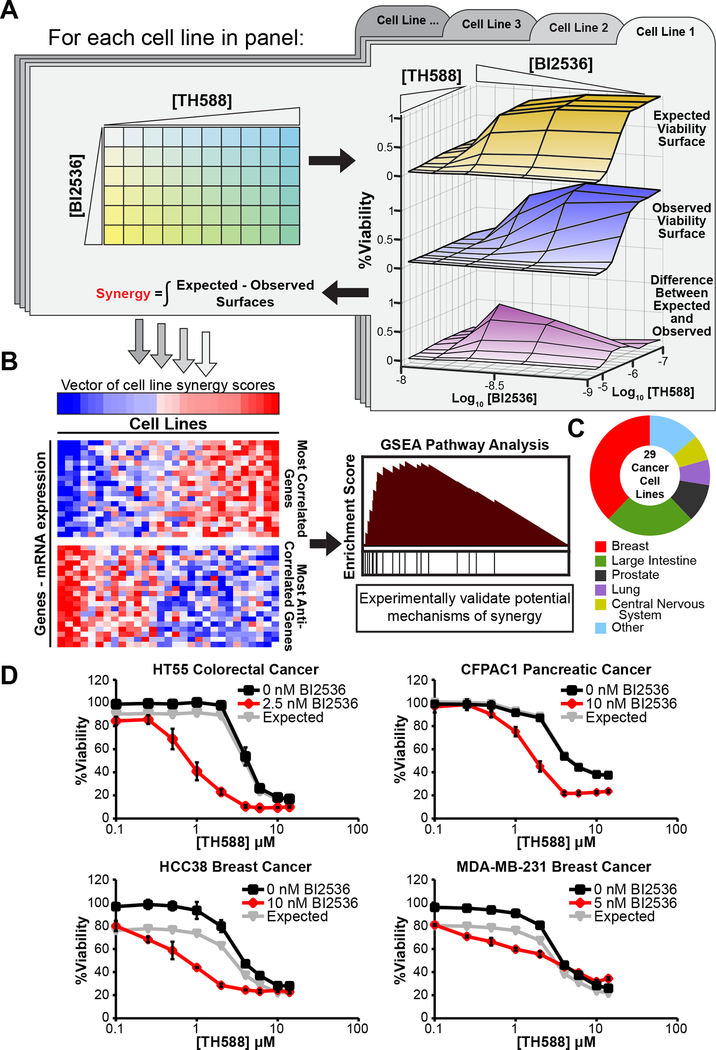
Figure 3. VISAGE – an approach for identifying mechanisms underlying synergistic drug combinations.
(A) Overview of the experimental component. In an iterative process each cell line is subjected to a 60-point dose matrix consisting of all pairwise combinations of 6 doses of BI2536 and 10 doses of TH588. The response at each point of this dose matrix is used to generate observed (middle surface) and expected (top surface) dose-response surfaces according to the Bliss independence model of drug additivity. The integrated difference between the observed and expected surfaces (bottom surface) is quantified as a volumetric measurement of synergy. This measure of synergy is analogous to a single drug AUC in that it represents the total effect observed in a particular dose space and is also known as the Bliss independence volume.
(B) Overview of the computational component. Cell lines are ranked according the quantity of synergy observed, and these data are used to calculate correlation coefficients between the synergy score vector and each gene’s mRNA expression profile across the CCLE cell lines included in the screen. Biologically coherent sets of genes whose basal expression are correlated with the presence or absence of synergy can be identified with GSEA, yielding insight and guiding validation of proposed synergistic mechanisms.
(C) Tissue of origin composition of cell lines included in the BI2536/TH588 VISAGE cell line screen for BI2536/TH588 synergy.
(D) Representative dose-response curves for TH588 (black) and TH588/BI2536 (red) co-treatment in cell lines that displayed a high (HT55), moderate (CFPAC1, HCC38), or very low (MDA-MB-231) degree of synergy. Relative viability was measured at 5 days. Expected viability (gray lines) was calculated using the Bliss independence model of drug additivity. Mean ± SEM (n = 3) is shown.
Twenty-nine cancer cell lines (Figure 3C) were subjected to a 60-point BI2536-TH588 dose matrix in triplicate, and viability was measured 5 days after treatment (Figures 3A,D and Table S1). These cell lines were selected for breadth of tissues of origin, and because of their inclusion in the Cancer Cell Line Encyclopedia (CCLE) (Barretina et al., 2012), which contains publicly-available transcriptomic data collected using a common platform. Sensitivities to treatment with BI2536 or TH588 alone, across our cell line panel, showed some correlation with each other as judged by area under the curve (AUC) (Figure 4A). In addition, nearly all cancer cell lines that we examined displayed some degree of synergy to combination treatment, the extent of which varied over a broad range (cf. panels in Figure 3D). The degree of synergy between the drugs, however, was not strongly correlated to the AUCs of either of the individual drugs alone (Figure 4A).
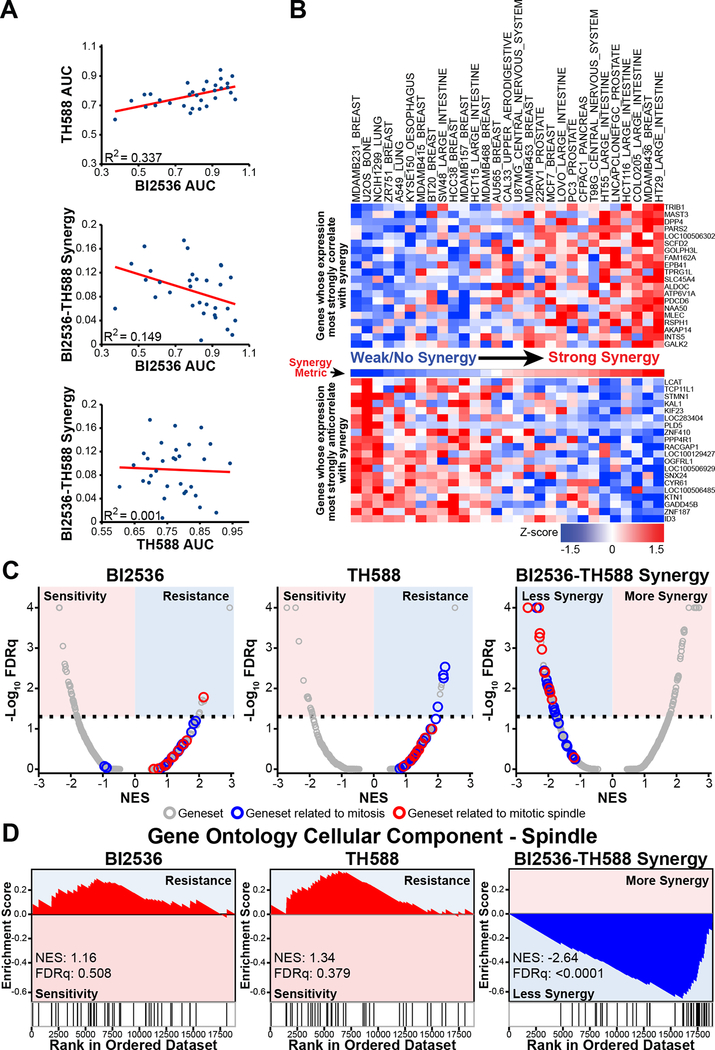
Figure 4. VISAGE predicts that the BI2536-TH588 drug combination synergistically targets mitosis and the mitotic spindle.
(A) Scatter plots depicting correlation between individual drug sensitivities (AUC) and their correlation with synergy observed across cell lines in our screen.
(B) Volumetric synergy scores were calculated as described in Figure 3A for each cell line (“Synergy Metric” between upper and lower heat maps). Cell lines were ranked according to synergy scores, increasing from left to right. Using CCLE data, genes were ranked based on the correlation between their expression and synergy across screened cell lines. Shown are the 20 genes whose expression profiles most strongly correlated (upper heat map) or anticorrelated (lower heat map) with the synergy scores. Rows were transformed to z-scores for ease of visualization.
(C) The expression levels of individual genes ranked by correlation with sensitivity to individual drugs (AUC; left and middle panels) or synergy (right panel) were analyzed by weighted GSEA using the Hallmark, KEGG, Reactome, BioCarta and GO gene sets. Plotted is the FDRq (false discovery rate q-value, a multiple-hypothesis corrected metric of significance) value versus the normalized enrichment score (NES) for each of these gene sets. The dotted line represents an FDRq value cut-off of 0.05. Gene sets related to mitosis are depicted in blue, and gene sets related specifically to the mitotic spindle are depicted in red. Colored backgrounds indicate gene set enrichment associated with drug sensitivity or resistance, or synergy, as indicated. Gene sets associated with mitosis and the mitotic spindle are more significantly associated with synergy than with sensitivity to either drug alone.
(D) The GO cellular component term ‘spindle’ was strongly enriched amongst genes with transcript levels that anticorrelated with the amount of synergy observed across various cell lines. This term was not significantly enriched amongst genes that correlated with resistance to the individual drugs alone.
To elucidate biological pathways and functions that potentially determine the observed synergy, we next combined our synergy measurements with gene expression data from the CCLE. For each gene, the Pearson correlation coefficient of the basal transcript level and the quantified synergy metric was calculated across the twenty-nine cell lines (Table S2). These correlation coefficients were then ranked (Figure 4B), and the ranked gene list used as input to GSEA (Subramanian et al., 2005), resulting in a list of biologically coherent gene sets whose elevated expression is associated with a greater or lesser degree of synergy. Identification of multiple gene sets related to mitotic progression, and more specifically to the mitotic spindle, readily emerged from this analysis. Members of these gene sets were enriched among genes whose increased expression correlated with decreased synergy between TH588 and BI2536. Of these, 11 of the 20 top-ranking gene sets were related to mitosis and 6 were specific to the mitotic spindle (Table S3). These mitosis and mitotic spindle gene sets were not observed to dominate the resistance to the individual drugs alone, and instead elevated expression of genes in these gene sets specifically correlated with reduced synergy of the combination (Figures 4C–D).
Plk1 inhibition and TH588 treatment synergistically disrupt mitotic spindle formation and metaphase chromosome alignment.
Our observation that genes whose expression anticorrelated with synergy were members of gene sets annotated as mitotic or spindle-related led us to hypothesize that the synergy between Plk1 inhibitors and TH588 results from synergistic disruption of certain key mitotic processes (Figures 4C–D). To explore this hypothesis, we examined the cell cycle profile of cells treated with the individual drugs or the combination using flow cytometry and phospho-histone H3 (pHH3) staining. As shown in Figure 5A, the BI2536/TH588 combination resulted in a marked synergistic mitotic arrest, with nearly 70% of cells staining positively for pHH3 16 hours post drug addition (Figure 5B). Plk1 inhibition alone with this dose of BI2536 resulted in a modest amount of mitotic arrest, as expected. TH588 treatment by itself resulted in the accumulation of mitotic cells to a similar extent to that seen with Plk1 inhibition (Figure 5B), rather than an accumulation of cells in G1 or S-phase, as might have been expected if the main cytotoxic target of TH588 was MTH1, an enzyme involved in maintaining the fidelity of DNA replication.
Figure 5. Plk1 inhibition and TH588 synergistically arrest cells in mitosis, disrupt spindle formation and prevent metaphase chromosome alignment.
(A) C4–2 CRPC cells were treated for 16 hours with 5 nM BI2536, 5 μM TH588, or both in combination, and mitotic arrest measured by flow cytometry using DAPI and pHH3 antibody staining. Red boxes indicate the mitotic population.
(B) Time course of pHH3+ cell accumulation caused by either BI2536 or TH588 alone, or their combination. C4–2 cells were fixed at the indicated time and stained as above. Mean ± SEM (n = 3).
(C) C4–2 cells were treated with a dose-response matrix of BI2536 and TH588 for 16 hours and defects in the assembly of the mitotic spindle were assessed by deconvolution microscopy. Pericentrin (red) and tubulin (green) were detected by immunofluorescence; DNA was stained with DAPI (blue). Shown are representative projections of deconvoluted Z-stacks from a subset of the dose-response matrix. (A 2.5 nM BI2536 column and 2.5 μM TH588 row are omitted for brevity, shown in Figure S4A). White arrows indicate the appearance of multipolar spindles and polar chromosomes. Scale bars at bottom right of each matrix indicate 10 μm. Top and bottom panels show identical fields of cells, with DAPI and pericentrin staining shown in top panels and DAPI, pericentrin, and tubulin staining shown in bottom panels.
(D) Mitotic spindle defects caused by each dose combination in the matrix described above were categorized based on phenotype. ‘Normal mitotic figures’ includes prophase, prometaphase, metaphase, anaphase and telophase cells. Mitotic cells with ‘polar or lagging chromosomes’ contained chromosomes that either overlapped the plane of a centrosome, or were positioned between the centrosome and the cell cortex. ‘Collapsed spindles’ were characterized by centrosomes surrounded by a cloud of chromosomes and tubulin polymers radiating from the center of the cell. The above three groups are mutually exclusive, but may co-occur with ‘excess pericentrin foci’. Between 61 and 134 mitotic cells were categorized per condition and their frequencies are depicted in heat maps (left). Representative micrographs for each of these categories are shown (right). Scale bar in the bottom right image represents 5 μm.
More specifically, the VISAGE approach pointed toward a particular role for the mitotic spindle in determining sensitivity to the combination of TH588 with Plk1 inhibition (Figure 4D). To further examine this, C4–2 cells were treated with a dose matrix of BI2536 and TH588 for 16 hours, fixed, and stained for tubulin, pericentrin, and DNA (Figures 5C and S4A). As expected, Plk1 inhibition led to an accumulation of prometaphase cells and collapsed spindles (Steegmaier et al., 2007; Sumara et al., 2004). When cells were treated with low dose TH588 (5 μM), the majority of mitotic cells (70.9%) contained supernumerary pericentriolar foci. Many of these supernumerary pericentrin foci, however, lacked centrosomes as judged by the absence of GFP-tagged centrin-1 colocalization (Figure S4B). Higher doses of TH588 caused additional spindle defects, including a clear defect in chromosome alignment along the metaphase plate, with a concomitant accumulation of polar chromosomes and astral microtubules. When the frequency of normal or aberrant mitotic spindles was accurately quantified using Z-stack image analysis (Figures 5D and S4C), we observed a synergistic induction of polar and lagging chromosomes at intermediate doses of each drug (4.9%, 12.9%, 9.3% and 41%, for DMSO, 2.5 nM BI2536, 2.5 μM TH588, and their combination, respectively).
It is important to note that the specific mitotic spindle phenotypes associated with the two individual drugs are distinct. Both drug treatments resulted in chromosome alignment defects accompanied by the appearance of polar chromosomes. However, strong Plk1 inhibition induced spindle collapse, whereas TH588 monotherapy resulted in a distorted spindle morphology characterized by more than two pericentrin foci, and unfocused or multiple poles (Figure 5D). The addition of a Plk1 inhibitor to TH588 nearly completely suppressed the supernumerary pericentrin foci induced by TH588 alone. Thus, at doses where strong cytotoxic synergy is observed, the prevailing phenotype is different from that of either drug alone, and is characterized by accumulation of polar chromosomes without excess pericentrin foci.
Synergistic cell killing after Plk1 inhibition and TH588 treatment occurs following prolonged mitosis and requires the spindle assembly checkpoint.
To determine whether these mitotic defects induced by TH588 were the cause of its toxicity and synergistic killing when combined with Plk1 inhibition, and further characterize the mechanism of cell death, we used live-cell microscopy to track cellular division and the fate of individual treated cells. C4–2 CRPC cells expressing histone H2B-mCherry and mEmerald-tubulin were imaged at 15-minute intervals over 40 hours using a combination dose matrix of BI2536 and TH588. A subset of drug-treated cells underwent a prolonged period of mitotic delay, followed by cell death during or immediately after mitotic exit (Figures 6A and S5A–S5E). A smaller percentage of cells underwent an abnormal cell division resulting in daughter cells with multiple decondensed chromatin bodies, or apparent exit from mitosis on the basis of chromosome decondensation without division (multinucleation and mitotic slippage, respectively (Vitale et al., 2011)), also generally followed by cell death (Figures S5B and S5C). These phenotypes were markedly enhanced by low doses of the drugs in combination, and resulted in a synergistic increase in the frequency of extended mitotic arrest with nearly uniform death of the arrested cells (Figures 6A–B). When viewed over time, the mitotic spindle defects caused by this drug combination were quite dynamic, with non-aligned chromosomes appearing to constantly alternate between the metaphase plate and the spindle poles, a situation that was commonly followed by spindle collapse and cell death (Figures S5D–E and Movies S1–S4). Based on our analysis of time-lapse microscopy, the combination of BI2536 and TH588 does not primarily induce a defect in chromosome congression, but instead results in a failure of stable chromosome alignment at the metaphase plate following initial congression similar to that seen in cells depleted of hNuf2 and HSET (Cai et al., 2009). Plk1 inhibition alone was associated with spindle collapse and death in mitosis, whereas TH588 treatment alone resulted in multinucleation, mitotic slippage, and death during or following mitosis. In cells treated with the drug combination, there was a synergistic increase in the fraction of cells killed overall. We found no indication of cell death caused by the individual drugs or the combination that was not associated with attempted mitosis (Figures 6A–B).
Figure 6. Cell death from TH588/BI2536 co-treatment occurs by prolonged mitotic arrest, requires the SAC, and is MTH1-independent.
(A) Quantification of phenotypic outcomes of C4–2 CRPC cells expressing mEmerald-tubulin and H2B-mCherry treated with a dose-response matrix of BI2536 and TH588, and imaged at 15 min intervals. Each bar represents a single cell, with color indicating its phenotype/cell cycle state over the time course of the experiment. Thirty cells that initiated and exited mitosis during the time-lapse were analyzed per condition.
(B) Violin plots depicting the distribution in duration of mitosis of C4–2 cells from panel (A) above. Position of horizontal lines on the y-axis indicates duration of mitosis of individual cells.
(C) Disruption of the SAC by MAD2L1 knockdown ablates sensitivity to TH588 and entirely prevents synergy with BI2536. Cells were treated with the indicated drugs 48 hours after control or MAD2L1 siRNA transfection and assessed for viability at 5 days. Bottom left inset on the right graph shows immunoblots of lysates from cells 48 hours after transfection of the indicated siRNA, confirming loss of MAD2. Mean ± SEM (n = 3) is shown.
(D) HCT116 cells, or single-cell clones expressing a control guide RNA (AAVS1-1), the MTH1 KO 9–4, clone, and the same clone transduced with constructs containing HA-MTH1 or HAMTH1 E56A were treated with the indicated drugs for 16 hours, fixed, stained with DAPI and antibodies against pHH3, and analyzed by flow cytometry to assess the percentage of mitotically arrested cells. Mean ± SEM (n = 3) is shown.
(E) Parental HCT116 cells, as well as the AAVS1-1 and MTH1 KO 9–4 clones were treated with 5 μM TH588 for 16 hours, fixed and stained for DNA (DAPI; blue), pericentrin (red) and tubulin (green). Representative deconvolved Z-stack projections are shown. Scale bar at bottom right indicates 10 μm. White arrowheads indicate cells undergoing abnormal mitosis.
The observation of dynamic spindle defects and polar chromosomes upon TH588 treatment or co-treatment with BI2536 suggested a failure to satisfy the spindle assembly checkpoint (SAC), resulting in cell death. The SAC delays mitotic progression until proper microtubule-kinetochore attachment and tension between opposite poles of the spindle are achieved. Loss of MAD2 (gene name MAD2L1) disables the SAC, and thus prevents the mitotic arrest normally induced by spindle defects (Musacchio and Salmon, 2007). Accordingly, we found that knockdown of MAD2L1 with siRNA in C4–2 cells almost completely abrogated both sensitivity to TH588 and the synergy between TH588 and the Plk1 inhibitor (Figure 6C). Furthermore, inhibition of Aurora B, a kinase required for initiation of the SAC and for sensing proper tension between paired centromeres (Saurin et al., 2011), with barasertib also abrogated TH588 cytotoxicity in C4–2 cells (Figure S5F). These data indicate that nearly all of the TH588 cytotoxicity, as well as its synergy with BI2536, is mediated through its disruptive effects on the mitotic spindle as monitored by the SAC.
The mitotic aberrations in cells treated with TH588 are completely independent of MTH1.
Since TH588 clearly binds directly to and inhibits MTH1 in cells (Figure S1C), we next examined whether the observed spindle morphology and chromosome alignment defects were in any way caused by acute inhibition of MTH1 and the associated accumulation of oxidized nucleotides. For example, incorporation of oxidized dNTPs into newly replicated chromosomes could alter mitotic chromatin structure, while increased levels of 8-oxo-GTP or 2-OH-ATP could impair mitotic ATPases or GTPases such as kinesins and tubulin. The former possibility was eliminated by demonstrating that progression through S-phase was not required for the TH588-induced mitotic defects (Figure S5G). The latter possibility was addressed by showing that 8-oxo-GTP did not block GTP-induced tubulin polymerization in vitro (Figure S5H), along with the observation that treatment of cells with another MTH1 inhibitor, (S)-crizotinib, did not result in mitotic arrest (Figure S5I). Finally, we looked for mitotic phenotypes in MTH1 knockout cells and those re-expressing either active HA-MTH1 or the catalytically inactive HA-MTH1 E56A mutant (Figure 2D–E). The presence or absence of MTH1 did not alter the accumulation of pHH3-positive cells either by TH588 treatment alone, or by co-treatment with TH588 and BI2536 (Figure 6D). Furthermore, while the parental cells, control clone AAVS1-1, and the MTH1 KO 9–4 clone all had normal mitotic spindles when left untreated, each of these cell types also accumulated the characteristic spindle defects described in Figure 5 when treated with TH588 (Figure 6E). Moreover, using live-cell microscopy we observed the same mitotic spindle and cell death phenotypes and synergistic disruption of mitotic progression with equal frequency in the MTH1 KO 9–4 clone, with or without expression of HA-MTH1 or the catalytically dead variant HA-MTH1 E56A (Figure S6A). MTH1 inhibition therefore makes no contribution to the observed mitotic arrest and spindle defects following TH588 treatment.
Cell killing by TH588 results from inhibition of tubulin polymerization through binding to the colchicine binding site.
TH588 is a modified aminopyrimidine analog (Figure 7A) that competes with dNTPs and NTPs for binding to MTH1 (Wang et al., 2017). This drug’s effects on the mitotic spindle might therefore arise from similar competitive inhibition of one or more NTPases. The kinesin motor CENP-E localizes to kinetochores and uses ATP hydrolysis to facilitate chromosome congression by capturing the plus end of kinetochore-associated microtubules. Loss of CENP-E activity causes an accumulation of polar chromosomes during mitotic spindle formation (Kapoor et al., 2006), and the spindle defects induced by a CENP-E inhibition (Wood et al., 2010) closely resemble those caused by TH588 treatment (Figure S6B). TH588, however, did not inhibit microtubule-dependent ATP hydrolysis by the purified CENP-E motor domain (Figure S6C), and combined treatment of cells with GSK923295 (CENP-E inhibitor) and BI2536 (Figure S6D) did not recapitulate the strong synergy between BI2536 and TH588, indicating that CENP-E is not likely to be the relevant target of TH588.
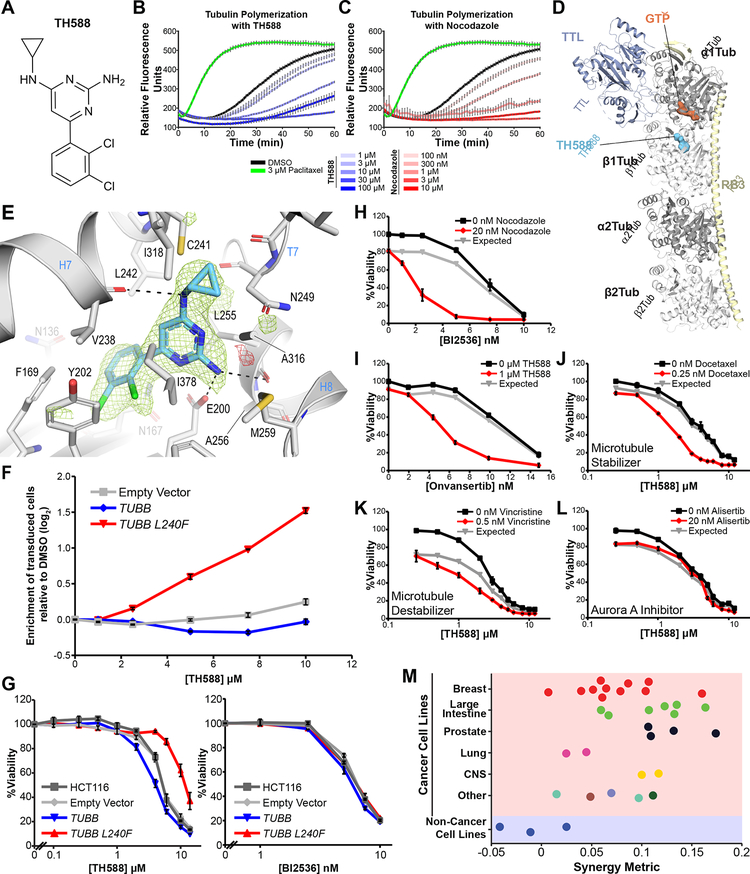
Figure 7. TH588 induces cell death by binding to the colchicine binding site in tubulin, inhibiting microtubule polymerization, and specifically synergizing with Plk1 inhibtors in cancer cells.
(A) Chemical structure of TH588.
(B, C) Microtubules were assembled from purified porcine a/b tubulin by incubation at 37°C in the presence of 1 mM GTP and varying doses of TH588 (B) or nocodazole (C), and tubulin polymerization measured by time-dependent fluorescence-enhancement of the microtubule indicator dye, 4’,6-Diamidino-2-phenylindole (Bonne et al., 1985). Paclitaxel was used a positive control for microtubule stabilization. Shown are time course experiments, where the relative fluorescent signal indicates the quantity of polymerized tubulin. Mean ± SEM (n = 3).
(D) Overall view of the T2R-TTL-TH588 complex structure at 2.3Å resolution. The a-tubulin and β-tubulin chains are in dark and light grey, TTL is in powder blue and RB3 is in pale yellow ribbon representation, respectively. The tubulin-bound TH588 in the colchicine binding site and the GTP molecule in the N-terminal binding domain of α-tubulin are shown in surface representation. The carbon atoms are colored in light blue (TH588) and dark orange (ATP), respectively.
(E) Close-up view of the atomic interaction network observed between TH588 (light blue) and tubulin (gray). Interacting residues of tubulin are shown in stick representation and are labeled. Oxygen and nitrogen atoms are colored red and blue, respectively; carbon atoms are in light blue (TH588) or gray (tubulin). Hydrogen bonds are depicted as black dashed lines. Secondary structural elements of tubulin are labeled in blue. The simulated annealing mFo-DFc omit map highlighting the shape of TH588 is contoured at +3.0 σ (green mesh) and −3.0 σ (red mesh), respectively.
(F) HCT116 colorectal cancer cells were transduced with empty vector pHR-SFFV-IRES-mCherry or the same vector containing either wildtype TUBB or the mutant TUBB L240F. These cells were then mixed one to one with untransduced HCT116 cells and subjected to increasing concentrations of TH588. After 72 hours the relative proportion of mCherry positive and negative cells was determined by flow cytometry. Enrichment was defined as the ratio of mCherry+/mCherry− for each sample normalized to the geometric mean of DMSO control samples and log2 transformed. Mean ± SEM (n = 3) is shown.
(G) HCT116 colorectal transduced as in panel (F) were subjected to increasing concentrations of the TH588 (left) or BI2536 (right) and assessed for viability after 5 days. Mean ± SEM (n = 3).
(H) C4–2 CRPC cells were treated with increasing doses of BI2536 in the presence (red lines) or absence (black lines) of the known microtubule polymerization inhibitor, nocodazole (top), or TH588 (bottom). Relative viability was measured at 5 days. The Bliss independence model of drug additivity was used to calculate expected relative viability (gray lines) and identify drug synergy. Mean ± SEM (n = 3) is shown.
(I) Onvansertib, a structurally distinct and highly specific Plk1 inhibitor, was combined with TH588 to assess synergy in C4–2 CRPC cells. Data was acquired, analyzed and plotted as in H.
(J-L) C4–2 CRPC cells were subjected to a dose-response matrix of the MTH1 inhibitor, TH588, and one of several antimitotic drugs (docetaxel (J), vincristine (K), or alisertib (L)) for 5 days. Shown are representative TH588 dose-response curves in the absence (black line) or presence (red line) of the indicated antimitotic drug. Gray line is expected viability according to the Bliss independence model of drug additivity. Mean ± SEM (n = 3) is shown. The dose of the antimitotic shown was chosen based on a 10–20% decrease in relative viability when that drug was used as a single agent.
(M) Swarm plot showing the degree of synergy observed in individual cell lines separated by tissue of origin and transformation status. On the top (red area), are cancer cell lines from the indicated tissues of origin, on the bottom are non-cancer cell lines (blue area).
The main structural constituent of the spindle, tubulin, is also a GTPase, raising the possibility that the observed TH588 effects on the mitotic spindle might arise from altered microtubule polymerization. Indeed, we found that increasing doses of TH588 markedly decreased the polymerization rate of purified α/β tubulin in vitro (Figure 7B) in a manner similar to that of nocodazole (Figure 7C), a known microtubule polymerization inhibitor. These findings are consistent with a recent report published while these studies were ongoing (Kawamura et al., 2016). To elucidate the molecular basis by which TH588 inhibits tubulin polymerization, and determine the binding mode of TH588 within the α/β-tubulin heterodimer, we soaked the compound into crystals formed by a protein complex composed of two bovine α/β-tubulin heterodimers, the stathmin-like protein RB3, and tubulin tyrosine ligase (termed T2R-TTL) (Prota et al., 2013a) and solved the structure of TH588 in complex with T2R-TTL by X-ray crystallography to 2.3 Å resolution (Figure 7D; Supplemental Table S4). We observed that TH588 binds to the colchicine binding site of tubulin, which is formed by residues of the strands βS8 and βS9 and helix βH8 of β-tubulin and the loop αT5 of α-tubulin (Figure 7E) at the intradimer interface.
The 2,3-dichlorophenyl ring of TH588 is buried into a hydrophobic pocket formed by the side chains of β-tubulin residues βPhe169, βTyr202, βVal238, βAsn167, βLeu242, βGlu200, βGln136, and βIle378 (Figure 7E). Moreover, the 2,4-amino-substituted pyrimidine core is stacked between the side chains of βLeu255 and βIle378, and is further stabilized by hydrogen bonds formed by the N3 nitrogen to OD1 of βAsn249 and by the 2-amino moiety to both the side chain of βGlu200 and the main chain carbonyl of βLeu255. Two additional hydrogen bonds are formed by the 4-amino moiety to both the main chain of βVal238 and the side chain of βCys241. The interaction is completed by hydrophobic contacts of the N4-cyclopropyl moiety to beta strand S8 and S9 residues βAla316, βIle318 and βAla354. The side chain of βTyr202 adopts a different orientation than the ones observed in all other structures of colchicine site binding agents (reviewed in (Steinmetz and Prota, 2018)), and in microtubules (PDB: 5SYF) (Supplemental figure S7A). Superpositions of the T2R-TTL-TH588 structure onto the T2R-TTL-colchicine (PDB: 4O2B, chains A,B, rmsd=0.267, 795 Cα-atoms), T2R-TTL-nocodazole (PDB: 5CA1, chains A,B, rmsd=0.251, 795 Cα-atoms) and the T2R-TTL-apo (PDB: 4I55, chains A,B, rmsd=0.268, 773 Cα-atoms) structures reveal a closer overlap of TH588 to nocodazole in zone 3 of the colchicine site (Massarotti et al., 2012) and almost no overlap with colchicine (Supplemental figure S7B). Moreover, both the aT5- and the βT7-loop conformations in the TH588 structure are closer to the ones observed in the T2R-TTL-apo structure (Supplemental figure S7C). These observations suggest that TH588 prevents tubulin straightening by perturbing the compaction in the deeply buried zone 3 of the colchicine site.
To validate that TH588 binding to the colchicine binding site of tubulin is responsible for its cell killing effects and its synergistic interaction with Plk1 inhibitors, we made use of a β-tubulin mutant, L242F, in the drug binding pocket. These studies were facilitated by a recent report from Jost et al. that elegantly demonstrated binding of the drug rigosertib to this T2R-TTL complex at the colchicine binding site and subsequently identified the specific L242F β-tubulin mutant (L240F in human TUBB) which, when expressed in cells, conferred resistance to rigosertib (Jost et al., 2017). Our T2R-TTL−TH588 complex structure shows that the side chain of βLeu242 lies in close proximity to the 2,4-diaminopyrimidine ring and the back of the 2,3-dichlorophenyl ring, and would be expected to result in modest steric hindrance of drug binding upon mutation of this leucine residue to phenylalanine.
We performed a competition assay for cell growth using parental HCT116 cells versus HCT116 cells expressing either an empty control vector, wild-type TUBB, or the TUBB L240F mutant in the presence of increasing concentrations of TH588. As shown in Figure 7F, there was a clear enrichment in net proliferation of cells expressing the mutant form of β-tubulin when grown in the presence of TH588. Furthermore, expression of wild-type TUBB resulted in a modest depletion of cells following TH588 treatment relative to cells transduced with empty vector alone, consistent with tubulin-TH588 complex being the active cytotoxic species. Moreover, direct assays of cell viability confirmed both a clear decrease in sensitivity to TH588 when TUBB L240F is expressed, and a modest increase in sensitivity when wild-type TUBB is expressed (Figure 7G, left panel). When synergy between BI2536 and TH588 was assessed in TUBB L240F-transduced cells, a synergistic response remained, but required higher doses of TH588 (Figure S7D). No change in sensitivity to the Plk1 inhibitor was observed in cells expressing either the wild-type or mutant forms of β-tubulin (Figure 7G, right panel). Together, these data demonstrate that the cellular effects of TH588, and its synergy with Plk1 inhibitors, results from inhibition of tubulin polymerization through binding to the colchicine binding site in β-tubulin.
Synergistic cell killing is specific to combined Plk1 inhibition and anti-microtubule drugs, and targets a cancer-specific vulnerability.
To further validate that inhibition of tubulin polymerization synergistically enhances the cytotoxicity of Plk1 inhibitors, we next examined the combination of BI2536 with low doses of nocodazole, a well-established microtubule poison. Indeed, the combination of BI2536 and nocodazole phenocopied the synergy observed between BI2536 and TH588 (Figure 7H), and nocodazole doses showing equivalent effects on viability resulted in spindle defects resembling those caused by TH588 (Figure S7E). Some synergy was also observed when BI2536 was combined with the microtubule stabilizing drug docetaxel, although the effect was not as pronounced as that seen when BI2536 was combined with tubulin polymerization inhibitors (Figure S7F). Moreover, the contribution of BI2536 to this synergy phenotype appears to be entirely mediated by the activity of BI2536 on Plk1, since TH588 also synergizes with the highly specific Plk1 inhibitor onvansertib/PCM-075 (Figure 7I), as well as volasertib and GSK461364, which represent a set of structurally distinct Plk1 inhibitors (Figure S7G–H). In contrast, when TH588 was combined with other antimitotic agents including the microtubule stabilizer docetaxel, the microtubule destabilizer vincristine, or the Aurora A inhibitor alisertib, synergy was markedly lower or absent (Figure 7J–L). Taken together, these results indicate that the synergistic cell killing effect is specific to the combination of Plk1 inhibition and anti-microtubule agents, particularly inhibitors of microtubule polymerization, and is not a general phenomenon of simply combining any two antimitotic agents.
When we evaluated the synergy between BI2536 and TH588 across a broad panel of cancer cell lines and three non-transformed lines, we observed a marked synergy in the majority of cancer cell lines, whereas there was minimal or no synergy in non-cancerous cells (Figure 7M). Thus, it appears that one or more features of the mitotic spindle unique to cancer cells makes them sensitive to the combination of Plk1 inhibitors and anti-microtubule drugs.
DISCUSSION
In a search for new synergistic drug treatments that could enhance tumor cell-specific killing by antimitotic drugs (Dominguez-Brauer et al., 2015), we attempted to leverage the observation that freely cycling mitotic cancer cells have elevated levels of ROS and biomolecule oxidation, which are even further elevated in mitotically-arrested cancer cells (Patterson et al., 2019). We focused on TH588, a well-publicized inhibitor of the nudix hydrolase MTH1, a derivative of which is currently in clinical trials (Gad et al., 2014) (NCT: ), in combination with inhibitors of the mitotic kinase Plk1. Although we observed very strong synergistic tumor cell killing by this drug combination in a large subset of cancer cell lines in vitro, and in a human tumor xenograft in vivo, the effect appeared to be completely independent of both ROS and MTH1 function, as shown by the persistent synergistic tumor cell killing despite the addition of antioxidants, or following the complete loss of MTH1 by CRISPR-mediated gene editing. Thus, while MTH1 may be important for the prevention of DNA damage-induced cancer (Speina et al., 2005; Tsuzuki et al., 2001), and degenerative neurological diseases (Yamaguchi et al., 2006), it seems that, despite previous reports (Rai et al., 2009), MTH1 is neither necessary for cancer cell proliferation, nor the critical target of TH588 responsible for tumor cell killing (Gad et al., 2014). Despite this lack of involvement of MTH1 in synergistic killing by TH588 in combination with Plk1 inhibitors, however, we still believe that co-targeting of other molecules involved in antioxidant or oxidative stress responses, such as OGG1 (Ba and Boldogh, 2018) or Nrf2 (Shim et al., 2009), may provide a mechanism for enhancing the ability of anti-mitotic drugs to specifically kill tumor cells, given the fact that cancer cells in general appear to have elevated levels of ROS (DeNicola et al., 2011; Szatrowski and Nathan, 1991), and our finding that tumor cell killing by Plk1 inhibitors depends, at least in part, on ROS (Figure 2A).
To contend with a synergistic drug combination in which both the drug target and the mechanism for synergy were unknown, we then devised a combined experimental and computational approach, VISAGE, that identifies gene sets associated with degree of synergy using a panel of cancer cell lines. In contrast to the limitations inherent to single gene correlation analyses, VISAGE uses the robustness of pathway-level analysis (Leiserson et al., 2015) to identify biologically coherent gene expression patterns associated with synergistic drug response. Furthermore, the approach uses a very generalized synergy metric that is readily scalable to cell line screens and incorporates all classes of greater-than-additive therapeutic efficacy for drug combinations (see STAR Methods and (Foucquier and Guedj, 2015)). What we believe is particularly compelling about this approach is the observation that quantitative phenotypic data from modestly sized cell line panels can yield insight about synergistic mechanisms. Equally important to understanding mechanisms that determine drug synergy is the ability to identify patients who will benefit from a particular drug combination. No cancer therapy will be universally effective, and without the ability to stratify patients prior to treatment, many potentially useful therapies will fail to show significant therapeutic effects (Sawyers, 2008). Genomic analysis of tumor samples for patient stratification, however, has identified specific actionable mutations that dictate treatment only in a minority of patients (Mendelsohn, 2013; Sparano et al., 2013). Because VISAGE identifies gene sets whose members’ expression levels are associated with the degree of synergy, without depending on the particular expression level of any individual gene, the VISAGE approach also has the potential to identify pathway-level biomarkers that might associate with efficacy of combination therapy in a clinical setting.
In the case of combined TH588-Plk1 inhibition, VISAGE clearly implicated the mitotic spindle as the mechanistic focal point responsible for synergistic cell killing. This was experimentally validated by demonstrating quantitative defects in metaphase chromosome alignment after combined drug treatment that resulted in prolonged mitotic arrest and SAC-dependent cell death. Our subsequent demonstration that TH588 directly inhibits tubulin polymerization in vitro, confirming a recent report (Kawamura et al., 2016), together with our X-ray crystal structure of the α/β-tubulin:TH588 complex showing that the drug binds directly to the colchicine binding site in tubulin, is consistent with this interpretation. The fact that mutation of the TH588 binding site in β-tubulin results in resistance to TH588 cytotoxicity definitively establishes that binding of the drug to tubulin is the predominant mechanism responsible for TH588-induced cell death.
It is important to note that the synergistic cell killing induced by TH588 and a variety of Plk1 inhibitors that we observed is not equivalent to a “double dose” of any single antimitotic agent, nor is it a generic effect of generalized combinations of any pair of antimitotic drugs. Instead, Plk1 inhibition and microtubule destabilization target independent processes in mitosis that appear to synergistically disrupt spindle formation and cell viability in cancer cells. We have made three observations that support this notion. First, these drugs in isolation elicit distinct spindle defects and mitotic phenotypes (see Figure 5D). Second, sensitivity to BI2536 was slightly correlated with sensitivity to TH588, and the amount of synergy observed did not correlate with TH588 sensitivity alone, and only weakly correlated with sensitivity to BI2536 alone (Figure 4A). This indicates that the component of TH588 cytotoxicity that correlates with sensitivity to Plk1 inhibition is separate from that which causes synergy, an observation that is exemplified by HUVECs, which are sensitive to each of the individual drugs but show no synergy. Third, TH588 did not synergize with the antimitotic drug alisertib, an Aurora A inhibitor, while BI2536 did not synergize with CENP-E inhibitors despite their induction of spindle defects that closely resemble those caused by TH588 treatment. Instead, substantial synergy occurs due to specific effects of TH588 and inhibitors of Plk1. Synergy between particular members of these two classes of anti-mitotic drugs - Plk1 inhibitors and microtubule poisons - has been recently observed in a few specific cancer types, including CCNE-amplified high grade serous ovarian cancer, neuroblastoma, rhabdomyosarcoma, and Ewing’s sarcoma (Czaplinski et al., 2016; Hugle et al., 2015; Noack et al., 2018; Weiss et al., 2016). Our results now demonstrate strong synergy specific to the combination of inhibitors of Plk1 and of microtubule polymerization that likely functions by targeting an inherent mitotic vulnerability present in a wide variety of cancer cells of disparate origin. The long-standing observation that tumor cells tolerate aneuploidy and are genetically unstable indicates that the fidelity of mitosis has been fundamentally compromised (Kops et al., 2005), which almost certainly contributes to this cancer-specific vulnerability.
It is interesting, in this regard, that docetaxel, a microtubule stabilizing agent that binds tubulin at an alternative site, did not induce the same extent of synergy with the Plk1 inhibitor as either nocodazole or TH588, both of which bind at the colchicine binding site and, like colchicine, almost certainly form drug-tubulin copolymers that display altered microtubule assembly kinetics (Lu et al., 2012; Sternlicht et al., 1980). The reason why this specific drug combination is particularly effective in cancer cells remains unclear, but may relate to the combination of altered dynamics of spindle microtubule polymerization, in combination with inhibition of Plk1-activated chromosome motor proteins responsible for metaphase congression (Grosstessner-Hain et al., 2011; Santamaria et al., 2011; Wynne and Funabiki, 2015). Taken together, these findings identify a targetable vulnerability that is specific to cancer cells, and for which future work exploring the details of this mechanism, and the combination’s clinical utility seem warranted.
STAR METHODS
CONTACTS FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Michael Yaffe (myaffe@mit.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
All cell lines were incubated at 37°C in a humidified incubator supplied with 5% CO2, maintained in subconfluent culture, and used for ≤ 20 passages. Thirty-two of thirty-four cell lines assayed for synergy in this study were authenticated by short tandem repeat genotyping (Genetica Cell Line Testing, LabCorp). HUVECs are pooled primary cells and have no reference STR profile. One of the cell lines used, KYSE150, was extinguished and not available for authentication. Re-analysis of the cell line screen data using VISAGE without data on KYSE150 cells did not significantly alter any of the results or conclusions of this study. All media was supplemented with 10% fetal bovine serum (FBS) and 2 mM glutamine, and lacked antibiotics unless otherwise specified for the selection of transduced cells. CCD-18co, MDA-MB-231, U-2 OS, A549, BT-20, MDA-MB-415, MDA-MB-157, U-87 MG, CFPAC1, HCT116, MDA-MB-468, T98G, MCF7, SW48, MDA-MB-453, MDA-MB-436, HT55, HT-29, HeLa, HEK293t and CCD-1112sk cells were grown in DMEM medium. C4–2, NCI-H1299, ZR-75–1, KYSE150, HCC38, HCT-15, 22RV1, LNCaP, AU565, CAL33, PC-3, LOVO and COLO205 cells were grown in RPMI-1640 medium. HUVEC cells were grown in EBM medium with EGM™−2 Bulletkit™.
METHOD DETAILS
Flow cytometry measurement of cell cycle stage and apoptosis
Cells were treated with drug or vehicle control as described and then collected by trypsinization. The media and PBS wash solutions were combined with the trypsinized cell pellet to retain any detached cells. Cells were fixed with 4% formaldehyde in PBS for 15 minutes, washed with PBS containing 1% bovine serum albumin (PBS-BSA), resuspended in −20°C methanol, washed twice with PBS-BSA containing 0.1% Tween-20, and incubated with primary antibodies overnight at 4°C. Cells were then washed with PBS-BSA 0.1% Tween-20 prior to a 1 hour-long room temp incubation with fluorescent dye-conjugated secondary antibodies diluted 1:200 (Alexa Fluor®, Molecular Probes). Finally, cells were washed twice with PBS-BSA 0.1% Tween-20, resuspended in PBS containing 1 μg/ml 4,6-diamidino-2-phenylindole (DAPI, Molecular Probes), and analyzed using a BD™ LSRII flow cytometer (Becton Dickson) and the FlowJo® software package.
Animal studies using the microwell tumor-implantable device
C4–2 prostate cancer xenograft tumors were grown in four- to six-week old castrated male NCR nude mice. Five million C4–2 cells were suspended in serum free media, mixed 1:1 with growth factor reduced matrigel (Invitrogen) to a total volume of 200 μl, and injected subcutaneously into the hind flank using a 23 gauge needle. Prior to injection, IMPACT rodent pathogen testing (IDEXX Bioreseach) was used to ensure cells were free of murine pathogens. Tumors took four to eight weeks to grow.
Microdose drug delivery devices were manufactured, implanted, and analyzed as previously described (Jonas et al., 2015). In brief, cylindrical microdevices (4 mm × 820 μm) were micromachined (CNC Micromachining Center) from medical-grade Delrin® acetal resin blocks (DuPont) to contain eighteen 200 μm (diameter) × 250 μm (depth) reservoirs. Drugs were mixed with PEG 1450, and 1 μg of the dry powder mixture then packed into a reservoir using a tapered metal needle (Electron Microscopy Science). One reservoir of each device contained doxorubicin, whose autofluorescence served as a fiduciary mark for orienting the rest of the wells. Implantation of the devices was performed using a 19-gauge spinal biopsy needle (Angiotech) with a retractable needle obturator. Tumors were excised 24 hours after device implantation, fixed in 10% formalin for 24 hours, and embedded in paraffin. Sections were stained with cleaved caspase-3 antibody (Cell Signaling Technologies, 9664) followed by detection with horseradish peroxidase conjugated secondary antibody and diaminobenzidine with hematoxylin used as a counterstain, following standard immunohistochemistry techniques. Images were viewed using a EVOS® Cell Imaging System (Invitrogen) microscope, and scored using ImageJ (Schindelin et al., 2015) in a blinded manner. Apoptotic index (AI) was calculated as the percentage of cells that were cleaved caspase-3–positive within 400 μm of the reservoir–tissue interface.
All mouse studies were approved by the Massachusetts Institute of Technology Committee for Animal Care or Beth Israel Deaconess Institutional Animal Care and Use Committee, and conducted in compliance with the Animal Welfare Act Regulations and other federal statutes relating to animals and experiments involving animals and adheres to the principles set forth in the Guide for the Care and Use of Laboratory Animals, National Research Council, 1996 (Institutional Animal Welfare Assurance #A-3125–01).
Viability measurements and cancer cell line screen
Cells were plated in 384-well plates at a density of 1000 cells per well in 22.5 μl of media containing 10% FBS. Rapidly growing cell lines, such as HCT116, were plated at 500 cells per well. The following day, 2.5 μl of drug, diluted in media, was added to each well. Following 5 days of incubation, viability was assessed using CellTiter-Glo™ (Promega) according to the manufacturer’s recommendations. Luminescence in each well was measured using an Infinite™ M200 Pro plate reader (Tecan Group Ltd.). Relative viability was determined by dividing the luminescent signal of each well to that of the vehicle-treated well for each replicate. To assess cell number and relative confluence, drug-treated cells were washed with PBS, fixed for one hour using 4% formaldehyde, stained with 1 μM SYTO 60 (Molecular Probes) and imaged using an Odyssey™ CLx scanner (LiCOR Biosciences).
VISAGE experimental-computational pipeline for identifying mechanistic determinants of drug combination synergy
The overall goal of our VISAGE strategy is to quantify the synergy observed in a dose-response matrix as a single value, and then to leverage inherent heterogeneity between cell lines in the basal state to make biologically coherent predictions about the mechanisms that underlie the synergy. The Bliss independence volume definition of synergy that we use is graphically straightforward and is defined as the volume between expected and observed dose-response surfaces in logarithmic space (Figure 3A). This approach has several advantages in the context of a drug-combination cell line screen. First, dose-effect definitions of synergy (in which combinations of drugs are always compared to individual drug concentrations of equivalent effectiveness) such as Loewe additivity (Greco et al., 1995) require accurate curve-fitting of the data. When screening cell lines, one must make assumptions about the appropriate dose regimes to be used. Accurate curve fitting is not possible when a fixed dose window does not capture the entire range of response of a particular cell line to a drug or drug combination. Expanding the dose range (and the distance between doses) may result in sparse data within the appropriate windows for accurately fitting individual response curves in addition to sampling the synergistic regime. The scale of a screen increases dramatically as the number of doses examined for each drug increases. Second, it is often the case that particular cell lines are entirely resistant to one or even both drugs until they are combined, precluding single drug dose-response curve fitting entirely. While this has often been considered a separate class of drug interaction from “synergy” (Foucquier and Guedj, 2015), these interactions represent greater-than-additive effects that ideally should be incorporated into a cell line screen using a single method for quantification of drug synergy. Finally, dose-effect synergy definitions in the Loewe additivity model assume that adding one drug to the dose-response curve of a second drug changes only the potency (EC50) of the second drug with no other effect. This assumption is often false and drug combinations frequently have a greater-than-additive effects that are not taken into account with Loewe additivity (Foucquier and Guedj, 2015). Overall effect-based definitions of synergy in which drug combinations are compared to sensitivity to individual drugs at the same dose, such as Bliss independence, however, do not rely on curve fitting and can readily combine greater than additive effects on both potency and efficiency (Figures S3A–D). Moreover, our method limits quantification of synergy to a particular dose response matrix with the aim of maintaining clinical relevance and ensuring that any synergy observed is due to the dominant cytotoxic target effects of the drugs (Figure S3E). Our method for quantifying synergy is consistent with the analyses of others (Foucquier and Guedj, 2015), i.e. that Bliss independence is preferable and more appropriate in certain contexts.
Indirect immunofluorescence and live cell microscopy
For indirect immunofluorescence microscopy, 105 cells were plated on poly-L-lysine coated glass coverslips (Corning-BioCoat #354085) in 12-well plates and dosed with the relevant drugs the following day. Fixation and staining for mitotic spindles were performed at room temperature as described in Ganem et al. (Ganem et al., 2009). Cells were washed for 1 minute with microtubule stabilization buffer (MTSB: 4 M glycerol, 100 mM PIPES pH 6.9, 1 mM EGTA, 5 mM MgCl2), then for 2 minutes with MTSB containing 0.5% Triton X-100, then for 2 minutes with MTSB, and then fixed with 1% glutaraldehyde in PBS for 10 minutes. After fixation, samples were aspirated and residual glutaraldehyde was quenched with 0.5 mg/ml of NaBH4 in ddH2O for 12 minutes twice. Fixed cells were blocked in TBS-BSA (10 mM Tris-HCl pH 7.5, 150 mM NaCl, 5% BSA) containing 0.5% Triton X-100 for 1 hour. All subsequent washes and 1 hour-long primary and 1:200 secondary antibody (Alexa Fluor®, Molecular Probes) incubations were performed in TBS-BSA. Samples were stained for DNA using 5 μg/ml DAPI in PBS for 3 minutes, washed with PBS, and mounted on glass slides using the antifade reagent ProLong Gold™ (Molecular Probes). For localization of centrosomes, cells expressing eGFP-centrin-1 were cultured, treated with the indicated drugs, fixed and stained as above, using fixation with −20°C methanol for 10 minutes at 4°C instead of glutaraldehyde, then immediately transferred to TBS-BSA for blocking. Images were acquired using a Deltavision™ Ultimate Focus microscope (Applied Precision) at 60X. Z-stacks were deconvolved and maximum intensity Z-stack projections were generated using softWoRx™ software (Applied Precision). All subsequent image analysis was done using the Fiji distribution of ImageJ (Schindelin et al., 2012; Schindelin et al., 2015; Schmid et al., 2010). For acquisition of movies, 6000 C4–2 cells expressing mEmerald-Tubulin and H2B-mCherry or 10,000 HCT116-derived MTH1 knockout cells expressing H2B-mCherry were plated in wells of a black optical bottom 96-well plate in phenol red-free media. The next day, the indicated drugs were added to wells, followed immediately by imaging using the IncuCyte ZOOM® Live Cell Analysis System (Essen Bioscience). Multiple images per well were acquired at 20X magnification every 15 minutes for a period of 40 hours. Each series of images was aligned using Adobe Photoshop CC (Adobe Systems). Violin plots were generated using the Seaborn statistical data visualization python package (version 0.7.1, http://seaborn.pydata.org).
siRNA knockdown
Silencer® Select siRNAs (Invitrogen) targeting MAD2L1 (s8392) and Negative Control No. 1 were transfected into C4–2 cells using Lipofectamine® RNAiMAX (Thermo Fisher Scientific) at a concentration of 10 nM, according to the manufacturer’s recommendations. After 24 hours, transfected cells were trypsinized and re-plated into both 6-well and 384-well plates. Cells in 6-well plates were lysed after an additional 24 hours and analyzed for MAD2 expression by immunoblotting. The indicated drugs were added to the 384-well plates 48 hours after transfection and analyzed for viability 5 days later using CellTiter-Glo®.
Measurement of tubulin polymerization kinetics in vitro.
Rates of microtubule polymerization were measured using a fluorescence-based porcine tubulin polymerization kit (Cytoskeleton, Inc., #BK011P) according to the manufacturer’s instructions. This assay contains a dye that fluoresces when incorporated in microtubules. Polymerization of tubulin (1.58 mg/ml) was initiated by addition of 1 mM GTP in the presence of either DMSO, or varying concentrations of TH588, nocodazole, 8-oxo-GTP or additional GTP, or 3 μM paclitaxel, and incubation at 37°C. The final concentration of DMSO was maintained at 1% for experiments comparing TH588 with nocodazole, or 0.14% for experiments with GTP or 8-oxo GTP. Fluorescence was measured using every 60 sec for 1 hour using an Infinite™ M200 Pro plate reader (Tecan Group Ltd.) with an excitation wavelength of 360 nm and an emission wavelength of 450 nm.
In vitro assay of CENP-E motor domain activity
The kinesin motor domain of CENP-E (GenBank: NM_001813.2, amino acids 1–340) was amplified from a cDNA library and cloned into the NdeI and SalI sites of pET-28a(+) (Novagen, Inc.), resulting in a C-terminal 6XHIS tag fusion. This fusion protein was expressed in the Rosetta 2™ (Novagen, Inc.) E. coli strain by induction with 1 mM Isopropyl β-D-1-thiogalactopyranoside for 16 hours at 20°C. Cells were centrifuged 7000 x g for 10 minutes and washed with 10 ml cold PBS. Pellets were then resuspended in lysis buffer (100 mM Tris HCl pH 8.0, 1 mM dithiothreitol, 0.5% Tween-20, 10% glycerol, 0.2 mg/ml lysozyme) containing EDTA-free Complete Protease Inhibitors (Roche Applied Sciences, Inc.), incubated on ice for 30 min and lysed by sonication. Lysates were clarified by centrifugation at 12,000 x g for 10 minutes, 0.5 ml Ni-NTA agarose beads (Qiagen, Inc.) was added and incubated for 1 hour at 4°C. Beads were washed with three times with 5 ml wash buffer (20 mM Tris-Cl pH 8.0, 300 mM NaCl, 10% Glycerol) and then eluted with 1 mL wash buffer containing 300 mM imidazole. This fusion was dialyzed against buffer containing 20 mM Tris HCl pH 7.5, 100 mM NaCl, 10% Glycerol and 1 mM DTT overnight. Microtubule-dependent CENP-E motor activity was assayed by incubating 25 nM CENP-E motor domain in 15 mM PIPES at pH 7.0, 1 mM MgCl2, 20 μM docetaxel and 0.5 mM ATP in the presence or absence of 5 μM tubulin dimers as pre-formed microtubules (Cytoskeleton, Inc.) with 1 μM TH588, 1 μM of the CENP-E inhibitor GSK923295, or a DMSO control. The reaction was stopped by the addition of EDTA to a final concentration of 3 mM, and motor-dependent ATPase activity measured by BIOMOL Green (Enzo Life Sciences) assay of released PO4, according to manufacturer’s instructions.
Preparation of lysates, SDS-PAGE and Immunoblotting
Lysates for immunoblotting were prepared by washing cells with PBS and lysing directly on the plate with the addition of lysis buffer (50 mM Tris-HCl pH 6.8, 2% SDS, 5% glycerol, 5 mM EDTA, 1 mM NaF, 10 mM beta-glycerophosphate, 1 mM phenylmethylsulfonyl fluoride, 1 mM Na3VO4, protease and phosphatase inhibitors (Roche EDTA-free complete protease inhibitor tablets and PhosSTOP tablets)). Media and PBS from washed plates were reserved and centrifuged, and then the pellet (visible or not) was combined with the lysate to prevent loss of loosely attached cells. After brief sonication and normalization for total protein concentration, 6X sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer (208 mM Tris-Hcl pH 6.8, 42% glycerol, 3 M beta-mercaptoethanol, 10% SDS, 5 mg/ml bromophenol blue) was added and samples were boiled for 5 minutes. Immunoblots were blocked with Odyssey™ blocking buffer (LiCOR Biosciences) membranes were incubated with primary antibody overnight at 4°C, secondary antibody for one hour. The anti-MTH1 H-159 antibody was diluted 1:100 in PBS 0.1% Tween-20 and blots were incubated overnight at room temperature. All other antibodies used for immunoblotting were diluted in a 1:1 mixture of PBS 0.1% tween and Odyssey™ blocking buffer. Immunoblots were imaged using an Odyssey™CLx scanner (LiCOR Biosciences).
Vector construction, virus production and clonal cell line selection
HEK293T cells were transiently transfected with viral vectors and packaging plasmids using the CalPhos™ mammalian transfection kit (Clontech Laboratories). Packaging and structural vectors expressed VSVG and GAG/POL or GAG/POL/Delta R8.2 were used for production of retroviruses and lentiviruses, respectively. Viral supernatants were passed through a 0.45 μm filter and used to transduce the target cells in the presence of 8 μg/ml polybrene for three 1-day rounds of infection. Drug concentrations used for selection of HCT116 transductants were 2 μg/ml puromycin, 500 μg/ml G418, 200 μg/ml hygromycin B. For selection of C4–2 transductants 2 μg/ml puromycin and 400 μg/ml G418 were used. Selection of transduced MDA-MB-231 cells was accomplished with 100 μg/ml hygromycin B or 2 μg/ml puromycin. To obtain MTH1 knockout cells, the HCT116 cell line was transduced with SpCas9, from Addgene #50661 cloned into pLVX-IRES-Hyg (Clontech Laboratories), and then subsequently transduced with Lentiguide-Puro (Addgene #52963) constructs containing the relevant guide RNA sequence. Guide RNAs targeted the first exon of MTH1 and were designed using the CRISPR Design Tool (http://crispr.mit.edu/, (Hsu et al., 2013)). Isolation of clonal cells after transduction with Cas9 guide RNAs was accomplished by plating cells in a 96 well plate using a dilution at which one in three wells would contain a single cell. After one week of growth, wells were visually inspected to identify wells that contained a single colony of cells. For rescue experiments, MTH1 (GenBank: NM_002452) was amplified from a cDNA library, N-terminally 2X-HA tagged and cloned into XhoI and HindIII sites of pLNCX2 (Clontech Laboratories). Silent mutations were introduced to prevent targeting by Cas9 and guide RNAs used to knock out MTH1. H2B-mCherry from Addgene #21044 was subcloned into the XhoI NotI sites of pLNCX2. The coding sequence for fluorescently tagged TUBA1B was removed from mEmerald-Tubulin-6 (Addgene #54291) using NheI and BamHI sites and inserted into the SpeI and BamH1 sites of pLVX-puro (Clontech Laboratories). Centrosomes were localized by transduction of cells with the plasmid pLVX-eGFP-C1-centrin-1 (Addgene #73331) and subsequent staining for GFP. All plasmids constructed during the course of this work and their sequences are available upon request.
EdU labeling and microscopy
To identify cells that had a history of DNA synthesis during a drug treatment time course we used the Click-iT® Plus EdU Alexa Fluor® 647 Kit (Molecular Probes, C10634). Cells were seeded onto poly-L-lysine coated coverslips and treated with 5 μM 5-ethynyl-2’-deoxyuridine (EdU) and the indicated drugs. Cells were washed, fixed, permeabilized and labeled with Alexa Fluor® 647 according to the manufacturer’s recommendations. Finally, fixed cells were stained with DAPI and mounted on microscope slides as above. EdU-labeled cells were imaged using an EVOS® FL Auto Cell Imaging System (Invitrogen) at 40X magnification.
Cellular thermal shift assay (CETSA)
CETSA was according to previously described protocols (Huber et al., 2015). Two million cells were plated in a 10 cm plate and incubated overnight. After incubation with the indicated drug for 3 hours, cells were trypsinized, washed, and separated into 10 μl aliquots in PBS. Cells were heated for 3 minutes at temperatures ranging from 40°C to 73°C in 3°C increments, and then incubated for 3 minutes at 25°C. Cells were lysed by addition of 40 μl lysis buffer (50 mM Tris-HCl pH 7.5, 100 mM NaCl, 0.2% NP-40, 5% glycerol, 1.5 mM MgCl2, 1 mM NaVO3, 25 mM NaF, 5 mM beta-glycerophosphate, 1 mM dithiothreitol, protease and phosphatase inhibitors (Roche EDTA-free complete protease inhibitor tablets and PhosSTOP tablets)) followed by three rounds of vortexing, freezing in liquid nitrogen and thawing on ice. Lysates were then centrifuged for 20 minutes at 17,000 x g at 4°C. Next, 30 μl of the supernatant was transferred to a new tube, combined with 6X SDS-PAGE loading buffer, boiled and analyzed by SDS-PAGE and immunoblotting.
Crystallization, Data Collection, and Structure Determination
Crystals of T2R-TTL were grown as described (Prota et al., 2013a; Prota et al., 2013b). Crystals were soaked for 31 hours at 20°C in reservoir solution (1% PEG 4K, 8% glycerol, 30 mM MgCl2, 30 mM CaCl2, 0.1 M MES/Imidazole pH 6.7) containing 25 mM TH588 and subsequently transferred into reservoir solutions supplemented with 16 or 20% glycerol prior to cryo-cooling in liquid nitrogen. All data were collected at beamline X06DA at the Swiss Light Source (Paul Scherrer Institut, Villigen, Switzerland). Images were indexed and processed using XDS (Kabsch, 2010). Structure solution using the difference Fourier method and refinement were performed using PHENIX (Adams et al., 2010). Model building was carried out iteratively using the Coot software (Emsley and Cowtan, 2004). Data collection and refinement statistics are given in Table S4. Molecular graphics and analyses were performed with PyMOL (The PyMOL Molecular Graphics System, Version 2.2.2 Schrödinger, LLC).
Competition assay to assess relative drug sensitivity
HCT116 cells were mixed one to one with HCT116 cells transduced with the mCherry-expressing pHR-SFFV empty vector or that same vector containing wild type TUBB or TUBB L240F. The following day this mixture of cells was treated with drug and 72 hours later cells were collected by trypsinization. The relative proportion of mCherry positive and negative cells was measured by flow cytometry using a BD™ LSRII flow cytometer (Becton Dickson) and the FlowJo® software package.
QUANTIFICATION AND STATISTICAL ANALYSIS
Single-drug AUC values were calculated directly as the area under single-drug viability curves normalized to vehicle control (Figure S3A). To calculate synergy and correct for different maximal killing efficiencies in different cell lines, observed viability across the dose-response matrix was linearly transformed such that 100% viability corresponded to vehicle control and 0% viability corresponded to the minimum observed viability within each cell line (see Figure S3F). Expected viability was calculated in this transformed space using the Bliss independence model as the product of the individual drugs’ effects on viability at the same doses. Synergy was calculated as the difference between the expected and observed surfaces. Robust multi-array average normalized gene centric CCLE data was analyzed and visualized using TIGR MeV version 4.9.0 (Saeed et al., 2006). Gene set enrichment analysis was performed using GSEA2 v2.1.0 and a subset of gene sets from MSigDB v5.1 with 1000 gene set permutations and the gene-to-synergy Pearson correlation coefficients as weights. Unless otherwise indicated, data is presented as the mean of triplicate experiments, and bars represent standard error of the mean.
DATA AND SOFTWARE AVAILABILITY
All cell line viability data and synergy calculations performed for this manuscript are in Supplemental Table 1 as a Microsoft Excel spreadsheet, along with a template for user-generated data (provided it conforms to a 10 × 6 matrix of drug doses). A more general implementation of VISAGE, for drug dose matrices of any dimension, is available as an R script at https://github.com/yaffelab/visage/. The cell line-specific gene expression data used in this manuscript was obtained from https://data.broadinstitute.org/ccle_legacy_data/mRNA_expression/CCLE_Expression_Entrez_2 012-10-18.res but is not redistributed with this manuscript due to CCLE licensing restrictions. Atomic coordinates and structure factors for the reported crystal structure have been deposited in the Protein Data Bank (www.rcsb.org) under the accession code 6QQN (T2R-TTL-TH588).
Supplementary Material
Movie S1, Related to Figure 6. Normal Cell Division. Multiple cells going through an apparently normal mitotic division.
Movie S2, Related to Figure 6. Death in Mitosis. A cell, which is in the center of the field, dies after attempting mitosis. During the first third of this movie, there is spindle with the characteristic an axis of symmetry, though with uncongressed chromosomes at the poles. The spindle then collapses with a concurrent loss of symmetry and the metaphase plate. Finally, the cell dies as indicated by a catastrophic loss of cell integrity and a large number of irregular chromatin bodies.
Movie S3, Related to Figure 6. Multinucleation. Attempted mitosis by the two cells near the center of the field results in the formation of multinucleate cells. Upon exit from mitosis, multiple decondensed chromatin bodies are formed without cell division.
Movie S4, Related to Figure 6. Mitotic Slippage. A cell near the center of the field unsuccessfully attempts mitosis (indicated by white arrow). After a prolonged period of failure to congress chromosomes and divide, this cell exits mitosis with an apparently unfragmented nucleus, essentially returning to an interphase-like state.
Table S1. Related to Figure 4. Cell line viability data and synergy calculations used for the VISAGE analysis. Sensitivity to BI2536 and TH588, and synergy between the two, for 31 cancerous and 3 non-cancerous cell lines, are presented in a supplemental Microsoft Excel file along with all of the viability measurements used to calculate these quantities for each cell line.
Table S2. Related to Figure 4. Pearson correlation coefficients for expression of each transcript with synergy.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Phospho S10 histone H3 [3H10] | EMD Millipore | Cat#05-806; RRID: AB_310016 |
| MTH1 [H-159] | Santa Cruz Biotechnology | Cat#sc-67291; RRID: AB_2153825 |
| MTH1 [D6V4O] | Cell Signaling Technologies | Cat#43918; RRID: AB_2799252 |
| Hemagglutinin tag HA.11 [16B12] | Covance | Cat#mms-101p; RRID: AB_2314672 |
| Tubulin [YL1/2] | Novus Biologicals | Cat#NB600-506; RRID: AB_10078394 |
| Pericentrin | Abcam | Cat#ab4448; RRID: AB_304461 |
| GFP [3E6] | Life Technologies Inc. | Cat#A-11120; RRID: AB_221568 |
| β-actin [AC-15] (mouse) | Sigma Aldrich | Cat#A5441; RRID: AB_476744 |
| MAD2 | Bethyl Labs Inc. | Cat#A300-300A; RRID: AB_309443 |
| Cleaved caspase-3 [C92-605] | BD Pharmingen | Cat#559565; RRID: AB_397274 |
| Cleaved caspase-3 [5A1E] | Cell Signaling Technologies | Cat#9664; RRID: AB_2070042 |
| β-actin (rabbit) | Cell Signaling Technologies | Cat#4967; RRID: AB_330288 |
| Bacterial and Virus Strains | ||
| Rosetta 2™ E. coli strain | Novagen | Cat#71402-3 |
| Biological Samples | ||
| None | ||
| Chemicals, Peptides, and Recombinant Proteins | ||
| DMEM | Corning CellGro™ | Cat#10-017-CV |
| RPMI-1640 | Gibco™ | Cat#11875-093 |
| EBM-2 | Lonza | Cat#CC-3156 |
| EGM™−2 Bulletkit™ | Lonza | Cat#CC-4176 |
| Fetal Bovine Serum (FBS) | Seradigm | Cat#1500-500 |
| TH588 | Thomas Helleday (Karolinska Institute) and Selleck Chemicals. | Cat#S7632 |
| BI2536 | Selleck Chemicals | Cat#S1109 |
| Volasertib | Selleck Chemicals | Cat#S2235 |
| GSK461364 | Selleck Chemicals | Cat#S2193 |
| Onvansertib/PCM-075/NMS-1286937 | Mark Erlander (Trovagene Inc.) | |
| (S)-crizotinib | Selleck Chemicals | Cat#S7505 |
| Barasertib | Selleck Chemicals | Cat#S1147 |
| GSK923295 / CENP-E inhibitor | Selleck Chemicals | Cat#S7090 |
| Alisertib | Selleck Chemicals | Cat#S1133 |
| N-acetyl cysteine | Sigma Aldrich | Cat#A9165 |
| Nocodazole | Sigma Aldrich | Cat#M1404 |
| Docetaxel | LC Laboratories | Cat#D-1000 |
| GTP | Jena Bioscience | Cat#NU-1012 |
| 8-oxo-GTP | Jena Bioscience | Cat#NU-11165 |
| 4,6-diamidino-2-phenylindole (DAPI) | Molecular Probes | Cat#D1306 |
| SYTO 60 | Molecular Probes | Cat#S11342 |
| ProLong Gold™ | Molecular Probes | Cat#P36934 |
| Lipofectamine® RNAiMAX | Invitrogen | Cat#13778-150 |
| PhosSTOP tablets | Roche | Cat#04 906 837 001 |
| cOmplete™, mini, EDTA-free protease inhibitor cocktail | Roche | Cat#11836170001 |
| CalPhos™ mammalioan transfection kit | Clontech Laboratories | Cat#631312 |
| Puromycin | Sigma Aldrich | Cat#P8833 |
| Hygromycin B | Invitrogen | Cat#10687-010 |
| G418 | Corning | Cat#30-234-CR |
| Tubulin protein (pre-formed microtubules): porcine brain | Cytoskeleton Inc. | Cat#MT002 |
| Critical Commercial Assays | ||
| CellTitreGlo™ | Promega | Cat#G7571 |
| Tubulin Polymerization assy | Cytoskeleton, Inc. | Cat#BK011 P |
| Click-iT® Plus EdU Alexa Fluor® 647 Kit | Molecular Probes | Cat#C10634 |
| BIOMOL® Green | ENZO Life Sciences | Cat#BML-AK111 |
| Deposited Data | ||
| CCLE Gene-centric RMA-normalized mRNA expression data (CCLE_Expression_Entrez_2012-10-18.res) | (Barretina et al., 2012) | https://portals.broadinstitute.org/cclelegacy/home |
| T2R-TTL-TH588 Crystal Structure | This paper | PDB: 6QQN |
| Experimental Models: Cell Lines | ||
| C4-2 | MD Anderson | RRID: CVCL_4782; Male |
| CCD-18co | ATCC | CRL-1459; RRID: CVCL_2379; Male |
| HUVEC | Lonza | C2519A |
| MDA-MB-231 | ATCC | HTB-26; RRID: CVCL_0062; Female |
| U-2 OS | ATCC | HTB-96; RRID: CVCL_0042; Female |
| NCI-H1299 | ATCC | CRL-5803; RRID: CVCL_0060; Male |
| ZR-75-1 | ATCC | CRL-1500; RRID: CVCL_0588; Female |
| A549 | ATCC | CCL-185; RRID: CVCL_0023; Male |
| KYSE-150 | DSMZ | ACC-375; RRID: CVCL_1348; Female |
| BT-20 | ATCC | HTB-19; RRID: CVCL_0178; Female |
| MDA-MB-415 | ATCC | HTB-128; RRID: CVCL_0621; Female |
| HCC38 | ATCC | CRL-2314; RRID: CVCL_1267; Female |
| MDA-MB-157 | ATCC | HTB-24; RRID: CVCL_0618; Female |
| HCT-15 | K. Haigis (Beth Israel Deaconess Medical Center) | RRID: CVCL_0292; Male |
| U-87 MG | ATCC | HTB-14; RRID: CVCL_0022; Male |
| 22Rv1 | ATCC | CRL-2505; RRID: CVCL_1045; Male |
| CFPAC1 | ATCC | CRL-1918; RRID: CVCL 1119; Male |
| HCT116 | ATCC | CCL-247; RRID: CVCL_0291; Male |
| LNCaP | ATCC | CRL-1740; RRID: CVCL_1379; Male |
| AU565 | ATCC | CRL-2351; RRID: CVCL_1074; Female |
| CAL-33 | DSMZ | ACC 447; RRID: CVCL 1108; Male |
| MDA-MB-468 | ATCC | HTB-132; RRID: CVCL_0419; Female |
| PC-3 | ATCC | CRL-1435; RRID: CVCL_0035; Male |
| T98G | ATCC | CRL-1690; RRID: CVCL 0556; Male |
| MCF7 | ATCC | HTB-22; RRID: CVCL_0031; Female |
| SW48 | K. Haigis (Beth Israel Deaconess Medical Center) | RRID: CVCL_1724; Female |
| MDA-MB-453 | ATCC | HTB-131; RRID: CVCL_0418; Female |
| MDA-MB-436 | ATCC | HTB-130; RRID: CVCL_0623; Female |
| LOVO | K. Haigis (Beth Israel Deaconess Medical Center) | RRID: CVCL_0399; Male |
| HT55 | K. Haigis (Beth Israel Deaconess Medical Center) | RRID: CVCL_1294; Gender Unknown |
| COLO 205DM | ATCC | CCL-229; RRID: CVCL_F402; Female |
| HT-29 | ATCC | HTB-38; RRID:CVCL_0320; Female |
| HeLa | ATCC | CCL-2; RRID:CVCL_0030; Female |
| HEK293t | ATCC | CRL-3216; RRID:CVCL_0063; Female |
| CCD-1112sk | ATCC | CRL-2429; RRID:CVCL_2769; Male |
| HCT116 AAVS1-1 | This paper | |
| HCT116 MTH1 KO 9-4 | This paper | |
| HCT116 MTH1 KO 10-2 | This paper | |
| Experimental Models: Organisms/Strains | ||
| Mouse: CrTac:NCr-Foxn1nu | Taconic | NCRNU |
| Oligonucleotides | ||
| Silencer select negative control #1 siRNA | Invitrogen | Cat#4390844 |
| MAD2L1 silencer select siRNA | Invitrogen | Cat#S8392 |
| AAVS1 targeting control guide RNA-GGGGCCACTAGGGACAGGAT | This paper | |
| MTH1 targeting guide RNA #7-GCCGGCCGGTGGAATGGCTT | This paper | |
| MTH1 targeting guide RNA #8-GCAAGAAGGAGAGACCATCG | This paper | |
| MTH1 targeting guide RNA #9-TTCGGGGCCGGCCGGTGGAA | This paper | |
| MTH1 targeting guide RNA #10-CATGAAAAAGCGAGGCTTCG | This paper | |
| Recombinant DNA | ||
| pLVX-IRES-Hyg | Clontech | Cat#632185 |
| pLVX-Puro | Clontech | Cat#632164 |
| pLNCX2 | Clontech | Cat#631503 |
| pCW-Cas9 | Addgene #50661 | (Wang et al., 2014) |
| pLVX-IRES-Hyg-S.p.Cas9 | This paper | |
| lentiGuide-Puro | Addgene #52963 | (Sanjana et al., 2014) |
| lentiGuide-Puro-sgAAVS1 | This paper | |
| lentiGuide-Puro-sgMTH1 #7 | This paper | |
| lentiGuide-Puro-sgMTH1 #8 | This paper | |
| lentiGuide-Puro-sgMTH1 #9 | This paper | |
| lentiGuide-Puro-sgMTH1 #10 | This paper | |
| pLNCX2-2XHA-MTH1 | This paper | GenBank: NM_002452 |
| pLNCX2-2XHA-MTH1 E56A | This paper | |
| pH2B-mCherry-IRES-neo3 | Addgene #21044 | (Steigemann et al., 2009) |
| pLNCX2-H2B-mCherry | This paper | |
| mEmerald-T ubulin-6 | Addgene #54291 | (Day and Davidson, 2009) |
| pLVX-Puro-mEmerald-TUBA1B | This paper | |
| pLVX-eGFP-C1-centrin-1 | Addgene #73331 | Manuel Thery |
| pHR-SFFV-IRES-mCherry | (Jost et al., 2017) | |
| pHR-SFFV-IRES-mCherry-TUBB | (Jost et al., 2017) | |
| pHR-SFFV-IRES-mCherry-TUBB L240F | (Jost et al., 2017) | |
| pET-28a(+) | Novagen | Cat#69864-3 |
| pET-28a(+) CENPE-6XHIS | This paper | GenBank: NM_001813.2 |
| Software and Algorithms | ||
| FlowJo version 7.6.5 | FlowJo, LLC | |
| TIGR MeV version 4.9.0 | (Saeed et al., 2006) | https://sourceforgenet/projects/mev-tm4/. |
| Gene Set Enrichment Analysis (GSEA) v2.1.0 | (Subramanian et al., 2005) | http://software.broadinstitute.org/gsea/ |
| MSigDB v5.1 | (Subramanian et al., 2005) | http://software.broadinstitute.org/gsea/ |
| softWoRx™ | Applied Precision | |
| Fiji distribution of ImageJ | (Schindelin et al., 2012; Schindelin et al., 2015; Schmid et al., 2010) | https://fiji.sc/ |
| Adobe Photoshop CC | Adobe Inc. | |
| Gnu Image Manipulation Program (GIMP) | https://www.gimp.org/ | |
| Seaborn statistical data visualization version 0.7.1 | http://seaborn.pyda.org | |
| CRISPR Design Tool | (Hsu et al., 2013) | http://crispr.mit.edu/ |
| XDS | (Kabsch, 2010) | http://xds.mpimf-heidelberg.mpg.de/ |
| PHENIX | (Adams et al., 2010) | https://www.phenix-online.org/ |
| Coot | (Emsley and Cowtan, 2004) | http://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/ |
| PyMOL Version 2.2.2 | Schrödinger, LLC | https://pymol.org/2/ |
| Excel | Microsoft | |
| VISAGE R Script | This paper | https://github.com/yaffelab/visage |
| Other | ||
Highlights.
The MTH1 inhibitor TH588 synergizes with Plk1 inhibition to drive cancer cell death
VISAGE implicates the mitotic spindle as the target of drug synergy, not MTH1
TH588 binds the colchicine binding site of β-tubulin blocking microtubule assembly
The cancer cell spindle is particularly vulnerable to Plk1 + microtubule inhibitors
ACKNOWLEDGEMENTS
We thank Dan Lim, Karl Merrick, Mun Kyung Hwang, Ian Cannell, Pau Creixell, Yi Wen Kong, Susanne Swartwout, and other members of the Yaffe and Lauffenburger laboratories, Neil Ganem, and Rebecca Heald for helpful discussion and advice, and Mark Erlander and Trovogene for onvansertib. We also thank Thomas Helleday and members of the Helleday laboratory for reagents and suggestions, David Freeman, and the Swanson Biotechnology Center including the Microscopy Core Facility (Eliza Vasile) and the Flow Cytometry Core Facility (Glenn Paradis). This research was supported by grants from the National Institute of Health (R35-ES028374 (M.B.Y.), R01-GM104047 (M.B.Y), R01-ES015339 (M.B.Y.), U54-CA112967 (D.A.L. and M.B.Y.), U54-CA217377 (D.A.L.), the Charles and Marjorie Holloway Foundation (M.B.Y.), the Ovarian Cancer Research Fund (M.B.Y.), the MIT Center for Precision Cancer Medicine, a Koch Institute - Dana-Farber/Harvard Cancer Center Bridge Project (M.B.Y. and S.P.B.), and an American Cancer Society Post-Doctoral Fellowship to J.C.P. Support was also provided in part by the Koch Institute Support Grant (P30-CA14051) from the National Cancer Institute and the Center for Environmental Health Support Grant (P30-ES002109).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DECLARATION OF INTERESTS
Michael B. Yaffe is a member of the Divisions of Surgical Oncology and Trauma and Critical Care, Department of Surgery, Beth Israel Deaconess Medical Center, Harvard Medical School, Boston, MA, and the Chief Academic Editor of the journal Science Signaling. The authors declare no competing interests.
REFERENCES
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. (2010). PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta crystallographica Section D, Biological crystallography 66, 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander J, Lim D, Joughin BA, Hegemann B, Hutchins JR, Ehrenberger T, Ivins F, Sessa F, Hudecz O, Nigg EA, et al. (2011). Spatial exclusivity combined with positive and negative selection of phosphorylation motifs is the basis for context-dependent mitotic signaling. Sci Signal 4, ra42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandre J, Batteux F, Nicco C, Chereau C, Laurent A, Guillevin L, Weill B, and Goldwasser F (2006). Accumulation of hydrogen peroxide is an early and crucial step for paclitaxel-induced cancer cell death both in vitro and in vivo. Int J Cancer 119, 41–48. [DOI] [PubMed] [Google Scholar]
- Archambault V, and Glover DM (2009). Polo-like kinases: conservation and divergence in their functions and regulation. Nat Rev Mol Cell Biol 10, 265–275. [DOI] [PubMed] [Google Scholar]
- Ba X, and Boldogh I (2018). 8-Oxoguanine DNA glycosylase 1: Beyond repair of the oxidatively modified base lesions. Redox Biol 14, 669–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, Wilson CJ, Lehar J, Kryukov GV, Sonkin D, et al. (2012). The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 483, 603–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bliss CI (1939). The toxicity of poisons applied jointly. Ann Appl Biol 26, 585–615. [Google Scholar]
- Bonne D, Heusele C, Simon C, and Pantaloni D (1985). 4’,6-Diamidino-2-phenylindole, a fluorescent probe for tubulin and microtubules. J Biol Chem 260, 2819–2825. [PubMed] [Google Scholar]
- Bruinsma W, Raaijmakers JA, and Medema RH (2012). Switching Polo-like kinase-1 on and off in time and space. Trends Biochem Sci 37, 534–542. [DOI] [PubMed] [Google Scholar]
- Cai S, O’Connell CB, Khodjakov A, and Walczak CE (2009). Chromosome congression in the absence of kinetochore fibres. Nat Cell Biol 11, 832–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crea F, Duhagon Serrat MA, Hurt EM, Thomas SB, Danesi R, and Farrar WL (2011). BMI1 silencing enhances docetaxel activity and impairs antioxidant response in prostate cancer. Int J Cancer 128, 1946–1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czaplinski S, Hugle M, Stiehl V, and Fulda S (2016). Polo-like kinase 1 inhibition sensitizes neuroblastoma cells for vinca alkaloid-induced apoptosis. Oncotarget 7, 8700–8711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day RN, and Davidson MW (2009). The fluorescent protein palette: tools for cellular imaging. Chem Soc Rev 38, 2887–2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeNicola GM, Karreth FA, Humpton TJ, Gopinathan A, Wei C, Frese K, Mangal D, Yu KH, Yeo CJ, Calhoun ES, et al. (2011). Oncogene-induced Nrf2 transcription promotes ROS detoxification and tumorigenesis. Nature 475, 106–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeVita VT Jr., and Chu E (2008). A history of cancer chemotherapy. Cancer Res 68, 8643–8653. [DOI] [PubMed] [Google Scholar]
- Dohner H, Lubbert M, Fiedler W, Fouillard L, Haaland A, Brandwein JM, Lepretre S, Reman O, Turlure P, Ottmann OG, et al. (2014). Randomized, phase 2 trial of low-dose cytarabine with or without volasertib in AML patients not suitable for induction therapy. Blood 124, 1426–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Döhner H, Symeonidis A, Sanz MA, Deeren D, Demeter J, Anagnostopoulos A, Esteve J, Fiedler W, Porkka K, Kim HJ, et al. (2016). Phase III randomized trial of volasertib plus low-dose cytarabine (LDAC) versus placebo plus LDAC in patients aged ≥65 years with previously untreated AML, ineligible for intensive therapy. Haematologica 93(s1), 185–186. [Google Scholar]
- Dominguez-Brauer C, Thu KL, Mason JM, Blaser H, Bray MR, and Mak TW (2015). Targeting Mitosis in Cancer: Emerging Strategies. Mol Cell 60, 524–536. [DOI] [PubMed] [Google Scholar]
- Elia AE, Cantley LC, and Yaffe MB (2003a). Proteomic screen finds pSer/pThr-binding domain localizing Plk1 to mitotic substrates. Science 299, 1228–1231. [DOI] [PubMed] [Google Scholar]
- Elia AE, Rellos P, Haire LF, Chao JW, Ivins FJ, Hoepker K, Mohammad D, Cantley LC, Smerdon SJ, and Yaffe MB (2003b). The molecular basis for phosphodependent substrate targeting and regulation of Plks by the Polo-box domain. Cell 115, 83–95. [DOI] [PubMed] [Google Scholar]
- Ellermann M, Eheim A, Rahm F, Viklund J, Guenther J, Andersson M, Ericsson U, Forsblom R, Ginman T, Lindstrom J, et al. (2017). Novel Class of Potent and Cellularly Active Inhibitors Devalidates MTH1 as Broad-Spectrum Cancer Target. ACS Chem Biol 12, 1986–1992. [DOI] [PubMed] [Google Scholar]
- Emsley P, and Cowtan K (2004). Coot: model-building tools for molecular graphics. Acta crystallographica Section D, Biological crystallography 60, 2126–2132. [DOI] [PubMed] [Google Scholar]
- Foucquier J, and Guedj M (2015). Analysis of drug combinations: current methodological landscape. Pharmacol Res Perspect 3, e00149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii Y, Shimokawa H, Sekiguchi M, and Nakabeppu Y (1999). Functional significance of the conserved residues for the 23-residue module among MTH1 and MutT family proteins. J Biol Chem 274, 38251–38259. [DOI] [PubMed] [Google Scholar]
- Gad H, Koolmeister T, Jemth AS, Eshtad S, Jacques SA, Strom CE, Svensson LM, Schultz N, Lundback T, Einarsdottir BO, et al. (2014). MTH1 inhibition eradicates cancer by preventing sanitation of the dNTP pool. Nature 508, 215–221. [DOI] [PubMed] [Google Scholar]
- Ganem NJ, Godinho SA, and Pellman D (2009). A mechanism linking extra centrosomes to chromosomal instability. Nature 460, 278–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greco WR, Bravo G, and Parsons JC (1995). The search for synergy: a critical review from a response surface perspective. Pharmacol Rev 47, 331–385. [PubMed] [Google Scholar]
- Grosstessner-Hain K, Hegemann B, Novatchkova M, Rameseder J, Joughin BA, Hudecz O, Roitinger E, Pichler P, Kraut N, Yaffe MB, et al. (2011). Quantitative phosphoproteomics to investigate the polo-like kinase 1-dependent phospho-proteome. Mol Cell Proteomics 10, M111 008540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D, and Weinberg RA (2011). Hallmarks of cancer: the next generation. Cell 144, 646–674. [DOI] [PubMed] [Google Scholar]
- Havens CG, Ho A, Yoshioka N, and Dowdy SF (2006). Regulation of late G1/S phase transition and APC Cdh1 by reactive oxygen species. Mol Cell Biol 26, 4701–4711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PD, Scott DA, Weinstein JA, Ran FA, Konermann S, Agarwala V, Li Y, Fine EJ, Wu X, Shalem O, et al. (2013). DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol 31, 827–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber KV, Olek KM, Muller AC, Tan CS, Bennett KL, Colinge J, and Superti-Furga G (2015). Proteome-wide drug and metabolite interaction mapping by thermal-stability profiling. Nat Methods 12, 1055–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber KV, Salah E, Radic B, Gridling M, Elkins JM, Stukalov A, Jemth AS, Gokturk C, Sanjiv K, Stromberg K, et al. (2014). Stereospecific targeting of MTH1 by (S)-crizotinib as an anticancer strategy. Nature 508, 222–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugle M, Belz K, and Fulda S (2015). Identification of synthetic lethality of PLK1 inhibition and microtubule-destabilizing drugs. Cell Death Differ 22, 1946–1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonas O, Landry HM, Fuller JE, Santini JT Jr., Baselga J, Tepper RI, Cima MJ, and Langer R (2015). An implantable microdevice to perform high-throughput in vivo drug sensitivity testing in tumors. Sci Transl Med 7, 284ra257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jost M, Chen Y, Gilbert LA, Horlbeck MA, Krenning L, Menchon G, Rai A, Cho MY, Stern JJ, Prota AE, et al. (2017). Combined CRISPRi/a-Based Chemical Genetic Screens Reveal that Rigosertib Is a Microtubule-Destabilizing Agent. Mol Cell 68, 210–223 e216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W (2010). XDS. Acta crystallographica Section D, Biological crystallography 66, 125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor TM, Lampson MA, Hergert P, Cameron L, Cimini D, Salmon ED, McEwen BF, and Khodjakov A (2006). Chromosomes can congress to the metaphase plate before biorientation. Science 311, 388–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura N (1960). Cytochemical and quantitative study of protein-bound sulfhydryl and disulfide groups in eggs of Arbacia during the first cleavage. Exp Cell Res 20, 127–138. [DOI] [PubMed] [Google Scholar]
- Kawamura T, Kawatani M, Muroi M, Kondoh Y, Futamura Y, Aono H, Tanaka M, Honda K, and Osada H (2016). Proteomic profiling of small-molecule inhibitors reveals dispensability of MTH1 for cancer cell survival. Sci Rep 6, 26521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keith CT, Borisy AA, and Stockwell BR (2005). Multicomponent therapeutics for networked systems. Nat Rev Drug Discov 4, 71–78. [DOI] [PubMed] [Google Scholar]
- Kettle JG, Alwan H, Bista M, Breed J, Davies NL, Eckersley K, Fillery S, Foote KM, Goodwin L, Jones DR, et al. (2016). Potent and Selective Inhibitors of MTH1 Probe Its Role in Cancer Cell Survival. J Med Chem 59, 2346–2361. [DOI] [PubMed] [Google Scholar]
- Kops GJ, Weaver BA, and Cleveland DW (2005). On the road to cancer: aneuploidy and the mitotic checkpoint. Nat Rev Cancer 5, 773–785. [DOI] [PubMed] [Google Scholar]
- Lehar J, Krueger AS, Avery W, Heilbut AM, Johansen LM, Price ER, Rickles RJ, Short GF 3rd, Staunton JE, Jin X, et al. (2009). Synergistic drug combinations tend to improve therapeutically relevant selectivity. Nat Biotechnol 27, 659–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leiserson MD, Vandin F, Wu HT, Dobson JR, Eldridge JV, Thomas JL, Papoutsaki A, Kim Y, Niu B, McLellan M, et al. (2015). Pan-cancer network analysis identifies combinations of rare somatic mutations across pathways and protein complexes. Nat Genet 47, 106–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liou GY, and Storz P (2010). Reactive oxygen species in cancer. Free Radic Res 44, 479–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Chen J, Xiao M, Li W, and Miller DD (2012). An overview of tubulin inhibitors that interact with the colchicine binding site. Pharm Res 29, 2943–2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macurek L, Lindqvist A, Lim D, Lampson MA, Klompmaker R, Freire R, Clouin C, Taylor SS, Yaffe MB, and Medema RH (2008). Polo-like kinase-1 is activated by aurora A to promote checkpoint recovery. Nature 455, 119–123. [DOI] [PubMed] [Google Scholar]
- Massarotti A, Coluccia A, Silvestri R, Sorba G, and Brancale A (2012). The tubulin colchicine domain: a molecular modeling perspective. ChemMedChem 7, 33–42. [DOI] [PubMed] [Google Scholar]
- Mazia D (1958). SH compounds in mitosis. I. The action of mercaptoethanol on the eggs of the sand dollar Dendraster excentricus. Exp Cell Res 14, 486–494. [DOI] [PubMed] [Google Scholar]
- Mendelsohn J (2013). Personalizing oncology: perspectives and prospects. J Clin Oncol 31, 1904–1911. [DOI] [PubMed] [Google Scholar]
- Musacchio A, and Salmon ED (2007). The spindle-assembly checkpoint in space and time. Nat Rev Mol Cell Biol 8, 379–393. [DOI] [PubMed] [Google Scholar]
- Noack S, Raab M, Matthess Y, Sanhaji M, Kramer A, Gyorffy B, Kaderali L, El-Balat A, Becker S, and Strebhardt K (2018). Synthetic lethality in CCNE1-amplified high grade serous ovarian cancer through combined inhibition of Polo-like kinase 1 and microtubule dynamics. Oncotarget 9, 25842–25859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson JC, Joughin BA, van de Kooij B, Lim DC, Lauffenburger DA, and Yaffe MB (2019). ROS and Oxidative Stress Are Elevated in Mitosis during Asynchronous Cell Cycle Progression and Are Exacerbated by Mitotic Arrest. Cell Syst 8, 163–167 e162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prota AE, Bargsten K, Zurwerra D, Field JJ, Diaz JF, Altmann KH, and Steinmetz MO (2013a). Molecular mechanism of action of microtubule-stabilizing anticancer agents. Science 339, 587–590. [DOI] [PubMed] [Google Scholar]
- Prota AE, Magiera MM, Kuijpers M, Bargsten K, Frey D, Wieser M, Jaussi R, Hoogenraad CC, Kammerer RA, Janke C, et al. (2013b). Structural basis of tubulin tyrosination by tubulin tyrosine ligase. J Cell Biol 200, 259–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pudelko L, Rouhi P, Sanjiv K, Gad H, Kalderen C, Hoglund A, Squatrito M, Schuhmacher AJ, Edwards S, Hagerstrand D, et al. (2017). Glioblastoma and glioblastoma stem cells are dependent on functional MTH1. Oncotarget 8, 84671–84684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai P, Onder TT, Young JJ, McFaline JL, Pang B, Dedon PC, and Weinberg RA (2009). Continuous elimination of oxidized nucleotides is necessary to prevent rapid onset of cellular senescence. Proc Natl Acad Sci U S A 106, 169–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt HC, and Yaffe MB (2013). Phospho-Ser/Thr-binding domains: navigating the cell cycle and DNA damage response. Nat Rev Mol Cell Biol 14, 563–580. [DOI] [PubMed] [Google Scholar]
- Saeed AI, Bhagabati NK, Braisted JC, Liang W, Sharov V, Howe EA, Li J, Thiagarajan M, White JA, and Quackenbush J (2006). TM4 microarray software suite. Methods Enzymol 411, 134–193. [DOI] [PubMed] [Google Scholar]
- Samaranayake GJ, Huynh M, and Rai P (2017). MTH1 as a Chemotherapeutic Target: The Elephant in the Room. Cancers (Basel) 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjana NE, Shalem O, and Zhang F (2014). Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods 11, 783–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santamaria A, Wang B, Elowe S, Malik R, Zhang F, Bauer M, Schmidt A, Sillje HH, Korner R, and Nigg EA (2011). The Plk1-dependent phosphoproteome of the early mitotic spindle. Mol Cell Proteomics 10, M110 004457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saurin AT, van der Waal MS, Medema RH, Lens SM, and Kops GJ (2011). Aurora B potentiates Mps1 activation to ensure rapid checkpoint establishment at the onset of mitosis. Nat Commun 2, 316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyers CL (2008). The cancer biomarker problem. Nature 452, 548–552. [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat Methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Rueden CT, Hiner MC, and Eliceiri KW (2015). The ImageJ ecosystem: An open platform for biomedical image analysis. Mol Reprod Dev 82, 518–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid B, Schindelin J, Cardona A, Longair M, and Heisenberg M (2010). A high-level 3D visualization API for Java and ImageJ. BMC Bioinformatics 11, 274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim GS, Manandhar S, Shin DH, Kim TH, and Kwak MK (2009). Acquisition of doxorubicin resistance in ovarian carcinoma cells accompanies activation of the NRF2 pathway. Free Radic Biol Med 47, 1619–1631. [DOI] [PubMed] [Google Scholar]
- Sparano JA, Ostrer H, and Kenny PA (2013). Translating genomic research into clinical practice: promise and pitfalls. Am Soc Clin Oncol Educ Book, 15–23. [DOI] [PubMed] [Google Scholar]
- Speina E, Arczewska KD, Gackowski D, Zielinska M, Siomek A, Kowalewski J, Olinski R, Tudek B, and Kusmierek JT (2005). Contribution of hMTH1 to the maintenance of 8-oxoguanine levels in lung DNA of non-small-cell lung cancer patients. J Natl Cancer Inst 97, 384–395. [DOI] [PubMed] [Google Scholar]
- Steegmaier M, Hoffmann M, Baum A, Lenart P, Petronczki M, Krssak M, Gurtler U, Garin-Chesa P, Lieb S, Quant J, et al. (2007). BI 2536, a potent and selective inhibitor of polo-like kinase 1, inhibits tumor growth in vivo. Curr Biol 17, 316–322. [DOI] [PubMed] [Google Scholar]
- Steigemann P, Wurzenberger C, Schmitz MH, Held M, Guizetti J, Maar S, and Gerlich DW (2009). Aurora B-mediated abscission checkpoint protects against tetraploidization. Cell 136, 473–484. [DOI] [PubMed] [Google Scholar]
- Steinmetz MO, and Prota AE (2018). Microtubule-Targeting Agents: Strategies To Hijack the Cytoskeleton. Trends Cell Biol 28, 776–792. [DOI] [PubMed] [Google Scholar]
- Sternlicht H, Ringel I, and Szasz J (1980). The co-polymerization of tubulin and tubulin chochicine complex in the absence and presence of associated proteins. J Biol Chem 255, 9138–9148. [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. (2005). Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 102, 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumara I, Gimenez-Abian JF, Gerlich D, Hirota T, Kraft C, de la Torre C, Ellenberg J, and Peters JM (2004). Roles of polo-like kinase 1 in the assembly of functional mitotic spindles. Curr Biol 14, 1712–1722. [DOI] [PubMed] [Google Scholar]
- Szatrowski TP, and Nathan CF (1991). Production of large amounts of hydrogen peroxide by human tumor cells. Cancer Res 51, 794–798. [PubMed] [Google Scholar]
- Tannock IF, de Wit R, Berry WR, Horti J, Pluzanska A, Chi KN, Oudard S, Theodore C, James ND, Turesson I, et al. (2004). Docetaxel plus prednisone or mitoxantrone plus prednisone for advanced prostate cancer. N Engl J Med 351, 1502–1512. [DOI] [PubMed] [Google Scholar]
- Tsuzuki T, Egashira A, and Kura S (2001). Analysis of MTH1 gene function in mice with targeted mutagenesis. Mutat Res 477, 71–78. [DOI] [PubMed] [Google Scholar]
- van Vugt MA, Gardino AK, Linding R, Ostheimer GJ, Reinhardt HC, Ong SE, Tan CS, Miao H, Keezer SM, Li J, et al. (2010). A mitotic phosphorylation feedback network connects Cdk1, Plk1, 53BP1, and Chk2 to inactivate the G(2)/M DNA damage checkpoint. PLoS Biol 8, e1000287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Heiden MG, and DeBerardinis RJ (2017). Understanding the Intersections between Metabolism and Cancer Biology. Cell 168, 657–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitale I, Galluzzi L, Castedo M, and Kroemer G (2011). Mitotic catastrophe: a mechanism for avoiding genomic instability. Nat Rev Mol Cell Biol 12, 385–392. [DOI] [PubMed] [Google Scholar]
- Wang JY, Jin L, Yan XG, Sherwin S, Farrelly M, Zhang YY, Liu F, Wang CY, Guo ST, Yari H, et al. (2016). Reactive Oxygen Species Dictate the Apoptotic Response of Melanoma Cells to TH588. J Invest Dermatol 136, 2277–2286. [DOI] [PubMed] [Google Scholar]
- Wang M, Zhou S, Chen Q, Wang L, Liang Z, and Wang J (2017). Understanding the molecular mechanism for the differential inhibitory activities of compounds against MTH1. Sci Rep 7, 40557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Wei JJ, Sabatini DM, and Lander ES (2014). Genetic screens in human cells using the CRISPR-Cas9 system. Science 343, 80–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warpman Berglund U, Sanjiv K, Gad H, Kalderen C, Koolmeister T, Pham T, Gokturk C, Jafari R, Maddalo G, Seashore-Ludlow B, et al. (2016). Validation and development of MTH1 inhibitors for treatment of cancer. Ann Oncol 27, 2275–2283. [DOI] [PubMed] [Google Scholar]
- Weichert W, Schmidt M, Gekeler V, Denkert C, Stephan C, Jung K, Loening S, Dietel M, and Kristiansen G (2004). Polo-like kinase 1 is overexpressed in prostate cancer and linked to higher tumor grades. Prostate 60, 240–245. [DOI] [PubMed] [Google Scholar]
- Weiss LM, Hugle M, Romero S, and Fulda S (2016). Synergistic induction of apoptosis by a polo-like kinase 1 inhibitor and microtubule-interfering drugs in Ewing sarcoma cells. Int J Cancer 138, 497–506. [DOI] [PubMed] [Google Scholar]
- Wood KW, Lad L, Luo L, Qian X, Knight SD, Nevins N, Brejc K, Sutton D, Gilmartin AG, Chua PR, et al. (2010). Antitumor activity of an allosteric inhibitor of centromere-associated protein-E. Proc Natl Acad Sci U S A 107, 5839–5844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wynne DJ, and Funabiki H (2015). Kinetochore function is controlled by a phospho-dependent coexpansion of inner and outer components. J Cell Biol 210, 899–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi H, Kajitani K, Dan Y, Furuichi M, Ohno M, Sakumi K, Kang D, and Nakabeppu Y (2006). MTH1, an oxidized purine nucleoside triphosphatase, protects the dopamine neurons from oxidative damage in nucleic acids caused by 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine. Cell Death Differ 13, 551–563. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Movie S1, Related to Figure 6. Normal Cell Division. Multiple cells going through an apparently normal mitotic division.
Movie S2, Related to Figure 6. Death in Mitosis. A cell, which is in the center of the field, dies after attempting mitosis. During the first third of this movie, there is spindle with the characteristic an axis of symmetry, though with uncongressed chromosomes at the poles. The spindle then collapses with a concurrent loss of symmetry and the metaphase plate. Finally, the cell dies as indicated by a catastrophic loss of cell integrity and a large number of irregular chromatin bodies.
Movie S3, Related to Figure 6. Multinucleation. Attempted mitosis by the two cells near the center of the field results in the formation of multinucleate cells. Upon exit from mitosis, multiple decondensed chromatin bodies are formed without cell division.
Movie S4, Related to Figure 6. Mitotic Slippage. A cell near the center of the field unsuccessfully attempts mitosis (indicated by white arrow). After a prolonged period of failure to congress chromosomes and divide, this cell exits mitosis with an apparently unfragmented nucleus, essentially returning to an interphase-like state.
Table S1. Related to Figure 4. Cell line viability data and synergy calculations used for the VISAGE analysis. Sensitivity to BI2536 and TH588, and synergy between the two, for 31 cancerous and 3 non-cancerous cell lines, are presented in a supplemental Microsoft Excel file along with all of the viability measurements used to calculate these quantities for each cell line.
Table S2. Related to Figure 4. Pearson correlation coefficients for expression of each transcript with synergy.
Data Availability Statement
All cell line viability data and synergy calculations performed for this manuscript are in Supplemental Table 1 as a Microsoft Excel spreadsheet, along with a template for user-generated data (provided it conforms to a 10 × 6 matrix of drug doses). A more general implementation of VISAGE, for drug dose matrices of any dimension, is available as an R script at https://github.com/yaffelab/visage/. The cell line-specific gene expression data used in this manuscript was obtained from https://data.broadinstitute.org/ccle_legacy_data/mRNA_expression/CCLE_Expression_Entrez_2 012-10-18.res but is not redistributed with this manuscript due to CCLE licensing restrictions. Atomic coordinates and structure factors for the reported crystal structure have been deposited in the Protein Data Bank (www.rcsb.org) under the accession code 6QQN (T2R-TTL-TH588).