Abstract
Several existing methods have been shown to consistently estimate causal direction assuming linear or some form of nonlinear relationship and no latent confounders. However, the estimation results could be distorted if either assumption is violated. We develop an approach to determining the possible causal direction between two observed variables when latent confounding variables are present. We first propose a new linear non-Gaussian acyclic structural equation model with individual-specific effects that are sometimes the source of confounding. Thus, modeling individual-specific effects as latent variables allows latent confounding to be considered. We then propose an empirical Bayesian approach for estimating possible causal direction using the new model. We demonstrate the effectiveness of our method using artificial and real-world data.
Keywords: structural equation models, Bayesian networks, estimation of causal direction, latent confounding variables, non-Gaussianity
1. Introduction
Aids to uncover the causal structure of variables from observational data are welcomed additions to the field of machine learning (Pearl, 2000; Spirtes et al., 1993). One conventional approach makes use of Bayesian networks (Pearl, 2000; Spirtes et al., 1993). However, these suffer from the identifiability problem. That is, many different causal structures give the same conditional independence between variables, and in many cases one cannot uniquely estimate the underlying causal structure without prior knowledge (Pearl, 2000; Spirtes et al., 1993).
To address these issues, Shimizu et al. (2006) proposed LiNGAM (Linear Non-Gaussian Acyclic Model), a variant of Bayesian networks (Pearl, 2000; Spirtes et al., 1993) and structural equation models (Bollen, 1989). Unlike conventional Bayesian networks, LiNGAM is a fully identifiable model (Shimizu et al., 2006), and has recently attracted much attention in machine learning (Spirtes et al., 2010; Moneta et al., 2011). If causal relations exist among variables, LiNGAM uses their non-Gaussian distributions to identify the causal structure among the variables. LiNGAM is closely related to independent component analysis (ICA) (Hyvärinen et al., 2001b); the identifiability proof and estimation algorithm are partly based on the ICA theory. The idea of LiNGAM has been extended in many directions, including to nonlinear cases (Hoyer et al., 2009; Lacerda et al., 2008; Hyvärinen et al., 2010; Zhang and Hyvärinen, 2009; Peters et al., 2011a).
Many causal discovery methods including LiNGAM make the strong assumption of no latent confounders (Spirtes and Glymour, 1991; Dodge and Rousson, 2001; Shimizu et al., 2006; Hyvärinen and Smith, 2013; Hoyer et al., 2009; Zhang and Hyvärinen, 2009). These methods have been used in various application fields (Ramsey et al., 2014; Rosenström et al., 2012; Smith et al., 2011; Statnikov et al., 2012; Moneta et al., 2013). However, in many areas of empirical science, it is often difficult to accept the estimation results because latent confounders are ignored. In theory, we could take a non-Gaussian approach (Hoyer et al., 2008b) that uses an extension of ICA with more latent variables than observed variables (overcomplete ICA) to formally consider latent confounders in the framework of LiNGAM. Unfortunately, current versions of the overcomplete ICA algorithms are not very computationally reliable since they often suffer from local optima (Entner and Hoyer, 2011).
Thus, in this paper, we propose an alternative Bayesian approach to develop a method that is computationally simple in the sense that no iterative search in the parameter space is required and it is capable of finding the possible causal direction of two observed vari-ables in the presence of latent confounders. We first propose a variant of LiNGAM with individual- specific effects. Individual differences are sometimes the source of confounding (von Eye and Bergman, 2003). Thus, modeling certain individual-specific effects as latent variables allows a type of latent confounding to be considered. A latent confounding vari-able is an unobserved variable that exerts a causal influence on more than one observed variables (Hoyer et al., 2008b). The new model is still linear but allows any number of latent confounders. We then present a Bayesian approach for estimating the model by integrating out some of the large number of parameters, which is of the same order as the sample size. Such a Bayesian approach is often used in the field of mixed models (Demidenko, 2004) and multilevel models (Kreft and De Leeuw, 1998), although estimation of causal direction is not a topic studied within it.
Granger causality (Granger, 1969) is another popular method to aid detection of causal direction. His method depends on the temporal ordering of variables whereas our method does not. Therefore, our method can be applied to cases where temporal information is not available, i.e., cross-sectional data, as well as those where it is available, i.e., time-series data.
The remainder of this paper is organized as follows. We first review LiNGAM (Shimizu et al., 2006) and its extension to latent confounder cases (Hoyer et al., 2008b) in Section 2. In Section 3, we propose a new mixed-LiNGAM model, which is a variant of LiNGAM with individual-specific effects. We also propose an empirical Bayesian approach for learning the model. We empirically evaluate the performance of our method using artificial and real-world sociology data in Sections 4 and 5, respectively, and present our conclusions in Section 6.
2. Background
In this section, we first review the linear non-Gaussian structural equation model known as LiNGAM (Shimizu et al., 2006). We then discuss an extension of LiNGAM to cases where latent confounding variables exist (Hoyer et al., 2008b).
In LiNGAM (Shimizu et al., 2006), causal relations between observed variables xl (l = 1, ..., d) are modeled as
| (1) |
where k(l) is a causal ordering of the variables xl. The causal orders k(l) (l = 1, ..., d) are unknown and to be estimated. In this ordering, the variables xl form a directed acyclic graph (DAG) so that no later variable determines, i.e., has a directed path to, any earlier variable in the DAG. The variables el are latent continuous variables called error variables, μl are intercepts or regression constants, and blm are connection strengths or regression coefficients.
In matrix form, the LiNGAM model in Eq. (1) is written as
| (2) |
where the vector μ collects constants μl, the connection strength matrix B collects regression coefficients (or connection strengths) blm, and the vectors x and e collect observed variables xl and error variables el, respectively. The zero/non-zero pattern of blm corresponds to the absence/existence pattern of directed edges (direct effects). It can be shown that it is always possible to perform simultaneous, equal row and column permutations on the connection strength matrix B to cause it to become strictly lower triangular, based on the acyclicity assumption (Bollen, 1989). Here, strict lower triangularity is defined as a lower triangular structure with the diagonal consisting entirely of zeros. Errors el follow non-Gaussian distributions with zero mean and non-zero variance, and are jointly independent. This model without assuming non-Gaussianity distribution is called a fully recursive model in conventional structural equation models (Bollen, 1989). The non-Gaussianity assumption on el enables the identification of a causal ordering k(l) and the coefficients blm based only on x (Shimizu et al., 2006), unlike conventional Bayesian networks based on the Gaussianity assumption on el (Spirtes et al., 1993).
To illustrate the LiNGAM model, the following example is considered, whose corresponding directed acyclic graph is provided in Fig. 1:
| (3) |
Figure 1:
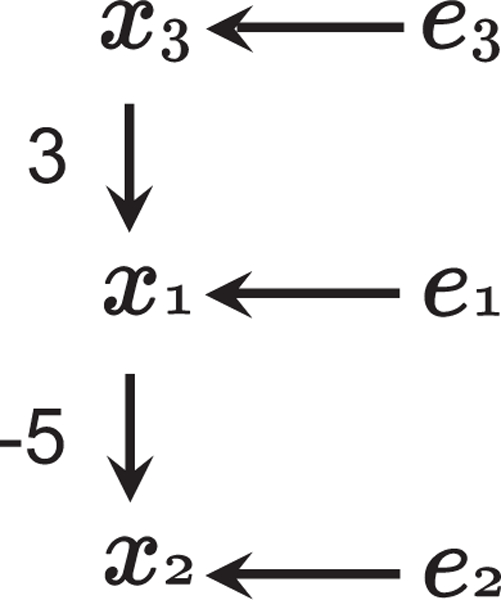
An example graph of LiNGAMs
In this example, x3 is equal to error e3 and is exogenous since it is not affected by either of the other two variables x1 and x2. Thus, x3 is in the first position of such a causal ordering such that B is strictly lower triangular, x1 is in the second, and x2 is the third, i.e., k(3) = 1, k(1) = 2, and k(2) = 3. If we permute the variables x1 to x3 according to the causal ordering, we have
| (4) |
It can be seen that the resulting connection strength (or regression coefficient) matrix is strictly lower triangular.
Several computationally efficient algorithms for estimating the model have been proposed (Shimizu et al., 2006, 2011; Hyvärinen and Smith, 2013). As with ICA, LiNGAM is identiable under the assumptions of non-Gaussianity and independence among error variables (Shimizu et al., 2006; Comon, 1994; Eriksson and Koivunen, 2003).1 However, for the estimation methods to be consistent, additional assumptions, e.g., the existence of their moments or some other statistic, must be made to ensure that the statistics computed in the estimation algorithms exist. The idea of LiNGAM can be generalized to nonlinear cases (Hoyer et al., 2009; Tillman et al., 2010; Zhang and Hyvärinen, 2009; Peters et al., 2011b).
The assumption of independence among el means that there is no latent confounding variable (Shimizu et al., 2006). A latent confounding variable is an unobserved variable that contributes to the values of more than one observed variable (Hoyer et al., 2008b). However, in many applications, there often exist latent confounding variables. If such latent confounders are completely ignored, the estimation results can be seriously biased (Pearl, 2000; Spirtes et al., 1993; Bollen, 1989). Therefore, in Hoyer et al. (2008b), LiNGAM with latent confounders, called latent variable LiNGAM, was proposed, and the model can be formulated as follows:
| (5) |
where fq are non-Gaussian individual-specific effects fq with zero mean and unit variance and λlq denote the regression coefficients (connection strengths) from fq to xl. This model is written in matrix form as follows:
| (6) |
where the difference from LiNGAM in Eq. (2) is the existence of a latent confounding variable vector f. The vector f collects fq. The matrix Λ collects λlq and is assumed to be of full column rank.
Another way to represent latent confounder cases would be to use dependent error variables. Denoting Λf + e in Eq. (6) by, , we have
| (7) |
| (8) |
Where are dependent due to the latent confounders fq. Observed variables that are equal to dependent errors, are connected by bi-directed arcs in their graphs. An example graph is given in Fig. 4. This representation can be more general since it is easier to extend it to represent nonlinearly dependent errors. In this paper, however, we use the aforementioned representation using independent errors and latent confounders since linear relations of the observed variables, latent confounders, and errors are necessary for our approach.
Figure 4:
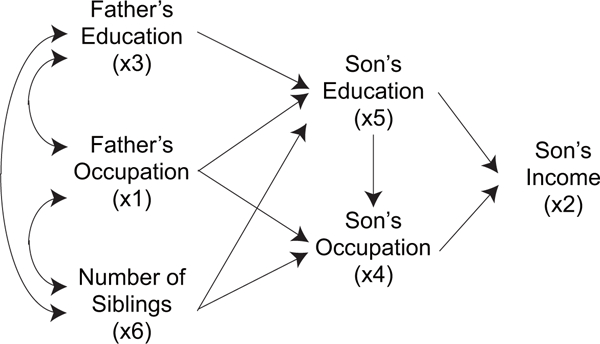
Status attainment model based on domain knowledge. Usually, the relations of x1, x3, and x6, represented by bi-directed arcs, are not modeled.
Without loss of generality, the latent confounders fq are assumed to be jointly independent since any dependent latent confounders can be remodeled by linear combinations of independent latent variables if the underlying model is linear acyclic and the error variables are independent (Hoyer et al., 2008b). To illustrate this, the following example model is considered:
| (9) |
| (10) |
| (11) |
| (12) |
| (13) |
| (14) |
where errors, , , and e1−e4 are non-Gaussian and independent. The associated graph is shown in Fig. 2. The relations of , , and x1−x4, are represented by a directed acyclic graph and latent confounders, and are dependent. In matrix form, this example model can be written as
| (15) |
Figure 2:
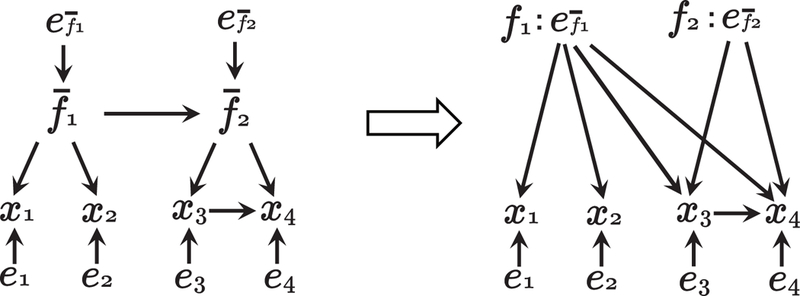
An example graph to illustrate the idea of independent latent confounders.
The relations of and to and in Eqs. (9)–(10):
| (16) |
we obtain
| (17) |
This is a latent variable LiNGAM in Eq. (6) taking, and since and , are non-Gaussian and independent.
Moreover, the faithfulness of xl and fq to the generating graph is assumed. The faithfulness assumption (Spirtes et al., 1993) here means that when multiple causal paths exist from one variable to another, their combined effect does not equal exactly zero (Hoyer et al., 2008b). The faithfulness assumption can be considered to be not very restrictive from the Bayesian viewpoint (Spirtes et al., 1993) since the probability of having exactly the parameter values that do not satisfy faithfulness is zero (Meek, 1995).
In the framework of latent variable LiNGAM, it has been shown (Hoyer et al., 2008b) that the following three models are distinguishable based on observed data2, i.e., the three different causal structures induce different data distributions:
| (18) |
| (19) |
| (20) |
where λ1qλ2q ≠ 0 due to the definition of latent confounders, that is, that they contribute to determining the values of more than two variables.
An estimation method based on overcomplete ICA (Lewicki and Sejnowski, 2000) explicitly modeling all the latent confounders fq was proposed (Hoyer et al., 2008b). However, in current practice, overcomplete ICA estimation algorithms often get stuck in local optima and are not sufficiently reliable (Entner and Hoyer, 2011). A Bayesian approach for estimating the latent variable LiNGAM in Eq. (6) has been proposed in Henao and Winther (2011). These previous approaches that explicitly model latent confounders (Hoyer et al., 2008b; Henao and Winther, 2011) need to select the number of latent confounders, and which can be quite large. This could lead to further computational difficulty and statistically unreliable estimates.
In Chen and Chan (2013), a simple approach based on fourth-order cumulants for estimating latent variable LiNGAM was proposed. Their approach does not need to explicitly model the latent confounders, however it requires the latent confounders fq to be Gaussian. The development of nonlinear methods that incorporate latent confounders is ongoing (Zhang et al., 2010).
None of these latent confounder methods incorporate the individual-specific effects that we model in the next section to consider latent confounders fq in the latent variable LiNGAM of Eq. (6).
3. Linear non-Gaussian acyclic structural equation model with individual-specific effects
In this section, we propose a new Bayesian method for learning the possible causal direction of two observed variables in the presence of latent confounding variables, assuming that the causal relations are acyclic, i.e., there is not a feedback relation.
3.1. Model
The LiNGAM (Shimizu et al., 2006) for observation i can be described as follows:
| (21) |
The random variables, are non-Gaussian and independent. The distributions of, are commonly assumed to be identical3 for every l. A linear non-Gaussian acyclic structural equation model with individual-specific effects for observation i is formulated as follows:
| (22) |
where the difference from LiNGAM is the existence of individual-specific effects, . The parameters, are independent of, and are correlated with through the structural equations in our Bayesian approach, introduced below. This means that the observations are generated from the identifiable LiNGAM, possibly with different parameter values of the means . We call this a mixed-LiNGAM, named after mixed models (Demidenko, 2004), as it has effects and that are common to all the observations and individual-specific effects . We note that causal orderings of variables k(l) (l = 1, ..., d) are identical for all the observations in the sample.
To use a Bayesian approach for estimating the mixed-LiNGAM, we need to model the distributions of error variables el and prior distributions of the parameters including individual-specific effects , unlike previous LiNGAM methods (Shimizu et al., 2006; Hoyer et al., 2008b). These individual-specific effects, whose number is of the same order as the sample size, are integrated out in the Bayesian method developed in Section 3.2, assuming an informative prior for them similar to the estimation of conventional mixed models (Demidenko, 2004). More details on the distributions of error variables and prior distributions of parameters are given in Section 3.2. These distributional assumptions were implied to be robust to some extent to their violations, at least in the artificial data experiments of Section 4.
We now relate the mixed-LiNGAM model above with the latent variable LiNGAM (Hoyer et al., 2008b). The latent variable LiNGAM in Eq. (6) for observation i is written as follows:
| (23) |
This is a mixed-LiNGAM taking . In contrast to the previous approaches for latent variable LiNGAM (Hoyer et al., 2008b; Henao and Winther, 2011), we do not explicitly model the latent confounders fq and rather simply include their sums in our model as its parameters since our main interest lies in estimation of the causal relation of observed variables xl and not in the estimation of their relations with latent confounders fq. Our method does not estimate λlq or the number of latent confounders Q.
3.2. Estimation of possible causal direction
We apply a Bayesian approach to estimate the possible causal direction of two observed variables using the mixed-LiNGAM proposed above. We compare the following two mixed- LiNGAM models with opposite possible directions of causation. Model 1 is
| (24) |
| (25) |
where b21 is non-zero. In Model 1, x2 does not cause x1. The second model, Model 2, is
| (26) |
| (27) |
where b12 is non-zero. In Model 2, x1 does not cause x2. The two models have the same number of parameters, but opposite possible directions of causation.
Once the possible causal direction is estimated, one can see if the common causal coefficient (connection strength) b21 or b12 is likely to be zero by examining its posterior distribution.4 We focus here on estimating the possible direction of causation as in many previous works (Dodge and Rousson, 2001; Hoyer et al., 2009; Zhang and Hyvärinen, 2009; Chen and Chan, 2013; Hyvärinen and Smith, 2013), and do not go to the computation of the posterior distribution5 since estimation of the possible causal direction of two observed variables in the presence of latent confounders has been a very challenging problem in causal inference and is the main topic of this paper.
We apply standard Bayesian model selection techniques to help assess the causal direction of x1 and x2. We use the log-marginal likelihood for comparing the two models. The model with the larger log-marginal likelihood is regarded as the closest to the true model (Kass and Raftery, 1995).
Let be the observed dataset , where . Denote Models 1 and 2 by M1 and M2. The log-marginal likelihoods of M1 and M2 are
| (28) |
| (29) |
| (30) |
where η1, η2 are the hyper-parameter vectors regarding the distributions of the parameters θ1and θ2, respectively. Since the last term log p() is constant with respect to Mr, we can drop it. To select suitable values for these hyper-parameters, we take an ordinary empirical Bayesian approach. First, we compute the log-marginal likelihood for every combination of the two models Mr and a number of candidate hyper-parameter values of ηr. Next, we take the model and hyper-parameter values that give the largest log-marginal likelihood, and finally estimate that the model with the largest log-marginal likelihood is better than the other model.
In basic LiNGAM (Shimizu et al., 2006), we have (Hyvärinen et al., 2010; Hoyer and Hyttinen, 2009)
| (31) |
Thus, in the same manner, the likelihoods under mixed-LiNGAM are given by
| (32) |
| (33) |
We model the parameters and their prior distributions as follows.6 The prior probabilities of M1 and M2 are uniform:
| (34) |
The distributions of the error variables and are modeled by Laplace distributions with zero mean and variances of var and var as follows:
| (35) |
| (36) |
Here, we simply use a symmetric super-Gaussian distribution, i.e., the Laplace distribution, to model and , as suggested in Hyvärinen and Smith (2013). Such super-Gaussian distributions have been reported to often work well in non-Gaussian estimation methods including independent component analysis and LiNGAM (Hyvärinen et al., 2001b; Hyvärinen and Smith, 2013). In some cases, a wider class of non-Gaussian distributions might provide a better model for and , e.g., the generalized Gaussian family (Hyvärinen et al., l 2 2001b), a finite mixture of Gaussians, or an exponential family distribution combining the Gaussian and Laplace distributions (Hoyer and Hyttinen, 2009).
The parameter vectors θ1 and θ2 are written as follows:
| (37) |
| (38) |
The prior distributions of common effects are Gaussian as follows:
| (39) |
| (40) |
| (41) |
| (42) |
| (43) |
| (44) |
where , , , , and are constants.
Generally speaking, we could use various informative prior distributions for the individual-specific effects and then compare candidate priors using the standard model selection approach based on the marginal likelihoods. Below we provide two examples.
If the data is generated from a latent variable LiNGAM, a special case of mixed-LiNGAM, as shown in Section 3.1, the individual-specific effects are the sums of many non-Gaussian independent latent confounders fq and are dependent. The central limit theorem states that the sum of independent variables becomes increasingly close to the Gaussian (Billingsley, 1986). Therefore, in many cases, it could be practical to approximate the non-Gaussian distribution of a variable that is the sum of many non-Gaussian and independent variables by a bell-shaped curve distribution (Sogawa et al., 2011; Chen and Chan, 2013). This motivates us to model the prior distribution of individual-specific effects by the multivariate t-distribution as follows:
| (45) |
where and are constants, and is a symmetric scale matrix whose diagonal elements are 1s. A random variable vector u that follows the multivariate t-distribution tv(0, Σ) can be created by where y follows the Gaussian distribution N(0, Σ), v follows the chi-squared distribution with v degrees of freedom, and y and v are statistically independent (Kotz and Nadarajah, 2004). Note that ui have energy correlations (Hyvärinen et al., 2001a), i.e., correlations of squares cov due to the common variable v. C is a diagonal matrix whose diagonal elements give the variance of elements of u, i.e., diag(Σ) for v > 2. The degree of freedom v is here taken to be six. The kurtosis of the univariate Student’s t-distribution with six degrees of freedom is three, the same as that of the Laplace distribution.
The hyper-parameter vectors η1 and η2 are
| (46) |
We want to take the constants , , , , and to be sufficiently large so that the priors for the common effects are not very informative. Whether these constants are sufficiently large depends on the scales of the variables. In the experiments in Sections 4–5, we set and so that they reflect the scales of the corresponding variables.
Moreover, we take an empirical Bayesian approach for the individual-specific effects. We test . That is, we uniformly vary the hyper-parameter value from that with no individual-specific effects, i.e., 0, to a larger value, i.e., , which implies very large individual differences. Further, we test σ12 = 0, ±0.3, ±0.5, ±0.7, ±0.9, i.e., the value with zero correlation and larger values with stronger correlations. This means that we test uncorrelated individual-specific effects as well as correlated ones. We take the ordinary Monte Carlo sampling approach to compute the log-marginal likelihoods with 1000 samples for the parameter vectors θr (r = 1,2).
The assumptions for our model are summarized in Table 1. Generally speaking, if the actual probability density function of individual-specific effects is unimodal and most often provides zero or very small absolute values and with few large values, i.e., many of the individual-specific effects are close to zero and many individuals have similar intercepts, the estimation is likely to work. If the individuals have very different intercepts, the estimation will not work very well.
Table 1:
Summary of the assumptions for our mixed-LiNGAM model
|
where blm are non-zero. |
| (l = 1, 2; i = 1, ..., n) are i.i.d.. |
| (l = 1, 2) are mutually independent. |
| (l = 1, 2) follow Laplace distributions with zero mean and standard deviations |
| Prior distributions: |
| μl, blm and hl (l = 1, 2; m = 1, 2; l ≠ m) follow Gaussian distributions with zero mean and variance |
| are the sum of latent confounders and are independent of . |
| (l = 1, 2; i = 1, ..., n) are i.i.d.. |
| μl (l = 1, 2) follow multivariate t-distributions with v degrees of freedom, zero mean, variances and correlation σ12 (here, v = 6). |
| Hyper-parameters: |
| (l = 1, 2; m = 1, 2; l ≠ m) are set to be large values so that the priors are not very informative. |
| (l = 1, 2) are uniformly varied from zero to large values. |
| σ12 are uniformly varied in the interval between −0.9 and 0.9. |
An alternative way of modeling the prior distribution of individual-specific effects would be to use the multivariate Gaussian distribution as follows:
| (47) |
where and are constants, and is a symmetric scale matrix whose diagonal elements are ones. Gaussian individual-specific effects or latent con- founders would not lead to losing the identifiability (Chen and Chan, 2013) since each observation still is generated by the identifiable non-Gaussian LiNGAM. However, if errors are Gaussian, there is no guarantee that our method can find correct possible causal direction. We could detect their Gaussanity by compariging our mixed-LiNGAM models with Gaussain error models based on their log-marginal likelihoods. If the errors are actually Gaussian or close to be Gaussian, Gaussian error models would provide larger log-marginal likelihoods. This would detect situations where our approach cannot find causal direction.
4. Experiments on artificial data
We compared our method with seven methods for estimating the possible causal direction between two variables: i) LvLiNGAM7 (Hoyer et al., 2008b); ii) SLIM8(Henao and Winther, 2011) iii) LiNGAM-GC-UK (Chen and Chan, 2013); iv) ICA-LiNGAM9 (Shimizu et al., 2006); v) DirectLiNGAM10 (Shimizu et al., 2011); vi) Pairwise LiNGAM11 (Hyvärinen and Smith, 2013); vii) Post-nonlinear causal model (PNL) 12 (Zhang and Hyvärinen, 2009). Their assumptions are summarized in Table 2. The first seven methods assume linearity, and the eighth allows a very wide variety of nonlinear relations. The last four methods assume that there are no latent confounders. We tested the prior t- and Gaussian distributions for individual-specific effects in our approach. LvLiNGAM and SLIM require to specify the number of latent confounders. We tested 1 and 4 latent confounder(s) for LvLiNGAM since its current implementation cannot handle more than four latent confounders, whereas we tested 1, 4 and 10 latent confounders(s) for SLIM. LiNGAM-GC-UK (Chen and Chan, 2013) assumes that errors are simultaneously super-Gaussian or sub-Gaussian and that latent confounders are Gaussian.
Table 2:
Summary of the assumptions of eight methods
| Functional form? | Latent confounders allowed? | Number of latent confounders necessary to be specified? | Iterative search in the parameter space required? | Distributional assumptions necessary? | |
|---|---|---|---|---|---|
| Our approach | Linear | Yes | No | No | Yes |
| LvLiNGAM | Linear | Yes | Yes | Yes | No13 |
| SLIM | Linear | Yes | Yes | No | Yes |
| LiNGAM-GC-UK | Linear | Yes | No | No | Yes |
| ICA-LiNGAM | Linear | No | N/A | Yes | No |
| DirectLiNGAM | Linear | No | N/A | No | No |
| Pairwise LiNGAM | Linear | No | N/A | No | No |
| PNL | Nonlinear | No | N/A | Yes | No |
We generated data using the following latent variable LiNGAM with Q latent confounding variables, which is a mixed-LiNGAM:
| (48) |
| (49) |
where μ1 and μ2 were randomly generated from N(0,1), and b21, λ1q, λ2q were randomly generated from the interval (−1.5, −0.5) ∪ (0.5, 1.5). We tested various numbers of latent confounders Q = 0, 1, 6, 12. The zero values indicate that there are no latent confounders. An example graph used to generate artificial data is given in Fig. 3.
Figure 3:
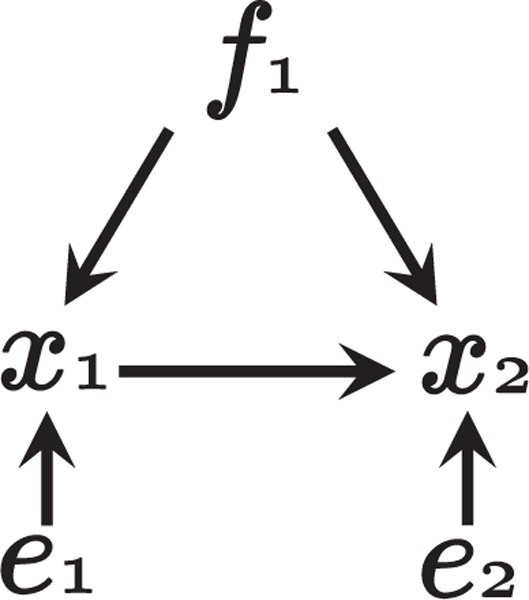
The associated graph of the model used to generate artificial data when the number of latent confounders Q = 1.
The distributions of the error variables e1, e2, and latent confounders fq were identical for all observations. The distributions of the error variables e1, e2, and latent confounders fq were randomly selected from the 18 non-Gaussian distributions used in Bach and Jordan (2002) to see if the Laplace distribution assumption on error variables and t- or Gaussian distribution assumption on individual-specific effects in our method were robust to different non-Gaussian distributions. These include symmetric/non-symmetric distributions, super-Gaussian/sub-Gaussian distributions, and strongly/weakly non-Gaussian distributions. The variances of e1 and e2 were randomly selected from the interval (0.52, 1.52). The variances of fq were 1s.
We permuted the variables according to a random ordering and inputted them to the eight estimation methods. We conducted 100 trials, with sample sizes of 50, 100, and 200. For the data with the number of latent confounders Q = 0, all the methods should find the correct causal direction for large enough sample sizes, as there were no latent confounders, which here means no individual-specific effects. The last four comparative methods should find the data with the number of latent confounders Q = 1,6,12 very difficult to analyze, because, unlike the other approaches, they assume no latent confounders.
To evaluate the performance of the algorithms, we counted the number of successful discoveries of possible causal direction and estimated their standard errors. We can calculate the 95% confidence intervals around the sample estimate of the number of successes based on the Gaussian approximation to the binomial distribution. If two approaches have overlapping confidence intervals, then they do not have a statistically significant difference at 5% level.
Table 3 shows the results. In the case with no latent confounders (Q = 0), the numbers of successes of linear methods that allow no latent confounders, ICA-LiNGAM, Di-rectLiNGAM, and Pairwise LiNGAM, were significantly larger or not significantly different from those of the methods that allow latent confounders. This would be because those latent confounder methods included many redundant parameters. In the cases that incorporated latent confounders (Q = 1, 6, 12), our two methods gave the largest numbers of successes for all the nine cases except the case of our method with Gaussian individual- specific effects with Q = 1 and sample size 200. In three of the nine cases, our two methods were significantly better than the others, although, in the other cases, they were not significantly different from LvLiNGAM or/and ICA-LiNGAM.
Table 3:
Number of successful discoveries (100 trials)
| Sample size | |||
|---|---|---|---|
| 50 | 100 | 200 | |
| Number of latent confounders Q = 0: | |||
| Our approach (t-distributed individual-specific effects) | 88 (3.25) | 91 (2.86) | 86 (3.47) |
| Our approach (Gaussian individual-specific effects) | 91 (2.86) | 87 (3.36) | 91 (2.86) |
| LvLiNGAM (1 latent confounder) | 73 (4.44) | 83 (3.76) | 83 (3.76) |
| LvLiNGAM (4 latent confounders) | 52 (5.00) | 68 (4.66) | 66 (4.74) |
| SLIM (1 latent confounder) | 29 (4.54) | 30 (4.58) | 25 (4.33) |
| SLIM (4 latent confounders) | 34 (4.74) | 31 (4.62) | 36 (4.80) |
| SLIM (10 latent confounders) | 30 (4.58) | 29 (4.54) | 30 (4.58) |
| LiNGAM-GC-UK | 33 (4.70) | 28 (4.49) | 35 (4.77) |
| ICA-LiNGAM | 93 (2.55) | 93 (2.55) | 96 (i.96) |
| DirectLiNGAM | 87 (3.36) | 95 (2.18) | 97 (1.71) |
| Pairwise LiNGAM | 89 (3.13) | 95 (2.18) | 95 (2.18) |
| Post-nonlinear causal model | 74 (4.39) | 71 (4.54) | 75 (4.33) |
| Number of latent confounders Q = 1: | |||
| Our approach t-distributed individual-specific effects) | 83 (3.76) | 80 (4.00) | 80 (4.00) |
| Our approach Gaussian individual-specific effects) | 79 (4.07) | 87 (3.36) | 69 (4.62) |
| LvLiNGAM (1 latent confounder) | 66 (4.74) | 71 (4.54) | 73 (4.44) |
| LvLiNGAM 4 latent confounders) | 63 (4.83) | 58 (4.94) | 67 (4.70) |
| SLIM (1 latent confounder) | 40 (4.90) | 47 (4.99) | 25 (4.33) |
| SLIM 4 latent confounders) | 40 (4.90) | 34 (4.74) | 44 (4.96) |
| SLIM (10 latent confounders) | 47 (4.99) | 39 (4.88) | 41 (4.92) |
| LiNGAM-GC-UK | 24 (4.27) | 32 (4.66) | 32 (4.66) |
| ICA-LiNGAM | 74 (4.39) | 71 (4.54) | 67 (4.70) |
| DirectLiNGAM | 48 (5.00) | 52 (5.00) | 54 (4.98) |
| Pairwise LiNGAM | 54 (4.98) | 58 (4.94) | 61 (4.88) |
| Post-nonlinear causal model | 55 (4.97) | 58 (4.94) | 57 (4.95) |
| Number of latent confounders Q = 6: | |||
| Our approach t-distributed individual-specific effects) | 88 (3.25) | 81 (3.92) | 87 (3.36) |
| Our approach Gaussian individual-specific effects) | 84 (3.67) | 85 (3.57) | 87 (3.36) |
| LvLiNGAM (1 latent confounder) | 58 (4.94) | 70 (4.58) | 70 (4.58) |
| LvLiNGAM 4 latent confounders) | 64 (4.80) | 61 (4.88) | 63 (4.83) |
| SLIM (1 latent confounder) | 50 (5.00) | 63 (4.83) | 47 (4.99) |
| SLIM 4 latent confounders) | 45 (4.97) | 47 (4.99) | 43 (4.95) |
| SLIM (10 latent confounders) | 58 (4.94) | 48 (5.00) | 58 (4.94) |
| LiNGAM-GC-UK | 29 (4.54) | 28 (4.49) | 21 (4.07) |
| ICA-LiNGAM | 74 (4.39) | 72 (4.49) | 47 (4.99) |
| DirectLiNGAM | 37 (4.83) | 48 (5.00) | 39 (4.88) |
| Pairwise LiNGAM | 48 (5.00) | 51 (5.00) | 37 (4.83) |
| Post-nonlinear causal model | 55 (4.97) | 42 (4.94) | 46 (4.98) |
| Number of latent confounders Q = 12: | |||
| Our approach t-distributed individual-specific effects) | 88 (3.25) | 86 (3.47) | 89 (3.13) |
| Our approach Gaussian individual-specific effects) | 91 (2.86) | 89 (3.13) | 91 (2.86) |
| LvLiNGAM (1 latent confounder) | 52 (5.00) | 55 (4.97) | 65 (4.77) |
| LvLiNGAM 4 latent confounders) | 65 (4.77) | 58 (4.94) | 64 (4.80) |
| SLIM (1 latent confounder) | 51 (5.00) | 55 (4.97) | 60 (4.90) |
| SLIM 4 latent confounders) | 45 (4.97) | 51 (5.00) | 63 (4.83) |
| SLIM 10 latent confounders) | 61 (4.88) | 54 (4.98) | 54 (4.98) |
| LiNGAM-GC-UK | 21 (4.07) | 25 (4.33) | 29 (4.54) |
| ICA-LiNGAM | 68 (4.66) | 72 (4.49) | 72 (4.49) |
| DirectLiNGAM | 37 (4.83) | 39 (4.88) | 38 (4.85) |
| Pairwise LiNGAM | 56 (4.96) | 42 (4.94) | 43 (4.95) |
| Post-nonlinear causal model | 51 (5.00) | 43 (4.95) | 46 (4.98) |
Largest numbers of successful discoveries were underlined.
Standard errors are shown in parentheses, which are computed assuming that the number of successes follow a binomial distribution.
Table 4 shows the average computational times. The computational complexity of the current implementation of our methods is clearly larger than that of the other linear methods ICA-LiNGAM, DirectLiNGAM, Pairwise LiNGAM, LvLiNGAM with 1 latent confounder, SLIM and LiNGAM-GC-UK and comparable to LvLiNGAM with 4 latent confounders and the nonlinear method PNL.
Table 4:
Average CPU time (s)
| Sample size | |||
|---|---|---|---|
| 50 | 100 | 200 | |
| Number of latent confounders Q = 0 | |||
| Our approach (t-distributed individual-specific effects) | 27.20 | 56.93 | 141.84 |
| Our approach (Gaussian individual-specific effects) | 35.48 | 69.59 | 117.10 |
| LvLiNGAM (1 latent confounder) | 2.41 | 2.55 | 9.91 |
| LvLiNGAM (4 latent confounders) | 22.25 | 30.12 | 87.96 |
| SLIM (1 latent confounder) | 5.89 | 6.25 | 6.81 |
| SLIM (4 latent confounders) | 7.60 | 8.14 | 9.13 |
| SLIM (10 latent confounders) | 10.88 | 12.02 | 13.96 |
| LiNGAM-GC-UK | 0.00 | 0.00 | 0.00 |
| ICA-LiNGAM | 0.04 | 0.03 | 0.02 |
| DirectLiNGAM | 0.00 | 0.01 | 0.01 |
| Pairwise LiNGAM | 0.00 | 0.00 | 0.00 |
| Post-nonlinear causal model | 19.59 | 27.68 | 57.37 |
| Number of latent confounders Q = 1: | |||
| Our approach (t-distributed individual-specific effects) | 35.87 | 65.55 | 131.25 |
| Our approach (Gaussian individual-specific effects) | 37.12 | 75.11 | 114.37 |
| LvLiNGAM (1 latent confounder) | 2.40 | 2.53 | 13.93 |
| LvLiNGAM (4 latent confounders) | 21.50 | 29.50 | 92.19 |
| SLIM (1 latent confounder) | 5.88 | 6.01 | 6.69 |
| SLIM (4 latent confounders) | 7.59 | 8.19 | 8.96 |
| SLIM (10 latent confounders) | 10.96 | 11.79 | 13.68 |
| LiNGAM-GC-UK | 0.00 | 0.00 | 0.00 |
| ICA-LiNGAM | 0.05 | 0.03 | 0.03 |
| DirectLiNGAM | 0.01 | 0.01 | 0.01 |
| Pairwise LiNGAM | 0.00 | 0.00 | 0.00 |
| Post-nonlinear causal model | 18.17 | 28.83 | 51.63 |
| Number of latent confounders Q = 6: | |||
| Our approach (t-distributed individual-specific effects) | 42.66 | 76.29 | 132.43 |
| Our approach (Gaussian individual-specific effects) | 33.13 | 69.07 | 104.83 |
| LvLiNGAM (1 latent confounder) | 2.40 | 2.56 | 9.38 |
| LvLiNGAM (4 latent confounders) | 22.17 | 30.12 | 83.01 |
| SLIM (1 latent confounder) | 5.89 | 6.22 | 6.77 |
| SLIM (4 latent confounders) | 7.58 | 8.18 | 9.11 |
| SLIM (10 latent confounders) | 11.03 | 12.02 | 13.91 |
| LiNGAM-GC-UK | 0.00 | 0.00 | 0.00 |
| ICA-LiNGAM | 0.06 | 0.05 | 0.05 |
| DirectLiNGAM | 0.01 | 0.01 | 0.01 |
| Pairwise LiNGAM | 0.00 | 0.00 | 0.00 |
| Post-nonlinear causal model | 18.71 | 29.62 | 52.21 |
| Number of latent confounders Q = 12: | |||
| Our approach (t-distributed individual-specific effects) | 29.16 | 59.30 | 134.89 |
| Our approach (Gaussian individual-specific effects) | 32.18 | 68.14 | 104.76 |
| LvLiNGAM (1 latent confounder) | 2.35 | 2.50 | 13.58 |
| LvLiNGAM (4 latent confounders) | 21.51 | 30.10 | 94.08 |
| SLIM (1 latent confounder) | 5.90 | 6.03 | 6.62 |
| SLIM (4 latent confounders) | 7.58 | 7.99 | 8.97 |
| SLIM (10 latent confounders) | 10.92 | 11.68 | 13.74 |
| LiNGAM-GC-UK | 0.00 | 0.00 | 0.00 |
| ICA-LiNGAM | 0.07 | 0.08 | 0.07 |
| DirectLiNGAM | 0.01 | 0.02 | 0.02 |
| Pairwise LiNGAM | 0.00 | 0.00 | 0.00 |
| Post-nonlinear causal model | 18.21 | 29.21 | 51.89 |
The MATLAB code for performing these experiments is available on our website.14
5. An experiment on real-world data
We analyzed the General Social Survey data set, taken from a sociological data repository (http://www.norc.org/GSS+Website/). The data consisted of six observed variables: x1: prestige of father’s occupation, x2: son’s income, x3: father’s education, x4: prestige of son’s occupation, x5: son’s education, and x6: number of siblings.15 The sample selection was conducted based on the following criteria: i) non-farm background; ii) ages 35–44; iii) white; iv) male; v) in the labor force at the time of the survey; vi) not missing data for any of the covariates; and vii) data taken from 1972–2006. The sample size was 1380.
The possible directions were determined based on the domain knowledge in Duncan et al. (1972), shown in Fig. 4. The causal relations of x1, x3, and x6 usually are not modeled in the literature since there are many other determinants of these three exogenous observed variables that are not part of the model. However, the possible causal directions among the three variables would be x1 ← x3, x6 ← x1, and x6 ← x3 based on their temporal orders.
Table 5 shows the results. Our mixed-LiNGAM approach with the t-distributed individual-specific effects gave the largest number of successful discoveries and achieved the highest precision. The second best method was our mixed-LiNGAM approach with the Gaussian individual-specific effects, which found one less correct possible directions than the t-distribution version. The third best method was LvLiNGAM with 1 latent confounder, which found two less correct possible directions than the t-distribution version. This would be mainly because our two methods allow individual-specific effects and the other methods do not, although these results are not significantly different for the number of trials 15.
Table 5:
Comparison of eight methods
| Possible directions | Our approach | LvLiNGAM | SLIM | ||||
|---|---|---|---|---|---|---|---|
| t-dist. | Gaussian | Num. | lat. conf. | Num. | lat. conf. | ||
| 1 | 4 | 1 | 4 | 10 | |||
| x1(FO) ← x3(FE) | ✓ | ✓ | ✓ | ✓ | |||
| x2(SI) ← x1(FO) | ✓ | ✓ | ✓ | ✓ | |||
| x2(SI) ← x3(FE) | ✓ | ✓ | ✓ | ✓ | ✓ | ||
| x2(SI) ← x4(SO) | ✓ | ✓ | ✓ | ✓ | |||
| x2(SI) ← x5(SE) | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | |
| x2(SI) ← x6(NS) | ✓ | ✓ | ✓ | ✓ | |||
| x4(SO) ← x1 (FO) | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
| x4(SO) ← x3(FE) | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | |
| x4(SO) ← x5(SE) | ✓ | ✓ | ✓ | ✓ | |||
| x4(SO) ← x6(NS) | ✓ | ✓ | ✓ | ✓ | ✓ | ||
| x5(SE) ← x1(FO) | ✓ | ||||||
| x5(SE) ← x3(FE) | ✓ | ✓ | ✓ | ✓ | ✓ | ||
| x5(SE) ← x6(NS) | ✓ | ✓ | ✓ | ✓ | ✓ | ||
| x6(NS) ← x1(FO) | ✓ | ✓ | |||||
| x6(NS) ← x3(FE) | ✓ | ✓ | ✓ | ✓ | |||
| Num. of successes | 12 | 11 | 10 | 9 | 9 | 7 | 8 |
| Standard errors | 1.55 | 1.71 | 1.83 | 1.90 | 1.90 | 1.93 | 1.93 |
| Precisions | 0.80 | 0.73 | 0.67 | 0.60 | 0.60 | 0.47 | 0.53 |
| Possible directions | LiNGAM-GC-UK | ICA | Direct | Pairwise | PNL | ||
| x1(FO) ← x3(FE) | ✓ | ✓ | |||||
| x2(SI) ← x1(FO) | ✓ | ✓ | ✓ | ||||
| x2(SI) ← x3(FE) | ✓ | ✓ | |||||
| x2(SI) ← x4(SO) | ✓ | ✓ | ✓ | ||||
| x2(SI) ← x5(SE) | ✓ | ✓ | |||||
| x2(SI) ← x6(NS) | ✓ | ✓ | |||||
| x4(SO) ← x1(FO) | ✓ | ✓ | |||||
| x4(SO) ← x3(FE) | ✓ | ✓ | |||||
| x4(SO) ← x5(SE) | ✓ | ✓ | |||||
| x4(SO) ← x6(NS) | ✓ | ||||||
| x5(SE) ← x1(FO) | ✓ | ✓ | ✓ | ||||
| x5(SE) ← x3(FE) | ✓ | ✓ | |||||
| x5(SE) ← x6(NS) | |||||||
| x6(NS) ← x1(FO) | ✓ | ✓ | |||||
| x6(NS) ← x3(FE) | ✓ | ✓ | ✓ | ||||
| Num. of successes | 3 | 8 | 9 | 2 | 9 | ||
| Standard errors | 1.55 | 1.93 | 1.90 | 1.32 | 1.90 | ||
| Precisions | 0.20 | 0.53 | 0.60 | 0.13 | 0.60 | ||
FO: Father’s Occupation
FE: Father’s Education
SI: Son’s Income
SO: Son’s Occupation
SE: Son’s Education
NS: Number of Siblings
ICA: ICA-LiNGAM (Shimizu et al., 2006)
Direct: DirectLiNGAM (Shimizu et al., 2011)
Pairwise: Pairwise LiNGAM (Hyvärinen and Smith, 2013)
PNL: Post-nonlinear causal model (Zhang and Hyvärinen, 2009)
Table 6 shows the estimated hyper-parameter values of our mixed-LiNGAM approach with the t-distributed individual-specific effects that performed best in the sociology data experiment. Either the estimated hyper-parameter and that represents the magnitudes of individual differences was non-zero in all pairs except (x4, x5). The non- ignorable influence of latent confounders was implied between the pairs (x2, x4), (x2, x6) and (x3, x6) since both or were non-zero for the pairs. In addition, for the pair (x2, x6), there might exist some nonlinear influence of latent confounders, since is zero, i.e., the individual-specific effects were linearly uncorrelated but dependent.16 If were larger, it would have implied a larger linear influence of the latent confounders on the pair (x2, x6). The estimates of the hyper-parameter were very large for the pairs (x2, x6) and (x4, x1), which implied very large individual differences regarding x2 and x4 respectively. This might imply that the estimated directions could be less reliable, although they were correct in this example.
Table 6:
Estimated hyper-parameter values of our method with t-distributed individual- specific effects
| Pairs analyzed | Possible directions |
Estimated directions |
|||
|---|---|---|---|---|---|
| (x1(FO), x3(FE)) | ← | ← | 0.42 (x1) | 0 | −0.7 |
| (x2(SI), x1(FO)) | ← | ← | 0.82 (x2) | 0 | 0.3 |
| (x2(SI), x3(FE)) | ← | ← | 0.82 (x2) | 0 | −0.5 |
| (x2(SI), x4(SO)) | ← | ← | 0.22 (x2) | 0.42 (x4) | −0.5 |
| (x2(SI), x5(SE)) | ← | ← | 0 | 0.42 (x5) | 0 |
| (x2(SI), x6(NS)) | ← | ← | 1.02 (x2) | 0.62 (x6) | 0 |
| (x4(SO), x1(FO)) | ← | ← | 1.02 (x4) | 0 | 0.9 |
| (x4(SO), x3(FE)) | ← | ← | 0 | 0.22 (x3) | −0.3 |
| (x4(SO), x5(SE)) | ← | ← | 0 | 0 | −0.3 |
| (x4(SO), x6(NS)) | ← | ← | 0.62 (x4) | 0 | −0.7 |
| (x5(SE), x1(FO)) | ← | → | 0 | 0.82 (x1) | 0.3 |
| (x5(SE), x3(FE)) | ← | ← | 0.62 (x5) | 0 | −0.5 |
| (x5(SE), x6(NS)) | ← | ← | 0.22 (x5) | 0 | −0.3 |
| (x6(NS), x1(FO)) | ← | → | 0.22 (x6) | 0 | −0.9 |
| (x6(NS), x3(FE)) | ← | → | 0.22 (x6) | 0.62 (x3) | 0.5 |
FO: Father’s Occupation
FE: Father’s Education
SI: Son’s Income
SO: Son’s Occupation
SE: Son’s Education
NS: Number of Siblings
represent the variances of the individual-specific effects for the variable pairs in the left-most column.
σ12 represents the correlation parameter value of the individual-specific effects for the variable pairs in the left-most column.
Another point to note is that both our methods with t-distributed and Gaussian individual-specific effects failed to find the possible direction x5 ← x1, although the causal relation is expected to occur from the domain knowledge (Duncan et al., 1972). This failure would be attributed to the model misspecification since the sample size was very large. Since the estimate of the hyper-parameter regarding x5 was zero, the influence of latent confounders might be small for this pair, although the estimate of was not small and the individual difference regarding x5 seemed substantial. Modeling both latent confounders and nonlinear relations and/or allowing a wider class of non-Gaussian distributions might lead to better performance. This is an important line of future research.
6. Conclusions
We proposed a new variant of LiNGAM that incorporated individual-specific effects in order to allow latent confounders. We further proposed an empirical Bayesian approach to estimate the possible causal direction of two observed variables based on the new model. In experiments on artificial data and real-world sociology data, the performance of our method was better than or at least comparable to that of existing methods.
For more than two variables, one approach would be to apply our method on every pair of the variables. Then, we can estimate a causal ordering of all the variables by integrating the estimation results. This approach is computationally much simper than trying all the possible causal orderings. Once a causal ordering of the variables is estimated, the remaining problem is to estimate regression coefficients or their posterior distributions. Then, one can see if there are direct causal connections between these variables. Although this could still be computationally challenging for large numbers of variables, the problem reduces to a significantly simpler one by identifying their causal orders. Thus, it is sensible to develop methods that can estimate causal direction of two variables allowing latent confounders.
Future work will focus on extending the model to allow cyclic and nonlinear relations and a wider class of non-Gaussian distributions as well as evaluating our method on various real-world data. Another important direction is to investigate the degree to which the model selection is sensitive to the choice of prior distributions.
Acknowledgments
S.S. was supported by KAKENHI #24700275. We thank Aapo Hyvärinen, Ricardo Silva and three reviewers for their helpful comments.
Footnotes
Comon (1994) and Erilsson and Koivunen (2003) established the identifiability of ICA based on the characteristic functions of variables. Moments of some variables may not exist, but their characteristic functions always exist.
If one or more error variables or latent confounders are Gaussian, it cannot be ensured that Models 3 to 5 will be distinguishable. Hoyer et al. (2008a) considered cases with one or more Gaussian error variables in the context of basic LiNGAM.
Relaxing this identically distributed assumption would lead to more general modeling of individual differences, howevers, however, this goes beyond the scope of the paper.
Chickering and Pearl (1996) considered a discrete variable model with known possible causal direction and proposed a Bayesian approach for computing the posterior distributions of causal effects in the presence of latent confounders
Point estimates of the parameters including the common causal connection strengths b12 and b12 can be obtained by taking their posterior means based on their posterior distributions, for example.
This is an example. The modeling method could depend on the domain knowledge.
Their current implementation of LvLiNGAM in Footnote 7 assumes a non-Gaussian distribution, which is a mixture of two Gaussian distributions.
Although x6 is discrete, it can be considered as continuous because it is an ordinal scale with many points.
Two variables that follow thw multivariate t-distribution are depedent, even when they are uncorrelated, as stated in section 3.2.
Contributor Information
Shohei Shimizu, The Institute of Scientific and Industrial Research Osaka University, Mihogaoka 8-1, Ibaraki, Osaka 567-0047, Japan.
Kenneth Bollen, Department of Sociology, CB 3210 Hamilton Hall University of North Carolina Chapel Hill, NC 27599-3210 U.S.A..
References
- Bach FR and Jordan MI Kernel independent component analysis. Journal of Machine Learning Research, 3:1–48, 2002. [Google Scholar]
- Billingsley P Probability and measure. Wiley-Interscience, 1986. [Google Scholar]
- Bollen K Structural Equations with Latent Variables. John Wiley & Sons, 1989. [Google Scholar]
- Chen Z and Chan L Causality in linear non Gaussian acyclic models in the presence of latent Gaussian confounders. Neural Computation, 25(6):1605–1641, 2013. [DOI] [PubMed] [Google Scholar]
- Chickering DM and Pearl J A clinician’s tool for analyzing non-compliance. In Proc. 13th National Conference on Artificial Intelligence (AAAI1996), pages 1269–1276, 1996. [Google Scholar]
- Comon P Independent component analysis, a new concept? Signal Processing, 36:62–83, 1994. [Google Scholar]
- Demidenko E Mixed models: Theory a,nd applications. Wiley-Interscience, 2004. [Google Scholar]
- Dodge Y and Rousson V On asymmetric properties of the correlation coefficient in the regression setting. The American Statistician, 55(1):51–54, 2001. [Google Scholar]
- Duncan OD, Featherman DL, and Duncan B Socioeconomic Background and Achievement. Seminar Press, New York, 1972. [Google Scholar]
- Entner D and Hoyer PO Discovering unconfounded causal relationships using linear non- gaussian models. In New Frontiers in Artificial Intelligence, Lecture Notes in Computer Science, volume 6797, pages 181–195, 2011. [Google Scholar]
- Eriksson J and Koivunen V Identifiability and separability of linear ICA models revisited. In Proc. Fourth International Conference on Independent Component Analysis and Blind Signal Separation (ICA2003), pages 23–27, 2003. [Google Scholar]
- Granger CWJ Investigating causal relations by econometric models and cross-spectral methods. Econometrica, 37(3):424–438, 1969. [Google Scholar]
- Henao R and Winther O Sparse linear identifiable multivariate modeling. Journal of Machine Learning Research, 12:863–905, 2011. [Google Scholar]
- Hoyer PO and Hyttinen A Bayesian discovery of linear acyclic causal models. In Proc. 25th Conference on Uncertainty in Artificial Intelligence (UAI2009), pages 240–248, 2009. [Google Scholar]
- Hoyer PO, Hyvärinen A, Scheines R, Spirtes P, Ramsey J, Lacerda G, and Shimizu S Causal discovery of linear acyclic models with arbitrary distributions. In Proc. 24th Conference on Uncertainty in Artificial Intelligence (UAI2008), pages 282–289, 2008a. [Google Scholar]
- Hoyer PO, Shimizu S, Kerminen A, and Palviainen M Estimation of causal effects using linear non-Gaussian causal models with hidden variables. International Journal of Approximate Reasoning, 49(2):362–378, 2008b. [Google Scholar]
- Hoyer PO, Janzing D, Mooij J, Peters J, and Scholkopf B Nonlinear causal discovery with additive noise models. In Advances in Neural Information Processing Systems 21, pages 689–696. 2009. [Google Scholar]
- Hyvärinen A and Smith SM Pairwise likelihood ratios for estimation of non-Gaussian structural equation models. Journal of Machine Learning Research, 14:111–152, 2013. [PMC free article] [PubMed] [Google Scholar]
- Hyvärinen A, Hoyer PO, and Inki M Topographic independent component analysis. Neural Computation, 13(7):1527–1558, 2001a. [DOI] [PubMed] [Google Scholar]
- Hyvärinen A, Karhunen J, and Oja E Independent component analysis. Wiley, New York, 2001b. [Google Scholar]
- Hyvärinen A, Zhang K, Shimizu S, and Hoyer PO Estimation of a structural vector autoregressive model using non-Gaussianity. Journal of Machine Learning Research, 11: 1709–1731, 2010. [Google Scholar]
- Kass RE and Raftery AE Bayes factors. Journal of the American Statistical Association, 90(430):773–795, 1995. [Google Scholar]
- Kotz Samuel and Nadarajah Saralees. Multivariate t-distributions and their applications. Cambridge University Press, 2004. [Google Scholar]
- Kreft IGG and De Leeuw J Introducing Multilevel Modeling. Sage, 1998. [Google Scholar]
- Lacerda G, Spirtes P, Ramsey J, and Hoyer PO Discovering cyclic causal models by independent components analysis. In Proc. 24th Conference on Uncertainty in Artificial Intelligence (UAI2008), pages 366–374, 2008. [Google Scholar]
- Lewicki M and Sejnowski TJ Learning overcomplete representations. Neural Computation, 12(2):337–365, 2000. [DOI] [PubMed] [Google Scholar]
- Meek C Strong completeness and faithfulness in Bayesian networks. In Proc. 11th Conference on Uncertainty in Artificial Intelligence, pages 411–418. Morgan Kaufmann Publishers Inc., 1995. [Google Scholar]
- Moneta A, Chlaß N, Entner D, and Hoyer P Causal search in structural vector autoregressive models. In Journal of Machine Learning Research: Workshop and Conference Proceedings, Causality in Time Series (Proc. NIPS2009 Mini-Symposium on Causality in Time Series), volume 12, pages 95–114, 2011. [Google Scholar]
- Moneta A, Entner D, Hoyer PO, and Coad A Causal inference by independent component analysis: Theory and applications. Oxford Bulletin of Economics and Statistics, 75 (5):705–730, 2013. [Google Scholar]
- Pearl J Causality: Models, Rea,soning, and Inference. Cambridge University Press, 2000. (2nd ed 2009). [Google Scholar]
- Peters J, Janzing D, and Scholkopf B Causal inference on discrete data using additive noise models. IEEE Transactions on Pattern Analysis and Machine Intelligence, 33(12): 2436–2450, 2011a. [DOI] [PubMed] [Google Scholar]
- Peters J, Mooij J, Janzing D, and Scholkopf B Identifiability of causal graphs using functional models. In Proc. 27th Conference on Uncertainty in Artificial Intelligence (UAI2011), pages 589–598, 2011b. [Google Scholar]
- Ramsey JD, Sanchez-Romero R, and Glymour C Non-Gaussian methods and high-pass filters in the estimation of effective connections. NeuroImage, 84(1):986–1006, 2014. [DOI] [PubMed] [Google Scholar]
- Rosenstrom T, Jokela M, Puttonen S, Hintsanen M, Pulkki-Raback L, Viikari JS, Raitakari OT, and Keltikangas-Jarvinen L Pairwise measures of causal direction in the epidemiology of sleep problems and depression. PloS ONE, 7(11):e50841, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu S, Hoyer PO, Hyvärinen A, and Kerminen A A linear non-Gaussian acyclic model for causal discovery. Journal of Machine Learning Research, 7:2003–2030, 2006. [Google Scholar]
- Shimizu S, Inazumi T, Sogawa Y, Hyvärinen A, Kawahara Y, Washio T, Hoyer PO, and Bollen K DirectLiNGAM: A direct method for learning a linear non-Gaussian structural equation model. Journal of Machine Learning Research, 12:1225–1248, 2011. [Google Scholar]
- Smith SM, Miller KL, Salimi-Khorshidi G, Webster M, Beckmann CF, Nichols TE, Ramsey JD, and Woolrich MW Network modelling methods for FMRI. NeuroImage, 54(2):875–891, 2011. [DOI] [PubMed] [Google Scholar]
- Sogawa Y, Shimizu S, Shimamura T, Hyvärinen A, Washio T, and Imoto S Estimating exogenous variables in data with more variables than observations. Neural Networks, 24 (8):875–880, 2011. [DOI] [PubMed] [Google Scholar]
- Spirtes P and Glymour C An algorithm for fast recovery of sparse causal graphs. Social Science Computer Review, 9:67–72, 1991. [Google Scholar]
- Spirtes P, Glymour C, and Scheines R Causation, Prediction, and Search. Springer Verlag, 1993. (2nd ed MIT Press; 2000). [Google Scholar]
- Spirtes P, Glymour C, Scheines R, and Tillman R Automated search for causal relations: Theory and practice In Dechter R, Geffner H, and Halpern J, editors, Heuristics, Probability, and Causality: A Tribute to Judea Pearl, pages 467–506. College Publications, 2010. [Google Scholar]
- Statnikov A, Henaff M, Lytkin NI, and Aliferis CF New methods for separating causes from effects in genomics data. BMC Genomics, 13(Suppl 8):S22, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillman RE, Gretton A, and Spirtes P Nonlinear directed acyclic structure learning with weakly additive noise models. In Advances in Neural Information Processing Systems 22, pages 1847–1855, 2010. [Google Scholar]
- von Eye A and Bergman LR Research strategies in developmental psychopathology: Dimensional identity and the person-oriented approach. Development and psychopathology, 15(3):553–580, 2003. [DOI] [PubMed] [Google Scholar]
- Zhang K and Hyvärinen A On the identifiability of the post-nonlinear causal model. In Proc. 25th Conference in Uncertainty in Artificial Intelligence (UAI2009), pages 647–655, 2009. [Google Scholar]
- Zhang K, Schölkopf B, and Janzing D Invariant Gaussian process latent variable models and application in causal discovery. In Proc. 26th Conference in Uncertainty in Artificial Intelligence (UAI2010), pages 717–724, 2010. [Google Scholar]


