Abstract
We present a scheme that propagates a reference skeletal model (s-rep) into a particular case of an object, thereby propagating the initial shape-related layout of the skeleton-to-boundary vectors, called spokes. The scheme represents the surfaces of the template as well as the target objects by spherical harmonics and computes a warp between these via a thin plate spline. To form the propagated s-rep, it applies the warp to the spokes of the template s-rep and then statistically refines. This automatic approach promises to make s-rep fitting robust for complicated objects, which allows s-rep based statistics to be available to all. The improvement in fitting and statistics is significant compared with the previous methods and in statistics compared with a state-of-the-art boundary based method.
Index Terms—: Correspondence propagation, medical imaging, skeletal model, statistical analysis, thin plate spline
I. Introduction
The ability to accurately and robustly represent sets of similar objects is an important and well-studied problem in computer vision [1], [2] and medical image analysis applications [3], [4]. Skeletal models for representing objects have shown particular strengths. As a result of their property of providing a shape-based coordinate system for the object interior and near exterior, they provide special capabilities for mechanical modeling [5]–[8] and for the image match term used in segmentation [9], [10]. Because they capture not only global boundary locations but also local object width properties and boundary directional properties, they have provided stronger statistical summaries of object populations, and these have led to improved prior terms needed for segmentation, which together with the advantages for the image match term have yielded superior segmentations [9].
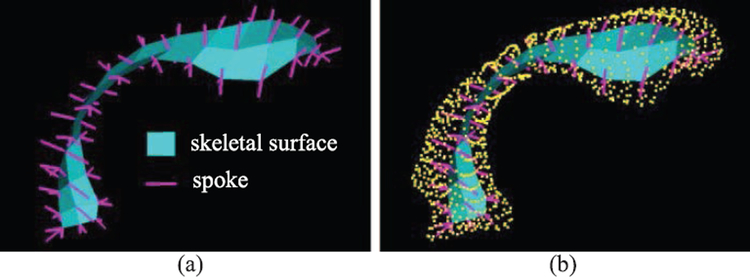
A particular form of quasi-medial skeletal model called the s-rep[11] has been shown in numerous recent papers to be more powerful for various statistical pattern recognition objectives, e.g., in classification (diagnosis) [12], hypothesis testing [13] and in the general probability distribution properties of specificity, generalization, and compactness [14] as compared to boundary point distribution models (PDMs) [15]. This s-rep consists of a grid of spoke vectors proceeding from the skeletal surface to the object boundary (Fig. 1).
Fig. 1.
(a) The template lateral ventricle s-rep; (b) that s-rep with its SPHARM boundary shown as yellow points. The magenta lines proceeding from the skeletal surface (cyan) to the object’s boundary are spokes.
To gain this expanded capability, s-reps must be fit tightly to the training objects in a way producing correspondence. Methods previously available [11], [13], [16] to obtain such fits can be summarized as 1) define a template s-rep (denoted as ); 2) solve an optimization problem that fits the interpolated form of to each target object; 3) compute the mean of the generated s-reps; 4) repeat this fitting process by replacing with the mean. This standard fitting process has been tedious to use and has required much manual intervention, leading to weaknesses in correspondence as well as limited use of this representation by others than those in or closely collaborating with our laboratory. Moreover, it performs poorly for more complex objects with variable bending and twisting.
Means of propagating a reference model into a particular object have been applied to PDMs. Cootes et al. presented such a method based on PDM statistics, active shape models [17]. Davies et al. [18] proposed a method for improving such statistical shape models by putting them into inter-object correspondence based on minimum description length. Styner et al. [19] demonstrated a thin plate spline (TPS) warping that maps objects to a common medial branching topology while matching their PDM boundaries perfectly.
In this letter we improve s-rep fitting by initializing the optimization in steps 2–4 above with a TPS-based propagation of into the target object. For complicated objects this leads to much more automatic fits with good correspondence. This promises to make the advantages of s-reps described above available to all users of shape statistics.
The propagation uses the spherical harmonics point distribution model (SPHARM-PDM) [3] representing both and the target object as the basis for computing the TPS warp and then applies that warp to the skeleton-to-boundary spoke vectors of .
Our main contribution is two-fold: 1) a novel scheme for fitting significantly improved s-reps via TPS warping; 2) an effective way to propagate the correspondence provided by the initial shape model.
The remainder of this letter is organized as follows. Section II describes the input and related formulations. Section III presents our proposed method. Experimental results are given in Section IV, followed by a discussion in Section V.
II. Initial Shape Model and SPHARM Surface
The input to the proposed method is a predefined template s-rep (see Fig. 1(a)), which is iteratively fitted to the object using the standard pipeline discussed in Section I under supervision; and a population of target PDMs sampled from each object. These PDMs can be those extended from any 3D surface detection method (e.g., [20], [21]). As aforementioned, here we use the SPHARM-PDM which is an up-to-date, open source, public available framework that has been extensively used in shape statistics [14], [18], [22]–[25] and medical image applications [26]–[29] to describe binary segmented magnetic resonance (MR) images. Spherical harmonics (SPHARM) describes a surface x(θ, φ) using
| (1) |
where the basis functions , −l ≤ m ≤ l and order m are defined on θ ∈ [0, π] × φ ∈ [0, 2π] and where the 3D coefficients are obtained by solving the least-squares problems in each spatial coordinate directions.
Every point pi on the surface is one-to-one mapped to a parameter vector (θi, φi) on the unit sphere. The bijective mapping of the surface to the sphere is done by modifying the parameter vectors in a constrained optimization procedure considering minimal quadrilateral distortion and area preservation that is used to force every object region to map to a region of proportional area in parameter space. Each object’s optimization is preceded by a setting of its axis and prime meridian using second moments of its {pi}.
A homogeneous sampling of the spherical parameter space uses a linear, uniform icosahedron subdivision along each edge of the original icosahedron. Suppose we get a set of parameter vectors (θi, φi) through the homogeneous sampling on the spherical parameter space. The PDM of the object surface can be obtained directly by putting the coefficients into Eq. (1), thus a sampled point at location (θi, φi) takes on the form:
| (2) |
where K is a linear subdivision level of the icosahedron, which was selected depending on the complexity of the objects.
In this letter each lateral ventricle was sampled by a linear subdivision level K = 10, which composes a PDM consisting of 1002 points. All PDMs were normalized to the unit space.
III. Method
The main issue addressed in this letter is the automatic and robust TPS-based propagation of a reference s-rep into unseen target objects. The following sections present the main components of our novel scheme: 1) get TPS deformations from to each of the target PDMs; 2) warp by each TPS deformation; 3) refine the warped target s-reps.
A. Thin Plate Spline Deformation
Given landmarks {pk = (xk, yk, zk), k = 1, …, m} that must map into target landmarks {pk′ = (xk′, yk′, zk′), k = 1, …, m}, the TPS [30] provides the deformation that minimizes the bending energy
| (3) |
where denotes the squares of the second-order partial derivatives. That deformation maps any point p = (x, y, z) into the target point p′ = (x′, y′, z′) by the equation
| (4) |
where U(s) = s2ln(s) and the three values in the translation , the nine values in the 3 × 3 matrix A, and the 3m values of the weights ωk of the warp basis functions U(|p − pk|) are computed by solving linear equations involving vectors connecting corresponding landmarks in {pk} and {pk′}.
B. Deriving the Warps for Target PDMs
The process starts from getting the SPHARM boundary for (Fig. 1(a)), which is achieved by applying the same formula as for the target PDMs presented in Section II. The resulting PDM acts as the template PDM (see Fig. 1(b)).
The calculation of the warp Tj for each target PDM j can be done by solving the linear equations discussed in Section III-A, where the landmarks {pk} are the boundary points in the SPHARM-PDM derived from and the landmarks {pk′} are analogous points in each of the target PDMs. Applying Tj to spoke’s two endpoints p yields the corresponding spoke endpoints p′.
For a population of N target objects, we get a set of mapping functions {Tj,j = 1,…,N} each defining a warp Tj that can be applied to deform the spoke endpoint pairs in to get its warped target s-rep.
C. Creating Initializing Warped S-Reps
The process of creating a warped target s-rep (denoted as ) is summarized in Table I. The set of transforms {Tj} are applied to the “landmark pairs” (the tail and tip of each spoke) of the template s-rep. The resulting “landmark pairs” are used to produce the jth target s-rep. Each spoke has a position (the coordinate of the spoke tail), a direction (a unit vector pointing from tail to tip) and a radius (the length of the spoke vector).
TABLE I.
Process for the Creation of Target S-Reps
| Input: Tsrep and {Tj, j = 1, … , N.} |
|---|
| Output: {} |
| for the jth TPS transformation Tj |
| for each spoke of Tsrep |
| tpsSpokeTail = applyTPS(Tj, spokeTail); |
| tpsSpokeTip = applyTPS(Tj, spokeTip); |
| spokeRadius = calculateSpokeRadius(tpsSpokeTail, tpsSpokeTip); |
| spokeDirection = calculateUnitDir(tpsSpokeTail, tpsSpokeTip); |
| saveNewSpoke(tpsSpokeTail, spokeRadiu, spokeDirection); |
| end |
| saveNewSrep(); |
| end |
These warped s-reps can be refined by slightly modifying each spoke’s length and direction to optimize the fit to the binary image. The refinement process is beyond the scope of this letter. The evaluations described in the next section are all based on the warped s-reps without refinement.
IV. Experimental Results
The proposed method was evaluated on a set of real world lateral ventricle objects semi-automatically segmented from MR images in neonate datasets [31]. We selected 94 lateral ventricles for our tests presented here. The program was implemented in C++, all experiments were done on a 64-bit 3.20 GHz Intel Quad Core PC with 8 GB RAM. It takes about 11 minutes to get the SPHARM-PDM surface, 5 minutes to get the TPS propagated s-rep, and 29 minutes to get the standardly fitted s-rep for one object.
We first investigated the smoothness of the surfaces implied by the propagated s-reps resulting from the proposed approach. Then we compared these s-reps with those from the standard method. Following this we evaluated the statistics of these s-reps via three commonly used measurements: generalization G(M), specificity S(M) and compactness C(M), which were first introduced by Davies [32] and have been widely used in previous literature [18], [26], [33]. Briefly, a lower value is desirable for all three metrics. Finally, we studied the shape variability captured by the proposed method and the baseline.
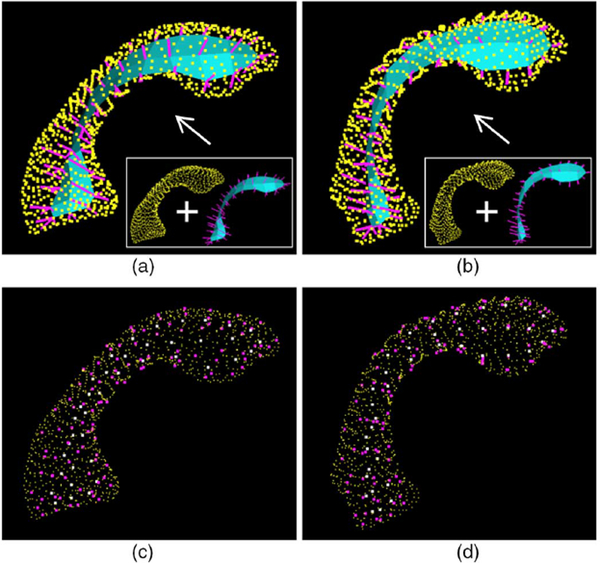
As described in Section II, our input objects were described by the SPHARM-PDMs, which also provide the ground truth to evaluate if our warped s-reps imply the correct object boundaries (similar to the input surfaces). Fig. 2 shows two example objects described by SPHARM-PDMs with the corresponding warped target s-reps shown inside.
Fig. 2.
Visualization of two example objects (each column is an object) with the propagated s-reps shown inside of their own baseline (SPHARM-PDM). (a) and (b) are the s-reps fitted into the baseline (yellow points); (c) and (d) are the spokes ends (spoke tail (white), spoke tip (magenta)) fitted into the baseline.
In Figs. 2(a),(b) we can see that all the spokes (magenta lines) located in the object interior (bounded by the yellow points). All the spoke tips (magenta points in Figs. 2(c), (d) lie approximately on the baseline surface (yellow points, the SPHARMPDM). These tell us that our warped target s-reps achieve a rather smooth surface.
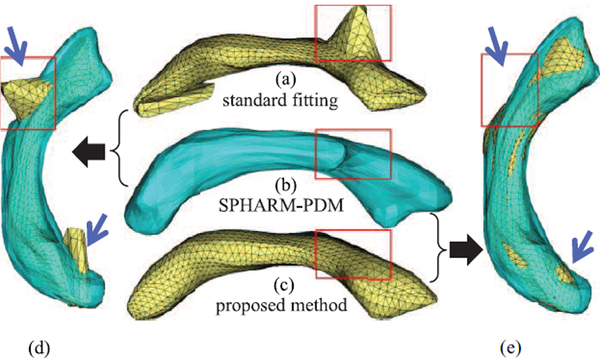
To further evaluate the propagated fitting, we compared the implied boundary of the s-reps resulting from the proposed method with those from the aforementioned standard fitting process that was extensively used for fitting relatively simple objects (e.g., [12], [13], [15], [34]). Results show that the proposed method achieves reasonable smooth surfaces with improved overlap with the target object, while there are error regions from their methods (e.g., Fig. 3).
Fig. 3.
(a) The surface for the standardly fitted s-rep; (b) the surface for the SPHARM-PDM (which is the baseline); (c) the surface for our propagated s-rep; (d) and (e) are the overlap of the baseline onto (a) and (c), respectively. The red frames indicate the approximate corresponding positions. The blue arrows indicate the significant differences of the two methods in comparison. (a) standard fitting (b) SPHARM-PDM (c) proposed method.
The erroneous bumps in Figs. 3(a), (d) show the surface implied by a standardly fitted s-rep. These bumps need to be adjusted manually followed by redoing of the fitting; even this doesn’t guarantee better fit for complicated objects. But our method (Figs. 3(c), (e)) automatically yields significantly improved s-reps with smooth surfaces. This is because the TPS warps are globally smooth and robust to narrow/thin regions, unlike the previous method for matching the s-rep model to the objects. Also, problems of poor convergence of the previous optimization method when initialized poorly are avoided.
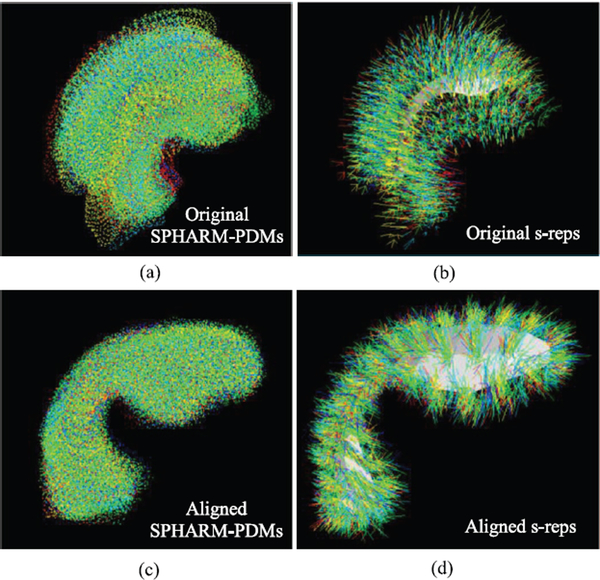
To evaluate the statistics of the resulting propagated s-reps, a Procrustes alignment was performed to remove the translation, scaling and rotation variances introduced by each model. Figs. 4(a), (b) display all the 94 samples in the population together; each was represented by the SPHARM-PDMs and the s-reps. The alignment brings the shapes closer (Figs. 4(c), (d)).
Fig. 4.
All the 94 training shapes overlaid on top of each other. Each shape is described by the SPHARM-PDM (a) and the s-rep (b). After applying the Procrustes alignment, the shapes described by both shape models get close and tight as (c) and (d). Colors indicate different shapes in the population.
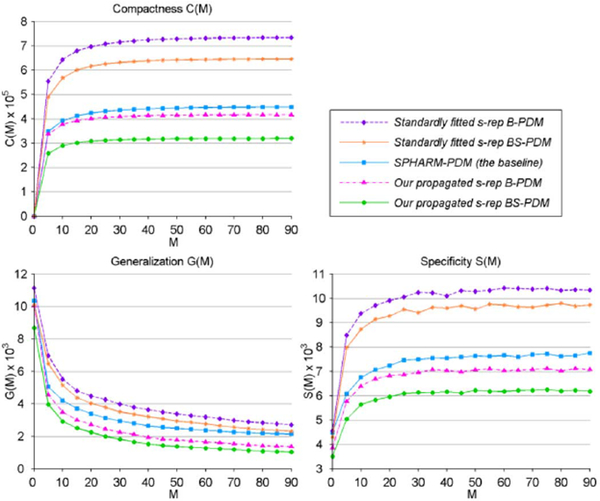
The correspondence quality among our propagated s-reps, the standardly fitted s-reps and the baseline are compared in Fig. 5. We collected two types of PDMs implied by s-rep spokes: B-PDM, which has 106 points (only spoke tip points) and BS-PDM, which has 212 points (spoke tail-and-tip points). Fig. 5 tells us that the proposed method achieves lower values than other methods in all three measurements, which means that our warped s-reps are superior.
Fig. 5.
Comparisons of correspondence quality among different PDMs. M is the shape parameters used for constructing new instances.
Table II lists the contribution of the first six eigenmodes for B-PDM and BS-PDM from our warped s-reps and the baseline; we can see that the total shape variances captured by these eigenmodes are 83.5%, 83.9% and 80.4% respectively. This suggests that the proposed model captures more shape variance even if we only consider the object boundaries implied by s-reps (B-PDM) and describes the object with lower dimension. On inspection, all three types of PDMs appear to be of good quality; each main eigenmode describes a plausible pattern of variation observed in the population (see Fig. 6 for a visualization of the first eigenmode).
TABLE II.
Shape Variances of Different Methods (%)
| Point set | λ1 | λ2 | λ3 | λ4 | λ5 | λ6 | Sum |
|---|---|---|---|---|---|---|---|
| B-PDM | 42.7 | 15.3 | 11.3 | 6.2 | 5.0 | 3.0 | 83.5 |
| BS-PDM | 37.8 | 18.6 | 11.8 | 7.1 | 5.5 | 3.1 | 83.9 |
| SPHARM-PDM | 40.4 | 15.7 | 10.5 | 6.0 | 5.1 | 2.9 | 80.4 |
Fig. 6.
From left to right column: B-PDM, BS-PDM and SPHARM-PDM. The middle row is the mean shape resulting from different point sets; the top and bottom rows are respectively.
V. Conclusion
We presented a novel scheme that propagates a reference skeletal model () to a set of biomedical objects to obtain their fitted s-reps. This is done by representing the surfaces of as well as the target objects by spherical harmonics and computing a thin plate spline warp between these, and applying this warp to . Experimental results proved that 1) this automatic scheme creates stable s-reps that are robust for complicated objects; 2) the propagated s-reps have significantly improved fitting and model properties as compared with the standardly fitted s-reps; 3) the propagated s-reps in the presence of considerable shape variability gain over the baseline. The resulting s-reps can be further statistically improved in spoke correspondence (e.g., [14]). In the future, we expect to obtain better fits by using shape change statistics in the refinement step. The resulting s-reps can be applied to achieve better results on classification, hypothesis testing and probability distribution estimation, as well as a variety of medical image applications dependent on these statistical analyses.
Acknowledgments
This work was supported in part by the National Institutes of Health (NIH) under Grants MH064065, MH070890, and MH091351, and in part by the National Natural Science Foundation of China under Grants 61173131, 91118005 and 11202249. The associate editor coordinating the review of this manuscript and approving it for publication was Prof. Edmund Lam.
Footnotes
Color versions of one or more of the figures in this paper are available online at http://ieeexplore.ieee.org.
Contributor Information
Liyun Tu, College of Computer Science, Chongqing University, Chongqing 400044 China, and also with the Department of Computer Science, University of North Carolina at Chapel Hill, NC 27599 USA..
Dan Yang, College of Computer Science, Chongqing University, Chongqing 400044 China.; School of Software Engineering, Chongqing University, Chongqing 400044 China, and also with the Key Laboratory of Dependable Service Computing in Cyber Physical Society Ministry of Education, Chongqing 400044 China
Jared Vicory, Department of Computer Science, University of North Carolina, Chapel Hill, NC 27599 USA..
Xiaohong Zhang, School of Software Engineering, Chongqing University, Chongqing 400044 China, and also with the Key Laboratory of Dependable Service Computing in Cyber Physical Society Ministry of Education, Chongqing 400044 China.
Stephen M. Pizer, Department of Computer Science, University of North Carolina, Chapel Hill, NC 27599 USA.
Martin Styner, Department of Computer Science and the Department of Psychiatry, University of North Carolina, Chapel Hill, NC 27599 USA.
References
- [1].Ruan Z, Wang G, Xue J-H, and Lin X, “Subcategory clustering with latent feature alignment and filtering for object detection,” IEEE Signal Process. Lett, vol. 22, no. 2, pp. 244–248, 2015. [Google Scholar]
- [2].Bai Y and Tang M, “Object tracking via robust multitask sparse representation,” IEEE Signal Process. Lett, vol. 21, no. 8, pp. 909–913, 2014. [Google Scholar]
- [3].Styner M, Oguz I, Xu S, Brechbühler C, Pantazis D, Levitt JJ, Shenton ME, and Gerig G, “Framework for the statistical shape analysis of brain structures using SPHARM-PDM,” in Open Science Workshop at MICCAI - Insight J, 2006, no. 1071, pp. 242–250. [PMC free article] [PubMed] [Google Scholar]
- [4].Gerig G, Styner M, and Szekely G, “Statistical shape models for segmentation and structural analysis,” in Proc. IEEE Int. Symp. Biomed. Imag, 2002, pp. 18–21. [Google Scholar]
- [5].Yushkevich PA, “Continuous medial representation of brain structures using the biharmonic PDE,” NeuroImage, vol. 45, pp. S99–S110, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Yushkevich PA, Zhang H, and Gee JC, “Continuous medial representation for anatomical structures,” IEEE Trans. Med. Imag, vol. 25, no. 12, pp. 1547–1564, 2006. [DOI] [PubMed] [Google Scholar]
- [7].Crouch JR, Pizer SM, Chaney EL, Hu Y-C, Mageras GS, and Zaider M, “Automated finite-element analysis for deformable registration of prostate images,” IEEE Trans. Med. Imag, vol. 26, no. 10, pp. 1379–1390, 2007. [DOI] [PubMed] [Google Scholar]
- [8].Yushkevich PA and Zhang HG, “Deformable modeling using a 3D boundary representation with quadratic constraints on the branching structure of the Blum skeleton,” in Proc. Inf. Proc. Med. Imag, Berlin, 2013, pp. 280–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Chaney EL and Pizer SM, “Autosegmentation of images in radiation oncology,” J. Amer. Coll. Radiol, vol. 6, no. 6, pp. 455–458, 2009. [DOI] [PubMed] [Google Scholar]
- [10].Stough JV, Broadhurst RE, Pizer SM, and Chaney EL, “Regional appearance in deformable model segmentation,” in Proc. Inf. Proc. Med. Imag, 2007, vol. 20, pp. 532–543. [DOI] [PubMed] [Google Scholar]
- [11].Pizer SM, Jung S, Goswami D, Vicory J, Zhao X, Chaudhuri R,Damon JN, Huckemann S, and Marron JS, “Nested sphere statistics of skeletal models,” in Innovations for Shape Analysis: Models and Algorithms, Part I, LNCS. Berlin, Germany: Springer, 2013, pp. 93–115. [Google Scholar]
- [12].Jung S and Qiao X, “A statistical approach to set classification by feature selection with applications to classification of histopathology images,” Biometrics, vol. 70, no. 3, pp. 536–545, 2014. [DOI] [PubMed] [Google Scholar]
- [13].Schulz J, Pizer SM, Marron JS, and Godtliebsen F, “Nonlinear hypothesis testing of geometric object properties of shapes applied to hippocampi,” J. Math. Imag. Vis, 2015, in press. [Google Scholar]
- [14].Tu L, Styner M, Vicory J, Paniagua B, Prieto JC, Yang D, and Pizer SM, “Skeletal shape correspondence via entropy minimization,” in Proc. SPIE Med. Imag, Florida, 2015, vol. 9413, p. 94130U. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Pizer SM, Hong J, Jung S, Marron JS, Schulz J, and Vicory J, “Relative statistical performance of s-reps with principal nested spheres vs. PDMs,” in Proc. Shape 2014- Symp. of Stat. Shape Models and Appl., Delémont, France, 2014, vol. 10, pp. 11–13. [Google Scholar]
- [16].Han Q, Merck D, Levy J, Villarruel C, Damon JN, Chaney EL, and Pizer SM, “Geometrically proper models in statistical training,” in Proc. Inf. Proc. Med. Imag, 2007, vol. 20, pp. 751–762. [DOI] [PubMed] [Google Scholar]
- [17].Cootes TF, Taylor CJ, Cooper DH, and Graham J, “Active shape models-their training and application,” Comput. Vis. Image Understand, vol. 61, no. 1, pp. 38–59, 1995. [Google Scholar]
- [18].Davies RH, Twining CJ, Cootes TF, and Taylor CJ, “Building 3-D statistical shape models by direct optimization,” IEEE Trans. Med. Imag, vol. 29, no. 4, pp. 961–981, 2010. [DOI] [PubMed] [Google Scholar]
- [19].Styner M and Gerig G, “Automatic and robust computation of 3d medial models incorporating object variability,” Int. J. Comp. Vis, vol. 55, no. 3, pp. 107–122, 2003. [Google Scholar]
- [20].Cates J, Fletcher PT, Styner M, Shenton M, and Whitaker R, “Shape modeling and analysis with entropy-based particle systems,” in Proc. Inf. Proc. Med. Imag, 2007, vol. 20, pp. 333–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Smith S and Williams I, “A statistical method for improved 3D surface detection,” IEEE Signal Process. Lett, vol. 22, no. 8, pp. 1045–1049, 2015. [Google Scholar]
- [22].Gerig G, Styner M, Shenton ME, and Lieberman JA, “Shape versus size: Improved understanding of the morphology of brain structures,” in Proc. Med. Image Comp. Comput.-Assist. Intervention, Utrecht, The Netherlands, 2001, vol. 2208, pp. 24–32. [Google Scholar]
- [23].Styner M, Lieberman JA, Pantazis D, and Gerig G, “Boundary and medial shape analysis of the hippocampus in schizophrenia,” Med. Image Anal, vol. 8, pp. 197–203, 2004. [DOI] [PubMed] [Google Scholar]
- [24].Shen L, Firpi HA, Saykin AJ, and West JD, “Parametric surface modeling and registration for comparison of manual and automated segmentation of the hippocampus,” Hippocampus, vol. 19, no. 6, pp. 588–595, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Paniagua B, Bompard L, Cates J, Whitaker R, Datar M, Vachet C, and Styner M, “Combined SPHARM-PDM and entropy-based particle systemsashape analysis framework,” in Proc. SPIE Med. Imag, 2012, vol. 8317, p. 83170L. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Xu R, Zhou X, Hirano Y, Tachibana R, Hara T, Kido S, and Fujita H, “Particle system based adaptive sampling on spherical parameter space to improve the MDL method for construction of statistical shape models,” Comput. Math. Method. M, pp. 1–9, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Oguz I, “Groupwise shape correspondence with local features,” Ph.D. dissertation, Dept. of Comput. Sci., Univ. North Carolina, Chapel Hill, NC, USA, 2009. [Google Scholar]
- [28].Paniagua B, Cevidanes L, Zhu H, Styner M, and Gerig G, “Outcome quantification using spharm-pdm toolbox in orthognathic surgery,” Int.J. Comput. Assist. Radiol. Surg, vol. 6, no. 5, pp. 617–626, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Meier D and Fisher E, “Parameter space warping: Shape-based correspondence between morphologically different objects,” IEEE Trans. Med. Imag, vol. 21, no. 1, pp. 31–47, 2002. [DOI] [PubMed] [Google Scholar]
- [30].Bookstein FL, “Principal warps: Thin-plate splines and the decomposition of deformations,” IEEE Trans. Patt. Anal. Mach. Intell, vol. 11, no. 6, pp. 567–585, 1989. [Google Scholar]
- [31].Paniagua B, Lyall A, Berger J-B, Vachet C, Hamer RM, Woolson S, Lin W, Gilmore J, and Styner M, “Lateral ventricle morphology analysis via mean latitude axis,” in Proc. SPIE Med. Imag, 2013, vol. 8672, p. 86720M. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Davies RH, “Learning shape: optimal models for analysing natural variability,” Ph.D. dissertation, Dept. Imag. Sci. Biomed. Eng., Univ. Manchester, Manchester, U.K, 2002. [Google Scholar]
- [33].Styner MA, Rajamani KT, Nolte L-P, Zsemlye G, Sźekely G. a.,Taylor CJ, and Davies RH, “Evaluation of 3D correspondence methods for model building,” in Proc. Inf. Proc. Med. Imag, 2003, vol. 18, pp. 63–75. [DOI] [PubMed] [Google Scholar]
- [34].Schulz J, Jung S, Huckemann S, Pierrynowski M, Marron JS, and Pizer SM, “Analysis of rotational deformations from directional data,” J. Comput. Graph. Statist, vol. 24, no. 2, pp. 539–560, 2015. [Google Scholar]