Abstract
Phase separation has emerged as a new paradigm currently revolutionizing our understanding of cell biology and intracellular organization. Disordered protein domains have recently been demonstrated as integral drivers of phase separation into condensed liquids with emergent material properties. Using in vitro model systems employing purified protein components is necessary to interrogate the molecular mechanisms underlying phase separation; however, these systems pose many experimental challenges. In this chapter we describe general strategies for purifying, handling, imaging, and characterizing the phase behavior of disordered proteins. We further outline methods for the purification of the model P granule protein LAF-1, the construction of phase diagrams, and the quantification of liquid droplet fusion or coalescence.
1. INTRODUCTION
Phase of biomolecules into condensed material states has recently emerged as a key principle underlying intracellular organization (Banani, Lee, Hyman, & Rosen, 2017; Boeynaems et al., 2018; Dolgin, 2018; Shin & Brangwynne, 2017). Intrinsically disordered proteins or domains have been identified as major drivers of phase separation into liquid droplets, including the P granule proteins LAF-1 (Elbaum-Garfinkle et al., 2015), PGL-3 (Saha et al., 2016) and MEG proteins (Smith et al., 2016; Wang et al., 2014), the germ granule protein DDX4 (Nott et al., 2015), the nucleolar protein Fib1 (Berry, Weber, Vaidya, Haataja, & Brangwynne, 2015; Feric et al., 2016), and stress granule proteins FUS (Burke, Janke, Rhine, & Fawzi, 2015; Murakami et al., 2015), TDP-43 (Conicella, Zerze, Mittal, & Fawzi, 2016), and hnRNPA1 (Lin, Protter, Rosen, & Parker, 2015; Molliex et al., 2015).
Reconstituting intracellular biomolecular condensates using in vitro model systems of purified disordered proteins or domains can offer significant insight into the mechanisms underlying phase separation and is a rapidly growing area of research. However, phase-separating proteins are often difficult to purify, handle, and characterize. This chapter outlines several methods and strategies for studying phase-separating disordered proteins, including protein purification, hunting for droplets, generating phase diagrams, and quantifying droplet morphology and coalescence.
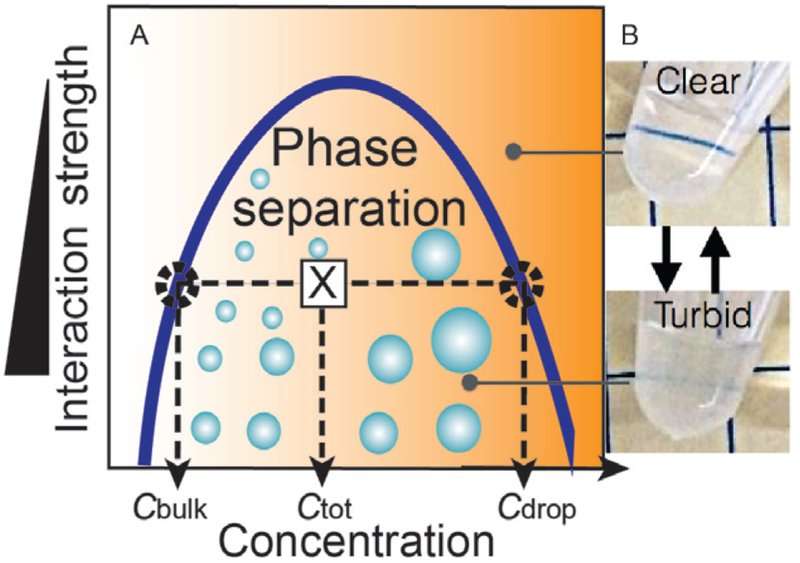
A liquid–liquid phase separation system in equilibrium can be quantitatively described by a phase diagram—a state diagram that delineates the regions where phase separation will and will not occur (Fig. 1A). The phase boundary, also known as a liquid–liquid coexistence curve or binodal, defines the two-phase or droplet forming region and the single-phase homogenous region where no phase separation occurs. The left arm of the binodal denotes the concentration of the bulk solution outside the droplets, i.e., the saturation concentration, while the right side denotes the significantly higher concentration of protein within the condensed phase droplets. Such diagrams are derived using the Flory–Huggins formalism and mean field theory approximation (Brady et al., 2017; Brangwynne, Tompa, & Pappu, 2015) where the enthalpy of interaction between molecules must be strong enough to counterbalance the entropy of mixing, for phase separation (i.e., demixing) to occur. In this framework, the Y-axis reflects the interaction strength, while the X-axis denotes protein concentration. Therefore, anything that tunes the interaction strength between molecules, including temperature, pH, ionic strength, etc., can influence their resulting phase separation behavior. Consideration of potential phase diagram parameters is important when strategizing the purification and characterization protocols for proteins suspected of phase separation.
Fig. 1.
Phase separation. (A) Phase diagram, binodal or liquid–liquid coexistence curve for a simplified 2-component system delineates the phase-separating region. Any starting total concentration/interaction strength pair that falls within that region—will result in phase separation. At equilibrium, the left side of the binodal denotes the concentration outside the droplets or the saturation concentration, while the right side denotes the concentration inside the droplets (B) LAF-1 protein solutions reversibly turn turbid upon phase separation.
2. METHODS
2.1. Purification Strategies
Phase separation of protein during the purification process often results in significant protein loss due in part to the increased viscosity of sticky condensed phases. To prevent phase separation all potential phase diagram interaction parameters, such as temperature, ionic strength, pH, or protein concentration, should be considered. Here, we briefly discuss general strategies for optimizing purification protocols of proteins prone to phase separation. These strategies are further illustrated in the detailed purification protocol for the P granule model protein LAF-1 in Section 2.2.
As with proteins that are prone to any concentration-dependent self-assembly, including aggregation and phase separation, care should be taken to avoid high local protein concentrations. During cell growth, such measures include careful monitoring of the OD600 of Escherichia coli cultures before induction—we find that induction at OD600 values of ~0.4 significantly reduce protein lost to the insoluble fraction compared to the more standard OD600 values of ~0.6—0.7. Similarly, overnight growth at cool temperatures (~18°C) after induction is often preferred. Additionally, purification column protocols may need to be adjusted to avoid drastic increases in local protein concentration that may induce phase separation on the column. Column modifications include, increasing column resin volumes or performing batch purifications by incubating dilute protein solutions with free resin. Furthermore, protein solutions should be buffer exchanged with extra care, especially if using centrifugal filter concentrators—spin times should be short and solutions should be frequently mixed inside the concentrator so that high local concentrations do not accumulate at the bottom of the centrifugal filter device.
If phase separation is suspected to have occurred buffer conditions should be systematically modified. A classic signature of liquid phase separation is the appearance of turbid solutions (Fig. 1B), resulting from the light scattered by liquid droplets in solution. Sudden and/or mysterious loss of large amounts of protein during a purification could also be a sign of phase separation, as the viscous phase-separated droplets tend to stick to tube walls and surfaces. Temperature is a very strong interaction parameter for phase separation and can be easily modified during purification. For example, while most proteins require purification at 4°C, proteins like LAF-1 more readily phase separate at cooler temperatures; therefore, we perform most of the purification at room temperature. Likewise, ionic strength, pH, and other buffer additives can be tuned to optimize protein yield. When working with a new protein, it is useful to pay attention to buffer changes that significantly alter yield, as this could indicate interaction parameters that will be useful when subsequently characterizing the protein’s phase behavior.
2.2. LAF-1 Purification Protocol
2.2.1. Buffers
All buffers, except lysis buffer, are stored and used at room temperature.
Lysis buffer (stored at 4°C)
20 mM Tris–HCl, pH 7.4
500 mM NaCl
10 mM imidazole
10% (vol/vol) glycerol
1% Triton-X
Added day of: 1 mg/mL lysozyme and protease inhibitor mixture one tablet EDTA-free per 50 mL (Roche Diagnostics), 14 mM β-mercaptoethanol, 1 mM PMSF
Nickel wash buffer
20 mM Tris–HCl, pH 7.4
500 mM NaCl
10% (vol/vol) glycerol
25 mM imidazole
Added day of: 14 mM β-mercaptoethanol
Nickel elution buffer
20 mM Tris–HCl, pH 7.4
500 mM NaCl
10% (vol/vol) glycerol
250 mM imidazole
Added day of: 14 mM β-mercaptoethanol
Heparin binding buffer
20 mM Tris–HCl, pH 7.4
50 mM NaCl
1% (vol/vol) glycerol
Added day of: 2 mM DTT
Heparin elution buffer
20 mM Tris–HCl, pH 7.4
1 M NaCl
1% (vol/vol) glycerol
Added day of: 2 mM DTT
2.2.2. Protocol
2.2.2.1. Cell Growth
LAF-1 plasmid, which has been codon optimized for Escherichia coli, synthesized and inserted into a Kanamycin resistant pET28a backbone with a N-terminal 6x-His tag (Elbaum-Garfinkle et al., 2015) is transformed and expressed in BL21 (DE3) cells according to standard protocols. LAF-1 protein yields are very sensitive to OD. We find it is essential to induce at relatively low OD of 0.3–0.4, followed by overnight growth at 18°C with 500 mM IPTG. Cell pellets can be flash frozen or immediately lysed and purified.
2.2.2.2. Cell Lysis
Add lysis buffer to pelleted (and/or thawed) cells to resuspend (~15–20 mL/L of culture). Gently swirl buffer over cell pellet until dissolved.
-
Transfer to 50-mL conical tubes. Incubate immersed in ice bucket for 30–60 min—swirl tube every so often.
Note: this is a good time to prepare the nickel beads for the purification (see later)
Sonicate solutions on ice to further disrupt/lyse cells—pulse on/off 10s, Amp: 40%, Time: 2min total.
Centrifuge balanced tubes at 20,000 × g for 30 min. Decant supernatant (cleared cell lysate) into fresh conical tube and discard cell pellet (cell debris).
2.2.2.3. Nickel Column Purification: “Batch” (Thermo Fisher Ni-NTA)
- Preparing nickel beads:
- Invert 50% nickel slurry several times before pipetting from the bottle to ensure it is well mixed.
- Remove desired amount and place in 15-mL Falcon tube. A good starting estimate is ~0.5 mL slurry for each Liter of original growth. (Note: growths are usually several liters; If you are doing a small prep and are only using 0.5 mL or less of slurry, then use an Eppendorf tube instead of a 15-mL conical tube to make sure you can get a nice pellet of the nickel resin).
- Add 10–12 mL of nickel wash buffer to the 15-mL conical tube. Invert several times to mix.
- Centrifuge at ~600 × g for 2min to pellet the nickel beads.
- Carefully pour or pipette off wash buffer into a waste bottle.
- Repeat steps c–e 3–4 × more times
-
Combine washed nickel beads with cell lysate. Tape to rocker at 4°C. Allow protein to bind the nickel for ~30 min with gentle rocking.
Note: from this point forward we work at room temperature.
Distribute lysate-bead mixture into several 15-mL Falcon tubes and centrifuge at 600 × g for 2min to collect the beads. Pour/pipette off supernatants and combine into 50-mL Falcon tube labeled “Flow Through.” Be very careful not to pour off any of the nickel beads.
Add ~10 mL nickel wash buffer to tubes and mix by inverting tube several times or gentle pipetting. Centrifuge at 600 × g for 2min. Pour/pipette off supernatant and combine into 50-mL Falcon tube labeled “Wash 1.” Repeat 2 × for “Wash 2” and “Wash 3.” Add additional 1–3 mL of wash buffer to each tube to resuspend nickel beads and combine all beads into a single 15 mL Falcon tube. Collect the beads by centrifugation. Remove supernatant, leaving minimal buffer to transfer beads to column.
Add nickel beads to a prepared, capped, disposable hand-held gravity flow column with filter and let the nickel beads sediment. Do not allow flow out of the column during this time! May use additional Nickel Wash Buffer to remove as much of the nickel beads as possible from the Falcon tube.
Once nickel beads have settled into a packed bed, open stopper and collect flow through. Do not let the bed dry out, stop flow just above top of bed.
Protein elution: Add nickel elution buffer carefully so as not to disturb the nickel bed. Collect elution fractions in Eppendorf tubes (0.5–1.0 mL fractions).
Measure protein concentration via absorption at 280-nm for each tube. Combine the tubes with protein and proceed with Heparin Column to further purify and remove any bound RNA. (Note: may stop here to run a gel).
2.2.2.4. Heparin Column Purification (GE HiTrap Heparin Columns, 1 or 5 mL)
Filter Heparin Buffers before use. Use Lueur lock syringes connected “drop-to-drop” with column to avoid introducing air to the column as per manufacturer instructions.
Equilibrate 1 or 5 mL column with 10× column volumes of Heparin Binding buffer
- Dilute protein obtained from nickel resin purification at least 5 × with room temperature Heparin Binding Buffer and load onto column.
- Note: If the protein concentration is sufficiently high, the solution will instantly become turbid as droplets form! Proceed quickly with loading onto the column. The protein still binds and is eluted as a clear solution with high salt buffer.
Load the protein solution on the column and collect flow through.
Wash with 10 × column volume Heparin Binding Buffer (i.e., 10 or 50 mL) and collect.
Elute with (~2 × column volume) Heparin Elution Buffer and collect in <~0.5 mL aliquots. Check concentrations of each fraction by absorbance at 280 nm. Pool protein-containing fractions, avoiding very dilute fractions so as not to reduce to the final protein concentration.
Add glycerol to pooled protein fractions to 10% (vol/vol), aliquot to desired volumes, and flash freeze with liquid nitrogen for storage in −80°C.
2.2.2.5. Thawing LAF-1 Protein Aliquots
LAF-1 aliquots are thawed by placing them on the bench top at room temperature. We find that both slow thawing on ice or fast thawing at 37°C promote aggregation. It is best to let aliquots thaw, untouched, on the bench top before proceeding. Typically, we perform a 2min high speed centrifugation step (maximum speed in a small table-top centrifuge) to pellet any aggregates, and then buffer exchange at room temperature into fresh storage buffer (20 mM Tris, pH 7.4, 1 M Nacl, 1 mM DTT).
2.3. Mapping Phase Diagrams
2.3.1. Hunting for Droplets/Identifying an Interaction Parameter
Determining whether or not your protein of interest phase separates, and/or identifying the conditions that promote or deter phase separation can be a laborious and often daunting process. Proteins may or may not require a binding partner to phase separate, including protein partners and/or RNA or other nucleotide analogs/small molecules. Additionally, macromolecular crowding agents may be required and can be sampled as necessary. Thus, the “hunt for droplets” is in many ways analogous to protein crystallography screens, where proteins of various concentrations are combined with a spectrum of buffer conditions and/or additives, incubated at multiple temperatures and screened via imaging or other forms of spectroscopy.
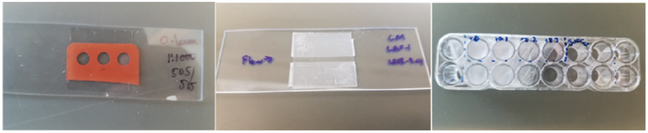
To determine phase separating conditions, we mix protein solutions in an imaging chamber with coverslip bottom (N = 1.5) and image over time using a high magnification brightfield, DIC or fluorescence inverted microscope. Different sized imaging chambers or wells can be used in order to accommodate varying sample conditions. For small, quick screens we make glass slide-coverslip chambers, either with rubber gaskets (Grace BioLabs) or with melted parafilm (Fig. 2). Larger culture wells (such as Nunc or Grace BioLabs) can be used for larger volumes (100–200 μL) and are easier to work with if solution components are added/mixed in at later times. For larger screens, we also use 96- or 384-well plates.
Fig. 2.
Imaging chambers glass slide-coverslip chambers made with Grace BioLabs spacers (left) and melted parafilm (middle). Grace BioLabs CultureWells with coverslip bottom (right).
Additional notes
One should be very cautious in evaluating the presence of droplets, especially if the protein is unlabeled. Condensation of any kind can give the appearance of droplet artifacts, including immersion oil on the bottom surface of the coverslip! Additionally, any evaporation or leakage, particularly problematic for small volume slides, can confound images and results. Sanity checks or controls for these artifacts include, always including a no-droplet control, cleaning the slide surfaces, and paying close attention to the Z-plane as you are imaging.
Fluorescently labeling protein, either through fusion to fluorescent proteins or via conjugation to fluorescent dyes following purification, can facilitate “droplet hunting” by improving image signal and minimizing nonprotein droplet artifacts. However, controls with unlabeled protein are strongly recommended as the phase behavior can be significantly influenced by fluorescent protein modifications.
To improve image contrast without fluorescence protein fusion or conjugation, small amounts of free fluorescent dyes can be added to the unlabeled protein solution. Many dyes will partition unequally between the condensed droplet phase and the dilute phase, depending on the dye–protein pair. Even here, care must be taken, as too much free dye, especially if it partitions into the droplet, may affect the phase behavior.
It is not always trivial to discern between the formation of liquid droplets and other types of assemblies or aggregates by imaging alone, especially if the assemblies are close to the resolution limit. For assemblies at least a few microns in size, liquid droplets appear spherical above the surface and may deform upon wetting the surface (see Section 2.4). Morphology and fusion events may be difficult to assess for assemblies closer to the resolution limit.
2.3.2. Mapping a Phase Boundary
As described above, a liquid–liquid phase diagram or coexistence curve delineates the conditions that do or do not give rise to phase separation. Mapping a full or partial phase diagram can be a useful first step in working with protein condensates, as it provides a framework for experimental setup and strategy. In addition to convenience, phase diagrams provide important information about the interactions that promote phase separation. LAF-1 droplets, for example, are very sensitive to salt concentration suggesting that electrostatics are very important. Other systems that are more driven by hydrophobic interactions may be less sensitive to salt, but may be more sensitive to temperature fluctuations. It is also insightful to see how certain additional perturbations to the system shift the phase diagram. For instance, macromolecular crowding agents often shift the phase diagram to the left, requiring less protein to phase separate. Experiments can be further designed to determine how sequence mutations and various binding partners or competitors influence the phase diagram and thus the interaction strengths of the system.
As depicted in Fig. 1A, a full phase diagram or binodal consists of left and right arms, denoting the concentrations of the bulk or dilute phase and droplet or condensed phase, respectively. Determination of full binodals has been recently described (Brady et al., 2017; Wei et al., 2017). Here, we outline complementary methods to determine the critical concentration for phase separation, or the left arm of the binodal/phase diagram.
2.3.3. Determining the Critical/Saturation Concentration Via Imaging
Optimize an imaging system for small volume samples for high throughput as described above. Prepare samples of varying protein concentration and interaction parameter (i.e., salt). It is best to mix directly within the imaging chamber, or immediately before placing samples into the imaging chamber. If condensed droplets form, they can stick to many surfaces and thus become lost during transfer.
Use a known droplet forming condition to determine the timescale of droplet formation and system equilibrium. For LAF-1, droplets form instantaneously for most conditions and settle down onto the coverslip roughly within 2h. For LAF-1 phase diagrams, mixed samples were observed after approximately 3h. Note: Some protein systems may phase separate on significantly longer timescales and the protocol should be adjusted accordingly.
Beginning with a previously determined droplet forming condition, decrease protein concentration until no droplets are observable. Then narrow down the phase boundary by testing intervening conditions. For any given set of conditions, measurements should be made a minimum of three times to ensure reproducibility. Any set of conditions is scored “yes/no” depending upon whether droplets are observed.
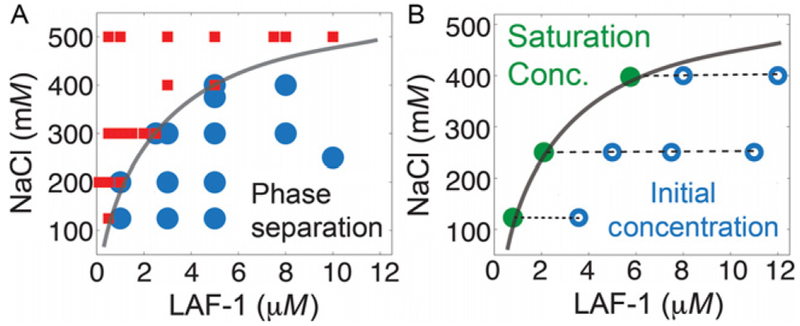
When close to the phase boundary, the system is very sensitive to small pipetting errors–thus conditions close to the boundary may result in inconsistent results for droplet formation upon repetition. A phase boundary can be drawn through these points to separate the region for phase separation which roughly corresponds to the saturation concentration (Fig. 3A).
Fig. 3.
Mapping the phase boundary. (A) LAF-1/NaCl conditions indicated were mixed, incubated, and scored for droplet formation via DIC microscopy. Positive and negative scoring for droplet formation are indicated by the blue circles and red squares, respectively. The phase boundary is drawn to distinguish between the two regions. (B) LAF-1/NaCl initial conditions within the phase separation region are incubated (open blue circles). The saturation concentration (i.e., critical concentration) is directly measured via absorbance at 280 nM of the solution after droplet removal via centrifugation. These values (green filled circles) are constant for increasing total protein concentration for a given NaCl concentration and overlay with the phase boundary determined in (A).
Scoring yes/no for droplets is naturally limited to droplets that are optically resolvable. A high magnification brightfield or a DIC microscope can be used to detect phase separation of unlabeled proteins (see additional notes in Section 2.3.1 for discussion of imaging tips and strategies).
2.3.4. Determining the Critical/Saturation Concentration: Complimentary Methods
Another method to measure the saturation concentration is a centrifugation assay. This method should be considered a complementary method, and droplets should always be confirmed by imaging. For a given interaction parameter value, create samples with increasing protein concentrations. Allow droplets to settle or slowly centrifuge for several hours to collect droplets. Then carefully measure the protein concentration in the supernatant via absorbance at 280 nm. The concentration of the supernatant corresponds to the critical or saturation concentration (Fig. 3B) and should be invariant for increasing total protein concentration at any single interaction parameter value.
One can also use turbidity measurements to report on phase separation. However, just as with centrifugation, many phenomena can result in turbidity increases, including aggregation or polymerization. Therefore, the presence of droplets in contrast to other assembled structures should be confirmed directly by imaging.
2.4. Droplet Shape: Surface Treatments
Pure viscous liquid droplets take on spherical shapes in solution, resulting from the energetic need to relax interfacial tension at the liquid–liquid interface (Hyman, Weber, & Julicher, 2014). Observance of spherical morphologies of protein assemblies has been used frequently to identify the presence of liquid material states (Banani et al., 2017; Shin & Brangwynne, 2017). However, liquids are not always spherical as they can deform upon wetting different surfaces, such as membranes in the case of P granules in the adult Caenorhabditis elegans germline (Brangwynne et al., 2009) or glass slides with varying surface treatments (Feric et al., 2016). Furthermore, when droplets approach the resolution limit, accurately resolving droplet morphology becomes increasingly challenging. Additionally, if a system transitions from a liquid to solid state, as has been suggested for Balbiani bodies (Boke & Mitchison, 2017), and centrosomal SPD-5 condensates (Woodruff et al., 2017), it may retain a spherical shape despite no longer being a pure viscous liquid.
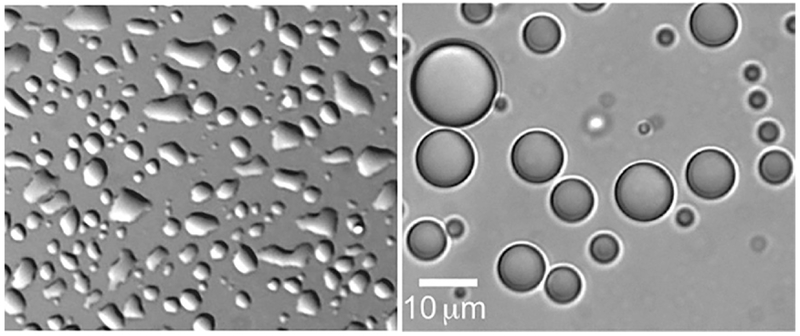
While further investigation is necessary to confirm the liquid material properties of a condensed protein phase (i.e., the coalescence assay described below, Fluorescence recovery after photobleaching (FRAP) or micro-rheology (Elbaum-Garfinkle et al., 2015; Zhang et al., 2015), varying surface treatments to promote spherical shapes can greatly assist with initial screening of droplet formation. Common surface treatments include Pluronics F-127, BSA, Sigmacote, and PEG. Protein droplets of varying content may interact uniquely with different surface treatments. LAF-1 droplets significantly wet untreated glass surfaces, but remain spherical upon treatment with Pluronics F-127 (Fig. 4). To treat glass surfaces, we add a filtered 10% solution of Pluronics F-127 to glass chambers/slides and incubate for ~1h. The chambers are rinsed 4–5 × with MilliQ water—and remain immersed in water until just before an experiment. Such treatments provide reproducible surfaces necessary for the quantification of droplet size and droplet fusion or coalescence.
Fig. 4.
Droplet shape and surfaces. Left: LAF-1 droplets wet the surface of untreated glass. Right: LAF-1 droplets do not wet the surface of Pluronics F127 treated glass and remain spherical.
2.5. Quantifying Droplet Coalescence
We can study the physical properties of our liquid droplets by taking advantage of their liquid-like behavior and spherical shape. Two spherical liquid droplets of length (ℓ) will collapse exponentially over time (τ) into a single spherical form. The time (τ) it takes for two liquid droplets to collapse into a single spherical shape is dictated by the relationship τ≈(η/γ)•ℓ, in which η is the droplet viscosity and γ is the surface tension (Eggers, Lister, & Stone, 1999). We can quantify the inverse capillary velocity, which is defined as the ratio of viscosity to surface tension (η/γ) by monitoring the coalescence of two droplets into a single sphere (Brangwynne et al., 2009; Brangwynne, Mitchison, & Hyman, 2011; Elbaum-Garfinkle et al., 2015).
2.5.1. Data Collection
To observe the coalescence events of LAF-1 liquid droplets the protein is incubated in CultureWell coverglass wells (GraceBioLabs) and visualized on an inverted microscope. The bottom of the well is treated with a Pluronics F-127 to prevent the liquid droplets from wetting the surface of the well, and to preserve their spherical shape. To visualize the liquid droplets, we mix unlabeled protein with ~1% labeled protein conjugated to DyLight NHS-Ester fluorophores (Thermo Fisher Scientific). We buffer exchange and concentrate the LAF-1 protein into a high salt (1 M NaCl) buffer along with reducing agent (1 mM DTT). The droplet size may vary depending on protein concentration, with higher concentrations resulting in larger droplets. We induce droplet formation by diluting into buffer with no or low NaCl depending on the experiment. We immediately observe the sample on the microscope, to image droplets and to capture fusion events as the liquid droplets coalesce and sink to the bottom of the well. Be mindful of the acquisition time per frame in each video as it will be important for the analysis.
2.5.2. Analysis
Once data collection is completed, time lapse images are first carefully reviewed in ImageJ. Ideal fusion events involve only two droplets that are similar in size. A single movie may contain several fusion events that can be useful for analysis, so it is important to look through the footage several times. We analyze each fusion event separately, noting the frames of each fusion event and saving each event with at least 10 frames prior to and after the event is completed. Each image sequence is saved as a .tif format file in a separate folder by making a unique “substack” folder for each fusion event to be analyzed.
Note: It is important to save several frames before and after the coalescence takes place to have a reference plateau and make sure the event is captured entirely. We look for liquid droplets in close proximity to each other, but which have not started the coalescence process. In this way we can track the aspect ratio change over time (Fig. 5A). The aspect ratio is defined as the ratio between the major and minor axis, given by the equation α=a/b in which a is the major axis and b is the minor axis. In our analysis, we assume the droplets to have a dimer shape because of their close proximity, with an aspect ratio of approximately 2. We monitor the collapse of the dimers into a sphere of aspect ratio equal to 1 after time (τ), which can be fit into an exponential decay graph. The MATLAB® analysis used to quantify coalescence is summarized in Figs. 5 and 6 and outlined here in detail:
Fig. 5.
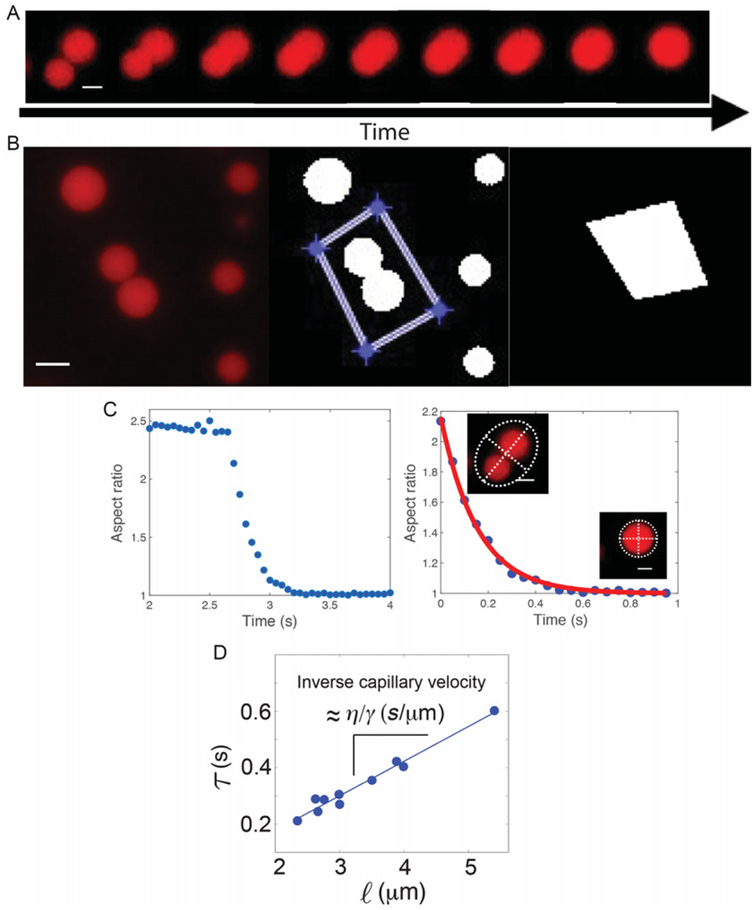
Coalescence assay. (A) Protein liquid droplets coalesce into a single sphere over time. Scale bar: 2 μm. (B) Single fusion events are selected and tracked over time in MATLAB®. The region corresponding to a single event (left) is selected (middle) in order to create a mask (right) to be applied to each frame in the time lapse. Scale bar: 5 μm. (C) Droplet coalescence raw data (left) are fit to an exponential decay (right). Inset: droplet ellipse with major/minor axis depicted by white dashed lines. (D)The fusion rate and droplet size are plotted and the slope is calculated to determine the inverse capillary velocity.
Fig. 6.
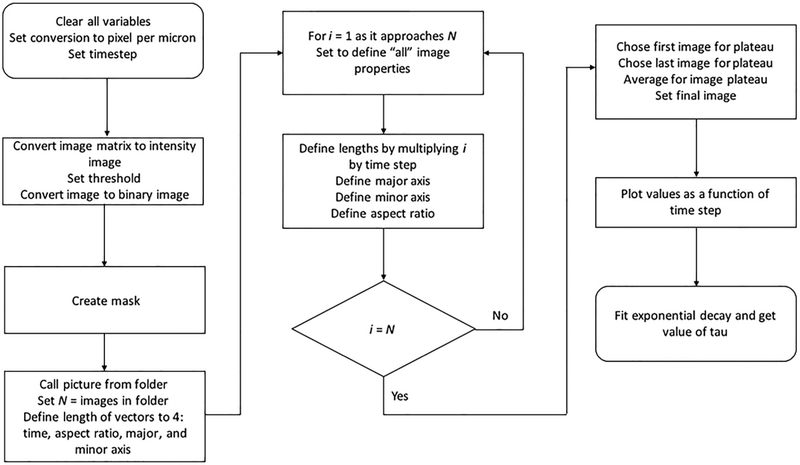
A flow chart of the image analysis used to quantify droplet coalescence.
-
Define parameters in your command window such as the pixels per micron in your images and the timestep in seconds of the video, i.e., the time interval of each image obtained.
Note: As with all time-sensitive imaging, be sure to confirm that the recorded frame rate corresponds to the frame rate set in acquisition settings!
Select the first image of the sequence and convert to double precision [double(x)] if the image does not already have this format.
Use ag=mat2gray(x) function to convert the intensities of the colors into ranges with values between 0 and 1.
Input this image into the command T=graythresh(image) to calculate the mean value of the image threshold. This will improve future visualization of the image.
Finally, convert the values of the image pixels to binary values of 0 and 1 using the function c=im2bw(ag,T), where T is the calculated threshold from step (d). Steps (c)–(e), should be input together and then observed using the imtool or imshow functions to see final product. Note that T is an automatic value calculated by MATLAB®. If the image looks too dark or too bright, you can decrease or increase the value of threshold to have a better contrast with the background.
-
Next draw a mask around the droplets to be analyzed. Call the modified image described earlier by using the command figure, imshow (c,[ ]). Input the command h = impoly to introduce a desired mask surrounding the droplets.
Note: It is important to avoid masking sections of droplets in the surrounding area. Take into consideration that droplets can move into your mask as the video progresses, making your analysis inaccurate. Introduce the commands BW=createMask (h) and figure, imshow (BW,[ ]) to return the mask created with the h=poly function. The mask shape is drawn by constructing line segments with the mouse, using a single click for each new line segment and a final double click once the mask shape is complete. A visual representation of this mask is shown in Fig. 5B.
Read in the full image sequence with the command pics=dir (fullfile(‘Name_of_file’,’*.tif’)); Define the number of elements in each folder; this will be the individual pictures in your folder (N = numel(pics);).
Run a “For” loop from image 1 to N (total) to: convert and threshold each image in the sequence as in steps (b)–(e); multiply mask created in step f to assign the area for analysis; quantify the major and minor axis and corresponding aspect ratio using the regionprops command.
After the loop is completed, create a graphical representation of the information. Set an average plateau for the graph by taking the average values of the matrix before the coalescence event takes place (command: ARi1=Image Number (initial), ARi2=image number (final), AR0=mean(AR(ARi1:ARi2)) (average between initial and final). The average will give an accurate starting point for the exponential decay of the droplets. Choose the final frame for the plot. Note: The final frame should be a few frames after the droplets have fused to ensure that the fusion event is captured in its entirety. Plot these values along with the timestep per frame. The resulting graph should look like the graph shown in Fig. 5C (left). Set the average plateau to equal the first data point in your graph.
Fit the decay of aspect ratio as it approaches 1 with the exponential decay form 1 + (AR0 − 1) * e−t/τ where AR0 is the initial aspect ratio and τ is the characteristic relaxation time, as shown in Fig. 5C (right). This will give a value for the decay represented by τ, which is the time of exponential decay as the two droplets fuse.
Plot the characteristic relaxation time (τ) against the length scale, or average size of the initial droplets. Since, τ≈(η/γ)•ℓ, the slope of τ v.s ℓ corresponds to the inverse capillary velocity, (η/γ), (Fig. 5D).
Additional notes
If the time step of the videos is too long relative to the fusion rate, the exponential decay may not have sufficient points to provide a reliable fit. (A minimum of 10 data points per decay is recommended).
If the aspect ratio decay cannot be fit to a single exponential, this may indicate divergence from a pure viscous liquid and/or contributions from interactions with the surface.
Try to avoid droplets that are too close to the surface, as any wetting of the droplets to the surface due to imperfect coating can inhibit fusion rates and compromise analysis.
3. CONCLUDING REMARKS
Here, we have outlined several methods and strategies for working with disordered proteins that are implicated in liquid phase separation. Optimizing reconstituted phase separation systems in vitro will provide a crucial foundation for uncovering the rules that govern disordered protein driven phase separation in and out of the cell.
REFERENCES
- Banani SF, Lee HO, Hyman AA, & Rosen MK (2017). Biomolecular condensates: Organizers of cellular biochemistry. Nature Reviews. Molecular Cell Biology, 18(5), 285–298. 10.1038/nrm.2017.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry J, Weber SC, Vaidya N, Haataja M, & Brangwynne CP (2015). RNA transcription modulates phase transition-driven nuclear body assembly. Proceedings of the National Academy of Sciences of the United States of America, 112(38), E5237–E5245. 10.1073/pnas.1509317112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeynaems S, Alberti S, Fawzi NL, Mittag T, Polymenidou M, Rousseau F, et al. (2018). Protein phase separation: A new phase in cell biology. Trends in Cell Biology, 28(6), 420–435. 10.1016/j.tcb.2018.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boke E, & Mitchison TJ (2017). The balbiani body and the concept of physiological amyloids. Cell Cycle, 16(2), 153–154. 10.1080/15384101.2016.1241605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady JP, Farber PJ, Sekhar A, Lin YH, Huang R, Bah A, et al. (2017). Structural and hydrodynamic properties of an intrinsically disordered region of a germ cell-specific protein on phase separation. Proceedings of the National Academy of Sciences of the United States of America, 114(39), E8194–E8203. 10.1073/pnas.1706197114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brangwynne CP, Eckmann CR, Courson DS, Rybarska A, Hoege C, Gharakhani J, et al. (2009). Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science, 324(5935), 1729–1732. 10.1126/science.1172046. [DOI] [PubMed] [Google Scholar]
- Brangwynne CP, Mitchison TJ, & Hyman AA (2011). Active liquid-like behavior of nucleoli determines their size and shape in Xenopus laevis oocytes. Proceedings of the National Academy of Sciences of the United States of America, 108(11), 4334–4339. 10.1073/Pnas.1017150108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brangwynne CP, Tompa P, & Pappu RV (2015). Polymer physics of intracellularphase transitions. Nature Physics, 11(11), 899–904. 10.1038/Nphys3532. [DOI] [Google Scholar]
- Burke KA, Janke AM, Rhine CL,& Fawzi NL (2015). Residue-by-residue view of in vitro FUS granules that bind the C-terminal domain of RNA polymerase II. Molecular Cell, 60(2), 231–241. 10.1016/j.molcel.2015.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conicella AE, Zerze GH, Mittal J, & Fawzi NL (2016). ALS mutations disrupt phase separation mediated by alpha-helical structure in the TDP-43 low-complexity C-terminal domain. Structure, 24(9), 1537–1549. 10.1016/j.str.2016.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolgin E (2018). What lava lamps and vinaigrette can teach us about cell biology. Nature, 555(7696), 300–302. 10.1038/d41586-018-03070-2. [DOI] [PubMed] [Google Scholar]
- Eggers J, Lister JR, & Stone HA (1999). Coalescence of liquid drops. Journal of Fluid Mechanics, 401, 293–310. 10.1017/S002211209900662x. [DOI] [Google Scholar]
- Elbaum-Garfinkle S, Kim Y, Szczepaniak K, Chen CC, Eckmann CR, Myong S, et al. (2015). The disordered P granule protein LAF-1 drives phase separation into droplets with tunable viscosity and dynamics. Proceedings of the National Academy of Sciences of the United States of America, 112(23), 7189–7194. 10.1073/pnas.1504822112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feric M, Vaidya N, Harmon TS, Mitrea DM, Zhu L, Richardson TM, et al. (2016). Coexisting liquid phases underlie nucleolar subcompartments. Cell, 165(7), 1686–1697. 10.1016/j.cell.2016.04.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman AA, Weber CA, & Julicher F (2014). Liquid-liquid phase separation in biology. Annual Review of Cell and Developmental Biology, 30, 39–58. 10.1146/annurev-cellbio-100913-013325. [DOI] [PubMed] [Google Scholar]
- Lin Y, Protter DS, Rosen MK, & Parker R (2015). Formation and maturation of phase-separated liquid droplets by RNA-binding proteins. Molecular Cell, 60(2), 208–219. 10.1016/j.molcel.2015.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molliex A, Temirov J, Lee J, Coughlin M, Kanagaraj AP, Kim HJ, et al. (2015). Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell, 163(1), 123–133. 10.1016/j.cell.2015.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami T, Qamar S, Lin JQ, Schierle GS, Rees E, Miyashita A, et al. (2015). ALS/FTD mutation-induced phase transition of FUS liquid droplets and reversible hydrogels into irreversible hydrogels impairs RNP granule function. Neuron, 88(4), 678–690. 10.1016/j.neuron.2015.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nott TJ, Petsalaki E, Farber P, Jervis D, Fussner E, Plochowietz A, et al. (2015). Phase transition of a disordered nuage protein generates environmentally responsive membraneless organelles. Molecular Cell, 57(5), 936–947. 10.1016/j.molcel.2015.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha S, Weber CA, Nousch M, Adame-Arana O, Hoege C, Hein MY, et al. (2016). Polar positioning of phase-separated liquid compartments in cells regulated by an mRNA competition mechanism. Cell, 166(6), 1572–1584, e1516. 10.1016/j.cell.2016.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin Y, & Brangwynne CP (2017). Liquid phase condensation in cell physiology and disease. Science, 357(6357). 10.1126/science.aaf4382. [DOI] [PubMed] [Google Scholar]
- Smith J, Calidas D, Schmidt H, Lu T, Rasoloson D, & Seydoux G (2016). Spatial patterning of P granules by RNA-induced phase separation of the intrinsically-disordered protein MEG-3. eLife, 5 10.7554/eLife.21337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JT, Smith J, Chen BC, Schmidt H, Rasoloson D, Paix A, et al. (2014). Regulation of RNA granule dynamics by phosphorylation of serine-rich, intrinsically disordered proteins in C. elegans. eLife, 3, e04591 10.7554/eLife.04591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei MT, Elbaum-Garfinkle S, Holehouse AS, Chen CC, Feric M, Arnold CB, et al. (2017). Phase behaviour of disordered proteins underlying low density and high permeability of liquid organelles. Nature Chemistry, 9(11), 1118–1125. 10.1038/nchem.2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodruff JB, Ferreira Gomes B, Widlund PO, Mahamid J, Honigmann A, & Hyman AA (2017). The centrosome is a selective condensate that nucleates microtubules by concentrating tubulin. Cell, 169(6), 1066–1077.e1010. 10.1016/j.cell.2017.05.028. [DOI] [PubMed] [Google Scholar]
- Zhang H, Elbaum-Garfinkle S, Langdon EM, Taylor N, Occhipinti P, Bridges AA, et al. (2015). RNA controls polyQ protein phase transitions. Molecular Cell, 60(2), 220–230. 10.1016/j.molcel.2015.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]