Abstract
Prokaryotic and eukaryotic Na+/Ca2+ exchangers (NCX) control Ca2+ homeostasis. NCX orthologs exhibit up to 104-fold differences in their turnover rates (kcat), whereas the ratios between the cytosolic (cyt) and extracellular (ext) Km values (Kint = KmCyt/KmExt) are highly asymmetric and alike (Kint ≤ 0.1) among NCXs. The structural determinants controlling a huge divergence in kcat at comparable Kint remain unclear, although 11 (out of 12) ion-coordinating residues are highly conserved among NCXs. The crystal structure of the archaeal NCX (NCX_Mj) was explored for testing the mutational effects of pore-allied and loop residues on kcat and Kint. Among 55 tested residues, 26 mutations affect either kcat or Kint, where two major groups can be distinguished. The first group of mutations (14 residues) affect kcat rather than Kint. The majority of these residues (10 out of 14) are located within the extracellular vestibule near the pore center. The second group of mutations (12 residues) affect Kint rather than kcat, whereas the majority of residues (9 out 12) are randomly dispersed within the extracellular vestibule. In conjunction with computational modeling-simulations and hydrogen-deuterium exchange mass-spectrometry (HDX-MS), the present mutational analysis highlights structural elements that differentially govern the intrinsic asymmetry and transport rates. The key residues, located at specific segments, can affect the characteristic features of local backbone dynamics and thus, the conformational flexibility of ion-transporting helices contributing to critical conformational transitions. The underlying mechanisms might have a physiological relevance for matching the response modes of NCX variants to cell-specific Ca2+ and Na+ signaling.
1. Introduction
Ca2+-transporting proteins (channels, pumps and transporters) exhibit nearly 107-fold differences in their transport rates, which are of very complex and dynamic physiological events (excitation-contraction coupling, action potential duration, hormone and neurotransmitter secretion, mitochondrial bioenergetics among many others) [1–3]. Among the Ca2+-transporting proteins, the Na+/Ca2+ exchanger (NCX) system plays a key role in controlling the Ca2+ homeostasis since the NCX-mediated Ca2+ extrusion rates must vary according to cell-specific patterns of Ca2+ oscillations in a given cell type [2–5]. The isoforms/splice variants of NCX are expressed in a tissue-and organelle-specific manner [1,4,5], where the NCX-mediated turnover rates of the ion transport cycle vary from 0.5 s−1 to 2500 s−1 among prokaryotic and eukaryotic NCXs [2,3,5]. Despite these differences in the transport rates, NCXs share a common stoichiometry (3Na+ :1Ca2+) of ion exchange [6–8], where the Na+ or Ca2+ bound species are transported in mutually exclusive (separate) steps along the transport cycle [3,5,9].
The NCX and other gene families (NCKX, NCLX, and CAX) belonging to the Ca2+/Cation antiporter (Ca/CA) superfamily contain highly conserved α1 and α2 repeats organized in an inverted two-fold “symmetry” to form an ion-passage pore (Fig. 1A and B) [1,10–15]. Despite their structural similarities [11–17], the five Ca/CA families display distinct selectivity for monovalent ion transport [3,5,10]. Prokaryotic and eukaryotic NCXs contain 10 transmembrane helices (TM1–10), with the cytosolic 5L6-loop between TM5 and TM6 forming 2 tightly packed hubs [12,17], TM2–5 and TM7–10 (colored white and blue, respectively, in Fig. 1A and B) [12], which can be superimposed upon rotation/inversion (Fig. 1B). The 5L6 loop (~520 residues) of eukaryotic NCXs contains two Ca2+-binding regulatory domains (CBD1 and CBD2), which play a critical role for tissue-specific regulation of NCXs [18–22]. Since the short 5L6-loop of NCX from Methanococcus jannaschii (NCX_Mj) lacks regulatory domains, its crystal structure provides an excellent basis for studying the ion-transport mechanisms [23–27,29,31]. In NCX_Mj, 12 ion-coordinating residues, located on TM2 (A47, T50, S51, and E54), TM3 (S77 and N81), TM7 (A206, T209, S210, and E213) and TM8 (S236 and D240) form an ion-passage pore with 4 binding sites, Sint, Smid, Sext, and SCa (Fig. 1C–F) [12]. The 4 sites exhibit distinct ion selectivity: Sint and Sext have high selectivity for Na+, SCa can bind either Na+ or Ca2+, whereas Smid cannot bind Na+ nor Ca2+ in the ground state [12,29,30].
Fig. 1.
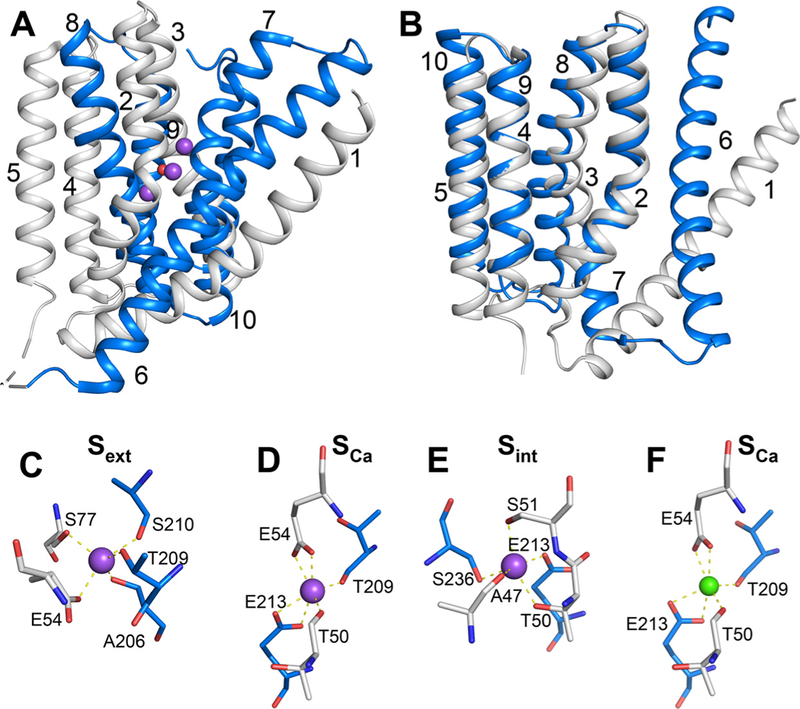
Structure of NCX_Mj. (A) Crystal structure of 3Na+-bound NCX_Mj (PDB 5HXE) in cartoon representation. The symmetry-related two halves are colored in gray (TM1–5) and blue (TM6–10), respectively. Purple and red spheres represent Na+ ions and water molecule, respectively. (B) Superposition of the symmetry-related two halves, as colored in panel A. (C–F) The ion-binding sites of NCX_Mj. The Smid site is not shown, since there is no experimental or computational evidence that this site can bind either Na+ or Ca2+. Ion-coordinating residues are shown as sticks. Purple and green spheres represent Na+ and Ca2+ ions, respectively.
During the transport cycle, NCX binds either 3Na+ ions at Sint, Sext, and SCa or 1Ca2+ ion at SCa, where the Na+ and Ca2+-bound species are transported in separate steps [9,28–30]. Recent studies with hydrogen-deuterium exchange mass-spectrometry (HDX-MS) show that NCX_Mj can adopt either the inward-facing (IF) or outward-facing (OF) conformation [31]. The recent crystallographic data obtained for the open, semi-open and occluded states of NCX_Mj in the OF conformation [12,30] are especially interesting in conjunction with HDX-MS and ion-flux analyses of mutants [23,26–31] for resolving the ion binding, occlusion and transport mechanisms. This might be of general interest since 11 (out of 12) ion-coordinating residues are identical between NCX_Mj and eukaryotic NCX variants. The only difference is at position 240 (Smid), occupied by an aspartate (D) or an asparagine (N) in NCX_Mj and eukaryotic NCXs, respectively. Interestingly, the D– >N replacement in NCX_Mj increases kcat by 5–10-fold, meaning that the D– > N replacement itself alone cannot fully account for the 104-fold differences in the kcat values among NCXs [27,29].
The mechanism of action of NCXs conforms to alternating access mechanism [32–34]. Under steady-state conditions NCX preferentially adopts the OF conformation [12,31] with Kint ≈ 0.15 [23,27], which might have a physiological relevance [25,26]. Like many other transporters, the structure-dependent matching of turnover rates and intrinsic asymmetry of NCX variants is physiologically relevant for attaining the relevant Ca2+-extrusion rates in distinct cell-types [23,27,33–35]. The structure-dynamics determinants of functional asymmetry are especially interesting in the case of NCXs (and similar proteins) since the cytosolic and extracellular vestibules, displaying the inverted two-fold symmetry, are exposed to symmetric substrates, like ions [27,34–37]. Thus, the question is: how the transport rates (kcat) and intrinsic asymmetry (Kint ≤ 0.15) are structurally controlled (predefined) in NCXs (and other transporters) exhibiting inverted two-fold topology?
The primary goal of the present work was to identify the key residues that predominantly determine the kcat and Kint values of NCX_Mj. For this purpose, we analyzed the bidirectional transport events in 55 mutants using established ion-flux assays [23–29,31,38] in conjunction with structure-based computational approaches [39–49]. We found that 14 pore-core residues limit the ion transport rates (kcat), whereas another set of 12 residues (randomly distributed within the pore and a nearby loop) alter the intrinsic asymmetry (Kint) of bidirectional Ca2+ movements. These data, analyzed in conjunction with HDX-MS analyses and computational modeling, elucidate important sites, which are located at highlighted TM segments showing the characteristic patterns of collective dynamics in controlling the ion exchange activities. Thus, the principle massage of the present work is that specific structural modules are involved in differential controlling of ion-transport rates (kcat) and intrinsic equilibrium (Kint) of bidirectional ion movements in NCX. The mechanistic and physiological significance of these findings is that, structurally predefined modules can diversify the ion transport rates, while retaining a comparable degree of functional asymmetry among NCXs. Alike structure-functional mechanisms may also exist in similar proteins, controlling the cell-specific Ca2+ homeostasis in prokaryotic and eukaryotic cell-types.
2. Materials and methods
2.1. DNA constructs and site-directed mutagenesis
DNA encoding the wild-type NCX_Mj was amplified by PCR from a Methanocaldococcus jannaschii cDNA library (DSMZ) and ligated between the NcoI and BamHI restriction sites of a pET-28a plasmid, as described before [23,27,29]. Single-point mutations were introduced by QuickChange mutagenesis (Stratagene) and were confirmed by sequencing, as outlined [27,29,31].
2.2. Isolation of E. coli-derived membrane vesicles overexpressing NCX_Mj and mutants thereof
Expression vectors were transformed into E. coli BL21 (DE3) pLysS competent cells [23,24,27]. E. coli cells were grown in 2xYT media with antibiotics and expression was induced when cell cultures reached OD600 = 0.5–0.6 at 16 °C by adding 0.4 mM IPTG. After 12–16 h incubation, cells were harvested and resuspended in buffer containing 50 mM Mops-Tris pH 7.4, 0.25 M sucrose, 1 mM EDTA, 1 mM DTT, 100 Units/ml DNase, and 1 mM PMSF. Before cell breakage, the suspension was supplemented with additives for cell disruption and subsequently, the cells were disintegrated in a French press (at 20,000 psi) or in a Microfluidizer (at 14,000 psi) to obtain right side out (RSO) or inside out (ISO) vesicles, respectively [23,24,27,29]. Cell lysates were centrifuged for 5 min at 6,000×g and the supernatant was pelleted at 200,000×g for 1 h. Next, the pellet was resuspended in buffer (50 mM Mops-Tris, pH 7.4, 1 mM EDTA, 1 mM DTT, and 0.25 M sucrose) and loaded onto a three-step sucrose gradient (2.02, 1.4, and 0.7 M, equilibrated with the same buffer). After centrifugation (for 15 h at 150,000×g, 4 °C), a brownish layer appearing between the 0.7 and 1.4 M sucrose layers was collected, diluted 4 times with buffer (50 mM Mops-Tris pH 7.4, 1 mM EDTA, and 1 mM DTT) and centrifuged at 200,000×g for 1 h. The pellet was resuspended in 50 mM Mops/Tris pH 7.4, 0.25 M sucrose, homogenized by passing it through a syringe needle, and subsequently washed by centrifugation (at 200,000×g for 1 h). The washed vesicles were resuspended in buffer (50 mM Mops-Tris pH 7.4 and 0.25 M sucrose), flash-frozen in liquid nitrogen, and stored at −80 °C until use. The total protein concentration was determined using the Lowry protein assay and the expression levels of the NCX_Mj mutants were evaluated using the GFP assay [23,24]. The orientation of NCX_Mj in E. coli-derived cell-membrane vesicles was evaluated by using an antibody (against the 6xHis-tag) assay, as previously outlined [23,24,27,29]. According to this test, both the ISO and RSO vesicles contained unidirectionally (90–95%) oriented NCX_Mj [23,27,29], where the orientation of vesicles is predefined by cell-breaking procedure (French-press or Micro-fluidizer, see above). No indication was found indicating any mutation-dependent alterations of NCX_Mj orientation in the E. coli-derived vesicles. For evaluating the expression levels of NCX_Mj (either in the case of WT or mutants), the GFP assay system was used, as previously described [23]. Limited number of mutants (exhibiting less than 20% of ion-exchange activity) were tested for the protein expression levels by using the GFP assay. The expression levels of tested mutants were comparable (if not identical) to WT NCX_Mj, where the GFP-labeled expression levels account for ~ 10–12% of total membrane protein in the preparations of E. coli-derived membrane vesicles.
2.3. Ion-exchange assays using E. coli-derived membrane vesicles
Initial rates (t = 5 s) of Na+/Ca2+ and Ca2+/Ca2+ exchange reactions were assayed at 35 °C by measuring the intravesicular 45Ca2+ content in E. coli-derived vesicles with a given overexpressed protein (WT or its mutant), as previously described [23,24,27,29]. The 45Ca2+-uptake was initiated by a rapid dilution of Na+ (160 mM) or Ca2+ (0.5 mM)-loaded vesicles into the assay medium (0.2–0.5 mL) containing 20 mM Mops/Tris, pH 6.5, 100 mM KCl, and 5–2000 μM 45CaCl2. The “de-calcified” buffers have been used for the 45Ca2+-uptake assays, where the stock solutions were passed through a Chelex column. The typical concentrations of endogenous free Ca2+ in the assay medium were in the range of 1–3 μM. Notably, without this treatment the endogenous free Ca2+ concentrations in the assay medium can easily reach 10–20 μM or even more. The free Ca2+ concentrations were measured according to previously established protocols by using the Arsenaso-III optical probe [9,23–25,52]. The 45Ca2+-uptake was quenched by rapid injection of cold EGTA-buffer (5 mL) into the assay medium. The intravesicular 45Ca2+ was measured by rapid filtration of the quenched solutions through a GF/C filter, followed by extensive washing (5×5 mL) of the filter. Importantly, the vesicles trapped on the filters were not allowed to dry during filtration, whereas after the filtration the filters were dried and placed in the scintillation vials [23,24,38]. For blank assays, E. coli-derived cell-membrane vesicles, lacking the NCX_Mj protein, were diluted, quenched, and filtrated under identical conditions. The blanks were taken for each concentration of 45Ca2+ (used in the assay medium) in the presence of 160 mM NaCl in the assay medium. Subsequently, nonspecific values of 45Ca2+ bound to the membrane vesicle and to the filters were subtracted from the samples as a background (nonspecific) signal. In the preparations of the E. coli-derived vesicles (containing WT NCX_Mj), the signal/background ratios of measured 45Ca2+-uptake vary from 10 to 50 (in proportion with 45Ca2+ concentrations used in the assay medium). For decreasing the nonspecific binding of 45Ca2+ to the filters, prior the filtration the GF/C filters were presoaked with 0.3% polyethylenimine as described before [9]. GraFit 7.0 software (Erithacus Software, Ltd.) was used for fitting the experimentally obtained values of 45Ca2+-uptake at varying ionic concentrations. The Km and Vmax values were derived from titration curves, where the experimentally measured levels of the observed signals reach at least 85% of the calculated Vmax values (this technical requirement is essential to obtaining accurate values for both Km and Vmax). The kinetic parameters were measured at least in three independent experiments (data are presented as mean ± SE). kcat values were calculated according to equation kcat = Vmax/[E]t, where Vmax was derived from the [Ca2+] titration curves and the [E]t values were evaluated by using the GFP assay [23].
2.4. Computer-aided structural modeling of the OF and IF NCX_Mj conformers and full-atomic simulations
A structural model for the inward-facing (IF) open conformation of NCX_Mj was generated using the inverted symmetry property of the two helical substructures TM2–5 and TM7–10 [12,23,27,31]. MODELLER [39] was used to generate the structure of each substructure of NCX_Mj, which were rigidly reoriented based on structural homology criteria for the symmetric parts of NCX_Mj [12,39]. The structure of each half of the protein was generated by homology to the opposite symmetric part and by sequence alignments of the helical segment TM1–3 against TM6–8, and TM4–5 against TM 9–10 [12,23,27,31]. A set of 100 homology models were generated to select the NCX_Mj IF open conformer with the best score (lowest MODELLER objective function). The OF conformer in the presence of three Na+ ions (PDB: 5HXE) [30] as well as the homology model generated for the IF NCX_Mj were further studied by molecular dynamics (MD) simulations. The missing 5L6 loop (S147 – N158) was constructed using MODELLER. The simulation systems were prepared in explicit membrane bilayer with 0.1 mM CaCl2 solution using CHARMM GUI [40]. We generated two sets of 100 ns full atomic MD trajectories for each conformer, using the simulation package NAMD [41]. For the OF and IF conformers, the root-mean-square deviations (RMSDs) of Cα-atoms from their initial positions attained plateaus at 2.3 ± 0.3 and 4.5 ± 0.3 Å, respectively, after ~50ns simulations, indicating that stable conformers were obtained.
2.5. Characterization of collective dynamics
The global dynamics of NCX_Mj was evaluated using the Gaussian network model (GNM) [42] and the anisotropic network model (ANM) [43], in the context of membrane lipids [44], as implemented in the DynOmics web server [45]. The dynamic features and predicted effects of mutated residues were calculated by the RhapSody interface [46]. We evaluated the following features [46–49]: (i) GNM-based root-mean-square fluctuations (RMSFs) of residues and (ii) the propensity of residues to act as sensors or effectors of allosteric signals based on per-turbation-response scanning (PRS) analysis.
3. Results
3.1. Mutational effects of pore-forming residues on the Vmax values of the Na+/Ca2+ exchange
The effects of single-point mutations on the initial rates of the Na+/Ca2+ and Ca2+/Ca2+ exchange were measured. For this assay, the E. coli-derived cell membrane vesicles containing the overexpressed NCX_Mj or its mutants were used [23,24,29]. In this assay system, the ion fluxes can be reliably monitored, since the overexpressed proteins account for 10–15% of the total membrane protein [23,24]. Moreover, independent biochemical tests have shown that overexpressed NCX_Mj proteins are uniformly (> 90%) oriented in the RSO (right-side out) or ISO (inside out) orientation, so that the Km and Vmax values of the Na+/Ca2+ and Ca2+/Ca2+ exchange rates can be measured (see Materials and Methods) by varying [Ca2+] or [Na+] at either side of the membrane [23,24,29,38]. In the present studies, the kinetic parameters (Km and Vmax) were measured in uniformly oriented vesicles (containing the overexpressed WT or mutated NCX_Mj) by measuring the initial rates (t = 5 s) of 45Ca2+-uptake (see Materials and Methods).
We tested the mutational effects of pore-forming residues located at TM2, TM3, TM7 and TM8 on the ion-transport activities. The Vmax values for 42 mutants of pore-forming residues were tested by examining the initial rates of Na+/Ca2+ exchange at saturating concentrations of [Ca2+] and [Na+] (Fig. 2A). According to observed values, the mutational effects can be divided into three major groups. In group 1 (in red), the mutations of 15 residues (out of 42) show < 20% of WT Vmax (Fig. 2A). Group 1 includes 6 ion-coordinating (S51, E54, S77, T209, E213 and D240) and 9 non-coordinating (G49, P53, G76, C78, C80, F202, P212, S217 and G235) residues, thereby underscoring the essential role of non-coordinating residues in ion transport. Notably, the most prominent effect (20–50-fold decrease in the Vmax) was observed for ion-coordinating and non-coordinating residues participating in the signature sequences, 49-GTSLPE-54 (TM2, α1) or 208-GTSLPE-213 (TM7, α2) at the center of the pore (Fig. 2A–C). Strikingly enough, all 15 residues strongly affecting the Na+/Ca+ exchange rates are located close to the ion-pore center (Fig. 2C). Interestingly, the C78 A and C80 A mutations have a devastating effect on the Na+/Ca2+ exchange activity (Fig. 2A), although the sulfhydryl group of these cysteine residues cannot be involved in ion coordination, since they cannot be reoriented to coordinate the ions within the pore. These are quite unexpected results since, instead of cysteine, the NCX and NCKX orthologs contain highly conserved alanine and phenylalanine residues at positions matching 78 and 80 [29,50,51].
Fig. 2.
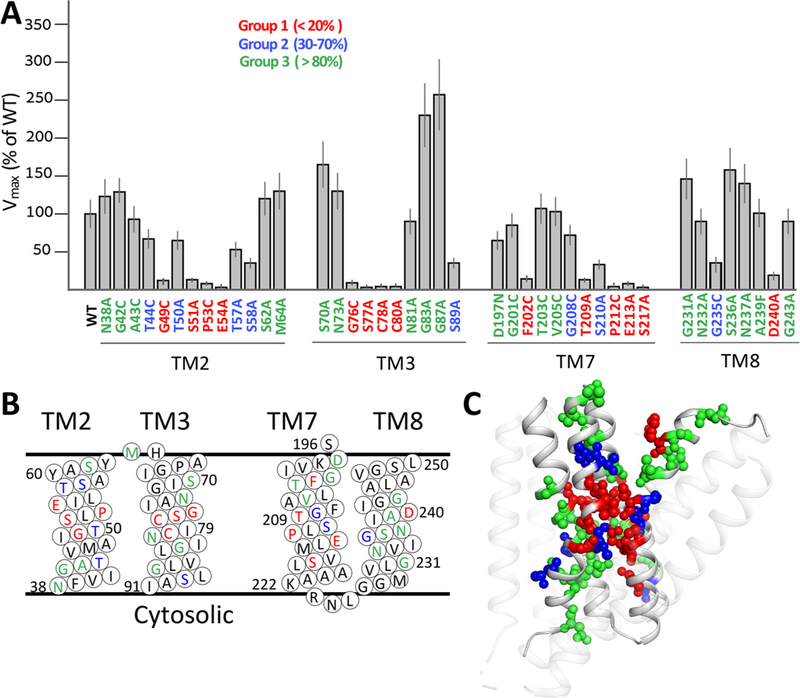
Mutational effects on the Vmax values of the Na+/Ca2+ exchange. (A) The initial rates (t = 5s) of Na+-dependent 45Ca2+-uptake were measured by using E. coli-derived vesicles containing the overexpressed protein of a given mutant or WT NCX_Mj (see Materials and Methods). The Vmax values of the Na+/Ca2+ exchange reaction were measured for NCX_Mj mutants as described in Materials and Methods. The Vmax values of the indicated mutants are presented in percentage values in comparison with WT Vmax (100%). Data are presented as bars (mean ± SE). The data were derived from at least 3 independent experiments. Residues are colored according to their mutational effects on Vmax, as indicated. (B) Topological positions of mutated residues are presented according to color assignments, described in panel A. (C) Cartoon representation of NCX_Mj. Mutated residues are shown as spheres and are displayed in color according to the mutational effect (see panel A). Note the specific distribution of residues belonging to group 1 (red) in the vicinity of the pore core.
Mutational effects of group 2 residues (in blue) represent a rather moderate decrease (30–70% of WT Vmax) in the Na+/Ca2+ exchange rates (Fig. 2A). This group includes two ion-coordinating (T50 and S210) and five non-coordinating (T44, T57, T58, S89, G208 and G235) residues (Fig. 2A). Group 3 (in green) includes 20 residues, of which 14 largely retain ion-transport activities. The remaining 6 residues even exhibited a 2–3-fold increase in their transport rates (Fig. 2A). Thus, these residues do not seem to be essential for ion-transport activities. Notably, 27 residues belonging to group 2 and group 3 are randomly distributed within the ion-pore, thereby showing no specific abundance within the pore (Fig. 2A and C). In sharp contrast with this, the group 1 residues are predominantly located within the pore core (Fig. 2A and C). Strikingly enough, the mutations of 3 ion-coordinating residues (as suggested by the crystal structure), T50, N81 and S236, do not reduce the Vmax values of Na+/Ca2+ exchange (Fig. 2A).
3.2. Mutational effects of pore-forming residues on the KmCyt (Ca2+) values of the Na+/Ca2+ exchange
Next, 32 pore-forming residue mutants were analyzed for their effects on the KmCyt values of Ca2+ transport by measuring the initial rates of Na+/Ca2+ exchange at varying concentrations of cytosolic (extravesicular) Ca2+ (5–2000 μM) and at a fixed, saturating concentration of extracellular (intravesicular) Na+ (160 mM) (see Materials and Methods). Notably, some mutants (P53C, G76C, P212C, and S217) were not included in this analysis, since the signal/noise ratios were insufficient for precise measurement of the Km values at very low values of ion-transport rates (< 10% of WT Vmax). Thus, the measurable KmCyt values, obtained for 28 mutants, can be divided into 2 groups. Group 1 residues (in blue) exhibit very small (if any) effect on the KmCyt values (50–200% of WT), whereas group 2 (in green) residues exhibit up to a 9-fold increase in the KmCyt values (Fig. 3A). Notably, 12 (out of 14) residues, showing the most prominent mutational effects on KmCyt, are located on TM7 and TM8 (Fig. 3B and C). The selected mutants (altering the KmCyt value of the Na+/Ca2+ exchange) were further examined for their effects on the KmCyt and KmExt values of the Ca2+/Ca2+ exchange (Fig. 4C). The primary goal of this approach was to identify the mutations significantly affecting the intrinsic equilibrium (Kint) of bidirectional Ca2+ movements (see below).
Fig. 3.
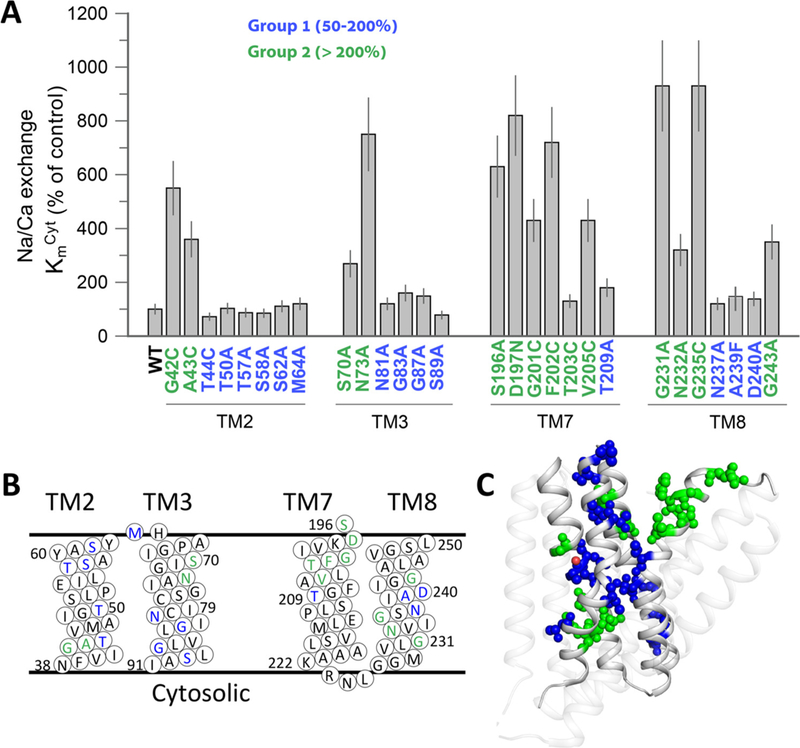
Mutational effects on the KmCyt values of the Na+/Ca2+ exchange. (A) The KmCyt values of the Na+/Ca2+ exchange reaction were measured for the indicated mutants, as described in Materials and Methods. The KmCyt values of the indicated mutants are presented in percentage values in comparison with the WT Kmcyt values. Data were obtained from at least 3 independent experiments and are presented as mean ± SE. Residues are colored according to their effects on Km, as indicated. (B) Topological positions of mutated residues are presented in color, as displayed in the panel A. (C) Cartoon presentation of NCX_Mj. Mutated residues are shown as spheres and colored according to the mutational effects, as in panel A.
Fig. 4.
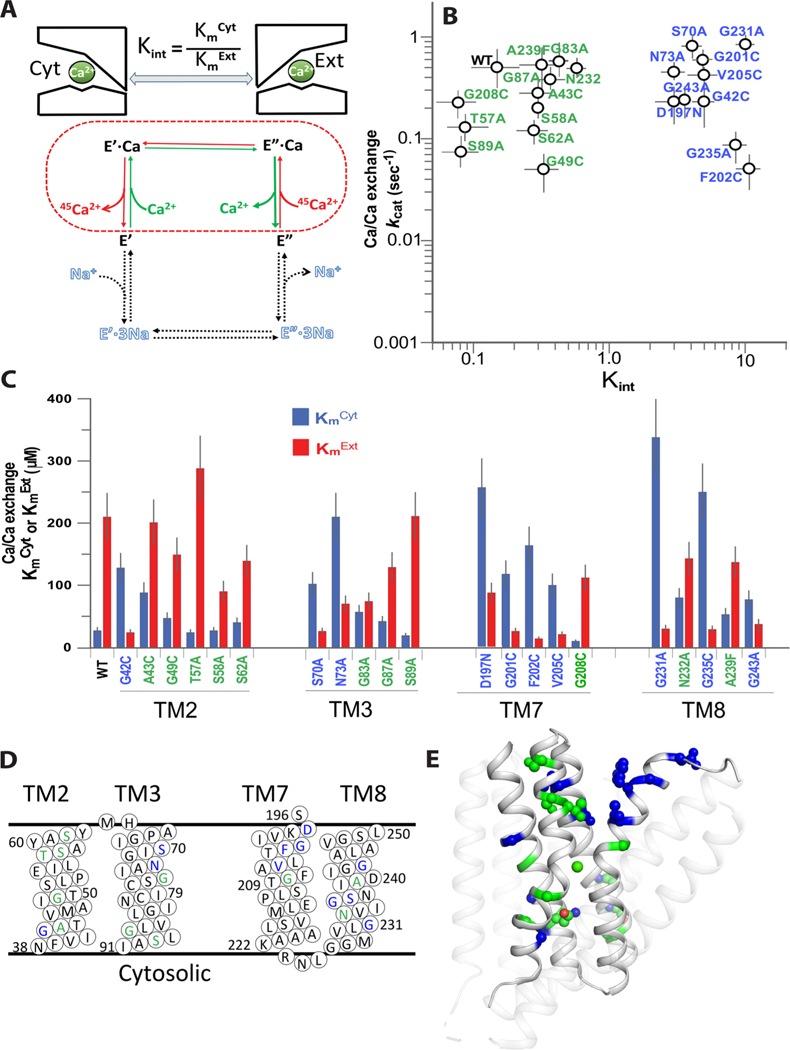
Mutational effects of pore-forming residues on the Kint and kcat values of the Ca2+/Ca2+ exchange. (A) Schematic representation of the Ca2+/Ca2+ exchange reaction for measuring the Kint and kcat values of bidirectional Ca2+ movements. The dashed line represents the partial reaction of the Ca2+/Ca2+ exchange, where the intrinsic equilibrium of bidirectional Ca2+ movements (Kint) is defined as the ratio of apparent affinities for Ca2+ at the cytosolic (KmCyt) and extracellular (KmExt) sides (Kint = KmCyt/KmExt). B) The initial rates (t = 5s) of Ca2+-dependent 45Ca2+-uptake were measured for determining the KmCyt and KmExt values. The KmCyt values were determined using varying [Ca2+]Cyt = 2–200 μM and saturating [Ca2+]Ext = 2 mM (blue bars), whereas the KmExt values were measured using varying [Ca2+]Ext = 10–2000 μM and fixed [Ca2+]cyt = 2mM (red bars). The kcat values were derived from the measured Vmax values of the Ca2+/Ca2+ exchange as described in Materials and Methods. Data are presented as mean ± SE of at least 3 independent experiments. Mutations resulting in Kint <1 or Kint >1 are in green and blue, respectively. (C) The KmCyt (blue bars) and KmExt (red bars) values of the Ca+/Ca2+ exchange reaction were measured as described in panel B. Data are presented as mean ± SE obtained at least 3 independent experiments (see Materials and Methods). Residues are colored according to their mutational effects on Kint, as shown in panel B. (D) Topological positions of residues are assigned according to their mutational effects on Kint, as colored in panel B. (E) Cartoon presentation of NCX_Mj. Mutated residues are shown as spheres, where they are colored according to mutational effects, described in panel B.
3.3. Mutational effects of pore-forming residues on the Kint and kcat values of the Ca2+/Ca2+ exchange
For Ca2+/Ca2+ exchange, the intrinsic equilibrium (Kint) of bidirectional Ca2+ movements can be defined as Kint = KmCyt/KmExt, whereas the turnover rates (kcat) can be presented as kcat= Vmax/[E]t (Figure S1) [23,37]. According to this formalism the Kint is derived from the measured values of KmCyt and KmExt, whereas kcat is derived by measuring the Vmax and [E]t (total enzyme concentration) values. The KmCyt values were measured at varying [Ca2+]Cyt (2–2000 μM) and saturating [Ca2+]Ext (2mM) (blue bars in Fig. 4C). In the complementary set of experiments the KmExt values were determined at varying [Ca2+]Ext and fixed [Ca2+]Cyt (red bars in Fig. 4C). Figure S2 describes the Ca2+-titration curves of Ca2+/Ca2+ exchange for three representative mutants (K156 A, D197 N and F202), showing up-to 100-fold differences in the Kint and Vmax values. The present analysis has identified 10 non-coordinating residues, the mutations of which result in concomitant changes in the KmCyt and KmExt values (Fig. 4C), thereby resulting in up to 50–70-fold changes in Kint (Fig. 4B). These mutational effects on the Kint values can be divided into 2 major groups (Fig. 4B). Group 1 mutations (in green) exhibit Kint < 1, which is consistent with stabilization of OF-state under steady-state conditions. Most interestingly, group 2 mutations (in blue) result in the increased Kint values (ranging from 2 to 10) although the effects on the kcat values are rather small if not negligible (Fig. 4B). Thus, group 2 residues consistently affect the intrinsic equilibrium of bidirectional movements, while having rather small (if any) effect on the ion transport rates. In light of present considerations, we posit that under steady-state conditions, the group 2 residues are capable of governing the relative stability of the IF and OF states, which may represent a structural basis for functional asymmetry of NCX_Mj.
3.4. Mutational effects of the loop-residues on the Kint and kcat values of the Ca2+/Ca2+ exchange
For elucidating the underlying structure-functional determinants of bidirectional Ca2+ movements, 13 loop-residues (located on the 1L2, 4L5, 5L6, 6L7 and 8L9 loops) were tested here for their effects on the kcat and Kint values (Fig. 5A and C). Interestingly, nearly all tested mutations either retain or increase the ion-transport rates (kcat), while the differential effects on the KmCyt and KmExt (Fig. 5C) lead to characteristic shifts in Kint (Fig. 5A). The mutational effects of loop-residues on Kint can be divided into 3 groups. Group 1 residues (in green) largely retain the WT Kint values upon mutation, which is compatible with insignificant contribution of these residues to relative stabilization of IF or OF species (Fig. 5A). The mutation of group 2 residues (in blue) results in 5–6-fold enhancement of Kint, thereby suggesting that these residues may have fairly small effect on relative stabilization of OF and IF states (Fig. 5A). Interestingly, group 2 residues are predominantly located on the 5L6 loop (Fig. 5B). Group 3 mutations (in red) result in 20–70-fold increase of the Kint values, thereby revealing a preferential stabilization of the IF state under steady-state conditions (Fig. 5A). Interestingly, the mutations of nearby residues, S196A, D197N and K198A (located at the interface of the 6L7 loop and TM7A) result in enhanced Kint values (Fig. 5A), whereas no significant changes are observed in the Kint of D194K, located on the 6L7 loop (Fig. 5A,C). These findings are consistent with existence of a functionally important module involving a hydrogen-bonding network at the 6L7/TM7A interface, which may play a key role in stabilizing the OF state and thus, in generating the functional asymmetry in NCX_Mj.
Fig. 5.
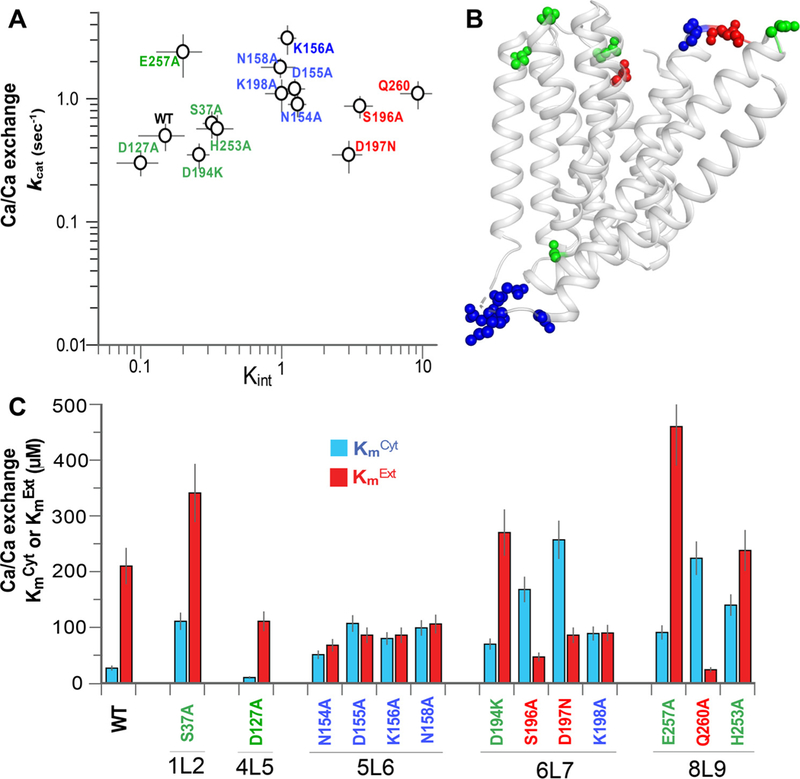
Mutational effects of loop-residues on the Kint and kcat values of the Ca2+/Ca2+ exchange. (A) Mutational effects on the Kint and kcat values of the Ca2+/Ca2+ exchange were analyzed as described in Fig. 4 (see also Materials and Methods). Data were derived from at least 3 independent experiments and are presented as mean ± SE. Mutations exhibiting Kint < < 1, Kint ≈ 1 and Kint > > 1 are in green, blue, and red, respectively. (B) Cartoon presentation of NCX_Mj. Mutated residues are shown as spheres, where the colored residues are assorted according to their mutational effects, as indicated in panel A. (C) The measured values of KmCyt (blue bars) and KmExt (red bars) of the Ca+/Ca2+ exchange reaction were derived from at least 3 independent experiments. Data are presented as mean ± SE. Residues are colored according to their mutational effects on Kint, as colored in panel A.
3.5. Mechanochemical role of selected residues revealed by computations
In order to interpret experimental results in light of the intrinsic dynamics of the exchanger, we first examined the conformational flexibility of residues. To this aim, we computed the RMSFs of NCX_Mj residues using the GNM [45]. Fig. 8A displays the results in comparison to those derived from experimental B-factors (PDB ID: 3v5u). A correlation coefficient of 0.79 between theory (GNM) and experiments (X-ray) is obtained, which supports further analysis using the GNM. Minima in the RMSF profile can be traced back to the hinge sites in the global modes of motions depicted in Fig. 8D. These regions indicate key mechanical sites that potentially act as hinges or anchors for supporting the collective dynamics of the exchanger. Strikingly, mutations of these sites (Fig. 8A) in TM2 (G42-S51), TM3 (G76-C80), TM7 (P212-S217) and TM8 (G231-D240) exhibited significant effects on the experimentally measured transport rates (see Fig. 2A) as well as KmCyt values (Fig. 3A). The majority of these key sites also coordinate the binding of ions (Fig. 1), implicating a mechanochemical role in regulating ion transport. This analysis thus suggests that mutations at putative hinge sites impact the ion-exchange kinetics.
Fig. 8.
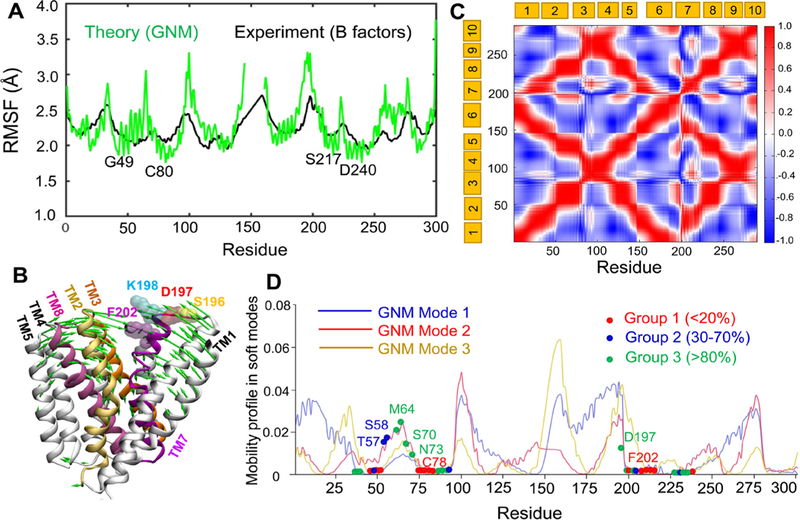
Conformational dynamics of NCX_Mj predicted by elastic network models. (A) Comparison of the theoretically predicted root-mean-square fluctuations (RMSFs) of residues (green) with the B-factors profile (black) observed in X-ray crystallographic experiments (PDB: 3V5S). (B) ANM mode 2 (green arrows) in the presence of lipid bilayer induce the closure of the extracellular vestibule, in favor of a transition from OF open state to an OF occluded state. (C) Cross-correlation map for the coupled fluctuations of all residues driven by the softest GNM modes 1–3. Red regions indicate pairs of residues undergoing correlated movements, blue regions indicate the pairs that undergo anticorrelated (coupled but opposite direction) fluctuations. Note that TM1 and TM6 are highly correlated, and they are anticorrelated with TM2–4 and TM8–9. (D) Residues in group 1 occupy minima in the mobility profile. The graph displays global mobilities of residues (corresponding to the soft modes 1, 2 and 3) obtained by GNM analysis of NCX_Mj embedded in a lipid bilayer. The blue, red and oranges curves represent the predictions from the respective three GNM modes. Red, blue and green spheres indicate the mutated residues belonging to those classified as Group 1 (Vmax < 20%), Group 2 (30 < Vmax < 70%), and Group 3 (> 80%), respectively in Fig. 2. The residues that show minimal (close to zero) mobility in at least one of the three soft modes are shown by their color-coded sphere. Those located in other regions, are indicated on the mode 2 curve. Minima refer to structural regions that serve as hinges or anchors in the global movements of the entire exchanger. All calculations and visualizations were performed using the DynOmics server.
Notably, C78, C80 and F202 do not coordinate ion binding, yet they exhibit minimal fluctuations typical of hinge sites in the slowest modes (Fig. 8D) and their mutations (C78A, C80A, and F202C) exhibit large reduction in Vmax (see Fig. 2), further highlighting the functional significance of hinge sites in the exchanger function. In contrast, residues S196-D197-K198 located around the loop connecting TM6 and TM7 show relatively high amplitude mobility by virtue of their exposure to the extracellular region (Fig. 8). Perturbation-response scanning analysis shows that these residues have a high propensity for acting as sensors, suggesting a potential role in relaying allosteric signals (Figure S3) [47]. Experiments conducted with mutations at these residues showed significant effects on Kint and a negligible effect on κcat values. A closer examination of collective motions (ANM mode 2; green arrows in Fig. 8B; and Supplemental Movie) revealed the correlated motions of TM1, TM6 and TM7 (block 1) and the strong coupling between TM2, TM3, TM4, TM5 and TM8 (block 2). These two blocks undergo anticorrelated motions with respect to each other (Fig. 8B and C). S196-D197-K198 lie within block 1, rather than the interface between those blocks. As such, mutations at those sites would not be expected to interfere with the collective dynamics of TM segments, which may explain the negligible effect on kcat.
4. Discussion
4.1. Mutational analysis of NCX-mediated ion fluxes
The effects of single-point mutations of α1/α2 repeat residues on Vmax and Km were previously reported for mammalian NCX1 [58,61,62] and NCKX2 [51,52,65] although at this time no structural information was available for any NCX protein. The determination of the crystal structure of NCX_Mj [12] and CAX [13–15] proteins, belonging to the Ca2+/CA superfamily of exchangers, provided a new opportunity for resolving the structure-functional basis of ion-transport mechanisms [22–31]. In general, NCX_Mj is an ideal model for studying the ion-transport mechanisms because this protein lacks any regulatory domains, where eleven ion-coordinating residues (out of twelve) are highly conserved among prokaryotic and eukaryotic NCX orthologs [12,22–25,30]. There is no doubt that a comparison of mutational effects obtained for NCX1 [58,61,62] and NCKX2 [51,52,65] with mutational effects obtained for NCX_Mj in the present study might be very compiling. However, it is worthwhile to mention that these kinds of comparisons are not trivial in the absence of exact structural information, since the structure-functional organization of four binding sites may differ in distinct protein, even though the ion-coordinating residues are highly conserved. For example, several lines of evidence suggest that NCX_Mj can bind 3Na+ ions without occupying the Smid site [23,26,27], whereas in NCKX2 (exhibiting the stoichiometry of 4Na+ :1Ca2+, 1K+) Smid is occupied either by Na+ or K+ [16,51,52,65]. More dedicated efforts are required for comparison of mutational effects observed in NCX1, NCKX2 and NCX_Mj in conjunction with structural data and MD simulations. Obviously, this kind of comparison is beyond the scope of the current study.
4.2. Mechanistic significance of the present analytical approaches
As a typical membrane transporter, NCX_Mj undergoes an alternative exposure of the ion-binding pocket at opposite sides of the membrane, thereby adopting either the IF or OF conformation during the transport cycle [12,28,30,31]. The kinetic analysis of bidirectional ion fluxes offers a quantitative evaluation of the kinetic (kcat) and equilibrium (Kint) parameters associated with bidirectional ion movements and thereby ascribes the overall rate-equilibrium relationships between the IF and OF states involving the apo and ion-bound species [23,24,38]. The previous kinetic analysis has shown that under steady-state conditions, NCX_Mj preferentially adopts the OF orientation, while showing Kint ≈ 0.15 for bidirectional Ca2+ movements [23,27,38]. Consistent with this, the crystallographic data, fluorescent labeling, and HDX-MS analyses have shown that the extracellular vestibule of NCX_Mj is preferentially exposed to the bulk phase either in the apo or ion-bound states, thereby revealing the conformational stability of the OF state [12,27,30,31]. Since the structure-dynamic determinants of the OF state stability are unknown, the present work was undertaken to resolve the partial contributions of individual residues in controlling the Kint and kcat parameters. Thus, the structure-based mutational effects of the pore-forming residues (located on TM2, TM3, TM7, and TM8) and the helix-loop residues were analyzed here with the goal of identifying residues affecting the functional asymmetry (Kint) and transport rates (kcat) of bidirectional Ca2+ movements.
The dynamic features of a protein have been shown to provide a metric for assessing the effect of missense mutations on the global dynamics [46–49]. In particular residues with the following properties were found to be highly pathogenic, if mutated [46]: high propensity to act as effector, low propensity to act as a sensor, low conformational flexibility (probed by RMSF), and high stiffness and mechanical bridging ability. Interestingly, NCX_Mj residues that displayed comparatively large effects upon mutation, broadly satisfied the aforementioned criteria. Comparison with experimental data shows that mutations at the hinge sites affect the transport/exchange rate kcat as well as Kint, while mutations of potential effectors or sensors may change the Kint.
4.3. Asymmetrically distributed deep-pore residues limit the ion-transport rates
Mutational analyses of pore-forming residues revealed that a limited number of ion-coordinating (S51, E54, S77, T209, S210, E21 and D240) and non-coordinating (G49, P53, G76, C78, C80, P212 and S217) residues limit (< 20% of WT Vmax) either the Na+/Ca2+ (Fig. 2A) or Ca2+/Ca2+ (Fig. 3B) exchange activity. Although these residues seem to be essential for mediating the ion-transport activities, the previous analysis has shown that the mutations of ion-coordinating residues have very small (if any) effect on Kint [27]. Thus, the pore-forming residues limiting the ion-transport rates have relatively insignificant effect on the intrinsic equilibrium of bidirectional ion movements. In contrast, our computational analysis shows that the pore-core residues play a key mechanical role in the collective dynamics (soft modes of motion) of the exchanger that enables its transition between OF and IF states (Fig. 8). Among these functionally important residues, four (G49, P212, E213 and S217) are located within the cytosolic vestibule, whereas the other key residues (S51, P53, E54, G76, S77, C78, C80, T209, S210 and D240) are located at the extracellular vestibule (Fig. 6A, B), and they all serve as hinges/anchors in the global motions (Fig. 8). Despite this “asymmetric” distribution of functionally important key residues between the extracellular and cytosolic vestibules, all these residues are located within ~ 10 Å from the pore center (Fig. 6A). Present analyses are consistent with previous findings suggesting that the 49-GTSLPE-54 (TM2, α1) and 208-GTSLPE-213 (TM7, α2) signature sequences (being a part of inverted two-fold symmetry and encompassed at the center of the ion-pocket) own to distinct conformational patterns, thereby suggesting an asymmetric preorganization of functional important key residues [27,31]. Compiling data supports a proposal according to which the ion binding may pull the “flexible” segments (TM7B, TM7C, TM2C and TM8A) toward the rigid TM2B (Fig. 7), which may initiate the OF/IF swapping. According to this proposal, a subtle conformational change involving the asymmetric compression of the pore-core forms a hydrophobic patch between TM2C and TM7C to allow the sliding of the gating bundle (TM1/TM6) on the protein surface [13,27,31]. This proposed model may serve as a common basis for ioncoupled alternating access in NCX and similar proteins [12–16,25–27,31]. A further resolution of underlying structure-dynamic details requires more dedicated and coordinated combination of experimental and computational approaches.
Fig. 6.
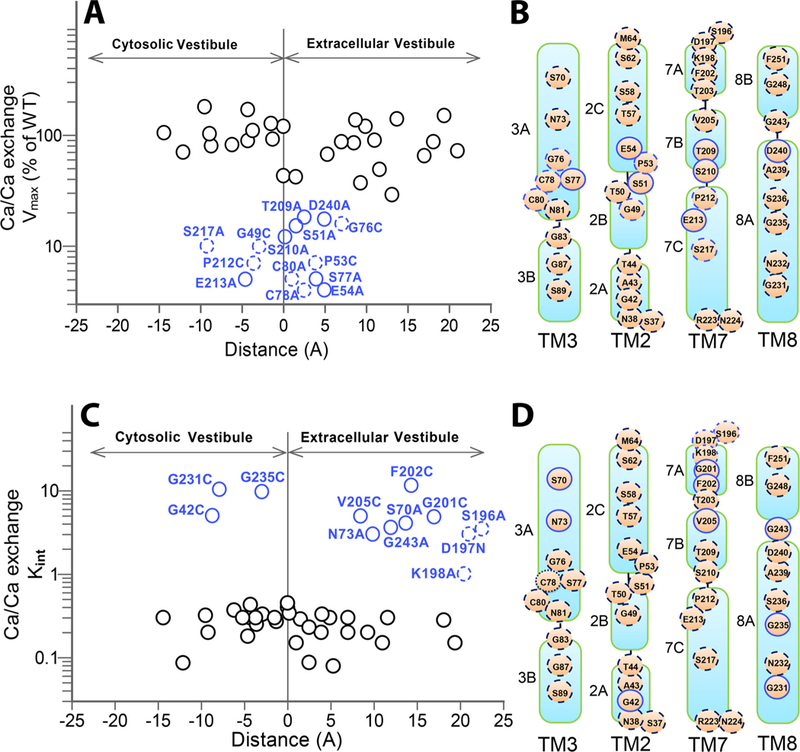
Spatial distribution of residues differentially affecting the Vmax and Kint values. (A) For each mutant, the Vmax value of the Ca2+/Ca2+ exchange was plotted vs. its position within the pore. The residue-positions are expressed as the distance from the pore center (the “zero” represents the position of the pore center). The residues significantly affecting the Vmax value (< 20% of WT Vmax) are shown in blue, where less significant residues affecting the ion-transport rates are shown as solid black circles. The solid and dashed blue circles represent the ion-coordinating and non-coordinating residues, respectively. (B) Topological presentation of mutated positions. The mutational effects of residues on Vmax are colored as in panel A. (C) For each mutant, the Kint value was plotted vs its distance from the pore center. The mutants exhibiting high Kint values are indicated in blue, whereas the solid and dashed circles represent the pore-forming and helix-loop residues, respectively. Solid black cycles represent mutations having an insignificant effect on Kit (D). Topological presentation of mutated positions are colored as in panel C.
Fig. 7.
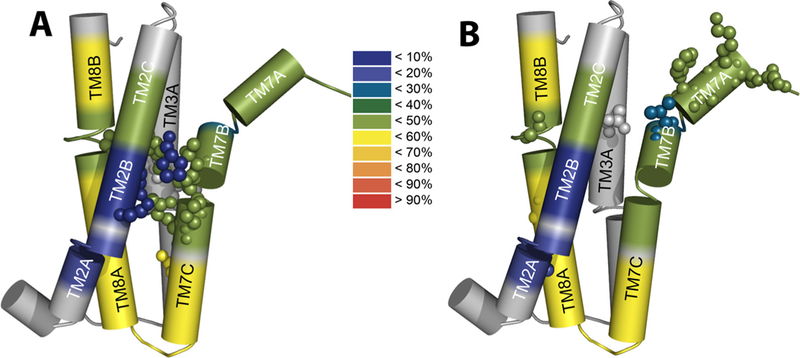
Structural-dynamics and functional relationships in NCX_Mj. Schematic heat maps of the hydrogen-deuterium exchange were overlaid on the pore-forming TMs by using the available HDX-MS data [31] and the crystal structural of NCX_Mj [12,30]. The color ruler represents the HDX levels (in %), thereby depicting the characteristic profiles of backbone dynamics in apo NCX_Mj (blue signifies the most rigid and water inaccessible segments in protein). (A) The key residues limiting the ion transport rates (shown as spheres) are located on the TM2B, TM3 A, TM7BC, and TM8 A segments within the pore core, while showing striking differences in the local backbone dynamics at respective locations. (B) Key residues affecting the intrinsic equilibrium (Kint) of bidirectional Ca2+ movements (shown as spheres) are distributed at peripheral locations on TM2 A, TM3 A, TM7AB, and TM8 AB, which is consistent with the known mechanisms of ion occlusion and also provides new clues for the mechanisms underlying ion-coupled alternating access.
4.4. Revisiting the structure-functional role of ion-coordinating residues
A large number of residues located within 10 Å from the ion porecore have very little (if any) mutational effects on the ion-transport rates (Fig. 2A and 6 A). Remarkably, the mutations in the ion-co-ordinating residues T50, N81, or S236 have no inhibitory effect on the Vmax values of the Na+/Ca2+ exchange (Fig. 2A). The questionable status of N81 (exclusively assigned to Smid) has been resolved in previous study, revealing that in the ground state the Smid site of NCX_Mj does not bind either Na+ or Ca2+ [27]. In contrast with NCX_Mj, however, the mutational studies of NCKX2 have demonstrated that the Smid site of NCKX can bind K+ [50,51]. Notably, the carbonyl group of T50 and T209 coordinate the Ca2+ ion at SCa, whereas the side chains of T50 and S236 ligate Na+ at Sint (Fig. 1D and F) [12,30]. Since the side-chain mutation of T50 or S236 does not inhibit the Na+/Ca2+ exchange (Fig. 2B), it is possible that some compensatory mechanisms are involved in Na+ ligation at Sint in the T50 and S236 mutants. For example, in the T50 or S236 mutant both oxygen atoms of the E213 carboxyl group may ligate Na+ at Sint, in contrast with NCX_Mj-WT, where according to the crystal structure, the Na+ ion at Sint is ligated through a single oxygen atom of the carboxyl group [12,30]. This kind of compensatory mechanism may not occur at Sext, since mutation of E54, S77, or T209 strongly inhibits the Na+/Ca2+ exchange (Fig. 2A). Thus, the mutational disparities between the Sint and Sext sites may represent the structure-dynamic asymmetry of matching residues (E54/E213, T50/T209, and S77/S236) at Sint and Sext in the context of inverted-twofold symmetry [27]. Consistent with this proposal, Sint of NCX_Mj has at least 10–20-fold higher affinity to Na+, compared with Sext [30]. Another interesting observation is that the mutation of S217 strongly reduces the Na+/Ca2+ exchange rates (Fig. 2A), although S217 is not an ion-coordinating residue according to the OF crystal structure of NCX_Mj. Since S217 is adjacent to Sint, we posit that this residue may interact with Sint. This is especially interesting in light of the fact that S217 is highly conserved among NCX and NCKX orthologs [16,29,50,51]. Moreover, the mutation of S552 in NCKX (the analog of S217 in NCX_Mj) dramatically increases the KmExt value for K+ [50,51].
Unfortunately, the measurement of the KmCyt and KmExt values for the S217 mutant is unavailable in the NCX_Mj system due to very low signal/background ratio at very low Vmax. Nonetheless, it is possible that S217 (TM7) forms hydrogen bonding with N232 (TM8), which may contribute to cytosolic gating. Interestingly, the MD simulations have shown that in the OF conformer, hydrogen bonding is intermittently formed between S217 and N232. But no hydrogen bond is formed between these two residues in the simulation of IF conformers. Notably, the mutation of S58 (matching S217) has no significant effects either on kcat or Kint (Figs. 2A and 4 B and C), meaning that S58 does not contribute to extracellular gating, neither controls the ion-transport rates.
4.5. Functional contributions of distinct glycine residues to the rate-equiHbrium relationships
Previous studies have shown that mutations of P53 or P212 have devastating effects on the ion-transport capacity [27]. These data, in conjunction with HDX-MS analyses [27,31], strongly support the notion that conformational flexibility of the signature repeats, 49-GTSLPE-54 and 208-GTSLPE-213 (located at the interface of TM2B/TM2C and TM7B/TM7C, respectively), plays a critical role in ion-transport activities [27]. Extending this analysis, we examined here the mutational effects of 10 glycine residues on the kcat and Kint values. Three glycine residues, G83 (TM3B), G87 (TM3B), and G208 (TM7) exhibit no appreciable effects on kcat and Kint even though G208 is a part of 208-GTSLPE-213. The mutations of G49 (TM2B) or G76 (TM3 A) dramatically reduce the Na+/Ca2+ exchange rates (Fig. 2A), thereby suggesting that these two glycine residues may control conformational dynamics of ion-bound intermediates involved in ion-transport catalysis and/or ion-occlusion events. Unfortunately, the Kint values for G49 and G76 mutations were unavailable since the signal/background ratios were low. In any case, it is quite clear that G49 and G208 (which are part of 49-GTSLPE-54 and 208-GTSLPE-213), exhibit very different mutational effects (Fig. 2A). These findings are especially interesting in light of the fact that according to HDX-MS analyses the backbone dynamics of 49-GTSLPE-54 is much more constrained in comparison with 208-GTSLPE-213 [27,31]. Interestingly, G76 is adjacent to S77, which ligates Na+ at Sext (Fig. 1C) and limits the ion-transport activity (Fig. 2A). Thus, the G76 controlled performance of the Sext site seems to be essential for ion-transport activity. Strikingly, the mutation of 5 glycine residues, G42 (TM2 A), G201 (TM7A), G231 (TM8 A), G235 (TM8 A), and G243 (TM8B) increase the Kint value 20–50-fold, whereas they have rather small effects on kcat (Fig. 4B,D). Thus, the observed Kint values for these mutations (Kint = 3–10) reveal the important role of the relevant glycine residues in the preferential stabilization of the OF state (Fig. 4B). These findings are consistent with a recent HDX-MS analysis revealing the characteristic differences in the backbone dynamics of the TM2 A, TM7A, TM8 A, and TM8B segments (Fig. 7). Since, TM2 A and TM2B exhibit very low flexibility and water accessibility either in the absence or presence of Na+ orCa2+ ions [31], G42, G201, G231, G235, and G243 can differentially govern the backbone dynamics at functionally important TM segments, which may effectively control the relative stability of the IF and OF states. Notably, G235 (inversely matching the position of G76) is next to S236, while ligating Na+ at Sint according to the crystal structure (Fig. 1D). Strikingly enough, the S236 A mutation activates, rather than inhibits, the Na+/Ca2+ (Fig. 2A) or Ca2+/Ca2+ exchange [27] activities. Notably, the S236 G mutation does not inhibit the ion-exchange activities as well [24], thereby supporting the notion that S236 is not an essential residue for mediating the Na+ transport activities in NCX. Collectively, the present data in conjunction with HDX-MS [27,31] and X-ray crystallography data [30] conclude that the Sext and Sint sites exhibit very different backbone dynamics and affinity for Na+ binding/transport.
4.6. Structure-based mechanistic insights for functional asymmetry
According to the X-ray data, the high affinity binding of 2 Na+ ions to Sint and SCa is followed by binding of the 3rd Na+ to Sext [30]. The binding at Sext is associated with the bending of the TM7A/TM7B segment, which subsequently results in occlusion of 3Na+ ions at the extracellular side [30]. Consistent with this, HDX-MS analysis of NCX_Mj identified hallmark changes in the backbone dynamics at the TM7A/TM7B interface upon Na+ binding [31]. The present analysis provides additional insights into the underlying mechanisms by identifying key functional residues. The mutations of G42, G231, and G235 (at the cytosolic vestibule) and of S70, N73, G201, F202, V205, and G243 (at the extracellular vestibule) result in up to 200-fold changes in Kint, where only 2 residues (G235 and F202) exhibit significantly reduced kcat values (Fig. 4B and 6 C). The mutations of 3 residues (G201, F202, and V205), located at the interface of TM7A and TM7B, result in increased Kint values (Fig. 4B). These findings are compatible with the contribution of these residues to extracellular ion occlusion. Notably, S70, N73, and G76 (at TM3 A) and their matching counterparts, G231, N232, and G235 (at TM8 A) alter either kCat (Fig. 2A) or Kint (Fig. 4B and C). These results are interesting from a structural perspective, revealing that the backbone carbonyls of T209 and T50 coordinate Ca2+, whereas the side chains of T209 and T50 may form hydrogen bonding with N73 and N232, respectively [12,30].
These interactions are likely to be unstable in the apo form, since the side chains of N73 and N232 in apo and Ca2+-bound NCX_Mj display different orientations, although the backbone folding at these areas is indistinguishable for the apo and ion-bound forms [12,30]. As suggested above, the interaction between N232 and S217 could be important for the conformational shaping and functioning of a “cytosolic gate”. Notably, S70, N73, and G76 (TM3A) and their inverted counterparts G231, N232, and G235 (TM8 A) are located at opposite sides of the ion-binding pocket, thereby suggesting that TM3 A (S70, N73, and G76) and TM8A (G231, N232, and G235) may differentially stabilize the ion-bound forms. Thus, according to the present findings, distinct residues located on TM2B (G49), TM3 A (S70, N73), TM7A (G201, F202), TM7B (V205), TM8A (G231, G235), and TM8B (G243) control the intrinsic equilibrium (Kint) of the bidirectional ion movements. The relevant structural arrangements may stabilize distinct ion-bound forms through specific interactions between distinct TM segments during the transport cycle. Further elucidation of the relevant mechanisms requires more dedicated and well-coordinated combinations of structural, biophysical, and computational approaches.
4.7. Loop-helix residues affecting the rate-equilibrium relationships of Ca2+ movements
Our previous studies have shown that elongation of the 5L6 loop of NCX_Mj by 8–14 residues results in increased kcat (up to 10-fold) and Kint (50–200-fold) values although the underlying mechanisms remain unclear [23,31]. This phenomenon is especially interesting in light of the fact that in contrast with NCX_Mj, the 5L6 loop of mammalian NCX orthologs contain the regulatory CBD domains, where Ca2+ binding to CBDs results in ~ 20-fold activation of ion-exchange rates under physiologically relevant conditions [52–57]. Interestingly, in our MD simulations of the IF NCX_Mj conformer, we observe that the 5L6 loop tends to swing into the intracellular vestibule and expose four acidic residues (E149, E151, E152, and D155), thus potentially facilitating the binding of Ca2+ ions from the cytosol (Fig. 9). Simulations suggest that this loop may play a role in distinguishing the asymmetric Ca2+ binding. Interestingly, the homologous 5L6 loop (assigned as the “acidic helix”) resolved in the IF-oriented structure of VCX1 (Ca2+/H+ exchanger, CAX family) is also oriented toward the intracellular vestibule (Fig. 9), according to the corresponding X-ray structure (PDB: 4K1C) [14]. In light of the present considerations, we posit that the global rearrangement of the TM helices during the OF-IF transition is accompanied by local rearrangements at the 5L6 loop. Interestingly, our MD simulations further showed that several acidic residues (D121, D127, E257, D194 and D197 on the extracellular surface and E28, E149, E151, D152, and D155 at the intracellular surface) transiently bound Ca2+ ions. Thus, in addition to the two acidic residues E54 and E213 that coordinate the binding of the Ca2+ ion in the central portion of the protein (Fig. 1F) as well as 3Na+ ions (Fig. 1C–E), several negatively charged residues on the extracellular and/or cytoplasmic loops may serve as cation attractors.
Fig. 9.
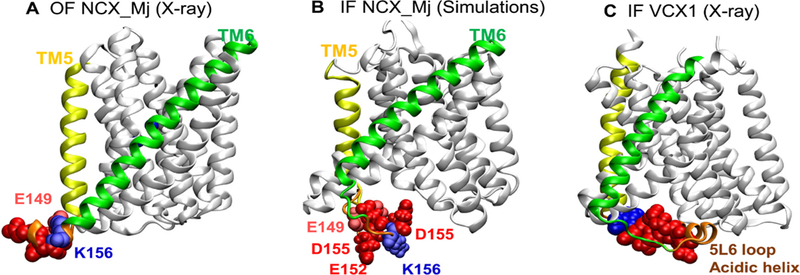
Conformational dynamics of NCX_Mj captured by full-atomic computations suggests the potential role of the 5L6 loop. (A–B) Simulations indicate that the 5L6 loop reorients differently in the outward-facing (OF) and inward-facing (IF) conformers of NCX_Mj as illustrated for (A) X-ray resolved NCX_Mj OF conformer (PDB: 5HWY), and (B) MD-refined IF conformer. (C) Structure of the IF homologous superfamily member VCX1 Calcium/Proton Exchanger resolved in the IF state (PDB: 4K1C). The simulations suggest that the 5L6 loop (orange) swings into the intracellular vestibule in the IF state. Notably, the homologous 5L6 loop (or acidic helix) of VCX1 (H+/Ca2+ exchanger) also exhibits a similar orientation in the IF state.
In search of underlying structure-functional mechanisms, we examined here the effect of single-point mutations, located at distinct loop-helix interfaces connecting TMs, on kcat and Kint of Ca2+/Ca2+ exchange (Fig. 5). The most interesting finding is that single-point mutations of three consecutive residues, D196, S197 and K198, located at the interface of TM7A and 6L7 loop (connecting TM6 and TM7), exhibit 8–25-fold higher Kint values as compared with WT, while having a negligible (if any) effect on kcat (Fig. 5A and 6 C). The crystal structure of NCX_Mj is compatible with possible interaction of these residues through hydrogen bonding network. Moreover, the 196-DSK-198 sequence is located in a close vicinity with G201, F202 and V205 (located on the TM7A and TM7B segments), which also exhibit comparable mutational effects on the Kint values (Fig. 4B and 6 C). Thus, the present functional studies are consistent with results from X-ray [30] and HDX-MS [27,31] studies, thereby underscoring the structure-functional significance of the TM7A/TM7B segment in controlling the extracellular ion occlusion. Notably, no significant mutational effects were observed for the cytosolic-facing residues S37 and N38 (Fig. 2A and 5 A), which represent inverted pair-residues for D196 and S197. Thus, the 196-DSK-198 module (at the TM7A/6L7 interface) stabilizes the OF state, which significantly contributes to functional asymmetry of WT NCX_Mj. Since the 6L7 loop connects TM7A with TM6 (which forms the gating bundle in association with TM1), the 196-DSK-198 module may shape the dynamic features of the TM1/TM6 cluster, which in turn may contribute to conformational dynamics of the TM1/TM6 movements toward the ion-coupled alternating access.
4.8. Physiological relevance of Kint and kcat values
The structure-based mutational analyses of prokaryotic NCX, described here, provide an excellent basis for interpreting mutational effects observed in mammalian NCXs and similar proteins. The resolution of mutational effects of Kint and kcat is essential for elucidating the structural determinants governing the physiologically relevant parameters of bidirectional ion movements [23–28]. The present work demonstrates that distinct structural elements can differentially affect the Kint and kcat values, where the underlying structure-functional mechanisms might be relevant not only for NCX_Mj, but also for eukaryotic NCXs and similar proteins belonging to the Ca2+/CA superfamily [12–15,25–27,53,54]. How the relevant mechanisms contribute to 104-fold differences in the kcat values while keeping the Kint values comparable remains to be discovered. Nevertheless, the present findings strongly support the notion that NCXs share a common structural basis for ion transport mechanisms, which is secondarily modified in mammalian isoform/splice variants [3,5,59–66]. In any case, the relevant mechanisms are essential for matching the NCX-mediated Ca2+-extrusion rates in cardiac, neuro-glia, kidney, pancreas and many other cell types although the different regulatory modules are involved in “secondary” modulation of tissue-specific NCXs [3,7,59,60,66]. The fine-tuning of NCX-mediated rates of ion-exchange seem to be especially important during the action potential, where the directionality of charge/ion fluxes reverses and ion-exchange rates permute up to 50-fold within a few milliseconds in cardiac [52,63,66] and neuroglia [52,59,60,64] systems. For example, in astrocytes the reversal potential of NCX (ENCX) is close to the resting membrane potential (Em ~ –80 mV), so even small changes in [Ca2+]i, [Na+]i and membrane potential can dynamically alternate the directionality and rates of NCX-mediated ion-exchange from the reverse (Ca2+-entry) to the forward (Ca2+-extrusion) mode and vice versa [59,60,64]. This in turn, may have a huge impact on the Ca2+ and Na+-dependent release/uptake of neurotransmitters among many other fundamental events controlled by Ca2+ [64].
5. Conclusions
In the present study, fifty-five mutants of NCX_Mj were analyzed with the goal of identifying the key residues controlling the turnover rates (kcat) and intrinsic asymmetry (Kint) of bidirectional Ca2+ movements. The mutations can be divided into two major groups based on the observed effects on the ion-exchange fluxes. The first group of mutations affect kcat rather than Kint. All the corresponding residues are located within ~ 10 Å of the pore center. The second group of mutations affects Kint rather than kcat. The relevant residues are dispersed either along the pore or are localized at specific sites on the extracellular and cytosolic loops, in this case. In conjunction with the present mutational analyses and previous HDX-MS studies [27,31], the simulations performed here reveal that distinct inter-residue interactions control the relative stability of the IF and OF states, as well as the opening or closure of the intracellular and extracellular vestibules. The corresponding structural and dynamic modules may serve as a basis for the intrinsic asymmetry of bidirectional Ca2+ movements, as documented in prokaryotic [23,27] or mammalian NCXs [38]. Thus, the combination of experimental and computational analyses described here is very effective for elucidating the structure-dynamics determinants at the single residue level in NCXs and similar proteins. This may provide a good basis for more systematic and extended computational modeling of NCX and similar proteins.
Supplementary Material
Acknowledgments
This work was partially funded by Israel Science Foundation grants 825/14 and 1351/18 to DK. The financial support of the Fields Estate foundation to DK is highly appreciated. Support from NIH awards P41 GM103712 and P30DA035778 is gratefully acknowledged by IB.
☆ This work was performed in partial fulfillment of the PhD thesis requirements of Liat van Dijk and Bosmat Refaeli at the Sackler Faculty of Medicine, Tel Aviv University.
Abbreviations:
- NCX
sodium–calcium exchanger
- CBD
calcium binding domain
- Mops
3-(N-morpholino)propanesulfonic acid
- Tris
tris(hydroxymethyl)-amino-methane
- HEPES
4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid
- MES
2-(N-morpholino)ethanesulfonic acid
- Fluo-3
N-[2-[2[bis-(carboxymethyl)-amino]-5-(2,7dichloro-6-hydroxy-3-oxy-3Hxanthen-9-yl) henoxy]ethoxy]-4methylphenyl]-N(carboxymethyl)glycine
- SDS-PAGE
sodium dodecyl sulfatepolyacrylamide gel electrophoresis
- EGTA
ethyleneglycoltetraacetic acid
- PMSF
phenylmethanesulfonyl fluoride
- HDX-MS
Hydrogen-deuterium exchange mass spectrometry
- DTT
1,4-dithiothreitol
Footnotes
Conflict of interest
All authors state no conflict of interests.
Appendix A. Supplementary data
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.ceca.2018.09.004.
References
- [1].Philipson KD, Nicoll DA, Sodium-calcium exchange: a molecular perspective, Annu. Rev. Physiol. 62 (2000) 111–133. [DOI] [PubMed] [Google Scholar]
- [2].Blaustein MP, Lederer WJ, Sodium/calcium exchange: its physiological implications, Physiol. Rev. 79 (1999) 763–854. [DOI] [PubMed] [Google Scholar]
- [3].Khananshvili D, The SLC8 gene family of sodium-calcium exchangers (NCX): structure, function, and regulation in health and disease, Mol. Asp. Med. 34 (2013) 220–235. [DOI] [PubMed] [Google Scholar]
- [4].Palty R, Silverman WF, Hershfinkel M, Caporale T, Sensi SL, Parnis J, Nolte C, Fishman D, Shoshan-Barmatz V, Herrmann S, Khananshvili D, Sekler I, NCLX is an essential component of mitochondrial Na+/Ca2+ exchange, Proc. Natl. Acad. Sci. U. S. A. 107 (2010) 436–441, 10.1073/pnas.0908099107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Khananshvili D, Sodium-calcium exchangers (NCX): molecular hallmarks underlying the tissue-specific and systemic functions, Plügers Arch. 466 (2014) 43–60. [DOI] [PubMed] [Google Scholar]
- [6].Reeves JP, Hale CC, The stoichiometry of the cardiac sodium-calcium exchange system, J. Biol. Chem. 259 (1984) 7733–7739. [PubMed] [Google Scholar]
- [7].Bers DM, Ginsburg KS, Na:Ca stoichiometry and cytosolic Ca-dependent activation of NCX in intact cardiomyocytes, Ann. N. Y. Acad. Sci. 1099 (2007) 326–338. [DOI] [PubMed] [Google Scholar]
- [8].Shlosman I, Marinelli F, Faraldo-Gomez JD, Mindell JA, The prokaryotic Na+/Ca2+ exchanger NCX_Mj transports Na+ and Ca2+ in a 3:1 stoichiometry, J. Gen. Physiol. 150 (2018) 51–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Khananshvili D, Distinction between the two basic mechanisms of cation transport in the cardiac Na+-Ca2+ exchange system, Biochemistry 29 (1990) 2437–2442. [DOI] [PubMed] [Google Scholar]
- [10].Cai X, Lytton J, The cation/Ca2+ exchanger superfamily: phylogenetic analysis and structural implications, Mol. Biol. Evol. 21 (2004) 1692–1703. [DOI] [PubMed] [Google Scholar]
- [11].Schntkamp PP, Jalloul AH, Liu G, Szerencsei RT, The SLC24 family of K+-dependent Na+-Ca2+ exchangers: structure-function relationships, Curr. Top. Membr. 73 (2014) 263–287. [DOI] [PubMed] [Google Scholar]
- [12].Liao J, Li H, Zeng W, Sauer DB, Belmares R, Jiang Y, Structural insight into the ion-exchange mechanism of the sodium/calcium exchanger, Science 335 (2012) 686–690. [DOI] [PubMed] [Google Scholar]
- [13].Nishizawa T, Kita S, Maturana AD, Furuya N, Hirata K, Kasuya G, Ogasawara S, Dohmae N, Iwamoto T, Ishitani R, Nureki O, Structural basis for the counter-transport mechanism of a H+/Ca2+ exchanger, Science 341 (2013) 168–172. [DOI] [PubMed] [Google Scholar]
- [14].Waight AB, Pedersen BP, Schlessinger A, Bonomi M, Chau BH, Roe-Zurz Z, Risenmay AJ, Sali A, Stroud RM, Structural basis for alternating access of a eukaryotic calcium/proton exchanger, Nature 499 (2013) 107–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Wu M, Tonga S, Walterspergerb S, Diederichsc K, Wang M, Zheng L, Crystal structure of Ca2+/H+ antiporter protein YfkE reveals the mechanisms of Ca2+ efflux and its pH regulation, Proc. Natl. Acad. Sci. U. S. A. 110 (2013) 11367–11372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Zhekova H, Zhao C, Schnetkamp PP, Noskov SY, Characterization of the cation binding sites in the NCKX2 Na+/Ca2+-K+ exchanger, Biochemistry 55 (2016) 6445–6455. [DOI] [PubMed] [Google Scholar]
- [17].Ren X, Philipson K, The topology of the cardiac Na+/Ca2+ exchanger, NCX1, J. Mol. Cell. Cardiol. 57 (2013) 68–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Hilge M, Aelen J, Vuister GW, Ca2+ regulation in the Na+/Ca2+ exchanger involves two markedly different Ca2+ sensors, Mol. Cell. 22 (2006) 15–25. [DOI] [PubMed] [Google Scholar]
- [19].Nicoll DA, Sawaya MR, Kwon S, Cascio D, Philipson KD, Abramson J, The crystal structure of the primary Ca2+ sensor of the Na+/Ca2+ exchanger reveals a novel Ca2+ binding motif, J. Biol. Chem. 281 (2006) 21577–21581. [DOI] [PubMed] [Google Scholar]
- [20].Giladi M, Sasson Y, Fang X, Hiller R, Buki T, Wang Y-X, Hirsch JA, Khananshvili D, Ca2+-driven interdomain switch of NCX: structural and bio-chemical studies of the two-domain Ca2+ sensor, PloS One 7 (6) (2012) e39985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Tal I, Kozlovsky T, Brisker D, Giladi M, Khananshvili D, Kinetic and equilibrium properties of regulatory Ca2+-binding domains in sodium-calcium exchangers 2 and 3, Cell Calcium 59 (2016) 181–188. [DOI] [PubMed] [Google Scholar]
- [22].Giladi M, SY Lee, Ariely Y, Teldan Y, Granit R, Strulovich R, Haitin Y, Chung KY, Khananshvili D, Structure-based dynamic arrays in regulatory domains of sodium-calcium exchanger (NCX) isoforms, Sci. Rep. 7 (1) (2017) 993, 10.1038/s41598-017-01102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Almagor L, Giladi M, van Dijk L, Buki T, Hiller R, Khananshvili D, Functional asymmetry of bidirectional Ca2+-movements in an archaeal sodium-calcium exchanger (NCX_Mj), Cell Calcium 56 (2014) 276–284. [DOI] [PubMed] [Google Scholar]
- [24].Refaeli B, Giladi M, Hiller R, Khananshvili D, Structure-based engineering of lithium-transport capacity in an archaeal sodium-calcium exchanger, Biochemistry 55 (2016) 1673–1676. [DOI] [PubMed] [Google Scholar]
- [25].Giladi M, Tal I, Khananshvili D, Structural features of ion transport and allosteric regulation in sodium-calcium exchanger (NCX) proteins, Front. Physiol. 7 (2016) 30, 10.3389/fphys.2016.00030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Giladi M, Shor R, Lisnyansky M, Khananshvili D, Structure-functional basis of ion transport in sodium-calcium exchanger (NCX) proteins, Int. J. Mol. Sci. 17 (11) (2016), 10.3390/ijms17111949 pii: E1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Giladi M, Almagor L, van Dijk L, Hiller R, Man P, Forest E, Khananshvili D, Asymmetric preorganisation of inverted pair residues in the sodium-calcium exchanger, Sci. Rep. 16 (20753) (2016) 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Khananshvili D, Shaulov G, Weil-Maslansky E, Rate-limiting mechanisms of exchange reactions in the cardiac sarcolemma Na+-Ca2+ exchanger, Biochemistry 34 (1995) 10290–10297. [DOI] [PubMed] [Google Scholar]
- [29].Marinelli F, Almagor L, Hiller R, Giladi M, Khananshvili D, Faraldo-Gómez JD, Sodium recognition by the Na+/Ca2+ exchanger in the outward-facing conformation, Proc. Natl. Acad. Sci. U. S. A. 111 (2014) E5354–E5362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Liao J, Marinelli F, Lee C, Huang Y, Faraldo-Gómez JD, Jiang Y, Mechanism of extracellular ion exchange and binding-site occlusion in a sodium/calcium exchanger, Nat. Struct. Mol. Biol. 23 (2016) 590–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Giladi M, van Dijk L, Refaeli B, Almagor L, Hiller R, Man P, Forest E, Khananshvili D, Dynamic distinctions in the Na+/Ca2+ exchanger adopting the inward-and outward-facing conformational states, J. Biol. Chem. 292 (2017) 12311–12323, 10.1074/jbc.M117.787168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Jardetzky O, Simple allosteric model for membrane pumps, Nature 211 (1966) 969–970. [DOI] [PubMed] [Google Scholar]
- [33].Stein WD, Transport and Diffusion Across Cell Membranes, Acad. Press, NY, 1986, pp. 55–120. [Google Scholar]
- [34].Forrest LR, Krämer R, Ziegler C, The structural basis of secondary active transport mechanisms, Biochem. Biophys. Acta 1807 (2011) 167–188. [DOI] [PubMed] [Google Scholar]
- [35].Forrest LR, (Pseudo-) symmetrical transport, Science 339 (2013) 399–341. [DOI] [PubMed] [Google Scholar]
- [36].Bai X, Moraes TF, Reithmeier RAF, Structural biology of solute carrier (SLC) membrane transport proteins, Mol. Membr. Biol. 13 (2018) 1–32. [DOI] [PubMed] [Google Scholar]
- [37].Duran AM, Meiler J, Inverted topologies in membrane proteins: a mini-review, Comput. Struct. Biotechnol. J. 8 (2013) e201308004, 10.5936/csbj.201308004eCollection2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Khananshvili D, Weil-Maslansky E, Baazov D, Kinetics and mechanism: modulation of ion transport in the cardiac sarcolemma sodium-calcium exchanger by protons, monovalent, ions, and temperature, Ann. N. Y. Acad. Sci. 779 (1996) 217–235. [DOI] [PubMed] [Google Scholar]
- [39].Marti-Renom MA, Stuart A, Fiser A, Sánchez R, Melo F, Sali A, Comparative protein structure modeling of genes and genomes, Annu. Rev. Biophys. Biomol. Struct. 29 (2000) 291–325. [DOI] [PubMed] [Google Scholar]
- [40].Jo S, Kim T, Iyer VG, Im W, CHARMM-GUI: a web-based graphical user interface for CHARMM, J. Comput. Chem. 29 (2008) 1859–1865. [DOI] [PubMed] [Google Scholar]
- [41].Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, Schulten K, Scalable molecular dynamics with NAMD, J. Comput. Chem. 26 (16) (2005) 1781–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Bahar I, Atilgan AR, Erman B, Direct evaluation of thermal fluctuations in protein, Fold. Des. 2 (1997) 173–181. [DOI] [PubMed] [Google Scholar]
- [43].Eyal E, Lum G, Bahar I, The anisotropic network model web server at 2015 (ANM 2.0), Bioinformatics 31 (2015) 1487–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Lezon TR, Bahar I, Constraints imposed by the membrane selectively guide the alternating access dynamics of the glutamate transporter GltPh, Biophys. J. 102 (2012) 1331–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Li H, Chang YY, Lee JY, Bahar I, Yang LW, DynOmics: dynamics of structural proteome and beyond, Nucleic Acids Res. 45 (2017) 374–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Ponzoni L, Bahar I, Structural dynamics is a determinant of the functional significance of missense variants, Proc. Natl. Acad. Sci. U. S. A. 115 (2018) 4164–4169, 10.1073/pnas.1715896115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].General IJ, Liu Y, Blackburn M, Mao W, Gierasch LM, Bahar I, ATPase sub-domain IA is a mediator of interdomain allostery in Hsp70 molecular chaperones, PLoS Comp. Biol. 10 (2014) e1003624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Atilgan C, Atilgan AR, Perturbation-response scanning reveals ligand entry-exit mechanisms of ferric binding protein, PLoS Comput. Biol. 5 (10) (2009) e1000544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Ponzoni L, Rossetti G, Maggi L, Giorgetti A, Carloni P, Micheletti C, Unifying view of mechanical and functional hotspots across class A GPCRs, PLoS Comput. Biol. 13 (2) (2017) e1005381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Jalloul AH, Cai S, Szerencsei RT, Schnetkamp PPM, Residues important for K+ ion transport in the K+-dependent Na+-Ca2+ exchanger (NCKX2), Cell Calcium 74 (2018) 61–72. [DOI] [PubMed] [Google Scholar]
- [51].Jalloul AH, Liu G, Szerencsei RT, Schnetkamp PPM, Residues important for Ca2+ ion transport in the neuronal K+-dependent Na+-Ca2+ exchanger (NCKX2), Cell Calcium 74 (2018) 187–197. [DOI] [PubMed] [Google Scholar]
- [52].Boyman L, Hagen BM, Giladi M, Hiller R, Lederer WJ, Khananshvili D, Proton-sensing Ca2+ binding domains regulate the cardiac Na+/Ca2+ exchanger, J. Biol. Chem. 286 (2011) 28811–28820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Khananshvili D, Structure-dynamic coupling through Ca2+-binding regulatory domains of mammalian NCX isoform/splice variants, Adv. Exp. Med. Biol. 981 (2018) 41–58. [DOI] [PubMed] [Google Scholar]
- [54].Khananshvili D, Long-range allosteric regulation of pumps and transporters: what can we learn from mammalian NCX antiporters, Adv. Biochem. Health Dis. 14 (2016) 93–115. [Google Scholar]
- [55].Giladi M, Hiller R, Hirsch JA, Khananshvili D, Population shift underlies Ca2+-induced regulatory transitions in the sodium-calcium exchanger (NCX), J. Biol. Chem. 288 (2013) 23141–23149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Lee SY, Giladi M, Bohbot H, Hiller R, Chung KY, Khananshvili D, Structure-dynamic basis of splicing dependent regulation in tissue-specific variants of the sodium-calcium exchanger (NCX1), FASEB J. 30 (2016) 1356–1366. [DOI] [PubMed] [Google Scholar]
- [57].Bode K, O’Halloran DM, NCX-DB: a unified resource for integrative analysis of the sodium calcium exchanger super-family, BMC Neurosci. 19 (1) (2018) 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].John SA, Liao J, Jiang Y, Ottolia M, The cardiac Na+-Ca2+ exchanger has two cytoplasmic ion permeation pathways, Proc. Natl. Acad. Sci. U. S. A. 110 (2013) 7500–7505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Verkhratsky A, Trebak M, Perocchi F, Khananshvili D, Sekler I, Crosslink between calcium and sodium signalling, Exp. Physiol. 103 (2018) 157–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Plattner H, Verkhratsky A, The remembrance of the things past: conserved signalling pathways link protozoa to mammalian nervous system, Cell Calcium 73 (2018) 25–39, 10.1016/j.ceca.2018.04.001. [DOI] [PubMed] [Google Scholar]
- [61].Nicoll DA, Hryshko LV, Matsuoka S, Frank JS, Philipson KD, Mutation of amino acid residues in the putative transmembrane segments of the cardiac sarcolemmal Na+-Ca2+ exchanger, J. Biol. Chem. 271 (1996) 13385–13391. [DOI] [PubMed] [Google Scholar]
- [62].Iwamoto T, Uehara A, Imanaga I, Shigekawa M, The Na+/Ca2+ exchanger NCX1 has oppositely oriented reentrant loop domains that contain conserved aspartic acids whose mutation alters its apparent Ca2+ affinity, J. Biol. Chem. 275 (2000) 38571–38580. [DOI] [PubMed] [Google Scholar]
- [63].Bers DM, Calcium cycling and signaling in cardiac myocytes, Annu. Rev. Physiol 70 (2008) 23–49. [DOI] [PubMed] [Google Scholar]
- [64].Rose CR, Verkhratsky A, Principles of sodium homeostasis and sodium signalling in astroglia, Glia 64 (2016) 1611–1627. [DOI] [PubMed] [Google Scholar]
- [65].Winkfein RJ, Szerencsei RT, Kinjo TG, Kang K, Perizzolo M, Eisner L, Schnetkamp PP, Scanning mutagenesis of the alpha repeats and of the transmembrane acidic residues of the human retinal cone Na/Ca-K exchanger, Biochemistry 42 (2003) 543–552. [DOI] [PubMed] [Google Scholar]
- [66].Chu L, Greenstein JL, Winslow RL, Modeling Na+-Ca2+ exchange in the heart: Allosteric activation, spatial localization, sparks and excitation-contraction coupling, J. Mol. Cell. Cardiol. 99 (2016) 174–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


