Abstract
Severe liver disease is the 12th leading cause of death in the USA, with organ transplantation often being the only viable option for treatment. However, due to the shortage of viable donor livers, it is estimated that over 1200 patients died in 2015 while waiting for liver transplantation. This highlights the need for alternative sources of viable organs. In this study, we describe a method that provides the groundwork for the development of functional liver grafts. The approach described here is for removal of cells from intact livers and subsequently repopulating them with functional liver cells. Briefly, rat livers are harvested and subjected to a series of perfusion decellularization steps using an anionic detergent such that an intact decellularized liver matrix (DLM) scaffold with preserved vascular architecture is obtained. Further, we describe methods to recellularize DLM scaffolds with adult primary hepatocytes, creating a liver graft that exhibits hepatic functions in vitro.
Keywords: Decellularization, Liver tissue engineering, Recellularization, Whole liver scaffold
1. Introduction
Whole liver engineering is a new approach to tissue engineering that has gained attention over the past few years [1]. It utilizes whole organs that are devoid of cells, thus creating scaffolds that retain structural, mechanical, and chemical attributes of the native tissue, and serving as an ideal material for development of engineered tissues. One major advantage of this approach is that it takes advantage of the natural tissue matrix by preserving the intact microvascular structure, which facilitates connection of the tissue directly with patient vasculature upon transplantation, allowing immediate circulation and instant delivery of nutrients and oxygen. This approach is ideal for engineering transplantable liver grafts where constant perfusion is required for nutrient and oxygen delivery and waste removal to maintain highly metabolically active liver cells. In addition, the well-preserved extracellular matrix (ECM) enhances the cell attachment and reorganization [2].
Perfusion decellularization is the first step in whole-organ engineering that aims to eliminate resident cells and cellular components from the target organ while retaining the native composition and structure of extracellular matrix. The next step is recellularization which is repopulating the decellularized organ with the cells of the native organ. This approach has been successfully applied for tissues and organs such as trachea [3], heart [4], lung [5,6], kidney [7], urinary bladder [8], and liver [9, 10]. Here, we describe decellularization, recellularization, and characterization techniques for liver engineering using rat livers as the model system. We also describe a nondestructive monitoring method during the decellu-larization that ensures the consistency of the scaffold’s quality, including the extent of cell removal and the preservation of extracellular matrix components, without disrupting the scaffold’s integrity using computed tomography and perfusate analysis. We recently reported that there is a strong correlation between DNA removal and Hounsfield unit (HU) where HU is a quantitative scale unit used in computed tomography for describing radiodensity. The methods described here are the first steps of creating transplantable liver grafts and can be scaled up to human size liver grafts. The recellularized liver grafts can be used as alternatives to donor livers for transplantation or can serve as a drug testing platform superior to in vitro hepatocyte cultures.
2. Materials
Solutions are prepared with ultrapure water which has a resistivity of 18 MΩ-cm at 25 °C. Reagents are subsequently stored at room temperature (unless indicated otherwise).
2.1. Liver Harvest
2.1.1. Animals
150–200 g Female Lewis rats (Charles River Laboratories, Wilmington, MA).
2.1.2. Chemicals and Surgical Tools
Phosphate buffered saline (PBS).
70% (v/v) Isopropanol.
Gas tank 100% O2.
Cotton tips (2×).
Gauze (2×) (Kendall Versalon 4 ply 2″ 1×2″).
3.0 and 5.0 polypropylene sutures (Ethicon).
18G catheter (Terumo Surflash IV catheters, Somerset, NJ).
Cell culture dish (100 mm × 20 mm, Corning, NY).
Tape (0.5″ wide Transpore 3 M).
Heparin (Hospira, Inc. Lake Forest, IL).
Large scissors (1×) (Aesculap, Roboz).
Micro dissecting rat tooth forceps (1×) (Aesculap, Roboz).
Large hemostats (1×) (Aesculap, Roboz).
Fine forceps, straight and curved (Aesculap, Roboz).
Curved micro forceps, stainless steel (Aesculap, Roboz).
Small blunt nosed dissecting scissors (1×) (Aesculap, Roboz).
30 ml syringe.
Isoflurane vaporizer (Colonial Medical Supply, Windham, NH).
2.2. Perfusion Decellularization
2.2.1. Reagents
Phosphate buffered saline (PBS).
Sodium dodecyl sulfate (SDS) (Sigma-Aldrich), 0.01% (w/v) in dH2O.
SDS, 0.1% (w/v) in dH2O.
SDS, 0.2% (w/v) in dH2O.
SDS, 0.5% (w/v) in dH2O.
Triton X-100 (Sigma-Aldrich), 1% (w/v) in dH2O.
Peracetic acid (Sigma-Aldrich).
Gentamicin (Sigma-Aldrich).
Amphotericin B (Sigma-Aldrich).
Distilled water.
Ethanol (200 proof).
2.2.2. Perfusion System Set-Up
L/S® standard digital drive (Cole-Parmer Cat # EW-07523–80).
Masterflex® standard pump head for L/S 15 (Cole-Parmer Cat # EW-07015–21).
Masterflex® platinum-cured silicone tubing, L/S® 14 (Cole-Parmer Cat # EW-96410–14).
8–1 containers.
Bubble trap (Radnoti part# 130149).
10-cm petri dishes.
30 cc syringes.
2.3. Characterization of Decellularized Liver Matrix
2.3.1. Computed Tomography (CT) Scanning of Decellularized Livers
Clear ziplock bag.
CereTom CT scanner (Neurologica, Danvers, MA) with Cere-Tom CT Scanner software.
AMIDE open source software (http://amide.sourceforge.net).
2.3.2. Immunoassay and Biochemical Assays
Total Protein Assay
Scienceware® Liquid Nitrogen Cooled Mini Mortar & Pestle Set (Thomas Scientific, Swedesboro, NJ).
RIPA Lysis and Extraction Buffer (Thermo Fisher Scientific, Waltham, MA).
Pierce BCA Assay Kit (Life Technologies, Grand Island, NY).
Western Blot
PBS.
JLA-20 antibody (Developmental Studies Hybridoma Bank, Iowa City, IA).
Laemmli buffer (BIO-RAD, Hercules, CA).
β-mercaptoethanol (Sigma-Aldrich, St. Louis, MO).
Tris-buffered saline, 0.1% Tween 20 (TBST).
Nitrocellulose membrane (BIO-RAD, Hercules, CA).
Protein ladder (Li-Cor, Lincoln, NE).
Blocking buffer (Li-Cor, Lincoln, NE).
Transfer buffer: 20 mM Tris, 190 mM glycine, 20% methanol.
Running buffer: 4% SDS, 10% 2-mercaptoethanol, 20% glycerol, 0.004% bromophenol blue, 0.125 M Tris-HCl.
Odyssey CLx (Li-Cor, Lincoln, NE).
DNA Assay
Purelink Genomic DNA kit (Life Technologies, Grand Island, NY).
Picogreen Assay kit (Life Technologies, Grand Island, NY).
Glycosaminoglycan (GAG) Assay
Lyophilizer (FTS SYSTEMS, Stone Ridge, NY).
6 N hydrochloric acid.
0.5 mg/ml chondroitin sulfate A (Sigma-Aldrich, St. Louis, MO).
Dimethylene blue (Sigma-Aldrich, St. Louis, MO).
Microplate spectrophotometer: Benchmark Plus (BIO-RAD, Hercules, CA).
Collagen Assay
Quickzyme Total Collagen assay kit (Cedarlane Labs, Burlington, NC).
2.3.3. Corrosion Casting
Batson’s 17 anatomic corrosion kit (Polysciences Inc. Cat # 07349–1).
1 N potassium hydroxide.
2.3.4. Histological Analyses
10% formalin.
70% ethanol.
2.3.5. Perfusate Analysis
Beakers (600 ml or 1000 ml).
Conical tubes (50 ml).
2.4. Perfusion Recellularization
2.4.1. Cells
Adult primary rat hepatocytes isolated from 150 to 200 g female Lewis rats using the two-step perfusion protocol as described by Dunn et al. [11] and used at a density of 10 million cells per ml.
2.4.2. Reagents
Seeding medium: High glucose Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum (Peak Serum), 100 U/ml penicillin, and 100 μg/ml streptomycin (Invitrogen).
Culture medium: High glucose Dulbecco’s modified Eagle’s medium with 10% fetal bovine serum (Peak Serum), 7 ng/ml glucagon (Bedford Laboratories), 7.5 μg/ml hydrocortisone (Pfizer), 0.5 U/ml insulin (Novolin), 20 ng/ml Epidermal Growth Factor (Invitrogen), 200 U/ml penicillin, and 200 μg/ml streptomycin (Invitrogen).
2.4.3. Engineering Parts
Cell Seeding Perfusion Setup
Masterflex® standard pump head for L/S® 15 (Cole-Parmer Cat # EW-07015–21).
Masterflex® platinum-cured silicone tubing, L/S® 14 (Cole-Parmer Cat # EW-96410–14).
Bubble trap (Radnoti part# 130149).
Three-way stopcocks with luer connections (Cole-Parmer cat # EW-30600–02).
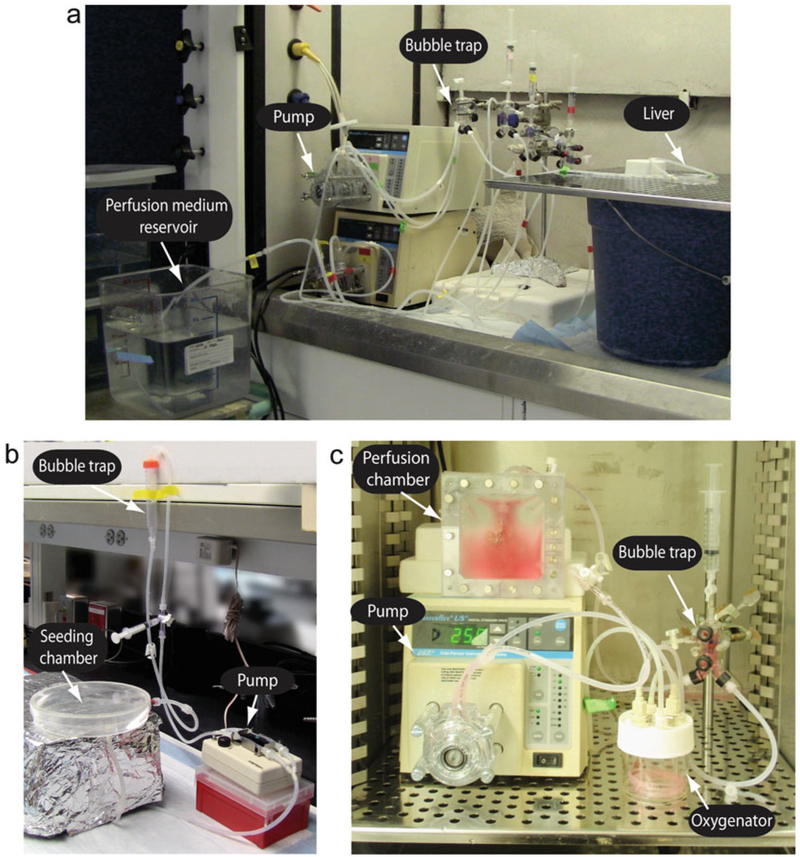
15-cm petri dish equipped with inflow and an outflow tubing (Fig. 1b).
Fig. 1.
(a) Perfusion decellularization system. The perfusion decellularization system is composed of a perfusion medium reservoir, a pump, and a bubble trap. Decellularization medium is pumped from the reservoir through the bubble trap and perfused into the liver graft which is placed in a 10-cm petri dish. (b) Cell seeding perfusion set-up. A 15-cm petri dish is served as the seeding chamber. The decellularized liver graft (not shown in the figure) is placed in the petri dish and connected to the system for seeding. A peristaltic pump facilitates seeding media flow from the chamber through a bubble trap into the graft. Cells are injected through a three-way valve prior to the bubble trap and thus perfused in the graft. (c) Perfusion culture system. The recellularized liver graft is placed in a perfusion chamber which is composed of two silicon sheets sealed by a frame and metallic screws. A three-way valve is added respectively at the inlet and the outlet of the perfusion chamber. The culture medium is circulated in the system from the chamber to an oxygenator and a bubble trap back to the liver. A pump is employed between the chamber and the oxygenator to facilitate the flow
In-Vitro Culture Perfusion Setup
L/S® standard digital drive (Cole-Parmer Cat # EW-07523–80).
Masterflex® standard pump head for L/S® 15 (Cole-Parmer Cat # EW-07015–21).
Masterflex® platinum-cured silicone tubing, L/S® 14 (Cole-Parmer Cat # EW-96410–14).
Oxygenator (Radnoti part# 130144).
Bubble trap (Radnoti part# 130149).
- Perfusion chamber (see Note 1):
- Two square polycarbonate frames (made to order).
- Two silicone sheets (McMaster-Carr Cat # 87315K62).
- Masterflex® platinum-cured silicone tubing, L/S® 14 (Cole-Parmer cat # EW-96410–14).
- 20G catheter.
- 1-way, 3-way/4-way stopcocks with luer connections (Cole-Parmer cat # EW-30600–01, # EW-30600–02, # EW-30600–03).
Syringes (5-cc, 30-cc)
95% Oxygen, 5% Carbon Dioxide Gas Tank
3. Methods
3.1. Hepatectomy
Ensure animals are maintained in accordance with National Research Council guidelines. Experimental protocols must be approved by relevant Committees on Research Animal Care.
Induce anesthesia in an induction chamber with isoflurane (see Note 2).
Put the animal on surgical table in supine position with continuous administration of isoflurane through nose cone (see Note 3).
Shave the animal’s thorax and abdomen ventrally.
Place animal in supine position on surgical table, and secure four limbs firmly on the table with tape.
Disinfect the shaved area with 70% ethanol. Check the leg reflex by pinching the foot of the animal. If there’s no reflex, proceed the surgery.
Make an incision at xiphoid process. Make large transverse abdominal incision with large scissors and rat tooth forceps.
Clamp the xiphoid process with straight hemostats, retract, exposing abdominal contents. Tape the hemostats’ handle.
The falciform ligament will be visible immediately inferior and deep to the xiphoid process. Wet two cotton tips in the solution reservoir. With a wet cotton bud and small scissors, divide this ligament without severing the superior vena cava or the diaphragm.
Inject 20 U heparin through the diaphragm into the heart to prevent blood coagulation.
Cut the ligaments between each lobe (see Note 4).
Free inferior caudate lobe (ICL) by gently rolling up the stomach and making an incision to the lesser omentum along the junction of the lobe and the posterior side of stomach. Using wet cotton tipped swabs and a gentle rolling motion, move the lobe superiorly to the stomach through the hole produced by these incisions (see Note 5).
Reveal the portal vein (PV) by tucking the stomach around the ribcage to the right and shifting the spleen, and the intestines downwards to the right side out of the body of the animal.
Identify the PV and the bile duct. Put a damped gauze on the small intestine and pull lightly aside to provide downward tension on the PV.
Prepare two sutures around the PV for cannulation: The proximal position is prepared superior to a small branch extending from the right side ofthe PV. With a wet cotton bud in the left hand and a pair of curved forceps in the right, create a window on each side of the aforementioned areas (see Note 6). Prepare the distal suture by repeating the same procedure in a position inferior to the branch of the PV. Insert the curved forceps underlying the PV with a clockwise rolling motion. Feed the forceps with the 3.0 or 5.0 sutures and gently pull the sutures in a counterclockwise rolling motion through the proximal and distal windows that were just created. Tie a loose ligature with each.
Insert an 18-gauge catheter. Retract the needle and simultaneously advance the catheter. Advance the catheter until it lies between the two sutures (see Note 7).
Connect the catheter with the 30 ml syringe filled with PBS.
Tie the proximal suture first by tightening the knot and securing with two more. Repeat with the distal suture.
Cut the inferior vena cava (IVC). Perfuse the liver by slowly pushing 20 ml PBS to clear the blood within the liver (see Note 8).
Excise the liver from the body cavity by ligating the superior hepatic vena cava (SHVC), bile duct, and the distal part of the PV, and freeing the ligaments around the liver (see Note 9).
Remove the liver into the petri dish containing PBS, and flush the liver with 10 ml PBS that left in the syringe. Place the liver at −80 °C prior to decellularization.
3.2. Perfusion Decellularization
Thaw harvested rat livers before perfusion while setting up the perfusion system (Fig. 1a).
Fill the perfusion system with PBS and keep it running for 10 min. Fill a 10-cm petri dish with PBS.
Reduce the flow rate to 1.2 ml/min.
Carefully transfer the thawed liver to the PBS filled 10-cm petri dish.
Remove any air bubbles in the tubing and the catheters. Make sure there is no air inside the catheters (see Note 10).
Carefully connect the liver to the perfusion system through the portal vein catheter while PBS is flowing through the tubing.
Record date and time. Continue with PBS perfusion overnight.
Stop the flow and transfer the tubing from PBS containing reservoir to 0.01% SDS containing reservoir. Resume the flow and perfuse with 0.01% SDS for 5 min.
Stop the flow and transfer the tubing from 0.01% SDS containing reservoir to PBS containing reservoir. Resume the flow and perfuse with PBS for 1 h.
Repeat SDS (0.01%) and PBS perfusion steps (steps 9 and 10) three more times increasing SDS perfusion time to 10,15, and 20 min.
Continue perfusion with 0.01% SDS for 24 h.
Start perfusion with 0.1% SDS for 24 h (see Note 11).
Perfuse with 0.2% SDS for 3 h. Go to step 16 if the effluent is clear and the liver is transparent (see Note 12).
Perfuse with 0.5% SDS for 3 h. Go to step 16 if the effluent is clear and the liver is transparent (see Note 13).
Perfuse with dH2O for 15 min.
Perfuse with 1% Triton X-100 for 30 min.
Perfuse with PBS for 1–2 h.
Store in a clean and sealed petri dish soaked in PBS at 4 °C until ready to use (see Note 14).
Sterilization: (a) Using a 30-ml syringe, flush the decellularized liver with sterile PBS containing 0.1% (v/v) peracetic acid and 4% (v/v) ethanol and incubate for 3 h at 4 °C in a sealed petri dish. (b) Transfer the liver into a new petri dish that contain sterile PBS and flush the decellularized liver twice with sterile PBS. (c) Wash with sterile PBS containing 2% penicillinstreptomycin, 10 μg/ml gentamicin, and 2.5 μg/ml amphotericin B. Store the decellularized liver in the same solution at 4 °C until ready to use for recellularization experiments.
3.3. Characterization of Decellularized Liver Matrix
3.3.1. Computed Tomography (CT) Scanning of Decellularized Livers
Calibrate CT scanner before each application.
Place liver in a sterile ziplock bag, take measurement of liver’s dimensions to ensure full scan.
Perform non-contrast CT scan through the decellularization process at 0 h, at the end of PBS/0.01% SDS alteration stage, 0.01% SDS perfusion stage, 0.1% SDS perfusion stage, and at the end of decellularization.
Use CereTom® CT scanner software to produce and construct Dicom 3 compliant images of the scans.
Determine Hounsfield units using AMIDE open source software.
3.3.2. Perfusate Collection (for Nondestructive Decellularization Analysis Method)
When setting up the decellularization, place a 600 ml or 1000 ml beaker under each liver graft ensuring perfusate is collected in the beaker.
Collect 50 or 100 ml of perfusate at appropriate time points for analysis.
Store at −80 °C until analysis is performed.
3.3.3. Immunoassay and Biochemical Assays
Whole liver weights prior to decellularization and after decellularization should be noted. Biopsies and perfusate samples for biochemical assays should be taken prior to starting decellularization [e.g., t = 0, 24, 48, 66 and 76 h (end)].
Total Protein Assay
Crush the tissue using the mortar and pestle set (see Note 15).
Once the tissue is crushed, add RIPA buffer and digest on a shaker at 4 °C for 2.5 h. Pipet the tissue digestion up and down several times after 2 h digestion.
Place the tissue digestion in centrifuge, spin at 3000 × g for 20 min and then 4000 × g for another 10 min.
Collected 1 ml of supernatant, ensuring no solid was left in the sample.
Proceed the Pierce BCA protein assay by following the manufacturer’s recommendations.
Western Blot
After determining the protein concentration with Pierce BCA assay, dilute the samples to the same concentration in PBS.
Add 50 μl of β-mercaptoethanol to 950 μl Laemmli, then take diluted samples and add 1:1 ratio of Laemmli buffer.
Boil mixed sample in dH2O for 5 min, centrifuge for 1 min at 13,000 × g.
Load sample into wells of the gel along with protein ladder.
Set-up the gel running system and run the samples on gel (see Note 16).
Transfer the protein bars from gel to film (see Note 17).
Incubate with primary antibody, followed by secondary antibody incubation (see Note 18).
Develop the film with Odyssey scanner (see Note 19).
DNA Assay (See Note 20)
Weigh 20 mg liver biopsy sample.
Follow the manufacturer’s instructions of Purelink Genomic DNA kit for DNA extraction and purification.
Use Picogreen Assay kit to determine the extracted DNA concentration.
GAG Assay
The GAG content of the DLM is determined according to Farndale et al. [12] (see Note 21).
Cut DLM samples into 100 mg small pieces.
Lyophilize the DLM samples.
Hydrolyze each sample in 0.25 –0.5 ml of 6 N HCl at 95 °C for 20 h.
Cool the samples to room temperature.
Mix 10 μl sample and 250 μl dimethylene blue and immediately measure the absorbance at 525 nm.
Collagen Assay
Quantify collagen content by applying Quickzyme Total Collagen assay kit. Follow the manufacturer’s recommendations.
3.3.4. Corrosion Casting
During the surgery, prepare the graft with cannulation of the IVC, portal vein, and common bile duct.
After decellularization, follow the manufacturer’s recommendations to proceed corrosion cast.
Inject 4 ml polymer mixture through the cannula.
Place the DLM in an ice bath for 2–3 h.
Macerate the tissue in 1 N KOH solution for 2 h at 50 °C or 24 h at room temperature.
3.3.5. Histological Analysis
Fix the DLM by immersing in 10% formalin for 24 h.
Transfer the DLM to 70% ethanol.
Proceed further histology steps by embedding the tissue in paraffin, sectioning, and staining with hematoxylin and eosin.
3.4. Perfusion Recellularization
Set-up the seeding perfusion system that is equipped with a peristaltic pump, bubble trap, and a perfusion chamber (Fig. 1b). Fill the perfusion system by pipetting 200 ml of seeding medium into the perfusion chamber.
Place the DLM into the perfusion chamber and connect it to the perfusion system through the portal vein cannula while the pump is running at 5 ml/min to avoid formation of any air bubbles. Allow the medium to perfuse the DLM for 30 min.
Stop the flow in the perfusion system and slowly inject 10–30 million hepatocytes through a 3-way stopcock situated before the bubble trap.
Restart the flow at 10 ml/min and recirculate the medium for 10 min.
Stop the flow of medium again, inject an additional 10–30 million hepatocytes. Resume the flow again to 10 ml/min. Repeat this procedure two more times until a total of 40–120 million hepatocytes have been introduced into the DLM.
Once addition of cells into the DLM is complete, cut the portal vein catheter at 45° angle to free the recellularized liver graft and transfer the graft to the in vitro perfusion chamber (see Note 22).
Aseptically, set-up sterile perfusion system (Fig. 1c).
Fill the perfusion system by pipetting 25 ml of culture media into the perfusion chamber. Start the pump at 10 ml/min and remove any air from the tubing.
Place the recellularized liver graft into the perfusion chamber and connect the DLM to the perfusion system through the portal vein cannula while the pump is running at 5 ml/min to avoid introducing any air bubbles into the recellularized liver graft.
Close the perfusion chamber and seal tightly to avoid any leakage during the culture.
Fill the chamber with additional 15–55 ml culture media using a sterile syringe through the 3-way/4-way stopcock till the total culture media reaches 40–80 ml.
Incubate the perfusion system at 37°C with 10% CO2 and keep the perfusion flow rate at 8–10 ml/min.
Connect the oxygenator to a 95% O2 and 5% CO2 gas mixture tank and set the gas flow rate to 0.5 l/min.
Continue the culture with daily changes of culture medium (see Note 23).
4. Notes
The in vitro perfusion chamber consists of two silicone sheets pressed together by plastic frames. The chamber has one inlet and one outlet port, each equipped with size 14 silicone tubing and 3-way/4-way stopcocks for medium change and sampling purposes. At the inlet, a 20G catheter that has been cut at a 45° angle at 1/2 in. length is placed to connect the recellularized graft.
Turn on the 100% O2 tank and make sure the regulator is at 50–55 psi. Turn up the flow rate to 0.8–1 LPM. Turn on the vaporizer to 5. When the breathing of the animal is slow and regular, and there is no muscle tone, turn off the vaporizer and flush the chamber by pressing the oxygen flush valve button for 10 s to ensure a safe working environment.
Before placing the animal in the nose cone, open the valve to the nose cone circuit, close the valve to the chamber circuit, and turn on the vaporizer to 5 again.
With two wet cotton buds move the medium lobe and the left lateral lobe (LLL) up in a gentle rolling motion against the diaphragm. Cut the hepatogastric ligament between LLL and superior caudate lobe (SCL). Roll SCL up against LLL and cut the ligament posterior to the SCL.
The ICL lies posterior to the stomach. This lobe is nestled in the lesser curvature of the body of the stomach and inferior to the esophagogastric junction. It is anchored to the stomach and the spleen by the lesser omentum, which must be severed to free this lobe. Be careful not to sever any of the vasculature of the lesser omentum.
To create windows, use a pair of forceps making initial incisions and conduct an open-close motion along the vessel with the forceps. When necessary, hold a cotton bud on the left to create a surface where the forceps can apply pressure on.
Grasp the 18G catheter in the right hand with thumb and middle finger. Create traction with left hand by gently pressing the most distal end of PV. Insert the cannula from as distal to the sutures as possible. Retract the needle by pushing the retraction button with index finger. Blood should flow out. The beveled edge of the catheter should lie directly superior to the proximal suture.
Keep the syringe at the same position by gently taping it.
Roll the anterior liver lobes back to the physiological position. Gently retract the liver away from the diaphragm and identify the connection with the SHVC. Sever the SHVC and continue to carefully sever all the ligaments that anchor the liver to the body and surrounding tissue. Cut the diaphragm. Cut the ligaments connecting the liver and the esophagus as well as the diaphragm. Grasp the bile duct and sever. Grasp the portal vein and cannula and cut away from the vascular system without dislodging from the liver. Using the cotton tips, lift the inferior right lobe (IRL) that lies directly superior to the IVC. Completely sever the IVC from it. Further lift to sever the ligaments that anchor the liver deep to this lobe.
If there’s any bubble in the catheter, fill the catheter with PBS using a syringe and a needle.
Alternative: in the shortened protocol [13], instead of perfusion for 24 h, perfuse 4 h with 0.1% SDS.
Alternative: in the shortened protocol [13], instead of perfusion for 3 h, perfuse 1 h with 0.2% SDS.
Alternative: in the shortened protocol [13], instead of perfusion for 3 h, perfuse 1 h with 0.5% SDS.
Optional: depending on the experimental purpose and utility, ligate and resect all lobes except for the median lobe.
To crush the tissue: fill the reservoir underneath the mortar with enough liquid nitrogen. The darker area in the metal cup is the coolest point in the cup. Crush the tissue in this exact area to prevent tissue from sticking to the cup. Be careful with the first initial crush to prevent loss of tissue. Hold spatula and sample tube in the liquid nitrogen to ensure the low temperature. Scrape crushed tissue and tap into sample tube which is on dry ice. Rinse spatula, pestle, and metal cup with water, repeat for next sample.
Put voltage lid on, making sure to match black to black, red to red. Bubbles should be observed once the power supply is turned on. Fill entire tank with running buffer and make sure buffer fills space between empty glass plates. Run at 200 Vuntil marker reaches bottom of plate.
Make 1 × transfer buffer and add methanol, place in −80 °C to chill before use. To set-up the transfer system: Remove gel plate with green spatula. Soak sponge in transfer buffer, put on black side of cassette. Soak filter paper: place on sponge then place gel on top. Soak membrane: cut to size first, soak, lay on top of gel. Soak another filter paper, place on membrane, and remove any bubbles. Soak another sponge. Close up cassette and place in chamber. Black side should face black side of chamber. Place chamber, add transfer buffer, magnetic stir bar, icepack, and then place the lid on top. Run for 90 min at 100 V.
Cut the film as needed. Indicate the reaction side. Wash the membrane with TBST for two times on orbital shaker. Block the membrane with blocking buffer for 1 h at room temperature on the shaker. Wash again with TBST after the blocking. Incubate with primary antibody (in this case, β-actin at 0.2 μg/ml) overnight at 4 °C on shaker. Wash three times with TBST after the primary antibody incubation. Apply secondary antibody at an appropriate concentration. Incubate at dark on shaker for 1 h. Wash the film with TBST for three times.
Wipe the surface of the scanner before reading. Lay the membrane facing down on the scanner. Determine the membrane size in the program per the ruler on the scanner. Start scanning.
- The quantification of DNA, GAG, and collagen content should be normalized accordingly. In our study, we weighed the sample, the whole liver weight of pre-and post-decellularization. The results were normalized to wet tissue weight and corrected by the ratio of post-to pre-decellularization whole liver weights at each time point. The normalization calculation is as follows:
Plot standard curve by applying serial dilution of 0.5 mg/ml chondroitin sulfate A in PBS. The preparation of standard samples is the same as the DLM samples.
Consequently, collect the perfusate into four 50-ml centrifuge tubes and centrifuge at 600 rpm for 10 min. Discard the supernatants and combine the pellets into a single tube. Determine the number of cells and their viability via trypan blue exclusion to establish the seeding efficiency.
In ideal situation, the culture can be kept for up to 10 days. Sample the medium daily for measurement of metabolites in the perfusate such as albumin, urea, and total bile acid. At the end of the culture period, sample the recellularized liver graft for molecular and histological analysis.
References
- 1.Badylak SF, Taylor D, Uygun K(2011) Whole-organ tissue engineering: decellularization and recellularization of three-dimensional matrix scaffolds. Annu Rev Biomed Eng 13:27–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gilbert TW, Sellaro TL, Badylak SF (2006) Decellularization of tissues and organs. Biomaterials 27:3675–3683 [DOI] [PubMed] [Google Scholar]
- 3.Macchiarini P, Jungebluth P, Go T et al. (2008) Clinical transplantation of a tissue-engineered airway. Lancet 372:2023–2030 [DOI] [PubMed] [Google Scholar]
- 4.Ott HC, Matthiesen TS, Goh S-K et al. (2008) Perfusion-decellularized matrix: using nature’s platform to engineer a bioartificial heart. Nat Med 14:213–221 [DOI] [PubMed] [Google Scholar]
- 5.Petersen TH, Calle EA, Zhao L et al. (2010) Tissue-engineered lungs for in vivo implantation. Science 329:538–541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ott HC, Clippinger B, Conrad C et al. (2010) Regeneration and orthotopic transplantation of a bioartificial lung. Nat Med 16:927–933 [DOI] [PubMed] [Google Scholar]
- 7.Sullivan DC, Mirmalek-Sani S-H, Deegan DB et al. (2012) Decellularization methods of porcine kidneys for whole organ engineering using a high-throughput system. Biomaterials 33:7756–7764 [DOI] [PubMed] [Google Scholar]
- 8.Bolland F, Korossis S, Wilshaw S-P et al. (2007) Development and characterisation of a fullthickness acellular porcine bladder matrix for tissue engineering. Biomaterials 28:1061–1070 [DOI] [PubMed] [Google Scholar]
- 9.Uygun BE, Soto-Gutierrez A, Yagi H et al. (2010) Organ reengineering through development of a transplantable recellularized liver graft using decellularized liver matrix. Nat Med 16:814–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Uygun BE, Izamis M-L, Jaramillo M et al. (2016) Discarded livers find a new life: engineered liver grafts using hepatocytes recovered from marginal livers. Artif Organs 13:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dunn JC, Tompkins RG, Yarmush ML (1991) Long-term in vitro function of adult hepato-cytes in a collagen sandwich configuration. Bio-technol Prog 7:237–245 [DOI] [PubMed] [Google Scholar]
- 12.Farndale RW, Buttle DJ, Barrett AJ (1986) Improved quantitation and discrimination of sulphated glycosaminoglycans by use of dimethylmethylene blue. Biochim Biophys Acta 883:173–177 [DOI] [PubMed] [Google Scholar]
- 13.Geerts S, Ozer S, Jaramillo M et al. (2016) Nondestructive methods for monitoring cell removal during rat liver decellularization. Tissue Eng Part C Meth 22:671–678 [DOI] [PMC free article] [PubMed] [Google Scholar]