Abstract
Small molecule inhibitors often only block a subset of the cellular functions of their protein targets. In many cases, how inhibiting only a portion of a multi-functional protein’s functions affects the state of the cell is not well understood. Therefore, tools that allow the systematic characterization of the cellular interactions that inhibitor-bound proteins make would be of great utility, especially for multi-functional proteins. Here, we describe a chemoproteomic strategy for interrogating the cellular localization and interactomes of inhibitor-bound kinases. By developing a set of orthogonal inhibitors that contain a trans-cyclooctene (TCO) click handle, we are able to enrich and characterize the proteins complexed to a drug-sensitized variant of the multi-domain kinase Src. We show that Src’s cellular interactions are highly influenced by the intermolecular accessibility of its regulatory domains, which can be allosterically modulated through its ATP-binding site. Furthermore, we find that the signaling status of the cell also has a large effect on Src’s interactome. Finally, we demonstrate that our TCO-conjugated probes can be used as a part of a proximity ligation assay to study Src’s localization and interactions in situ. Together, our chemoproteomic strategy represents a comprehensive method for studying the localization and interactomes of inhibitor-bound kinases and, potentially, other druggable protein targets.
Graphic abstract
(TOC) A Chemoproteomic Strategy for Profiling Inhibitor-Bound Kinase Complexes
Introduction
The phenotypic effects of pharmacologically inhibiting a protein target can significantly differ compared to pre-translational disruption of function.1–3 The slow onset of pre-translational disruption can often lead to compensatory changes within the cell. In addition, pre-translational disruption methods usually lead to the complete loss of a target protein from a cell, while most small molecules do not lead to degradation of their inhibited protein targets. As most small molecule inhibitors block only one or two interaction sites, additional functional sites are left intact and are capable of participating in various cellular processes. In many cases, how an inhibited protein target influences cellular function is not well understood.
Kinases are of particular interest in terms of understanding how inhibitor binding affects cellular target function because almost 50% of the ~540 kinases in humans contain separate domains outside of the catalytic domain.4 Even kinases composed of a single catalytic domain often contain multiple binding interaction sites that are distal to their active sites. These separate domains and catalytic domain binding sites participate in numerous interactions that can influence cellular signaling. Furthermore, for many kinases, binding interactions distal to their active sites mediate a number of phosphotransferase-independent functions.5–7 The abilities of inhibited kinases to participate in and to modulate phosphotransferase-independent functions are not well understood. Determining how inhibitor binding affects the interactions and phosphotransferase-independent functions of protein kinase is especially important because ATP-competitive inhibitors can allosterically modulate interaction surfaces and domains that are distal to the active site.8–10 Furthermore, because kinase inhibitors can differentially modulate distal interaction surfaces and binding domains depending on the ATP-binding site conformation that they stabilize, the interactomes and cellular localization of probe-bound kinases can vary greatly. Therefore, we sought to develop a methodology that would allow the interactions and cellular context of an inhibited protein kinase to be interrogated.
To develop a general strategy for co-enriching proteins that are non-covalently, and often weakly, associated with a probe-bound target, we required a mild and rapid bioorthogonal click reaction. Numerous chemical proteomics strategies, such as activity-based protein profiling,11–13 rely on the use of probes containing bioorthogonal click handles for target enrichment. The most dominant click reaction for this purpose is the copper-catalyzed alkyne-azide cycloaddition (CuAAC).14 The robust nature of the CuAAC reaction make it attractive for enriching low abundance targets. However, we were concerned that the high concentrations of copper used in the CuAAC reaction, and the copper-mediated protein oxidation that can occur,15–20 may result in protein complex dissociation. Strain promoted copper-free alkyne-azide cycloaddition (SPAAC)21 have been developed but we felt its moderate reaction rate would require prolonged incubation time for enrichment, which reduces the likelihood that intact protein complexes can be captured. For these reasons, we were attracted to the inverse electron demand Diels-Alder (iEDDA) reaction between a tetrazine (Tz) and trans-cyclooctene (TCO).12, 22–23 The iEDDA reaction has been demonstrated to be rapid (~105 M−1S−1), mild, and does not require additives or catalysts.
Here, we demonstrate that selective inhibitors conjugated to a TCO click handle can be used to interrogate the interactomes and cellular context of protein kinases (Figure 1). We show that the rapid, mild, and selective reaction between TCO-conjugated probes and tetrazines is ideal for interrogating the interactomes of kinases. Using a set of TCO-conjugated probes that stabilize different ATP-binding site conformations of Src kinase, we were able to obtain a comprehensive overview of how modulating the global conformation of this multi-domain protein influences its interactome. Furthermore, we were able to demonstrate the cellular context of Src profoundly influences its cellular interaction partners. Finally, we showed that our TCO-conjugated probes can be used as part of a proximity ligation assay (PLA) to study the in situ localization and interactions of Src. Our chemical proteomic method represents an inhibitor-focused strategy for studying the localization and interactomes of inhibitor-bound kinases and potentially other druggable protein targets.
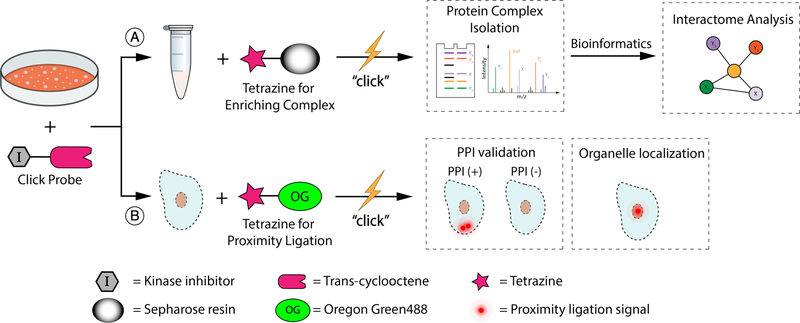
Figure 1.
A chemoproteomic platform for characterizing inhibitor-bound protein complexes. (A) Schematic for isolating and analyzing inhibitor-bound protein complexes. (B) Schematic for validating intracellular target interactions and visualizing inhibitor-bound target localization using a proximity ligation assay.
Results and Discussion
Probe Design and Rapid Enrichment of Probe-bound Target.
We pursued a strategy that involves derivatizing selective inhibitors with a TCO moiety that can be directly enriched with Tetrazine-conjugated sepharose beads (Tz-beads). To do this, we explored whether our inhibitor-bound protein complex enrichment strategy could be integrated with a chemical genetic strategy we recently developed called Cysteine Installation for Modulating Allostery and Targeted Inhibition of Kinases (CystIMATIK).8 CystIMATIK involves sensitizing specific kinases to electrophile-containing inhibitors, which are orthogonal to most wild-type (WT) kinases (Supporting Information. Figure S1), through the introduction of a cysteine residue into the N-terminal lobes of their catalytic domains.24–27 By generating a TCO-conjugated version of our orthogonal inhibitors, this strategy should allow multiple sensitized kinases to be selectively targeted.
Our first target was a TCO-conjugated analog of pyrrolo[2,3]pyrimidine-based CystIMATIK probe 1 (Figure 2A, left). Probe 1 contains a reversible covalent electrophile at the C-6 position and a 4-chlorophenyl moiety at the C-5 position of the pyrrolopyrimidine scaffold, which makes it highly selective for kinases that contain a threonine gatekeeper residue and a cysteine residue in the β2 strand of the N-terminal lobe that lines the roof of the ATP-binding cleft of the catalytic domain. Because only the C-terminal kinase domain of RSK2 has both of these residues at these positions, inhibitor 1 minimally interacts with most wild-type kinases (Supporting Information. Figure S1), including Src WT. Introduction of a cysteine residue into Src, which contains a threonine gatekeeper residue, sensitizes it to probe 1 (Supporting Information. Figure S2). Based on a co-crystal structure of 1 bound to a cysteine-containing Src mutant (Src V284C; PDB ID: 5SWH), we designed and generated 1-TCO (Figure 2A, right), which contains TCO conjugated to a position directed outside of the ATP-binding pocket (Figure 2B). Like 1, 1-TCO potently inhibits Src V284C (Figure 2C).
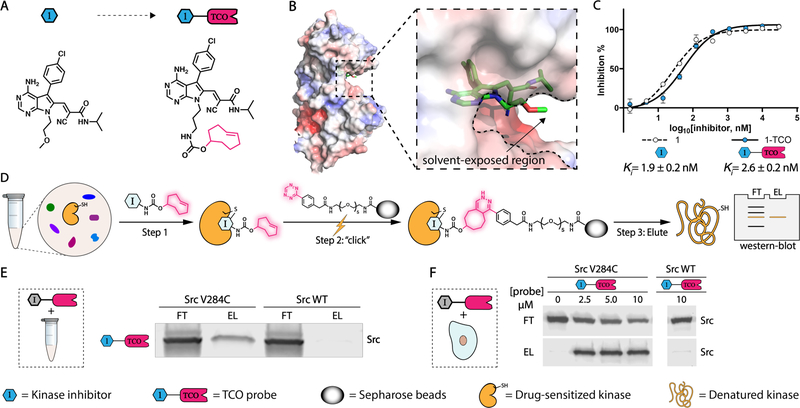
Figure 2.
Rapid and mild enrichment of protein kinases with TCO-conjugated probes. (A) Schematic for conversion of 1 (left) into 1-TCO (right). (B) Co-crystal structure of 1 bound to Src V284C (PDB ID: 5SWH). (C) Inhibition curves and Ki values of 1 and 1-TCO against recombinant Src V284C activity. Values shown are mean ± SEM, n = 3. (D) Schematic representation of kinase enrichment with TCO-conjugated probes. (E) 1-TCO selectively enriches a drug-sensitized Src mutant (Src V284C) from cell lysates. HEK293 cell lysates containing either Src V284C or Src WT were treated with 1-TCO (5 μM), incubated with tetrazine-conjugated sepharose beads (Tz-beads), and captured proteins were eluted under reducing and denaturing conditions. A Src immunoblot of the flow through (FT) and elution (EL) are shown. (F) 1-TCO selectively enriches Src V284C from cells. HEK293 cells expressing either Src V284C or Src WT were treated with the indicated concentrations of 1-TCO, lysed, and then incubated with Tz-beads. Captured proteins were eluted under reducing and denaturing conditions. A Src immunoblot of the flow through (FT) and elution (EL) are shown.
Next, we investigated whether our 1-TCO probe can be used to directly enrich Src V284C from complex proteomes. To do this, we incubated 1-TCO with HEK293T cell lysate containing either recombinant drug-sensitized Src V284C or Src WT constructs, followed by direct enrichment with Tz-beads (Figure 2D). Western blot analysis of bound proteins eluted under reducing and denaturing conditions demonstrated that a significant percentage of Src V284C was captured under these conditions, while Src WT was not (Figure 2E). We found that 1-TCO was also capable of efficiently enriching Src V284C from live mammalian cells. Treatment of HEK293 cells stably expressing Src V284C with 1-TCO, followed by incubation with Tz-beads immediately after cell lysis led to significant enrichment of Src V284C (Figure 2F). Thus, the click reaction between TCO and tetrazine is capable of rapidly and efficiently enriching probe-bound targets.
We also determined whether additional kinases can be sensitized to CystIMATIK probes 1 and 1-TCO (Supporting Information, Figure S2). We found that EphA2 (EphA2 V627C), JNK2 (JNK2 V40C/M108T), and PAK1 (PAK1 V286C/M346T) can be sensitized to probe 1 by introducing Cys at the residue homologous to V284 of Src and, if required, threonine at the gatekeeper position (Supporting Information, Figure S3A). As expected, WT JNK2, EphA2 and PAK1 were insensitive to 1. Like Src V284C, we observed that 1-TCO was capable of selectively enriching diverse drug-sensitized kinases from lysates (Supporting Information. Figure S3B, S3C) or live mammalian cells (Supporting Information. Figure S3D). Thus, 1-TCO should be of utility for the chemical proteomic interrogation of a number of kinases beyond Src.
Conformation-Selective TCO Conjugates.
We and others have demonstrated that there is an allosteric, bi-directional relationship between the ATP-binding sites and regulatory domains of protein kinases.5, 28–31 Intramolecular engagement of kinase regulatory domains can stabilize ATP-binding site conformations that either inhibit or enhance phosphotransferase activity. Conversely, small molecule inhibitors that stabilize different ATP-binding site conformations can divergently modulate the regulatory domains of multi-domain protein kinases, which can greatly influence intracellular protein-protein interactions.29 To facilitate studies of how ATP-binding site conformation and regulatory domain engagement influence the intra-cellular functions of kinases, we developed analogs of CystIMATIK probe 1 that stabilize different ATP-binding site conformations. Inhibitor 2 stabilizes an ATP-binding site conformation that involves displacement of a catalytically important helix–helix αC–in the N-terminal lobe of the catalytic domain from an active position (Figure 3A, top left). Stabilizing this inactive conformation, which is often referred to as the helix αC-out conformation, with ATP-competitive inhibitors leads to enhanced intramolecular engagement of Src’s regulatory domains and a closed, autoinhibited global conformation. CystIMATIK probe 3 stabilizes an ATP-binding site conformation that involves flipping of the DFG-motif in the activation loop of the catalytic domain to an inactive conformation–often referred to as the DFG-out conformation (Figure 3A, bottom left). We have demonstrated that the stabilization of the DFG-out conformation of Src leads to intramolecular disengagement of its regulatory domains and an open global conformation. TCO-conjugated versions of 2 and 3–2-TCO and 3-TCO (Figure 3A)–were generated in an analogous manner as 1-TCO.
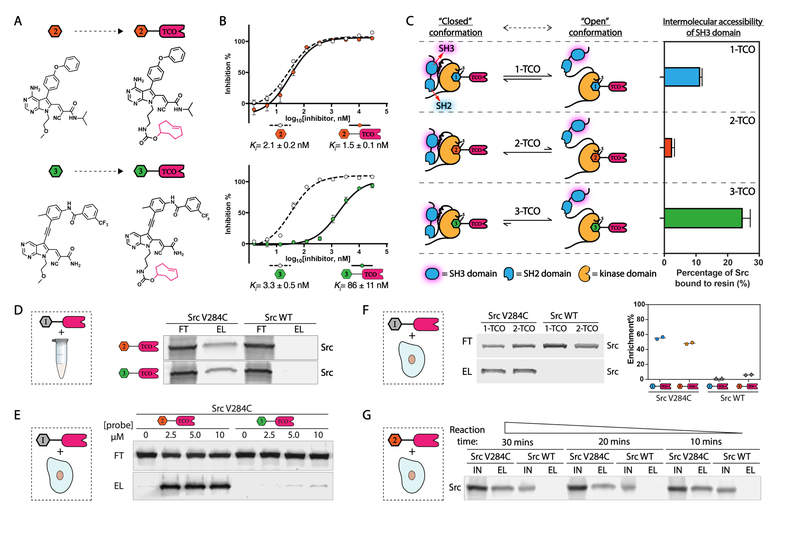
Figure 3.
Conformation-selective versions of TCO-conjugated probes. (A) Schematic for conversion of 2 and 3 (left) into 2-TCO and 3-TCO (right), respectively. (B) Inhibition curves and Ki values of 2, 2-TCO, 3, and 3-TCO against recombinant Src V284C activity. Values shown are mean ± SEM, n = 3. (C) 1-TCO, 2-TCO and 3-TCO promote expected levels of intermolecular SH3 domain accessibility of Src V284C. Schematic of how conformation-selective probes are predicted to modulate the intramolecular regulatory interactions and global confomrational state of Src (left). Percentage retained Src in an SH3 pulldown assay with recombinant Src V284C treated with 1-TCO, 2-TCO or 3-TCO (right). Values shown are mean ± SEM, n = 3. (D) 2-TCO and 3-TCO selectively enrich Src V284C from cell lysates. Cell lysates containing either Src V284C or Src WT were treated with 2-TCO or 3-TCO (5 μM), incubated with Tz-beads, and captured proteins were eluted under reducing and denaturing conditions. A Src immunoblot of the flow through (FT) and elution (EL) are shown. (E) 2-TCO–but not 3-TCO–selectively enriches Src V284C from cells. HEK293 cells expressing either Src V284C or Src WT were treated with the indicated concentrations of 2-TCO or 3-TCO, lysed, and then incubated with Tz-beads. Captured proteins were eluted under reducing and denaturing conditions. A Src immunoblot of the flow through (FT) and elution (EL) are shown. (F) 1-TCO and 2-TCO selectively enrich Src V284C from HeLa cells. HeLa cells expressing either Src V284C or Src WT were treated with 1-TCO (5 μM) or 2-TCO (2.5 μM), lysed, and then incubated with Tz-beads. Captured proteins were eluted under reducing and denaturing conditions. A Src immunoblot (left) and qunatification (right) of the flow through (FT) and elution (EL) are shown. (G) Kinetics of probe-bound target capture with Tz-beads. HeLa cells expressing either Src V284C or Src WT were treated with 2-TCO (2.5 μM), lysed, and then incubated with Tz-beads at 4 °C for the reaction time indicated. Immunoblots of the total lysate input (IN) and eluted proteins (EL) are shown.
We tested 2-TCO and 3-TCO for the inhibition of Src V284C and found that both probes potently inhibited this drug-sensitized kinase (Figure 3B), though conjugation of probe 3 to TCO led to a significant decrease in potency. Next, we validated that each TCO-conjugated probe biochemically modulated intramolecular SH3 domain engagement like their underivatized counterparts by performing pulldown assays of probe-bound Src V284C complexes with an immobilized SH3 domain ligand (Figure 3C). As expected, we found that the SH3 domain of the Src V284C/2-TCO complex was largely inaccessible to intermolecular ligand engagement, which is consistent with 2-TCO promoting a closed, autoinhibited global conformation of Src. In contrast, the Src V284C/3-TCO complex was efficiently pulled down by the immobilized SH3 domain ligand, which is consistent with 3-TCO promoting a regulatory domain disengaged conformation of Src. Finally, we observed an intermediate amount of pulldown of the Src V284C/1-TCO complex, which is consistent with the pharmacophore of 1 minimally influencing the ATP-binding site conformation of Src, resulting in a moderately open global conformation.8, 29, 31
Next, we validated that 2-TCO and 3-TCO were also capable of enriching Src V284C from complex proteomes. Like 1-TCO, 2-TCO and 3-TCO significantly enriched recombinant Src V284C but not Src WT from a HEK293 lysate (Figure 3D). Treatment of HEK293 cells expressing Src V284C with 2-TCO followed by lysis and incubation with Tz-beads led to efficient Src V284C enrichment. Unfortunately, very little Src V284C was captured with Tz-beads following lysis of 3-TCO-treated HEK293 cells expressing this mutant. The low level of captured Src V284C observed with 3-TCO is likely due to its poor cell permeability, as this probe worked efficiently in lysates (Figure 3D,E).
Next, we investigated whether our conformation-selective CystIMATIK probes are compatible with the diverse set of kinases that can be sensitized to 1 and 1-TCO. Like 1, we found that probes 2 and 3 inhibited drug-sensitized EphA2 and JNK2 selectively over their WT variants (Supporting Information. Figure S4A, S4B). The V286C/M346T variant of PAK1 was inhibited by probe 2 but was almost completely insensitive to probe 3. The inability to sensitize PAK1 to DFG-out-stabilizing probe 3 reflects the fact that some kinases may be incompatible with certain conformation-selective probes. We also observed that 2-TCO and 3-TCO were similarly efficient as 1-TCO in enriching drug-sensitized kinases from lysates (Supporting Information. Figure S4C), while only 2-TCO was effective in mammalian cells (Supporting Information. Figure S4D). The ability to sensitize diverse kinases to one or more conformation-selective TCO conjugates will allow broader interrogation of how ATP-binding site conformation influences the interactomes of kinases.
Co-clickable precipitation for interrogating Src’s interactome.
Having demonstrated that the TCO/tetrazine reaction is suitable for capturing inhibitor-bound kinase targets, we next developed cell lines suitable for studying Src’s interactome under different signaling conditions. To do this, we generated HeLa cells lines stably expressing either exogenous Src WT or Src V284C. Development of HeLa lines that express equal amounts of a Src variant–Src V284C–that is sensitive to our TCO probes and a probe-resistant variant–Src WT–are valuable for performing comparative proteomics of proteins that are enriched through their interactions with Src. Like in HEK293 cells, treatment of Src V284C-expressing HeLas with 1-TCO or 2-TCO followed by lysis and Tz-beads capture led to a significant amount of captured Src (Figure 3F). We observed almost no enrichment of Src WT when HeLa cells expressing this variant were treated with either probe. Prior to performing coclickable precipitation experiments to study Src’s cellular interactome, we determined the mildest conditions that could be used for capturing probe-bound Src after lysis, while maintaining sufficiently stringent washing conditions for removing proteins non-specifically bound to the sepharose matrix. A time-course experiment revealed that the click reaction between probe-bound Src and Tz-beads was almost complete within 10 mins at 4 °C (Figure 3G). We felt that these fast and mild capture kinetics would be advantageous for enriching protein complexes with minimal dissociation following cell lysis.
Co-clickable precipitation for mapping Src’s cellular interactome.
Prior to performing proteomic experiments, we verified that our co-clickable precipitation method was capable of co-enriching proteins that are likely to be associated with Src in growth factor-stimulated cells, such as tyrosine-phosphorylated signaling proteins. To do this, HeLa cells expressing Src V284C or WT were pre-incubated with either 1-TCO or 2-TCO, stimulated with growth factor for 15 minutes, and subsequently lysed (Figure 4A). Lysates were immediately incubated with Tz-beads and subjected to Western blot analysis with an anti-phosphotyrosine antibody following elution. Consistent with 1-TCO stabilizing a global conformation with an SH2 domain accessible to intermolecular interactions, we observed that treatment of Src V284C-expressing HeLas with this probe resulted in capture of several phosphotyrosine-containing proteins (Figure 4B).36 In contrast, treatment of Src V284C-expressing HeLas with 2-TCO, which stabilizes a closed global conformation with an inaccessible SH2 domain, led to capture of only tyrosine-phosphorylated Src. Thus, our co-clickable precipitation method is capable of co-enriching proteins interacting with Src and TCO-conjugated probes are able to differentially modulate the accessibility of Src’s SH2 domain through its ATP-binding site.
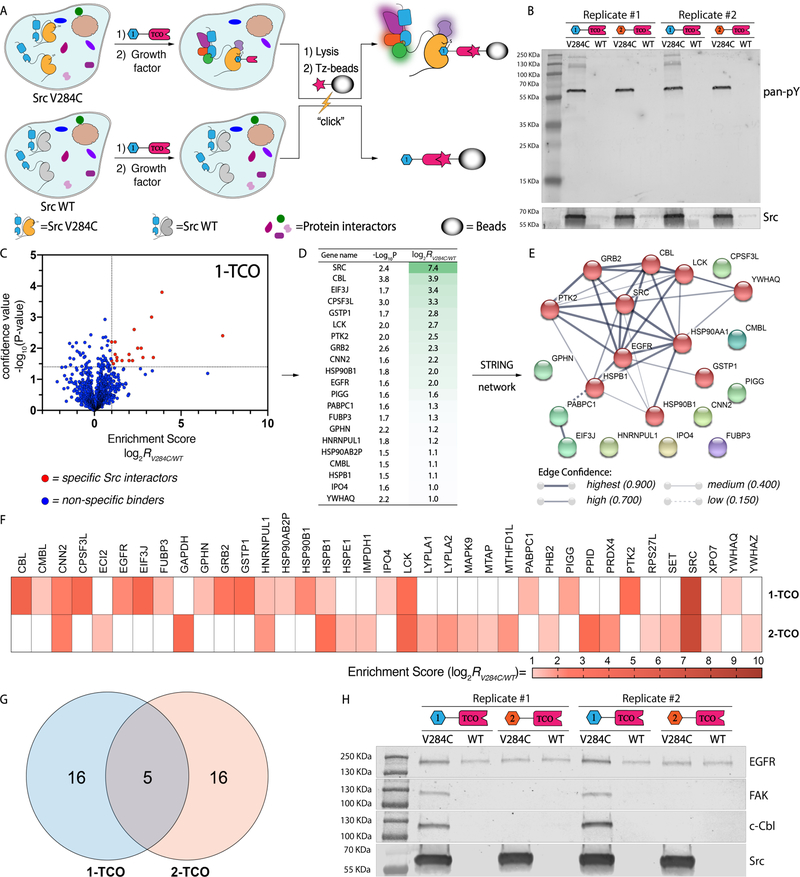
Figure 4.
Interrogation of Src’s interactome with co-clickable precipitation. (A) A schematic representation of the co-clickable precipitation strategy for identifying Src’s interactome. Cells stably expressing drug-sensitized Src V284C or drug-resistant Src WT are treated with a TCO-conjugated probe. Cells are then lysed, incubated with Tz-beads, and captured proteins are analyzed by Western blot or LC-MS/MS. Proteins that are enriched from cells expressing drug-sensitized Src V284C and not Src WT are classified as specific Src interactors. (B) Phosphotyrosine immunoblot of proteins enriched from growth factor-stimulated HeLa cells with TCO-conjugated probes. Cells expressing either Src V284C or Src WT were treated with 1-TCO or 2-TCO, lysed, and then incubated with Tz-beads. Captured proteins were eluted under reducing and denaturing conditions. Replicates of phosphotyrsosine (top) and Src (bottom) immunoblots of elutions are shown. (C-E) Interactome of 1-TCO-bound Src V284C in growth factor-stimulated HeLa cells. (C) Volcano plot showing the log2 transformed MaxQuant intensities of the label-free comparison of 1-TCO-treated, growth factor-stimulated HeLa cells expressing Src V284C and Src WT (n=3). Statistically significant interactors for drug-sensitized Src were defined as protein hits with an enrichment score = log2[intensity (drug-sensitized target)/intensity (WT target)] ≥ 1.0 and a confidence value = −log10(P-value) ≥ 1.4. The calculation was based on intensity values computed by MaxQuant. Missing protein intensity values were imputed by Perseus with a distribution downshift of 1.3 and a width of 0.2.32 Proteins that fit the classification as statistically significant Src V284C interactors are shown in red. (D) Proteins that show statistically significant enrichment with 1-TCO from Src V284C-expressing, growth factor-stimulated HeLa cells. The second column shows confidence values (-log10(P-value)) and the third column shows enrichment scores. (E) STRING network of the interactome of 1-TCO-bound Src V284C. The STRING database was used to generate a network with significant interactors. Each circle (node) represents a member of the interactome. Each line between the nodes (edge) represents a detected protein-protein interaction (PPI) in the STRING database.33 The edge weight denotes the confidence of the PPI.34 (F-H) Comparison of the interactomes between 1-TCO- and 2-TCO-bound Src V284C. (F) Heat map showing the enrichment score for each statistically significant interactor with 1-TCO- and 2-TCO-bound Src V284C (n=3). The panel’s color scale indicates the mean enrichment score from three biological replicates. Cells colored in white represent proteins that are not classified as significant interactors for the Src V284C-probe complex shown. (G) Venn diagram35 of common interactors between 1-TCO- and 2-TCO-bound Src V284C complexes. (H) Western blot confirmation of probe-bound Src V284C complex interactions.
Next, we established a quantitative proteomic analysis protocol to identify Src’s intracellular interactome. To differentiate proteins that were co-enriched with Src V284C from non-specific interactors, we performed quantitative label free comparisons of TCO probe-enriched proteins from HeLa cells expressing Src V284C to HeLa cells expressing Src WT. Proteomic experiments were performed like the Western blot analysis experiments described in Figure 4B, except Tz-bead-captured proteins were subjected to on-bead tryptic digestion and resultant peptides were identified and quantified by tandem mass spectrometry. Raw MS data were processed with MaxQuant37 across three biological replicates and analyzed with Perseus32 (Supporting Information). Enrichment scores (log2[intensity (Src V284C-expressing)/intensity (Src WT-expressing)]) were calculated using the relative intensity values from Src V284C- and WT-expressing cells with confidence values (-log10(P-value)) for each identified protein (Supporting Information, Table 1–2). We defined significant interactors as protein hits with an enrichment score ≥ 1.0 and a confidence value ≥ 1.4.
We first applied our quantitative proteomic analysis protocol to growth factor-stimulated HeLa cells. Our analysis showed that 21 high-confidence proteins are more than 2-fold enriched by 1-TCO from Src V284C-expressing cells relative to Src WT-expressing cells (Figure 4C,D). Consistent with the orthogonality of the 1-TCO probe, Src was the most highly enriched protein. The 20 putative Src interactors were enriched in the expected Gene Ontology (GO) terms38–39 “EGFR signaling pathway” (Biological Process), “SH2 domain binding” (Molecular Function), and “cell junction” (Cellular Component). Among the 20 identified Src interactions, 11 (55%) have not been previously reported in GO and Kegg repositories. Highly co-enriched proteins include the tyrosine kinase PTK2 (FAK) and the E3 ligase c-Cbl, which have previously been demonstrated to interact with the SH2 and SH3 domain of Src,40–41 respectively. These interactions are consistent with 1-TCO stabilizing a moderately open global conformation of Src with intermolecularly accessible regulatory domains. Another notably co-enriched protein includes the receptor tyrosine kinase EGFR, which has been demonstrated to be a substrate of Src in growth factor-stimulated cells.42 To better visualize Src’s interactome in growth factor-stimulated cells, we generated a STRING interaction network of co-enriched proteins (Figure 4E). Overall, these results are consistent with Src participating in signaling complexes directly downstream of activated EGFR.
We observed that a similar number of proteins were selectively enriched from growth factor-stimulated, Src V284C-expressing HeLa cells treated with 2-TCO (Figure 4F and Supporting Information, Figure S6). Like 1-TCO, the most highly enriched protein by 2-TCO was Src. Of the 20 putative interactors with the Src/2-TCO complex, five have previously been described in GO and Kegg repositories. Consistent with 2-TCO promoting intramolecular SH2 and SH3 engagement, most co-enriched proteins are different than those enriched from 1-TCO-treated cells (Figure. 4F,G). The small heat shock protein HSPB1 and the SFK Lck are the only two proteins that are enriched by both 1-TCO and 2-TCO. Proteins that have been characterized to interact with the SH2 or SH3 domains of Src are notably absent in the list of proteins enriched by 2-TCO, which further confirms that stabilizing the helix aC-out conformation prevents intermolecular interactions with these domains. Therefore, modulating the global conformational state of Src with conformation-selective, ATP-competitive inhibitors has a dramatic effect on its intracellular interactome. Prior to performing more expansive proteomic studies, we validated the enrichment of several representative protein targets with Western blot analysis. We found that only 1-TCO enriched EGFR, FAK and c-Cbl, three key players in the EGFR pathway (GO:0007173),38–39 from HeLa cells expressing the Src V284C mutant relative to HeLa cells expressing drug-resistant Src WT (Figure 4H). Only 2-TCO detectably enriched PPID and PRDX4 from cells expressing drug-sensitized Src V284C relative to drug-resistant Src WT (Supporting Information, Figure S5).
Interrogation of Src’s interactome under diverse cellular conditions.
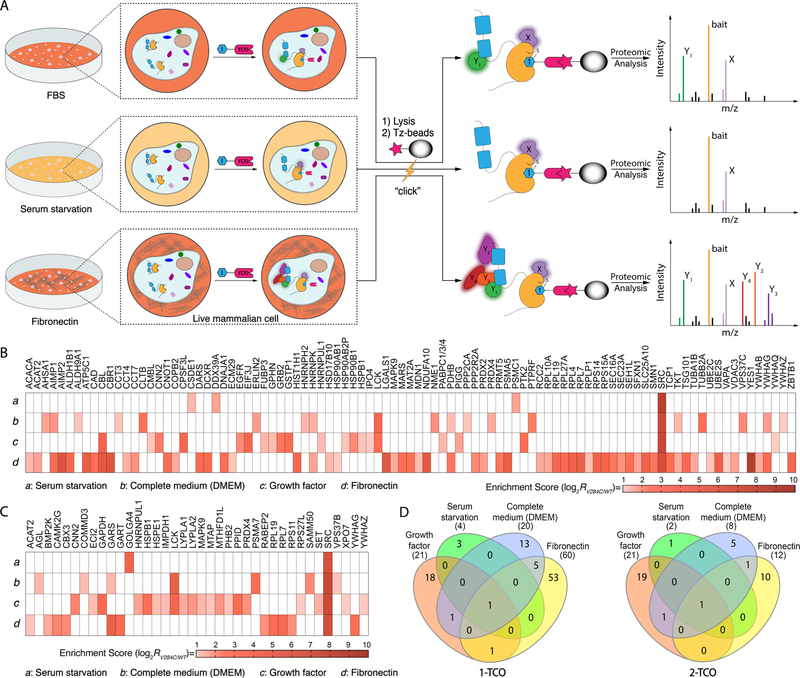
We next subjected HeLa cells to a number of conditions to assess how cellular state affects the interactome of inhibitor-bound Src (Figure 5A). Src V284C- and Src WT-expressing Hela cells were first treated with 1-TCO or 2-TCO and then either serum-starved, incubated in complete medium (DMEM), or plated on fibronectin. Cells were then lysed, and lysates were subjected to the same proteomic workflow as growth-factor-stimulated HeLas described in Figure 4C. Heatmaps showing the proteins that were specifically enriched by 1-TCO and 2-TCO under the four cellular states, which includes growth factor stimulated HeLas, that were probed are shown in Figures 5B,C. For all four conditions, Src was the most highly enriched cellular target and a total of 94 and 37 co-enriched proteins were identified for 1-TCO and 2-TCO, respectively. Consistent with 1-TCO-bound Src promoting a global conformation that is more capable of interacting with intracellular targets, more total proteins were enriched under all conditions with this probe than with 2-TCO.
Figure 5.
Src’s interactome in different cellular states. (A) A schematic representation of co-clickable precipitation experiments performed in three distinct cellular contexts. Hela cells stably overexpressing Src V284C or Src WT were serum-starved, cultured in complete medium (DMEM) or plated on fibronectin. Prior to lysis, cells were incubated with 1-TCO or 2-TCO. Probe-bound Src complexes were enriched with Tz-beads and captured proteins were analyzed using LC-MS/MS. (B, C) Heat maps showing the enrichment score for each statistically significant interactor with 1-TCO- and 2-TCO-bound Src V284C under four different cellular conditions: (a) serum-starvation, (b) complete medium (DMEM), (c) growth factor stimulation, or (d) plating on fibronectin. The panel’s color scale indicates the mean enrichment score from three biological replicates. Cells colored in white are proteins that were not classified as significant interactors with the Src V284C-probe complex shown. (D) Venn diagrams representing common interactors in four cellular states for 1-TCO- (left) and 2-TCO-bound (right) Src.
Our proteomic data with 1-TCO shows that the cellular state of a cell can dramatically affect Src’s interactome (Figure 5B). Consistent with serum starvation inactivating basal signaling pathways that Src participates in, only three proteins were co-enriched under this condition (Figure 5B and Supporting Information, Figure S7A–C). These three proteins have not previously been reported to interact with Src and no significant functional enrichments were detected in GO terms. Culturing cells in complete medium (DMEM) activates basal cell signaling and increases the number of interactions with 1-TCO-bound Src (Figure 5B and Supporting Information, Figure S8A–C). 19 proteins co-enriched with 1-TCO-bound Src from HeLa cells cultured in complete medium, with the GO terms “cellular metabolic process” (Biological process) and “SH2 domain binding” (Molecular function) being the most enriched. Growth factor stimulation further enhances the complexity of Src’s interactome. Despite resulting in a similar number of Src interactors, growth factor stimulation led to 3.0-fold, 2.0-fold and 2.4-fold increases in involvement of GO terms “Biological Process”, “Cellular Component” and “Molecular Function”, respectively. Finally, we observed that activation of integrin signaling by plating cells on fibronectin led to by far the most complex Src interactome (Figure 5B and Supporting Information, Figure S9A–C). In total, 59 proteins were co-enriched by 1-TCO-bound Src V284C under this condition. This interconnected interaction network displayed a high degree of complexity, highly enriching in several GO terms (104 of “Biological Process” terms, 22 of “Molecular Function” terms, and 29 of “Cellular Component” terms).
Similar to 1-TCO, our proteomic data with 2-TCO shows significant differences in Src’s interactome in different cellular states (Figure 5C). In serum starved HeLa cells, only one protein was significantly co-enriched with 2-TCO-bound Src V284C (Figure 5C and Supporting Information, Figure S7D–F). In HeLa cells with basal signaling activated by culturing in complete medium (DMEM), 7 proteins were enriched by 2-TCO-bound Src V284C, only 1 of which, Lck, was a mutual interactor shared with 1-TCO-bound Src V284C (Figure 5C, and Supporting Information, Figure S8D–F). Consistent with growth factor stimulation further activating cellular signaling, the 2-TCO/Src V284C complex interacts with 1.6-fold more proteins under this condition than in HeLas cultured in complete medium (Figure 5D). Notably, activation of integrin signaling by plating cells on fibronectin didn’t further enhance the size and complexity of the 2-TCO/Src V284C complex interactome (Figure 5C and Supporting Information, Figure S9D–F). Only 11 proteins were significantly co-enriched with 2-TCO-bound Src V284C. This loss of sensitivity towards stimulation with fibronectin is likely due to 2-TCO’s promotion of a closed global conformation of Src with reduced intermolecular accessibility of regulatory SH2 and SH3 domains. While 2-TCO reduced the number of Src interactors compared to 1-TCO under all of the cellular states tested, the largest difference between the two probes was in cells plated on fibronectin. Src forms 48 more interactions when bound to 1-TCO compared to 2-TCO under integrin stimulation, with only YWHAG–a 14-3-3 protein–commonly co-enriched. Thus, stabilizing a closed global conformation of Src has the most dramatic effect on Src’s interactome in cells.
TCO-conjugated probes for performing in situ proximity ligation assays.
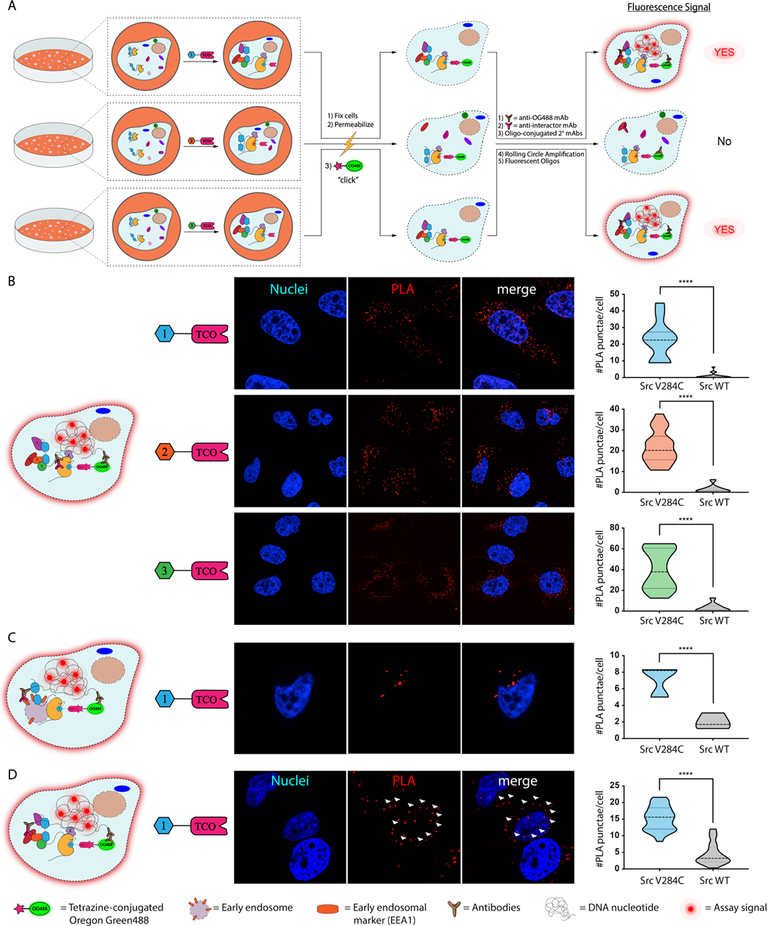
Beyond allowing the proteomic interrogation of drug-bound kinase complexes, we reasoned that our TCO conjugates could be used to characterize the in situ localization and interactions of probe-bound kinases. To do this, we developed a click chemistry-mediated proximity ligation assay with TCO probe-bound cellular targets (Figure 6A).43–46 In this method, cells are treated with a TCO-conjugated probe and then fixed and detergent-permeabilized. The TCO moiety is used to deliver a tetrazine-containing Oregon Green488 (OG488) dye, which allows visualization and serves as an exogenous epitope for an anti-OG488 antibody, to an inhibitor-bound target. Cells are then incubated with primary antibodies for OG488 and potential interactors or cellular organelle markers, which are recognized by secondary antibodies conjugated to single-stranded oligonucleotides that only template the formation of a circular DNA when two binding epitopes are <40 nm in proximity.43, 47 A rolling circle amplification (RCA) is then performed, followed by hybridization with complementary oligonucleotides labeled with fluorophores, which are imaged using confocal microscopy.
Figure 6.
Clickable proximity ligation assays (PLAs) with TCO-conjugated probes. (A) A schematic representation of PLAs performed with TCO-conjugated probes. Cells are treated with a TCO-conjugated probe, fixed, permeabilized, and then treated with a tetrazine-containing Oregon Green488 (OG488) dye. Fixed cells are then subjected to a standard proximity ligation assay protocol. (B) PLA for visualization of a probe-bound target. Representative images (nuclei in blue and PLA signal in red) of cells from three biological replicates are shown (middle). Quantified signals for the number of PLA-mediated fluorescent puncta observed per cell are shown in the violin plot (right). Values shown are the means of fluorescent puncta per cell from three biological replicates. (number of total cells quantified: n=97 for 1-TCO/Src V284C, and n=215 for 1-TCO/Src WT; n=148 for 2-TCO/Src V284C, and n=171 for 2-TCO/Src WT; n=142 for 3-TCO/Src V284C, and n=179 for 3-TCO/Src WT). (C) Clickable PLA for visualizing the cellular localization of TCO probe-bound Src in HeLa cells incubated in complete medium (DMEM). Representative images (nuclei in blue and PLA signal in red) of TCO probe-treated Src V284C-expressing HeLa cells are shown (middle). Quantified signals for the number of PLA-mediated fluoscent puncta observed per cell are shown in the violin plot (right). Values shown are the means of fluorescent puncta per cell from three biological replicates (number of total cells quantified: n=66 for Src V284C, and n=39 for Src WT). (D) Clcikable PLA for visualizing the interaction between Src and FAK. Representative images (nuclei in blue and PLA signal in red) of TCO probe-treated Src V284C-expressing HeLa cells plated on fibronectin are shown. The arrows (white) indicated examples of plasma membrane localization. Quantified signals for the number of PLA-mediated fluoscent puncta observed per cell are shown in the violin plot (right). Values shown are means of PLA puncta per cell from three biological replicates (number of total cells quantified: n= 375 for Src V284C, and n=314 for Src WT).
Prior to performing PLA experiments, we validated that Src V284C and Src WT have the same overall expression level (Supporting Information, Figure S10) and cellular localization in HeLa cells (Supporting Information, Figure S11–S12). Next, we confirmed that probe-bound Src V284C could selectively be visualized with an in situ PLA (Figure 6B). Src V284C- and Src WT-expressing HeLa cells were treated with TCO probes, fixed, detergent-permeabilized and reacted with tetrazine-conjugated Oregon Green488 dye. Primary antibodies for Oregon Green488 and Src were then added prior to performing RCA with single-strand oligonucleotide-conjugated secondary antibodies and hybridization with complementary oligonucleotides (Figure 6B, left). We observed that fluorescent puncta formation required the presence of both the OG488 and Src antibodies (Supporting Information, Figure S13). Consistent with our probes only selectively engaging drug-sensitized Src V284C in situ, significantly more fluorescent puncta were detected in Src V284C-expressing Hela cells versus cells expressing Src WT. Similar to the 1-TCO probe, significantly more signal was observed in Src V284C-expressing HeLa cells treated with 2-TCO and 3-TCO than in cells expressing Src WT.
Next, we used an organelle marker to determine if PLA can be used to provide information on the localization of probe-bound Src complexes (Figure 6C). Specifically, we determined if probe-bound Src is localized to early endosomes in Src V284C-expressing Hela cells cultured in complete medium (DMEM). Src has been shown to traffic through early endosomes during specific signaling events.48 Antibodies for the early endosomal marker EEA149 and OG488 were added to fixed and detergent-permeabilized cells following treatment with 1-TCO. Consistent with 1-TCO bound Src V284C localizing to early endosomes, fluorescent puncta were observed in the perinuclear region of Src V284C-expressing HeLas only with the presence of both primary antibodies (Figure 6C and Supporting Information, Figure S13).
Finally, we used PLA to validate kinase interactors in situ (Figure 6D). To do this, we probed the well-characterized interaction between FAK and Src in HeLa cells plated on fibronectin. As expected, fluorescent puncta were specifically observed at the plasma membrane–the site of focal adhesions–in Src V284C-expressing HeLa cells that were treated with 1-TCO followed by FAK and OG488 antibodies after fixation and cell permeabilization (Figure 6D, left). Interestingly, fluorescent signal was also observed in the perinuclear region, suggesting that Src and FAK interact in this subcellular region as well. Thus PLA, can validate probe-bound kinase interactions in situ and provide information on where these events occur.
Conclusions
In sum, we have explored the utility of click handle-conjugated probes for studying the interactomes and cellular localization of inhibitor-bound targets. We found that the rapid kinetics and mild reaction conditions of the iEDDA click reaction are well suited for capturing transient protein complexes associated with inhibitor-bound Src kinase. While click handle-conjugated probes have become important reagents for profiling and enriching direct cellular targets, our study, to the best of our knowledge, is the first to use this strategy to identify the proteins complexed to an inhibitor-bound cellular target. By developing TCO-conjugated probes that are able to stabilize different ATP-binding site conformations and divergently modulate the regulatory domains of Src, we are able to interrogate the cellular interactome of Src in various global conformations. We find that when the regulatory domains of Src are intramolecularly disengaged that its binding partners are dominated by SH2 and SH3 domain interactors. In contrast, when the regulatory domains of Src are fully intramolecularly engaged it interacts with a complementary set of proteins. We also find that cellular context has a dramatic effect on probe-bound Src’s interactome. As expected, Src interacts with very few proteins in cells with low basal signaling. As the number of active signaling pathways increases, the number of proteins comprising Src’s interactome expands. Our observation that Src forms the largest number of intermolecular interactions under prolonged integrin activation with fibronectin is consistent with the centrality of Src in downstream signaling events. More generally, our studies demonstrate that both kinase global conformation and cellular context greatly influence the interactions that kinases form within the cell.
In our study, we used a drug-sensitized Src mutant to obtain selectivity for our TCO-conjugated probes. We demonstrate that an advantage of this strategy is that the same TCO-conjugated probes can be used to target multiple drug-sensitized kinases. While the generality of our chemical genetic method throughout the broader kinome remains to be determined, the ability to selectively sensitize diverse kinases to our probes suggests that it should be of utility for the chemical proteomic interrogation of a number of kinases beyond the Src family. Our chemical proteomic strategy should also be applicable to any WT kinase or druggable protein target for which a selective inhibitor that contains a suitable site of chemical derivatization is available. Our chemoproteomic method is an alternative to the co-immunoprecipitation of proteins from inhibitor-treated cells. We feel that the rapid capture of inhibitor-bound targets that our method facilitates will likely provide a better representation of protein complexes present upon cell lysis. Furthermore, our strategy does not require the introduction of an epitope tag into a protein target, which can interfere with the regulation of proteins like Src that contain regulatory interactions at both termini, or the use of a target-specific antibody that can potentially disrupt the protein-protein interactions of an inhibitor-bound target. Finally, the ability to perform direct on-bead digestion of inhibitor-bound targets decreases the number of MS runs that need to be performed and increases the diversity of conditions that can be profiled. Going forward, we envision that our chemoproteomic strategy can be applied to a number of kinases and other druggable protein targets.
Supplementary Material
ACKNOWLEDGMENT
We thank Nathaniel Peters at the W.M. Keck Center for Advanced Studies in Neural Signaling for microscopy assistance. We thank Priska D. von Haller at University of Washington Proteomics Resource (UWPR) and Martin Sadilek for mass spectrometry assistance. We thank Stephen Taylor for providing Flp-In T-Rex HeLa cell line. This work was supported by the National Institute of General Medical Sciences (R01GM086858 to D.J.M.). E.M.D. was supported by a National Science Foundation Graduate Research Fellowship.
Funding Sources
National Institute of General Medical Sciences (R01GM086858 to D.J.M.) and National Institute of Diabetes and Digestive and Kidney Diseases (R01DK116064 to D.J.M.)
ABBREVIATIONS
- Tz
tetrazine
- TCO
trans-cyclooctene
- PLA
Proximity Ligation Assay
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website.
Supplementary figures, experimental details and synthesis of all compounds used in the study (PDF)
Supplementary table 1. Mass spectrometry proteomics datasets for all identified proteins (XLSX)
Supplementary table 2. Mass spectrometry proteomics datasets for all identified interactors (XLSX)
Notes
The authors declare no competing financial interest.
REFERENCES
- (1).Housden BE; Muhar M; Gemberling M; Gersbach CA; Stainier DY; Seydoux G; Mohr SE; Zuber J; Perrimon N, Nat Rev Genet 2017, 18 (1), 24–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Weiss WA; Taylor SS; Shokat KM, Nat Chem Biol 2007, 3 (12), 739–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Knight ZA; Shokat KM, Cell 2007, 128 (3), 425–30. [DOI] [PubMed] [Google Scholar]
- (4).Manning G; Whyte DB; Martinez R; Hunter T; Sudarsanam S, Science 2002, 298 (5600), 1912–34. [DOI] [PubMed] [Google Scholar]
- (5).Kung JE; Jura N, Structure 2016, 24 (1), 7–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Hatzivassiliou G; Song K; Yen I; Brandhuber BJ; Anderson DJ; Alvarado R; Ludlam MJ; Stokoe D; Gloor SL; Vigers G; Morales T; Aliagas I; Liu B; Sideris S; Hoeflich KP; Jaiswal BS; Seshagiri S; Koeppen H; Belvin M; Friedman LS; Malek S, Nature 2010, 464 (7287), 431–5. [DOI] [PubMed] [Google Scholar]
- (7).Murphy JM; Nakatani Y; Jamieson SA; Dai W; Lucet IS; Mace PD, Structure 2015, 23 (11), 2111–21. [DOI] [PubMed] [Google Scholar]
- (8).Ahler E; Register AC; Chakraborty S; Fang L; Dieter EM; Sitko KA; Vidadala RSR; Trevilli-an BM; Golkowski M; Gelman H; Stephany JJ; Rubin AF; Merritt EA; Fowler DM; Maly DJ, Molecular Cell 2019, 74, 393–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Gonfloni S; Weijland A; Kretzschmar J; Superti-Furga G, Nat Struct Biol 2000, 7 (4), 281–6. [DOI] [PubMed] [Google Scholar]
- (10).Wang L; Perera BG; Hari SB; Bhhatarai B; Backes BJ; Seeliger MA; Schurer SC; Oakes SA; Papa FR; Maly DJ, Nat Chem Biol 2012, 8 (12), 982–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Moellering RE; Cravatt BF, Chem Biol 2012, 19 (1), 11–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Kang K; Park J; Kim E, Proteome Sci 2016, 15, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Cravatt BF; Wright AT; Kozarich JW, Annu Rev Biochem 2008, 77, 383–414. [DOI] [PubMed] [Google Scholar]
- (14).Speers AE; Adam GC; Cravatt BF, J Am Chem Soc 2003, 125 (16), 4686–7. [DOI] [PubMed] [Google Scholar]
- (15).Hong V; Steinmetz NF; Manchester M; Finn MG, Bioconjug Chem 2010, 21 (10), 1912–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Khossravi M; Borchardt RT, Pharm Res 2000, 17 (7), 851–8. [DOI] [PubMed] [Google Scholar]
- (17).Cheng RZ; Uchida K; Kawakishi S, Biochem J 1992, 285 (Pt 2), 667–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Houghton EA; Nicholas KM, J Biol Inorg Chem 2009, 14 (2), 243–51. [DOI] [PubMed] [Google Scholar]
- (19).Khossravi M; Borchardt RT, Pharm Res 1998, 15 (7), 1096–102. [DOI] [PubMed] [Google Scholar]
- (20).Ramirez DC; Mejiba SE; Mason RP, J Biol Chem 2005, 280 (29), 27402–11. [DOI] [PubMed] [Google Scholar]
- (21).Baskin JM; Prescher JA; Laughlin ST; Agard NJ; Chang PV; Miller IA; Lo A; Codelli JA; Bertozzi CR, Proc Natl Acad Sci U S A 2007, 104 (43), 16793–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Rutkowska A; Thomson DW; Vappiani J; Werner T; Mueller KM; Dittus L; Krause J; Muelbaier M; Bergamini G; Bantscheff M, ACS Chem Biol 2016, 11 (9), 2541–50. [DOI] [PubMed] [Google Scholar]
- (23).Blackman ML; Royzen M; Fox JM, J Am Chem Soc 2008, 130 (41), 13518–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Serafimova IM; Pufall MA; Krishnan S; Duda K; Cohen MS; Maglathlin RL; McFarland JM; Miller RM; Frodin M; Taunton J, Nat Chem Biol 2012, 8 (5), 471–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Bradshaw JM; McFarland JM; Paavilainen VO; Bisconte A; Tam D; Phan VT; Romanov S; Finkle D; Shu J; Patel V; Ton T; Li X; Loughhead DG; Nunn PA; Karr DE; Gerritsen ME; Funk JO; Owens TD; Verner E; Brameld KA; Hill RJ; Goldstein DM; Taunton J, Nat Chem Biol 2015, 11 (7), 525–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Krishnan S; Miller RM; Tian B; Mullins RD; Jacobson MP; Taunton J, J Am Chem Soc 2014, 136 (36), 12624–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Miller RM; Paavilainen VO; Krishnan S; Serafimova IM; Taunton J, J Am Chem Soc 2013, 135 (14), 5298–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Feldman HC; Tong M; Wang L; Meza-Acevedo R; Gobillot TA; Lebedev I; Gliedt MJ; Hari SB; Mitra AK; Backes BJ; Papa FR; Seeliger MA; Maly DJ, ACS Chem Biol 2016, 11 (8), 2195–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Leonard SE; Register AC; Krishnamurty R; Brighty GJ; Maly DJ, ACS Chem Biol 2014, 9 (8), 1894–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Hari SB; Merritt EA; Maly DJ, Chem Biol 2014, 21 (5), 628–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Krishnamurty R; Brigham JL; Leonard SE; Ranjitkar P; Larson ET; Dale EJ; Merritt EA; Maly DJ, Nat Chem Biol 2013, 9 (1), 43–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Tyanova S; Temu T; Sinitcyn P; Carlson A; Hein MY; Geiger T; Mann M; Cox J, Nat Methods 2016, 13 (9), 731–40. [DOI] [PubMed] [Google Scholar]
- (33).Szklarczyk D; Franceschini A; Wyder S; Forslund K; Heller D; Huerta-Cepas J; Simonovic M; Roth A; Santos A; Tsafou KP; Kuhn M; Bork P; Jensen LJ; von Mering C, Nucleic Acids Res 2015, 43 (Database issue), D447–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Szklarczyk D; Morris JH; Cook H; Kuhn M; Wyder S; Simonovic M; Santos A; Doncheva NT; Roth A; Bork P; Jensen LJ; von Mering C, Nucleic Acids Res 2017, 45 (D1), D362–D368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Heberle H; Meirelles GV; da Silva FR; Telles GP; Minghim R, BMC Bioinformatics 2015, 16, 169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Liu BA; Engelmann BW; Nash PD, FEBS Lett 2012, 586 (17), 2597–605. [DOI] [PubMed] [Google Scholar]
- (37).Cox J; Mann M, Nat Biotechnol 2008, 26 (12), 1367–72. [DOI] [PubMed] [Google Scholar]
- (38).The Gene Ontology C, Nucleic Acids Res 2017, 45 (D1), D331–D338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Ashburner M; Ball CA; Blake JA; Botstein D; Butler H; Cherry JM; Davis AP; Dolinski K; Dwight SS; Eppig JT; Harris MA; Hill DP; Issel-Tarver L; Kasarskis A; Lewis S; Matese JC; Richardson JE; Ringwald M; Rubin GM; Sherlock G, Nat Genet 2000, 25 (1), 25–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Xing Z; Chen HC; Nowlen JK; Taylor SJ; Shalloway D; Guan JL, Mol Biol Cell 1994, 5 (4), 413–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Tanaka S; Neff L; Baron R; Levy JB, J Biol Chem 1995, 270 (24), 14347–51. [DOI] [PubMed] [Google Scholar]
- (42).Bromann PA; Korkaya H; Courtneidge SA, Oncogene 2004, 23 (48), 7957–68. [DOI] [PubMed] [Google Scholar]
- (43).Gao X; Hannoush RN, J Am Chem Soc 2014, 136 (12), 4544–50. [DOI] [PubMed] [Google Scholar]
- (44).Weibrecht I; Leuchowius KJ; Clausson CM; Conze T; Jarvius M; Howell WM; Kamali-Moghaddam M; Soderberg O, Expert Rev Proteomics 2010, 7 (3), 401–9. [DOI] [PubMed] [Google Scholar]
- (45).Gao X; Hannoush RN, Nat Chem Biol 2014, 10 (1), 61–8. [DOI] [PubMed] [Google Scholar]
- (46).Gao X; Hannoush RN, Nat Protoc 2014, 9 (11), 2607–23. [DOI] [PubMed] [Google Scholar]
- (47).Soderberg O; Gullberg M; Jarvius M; Ridderstrale K; Leuchowius KJ; Jarvius J; Wester K; Hydbring P; Bahram F; Larsson LG; Landegren U, Nat Methods 2006, 3 (12), 995–1000. [DOI] [PubMed] [Google Scholar]
- (48).Donepudi M; Resh MD, Cell Signal 2008, 20 (7), 1359–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Stenmark H; Aasland R; Toh BH; D’Arrigo A, J Biol Chem 1996, 271 (39), 24048–54. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.