Abstract
Hypoxia is a common feature in solid tumors. Clinical samples show a positive correlation between the expression of the hypoxia-inducible factor HIF-1α and estrogen receptor alpha (ERα) and a negative correlation between HIF-1α and hormone sensitivity. Results from monolayer cultures are in contention with clinical observations, showing that ER (+) cell lines no longer express ERα under hypoxic (1% O2) conditions. Here, we compared the impact of hypoxia on the ERα signaling pathway for T47D cells in a 2D and 3D culture format. In the 2D format, the cells were cultured as monolayers. In the 3D format, paper-based scaffolds supported cells suspended in a collagen matrix. Using ELISA, western blot, and immunofluorescence measurements, we show that hypoxia differentially regulates ERα protein levels. In the 2D format, the protein levels are significantly decreased in hypoxia. In the 3D format, the protein levels are maintained in hypoxia. Hypoxia reduced ERα transcriptional activation in both culture formats. These results highlight the importance of considering tissue dimensionality for in vitro studies. They also show that ERα protein levels in hypoxia are not an accurate indicator of ERα transcriptional activity, and confirm that a positive stain for ERα in a clinical sample may not necessarily indicate hormone sensitivity.
Keywords: estrogen signaling, hypoxia-inducible factor (HIF), breast cancer
Graphical Abstract:
1. Introduction
Breast cancer is the most commonly occurring cancer in women worldwide [1]. Approximately 70% of breast cancers stain positively for estrogen receptor alpha (ERα) [2] and rely on estrogen to regulate genes required for proliferation, differentiation, and motility [3-5]. Endocrine therapies—including aromatase inhibitors, selective estrogen receptor modulators, and selective estrogen receptor disruptors—are used as adjuvant therapies, preventing breast tumor growth and recurrence by targeting proteins involved in the ERα signaling pathway [6-8].
Nearly 40% of estrogen-sensitive ER (+) tumors become hormone-insensitive [7, 8]. A number of factors have been implicated in this progression, including the tumor microenvironment [6, 8]. Solid tumor masses are often poorly vascularized, resulting in regions of insufficient oxygen tension to support normal cellular functions (hypoxia) [9]. In healthy breast tissue, the average partial pressure of oxygen (pO2) is 52.0 mmHg (6.8%). The average pO2 for 212 breast tumors ranging in size and stage was hypoxic at 10.0 mmHg (1.3%) [10, 11]. Hypoxia is associated with both tumor recurrence and aggressiveness in breast cancer [9, 12].
Hypoxia-inducible factor 1 (HIF-1) is a master regulator of cellular responses in hypoxic conditions [12], regulating the expression of more than 70 target genes involved in angiogenesis, tissue remodeling, metabolism, and proliferation [13-15]. This heterodimeric transcription factor consists of an oxygen-sensitive α subunit and a stable β subunit. Under normoxic conditions, HIF-1α is readily targeted for degradation by prolyl-4-hydroxylases (PHDs) and the von Hippel-Lindau tumor suppressor. Under hypoxic conditions, HIF-1α is stabilized and able to translocate to the nucleus, where it forms an active transcription factor complex.
HIF-1α protein stabilization has been associated with endocrine therapy insensitivity in ER (+) breast cancer patients and its expression is positively correlated with ERα expression in tissue samples [16-18]. Interestingly, previous in vitro studies exposing monolayer cultures of ER (+) breast cell lines to hypoxic conditions (1% O2, 24 – 48 h) had significantly reduced ERα protein levels [19, 20]. The discrepancy between clinical and in vitro studies led us to question if the 3D tumor environment alters the interplay between the hypoxia and ERα signaling pathways within in vitro models.
Three-dimensional (3D) culture models emulate key aspects of the tumor microenvironment [21-24]. Both ERα and HIF-1α signaling pathways are sensitive to the culture environment. Vantangoli et al. showed that transcriptional regulation in the ER (+) MCF-7 cell line was markedly different for monolayers or microtissues in an agarose gel exposed to 17β-estradiol (E2) [25]. After 24 h, five gene transcripts increased above basal levels in the 2D cultures, whereas 22 transcripts were either increased or decreased in the 3D cultures. In another study, DelNero et al. found that 214 genes were differentially regulated when OSCC-3 cells in monolayers or suspended in alginate discs were exposed to hypoxia for six days [26]. In particular, they observed increased expression of pro-inflammatory genes in 3D culture, compared to levels seen in 2D culture.
There is not a study to our knowledge that compares how the transition from 2D to a 3D culture format affects the interplay between the HIF-1α and ERα signaling pathways. In this work, we compared the impact of 24 h of hypoxia on the expression and transcriptional activity of ERα in 2D and 3D culture formats. The 2D cultures were monolayers on plasticware and the 3D cultures were cell-containing collagen suspensions in wax-patterned paper scaffolds. The paper scaffolds, which allow thin gel slabs (40 microns thick) to be easily manipulated without fear of cracking or breaking, have been used to generate 3D models of breast [27-30], lung [31, 32], colon [33], ovarian and cervical [34], and head and neck tumors [35]. Our studies show that both the HIF-1α and the ERα signaling pathways of the T47D-KBluc cell line are differentially regulated in different culture formats. In particular, ERα levels in 3D cultures are not impacted by hypoxia, yet ERα transcriptional activity is decreased under hypoxia in both 2D and 3D culture formats.
2. Materials and methods
2.1. Materials
All reagents were used as received unless otherwise stated. 17β-estradiol (E2) and MG-132 were purchased from Sigma-Aldrich. Dimethyloxalylglycine (DMOG) was purchased from Frontier Scientific. Cell culture medium and additives were purchased from Gibco, except for collagen I (rat tail, Corning), DMSO (Fisher Scientific), ethanol (Fisher Scientific), and fetal bovine serum (FBS, VWR). CellTiter-Glo, ONE-Glo, and Reporter Lysis 5X Buffer were purchased from Promega and used according to the manufacturer’s protocols.
2.2. Cell culture
The T47D-KBluc (T47D) cell line was kindly provided by Dr. Vickie Wilson at the EPA. These cells are an engineered variant of the ER (+) T47D cell line that expresses luciferase, in a dose-dependent manner, in the presence of estrogenic agonists [36]. The cells were cultured as monolayers in phenol red-free DMEM supplemented with 10% FBS, 4.5 g/L D-glucose, 4 mM L-glutamine, 25 mM HEPES, 1 mM sodium pyruvate, 0.1 mM nonessential amino acids, 0.5 mg/mL Geneticin, and 0.05 mg/mL gentamicin. Cells were maintained at 37 °C in a 5% CO2 environment. Culture medium was exchanged every 2-3 days, and the cells were passed at a 1:10 dilution upon confluency. Unless otherwise stated, the cells were placed in withdrawal medium 3 d prior to use. Withdrawal medium consisted of phenol red-free DMEM supplemented with 10% charcoal-stripped FBS, 4.5 g/L D-glucose, 4 mM L-glutamine, 25 mM HEPES, 1 mM sodium pyruvate, 0.1 mM nonessential amino acids, and 0.05 mg/mL gentamicin.
For 2D cultures, the T47D cells were seeded at a density of 40,000 cells/well in tissue culture-treated 96-well plates. For 3D cultures, the T47D cells were suspended in 1.2 mg/mL collagen I at a density of 40,000 cells/μL. Paper scaffolds containing a single zone surrounded by a wax-patterned border were seeded with 1 μL of the cell-containing suspension, a final density of 4 × 107 cells/cm3. The paper scaffolds are detailed in Fig. S1. Their preparation and characterization have been previously detailed [27].
For normoxia studies, cells were maintained at 21% oxygen in a standard cell culture incubator. For hypoxia studies, cells were maintained at 1% oxygen in an O2 Control In Vitro Glove Box (Coy Laboratory Products). Both culture environments maintained the cells at 37 °C in a 5% CO2 environment.
2.3. ELISA and western blot
Prior to ELISA and western blot analysis, cells in both the 2D and 3D cell culture formats were lysed in ice-cold RIPA buffer (20 min, frequent agitation, 4 °C, 2 × 106 cells per mL of lysis buffer). The resulting lysates were clarified by centrifugation at 10,000 ×g at 4 °C for 10 min. For ELISA, ERα and HIF-1α concentrations were determined with commercial kits (R&D Systems, DYC-5715 and DYC1935); β-actin concentration was determined with a sandwich assay consisting of a β-actin capture antibody (R&D Systems, AF4000) and an HRP-linked detection antibody (SantaCruz Biotechnology, sc-47778).
For western blot analyses, clarified lysates were separated by SDS-PAGE and transferred to PVDF membranes. Membranes were probed with antibodies for ERα (1:500), HIF-1α (1:200), and β-actin (1: 500), detected with HRP-conjugated secondary antibodies (1: 1000 m-IgGº BP-HRP), and visualized with the Pierce ECL Plus Western blotting substrate. Antibodies for ERα (sc-8002), β-actin (sc-47778), and mIgGº BP-HRP (sc-516102) were purchased from Santa Cruz Biotechnology; the HIF-1α antibody (610958) was purchased from BD Transduction Laboratories.
2.4. Immunofluorescence staining
Withdrawn cells were seeded on glass coverslips or in paper scaffolds. The cells were incubated for 24 h under normoxic or hypoxic conditions, in the presence of either vehicle (0.1% ethanol) or 10 nM E2. The coverslips and paper scaffolds were washed with 1X PBS, fixed with 3.2% paraformaldehyde in 1X PBS for 15 min, blocked for 1 hr in 1X PBS containing 5% normal goat serum and 0.3% Triton X-100, and incubated overnight in anti-ERα (1:100, Santa Cruz Biotechnology sc-8002). The stained samples were washed with 1X PBS containing 0.3% Triton X-100, and incubated with IgG-CFL 555 (1:200, goat anti-mouse, Santa Cruz Biotechnology sc-362267) and DAPI (1:1000) for 3 h. Stained samples were washed with 1X PBS containing 0.3% Triton X-100 and with 1X PBS before mounting on glass microscope slides with ProLong Gold Antifade Mountant (Invitrogen). Mounted samples were sealed with clear nail polish and allowed to cure overnight before image acquisition.
2.5. Fluorescence image acquisition and analysis
Fluorescence micrographs were obtained on a Zeiss LSM 710 confocal laser scanning microscope using a 25x/0.8 LD LCI Plan-Apochromatic objective. For 2D cultures, a single confocal image was acquired; for 3D cultures, z-stack images were taken every 5 μm. Each image (512 × 512 pixels) is the average of six scans obtained at an acquisition speed of 1.27 μs/pixel, using a 36 μm pinhole. ERα was imaged with a 543/604 nm filter set and DAPI with a 405/478 filter set.
Single-slice images from each z-stack were analyzed with FIJI using the following procedure. First, the DAPI images were automatically thresholded using the Otsu method [37]. Next, we generated a binary mask for each nucleus. Regions of interest (ROIs) were then generated from each nuclear mask using the analyze particles function (parameters: size = 20 – 250 μm and circularity = 0.2 – 1.0). These ROIs were applied to the ERα images to measure the intensity of nuclear ERα staining. The raw intensity data for each culture format was normalized to the intensity signal for cultures incubated under normoxia in E2-deprived medium.
2.6. RNA isolation and RT-qPCR
Total RNA was isolated from both 2D and 3D cultures after a 24 h treatment using the TRIzol Plus RNA Purification Kit (Thermo Fisher Scientific); 1 μg of RNA was reverse transcribed with the High-Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific). Each qPCR reaction (10 μL total volume) was prepared in a 384-well plate with 0.3 μM primers and PowerUp SYBR Green PCR Master Mix (Applied Biosystems). qPCR was performed with a QuantStudio 6 Flex Real-Time PCR System (Applied Biosystems) using the following amplification protocol: initial denaturation at 95.0 °C for 20 seconds followed by 40 cycles of 2 seconds at 95 °C and 30 seconds at 60 °C. The primers for estrogen receptor α (ESR1), progesterone receptor (PGR), hypoxia-inducible factor 1α (HIF1A), vascular epidermal growth factor A (VEGFA) and β-actin (ACTB) are listed in Table S1. Standard curves for each primer pair were generated and the efficiency of each qPCR reaction was calculated (Fig. S2). Relative quantification of target genes was performed in triplicate and analyzed with the ΔΔCt method using β-actin (ACTB) as the reference gene [38].
2.7. ER transcriptional activation assay
ERα transcriptional activation was measured as described previously [27]. Withdrawn cells were seeded at a density of 40,000 cells per 100 μL in a 96-well plate or seeded in paper scaffolds at 40,000 cells per zone. Luciferase activity and cellular viability were assessed with the One-Glo and CellTiter-Glo reagents, respectively. For each assay, cells were lysed in ambient oxygen conditions for 30 min. All of the luciferase activity reactions were also performed under ambient oxygen levels. Luminescence values were recorded on a SpectraMax M5 plate reader and are reported as a fold-change relative to the cultures incubated in E2-deprived medium under normoxia.
2.8. In vitro proteasome activity assay
In vitro proteasome activity was assessed using a fluorescence-based assay (ab107921, Abcam). Briefly, 2 × 106 cells were lysed in 400 μL NP-40 lysis buffer (20 min, frequent agitation, 4 °C). The lysate was clarified by centrifugation. Activity reactions were prepared by adding 20 μL of sample to 80 μL of assay buffer in either the absence or presence of the proteasome inhibitor MG-132 (100 μM). The reactions were initiated by the addition of 1 μL of fluorogenic proteasome substrate, incubated at 37 °C for 30 min, and then the fluorescence intensity recorded for an additional 30 min (Ex350/440). Proteasome activity was normalized to the total protein from each reaction, determined with a BCA assay (Pierce).
2.9. Statistics
Unless otherwise specified, each dataset represents the average and standard error of the mean (SEM) of at least six replicates. GraphPad Prism v7.0b was used for statistical analyses. Two groups were compared using an unpaired Student’s t-test with Welch’s correction; multiple comparisons were made using one-way ANOVA. A p value of < 0.05 was considered significant.
3. Results
3.1. ERα levels are insensitive to hypoxia in a 3D culture format
To evaluate the impact of decreased oxygen tension on ERα protein expression, we exposed both monolayers and 3D cultures containing 40,000 T47D cells to either normoxic (~21% O2) or hypoxic (1% O2) conditions for 24 h. Schematics of both culture formats are shown in Fig. 1A. We quantified ERα levels, after lysis, with an ELISA.
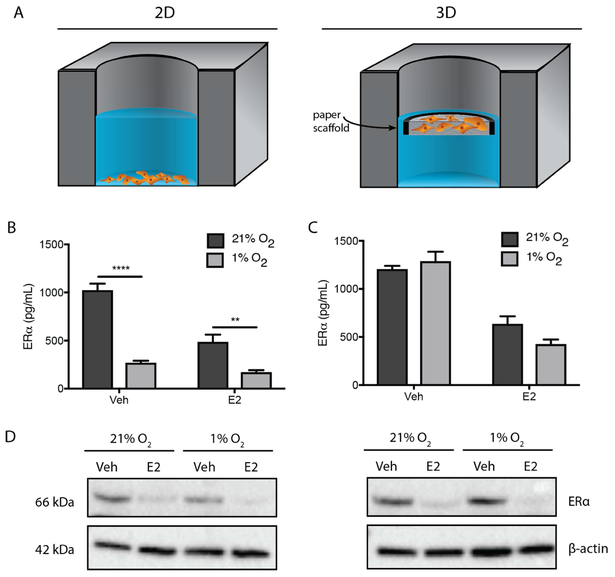
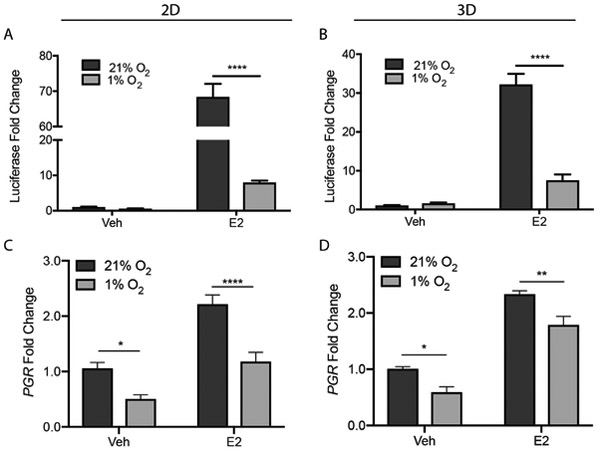
Fig. 1.
(A) Schematics of the 2D and 3D culture formats used throughout this work. Both formats contained 40,000 T47D cells that were either plated directly in a commercial 96 well plate or suspended in collagen I and seeded into a paper-based scaffold with a 1 × 10−3 cm3 culture zone. Once seeded, the paper-based scaffolds were also placed in a commercial 96 well plate. All experiments were incubated under normoxic (21% O2) or hypoxic (1% O2) conditions in E2-deprived (Veh) or -supplemented (E2) medium for 24 h. (B, C) Average ± SEM of ERα protein levels for each experimental condition determined by ELISA. Protein levels were normalized to β-actin. Each bar represents n ≥ 12 replicate cultures from three different cell passages. *p ≤ 0.05, ** p ≤ 0.01, *** p ≤ 0.001, **** p ≤ 0.0001. (D) A representative western blot of ERα protein levels with a β-actin loading control.
In both culture formats under normoxic conditions, ERα significantly decreased upon exposure to E2, consistent with previous reports using monolayer cultures [39-41]. In 2D, the ERα decrease was greater (1022 to 484 pg/mL, Fig. 1B) than in 3D (1204 to 634 pg/mL, Fig. 1C). Hypoxia significantly reduced ERα expression in 2D cultures in both E2-deprived (3.8-fold decrease) and E2-supplemented (2.9-fold decrease) medium. These findings agree with previous in vitro studies [19, 20, 42]. Surprisingly, ERα expression levels were not affected by hypoxia in the 3D cultures. Western blots confirmed the ELISA results, showing that the fold-changes were representative of full-length ERα and not artifacts caused by ERα truncates or degradation products. Fig. 1D is a representative set of western blots; Fig. S3 summarizes relative band intensities from three biological replicates.
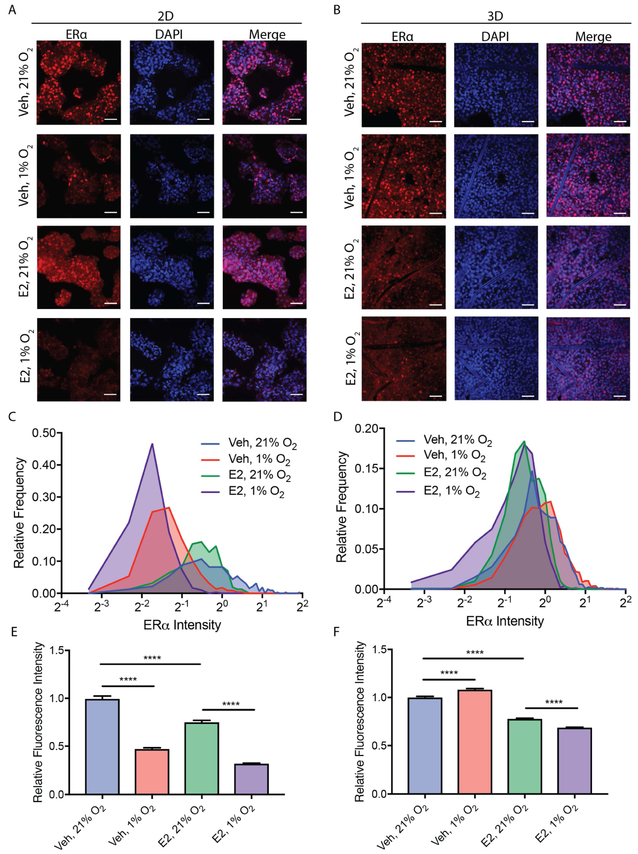
To determine if the ELISA results were representative of all cells in the culture, we assessed nuclear localization of ERα with immunofluorescence. Representative single-plane images of cells in both culture formats are displayed in Fig. 2A and 2B. To quantify nuclear localization of ERα, we used DAPI-stained nuclei to generate co-localized regions of interest, as described previously [43]. Fig. 2C and 2D contain frequency distributions of ERα nuclear staining for the 2D and 3D culture formats, respectively. The fluorescence intensity of nuclear ERα staining for each condition was normalized to the average fluorescence intensity of cultures incubated in E2-deprived medium under normoxic conditions. Each frequency distribution could be fit to a single Gaussian (Fig. S4) with R2 values above 0.9 (Table S2). These results indicate a unimodal response and demonstrate that the average changes in ERα levels are representative of the entire population of cells rather than a subpopulation of strongly expressing cells. The average fluorescence intensity of nuclear ERα staining in 2D (Fig. 2E) and 3D (Fig. 2F) culture formats are an alternative means of displaying this dataset, and further confirm that ERα is sensitive to E2 in both culture formats and that ERα is sensitive to hypoxia in the 2D culture format but not in the 3D culture format.
Fig. 2.
Representative single-plane confocal fluorescence micrographs of T47D cells in (A) 2D and (B) 3D culture formats after a 24 h incubation under normoxic (21% O2) or hypoxic (1% O2) conditions in E2-deprived (Veh) or -supplemented (E2) medium. Cells were fixed, immunostained for ERα (red), and the nuclei co-stained with DAPI (blue). The scale bar in each image represents 50 μm. Nuclear ERα values were normalized to the normoxic vehicle control for the 2D and 3D cultures. Frequency distributions of ERα staining, plotted for both (C) 2D and (D) 3D culture formats. ERα staining intensity, plotted as average ± SEM for both (E) 2D and (F) 3D culture formats. Each data is n > 150 nuclei from four replicate cultures from a total of two passages of cells. *p ≤ 0.05, ** p ≤ 0.01, *** p ≤ 0.001, **** p ≤ 0.0001.
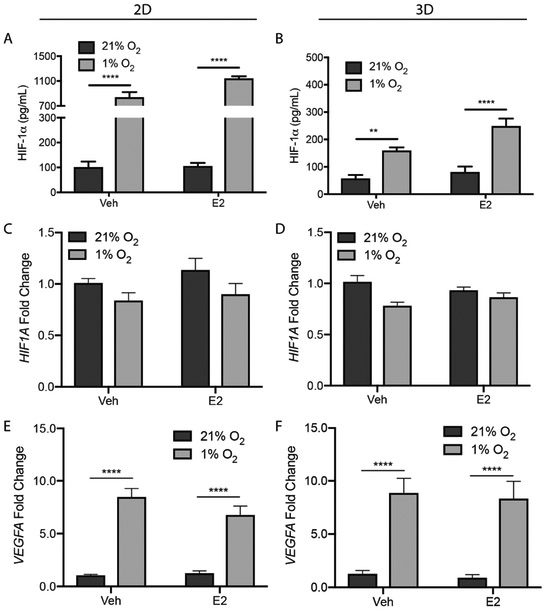
Fig. 3.
2D and 3D T47D cultures under normoxic (21% O2) or hypoxic (1% O2) conditions in E2-deprived (Veh) or E2-supplemented (E2) medium for 24 h, and then probed with ELISA to quantify HIF-1α protein levels (A, B). Protein levels were normalized to β-actin. Data represent the average ± SEM, from n ≥ 12 replicate cultures from three different cell passages. Total RNA was extracted, and the relative expression of HIF1A (C, D) and VEGFA (E, F) for each experimental condition determined using the ΔΔCt method; β-actin served as the reference gene. Data represent the average ± SEM, from n = 9 replicate cultures from a total of three passages of cells. *p ≤ 0.05, ** p ≤ 0.01, *** p ≤ 0.001, **** p ≤ 0.0001.
3.2. Cells in both culture formats undergo classic responses to hypoxia
To confirm that cells in both culture formats underwent a canonical response to hypoxia, we repeated the experimental setups detailed above and quantified HIF-1α levels with both ELISA (Fig. 3A and 3B) and western blot (Fig. S5). The basal levels of HIF-1α were the same in both culture formats, suggesting that the cells in the scaffolds were well oxygenated. HIF-1α levels increased significantly after a 24 h exposure to hypoxia, with a culture format-dependent stabilization: a 9.5-fold increase in 2D and a 3.0-fold increase in 3D.
Previous studies showed that hypoxia does not have an impact on HIF1A expression [44], and that increased levels of HIF-1α are caused by a decrease in its rate of degradation [15]. We quantified the relative abundance of both HIF1A and VEGFA, a classic HIF-responsive gene, using RT-qPCR. As expected, hypoxia had no impact on relative HIF1A abundance in any of the setups (Fig. 3C and 3D). We observed a significant increase in VEGFA mRNA expression under hypoxia in both 2D (Fig. 3E) and 3D (Fig. 3F) culture formats, confirming HIF-1α transcriptional activity.
3.3. ERα is both transcriptionally and post-translationally regulated under hypoxia
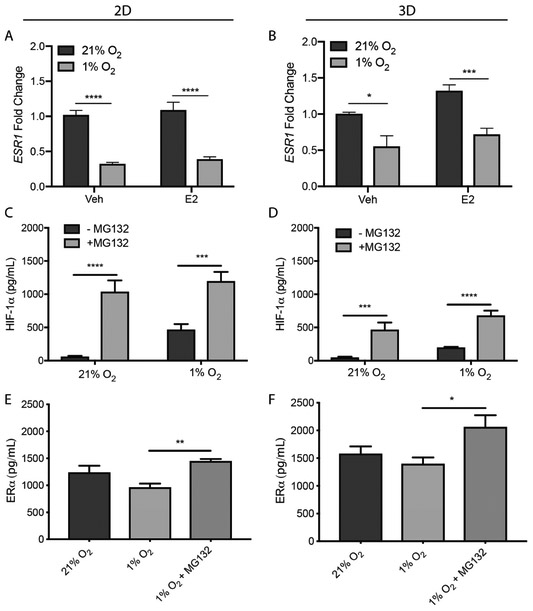
Despite an agreement that hypoxia decreases ERα protein expression in 2D, its impact on ESR1 gene expression is contended. Two separate studies found that hypoxia decreased ESR1 gene expression in MCF-7 and T47D cells [19, 45]. A third study found hypoxia did not affect ESR1 in a panel of 10 ER (+) cell lines that included MCF-7 and T47D [20]. We observed a significant reduction in ESR1 in both culture formats (Fig. 4A and 4B). These results highlight that transcriptional and translational regulation of ERα is not directly related in the 3D cultures.
Fig. 4.
2D and 3D T47D cultures exposed to normoxic or hypoxic conditions in E2-deprived (Veh) or -supplemented (E2) medium for 24 h. Total RNA was extracted and relative expression of ESR1 (A, B) was determined using the ΔΔCt method; β-actin served as the reference gene. Data represent the average ± SEM, from n = 9 replicate cultures from three different cell passages. 2D and 3D T47D cultures were exposed to normoxic or hypoxic conditions in estrogen-deprived medium in the presence or absence of MG-132 (10 μM) for 8 h. HIF-1α (C, D) and ERα (E, F) protein levels were quantified with ELISA. Data represent the average ± SEM, from n ≥ 6 replicate cultures from two different cell passages. *p ≤ 0.05, ** p ≤ 0.01, *** p ≤ 0.001, **** p ≤ 0.0001.
Previous studies using monolayer cultures determined ERα loss under hypoxic conditions was mediated by the ubiquitin-proteasome system (UPS) [46, 47]. To confirm this finding, we treated both culture formats with MG-132 (10 μM), a selective proteasome inhibitor, for 8 h. Under normoxia, HIF-1α levels were significantly increased in both 2D (Fig. 4C) and 3D (Fig. 4D) formats, confirming proteasome inhibition. Under hypoxic conditions, ERα protein levels increased significantly in both 2D (Fig. 4E) and 3D (Fig. 4F) formats, confirming that ERα is, at least partially, regulated by the UPS in hypoxia.
Given the role of the UPS in regulating ERα under hypoxic conditions, we hypothesized that altered proteasome activity could explain the differences in ERα levels between the two culture formats. Liu et al. showed that proteasome activity in monolayer cultures of HUVECs and in lung tissue from a mouse model increased after exposure to hypoxia [48]. We measured 26S proteasome activity in cell lysates collected from both culture formats with a fluorescence-based assay. Hypoxia had no impact on proteasomal activity in either culture format, suggesting that the constant levels of ERα in 3D cultures are not caused by altered UPS activity (Fig. S6).
3.4. Hypoxia reduces ERα transcriptional activity in both 2D and 3D cultures
To determine if ERα protein levels were an accurate indicator of its transcriptional activity, we quantified luciferase activity in both culture formats with the ONE-Glo assay. Under normoxic and hypoxic conditions, the T47D cells had low basal luciferase activity in both culture formats (Fig. 5A and 5B). Upon treatment with a saturating concentration of E2 (10 nM), luciferase activity increased significantly in each setup. Under normoxic conditions, there was a 68-fold increase in 2D and a 32-fold increase in 3D. Under hypoxic conditions, the increases were muted with an 8-fold increase in 2D and a 7-fold increase in 3D. These results indicate that hypoxia significantly reduced ERα transcriptional activity in both culture formats.
Fig. 5.
ERα transcriptional activation is reduced under hypoxic conditions in both 2D and 3D culture formats. Both culture formats were incubated in E2-deprived (Veh) or -supplemented (E2) medium under normoxic or hypoxic conditions for 24 h. Luciferase activity was quantified using the ONE-Glo luciferase assay; the fold-change in luminescence relative to E2-deprived cultures under normoxia was plotted for 2D (A) and 3D (B) culture formats. Figures represent the average ± SEM, from n ≥ 6 replicates from three different cell passages. Total RNA was extracted and the relative expression of PGR mRNA (C, D) was determined using the ΔΔCt method; β-actin served as the reference gene. Data represent the average ± SEM, from n = 9 replicate cultures from three different cell passages *p ≤ 0.05, ** p ≤ 0.01, *** p ≤ 0.001, **** p ≤ 0.0001.
We also quantified the expression of the progesterone receptor (PGR), a canonical estrogen receptor target gene. Under normoxic conditions, basal PRG levels were similar in both culture formats (Table S3) and increased upon addition of E2 (Fig. 5C and 5D). In both culture formats, hypoxia significantly decreased PGR expression in E2-deprived and -supplemented medium, mirroring trends observed using the luciferase transcriptional activation assay.
3.5. ERα transcriptional activity is decreased by chemical stabilization of HIF-1α
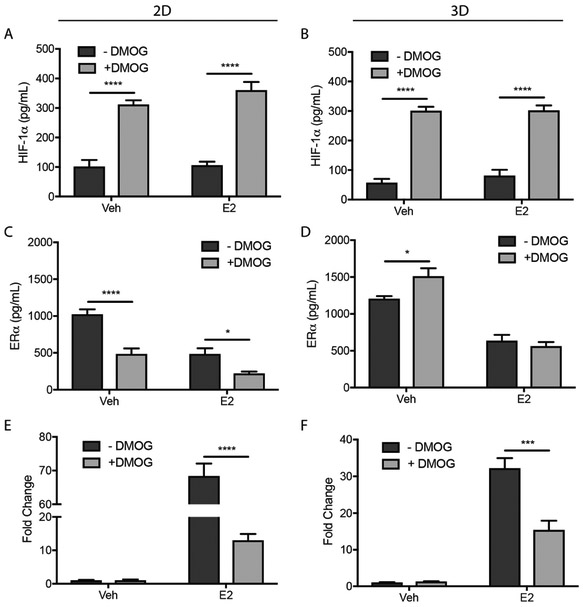
To determine whether ERα expression levels and transcriptional activity were mediated by HIF-1α or a different hypoxia-mediated mechanism (e.g., increased mitochondrial reactive oxygen species), we treated cultures with DMOG (1 mM). This small molecule stabilizes HIF-1α under normoxia by inhibiting PHD activity. ERα and HIF-1α protein levels in cell lysates were quantified with an ELISA; ERα transcriptional activity was measured with the ONE-Glo assay.
In both culture formats, DMOG stabilized HIF-1α levels in normoxia (Fig. 6A and 6B), confirming inhibition of PHD. The DMOG had culture-dependent effects, with significant decreases in ERα for 2D cultures in the presence and absence of E2 (Fig. 6C); a small but significant increase in ERα for 3D cultures in the absence of E2 (Fig. 6D); and no change in ERα in E2-treated cultures (Fig. 6D). In both culture formats DMOG decreased ERα transcriptional activation in the presence of E2 (Fig. 6E and 6F).
Fig. 6.
T47D cells in 2D and 3D culture formats were incubated in the absence or presence of 1 mM DMOG for 1 h before culture in E2-deprived (Veh) or -supplemented (E2) medium (± DMOG) for 24 h. ELISA was used to quantify HIF-1α (A, B) and ERα (C, D) protein levels which were normalized to β-actin. ERα transcriptional activity was measured using the ONE-Glo luciferase activity assay (E, F). Data represent the average ± SEM from n ≥ 6 replicate cultures from at least two different cell passages.
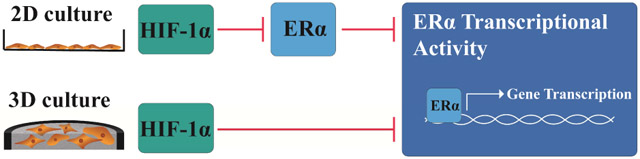
Together, these results suggest that HIF-1α is responsible for the significant reduction of ERα transcriptional activity observed under hypoxic conditions in both culture formats. HIF-1α is also responsible for the loss of ERα protein expression in 2D culture formats. Neither hypoxia nor stabilization of HIF-1α significantly impacted ERα protein expression in 3D culture formats.
4. Discussion
The dysregulated expression of ERα is common in tumors of hormone-sensitive tissues, including breast, ovary, and thyroid [4, 49-51]. Progression of ER (+) breast tumors from hormone-responsive to hormone-insensitive is associated with a poor prognosis, as the cancers no longer respond to adjuvant endocrine therapies [8, 52]. Clinical samples of hormone-insensitive ER (+) tumors co-stain for HIF-1α and ERα, suggesting that hypoxia has a role in promoting this transition [16, 17, 53]. Hypoxia also decreases hormone sensitivity in monolayer cultures. This decrease is coupled with decreased ERα protein levels, suggesting that activity is directly related to protein availability. In our experimental setup, we determined that culture format significantly altered ERα regulation in the T47D cell line, when exposed to hypoxic conditions.
In both formats, the cells underwent a canonical HIF-1α stabilization upon exposure to hypoxic conditions [9, 30, 54, 55]. The extent of this stabilization was four-fold greater in the 2D format, but was not reflected in overall transcriptional activity as both culture formats had a nearly six-fold increase in VEGF expression. We are unaware of a single study that directly compared HIF-1α stabilization between 2D and 3D culture formats, but discrepancies in transcriptional activity have been noted previously [26]. Cells in both formats had similar levels of HIF-1α under normoxic conditions, suggesting the cells in the 40-μm thick scaffolds were not hypoxic. Our previous characterization of a paper-based colon tumor model showed that scaffolds containing similar cell densities had oxygen tensions of less than 21% but were still within the clinically accepted values of normoxia [10, 33]. These results are also supported by previous work, which showed that hypoxia occurs in a tissue, multicellular aggregate, or collagen I suspension when the distance between the cells and the oxygen source exceeds 150 μm [10, 56, 57]. Based on previous comparisons of oxygen and glucose gradients in spheroids, we can also conclude that the cells in the paper scaffolds were not experiencing nutrient starvation [58].
In normoxia, the basal ERα levels and transcriptional activity were similar in both culture formats. The changes in ERα levels caused by hypoxia were, however, markedly different when moving from a 2D to a 3D culture format. Hypoxia decreased ESR1 levels in both culture formats, which is supported by previous studies in monolayers [19, 45], and suggests ERα is regulated at either the translational or post-translational level.
When exposed to hypoxic conditions, ERα levels were significantly reduced in the 2D format. A similar reduction also occurred in the presence of E2. These results agree with previous in vitro experiments [19, 20, 42]. The combination of hypoxia and E2 decreased ERα in monolayer cultures, but did not have an obvious additive or cooperative response. The use of a PHD inhibitor, which stabilized HIF-1α under normoxia, confirmed a HIF-1α-dependent process caused these changes in ERα. While E2 decreased ERα in the 3D cultures, neither hypoxia nor the stabilization of HIF-1α altered its expression.
While there are no direct comparisons of ERα expression between 2D and 3D cultures, others have evaluated ERα in spheroids. Truchet et al. found uniform levels of ERα across MCF-7 spheroids (~ 400 μm in diameter) that possessed proliferative gradients [59]. The addition of E2 decreased these levels across the spheroid, with no differences between the oxygen-rich periphery and the oxygen-poor core. These results support our observations that hypoxia had no impact on ERα expression in 3D culture. Counter to our findings, Munoz et al. showed that larger MCF-7 spheroids (~ 800 μm in diameter) lost ERα expression in all but a few cells along the periphery [60]. These studies, however, make it difficult to isolate the impacts of hypoxia on ERα expression from the multiple gradients known to form across spheroids of greater than 150 - 200 microns [57].
The observed decreases in ERα upon addition of E2 was expected because its transcriptional activation enables the subsequent proteolysis by the 26S proteasome [41]. Our proteasome inhibition studies confirmed that ERα was degraded by the UPS under normoxic and hypoxic conditions. Proteasomal activity was similar in both culture formats and unaffected by hypoxia, suggesting that the increased stability of ERα in the 3D culture environment is a result of differential post-translational regulation.
The insensitivity of ERα protein levels to hypoxia in the 3D culture format raises questions about the environmentally mediated regulation of the receptor at either the transcriptional or post-translational level. This increased stability of ERα in the 3D culture formats may be a result of transcriptional repression of E3 ubiquitin ligases [61-64]; increased levels of proteins that directly interact with ERα and increase its stability [65-69]; or altered kinase activities, which can alter the affinity of ERα to other proteins through an altered phosphorylation status [65, 66]. Numerous studies have observed altered kinase activities between 2D and 3D culture formats [23, 70]. It is also possible that the availability of shared E3 ubiquitin ligases or stabilizing proteins is different between 2D and 3D cultures, impacting the stability of ERα under hypoxic conditions [71].
Despite differential regulation of ERα protein levels in the 2D and 3D cultures, hypoxia decreased its transcriptional activity in both culture formats. We attribute the decreased luciferase activity in hypoxia to reduced transcriptional activity of ERα specifically and not to changes in overall translation efficiency. Previous reports support this assumption and show that translation efficiency in breast cancer cell lines is unaffected by moderate hypoxia [20, 72]. Using a selective inhibitor of PHD, we showed that this decrease is mediated by HIF-1α. One possibility for this decrease in activity is the competition for shared transcriptional coactivators (e.g., p300/CPB and SRC3) [73-75]. Alternatively, decreases in the amount of ERα relative to ERβ could result in a decrease in ERα transcriptional activity independent of absolute ERα protein levels [19, 76].
In summary, we highlight the importance of incorporating a tissue-like microenvironment when predicting cellular responses to hypoxia. By simply placing cells in a collagen matrix, we found a differential regulation of ERα expression levels. Hypoxia decreased the transcriptional activity of ERα in both culture formats. The significant decrease in ERα in 2D cultures suggested that loss of activity required loss of protein. The stabilization of ERα in 3D cultures, however, can explain both co-staining for HIF-1α and ERα and hormone insensitivity in clinical samples. Future studies on the hypoxia-mediated progression of breast cancer using a 3D culture platform will likely reveal novel insights into the role of hypoxia in regulating ERα protein levels and transcriptional activity, along with de novo and acquired resistance to endocrine therapies used for treatment.
Supplementary Material
Highlights:
ERα levels in 3D cultures are insensitive to hypoxia.
Hypoxia decreases ERα transcriptional activity in 2D and 3D culture formats.
HIF-1α mediates the observed hypoxic responses in both culture formats.
Acknowledgments
This work was supported by funds provided by the National Center for Advancing Translational Science (NCATS) through Grant Award Number UL1TR001111 and the National Institute of General Medical Science through Grant Award Number R35GM128697. NAW and RMK thank the Graduate School at UNC for support through Dissertation Completion Fellowships. ZWL and LA thank the NC Louis Stokes Alliance for Minority Participation, NSF Grant Number 1202467, for summer research support. We thank Professor Qing Zhang for access to his hypoxic culture chamber and Melanie Sinanian for helpful feedback and discussion.
Abbreviations:
- 2D
two dimensional
- 3D
three dimensional
- ERα
estrogen receptor alpha
- ER (+)
estrogen receptor positive
- ELISA
enzyme-linked immunosorbent assay
- HIF-1α
hypoxia inducible factor 1 alpha
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Cancer Facts & Figures, in: A.C. Society (Ed.) American Cancer Society, Atlanta, 2019. [Google Scholar]
- [2].Castoria G, Migliaccio A, Giovannelli P, Auricchio F, Cell proliferation regulated by estradiol receptor: Therapeutic implications, Steroids 75(8-9) (2010) 524–527. [DOI] [PubMed] [Google Scholar]
- [3].Heldring N, Pike A, Andersson S, Matthews J, Cheng G, Hartman J, Tujague M, Strom A, Treuter E, Warner M, Gustafsson JA, Estrogen receptors: How do they signal and what are their targets, Physiol. Rev 87(3) (2007) 905–931. [DOI] [PubMed] [Google Scholar]
- [4].Deroo BJ, Korach KS, Estrogen receptors and human disease, J. Clin. Invest 116(3) (2006) 561–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Bouris P, Skandalis SS, Piperigkou Z, Afratis N, Karamanou K, Aletras AJ, Moustakas A, Theocharis AD, Karamanos NK, Estrogen receptor alpha mediates epithelial to mesenchymal transition, expression of specific matrix effectors and functional properties of breast cancer cells, Matrix Biol. 43(1) (2015) 42–60. [DOI] [PubMed] [Google Scholar]
- [6].De Marchi T, Foekens JA, Umar A, Martens JWM, Endocrine therapy resistance in estrogen receptor (ER)-positive breast cancer, Drug Discov. Today 21(7) (2016) 1181–1188. [DOI] [PubMed] [Google Scholar]
- [7].Osborne CK, Schiff R, Mechanisms of endocrine resistance in breast cancer, Annu. Rev. Med 62(1) (2011) 233–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Musgrove EA, Sutherland RL, Biological determinants of endocrine resistance in breast cancer, Nat. Rev. Cancer 9(9) (2009) 631–643. [DOI] [PubMed] [Google Scholar]
- [9].Semenza GL, The hypoxic tumor microenvironment: A driving force for breast cancer progression, Biochim. Biophys. Acta, Mol. Cell Res 1863(3) (2016) 382–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].McKeown SR, Defining normoxia, physoxia and hypoxia in tumours: Implications for treatment respon, Br. J. Radiol 87(1035) (2014) 20130676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Vaupel P, Hockel M, Mayer A, Detection and characterization of tumor hypoxia using pO(2) histography, Antioxid. Redox. Signal 9(8) (2007) 1221–1235. [DOI] [PubMed] [Google Scholar]
- [12].Gilkes DM, Semenza GL, Role of hypoxia-inducible factors in breast cancer metastasis, Future Oncol. 9(11) (2013) 1623–1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Hickey MM, Simon MC, Regulation of angiogenesis by hypoxia and hypoxia-inducible factors, Curr. Top. Dev. Biol 76(1) (2006) 217–257. [DOI] [PubMed] [Google Scholar]
- [14].Chiche J, Brahimi-Horn MC, Pouyssegur J, Tumor hypoxia induces a metabolic shift causing acidosis: A common feature in cancer, J. Cell. Mol. Med 14(4) (2010) 771–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Semenza GL, HIF-1 and mechanisms of hypoxia sensing, Curr. Opin. Cell Biol 13(2) (2001) 167–171. [DOI] [PubMed] [Google Scholar]
- [16].Generali D, Berruti A, Brizzi MP, Camp L, Bonardi S, Wigfield S, Bersiga A, Allevi G, Milani M, Aguggini S, Gandolfi V, Dogliotti L, Bottini A, Harris AL, Fox SB, Hypoxia-inducible factor-1 alpha-expression predicts a poor response to primary chemoendocrine therapy and disease-free survival in primary human breast cancer, Clin. Cancer Res 12(15) (2006) 4562–4568. [DOI] [PubMed] [Google Scholar]
- [17].Bos R, van der Groep P, Greijer AE, Shvarts A, Meijer S, Pinedo HM, Semenza GL, van Diest PJ, van der Wall E, Levels of hypoxia-inducible factor-1 alpha independently predict prognosis in patients with lymph node negative breast carcinoma, Cancer 97(6) (2003) 1573–1581. [DOI] [PubMed] [Google Scholar]
- [18].Bos R, Zhong H, Hanrahan CF, Mommers ECM, Semenza GL, Pinedo HM, Abeloff MD, Simons JW, van Deist PJ, van der Wall E, Levels of hypoxia-inducible factor-1a during breast carcinogenesis, JNCI, J. Natl. Cancer Inst 93(4) (2001) 309–314. [DOI] [PubMed] [Google Scholar]
- [19].Wolff M, Kosyna FK, Dunst J, Jelkmann W, Depping R, Impact of hypoxia inducible factors on estrogen receptor expression in breast cancer cells, Arch. Biochem. Biophys 613 (2017) 23–30. [DOI] [PubMed] [Google Scholar]
- [20].Padro M, Louie RJ, Lananna BV, Krieg AJ, Timmerman LA, Chan DA, Genome-independent hypoxia repression of estrogen receptor alpha in breast cancer cells, BMC Cancer 17 (2017) 203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Yamada KM, Cukierman E, Modeling tissue morphologies and cancer in 3D, Cell 130(4) (2007) 601–610. [DOI] [PubMed] [Google Scholar]
- [22].Vantangoli MM, Madnick SJ, Huse SM, Weston P, Boekelheide K, MCF-7 human breast cancer cells form differentiated microtissues in scaffold-free hydrogels, PLoS Med. 10(8) (2015) e0135426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Breslin S, O'Driscoll L, The relevance of using 3D cell cultures, in addition to 2D monolayer cultures, when evaluating breast cancer drug sensitivity and resistance, Oncotarget 7(29) (2016) 45745–45756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Barcellos-Hoff MH, Aggeler J, Ram TG, Bissell MJ, Functional differentiation and alveolar morphogenesis of primary mammary cultures on reconstituted basement membrane, Development 105(2) (1989) 223–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Vantangoli MM, Madnick SJ, Wilson S, Boekelheide K, Estradiol exposure differentially alters monolayer versus microtissue MCF-7 human breast carcinoma cultures, PLoS One 11(7) (2016) e0157997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].DelNero P, Lane M, Verbridge SS, Kwee B, Kermani P, Hempstead B, Stroock A, Fischbach C, 3D culture broadly regulates tumor cell hypoxia response and angiogenesis via pro-inflammatory pathways, Biomaterials 55(1) (2015) 110–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Whitman NA, Lin ZW, DiProspero TJ, McIntosh JC, Lockett MR, Screening estrogen receptor modulators in a paper-based breast cancer model, Anal. Chem 90(20) (2018) 11981–11988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Kenney RM, Boyce MW, Truong AS, Bagnell CR, Lockett MR, Real-time imaging of cancer cell chemotaxis in paper-based scaffolds, Analyst 141(2) (2016) 661–8. [DOI] [PubMed] [Google Scholar]
- [29].Kenney RM, Boyce MW, Whitman NA, Kromhout BP, Lockett MR, A pH-sensing optode for mapping spatiotemporal gradients in 3D paper-based cell cultures, Anal. Chem 90(3) (2018) 2376–2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Truong AS, Lockett MR, Oxygen as a chemoattractant: confirming cellular hypoxia in paper-based invasion assays, Analyst 141(12) (2016) 3874–3882. [DOI] [PubMed] [Google Scholar]
- [31].Camci-Unal G, Newsome D, Eustace BK, Whitesides GM, Fibroblasts enchance migration of human lung cancer cells in paper-based coculture system, Adv. Healthcare Mater 5(6) (2016) 641–647. [DOI] [PubMed] [Google Scholar]
- [32].Simon KA, Mosadegh B, Minn KT, Lockett MR, Mohammady MR, Boucher DM, Hall AB, Hillier SM, Udagawa T, Eustace BK, Whitesides GM, Metabolic response of lung cancer cells to radiation in a paper-based 3D cell culture system, Biomaterials 95(1) (2016) 47–59. [DOI] [PubMed] [Google Scholar]
- [33].Boyce MW, LaBonia GJ, Hummon AB, Lockett MR, Assessing chemotherapeutic effectiveness using a paper-based tumor model, Analyst 142(15) (2017) 2819–2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Rodenhizer D, Gaude E, Cojocari D, Mahadevan R, Frezza C, Wouters BG, McGuigan AP, A three-dimensional engineered tumour for spatial snapshot analysis of cell metabolism and phenotype in hypoxic gradients, Nat. Mater 15(2) (2016) 227–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Young MK, Rodenhizer D, Dean T, D'Arcangelo E, Xu B, Ailles L, McGuigan AP, A TRACER 3D co-culture tumour model for head and neck cancer, Biomaterials 164(1) (2018) 54–69. [DOI] [PubMed] [Google Scholar]
- [36].Wilson VS, Bobseine K, Gray LE, Development and characterization of a cell line that stably expresses an estrogen-responsive luciferase reporter for the detection of estrogen receptor agonist and antagonists, Toxicol. Sci 81(1) (2004) 69–77. [DOI] [PubMed] [Google Scholar]
- [37].Otsu N, Threshold selection method from gray-level histograms, IEEE Trans. Syst. Man. Cybern 9(1) (1979) 62–66. [Google Scholar]
- [38].Livak KJ, Schmittgen TD, Analysis of relative gene expression data using real-time quantitative PCR and the 2(T)(-Delta Delta C) method, Methods 25(4) (2001) 402–408. [DOI] [PubMed] [Google Scholar]
- [39].Alarid ET, Bakopoulos N, Proteasome-mediated proteolysis of estrogen receptor: A novel component in autologous down-regulation, Mol. Endocrinol 13(9) (1999) 1522–1534. [DOI] [PubMed] [Google Scholar]
- [40].Eckert RL, Mullick A, Rorke EA, Katzenellenbogen BS, Estrogen-receptor syntehsis and turnover in MCF-7 breast-cancer cells measured by a density shift technique, Endocrinology 114(2) (1984) 629–637. [DOI] [PubMed] [Google Scholar]
- [41].Valley CC, Solodin NM, Powers GL, Ellison SJ, Alarid ET, Temporal variation in estrogen receptor-alpha protein turnover in the presence of estrogen, J. Mol. Endocrinol 40(1-2) (2008) 23–34. [DOI] [PubMed] [Google Scholar]
- [42].Cho JY, Kim D, Lee SK, Lee YJ, Cobalt chloride-induced estrogen receptor alpha down-regulation involves hypoxia-inducible factor-1 alpha in MCF-7 human breast cancer cells, Mol. Endocrinol 19(5) (2005) 1191–1199. [DOI] [PubMed] [Google Scholar]
- [43].Lang JD, Berry SM, Powers GL, Beebe DJ, Alarid ET, Hormonally responsive breast cancer cells in a microfluidic co-culture model as a sensor of microenvironmental activity, Integ. Biol 5(5) (2013) 807–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Doe MR, Ascano JM, Kaur M, Cole MD, Myc posttranscriptionally induces HIF1 protein and target gene expression in normal and cancer cells, Cancer Res. 72(4) (2012) 949–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Ryu K, Park C, Lee Y, Hypoxia-inducible factor 1 alpha represses the transcription of the estrogen receptor alpha gene in human breast cancer cells, Biochem. Biophys. Res. Commun 407(4) (2011) 831–836. [DOI] [PubMed] [Google Scholar]
- [46].Stoner M, Saville B, Wormke M, Dean D, Burghardt R, Safe S, Hypoxia induces proteasome-dependent degradation of estrogen receptor a in ZR-75 breast cancer cells, Mol. Endocrinol 16(10) (2002) 2231–2242. [DOI] [PubMed] [Google Scholar]
- [47].Cooper C, Liu GY, Niu YL, Santos S, Murphy LC, Watson PH, Intermittent hypoxia induces proteasome-dependent down-regulation of estrogen receptor in human breast carcinoma, Clin. Cancer Res 10(24) (2004) 8720–8727. [DOI] [PubMed] [Google Scholar]
- [48].Liu H, Wang Z, Yu S, Xu J, Proteasomal degradation of O-GlcNAc transferase elevates hypoxia-induced vascular endothelial inflammatory response, Cardiovasc. Res 103(1) (2014) 131–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Di Vito M, De Santis E, Perrone GA, Mari E, Giordano MC, De Antoni E, Coppola L, Fadda G, Tafani M, Carpi A, Russo MA, Overexpression of estrogen receptor-alpha in human papillary thyroid carcinomas studied by laser-capture microdissection and molecular biology, Cancer Sci. 102(10) (2011) 1921–1927. [DOI] [PubMed] [Google Scholar]
- [50].Tafani M, Russo A, Di Vito M, Sale P, Pellegrini L, Schito L, Gentileschi S, Bracaglia R, Marandino F, Garaci E, Up-regulation of pro-inflammatory genes as adaptation to hypoxia in MCF-7 cells in human mammary invasive carcinoma microenvironment, Cancer Sci. 101(4) (2010) 1014–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Chan KKL, Siu MKY, Jiang YX, Wang JJ, Wang Y, Leung THY, Liu SS, Cheung ANY, Ngan HYS, Differential expression of estrogen receptor subtypes and variants in ovarian cancer: Effects on cell invasion, proliferation, and prognosis, BMC Cancer 17(1) (2017) 606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Lopez-Tarruella S, Schiff R, The dynamics of estrogen receptor status in breast cancer: Re-shaping the paradigm, Clin. Cancer Res 13(23) (2007) 6921–6925. [DOI] [PubMed] [Google Scholar]
- [53].Generali D, Buffa FM, Berruti A, Brizzi MP, Campo L, Bonardi S, Bersiga A, Allevi G, Milani M, Aguggini S, Papotti M, Dogliotti L, Bottini A, Harris AL, Fox SB, Phosphorylated ERalpha, HIF-1alpha, and MAPK signaling as predictors of primary endocrine treatment response and resistance in patients with breast cancer, J. Clin. Oncol 27(2) (2009) 227–234. [DOI] [PubMed] [Google Scholar]
- [54].Lehmann S, Te Boekhorst V, Odenthal J, Bianchi R, van Helvert S, Ikenberg K, Ilina O, Stoma S, Xandry J, Jiang L, Grenman R, Rudin M, Friedl P, Hypoxia induces a HIF-1-dependent transition from collective-to-amoeboid dissemination in epithelial cancer cells, Curr. Biol 27(3) (2017) 392–400. [DOI] [PubMed] [Google Scholar]
- [55].Daster S, Amatruder N, Calabrese D, Ivanek R, Turrini E, Droesser RA, Zajac P, Fimognari C, Spagnoli GC, Iezzi G, Mele V, Muraro MG, Induction of hypoxia and necrosis in multicellular tumor spheroids is associated with resistance to chemotherapy treatment, Oncotarget 8(1) (2017) 1725–1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Szot CS, Buchanan CF, Freeman JW, Rylander MN, 3D in vitro bioengineered tumors based on collagen I hydrogels, Biomaterials 32(31) (2011) 7905–7912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Hirschhaeuser F, Menne H, Dittfeld C, West J, Mueller-Klieser W, Kunz-Schughart LA, Multicellular tumor spheroids: An underestimated tool is catching up again, J. Biotechnol 148(1) (2010) 3–15. [DOI] [PubMed] [Google Scholar]
- [58].Milotti E, C. R., Emergent properties of tumor microenvironment in a real-life model of multicell tumor spheroids, PLoS One 5(11) (2010) e13942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Truchet I, Jozan S, Baron S, Frongia C, Balaguer P, Richard-Foy H, Valette A, Estrogen and antiestrogen-dependent regulation of breast cancer cell proliferation in multicellular spheroids: Influence of cell microenvironment, Int. J. Oncol 32(5) (2008) 1033–1039. [DOI] [PubMed] [Google Scholar]
- [60].Munoz L, Espinosa M, Quintanar-Jurado V, Hildago A, Melendez-Zajgla J, Maldonado V, Paradoxial changes in the expression of estrogen receptor alpha in breast cancer multicellular spheroids, Tissue Cell 42(5) (2010) 344–347. [DOI] [PubMed] [Google Scholar]
- [61].Bhatt S, Xiao Z, Meng Z, Katzenellenbogen BS, Phosphorylation by p38 mitogen-activated protein kinase promotes estrogen receptor alpha turnover and functional activity via the SCF(Skp2) proteasomal complex, Mol. Cell Biol 32(10) (2012) 1928–1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Eakin CM, Maccoss MJ, Finney GL, Klevit RE, Estrogen receptor alpha is a putative substrate for the BRCA1 ubiquitin ligase, Proc. Natl. Acad. Sci. USA 104(14) (2007) 5794–5799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Fan M, Park A, Nephew KP, CHIP (carboxyl terminus of Hsc70-interacting protein) promotes basal and geldanamycin-induced degradation of estrogen receptor-alpha, Mol. Endocrinol 19(12) (2005) 2901–2914. [DOI] [PubMed] [Google Scholar]
- [64].Sun J, Zhou W, Kaliappan K, Nawaz Z, Slingerland JM, ERα phosphorylation at Y537 by Src triggers E6-AP-ERα binding, ERα ubiquitylation, promoter occupancy, and target gene expression, Mol. Endocrinol 26(9) (2012) 1567–1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Wei X, Xu H, Kufe D, MUC1 oncoprotein stabilizes and activates estrogen receptor alpha, Mol. Cell 21(2) (2006) 295–305. [DOI] [PubMed] [Google Scholar]
- [66].Rajbhandari P, Schalper KA, Solodin NM, Ellison-Zelski SJ, Ping Lu K, Rimm DL, Alarid ET, Pin1 modulates ERα levels in breast cancer through inhibition of phosphorylation-dependent ubiquitination and degradation, Oncogene 33(11) (2014) 1438–1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Grisouard J, Medunjanin S, Hermani A, Shukla A, Mayer D, Glycogen synthase kinase-3 protects estrogen receptor alpha from proteasomal degradation and is required for full transcriptional activity of the receptor, Mol. Endocrinol 21(10) (2007) 2427–2439. [DOI] [PubMed] [Google Scholar]
- [68].Giamas G, Filipovic A, Jacob J, Messier W, Zhang H, Yang D, Zhang W, Shifa BA, Photiou A, Tralau-Stewart C, Castellano L, Green AR, Coombes RC, Ellis IO, Ali S, Lenz HJ, Stebbing J, Kinome screening for regulators of the estrogen receptor identifies LMTK3 as a new therapeutic target in breast cancer, Nat. Med 17(6) (2011) 715–719. [DOI] [PubMed] [Google Scholar]
- [69].He X, Zheng Z, Song T, Wei C, Ma H, Ma Q, Zhang Y, Xu Y, Shi W, Ye Q, Zhong H, c-Abl regulates estrogen receptor alpha transcription activity through its stabilization by phosphorylation, Oncogene 29(15) (2010) 2238–2251. [DOI] [PubMed] [Google Scholar]
- [70].Weigelt B, Lo AT, Park CC, Gray JW, Bissell MJ, HER2 signaling pathway activation and response of breast cancer cells to HER2-targeting agents is dependent strongly on the 3D microenvironment, Breast Cancer Res. Treat 122(1) (2010) 35–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Jung YS, Lee SJ, Yoon MH, Ha NC, Park BJ, Estrogen receptor α is a novel target of the Von Hippel-Lindau protein and is responsible for the proliferation of VHL-deficient cells under hypoxic conditions, Cell Cycle 11(23) (2012) 4462–4473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Connolly E, Braunstein S, Formenti S, Schneider RJ, Hypoxia inhibits protein synthesis through 4E-BP1 and elongation factor 2 kinase pathway controlled by mTOR and uncoupled in breast cancer cells, Mol. Cell Biol 26(10) (2006) 3955–3965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Klinge CM, Estrogen receptor interaction with co-activators and co-repressors, Steroids 65(6) (2000) 2271–2251. [DOI] [PubMed] [Google Scholar]
- [74].Arany Z, Huang LE, Eckner R, Bhattacharya S, Jiang C, Goldberg MA, Bunn HF, Livingston DM, An essential role for p300/CBP in the cellular response to hypoxia, Proc. Natl. Acad. Sci. USA 93(23) (1996) 12969–12973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Zhao W, Chang C, Cui Y, Zhao X, Yang J, Shen L, Zhou J, Hou Z, Zhang Z, Ye C, Hasenmayer D, Perkins R, Huang X, Yao X, Yu L, Huang R, Zhang D, Guo H, Yan J, Steroid receptor coactivator-3 regulates glucose metabolism in bladder cancer cells through coactivation of hypoxia inducible factor 1α, J. Biol. Chem 289(16) (2014) 11219–11229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Sotoca AM, van den Berg H, Vervoort J, van der Saag P, Strom A, Gustafsson JA, Rietjens I, Murk AJ, Influence of cellular ERalpha/ERbeta ratio on the ERalpha-agonist induced proliferation of human T47D breast cancer cells, Toxicol. Sci 105(2) (2008) 303–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.