Abstract
Positron Emission Tomography (PET) is an important neuroimaging tool to quantify the distribution of specific molecules in the brain. The quantification is based on a series of individually designed data preprocessing steps (pipeline) and an optimal preprocessing strategy is per definition associated with less noise and improved statistical power, potentially allowing for more valid neurobiological interpretations. In spite of this, it is currently unclear how to design the best preprocessing pipeline and to what extent the choice of each preprocessing step in the pipeline minimizes subject-specific errors.
To evaluate the impact of various preprocessing strategies, we systematically examined 384 different pipeline strategies in data from 30 healthy participants scanned twice with the serotonin transporter (5-HTT) radioligand [11C]DASB. Five commonly used preprocessing steps with two to four options were investigated: (1) motion correction (MC) (2) co-registration (3) delineation of volumes of interest (VOI’s) (4) partial volume correction (PVC), and (5) kinetic modeling. To quantitatively compare and evaluate the impact of various preprocessing strategies, we used the performance metrics: test-retest bias, within- and between-subject variability, the intraclass-correlation coefficient, and global signal-to-noise ratio. We also performed a power analysis to estimate the required sample size to detect either a 5% or 10% difference in 5-HTT binding as a function of preprocessing pipeline.
The results showed a complex downstream dependency between the various preprocessing steps on the performance metrics. The choice of MC had the most profound effect on 5-HTT binding, prior to the effects caused by PVC and kinetic modeling, and the effects differed across VOI’s. Notably, we observed a negative bias in 5-HTT binding across test and retest in 98% of pipelines, ranging from 0–6% depending on the pipeline. Optimization of the performance metrics revealed a trade-off in within- and between-subject variability at the group-level with opposite effects (i.e. minimization of within-subject variability increased between-subject variability and vice versa). The sample size required to detect a given effect size was also compromised by the preprocessing strategy, resulting in up to 80% increases in sample size needed to detect a 5% difference in 5-HTT binding.
This is the first study to systematically investigate and demonstrate the effect of choosing different preprocessing strategies on the outcome of dynamic PET studies. We provide a framework to show how optimal and maximally powered neuroimaging results can be obtained by choosing appropriate preprocessing strategies and we provide recommendations depending on the study design.
In addition, the results contribute to a better understanding of methodological uncertainty and variability in preprocessing decisions for future group- and/or longitudinal PET studies.
Keywords: Positron Emission Tomography, preprocessing, head motion, optimization, partial volume correction, kinetic modeling, test-retest, [11C]DASB
INTRODUCTION
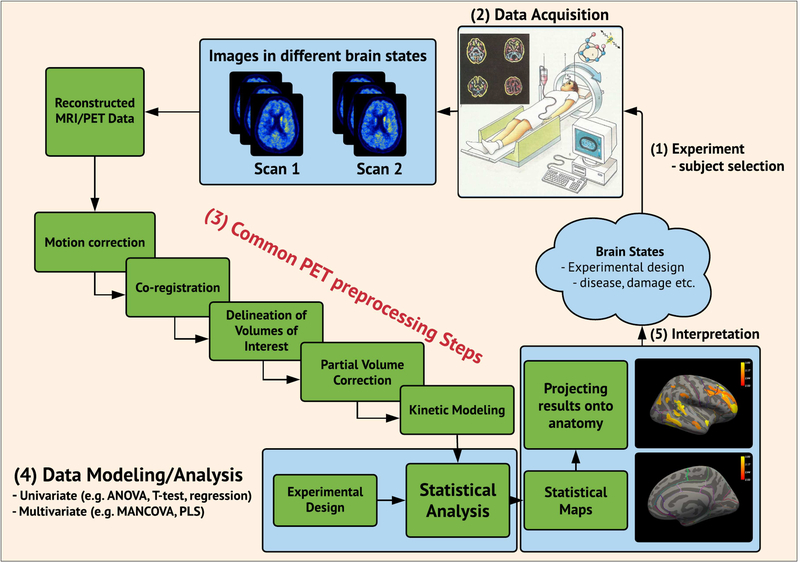
Positron Emission Tomography (PET) is a state-of-the-art neuroimaging tool for quantification of the in vivo spatial distribution of specific molecules in the brain. It has long been recognized that precise quantification using a series of individually designed preprocessing steps (“pipeline”) is a critical part of a PET analysis framework, and as part of the validation of new PET radioligands, these are often preprocessed with different kinetic models and at different scan lengths. The outcomes are often examined in a test-retest setting (Parsey et al. 2000, Ginovart et al. 2001) under the implicit assumption that test and retest should generate similar outcomes. However, despite the importance and usefulness of validating kinetic models and scan length, the impact of several other important choices such as preprocessing strategies for delineating volumes of interest (VOI), whether to apply motion correction (MC), how to accurately perform co-registration, and whether to use partial volume correction (PVC), remain unresolved (Figure 1).
Figure 1:
Flowchart depicting a common pipeline for neuroimaging studies (multimodal PET and MRI) and its multiple stages ranging from (1) experimental design / subject selection, (2) data acquisition, (3) preprocessing, (4) data modeling/analysis, and (5) interpretation.
As a result, centres or even individual scientists often design their own unique preprocessing strategy (Nørgaard et al. 2019), with each choice potentially compromising one another to affect performance (e.g. NRM Grand Challenge 2018, www.petgrandchallenge.com).
However, in the absence of a “ground truth”, it remains a major challenge in PET to optimize preprocessing strategies, and it may take alternative performance metrics to quantitatively evaluate and compare various preprocessing strategies (Strother et al. 2002, Churchill et al. 2015). The uptake of radioligand in the brain varies across both regions and subjects, but also between scan sessions (Frankle et al. 2005). Therefore, there exists no unifying pattern of radioligand uptake that can be predicted and generalized to the population. Simulations can overcome these latter limitations of real data by providing the “ground truth”, having knowledge about the true underlying data generating process (Ichise et al. 2003). However, while simulations can be instructive, it is obviously very difficult to simulate the complex spatio-temporal noise patterns arising from a PET scanner. Simulations therefore provide only limited information on preprocessing effects.
In this study, we extend the work of previous validation studies of the radioligand [11C]DASB, which binds to the serotonin transporter (5-HTT), a target for many anti-depressive drugs (Houle et al. 2000, Meyer et al. 2001). Previously published [11C]DASB PET papers have mainly used five preprocessing steps with multiple levels of options within each preprocessing step (Figure 1). However, while there is some consensus on the main preprocessing steps (MC, co-registration, VOI, PVC, and kinetic modeling), there is less consensus on the details within each step. In addition, with new methodological improvements continually being developed and refined (Zanderigo et al. 2017, Gryglewski et al. 2017) it may also be difficult to establish an optimal pipeline, with each choice potentially compromising one another. Nevertheless, for scientific, ethical and economical reasons it is important to know how the choice of preprocessing strategy influences the noise levels and thereby the sample size required to establish e.g. group differences. Inspired by the previously published preprocessing strategies for the radioligand [11C]DASB (Nørgaard et al. 2019), in this work we will focus on three key questions:
are measures of 5-HTT BPND using [11C]DASB robustly determined across a wide range of preprocessing strategies? The robustness will be estimated using the performance metrics; test-retest bias, within- and between-subject variability, global signal-to-noise ratio (gSNR), and intraclass correlation coefficient (Kim et al. 2006, Strother et al. 2002).
does optimization of the performance metrics result in a detectable tradeoff in within- and between-subject variability at the group level?
can study power be enhanced by optimized preprocessing of [11C]DASB?
We specifically chose to focus on the PET radioligand [11C]DASB because of its widespread use to study various aspects of brain function, but more importantly because the foundation for selecting a given preprocessing strategy seems to be an overlooked aspect in modern PET neuroscience (Nørgaard et al. 2019).
MATERIALS AND METHODS
1.1. Participants
A total of N=30 healthy women (mean age: 25 ± 5.9 years, range: 18 – 37) were recruited from a previous randomized, placebo-controlled and double-blind intervention study investigating the role of 5-HTT changes in depressive responses to sex-steroid hormone manipulation (Frokjaer et al. 2015). The women served as a control group receiving placebo treatment only (a saline injection), i.e., the data is considered to represent test-retest without any expected changes in [11C]DASB binding. All participants were blinded and the blinding was successful as specified in detail in Frokjaer et al. 2015. The study by Frokjaer et al. 2015 was designed to capture brain chemistry in two consecutive follicular phases of the menstrual cycle and participants were therefore planned to be re-scanned 23–35 days after their baseline cycle scan (depending on their follow-up cycle). Three participants were scanned one cycle-period later (61 days, 70 days, 56 days), one participant two periods later (92 days), and one participant three periods later (122 days). The midfollicular timing of the scan was kept in all participants. All the remaining 25 participants were scanned in a cycle-period ranging between 27 and 37 days. In addition, participants were scanned at similar time of the day in scan 1 and scan 2, eliminating potential diurnal effects. Additional information can be found in Frokjaer et al. 2015. The study was registered and approved by the local ethics committee (protocol-ID: H-2–2010-108). All participants gave written informed consent.
1.2. Magnetic Resonance Imaging Acquisition
An anatomical 3D T1-weighted MP-RAGE sequence with matrix size = 256 × 256 × 192; voxel size = 1 × 1 × 1 mm; TR/TE/TI = 1550/3.04/800 ms; flip angle = 9° was acquired for all patients using a Siemens Magnetom Trio 3T MR scanner or a Siemens 3T Verio MR scanner. In addition, a 3D T2-weighted isotropic sagittal sequence with matrix size 256 × 256 × 176; voxel size = 1 × 1 × 1 mm; TR/TE = 3200/409 ms; flip angle = 120° was also acquired for all subjects. All single-subject MRI sequences were corrected for gradient nonlinearities according to Jovicich et al. 2006, in order to correct for spatial distortions and achieve optimal PET-MR co-registration. All the acquired MR images were examined for structural abnormalities, as a criterion for subject inclusion.
1.3. Positron Emission Tomography using [11C]DASB
All patients were scanned using a Siemens ECAT High-Resolution Research Tomography (HRRT) scanner operating in 3D list-mode and with the highly selective radioligand [11C]DASB. The imaging protocol consisted of a single-bed, 90 minutes transmission acquisition post injection of 587 ± 30 (mean ± SD) MBq, range 375–612 MBq, bolus into an elbow vein. PET data was reconstructed into 36 frames (6×10, 3×20, 6×30, 5×60, 5×120, 8×300, 3×600 seconds) using a 3D-OSEM-PSF algorithm with TXTV based attenuation correction (image matrix, 256 × 256 × 207; voxel size, 1.22 × 1.22 × 1.22 mm) (Sureau et al. 2008, Keller et al. 2013).
1.4. Preprocessing Steps for PET and MRI
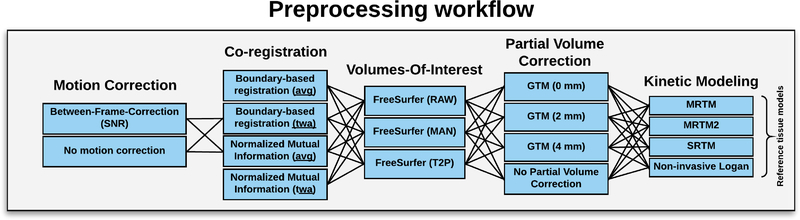
Here we establish a 5-step pipeline, each step with two to four options, to estimate the outcome measure BPND. All the individual procedures have previously been used in published [11C]DASB PET studies, except for PVC using the GTM. The steps are listed below in the order in which they were applied. Specific rationales for including/excluding each unique preprocessing step and their options are listed below.
Step 1 – Motion correction (2 options)
Within-scan PET motion correction was executed using a data-driven automated image registration (AIR v. 5.2.5, http://loni.usc.edu/Software/AIR). Prior to alignment, each frame was smoothed using a 10 mm Gaussian 3D kernel and thresholded at the 20-percentile level to boost SNR. Alignment parameters were estimated for the smoothed PET frames 10–36 to a reference frame with high SNR (frame 26) using a scaled least squares cost-function in AIR. Subsequently, the non-smoothed frames were transformed using the estimated alignment parameters and resliced into a 4D motion corrected data set (e.g., as applied in Frokjaer et al. 2015 and Beliveau et al. 2017). The motion correction estimation for frame 10 was applied to the first 9 frames. We chose to register frames 10–36 only, because the first 9 time frames (10/20 sec) have low count statistics, high noise levels and have shown to produce highly variable alignment parameters.
Criterion for acceptable motion (quality control) was a median movement less than 3 mm across frames, as estimated by the median of the sum of the squared translations (x,y,z) across all voxels. All 30 participants had acceptable median motion below 3 mm. The MC process was fully automated according to parameters defined in previous work (e.g. Frokjaer et al. 2015, Beliveau et al. 2017). The rationale for testing the effect of MC in the pipeline is because motion artefacts vary by dataset. Furthermore, MC should ultimately control motion artefacts, but may also impose unwanted biases on the data or reduce experimental power, especially in cases of minor or no head movement (Churchill et al. 2012). In addition, Nørgaard et al. 2019 showed that MC lowers between-subject variability in striatum, resulting in 26% fewer subjects needed in a group analysis to achieve similarly power statistical tests. It is therefore of interest to validate this observation in an independent dataset.
Step 2 – Co-registration (4 options)
All single-subject PET frames were initially either summed (according to their frame length i.e. integral) or averaged over all time frames to estimate a time-weighted (twa) or averaged (avg) 3D image for rigid-body transformation co-registration. Two different co-registration techniques were subsequently applied to either the twa or the avg image, namely Normalized Mutual Information (NMI, Studholme et al. 1999) or Boundary-Based Registration (BBR, Greve et al. 2009). Each co-registration technique depends on a cost-function, seeking to minimize the registration error of the PET and MRI image alignment. Both NMI and BBR use mutual information as their cost function, but BBR puts an additional cost on the boundaries being aligned.
The co-registation step is explicitly evaluated in this work, as its effects may vary by dataset and as a function of the spatial and temporal distribution of the PET signal. For example, 5-HTT is only modestly expressed in the neocortex, and BBR may therefore not be the best algorithm to align cortical folding patterns, especially not if the resolution of the PET scanner is limited.
Step 3 – Delineation of Volumes of Interest (3 options)
All MRI scans were processed using FreeSurfer (http://surfer.nmr.mgh.harvard.edu, version 5.3). FreeSurfer contains a fully automatic structural imaging pipeline for processing of cross-sectional as well as longitudinal data. Furthermore, it includes several features such as skull stripping, B1 bias field correction, non-linear registration to a stereotaxic atlas, statistical analysis of morphometric differences, and probabilistic labeling of cortical/subcortical brain structures based on the Desikan-Killiany atlas (Fischl et al. 2004). A total of 28 subcortical and cortical regions were extracted, and averaged across hemispheres producing a final sample of 14 regions pr. subject/pipeline. The volumetric regions included the amygdala, thalamus, putamen, caudate, anterior cingulate cortex (ACC), hippocampus, orbital frontal cortex, superior frontal cortex, occipital cortex, superior temporal gyrus, insula, inferior temporal gyrus, parietal cortex, and entorhinal cortex. We chose these regions because they largely cover the entire brain, but also because many of the regions have been used in previously published DASB PET studies. Out of more than 100 published DASB PET studies (Nørgaard et al. 2019), each region is mentioned N times: amygdala (N=72), thalamus (N=105), putamen (N=88), caudate (N=82), ACC (N=74), hippocampus (N=71), frontal cortex (N=66), occipital cortex (N=48), temporal cortex (N=58), parietal cortex (N=34), entorhinal cortex (N=16). Subsequently to running the FreeSurfer pipeline, the researcher can choose to perform user-dependent manual edits to the FreeSurfer output, to correct for errors mostly located in the white matter (WM), cerebrospinal fluid (CSF) or on the pial surface. The manual editing was carried out according to FreeSurfer recommendations (https://surfer.nmr.mgh.harvard.edu/fswiki/Edits). To minimize eventual rater bias, the manual edits were carried out by two experts (Beliveau et al. 2017). For each MRI, manual edits were initially carried out by the two experts independently. Next, the manual edits from each expert were evaluated jointly by the two experts and a consensus was reached on the optimal edits. If a T2-weighted MRI is also available, semi user-independent edits can also be made to the FreeSurfer output by re-running the FreeSurfer reconstruction including the T2-weighted MRI. We examined all three pipelines in this study and now refer to these as FS-RAW (standard output from FreeSurfer), FS-MAN (output from FreeSurfer with manual edits) and FS-T2P (output from FreeSurfer with the T2 stream). Only the first test-scan MRI was used for the analysis. Different FreeSurfer options are tested, as the optimal correction has been reported to vary as a function of subject and scanner (McCarthy et al. 2015). Although choice of atlas (e.g. PVElab, AAL or MNI305) may have an impact on the outcome, we considered assessment of various atlas choices to be beyond the scope of the current work and we consistently applied the Desikan-Killiany atlas provided in FreeSurfer. However, as it is also common to include a normalization step to standard space and subsequently extract VOIs using a volumetric atlas, an evaluation and comparison of such a pipeline can be found in the supplemental material.
Step 4 – Partial Volume Correction (4 options)
The data were analyzed either without or with three partial volume correction (PVC) approaches. The VOI-based PVC technique, Geometric Transfer Matrix (GTM), by Rousset et al. 1998 was applied using PETsurfer (https://surfer.nmr.mgh.harvard.edu/fswiki/PetSurfer), establishing a forward linear model relating [11C]DASB intensities to the VOI means, as described in Greve et al. 2016. Because the PSF for a HRRT scanner reconstructed with a OP-OSEM-PSF algorithm varies from 1–2.5 mm in radial orientation depending on the distance from the centre of the field of view (Olesen et al. 2009), we ran the analyses with the PSF settings; 0 mm and 2 mm. However, because motion, inhomogeneous tracer uptake and varying uptake across frames are likely to further decrease the spatial resolution as compared to a point source in Olesen et al. 2009, we also ran the PVC analyses with 4 mm, as used in Greve et al. 2014. The PVC step is evaluated, because it has been suggested to be the optimal solution for VOI analysis, given that assumptions about the PSF, accurate delineation of regions, correct PET-MRI registration, and constant uptake within each VOI are satisfied (Greve et al. 2016). In addition, a homogeneous CSF and WM segmentation is important (provided in FreeSurfer), as these are primary regions to compensate for in gray matter uptake of the tracer. When the assumptions are satisfied (and under noiseless conditions), the GTM will provide the exact mean in each VOI.
Step 5 – Kinetic Modeling (4 options)
The Multilinear Reference Tissue Model (MRTM) was applied as described by Ichise et al. 2003 with cerebellum (excluding vermis) as a reference region, allowing for estimation of three parameters from which the non-displaceable binding potential (BPND) can be derived.
The second model applied was the Multilinear Reference Tissue Model 2 (MRTM2) (Ichise et al. 2003) with cerebellum (excluding vermis) as a reference region (Ganz et al. 2017), and thalamus, putamen and caudate were averaged to represent a single less noisy high-binding region for estimation of k2’, the clearance rate constant from reference region to plasma (Beliveau et al. 2017). The MRTM2 is similar to MRTM, except that k2’ is determined after the first iteration of MRTM and its value is subsequently entered into the two-parameter MRTM2 model. This approximates a linear kinetic analysis, but is executed in only a fraction of the computational time. The simplified reference tissue model, SRTM, was applied as described by Lammertsma and Hume, 1996. SRTM allows for nonlinear least squares estimation of 3 parameters (R1, k2’ and k2a) from the full dataset, and the BPND can be estimated from . R1 is the relative radioligand delivery and k2a is the apparent rate constant.
The non-invasive Logan reference tissue model was applied as described in Logan et al. 1996 with t* = 35 minutes for all regions and subjects. It also assumes the existence of a valid reference region and an average tissue-to-plasma clearance k2’, and the distribution volume ratio can estimated as DVR = BPND − 1. All kinetic models applied in this work were implemented in MATLAB v. 2016b as specified in their original paper. The implementation in MATLAB was validated with PMOD v. 3.0 (10 subjects < 0.1% difference in BPND), but was carried out in MATLAB for parallel execution purposes to substantially reduce processing time.
Different kinetic modeling approaches are tested in this study, as the optimal estimation of 5-HTT binding may vary as a function of scanner (i.e. resolution), subject and region.
From this 5-step list of preprocessing choices, we can quantify BPND for 3×2×4×4×4 = 384 different pipelines per subject (Figure 2) and subsequently examine their impact on a set of chosen performance metrics (Section 1.5).
Figure 2:
Schematic overview of the various preprocessing steps applied for the [11C]DASB quantification. Abbreviations; average (avg), time-weighted average (twa), signal-to-noise ratio (SNR).
1.5. Analysis and pipeline performance metrics
To evaluate the effects of different PET preprocessing choices, we tested their performance on a set of common performance metrics, namely the test-retest bias, within-subject variability, between-subject variability, intraclass correlation, power calculation, and failure rate. While these analyses were applied for each region j individually and summarized over all subjects i, we also adopted a global reproducibility metric from the fMRI literature, producing a single reproducibility measure for each subject i and pipeline k, taking the information from all regions into account (Strother et al. 2002). This sums to a total of 7 performance metrics that serve to assess the individual pipelines against each other.
Unless otherwise stated, we used statistical subsampling to test several sample sizes of either ñ = 10 or 20 subjects randomly selected without replacement from the 30 subjects, and this was repeated 1000 times, to produce a mean estimate and a 95% confidence interval (CI). The sample sizes of 10 or 20 subjects were chosen to reflect the commonly used sample sizes in [11C]DASB PET studies (Nørgaard et al. 2019). Notation-wise, ñ indicates a resampling analysis, whereas N = 30 indicates that all subjects were included in the analysis.
Statistical differences in pipeline choice (e.g., motion correction vs. no motion correction) for each performance metric was determined across 1000 resamples (subsampling 20 subjects without replacement), and then using the empirical distribution of the differences of the performance metric. This provides an empirical P-value for the difference between pipeline choices for each performance metric. Correction for multiple comparisons across regions was carried out using False-Discovery Rate (FDR), at FDR=0.05. The rationale for choosing these 7 performance metrics is to provide a quantitative estimate of what can be expected of biases and variability as a function of preprocessing pipeline choice and sample size.
Global Within-Subject Reproducibility Metric (FIX)
A global within-subject reproducibility metric over all regions was estimated by generating global signal-to-noise (gSNR) metrics for each subject i and pipeline k, as described in (Strother et al. 2002, Churchill et al. 2012, Churchill et al. 2015). The fourteen brain regions, described in section 1.4 step 3, were selected for analysis, and a pairwise linear correlation based on the Pearson linear correlation coefficient, R, was estimated based on the test and retest BPND’s.
The gSNR for each subject and pipeline was estimated as
Subsequently, we identified the pipeline that maximized the median-rank across all subjects, as described in (Churchill et al. 2012), and described in Supplemental Text 1. This pipeline is defined as the optimal fixed pipeline (FIX) across all subjects and regions.
The rationale for choosing the gSNR as performance metric is because it captures the test-retest correlation between BPND estimates across all brain regions for each subject. Furthermore, it is less prone to real (physiological) second-scan effects, as captured by the test-retest bias metric presented below.
Test-retest Bias
The test-retest bias was estimated as the difference between the two measurements for subject i, region j, and pipeline k, and expressed as a percentage of the first measurement (Kim et al. 2006). This is given by
In the estimation of an average group-level bias (i.e. ), BPND’s that were ≤ 0 or ≥ 10 in either test or retest were excluded in the estimation to avoid the influence of outliers. To account for this exclusion, we estimated a “failure rate” for a given region j and pipeline k, defined as the number of outliers divided by the number of subjects × 100.
Failure rate information is available in the supplementary material.
Within-Subject Variability Metric (WSV)
The within-subject variability (WSV) was estimated as the standard deviation across regions of the difference between test and retest. To normalize the metric to a coefficient of variation (CV) %, we divided the WSV by the average BPND’s over test and retest for all 30 subjects (outliers excluded). This can mathematically be expressed as
where di,j,k = testi,j,k − reseti,j,k, and S is the number of subsamples (i.e. without outliers). BPND’s that were ≤ 0 or ≥ 10 in either test or retest were excluded in the estimation to avoid the influence of outliers.
The rationale for choosing WSV as performance metric is because it reflects the variability between measurements of the BPND estimate for individuals in a sample. For example if the WSV is too high it becomes infeasible to perform longitudinal studies applying a pharmacological intervention, e.g. if the expected within-subject variability is larger than the effect of the intervention.
Between-Subject Variability Metric (BSV)
Between-subject variability (BSV) was captured by identifying the pipeline that minimized the mean standard deviation across all regions and across subjects at baseline (i.e. test). To compare regions, we estimated the CV by dividing the standard deviation, σ, for ñ = 10 or 20 subjects for a given region by the mean, μ, estimated from all subjects at baseline and re-scan (outliers excluded). This is our final between-subject variability measure.
where and . BPND’s that were ≤ 0 or ≥ 10 in either test or retest were excluded in the estimation to avoid the influence of outliers.
The rationale for choosing BSV as performance metric is because it reflects the differences in BPND between individuals and our ability to detect group differences. For example, if the BSV is too high it may require an unreasonable number of subjects to establish group differences.
Intraclass-Correlation Reproducibility Metric (ICC)
We estimated the intraclass-correlation coefficient (ICC(3,1)) across test-retest for all regions and for each pipeline, as given below
where q is the number of repeated measurements (i.e. q = 2), MSBS is the between-subjects’ sum of squares, and MSE is the error mean square. We chose ICC(3,1), as this measure eliminates possible systematic test-retest effects due to the scan-order, by treating repeated measurements as fixed instead of random. In the estimation of the ICC metric, BPND’s that were ≤ 0 or ≥ 10 in either test or retest were excluded to avoid the influence of outliers. We subsequently chose the pipeline, that maximized the ICC(3,1) and from now on we refer to this pipeline as ICC.
Power Analysis
Power analysis involves determining the number of subjects needed to show a given effect size, based on the variability of the data (Whitley et al. 2002). An example of such an estimation, can be expressed as
where is the number of subjects needed to show an effect E, 1.96 corresponds to a 95% confidence interval, and σ is the group-level standard deviation i.e. the BSV. We estimated the average number of subjects needed to show an effect of either 5% or 10% change in BPND based on the previously estimated between-subject variabilities for 10 and 20 subjects, including a 95% confidence interval. The effects of either 5% or 10% were estimated as the percent change from the average BPND for a given region j and pipeline k. Outliers (BPND’s that were ≤ 0 or ≥ 10) were excluded in the estimation of both σ and E. We note, that there are different ways to estimate the needed sample size depending on the experimental setup, however, as we are mainly interested in the relative differences in sample size between pipelines, this procedure should be sufficient.
RESULTS
An optimal pipeline across subjects and regions (Figure 3)
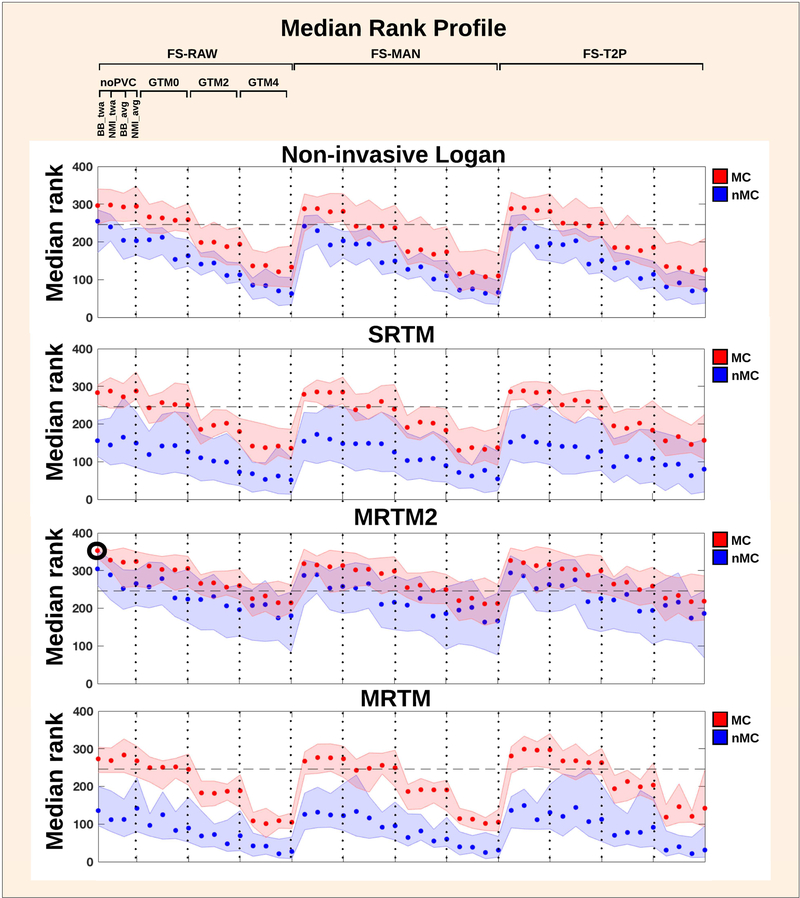
Figure 3:
Median rank profile for all pipelines across all subjects. The shaded errorbars indicate 95% confidence intervals. The optimal pipeline across subjects and regions (FIX) is visualized by the black bold circle. The horizontal dotted line indicates that pipelines below this line are significantly different from FIX. The pipelines above the cut-off are not significantly different from each other.
The evaluation of a median-rank profile for relative pipeline performance for each pipeline and across all subjects (N = 30) and regions (N = 14) is shown in Figure 3, based on the gSNR. Higher median rank indicates a higher gSNR, and better test-retest performance across subjects and regions for a given preprocessing pipeline. We found a significant pipeline performance effect across subjects (P < 0.0001, Friedman test), suggesting the existence of an optimal fixed pipeline. The highest median rank across subjects (Rmax = 0.995), was achieved with the following preprocessing pipeline (FIX): without manual edits in FreeSurfer (FS-RAW), with motion correction (MC), boundary-based co-registration with the time-weighted average image, without partial volume correction (noPVC), and with MRTM2 as preferred kinetic modeling approach. We also identified a subset of several other pipeline choices, that statistically performed equally well as FIX, based on a Dunn-Sidak test, correcting for multiple comparisons for all possible pairwise combinations (P = 0.05). The horizontal dotted line in Figure 3 indicates that pipelines below this line are significantly different from FIX (Rcut-off = 0.989). The pipelines above the cut-off are not significantly different from each other (Rmin = 0.857).
MC was the factor that influenced pipeline performance most; it consistently increased the median rank when applying MC. The effect of MC also depended on which kinetic model was subsequently applied: whereas the rank for MRTM2 with either MC or nMC were not significantly different from each other (overlapping CI’s), the Non-invasive Logan, SRTM and MRTM performed significantly better after MC.
PVC with GTM generally decreased the median rank with increasing PSF (i.e. 0 mm, 2 mm, 4 mm), but the effects were most evident when MC was applied.
The application of GTM with all PSF options and in combination with MRTM2 showed comparable performance to the noPVC, whereas PVC combined with other kinetic models significantly lowered the test-retest performance, as measured by the gSNR.
The co-registration with the time-weighted PET image for both BBR and NMI marginally outperformed the average image, when no MC was applied. This was particularly evident for the higher-rank cases where either Non-invasive Logan or MRTM2 were applied. Only minor effects of co-registration were evident when MC was applied.
The three different FreeSurfer approaches to delineate brain regions did not cause any consistent differences in median rank performance.
Test-retest bias (Figure 4)
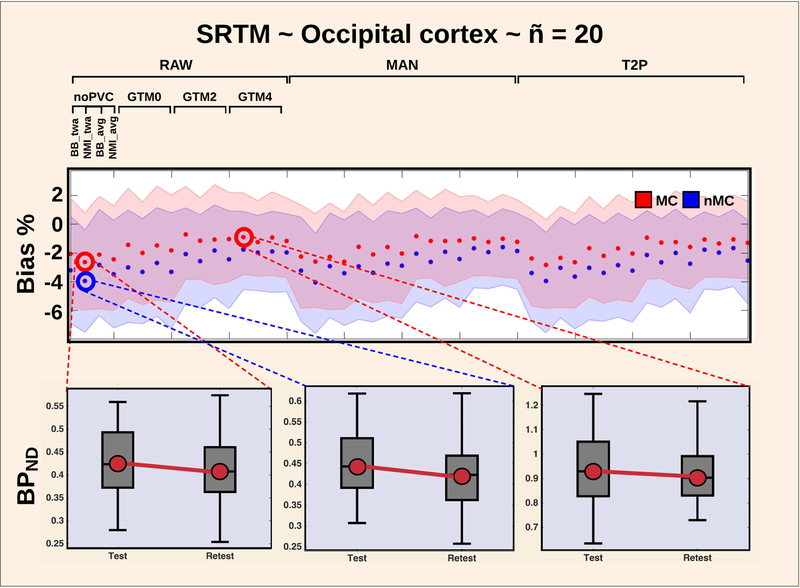
Figure 4:
Test-retest bias (%) as a function of pipeline for the occipital cortex, when SRTM is applied. The use of motion correction generally decreases the bias, and ranges from −1% to −4%. This is highlighted by three plots in the bottom, showcasing the test-retest effect on BPND.
98% of the pipelines revealed a negative test-retest bias (range: −6% to 0%), meaning that the regional BPND’s were lower on the second scan compared to the first scan. Motion correction had only minor effects on the mean bias for the high-binding regions thalamus, putamen and caudate ranging from 1–2% (Figure S1).
In contrast, when applying SRTM to the occipital cortex, the bias was reduced to −2% when using MC, whereas it was −4% without MC (Figure 4). The bias for the occipital cortex was reduced to −1% when combining MC and GTM with either 2 or 4 mm. For the superior frontal cortex and entorhinal cortex, MRTM and SRTM created some spurious outliers producing up to 15% bias. The amygdala created the highest consistent test-retest bias over all pipelines for both MRTM and SRTM, ranging from −4% when MC was applied, to −6% when no MC was applied (Figure S2). The amygdala bias was reduced to negative 1–2% when either Non-invasive Logan or MRTM2 was used.
Figures of all estimated biases as a function of sample size (ñ = 10 or ñ =20), region and preprocessing pipeline are available through the CIMBI database (Knudsen et al. 2016).
Tradeoff in within- and between subject variability at the group level (Figure 5)
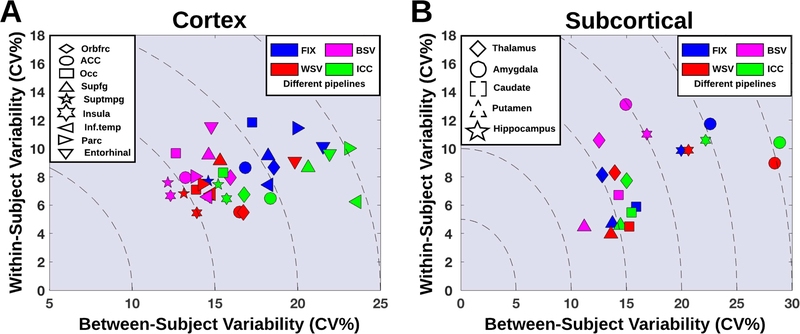
Figure 5:
Between-subject variability as a function of within-subject variability for different pipeline optimization schemes, and for both cortical (A) and subcortical regions (B).
The within- and between subject variability were assessed for four different optimization schemes according to the pipelines that for 20 subjects (subsampled 1000 times without replacement) and region j, (1) minimized the between-subject variability (BSV), (2) minimized the within-subject variability (WSV) across test and retest, (3) maximized the ICC(3,1) across test and retest, and (4) the fixed optimal pipeline (FIX).
Figure 5 shows the between-subject variability as a function of within-subject variability for these four pipelines, depicted for subcortical and cortical regions. For all regions, we observed a trade-off in between- and within-subject variability, meaning that e.g. minimization of within-subject variability increased between-subject variability, and vice versa (Figure 5). The worst case was the hippocampus showing stable WSV across the pipelines WSV, BSV, FIX and ICC at 10–11%, whereas between-subject variability decreased from 22% to 17% when using the BSV pipeline instead of the ICC pipeline. This translates into 30 fewer subjects needed in a group analysis to detect a 5% difference in BPND and to achieve similarly powered statistical results (approximately 6 more subjects needed per % increase in BSV).
Table 1 lists the optimal preprocessing pipelines for the 14 regions. Across all regions, use of either MRTM or MRTM2 consistently minimized WSV (Table 1). Notably, the application of GTM 4 mm minimized the between-subject variability in all regions except for the amygdala, thalamus and hippocampus, and the within-subject variability was similarly minimized in all cortical regions, except from the regions insula and entorhinal cortex (Table 1).
Table 1:
Overview of optimal pipelines for the brain regions amygdala, thalamus, putamen, caudate, anterior cingulate, hippocampus, orbital frontal cortex (FC), occipital, superior FC, superior temporal gyrus (TG), insula, medial-inferior TG, parietal cortex and entorhinal, when optimized by median-rank (FIX), within-subject variability (WSV), between-subject variability (BSV) and intra-class correlation (ICC). 1st letter (Motion Correction (MC); A=MC, B=noMC), 2nd letter (Co-registration; A=BBTWA, B=NMITWA, C=BBAVG, D=NMIAVG), 3rd letter (Delineation of regions; A=FS-raw, B=FS-man, C=FS-T2p), 4th letter (Partial Volume Correction (PVC); A=noPVC, B=Geometrix Transfer Matrix (GTM) 0 mm, C=GTM 2 mm, D=GTM 4 mm), 5th letter (Kinetic modeling; A=MRTM, B=MRTM2, C=SRTM, D=Non-invasive Logan).
| FIX | WSV | BSV | ICC | |
|---|---|---|---|---|
| Amygdala | AAAAB | BBCCB | AABAD | ABACB |
| Thalamus | AAAAB | AABAA | BBAAD | BABDA |
| Putamen | AAAAB | AACAA | DACDA | BAADA |
| Caudate | AAAAB | AACDB | DACDA | AAADB |
| Anterior Cingulate | AAAAB | ABBDB | DBADD | ABCDB |
| Hippocampus | AAAAB | BBBAB | BBAAD | BBCCB |
| Orbital FC | AAAAB | BBBDB | DBCDD | ABBDB |
| Occipital | AAAAB | BABDB | DBADC | BACDA |
| Superior FC | AAAAB | CBADB | BBADA | ABCDC |
| Superior TG | AAAAB | BBBDB | BAADD | ABBBB |
| Insula | AAAAB | BACBA | BABDD | BBCDB |
| Medial-Inferior TG | AAAAB | BBBDB | BABDB | BBCDB |
| Parietal C | AAAAB | ABADA | CBADB | ABBBC |
| Entorhinal | AAAAB | BACAB | BBCDD | BABDB |
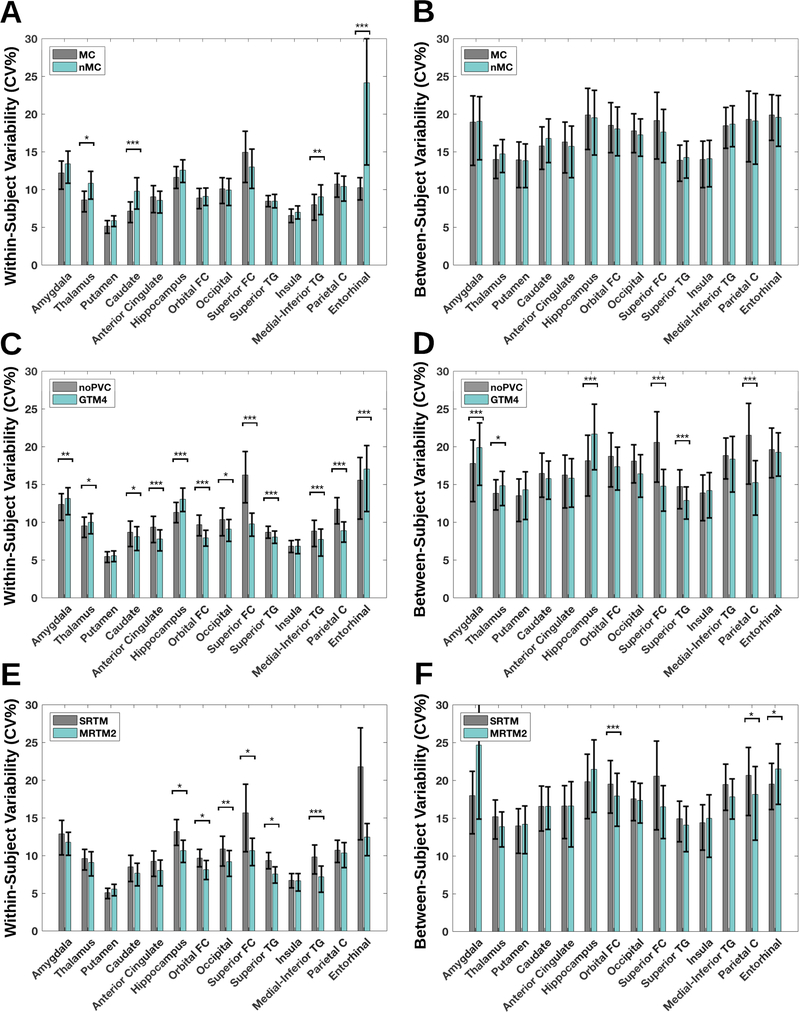
Figure 6A and 6B display the within- and between-subject variability as a function of region, and with or without application of MC. In Figure 6C and 6D, the within- and between subject variability is displayed for noPVC vs. GTM 4 mm. Figure 6 shows the impact on within- (E) and between-subject (F) variability of choosing SRTM versus MRTM2.
Figure 6:
(A) within-subject variability for 14 regions with or without motion correction, including a 95% confidence interval (B) between-subject variability for 14 regions with or without motion correction, including a 95% confidence interval (C-D) similar to A and B, but with either no partial volume correction (noPVC) or with the Geometric Transfer Matrix (GTM) and a point spread function assumption of 4 mm (E-F) similar to A and B, but with the application of either the Simplified Reference Tissue Model (SRTM) or the Multilinear Reference Tissue Model 2 (MRTM2) as kinetic modeling choice. * P < 0.05, ** P < 0.01, *** P < 0.001, FDR corrected for multiple comparisons (FDR=0.05).
Figures of all estimated variabilities as a function of sample size ( ñ =10 or ñ =20), region and preprocessing pipeline are avilable through the CIMBI database (Knudsen et al. 2016).
Power analysis across preprocessing pipeline choices (Figure 7)
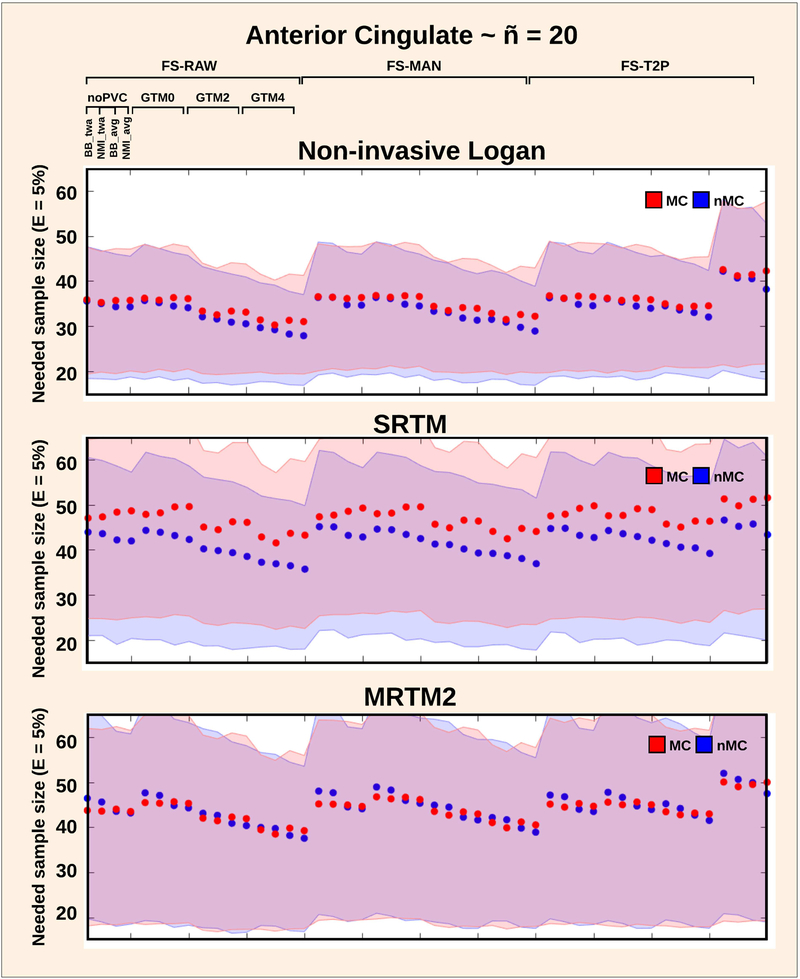
Figure 7:
Sample size required to detect a group differece of 5% in BPND for the anterior cingulate cortex, depending on the kinetic modeling approach (Non-invasive Logan, SRTM and MRTM), and for all other pipeline choices. The blue is without motion correction (nMC) and the red line is with motion correction (MC). The shaded error bars indicate the 95% confidence interval, estimated by randomly choosing 20 subjects over 100 resampling’s.
The effects of pipeline choice on the sample size required to show a given effect size were also examined. Figure 7 shows the sample size required to detect a 5% difference from the anterior cingulate mean BPND. A 95% CI is plotted for all pipeline combinations for Non-invasive Logan, SRTM and MRTM2 for ñ = 20.
The greatest reduction in sample size was seen with the choice of kinetic modeling: the Non-invasive Logan in combination with GTM 4 mm and no MC was associated with a sample size of 27 subjects [CI: 16 subjects – 36 subjects] (Figure 7). Notably, when combined with GTM 4 mm, the FS-T2P stream resulted in substantially higher sample size (Figure 7). This result was driven by a single subject producing a substantially higher BPND after FS-T2P correction, consequently increasing the between-subject variability.
Figures for sample size as a function of pipelines, regions, subjects and effect sizes are available through the CIMBI database (Knudsen et al. 2016).
All average BPND estimates, standard deviation, min and max values for each region and preprocessing strategy is available in the Supplementary. In addition, average volume, standard deviation, min and max value for each region for the VOI choices FS-RAW, FS-MAN and FS-T2p is available in the supplementary material.
DISCUSSION
In a comprehensive preprocessing framework, we report the evaluation of the impact of 384 different preprocessing pipelines on a set of common performance metrics, based on test-retest [11C]DASB neuroimaging data. Our findings suggest that the observed complex interaction between various preprocessing steps and brain regions necessitates careful consideration of the performance of a chosen preprocessing pipeline and the final outcome measure BPND.
Test-retest bias on [11C]DASB binding
Whereas test-retest studies are generally considered to provide valuable information about the repeatability of PET measures, variability may not only arise from measurement errors but also from biological variations between scans. Independent of the chosen preprocessing pipeline, we consistently found lower [11C]DASB BPND at the second compared to the first scan.
This observation was also made by Kim et al. 2006 who reported a negative bias of 2.5% - 7.5% between first and second scan. Two other test-retest studies by Frankle et al. 2005 and Ogden et al. 2006 did not apply any test-retest bias metric in their evaluation.
Regardless, if the negative test-retest bias is a true biological effect or if it is introduced in the data-acquisition and/or preprocessing stage, care must be taken in analysis of longitudinal data to avoid attributing an effect to a treatment/condition that was actually due to the retest alone. Further, test-retest studies with a biologically determined bias means that attempts to define a pipeline that minimizes the bias may be counterproductive.
Extensive research in humans support a number of factors affecting cerebral [11C]DASB binding. Diurnal variation has been reported to affect 5-HTT binding (Meyerson et al. 1989) causing an increase in measurement variability if test and retest scans are executed in the morning and afternoon. The effects of having repeated tracer injections may introduce carry-over effects, or induce internalization or conformational changes of the 5-HTT to a different state (Zhang et al. 2016). The data used in the present study were acquired with an interval of 5 weeks, at the same phase in the menstrual cycle and at the same time of the day, which makes it unlikely that carry-over effects, hormonal or diurnal changes explain our observation. Kim et al. 2006 also discussed the possibility of increased stress levels at the first scan, elevating the circulation of cortisol, consequently increasing 5-HTT synthesis and thereby potentially lowering 5-HTT binding observed at retest. While increased stress levels at the first scan may be causing the negative bias, it may also be attributed to a change in levels of motion between test and retest, with less motion contributing to an increase in SNR (if subjects are more calm at retest). In contrast, high levels of motion may have substantial impact on the scanner reconstruction, potentially either over- or underestimating the true uptake by affecting attenuation- and scatter correction (Van den Heuvel et al. 2013).
Irrespective whether the bias appears from biological variations and/or is caused by data acquisition and/or preprocessing, it should be taken into account if it is likely to have an impact on the scientific question. More specifically, scientific question could depend on e.g. 1) region 2) one or more scans 3) structural abnormalities such as atrophy, and 4) disorder related head motion. In contrast, bias may trade off with true between subject variability where greater variability may reflect more accurate between-subject biological variation. This is contrary to focusing only on group mean differences which we have mainly focused on in this study.
Nevertheless, depending on the ability to remove potential biases and depending on the size of their individual contributions, both the within- and between-subject variability should subsequently be assessed to decide whether an estimated effect can be considered a true biological effect or not.
Impact of preprocessing pipeline strategy
Using our evaluation framework, we identified a set of optimal pipelines across subjects and regions showing significant effects for MC, co-registration, PVC and kinetic modeling (Figure 3). Although it is well-known that MC can have a significant impact on PET results (Montgomery et al. 2006), about 40% of published [11C]DASB PET neuroimaging studies leave out MC (Nørgaard et al. 2019).
One of the most consistent outcomes of our analysis was that MC had an impact on the pipeline performance. Given that we only included scans with < 3 mm median movement, MC is likely to have an even larger impact in people with larger head motion. Conversely, in the absence of motion, MC will lead to some degree of smoothing due to interpolation which might have improved the performance. The improved performance with MC could also result from higher noise-levels at the end of the scan where the distribution of radioactivity is lower producing less true counts, or by a re-distribution of the tracer. Freire and Mangin 2001, and Orchard and Atkins 2003, demonstrated that least-squares cost functions may be susceptible to fMRI activation biases, which for PET means that the MC algorithm may attempt to incorrectly account for motion if the VOI has low SNR, or if the tracer distribution in the target volume changes significantly over time compared to the reference volume. While the registration to one single frame is possible for [11C]DASB, this may turn out to be more complicated for other radiotracers, e.g., with heavily changing uptake over long acquisition times. This limitation could be overcome by using time dependent changing reference frames. One such approach is the AIR reconcile function (http://air.bmap.ucla.edu/AIR5/reconcile_air.html) which attempts to reconcile discrepancies between frames that represent all pairwise registrations. However, while this approach may be more optimal in cases with heavily changing uptake over time, it comes at a significantly increased computional cost and execution time. Nevertheless, given the impact of the motion corrections step this issue is an important topic for future research. While we identified an overall impact of MC on pipeline ranking using the performance metric gSNR (Figure 3), we also found that particularly the thalamus, caudate, medial-inferior TG and entorhinal cortex (Figure 6A) contributed to the within-subject variability. This is important, because thalamus and caudate are often used as high-binding regions in MRTM2, affecting the estimation of k2’ and consequently the BPND. While choice of reference region will impact all RTM’s, MRTM2 and Non-invasive Logan are particularly sensitive to the choice of an adequate high-binding region for estimation of k2’ (Ichise et al. 2003, Mandeville et al. 2016). In previous studies, different high-binding regions have been used, without any particular justification, e.g., raphe, thalamus and striatum (Kim et al. 2006); midbrain, thalamus and striatum (Matsumoto et al. 2010); raphe (Hesse et al. 2011); occipital cortex (Brown et al. 2007); or thalamus (James et al. 2017). The same group may even choose different high-binding regions across studies, e.g. Gryglewski et al. 2017 (striatum) and James et al. 2017 (thalamus), and some studies do not mention which high-binding region was chosen (e.g. Zientek et al. 2016). A few studies also cite other studies as justification for using a high-binding region, but then use another high-binding region than the cited study (e.g. Kupers et al. 2011 & Frokjaer et al. 2009). Based on previous literature (e.g. Beliveau et al. 2017), we rather arbitrarily decided to use thalamus and striatum as high-binding regions for estimation high-binding regions for estimation of k2’. However, as displayed in Figure 6, this choice may not be optimal, as the putamen not only minimizes the within- and between-subject variability relative to thalamus and caudate, it is also the region least affected by preprocessing strategy; MC, PVC and kinetic modeling. The putamen delineation in FreeSurfer is a more homogeneous gray-matter region compared to thalamus (see supplementary text 3 for evaluation), and does not suffer from the same severe partial volume effects as caudate does because of its proximity to CSF. Therefore, one could consider the putamen to be the optimal choice of high-binding region to minimize potential biases originating from subject-dependent differences. In a post-hoc test we evaluated the use of putamen as high-binding region, and this indeed lowered the between-subject variability with 1–10% depending on the VOI, at the expense of bias in group mean. As this post-hoc test is a circular analysis, our observation needs to be tested in an independent cohort.
The performance of the optimal pipeline was also largely dependent on the use of noPVC or GTM with either 0 mm, 2 mm or 4 mm, with the latter contributing negatively to the overall pipeline rank, as highlighted by the performance metric gSNR (Figure 3). At first sight, this effect would seem to be caused by violations of the GTM assumptions, presumably the PSF and the constant uptake within each VOI. For subcortical regions, the thalamus delineation and consequently the [11C]DASB uptake homogeneity has been shown to vary substantially between atlases (Nørgaard et al. 2015), which may make the estimate more noisy. For cortical regions where the 5-HTT density is relatively low and the average cortical thickness only 3 mm (Fischl et al. 2000), the voxel-wise noise level may be higher than in the subcortical regions.
However, in a post-hoc analysis on variability (Figure 6C and 6D), we identified a distinct difference in PVC performance across subcortical and cortical regions. While the application of GTM 4 mm caused a significant decrease in within-subject variability in all cortical regions except for the insula and entorhinal cortex, it significantly increased it in the amygdala, thalamus and hippocampus. More specifically, the amygdala and hippocampus were critically affected by this preprocessing step, increasing both within-subject and between-subject variability. This observation may be attributed to partial volume effects being similar in the amygdala, hippocampus, and cerebellum, resulting in more unstable estimates due to the correlation between regions. Regardless of the contribution to noise of the GTM, PVC is still highly recommended in studies where brain atrophy interacts with an effect of interest (e.g., age or diagnosis). Failure to properly account for partial volume effects in these cases can falsely inflate or degrade the effect of interest (Greve et al. 2016).
Amygdala and hippocampus have medium to high 5-HTT density and with long uptake times, the TACs tend to reflect irreversible binding, which may compromise the identification of stable model parameters, resulting in noisy estimates. For the pipeline-rank performance metric (Figure 3), if within-subject variability increases when GTM 4 mm is applied, this will have a negative impact on the pipeline-rank metric, as it largely depends on the Pearson’s correlation coefficient.
This variability was reduced after MC, but the remaining difference in pipeline performance was significantly affected by the choice of kinetic modeling. Depending on the difference in noise-levels at test and retest due to e.g. motion, this may be caused by a bias in the BPND estimates from kinetic models using non-invasive Logan, SRTM and MRTM, consequently reducing the test-retest performance. This is because BPND estimates from kinetic models are subject to noise-dependent bias, meaning that as the noise-level increases, the estimated BPND deviates from the true value (Ichise et al. 2003). The MRTM2 has no noisy term as independent variable when fitting the kinetic model parameters with multi-linear regression, thus effectively reducing the noise-induced bias and improving overall performance (Ichise et al. 2003).
Trade-off in within- and between-subject variability at the group level
The within-subject and between-subject variability analysis revealed important trade-offs in pipeline performance as a function of region (Figure 5). Minimization of between-subject variability increased within-subject variability relative to the fixed pipeline, particularly for the amygdala, thalamus and hippocampus quantified with the non-invasive Logan model.
Quantification with the non-invasive Logan method is often preferred due to it having the lowest between-subject coefficient of variation (Tyrer et al. 2016, Logan et al. 1996), however, our analyses indicate that this comes at the expense of a 3–5% increase in within-subject variability (range: 10% – 14%), as shown in Figure 5. Consequently, depending on the experimental setup (i.e. group or longitudinal study) the choice of preprocessing should be selected with caution and consideration of the study goals and design.
The analysis on the effects of spatial normalization on BSV and WSV (supplemental material), showed only a small difference in terms of average BPND compared to without normalization, but the within- and between-subject variability were substantially increased by a factor of 2–4. This effect is likely to be caused by contamination of CSF and white-matter in the VOI in standard space, requiring a substantial increase in number of subjects needed to obtain similar statistical power.
The within-subjects design captures the difference among conditions (i.e., test and retest) and has the clear advantage that fewer subjects are required. However, the within-subjects design is subject to learning effects across conditions if the design is not placebo vs active, which is not the case for between-subject designs. Care must therefore be taken in the analysis of longitudinal data to avoid attributing an effect to a treatment/condition that was actually due to a potential retest bias. From a physiological point-of-view, the minimization of BSV might be misleading given that the potentially higher and true underlying BSV is due to real physiological differences. However, from a statistical and experimental point-of-view it is of interest to minimize the BSV because it requires fewer subjects to detect group differences.
Moreover, by including participants that are thought to be similar (age, gender etc.), we expect the BSV to be smaller by minimizing the influence of other variables on the measurement. This assumption is, however, only valid under certain assumptions. For example, methods that introduce noise (inclusion of WM and CSF in the VOI) will have an impact on the BSV metric, but assuming these effects are independent of the true physiological variability the joint effects are additive and minimizing the methodological noise through BSV seems reasonable. However, there might be interactions between the true physiological signal and associated methodological effects leading to the major unsolved problem with BSV that it will be extremely difficult to distinguish between what is true biological variability and what is variability coming from the method. This is largely why we design preprocessing strategies to remove noise unrelated to the true underlying signal. In the absence of a “ground truth”, it remains a challenge to select the optimal preprocessing pipeline, and it may take alternative performance metrics to quantitatively evaluate and compare various pipelines (Strother et al. 2002, Churchill et al. 2015). In this work, we argue that explicit knowledge about the impact of preprocessing choices will help researcher’s to understand the variability in their data, so they are able to correctly identify and remove the noise sources affecting their measurements. Such understanding will help to ensure that noise is not mistakenly characterized as a true biological effect.
We want to emphasize that the aim of this study was not to identify a definitive preprocessing pipeline for [11C]DASB data, but to quantify the impact of the preprocessing choices selected in this study and their uncertainty on BPND.
Enhancement of study power with optimal preprocessing pipelines
The comparison of subjects needed to show a given effect size, provided insight into the effect of preprocessing pipeline choice on sample size and as a function of region, based on the between-subject variability performance metric. The test-retest studies published so far for [11C]DASB included between 8 and 11 volunteers (Ogden et al. 2007, Frankle et al. 2006, Kim et al. 2005) (the present study includes 30 subjects) and sample sizes in published [11C]DASB PET studies range from 5 (Ogawa et al. 2014) to 83 subjects (Miller et al. 2013), but with approximately 20 subjects being the most common (Nørgaard et al. 2019). However, while the sample size required to show an effect should ultimately be determined by the variability of the measured random variable (i.e. BPND), power analyses may become biased if incorrect variability measures are used. Therefore, here we provide an estimate of what sample size is needed to show an effect of either 5% og 10% difference in BPND as a function of pipeline choice and for a specific hypothesis related to a given region (available through the CIMBI database (Knudsen et al. 2016)).
As highlighted previously, there exist a trade-off between the optimization of within- and between subject variation as a function of VOI and preprocessing pipeline. This ultimately affects our recommendation of preprocessing strategy to maximize power. For example, given no apriori hypothesis related to a specific region, we recommend the pipeline from the rank analysis using gSNR as performance metric. However, as the gSNR metric is mostly sensitive to within-subject variance and because the power estimation is largely driven by between-subject variance, there will exist other pipelines that maximizes power by minimizing between-subject variance at the expense of increased within-subject variance.
We strongly suggest that researchers take the reported biases and variations into account when they conduct power analyses prior to a study. In addition, we recommend choosing a fixed preprocessing pipeline prior to data acquisition depending on the researcher’s biological question, as this should help to avoid underpowered studies.
While it is quite common in the PET community to perform regional analyses, several attempts have also been extended to both voxel- and surface based analyses. The effects of bias and variance trade-offs as a function of various tracers and preprocessing pipelines are thus largely unknown for these types of analyses, and only a few papers have attempted to address some of these challenges for PET (e.g. Greve et al. 2014) and fMRI (e.g. Churchill et al. 2015).
All the reported results and analyses are available through the CIMBI database (Knudsen et al. 2016).
Generalizability to other PET radioligands
Nørgaard et al. 2019 found that across 21 PET centres in 105 studies for [11C]DASB only, the heterogeneous range of preprocessing steps used allowed for a total of approximately 21.000.000 different workflows. We have no reason to believe that the preprocessing steps used for other radioligands and across PET centers are different. By contrast, the number of possible preprocessing strategies may even be larger with more available radioligands. Further, many of the noise sources such as subject-motion and partial volume effects are inherent to the PET signal, independently of the radioligand.
In the current work, we found that MC had the most profound effect on the outcome, followed by PVC and kinetic modeling. MC has also been found to have an impact for other radioligands such as [11C]raclopride (Montgomery et al. 2006) and [11C]PHNO (Jin et al. 2013), generally showing decreased variability with the application of MC. For PVC, Rousset et al. 2008 showed that for [11C]raclopride, average BPND and its variance increased with the application of PVC, and Yang et al. 2017 showed that the application of PVC for imaging of beta-amyloid plaques using the radioligand [18F]AV-45 increased the ability to detect the progression of Alzheimer’s. Generalization of such results across radioligands may be increased by expanding on the interactions for techniques of VOI definition and related characteristics.
While we are not in the position to evaluate the exact impact of preprocessing on other radioligands, the framework that we are proposing is intended to be used in the early test-retest stage for a new radioligand, where recommendations are made to the community for subsequent studies. This should help scientists to make informed choices about pipeline, and thereby increase the likelihood of producing reproducible outcomes across PET studies. We believe that providing such preliminary guidelines for PET receptor results produced by different preprocessing processes is an important part of the neuroimaging community’s response to the problem of producing more reproducible results for human PET receptor studies. In addition, by demonstrating potentially significant increases in study power we are helping to optimize the value of the research money spent on such studies.
LIMITATIONS AND FUTURE CONSIDERATIONS
Our study is not without limitations. The results were derived from the radiotracer [11C]DASB measured in the HRRT scanner; however, as stated above we expect the results to generalize to other radiotracers and scanners, with a possible exception of PVC. Inclusion of PVC only had minor effects on most performance metrics. While this may be a specific finding in the context of using the HRRT with the PSF-OSEM reconstruction, it may be questionable whether PVC would generably be favourable in cases of PET images obtained with a conventional PET scanner offering a resolution of 4–5 mm.
We observed only minor differences in performance between FS-RAW, FS-MAN and FS-T2P, but substantial differences in regional binding estimates could presumeably be obtained if a different brain atlas (e.g. PVElab, Svarer et al. 2005) is used. We deliberately abstained from testing other segmentation atlases since the outcome was likely to be influenced by differences in volumes, etc.
FreeSurfer returns 41 regions per hemisphere and to make the results more comparable to other atlases, we chose to extract only a subset of 14 regions covering all major parts of the brain. The results and interpretations of this study can therefore not be generalized to the remaining 27 regions, with the tradeoff being that the results are more comparable to regions from other atlases. Furthermore, the merging of regions between hemispheres is also a limitation if lateralized effects are present. On the other hand, averaging across hemispheres is commonly done in PET studies because it reduces the number of statistical tests.
However, even though the extent to which a change in brain atlas affects regional binding is quantifiable, it is not trivial to determine whether a decrease/increase in the performance metrics suggests a better choice of atlas. Further investigation is therefore needed in order to understand this question.
With respect to kinetic modeling we decided not to include SRTM2 because SRTM2 and MRTM2 perform similarly well. Optimally, the current framework should be expanded to include data from different scanners and other acquisition parameters to evaluate inter-site differences, however data sharing initiatives are needed to accomplish this task (Knudsen et al., 2016). This is beyond the scope of this paper.
Last but not least, the chosen steps of preprocessing in this study is also a limitation. Our future goal is to make the data publicly available, so researchers can download the data and benchmark their own preprocessing pipeline using the same performance metrics and the same data. The results of the benchmark can subsequently be made publicly available on a website or in a database. The effort aligns well with current interests in the PET community, as was highlighted at the NRM2018 PET Grand Challenge (www.petgrandchallenge.com).
CONCLUSION
In summary, we provide evidence that preprocessing pipeline choices have significant impact on [11C]DASB BPND in a distributed set of brain regions, as evaluated by 7 performance metrics. Given that no apriori hypothesis exist, we recommend researchers use the FIX pipeline (with MC, co-registration BBR and the time-weighted PET image, no PVC, and kinetic modeling using MRTM2). Given a specific clinical hypothesis (e.g. change in binding in putamen), we recommend researchers to use Table 1 as a guideline, with longitudinal studies using the WSV column, as this measure ensures minimum test-retest variability between scan sessions. For cross-sectional studies, we recommend researchers choose a pipeline that minimizes both within- and between subject variability (i.e. either the BSV or ICC column in Table 1), as this should ensure a compromise between low within-subject variability and low between-subject variability.
Systematic evaluation of preprocessing choices using data-driven performance metrics can identify strategies that result in 80% fewer subjects needed in a group analysis to achieve similar statistical results. The use of MC had the most profound effect on BPND, prior to the effects caused by PVC and kinetic modeling. MC increases reproducibility because it respects the structural geometry by aligning the PET data over time, and so does not contaminate the GM signal with that from other tissue types. Thus, a systematic evaluation of preprocessing strategies in PET studies should result in improvements in reproducibility and reliability of study outcomes, allowing for better understanding of human brain function.
Supplementary Material
ACKNOWLEDGMENTS
We wish to thank all the participants for kindly joining the research project. We thank the John and Birthe Meyer Foundation for the donation of the cyclotron and PET scanner. Peter Steen Jensen, Vincent Beliveau and Patrick M. Fisher are gratefully acknowledged. Finally, we express our gratitude to the anonymous reviewers for the many suggestions that helped improve this paper.
FUNDING
MN was supported by the National Institutes of Health (Grant 5R21EB018964-02), the Lundbeck Foundation (Grant R90-A7722), and the Independent Research Fund Denmark (DFF-1331-00109 & DFF-4183-00627).
Footnotes
DISCLOSURE/CONFLICT OF INTEREST
The authors declare no conflict of interest or financial disclosures. SCS is the consulting Chief Scientific Officer at ADMdx, Inc.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Beliveau V, Feng L, Svarer C, Knudsen GM, Fisher PM, Ozenne B, … Højgaard L (2017). A High-Resolution In Vivo Atlas of the Human Brain’s Serotonin System. The Journal of Neuroscience, 37(1), 120–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown AK, George DT, Fujita M, Liow JS, Ichise M, Hibbeln J, Ghose S, Sangare J, Hommer D, Innis RB. PET [11C]DASB imaging of serotonin transporters in patients with alcoholism. Alcohol Clin Exp Res. 2007. January;31(1):28–32. [DOI] [PubMed] [Google Scholar]
- Churchill NW, Oder A, Abdi H, Tam F, Lee W, Thomas C, … Strother SC (2012). Optimizing preprocessing and analysis pipelines for single-subject fMRI. I. Standard temporal motion and physiological noise correction methods. Human Brain Mapping, 33(3), 609–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchill NW, Spring R, Afshin-Pour B, Dong F, & Strother SC (2015). An automated, adaptive framework for optimizing preprocessing pipelines in task-based functional MRI. PLoS ONE, 10(7), 1–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl B, Dale AM. Measuring the thickness of the human cerebral cortex from magnetic resonance images. Proc Natl Acad Sci U S A. 2000. September 26;97(20):11050–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl B, van der Kouwe A, Destrieux C, Halgren E, Ségonne F, Salat DH, Busa E, Seidman LJ, Goldstein J, Kennedy D, Caviness V, Makris N, Rosen B, Dale AM. Automatically parcellating the human cerebral cortex. Cereb Cortex. 2004. January;14(1):11–22. [DOI] [PubMed] [Google Scholar]
- Frankle WG, Slifstein M, Gunn RN, Huang Y, Hwang DR, Darr EA, Narendran R, Abi-Dargham A, and Laruelle M (2006). Estimation of serotonin transporter parameters with 11C-DASB in healthy humans: reproducibility and comparison of methods. J Nucl Med, 47:815–826. [PubMed] [Google Scholar]
- Frokjaer VG, Pinborg A, Holst KK, Overgaard A, Henningsson S, Heede M, Larsen EC, Jensen PS, Agn M, Nielsen AP, Stenbæk DS, Da Cunha-Bang S, Lehel S, Siebner HR, Mikkelsen JD, Svarer C, and Knudsen GM (2015). Role of serotonin transporter changes in depressive responses to sex-steroid hormone manipulation: A positron emission tomography study. Biological Psychiatry, 78(8):534–543. [DOI] [PubMed] [Google Scholar]
- Frokjaer VG, Vinberg M, Erritzoe D, Svarer C, Baaré W, Budtz-Joergensen E, Madsen K, Madsen J, Kessing LV, and Knudsen GM (2009). High familial risk for mood disorder is associated with low dorsolateral prefrontal cortex serotonin transporter binding. NeuroImage, 46(2):360–366. [DOI] [PubMed] [Google Scholar]
- Ganz M, Feng L, Hansen HD, Beliveau V, Svarer C, Knudsen GM, and Greve DN (2017). Cerebellar heterogeneity and its impact on PET data quantification of 5-HT receptor radioligands. Journal of Cerebral Blood Flow & Metabolism, page 0271678X1668609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginovart N, Wilson a. a., Meyer JH, Hussey D, and Houle S (2001). Positron emission tomography quantification of [(11)C]-DASB binding to the human serotonin transporter: modeling strategies. Journal of cerebral blood flow and metabolism : official journal of the International Society of Cerebral Blood Flow and Metabolism, 21(11):1342–1353. [DOI] [PubMed] [Google Scholar]
- Greve D, Fischl B (2009). Accurate and robust brain image alignment using boundary-based registration. Neuroimage, 48, 63–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greve DN, Salat DH, Bowen SL, Izquierdo-Garcia D, Schultz AP, Catana C, Becker JA, Svarer C, Knudsen GM, Sperling RA, and Johnson KA (2016). Different partial volume correction methods lead to different conclusions: An 18F-FDG-PET study of aging. NeuroImage, 132:334–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greve DN, Svarer C, Fisher PM, Feng L, Hansen AE, Baare W, Rosen B, Fischl B, and Knudsen GM (2014). Cortical surface-based analysis reduces bias and variance in kinetic modeling of brain PET data. NeuroImage, 92:225–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gryglewski G, Rischka L, Philippe C, Hahn A, James GM, Klebermass E, Hienert M, Silberbauer L, Vanicek T, Kautzky A, Berroterán-Infante N, Nics L, Traub-Weidinger T, Mitterhauser M, Wadsak W, Hacker M, Kasper S, Lanzenberger R. Simple and rapid quantification of serotonin transporter binding using [11C]DASB bolus plus constant infusion. Neuroimage. 2017. January 22;149:23–32. [DOI] [PubMed] [Google Scholar]
- Hesse S, Stengler K, Regenthal R, Patt M, Becker GA, Franke A, Knüpfer H, Meyer PM, Luthardt J, Jahn I, Lobsien D, Heinke W, Brust P, Hegerl U, Sabri O. The serotonin transporter availability in untreated early-onset and late-onset patients with obsessive-compulsive disorder. Int J Neuropsychopharmacol. 2011. June;14(5):606–17. [DOI] [PubMed] [Google Scholar]
- Houle S, Ginovart N, Hussey D, Meyer JH, Wilson AA. Imaging the serotonin transporter with positron emission tomography: initial human studies with [11C]DAPP and [11C]DASB. Eur J Nucl Med. 2000. November;27(11):1719–22. [DOI] [PubMed] [Google Scholar]
- Ichise M, Liow J-S, Lu J-Q, Takano A, Model K, Toyama H, … Carson RE (2003). Linearized reference tissue parametric imaging methods: application to [11C]DASB positron emission tomography studies of the serotonin transporter in human brain. Journal of Cerebral Blood Flow and Metabolism : Official Journal of the International Society of Cerebral Blood Flow and Metabolism, 23(9), 1096–1112. [DOI] [PubMed] [Google Scholar]
- James GM, Baldinger-Melich P, Philippe C, Kranz GS, Vanicek T, Hahn A, Gryglewski G, Hienert M, Spies M, Traub-Weidinger T, Mitterhauser M, Wadsak W, Hacker M, Kasper S, Lanzenberger R. Effects of Selective Serotonin Reuptake Inhibitors on Interregional Relation of Serotonin Transporter Availability in Major Depression. Front Hum Neurosci. 2017. February 6;11:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin X, Mulnix T, Gallezot JD, & Carson RE (2013). Evaluation of motion correction methods in human brain PET imaging-A simulation study based on human motion data. Medical Physics, 40(10), 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovicich J, Czanner S, Greve DN, Haley E, van der Kouwe A, Gollub R, Kennedy D, et al. Reliability in Multi–Site Structural MRI Studies: Effects of Gradient Non–Linearity Correction on Phantom and Human Data. NeuroImage, 2006; 30(2): 436–43. [DOI] [PubMed] [Google Scholar]
- Keller SH, Svarer C, Sibomana M. Attenuation correction for the HRRT PET-scanner using transmission scatter correction and total variation regularization. IEEE Trans Med Imaging, 2013; 32(9): 1611–21. [DOI] [PubMed] [Google Scholar]
- Kim JS, Ichise M, Sangare J, and Innis RB (2006). PET Imaging of Serotonin Transporters with [11C]DASB: Test- Retest Reproducibility Using a Multilinear Reference Tissue Parametric Imaging Method. J. Nucl. Med, 47(2):208–214. [PubMed] [Google Scholar]
- Knudsen GM, Jensen PS, Erritzoe D, et al. (2015). The Center for Integrated Molecular Brain Imaging (Cimbi) database. NeuroImage, 124, 1213–1219. [DOI] [PubMed] [Google Scholar]
- Kupers R, Frokjaer VG, Erritzoe D, Naert A, Budtz-Joergensen E, Nielsen FA, … Knudsen GM (2011). Serotonin transporter binding in the hypothalamus correlates negatively with tonic heat pain ratings in healthy subjects: A [11C]DASB PET study. NeuroImage, 54(2), 1336–1343. [DOI] [PubMed] [Google Scholar]
- Lammertsma AA, Hume SP, 1996. Simplified reference tissue model for PET receptor studies. Neuroimage 4, 153–158. [DOI] [PubMed] [Google Scholar]
- Logan J, Fowler JS, Volkow ND, Wang GJ, Ding YS, Alexoff DL: Distribution volume ratios without blood sampling from graphical analysis of PET data. J Cereb Blood Flow Metab 1996, 16(5):834–840. [DOI] [PubMed] [Google Scholar]
- Mandeville JB, Sander CYM, Wey H, Hooker JM, Hansen HD, Svarer C, … Rosen BR (2016). NeuroImage A regularized full reference tissue model for PET neuroreceptor mapping. NeuroImage, 139, 405–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto R, Ichise M, Ito H, et al. (2010). Reduced serotonin transporter binding in the insular cortex in patients with obsessive-compulsive disorder: A [11C]DASB PET study. NeuroImage, 49(1):121–126. [DOI] [PubMed] [Google Scholar]
- McCarthy CS, Ramprashad A, Thompson C, Botti JA, Coman IL, & Kates WR (2015). A comparison of FreeSurfer-generated data with and without manual intervention. Frontiers in Neuroscience, 9(OCT), 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer JH, Wilson AA, Ginovart N, Goulding V, Hussey D, Hood K, Houle S. Occupancy of serotonin transporters by paroxetine and citalopram during treatment of depression: a [(11)C]DASB PET imaging study. Am J Psychiatry. 2001. November;158(11):1843–9. [DOI] [PubMed] [Google Scholar]
- Meyerson LR, Strano R, Ocheret D. Diurnal concordance of human platelet serotonin content and plasma alpha-1-acid glycoprotein concentration. Pharma- col Biochem Behav. 1989;32:1043–1047. 32. [DOI] [PubMed] [Google Scholar]
- Miller JM, Hesselgrave N, Ogden RT, Sullivan GM, Oquendo MA, Mann JJ, & Parsey RV (2013). Positron emission tomography quantification of serotonin transporter in suicide attempters with major depressive disorder. Biological Psychiatry, 74(4), 287–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery AJ, Thielemans K, Mehta MA, Turkheimer F, Mustafovic S, & Grasby PM (2006). Correction of Head Movement on PET Studies: Comparison of Methods. J. Nucl. Med, 47(12), 1936–1944. [PubMed] [Google Scholar]
- Nørgaard M, Ganz M, Fisher PM, et al. (2015). Estimation of regional seasonal variations in SERT-levels using the FreeSurfer PET pipeline: A reproducibility study. In: Proceedings of the MICCAI workshop on computational methods for molecular imaging 2015. [Google Scholar]
- Nørgaard M, Ganz M, Svarer C, Feng L, Ichise M, Lanzenberger R, Lubberink M, Parsey RV, Politis M, Rabiner EA, Slifstein M, Sossi V, Suhara T, Talbot PS, Turkheimer F, Strother SC, Knudsen GM. Cerebral Serotonin Transporter Measurements with [11C]DASB: A Review on Acquisition and Preprocessing across 21 PET Centres. J Cereb Blood Flow Metab. 2019. February;39(2):210–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa K, Tateno A, Arakawa R, Sakayori T, Ikeda Y, Suzuki H, & Okubo Y (2014). Occupancy of serotonin transporter by tramadol: a positron emission tomography study with [11C]DASB. The International Journal of Neuropsychopharmacology / Official Scientific Journal of the Collegium Internationale Neuropsychopharmacologicum (CINP), 17(6), 845–50. [DOI] [PubMed] [Google Scholar]
- Ogden RT, Ojha A, Erlandsson K, Oquendo MA, Mann JJ, and Parsey RV (2007). In vivo Quantification of Serotonin Transporters Using [11C]DASB and Positron Emission Tomography in Humans: Modeling Considerations. Journal of Cerebral Blood Flow & Metabolism, 27(1):205–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olesen OV, Sibomana M, Keller SH, Andersen F, Jensen J, Holm S, … Højgaard L (2009). Spatial resolution of the HRRT PET scanner using 3D-OSEM PSF reconstruction. IEEE Nuclear Science Symposium Conference Record, 3789–3790. [Google Scholar]
- Parsey RV, Slifstein M, Hwang DR, Abi-Dargham A, Simpson N, Mawlawi O, Guo NN, Van Heertum R, Mann JJ, Laruelle M: Validation and reproducibility of measurement of 5-HT1A receptor parameters with [carbonyl-11C]WAY-100635 in humans: comparison of arterial and reference tissue input functions. J Cereb Blood Flow Metab 2000, 20(7):1111–1133. [DOI] [PubMed] [Google Scholar]
- Rousset O, Rahmim A, Alavi A, & Zaidi H (2007). Partial Volume Correction Strategies in PET. PET Clinics, 2(2), 235–249. [DOI] [PubMed] [Google Scholar]
- Strother SC, Anderson J, Hansen LK, Kjems U, Kustra R, Sidtis J, Frutiger S, Muley S, Laconte S, and Rottenberg D. (2002). The quantitative evaluation of functional neuroimaging experiments: the NPAIRS data analysis framework. NeuroImage, 15, 747–71. [DOI] [PubMed] [Google Scholar]
- Studholme C, Hill DLG, and Hawkes DJ. An overlap invariant entropy measure of 3d medical image alignment. Pattern Recogn. 32, pp. 71–86, 1999. [Google Scholar]
- Sureau FC, Reader AJ, Comtat C, Leroy C, Ribeiro MJ, Buvat I, Trébossen R. Impact of image-space resolution modeling for studies with the high-resolution research tomograph. J Nucl Med, 2008; 49(6): 1000–8. [DOI] [PubMed] [Google Scholar]
- Svarer C, Madsen K, Hasselbalch SG, Pinborg LH, Haugbøl S, Frøkjær VG, Holm S, Paulson OB, Knudsen GM. MR-based automatic delineation of volumes of interest in human brain PET-images using probability maps. NeuroImage 2005;24:969–79. [DOI] [PubMed] [Google Scholar]
- Tyrer AE, Levitan RD, Houle S, Wilson AA, Nobrega JN, Rusjan PM, & Meyer JH (2016). Serotonin transporter binding is reduced in seasonal affective disorder following light therapy. Acta Psychiatrica Scandinavica, 134(5), 410–419. [DOI] [PubMed] [Google Scholar]
- van den Heuvel OA, Boellaard R, Veltman DJ, et al. Attenuation correction of PET activation studies in the presence of task-related motion. Neuroimage. 2003. August;19(4):1501–9. [DOI] [PubMed] [Google Scholar]
- Yang J, Hu C, Guo N, Dutta J, Vaina LM, Johnson KA, … Li Q (2017). Partial volume correction for PET quantification and its impact on brain network in Alzheimer’s disease. Scientific Reports, 7(1), 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitley E & Ball J. (2002). Statistics review 4: Sample size calculations. Crit Care, 6(4);335–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanderigo F, Mann JJ, & Ogden RT (2017). A hybrid deconvolution approach for estimation of in vivo non-displaceable binding for brain PET targets without a reference region. PLoS One. 2017 May 1;12(5):e0176636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YW, Turk BE, Rudnick G. Control of serotonin transporter phosphorylation by conformational state. Proc Natl Acad Sci U S A. 2016. May 17;113(20):E2776–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zientek F, Winter K, … Hesse S (2016). Effortful control as a dimension of temperament is negatively associated with prefrontal serotonin transporter availability in obese and non-obese individuals. European Journal of Neuroscience, 44(7), 2460–2466. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.