Abstract
Background:
Previous research has demonstrated significant relationships between obesity and brain structure. Both phenotypes are heritable, but it is not known whether they are influenced by common genetic factors. We investigated the genetic etiology of the relationship between individual variability in brain morphology and BMIz using structural MRI in adolescent twins.
Method:
The sample (n=258) consisted of 54 monozygotic and 75 dizygotic twin pairs (mean(SD) age = 13.61(0.505), BMIz = 0.608(1.013). Brain structure (volume and density of gray and white matter) was assessed using VBM. Significant voxelwise heritability of brain structure was established using the Accelerated Permutation inference for ACE models (APACE) program, with structural heritability varying from 15 to 97%, depending on region. Bivariate heritability analyses were carried out comparing additive genetic and unique environment models with and without shared genetics on BMIz and the voxels showing significant heritability in the APACE analyses.
Results:
BMIz was positively related to gray matter volume in the brainstem and thalamus and negatively related to gray matter volume in the bilateral uncus and medial orbitofrontal cortex, gray matter density in the cerebellum, prefrontal lobe, temporal lobe, and limbic system, and white matter density in the brainstem. Bivariate heritability analyses showed that BMIz and brain structure share ~1/3 of their genes and that ~95% of the phenotypic correlation between BMIz and brain structure is due to shared additive genetic influences. These regions included areas related to decision-making, motivation, liking vs. wanting, taste, interoception, reward processing/learning, caloric evaluation, and inhibition.
Conclusion:
These results suggested genetic factors are responsible for the relationship between BMIz and heritable BMIz related brain structure in areas related to eating behavior.
Keywords: VBM, Heritability, Obesity, Volume, Density, Adolescence
1. Introduction
Obesity has become a major health concern worldwide (James et al., 2001). Child and adolescent obesity rates have significantly increased since the 1970s (Sassi et al., 2009), with worldwide obesity rates in children and adolescents having risen from 0.7% to 5.6% in girls and 0.9% to 7.8% in boys in this time period (NCD-RisC, 2017). Early life obesity is typically measured through Body Mass Index Z score (BMIz) or percentile, a measure examining where a person’s weight falls on a distribution for people their age, sex, and height with a Z score above 1.645 (the 95th percentile) considered obese. Early life obesity increases the risk of later health problems, even if the person is no longer obese (Reilly and Kelly, 2011). A better understanding of etiological mechanisms of obesity and its neural correlates can facilitate prevention and intervention efforts. Obesity is a complex phenotype influenced by both genetic and environmental factors. Twin studies can estimate the heritability of a trait (e.g. BMI or brain structure), which is a proportion of the total phenotypic (observed) inter-individual variation that is attributable to genetic differences, and genetic correlation between traits (rG), i.e. the proportion of genes two variables share by comparing the covariances of a phenotype between people with differing degrees of genetic (e.g. 100% for identical twins, on average 50% for fraternal twins, etc.) and environmental similarity (e.g. 100% for twins raised together, 0% for unrelated individuals raised apart; Neale and Maes, 2004). Twin and family studies have shown that the risk for obesity is strongly influenced by genetic factors, with ~80% of adolescent BMIz variance being genetic (Elks et al., 2012), though the exact mechanisms by which genetic factors contribute to BMIz are not well understood. Research has found that eating behaviors predict weight and weight gain (Gallant et al., 2010; Pothos et al., 2009; Sung et al., 2009), are significantly heritable (Carnell et al., 2008; Keskitalo et al., 2008; Sung et al., 2010; Tholin et al., 2005), and share genetic influences with obesity (Keskitalo et al., 2008). Both eating behavior and obesity have been linked to brain structure in regions related to reward, caloric evaluation, taste, decision making, and inhibition, reflecting that brain structure and BMIz are linked through eating behavior, as either a cause or a consequence of overeating behaviors (Kennedy et al., 2016; Maayan et al., 2011; Su et al., 2017; van der Laan et al., 2016). Given that BMIz, eating behavior, and brain structure are heritable and phenotypically related (Blokland et al., 2012; Carnell et al., 2008; Elks et al., 2012; Jansen et al., 2015; Kennedy et al., 2016; Keskitalo et al., 2008; Maayan et al., 2011; Su et al., 2017; Sung et al., 2010; Tholin et al., 2005; van der Laan et al., 2016; van der Lee et al., 2017), it is reasonable to expect that there are shared genetic factors between BMIz and brain structure, possibly mediated by variance in eating behavior related to heritable brain structure. It is important to note that both heritability and genetic correlations between traits can change with age, particularly during development.
Understanding the heritable relationship between BMIz and brain structure during adolescence is particularly important as adolescence is a significant period in the development of the frontostriatal reward and inhibition regions (DePasque and Galvan, 2017; Galvan 2005; Sowell 2001) which are related to the overeating behaviors associated with obesity (Maayan et al., 2011; Su et al., 2017; van der Laan et al., 2016). Obesity is related to striatal volumes and response to food rewards (Kennedy et al., 2016; Stice et al., 2008) in a manner consistent with a reward deficit hypothesis that states individuals with a weaker reward response tend to overeat in order to reach a reward threshold (Blum 2014). Prefrontal regions involved in executive function and inhibition and insular regions involved in interoception have also been implicated in obesity and eating behavior (Kennedy et al., 2016; Kishinevsky et al., 2012; Le et al., 2007; Maayan et al., 2011; Pannacciulli et al., 2006; Tuulari et al., 2016). In particular, Kennedy et al (2016) found that BMIz was negatively related to gray matter volume in the medial prefrontal cortex/anterior cingulate, frontal pole, caudate, and uncus, as well as white matter volume in the anterior limb of the internal capsule, adjacent to the caudate. These regions are involved in decision-making, motivation, conflict monitoring, emotion processing, and reward processing (Gilbert et al., 2006; Kerns et al., 2004; Kim, 2013; Rushworth, 2008; Stice et al., 2008), suggesting that atypical forms of related behaviors may be involved in the overeating that leads to increased BMIz. Examining the relationship between these regions and obesity using a twin dataset can help inform to what extent BMIz and the brain structures linked to these behaviors are genetically related.
Another line of research into the etiology of obesity established heritable relationships between brain structure and obesity. In a diffusion tensor imaging (DTI) study of adult obesity in a sample of Mexican-American families, Spieker et al. (2015) found genetic correlations between average fractional anisotropy and obesity in the corpus callosum, internal capsule, thalamic radiations, and superior fronto-occipital fasciculus, ranging from −0.25 to −0.39. Surface and subcortical volume analyses found genetic correlations with cortical surface area in most lobes of the brain (range −0.013 to −0.385) and with regional subcortical volume (range −0.085 to −0.274). Curiously, the phenotypic correlations (i.e. Pearson’s) between obesity and brain structure were small (all below +/−0.1; Curran et al., 2013). Significance values were not reported for the surface and subcortical analyses, making interpretations difficult. These analyses were carried out in adults with a very wide age range (18-81 years old) and a lower obesity heritability (58%) than reported in adolescents. They also used summary measures (e.g. average surface area of the superior frontal gyrus), which are insensitive to local variation in what are often large structures encompassing multiple functionally distinct regions. It is important to note that estimates of heritability and genetic correlations in the above studies were based on family pedigree data, where genetic and common environmental factors are difficult to disentangle because the degree of gene sharing and environment sharing is strongly correlated within extended family structures (except special cases such as adoption).
Two major gaps in knowledge remain. First, while brain structure heritability, obesity heritability, and the association between brain structure and obesity have been separately examined (Blokland et al., 2012; Elks et al., 2012; Janowitz et al., 2015; Kennedy et al., 2016; Lenroot et al., 2009; Swagerman et al., 2014), it remains unclear whether genetic influences on brain volume and density significantly overlap with genetic influences on BMIz. Here we address this gap in knowledge by conducting a series of bivariate genetic analyses to estimate the degree of genetic overlap between BMIz and brain structure. Second, most of the previous research has been conducted in adult samples, whereas the frontostriatal BMIz-related brain structures involved in reward, decision-making, evaluation, and inhibition are largely shaped during adolescence, making adolescence a critical developmental stage for the study of the relationships between structural variation in the brain and BMIz. The present study addressed this problem by utilizing a sample of adolescent twins, allowing the estimation of shared genetics in a cohort where obesity is most heritable (Elks et al., 2012). It is not clear whether genetic influences on BMIz and brain structure are independent or shared, and whether phenotypic correlations between them can be attributed, at least in part, to overlapping genetic influences. Accordingly, the aim of this study was to address these questions through the assessment of genetic correlations between brain structure and BMIz using bivariate heritability analyses in a sample of adolescent twins.
We hypothesized that there are shared genetic influences for BMIz and brain regions implicated in eating behavior that account for a significant portion of the phenotypic relationship between BMIz and brain structure. Based on our previous study using similar methods in a sample of unrelated adolescents (Kennedy et al., 2016), we hypothesized that the volume of brain regions involved in regulation of eating behavior, including striatal, medial temporal, medial prefrontal, frontal pole, and anterior cingulate gray matter and peristriatal white matter volumes, would be negatively related to BMIz. Based on previous twin research, we expected that BMIz would have a high heritability (~80%; Elks et al., 2012). While we were not aware of previous voxel-based morphometry (VBM) heritability studies in adolescents, meta-analyses of adult regional structural volumes (as derived from lobular volume and FreeSurfer subcortical segmentations) suggested that the hypothesized regions should have univariate heritabilities in the 65-75% range (Blokland et al., 2012).
2. Materials and Methods
2.1. Participants
Monozygotic (MZ) and dizygotic (DZ) twin pairs were recruited through the Missouri Family registry. Participants were excluded based on history of neurological or psychiatric illness, physical or intellectual impairment, history of substance use, pregnancy, or non-removable metal in their body. The study protocol was approved by Washington University School of Medicine’s Human Research Protection Office. Informed consent was obtained from parents or legal guardians and an informed assent was obtained from child participants. The final sample used for these analyses consisted of 129 twin pairs (54 monozygotic and 75 dizygotic), ages 12 to 14 years old (mean(SD) 13.61(0.505) years old). Zygosity was determined using a modified version of the twin zygosity questionnaire (TZQ; Goldsmith, 1991) completed by parents of twins and included ratings of physical similarity and differences between twins, difficulty of telling the twins apart by family members, friends, and other parties. When TZQ scores were unavailable for both twins (two pairs), zygosity determination was based on an analogue scale measures of facial similarity completed by the study coordinator and research assistants who interacted with both twins during their laboratory visit. All twin pairs were the same sex, with 63 male (25 MZ) and 66 female (29 MZ) twin pairs.
2.2. BMI measurement
BMI z-score (BMIz) was calculated based on height, weight, and age obtained during a laboratory visit, typically the same day as the scan, using formulas derived from the 2000 Center for Disease Control growth charts (Kuczmarski et al., 2002). As child and adolescent obesity rates have risen since 2000 (Sassi et al., 2009), BMIz for this sample did not have a mean of 0. Z-scores derived by standardizing our sample’s BMIz values were used to identify outliers. Individuals with a standardized Z outside +/− 3 were considered outliers and they and their twin were removed from the dataset, resulting in two twin pairs being excluded. Post-hoc analyses including outliers did not change the phenotypic or genetic relationship between BMIz and brain structure. The mean(SD) BMIz was 0.608(1.013) and had a range of −2.36 to 2.76.
2.3. MRI Acquisition and Preprocessing
All participants were scanned on a Siemens 3T Prisma Fit at Washington University Medical School’s Neuroimaging Laboratories. A new navigator-guided T1 MPRAGE scan, developed to compensate for movement (Tisdall et al., 2012), was obtained using an axial acquisition in a 32-channel head coil array with a TR of 2500ms, TE of 2.88ms, TI of 1060ms, 176 slices, no gap, and field of view of 256mm. VBM preprocessing was carried out using Statistical Parametric Mapping 12’s Diffeomorphic Anatomical Registration Through Exponentiated Lie Algebra (Ashburner, 2007; Friston et al., 2007) package to warp segmented whole brain gray and white matter volumes into 1mm3 Montreal Neurological Institute space. Spatial smoothing was carried out using an 8mm full width at half maximum. Images were inspected for failed warping, ultimately no one was excluded. Brain masks were created by averaging all subject’s tissue maps and filtering out voxels with values under 0.3; the gray matter mask was then manually edited to exclude some para-orbitofrontal non-gray matter.
2.4. Statistical Analyses
Preliminary statistical analyses were carried out using the Permutation Analysis of Linear Models (PALM; Winkler et al., 2014) package as it can control for familial non-independence. The relationship between BMIz and gray and white matter volume and density was evaluated while controlling for age, sex, ethnicity (Caucasian/non-Caucasian), the interaction between BMIz and age, and total tissue volume. Clusters surviving a Threshold Free Cluster Enhancement family wise error rate of 0.05 (1000 permutations) were used for further analyses examining univariate brain structural heritability.
Univariate heritability of BMIz (controlling for age, sex, ethnicity and the interaction between BMIz and age) was carried out using OpenMx (Neale et al., 2016) implemented in R-3.4.4. Models tested were additive genetics (A), non-additive genetics (D), and unique environment (E; ADE), A, shared environment (C), and E (ACE), A and E, C and E, and only E. Model comparisons were performed to identify the best fitting model. This was repeated for the total gray and white matter volumes. Voxelwise brain structure heritability was examined using the Accelerate Permutation inference for ACE models (APACE; Chen et al., 2014) script on the significant clusters from the PALM analyses. APACE takes the voxelwise heritability and compares it to a null created by randomizing zygosity 1000 times. All voxels surviving a False Discovery Rate of 0.05 were used for bivariate analyses.
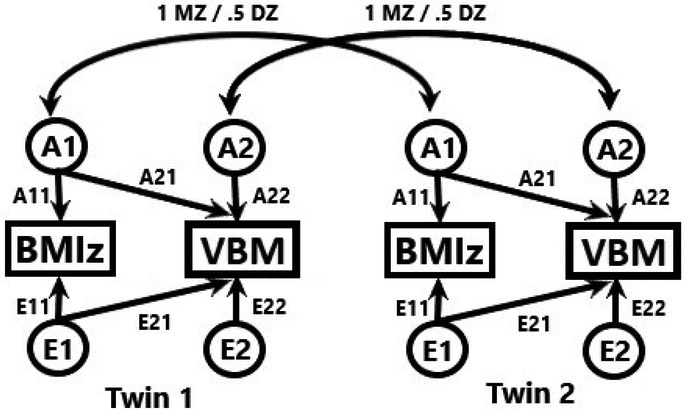
Each significantly heritable voxel was extracted and entered into a bivariate heritability analysis with BMIz in a lower Cholesky model (see Figure 1) in OpenMx. This was done after controlling for the effects of age, sex, ethnicity and the interaction of BMIz and age on BMIz and the effects of age, sex, ethnicity, the interaction between BMIz and age, and total tissue volume on brain structure. Three models were tested: A and E with paths allowing common and unique genetics and error, A and E with no common genetics between BMIz and brain structure (AEdA21), and an E only model. The last two models were tested against the AE model in likelihood ratio tests. A false discovery rate of 0.05 was applied to the voxelwise AE vs. AEdA21 model to control for multiple comparisons when testing if there were significant shared genetics between BMIz and brain structure. Clusters greater than 100 voxels were reported here. For summary purposes, this was repeated on average voxel values for each cluster. This was repeated (save for multiple comparison corrections) to examine the shared genetics of BMIz, total gray matter volume, and total white matter volume.
Figure 1:
Bivariate additive genetic non-shared environment heritability model. MZ = monozygotic, DZ = Dizygotic. A/E11/22 = Unique genetics/non-shared environment on BMIz/VBM. A/E21 = Shared genetics/non-shared environment between BMIz and VBM.
Data from this ongoing project can be made available by request of the principal investigator (A.P.A.) after de-identification and documentation is completed. Upon the completion of the study, the data will be shared with the scientific community, pending availability of appropriate resources. All code can be supplied upon request and can be freely shared or reused. This is in compliance with the funding body and institutional review board.
3. Results
3.1. Regression Results
3.1.1. Volume
BMIz was positively related to gray matter volume in the brainstem and in the bilateral thalamus. BMIz was negatively related to gray matter volumes in the bilateral uncus, extending into the orbitofrontal cortex (OFC). BMIz did not show a significant relationship with white matter volume. See Table 1 and Figure 2 for more detail.
Table 1:
TFCE results for the association of BMIz with gray and white matter volume and density
| Volume | ||||||||
|---|---|---|---|---|---|---|---|---|
| Gray Matter | ||||||||
| BMIz+ | ||||||||
| Area | k | X | Y | Z | TFCE p | Sig h2/TFCE | AEpD21/TFCE | AEpD21/Sig h2 |
| Brainstem | 1399 | −4 | −35 | −13 | 0.019 | 0.77 | 0.77 | 1.00 |
| L Thalamus | 1152 | −16 | −28 | 2 | 0.037 | 1.00 | 1.00 | 1.00 |
| R Thalamus | 1919 | 9 | −31 | −3 | 0.029 | 0.71 | 0.71 | 1.00 |
| BMIz− | ||||||||
| Area | k | X | Y | Z | TFCE p | Sig h2/TFCE | AEpD21/TFCE | AEpD21/Sig h2 |
| L Uncus/medial OFC | 5897 | −17 | 6 | −32 | 0.001 | 0.30 | 0.30 | 1.00 |
| R Uncus/medial OFC | 5727 | 19 | 8 | −35 | 0.001 | 0.17 | 0.17 | 1.00 |
| Density | ||||||||
| Gray Matter | ||||||||
| BMIz− | ||||||||
| Area | k | X | Y | Z | TFCE p | Sig h2/TFCE | AEpD21/TFCE | AEpD21/Sig h2 |
| L Cerebellum | 6448 | −32 | −58 | −51 | 0.025 | 0.10 | 0.09 | 0.99 |
| R Cerebellum | 8392 | 32 | −55 | −51 | 0.024 | 0.20 | 0.20 | 1.00 |
| R Uncus/OFC/inferior temporal/fusiform/ amygdala/hippocampus/parahippocampal/ inferior, middle and superior frontal/insula/ B cingulate/medial frontal/frontal pole | 127507 | 15 | 48 | −22 | 0.012 | 0.12 | 0.08 | 0.73 |
| L Uncus/OFC/inferior temporal/insula/ supramarginal/pallidum/amygdala/ hippocampus | 29978 | −27 | 9 | −35 | 0.023 | 0.20 | 0.16 | 0.82 |
| R Inferior occipital | 127 | 46 | −80 | 2 | 0.046 | 0.00 | 0.00 | - |
| White Matter | ||||||||
| BMIz− | ||||||||
| Area | k | X | Y | Z | TFCE p | Sig h2/TFCE | AEpD21/TFCE | AEpD21/Sig h2 |
| Brainstem | 1976 | −2 | −30 | −15 | 0.019 | 0.55 | 0.22 | 0.40 |
Table 1: Regions significantly associated with BMIz. TFCE = Threshold Free Cluster Enhancement analyses. BMIz+/− = Structures positively/negatively associated with BMIz. k = Volume, XYZ = MNI peak coordinate, TFCE p = peak TFCE significance, Sig h2/TFCE = proportion of TFCE cluster voxels significantly heritable, AEpD21/TFCE = proportion of TFCE cluster voxels with significant shared genetics, AEpD21/Sig h2 = proportion of significantly heritable voxels with significant shared genetics. L = Left, R = Right, B = Bilateral. / = cluster covers multiple regions. OFC = Orbitofrontal cortex.
Figure 2:
Brain regions significantly related with BMIz. A: Gray and white matter density. B: Gray matter volume. Color scales indicate the significance size of the correlations. Yellow-red indicates significant positive correlations, whereas light blue to dark blue indicate negative correlations. Lighter colors indicate higher significance level (lower p-value).
3.1.2. Density
BMIz was negatively related to gray matter density in the cerebellum, scattered clusters in all lobes, the insula, cingulate, and some subcortical structures. BMIz was negatively related to white matter density in the brainstem. See Table 1 and Figure 2 for a full list of regions.
3.2. Univariate Heritability
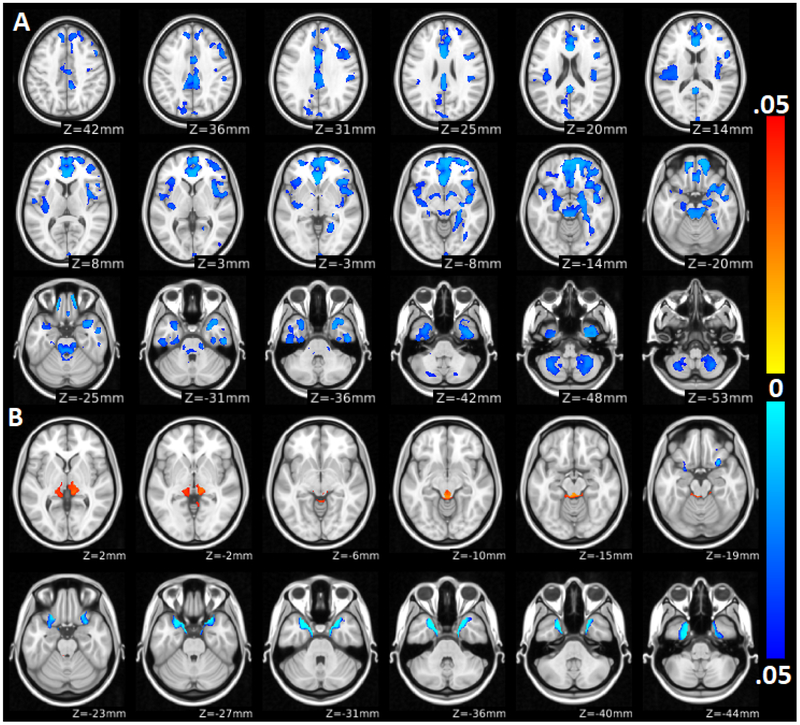
The AE model had the best fit for BMIz with a heritability of 89%. For total gray matter volume, an ACE model showed the best fit (A2 = 0.43, C2 = 0.47, E2 = 0.1, probability ACE is not a better fit than AE (p ACE < AE) = 0.002, p ACE < CE < 0.001, p ACE < E < 0.001). For total white matter volume, the AE model had the best fit (A2 = 0.93, E2 = 0.07, p ACE < AE = 0.074, p ACE < CE < 0.001, p ACE < E < 0.001). See Table 2 for more information. APACE analyses of clusters significantly associated with BMIz found significantly heritable brain gray matter volumes (36 to 89%) in parts of all regions associated with BMIz. Significantly heritable gray matter density (15 to 97%) was observed in most regions where a relationship between BMIz and brain structure were observed. White matter densities were heritable (56 to 97%) in the brainstem in the superior cerebellar peduncle and spinothalamic tract. See Table 3 and Figure 3 for the full list of significant regions.
Table 2:
Univariate heritability analyses
| Analysis | ADE A2 |
ADE D2 |
ADE E2 |
ACE A2 |
ACE C2 |
ACE E2 |
AE A2 |
AE E2 |
CE C2 |
CE E2 |
rMZ | rDZ | LL_ADE | LL_ACE | LL_AE | LL_CE | LL_E |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BMIz | 0.66 | 0.23 | 0.11 | 0.90 | 0.00 | 0.10 | 0.90 | 0.10 | 0.60 | 0.40 | 0.89 | 0.40 | 640.67 | 641.08 | 641.08 | 682.12 | 738.68 |
| Gray Volume | 0.90 | 0.00 | 0.10 | 0.43 | 0.47 | 0.10 | 0.90 | 0.10 | 0.78 | 0.22 | 0.90 | 0.70 | −1165.56 | −1175.63 | −1165.56 | −1156.74 | −1038.28 |
| White Volume | 0.93 | 0.00 | 0.07 | 0.65 | 0.28 | 0.07 | 0.93 | 0.07 | 0.74 | 0.26 | 0.92 | 0.62 | −1194.88 | −1198.07 | −1194.88 | −1159.99 | −1058.87 |
Table 2: Standardized univariate heritability values for ADE, ACE, AE, and CE values. rMZ = monozygotic twin correlations, rDZ = dizygotic twin correlations. LL_ = −2 log likelihood values.
Table 3:
Significantly heritable brain regions
| Volume | ||||
|---|---|---|---|---|
| Gray Matter | ||||
| BMI+ | ||||
| Area | k | X | Y | Z |
| R Thalamus | 1365 | 9 | −31 | −3 |
| L Thalamus | 1152 | −16 | −28 | 2 |
| Brainstem | 1073 | −4 | −36 | −16 |
| BMI− | ||||
| Area | k | X | Y | Z |
| L Uncus/medial OFC | 1775 | −18 | 7 | −32 |
| R Uncus/medial OFC | 428 | 19 | 8 | −32 |
| R Temporal pole/OFC | 320 | 28 | 14 | −28 |
| R Inferior temporal | 224 | 23 | −7 | −49 |
| Density | ||||
| Gray Matter | ||||
| BMI− | ||||
| Area | k | X | Y | Z |
| B Anterior cingulate/medial prefrontal | 3753 | −2 | 28 | −5 |
| L Hippocampus/pallidum/posterior insula | 3100 | −16 | −12 | −14 |
| R OFC/insula/temporal pole | 2974 | 44 | 25 | −4 |
| L Insula | 1533 | −45 | 5 | −13 |
| R medial OFC/medial prefrontal/frontal pole | 1408 | 13 | 49 | −21 |
| R Insula | 1360 | 34 | 15 | 4 |
| R Cerebellum | 1267 | 31 | −54 | −49 |
| R Lingual | 971 | 27 | −39 | −12 |
| R Fusiform/inferior temporal | 879 | 24 | −7 | −49 |
| L Cerebellum | 618 | −35 | −55 | −49 |
| R OFC | 587 | 33 | 39 | −12 |
| R Fusiform/inferior temporal | 516 | −27 | −11 | −48 |
| R Middle cingulate | 470 | 3 | 3 | 31 |
| R Cerebellum | 417 | 27 | −58 | −58 |
| B Middle cingulate | 395 | 3 | −27 | 32 |
| B Posterior cingulate | 372 | 1 | −39 | 27 |
| L Anterior insula | 336 | −31 | 25 | −1 |
| R Fusiform | 306 | 35 | −55 | −18 |
| L Posterior cingulate | 286 | −10 | −56 | 28 |
| R Posterior operculum | 272 | 54 | 12 | 13 |
| L Inferior temporal | 271 | −44 | −14 | −38 |
| L Medial superior frontal | 190 | −5 | 49 | 27 |
| L OFC | 166 | −23 | 33 | −14 |
| R Hippocampus/parahippocampal gyrus | 166 | 27 | −26 | −14 |
| L Parietal operculum | 149 | −38 | −27 | 21 |
| R Medial superior frontal | 138 | 10 | 40 | 41 |
| R Medial superior frontal | 135 | 13 | 41 | 40 |
| White Matter | ||||
| BMI− | ||||
| Area | k | X | Y | Z |
| R Superior cerebellar peduncle | 303 | 8 | −35 | −25 |
| L Superior cerebellar peduncle | 243 | −3 | −33 | −13 |
| L Superior cerebellar peduncle | 208 | −1 | −32 | −13 |
| L Superior cerebellar peduncle | 176 | −5 | −46 | −24 |
| R Spinothalamic tract | 150 | 15 | −22 | −19 |
Table 3: Significantly heritable regions associated with BMIz. BMIz+/− = Structures positively/negatively associated with BMIz. k = Volume, XYZ = MNI peak coordinate. L = Left, R = Right, B = Bilateral. / = cluster covers multiple regions. OFC = Orbitofrontal cortex.
Figure 3:
Voxelwise heritability of brain structure related to BMIz. Color scales indicate the proportion of phenotypic variance attributable to genetic factors. The yellow-red scale indicates heritability of density, and the blue scale indicates heritability of volume. Lighter colors represent higher heritability.
3.3. Bivariate Heritability and Genetic Correlations
Bivariate heritability analyses found significant shared genetic influences between BMIz and brain structure. For the gray matter analyses, significant genetic correlations were observed in clusters in most regions that were both related to BMIz and heritable. The mean(SD) genetic correlation (rG) was 0.33(0.04) for gray matter volumes positively related to BMIz, −0.42(0.09) for volumes negatively related to BMIz, and was −0.3(0.07) for densities negatively related to BMIz. The mean(SD) percentage of the phenotypic correlation explained by shared genetics (%A) was 99(5.5)% for gray matter volumes positively related to BMIz, 94(6.8)% for volumes negatively related to BMIz, and 101(12)% for density (%A can exceed 100% if the variance due to error reduces the relationship between two variables). For white matter density, shared genetic influences were observed in the superior cerebellar peduncle with a mean(SD) rG of −0.25(0.02) and %A of 94(7.3)%. There were no voxels where the model with no genetics fit best. See Table 4 and Figure 4 for the full list of regions. BMIz was not related to total gray matter volume (probability the AE model (p AE) is a better fit than the AE model with no shared genetics (AE dA21) = 0.81) but was a trend for total white matter volume (p AE better fit than AE dA21 = 0.086). Total gray and white matter volume were significantly related (p AE better fit than AE dA21 < 0.001) with an rG of 0.71 and %A of 87.9%.
Table 4:
Regions with significant shared genetics between BMIz and gray or white matter volume or density
| Volume | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gray Matter | ||||||||||||||||
| BMI+ | ||||||||||||||||
| Area | k | x | y | z | SA2 | LL_AE | LL_AEdA21 | LL_E | pAEdA21 | PE | rG | rE | %A | %E | SrMZ | SrDZ |
| R Thalamus | 1365 | 9 | −31 | −3 | 0.67 | −359.04 | −345.99 | −228.06 | <0.001 | <0.001 | 0.34 | 0.04 | 97.26 | 2.74 | 0.63 | 0.39 |
| L Thalamus | 1152 | −16 | −28 | 2 | 0.73 | −345.87 | −333.46 | −214.69 | <0.001 | <0.001 | 0.33 | 0.00 | 99.94 | 0.06 | 0.68 | 0.31 |
| PAG | 1073 | −4 | −36 | −16 | 0.66 | −619.57 | −594.39 | −486.03 | <0.001 | <0.001 | 0.46 | 0.00 | 99.74 | 0.26 | 0.66 | 0.55 |
| BMI− | ||||||||||||||||
| Area | k | x | y | z | SA2 | LL_AE | LL_AEdA21 | LL_E | pAEdA21 | pE | rG | rE | %A | %E | SrMZ | SrDZ |
| L Uncus/temporal pole | 1775 | −18 | 7 | −32 | 0.64 | −310.63 | −288.03 | −190.76 | <0.001 | <0.001 | −0.47 | −0.17 | 91.48 | 8.52 | 0.65 | 0.36 |
| R Uncus/temporal pole | 428 | 19 | 8 | −32 | 0.61 | −378.36 | −352.15 | −260.34 | <0.001 | <0.001 | −0.51 | −0.13 | 93.33 | 6.67 | 0.65 | 0.31 |
| R Temporal pole | 320 | 28 | 14 | −28 | 0.62 | −338.56 | −320.58 | −218.83 | <0.001 | <0.001 | −0.43 | −0.02 | 99.02 | 0.98 | 0.72 | 0.22 |
| R Fusiform | 224 | 23 | −7 | −49 | 0.57 | −324.31 | −306.27 | −206.45 | <0.001 | <0.001 | −0.44 | 0.00 | 99.88 | 0.12 | 0.58 | 0.29 |
| Density | ||||||||||||||||
| Gray Matter | ||||||||||||||||
| BMI− | ||||||||||||||||
| Area | k | x | y | z | SA2 | LL_AE | LL_AEdA21 | LL_E | pAEdA21 | pE | rG | rE | %A | %E | SrMZ | SrDZ |
| L Hippocampus/pallidum/insula | 2672 | −16 | −12 | −14 | 0.66 | −401.13 | −388.95 | −268.28 | <0.001 | <0.001 | −0.33 | 0.07 | 105.21 | −5.21 | 0.71 | 0.21 |
| B Anterior cingulate/L medial prefrontal | 2554 | −2 | 28 | −5 | 0.83 | −802.19 | −780.33 | −635.36 | <0.001 | <0.001 | −0.39 | 0.19 | 108.36 | −8.36 | 0.85 | 0.25 |
| R Insula/uncus | 2250 | 44 | 25 | −4 | 0.53 | −321.02 | −313.55 | −209.55 | 0.006 | <0.001 | −0.31 | 0.06 | 106.21 | −6.21 | 0.60 | 0.00 |
| R Cerebellum | 1262 | 31 | −54 | −49 | 0.73 | −644.71 | −637.46 | −498.63 | 0.007 | <0.001 | −0.25 | −0.12 | 90.49 | 9.51 | 0.78 | 0.24 |
| R Medial OFC/frontal pole | 1156 | 13 | 49 | −21 | 0.66 | −540.79 | −534.50 | −412.27 | 0.012 | <0.001 | −0.25 | 0.06 | 106.25 | −6.25 | 0.68 | 0.23 |
| L Insula/temporal pole | 982 | −45 | 5 | −13 | 0.69 | −539.70 | −530.14 | −404.75 | 0.002 | <0.001 | −0.29 | −0.02 | 98.54 | 1.46 | 0.71 | 0.28 |
| R Fusiform/inferior temporal | 860 | 24 | −7 | −49 | 0.57 | −513.65 | −502.82 | −392.49 | 0.001 | <0.001 | −0.35 | 0.11 | 110.97 | −10.97 | 0.67 | 0.01 |
| R Lingual/parahippocampal | 859 | 27 | −39 | −12 | 0.67 | −429.45 | −421.37 | −292.86 | 0.004 | <0.001 | −0.27 | 0.05 | 104.80 | −4.80 | 0.72 | 0.30 |
| L Cerebellum | 612 | −35 | −55 | −49 | 0.72 | −315.82 | −307.66 | −175.57 | 0.004 | <0.001 | −0.27 | −0.12 | 91.02 | 8.98 | 0.77 | 0.10 |
| L Medial prefrontal | 596 | −4 | 56 | −12 | 0.62 | −655.29 | −646.17 | −531.51 | 0.003 | <0.001 | −0.31 | 0.02 | 101.62 | −1.62 | 0.68 | 0.18 |
| L Fusiform/inferior temporal | 464 | −27 | −11 | −48 | 0.64 | −767.09 | −744.27 | −643.80 | <0.001 | <0.001 | −0.47 | 0.04 | 102.33 | −2.33 | 0.72 | 0.01 |
| R Anterior insula | 447 | 35 | 15 | 4 | 0.63 | −580.61 | −567.99 | −459.81 | <0.001 | <0.001 | −0.36 | 0.13 | 111.05 | −11.05 | 0.63 | 0.19 |
| R Cerebellum | 417 | 27 | −58 | −58 | 0.62 | −450.65 | −441.78 | −324.63 | 0.003 | <0.001 | −0.30 | 0.03 | 102.61 | −2.61 | 0.63 | 0.19 |
| B Middle cingulate | 377 | 3 | 3 | 31 | 0.75 | −596.90 | −578.41 | −456.11 | <0.001 | <0.001 | −0.38 | −0.02 | 98.87 | 1.13 | 0.75 | 0.29 |
| L Anterior insula | 336 | −31 | 25 | −1 | 0.74 | −647.77 | −633.27 | −504.18 | <0.001 | <0.001 | −0.34 | −0.09 | 94.68 | 5.32 | 0.80 | 0.28 |
| R Posterior insula | 328 | 40 | −18 | 18 | 0.68 | −451.83 | −444.71 | −324.83 | 0.008 | <0.001 | −0.26 | 0.15 | 115.72 | −15.72 | 0.73 | 0.11 |
| R Fusiform | 292 | 35 | −55 | −18 | 0.69 | −600.26 | −591.89 | −458.94 | 0.004 | <0.001 | −0.27 | −0.03 | 97.79 | 2.21 | 0.72 | 0.37 |
| B Posterior cingulate | 280 | 3 | −27 | 32 | 0.80 | −471.84 | −459.20 | −315.67 | <0.001 | <0.001 | −0.31 | −0.03 | 98.20 | 1.80 | 0.82 | 0.27 |
| L Inferior temporal | 267 | −44 | −14 | −38 | 0.80 | −692.90 | −678.58 | −528.03 | <0.001 | <0.001 | −0.32 | −0.15 | 92.71 | 7.29 | 0.84 | 0.34 |
| B Posterior cingulate | 262 | 1 | −37 | 27 | 0.74 | −670.98 | −651.56 | −523.86 | <0.001 | <0.001 | −0.39 | −0.01 | 99.61 | 0.39 | 0.79 | 0.32 |
| R Inferior frontal | 168 | 45 | 43 | −12 | 0.54 | −489.75 | −479.10 | −367.94 | 0.001 | <0.001 | −0.35 | 0.13 | 113.95 | −13.95 | 0.62 | 0.14 |
| L Posterior cingulate | 144 | −10 | −56 | 28 | 0.83 | −616.39 | −598.11 | −443.33 | <0.001 | <0.001 | −0.35 | 0.04 | 101.78 | −1.78 | 0.83 | 0.45 |
| L Post Insula | 143 | −38 | −27 | 21 | 0.71 | −511.30 | −502.23 | −368.40 | 0.003 | <0.001 | −0.28 | −0.02 | 98.42 | 1.58 | 0.76 | 0.27 |
| R Hippocampus/parahippocampal | 140 | 28 | −29 | −22 | 0.53 | −634.55 | −625.37 | −520.62 | 0.002 | <0.001 | −0.33 | −0.10 | 91.13 | 8.87 | 0.57 | 0.22 |
| L Medial OFC | 123 | −23 | 33 | −14 | 0.75 | −713.37 | −694.35 | −569.74 | <0.001 | <0.001 | −0.39 | 0.05 | 102.52 | −2.52 | 0.79 | 0.26 |
| White Matter | ||||||||||||||||
| BMI- | ||||||||||||||||
| Area | k | x | y | z | SA2 | LL_AE | LL_AEdA21 | LL_E | pAEdA21 | pE | rG | rE | %A | %E | SrMZ | SrDZ |
| L Superior cerebellar peduncle | 163 | −3 | −33 | −13 | 0.67 | −470.25 | −462.92 | −328.18 | 0.007 | <0.001 | −0.25 | −0.12 | 89.84 | 10.16 | 0.72 | 0.34 |
| R Superior cerebellar peduncle | 161 | 8 | −35 | −25 | 0.59 | −631.26 | −624.33 | −503.69 | 0.008 | <0.001 | −0.27 | −0.08 | 91.79 | 8.21 | 0.65 | 0.18 |
| L Superior cerebellar peduncle | 112 | −5 | −46 | −24 | 0.67 | −620.60 | −613.22 | −483.09 | 0.007 | <0.001 | −0.26 | 0.01 | 100.81 | −0.81 | 0.70 | 0.27 |
Table 4: Regions with significant shared genetics. BMIz+/− = Structures positively/negatively associated with BMIz. k = Volume, XYZ = MNI peak coordinate. SA2 = standardized heritability of the selected structure. LL_ = −2 log likelihood. pAEdA21 = probability the AE model fits better than the model with no shared genetics. pE = probability the AE model is better than the E model. rG = genetic correlation. rE = error correlation. SrMZ = Structure’s monozygotic twin correlation. SrDZ = Structure’s dizygotic twin correlation. L = Left, R = Right, B = Bilateral. / = cluster covers multiple regions. PAG = Periaqueductal gray, OFC = Orbitofrontal cortex.
Figure 4:
Genetic correlations between BMIz and brain structure. A: Gray and white matter density. B: Gray matter volume. Yellow-red = positive correlations, light blue-blue = negative correlations.
4. Discussion
4.1. Accuracy of Hypotheses and Replication of Previous Research
Our hypotheses that BMIz would be heritable as well as phenotypically and genetically related to brain structure in adolescents were partially supported. BMIz heritability was high, as were gray and white matter volumes and densities in many of the areas associated with BMIz. BMIz was significantly associated with gray matter volume in the uncus/orbitofrontal cortex and with gray matter density in the medial prefrontal, frontal pole, and anterior cingulate. While the expected caudate volume differences were not observed, BMIz was associated with pallidum density, another region involved in reward processing. Most heritable clusters shared genetic influences with BMIz. Our findings suggest that the observed phenotypic correlations between BMI and heritable brain structure can be attributed almost entirely (>90%) to common genetic influences.
Brain structural correlates of BMIz were largely consistent with previous research. Structural BMI analyses typically reported reduced volume or density in subcortical, insula, cerebellar, and prefrontal structures that were observed in our density analyses (Kennedy et al., 2016; Pannacciulli et al., 2006; Willette and Kapogiannis, 2015). The positive correlations with gray matter volume and negative with white matter density measures appeared to be novel. This may be due to the use of the permutation based Threshold Free Cluster Enhancement significance approach, as previous research used the random field theory based clustering approach which is less accurate (Eklund et al., 2016) and less sensitive to small, highly significant regions (Hayasaka and Nichols, 2003). VBM based volume and density heritabilities appeared similar or higher than volume heritabilities previously reported (van der Lee et al., 2017; Weise et al., 2017). This may be because previous VBM heritability research was based on adult samples while ours was from a younger age group and may be subject to developmental effects on heritability (Peper et al., 2009; van den Berg 2006). Genetic correlations between brain structure and BMIz were higher in our analyses than those previously reported (Curran et al., 2013; Spieker et al., 2015), possibly due to the previous study’s use of pedigree data which conflates genetic and common environmental effects and use of summary measures from different structural modalities.
The direction of causation in the relationship between brain structure and BMIz is difficult to resolve in a cross-sectional analysis. One plausible hypothesis is that genetically determined differences in brain structure contribute to individual differences in the regulation of eating behavior which in turn influences BMIz. This causal interpretation is indirectly supported by the fact that the majority of brain regions associated with BMIz in the present study have been previously linked to eating behaviors (Kishinevsky et al., 2012; Maayan et al., 2011; van der Laan et al., 2016). An alternative model would be that differences in BMIz lead to differences in brain structure, which is supported by evidence that weight loss surgery resulted in density recovery (Tuulari et al., 2016). The issue of causal relationships between BMIz and brain structure can be clarified by longitudinal or co-twin control studies.
4.2. Functional Associations of Structures Phenotypically Correlated with BMIz
Gray matter volume or density reductions were observed in structures in some way related to food consumption, including systems related to sensation, reward evaluation, and decision-making. As we did not evaluate eating behavior directly any inference on the behavioral significance of structural variances related to BMIz in these analyses is speculative. Cingulate and prefrontal volume reductions have been linked to increased self-reported dietary restraint, though restrainers generally do not eat less than non-restrainers and are likely to be overweight, suggesting self-reported restraint is more linked to intention than behavior (van der Laan et al., 2016). Reduced orbitofrontal cortex (OFC) volume has been linked to disinhibited eating behavior, where someone eats impulsively to environmental food cues (Maayan et al., 2011). Anterior cingulate cortex activity has also been negatively linked to food related disinhibition, but only in the obese (Martin et al., 2010). In cases where someone has both high dietary restraint and high disinhibition, people do not update their eating behaviors based on earlier food exposure, causing them to overeat when new eating opportunities appear (Westenhoefer et al., 1994). The volume and density reductions observed in our analyses in the prefrontal and cingulate cortices support the high food disinhibition/restraint model of overeating in obesity. We also observed structural decreases in the insula, hippocampus, and spinothalamic tract, which are involved in interoceptive awareness (Davidson et al., 2007; Kennedy and Dimitropoulos, 2014; Krames and Foreman, 2007; Rolls, 2016). This may result in reduced awareness of bodily signals of hunger and satiety, leading to overeating and disinhibition related eating to external food cues. The reduced pallidum and posterior cingulate density may also contribute to overeating as the pallidum is involved in controlling reward based on interoceptive need (Nangunoori et al., 2016, Smith and Berridge, 2005, Tindell et al., 2006) and the posterior cingulate has been linked to craving (Brewer et al., 2013) and emotional salience of food in a sensory specific satiety task (Small et al., 2001). Structural reductions here may inhibit the updating of reward values as people reach satiety. Observed reductions in the primary taste and smell cortices of the brain, namely the insula (Rolls, 2016) and uncus (Kiernan, 2012), may reflect reduced sensory input from consumption, which may lead to less sensory reward and overeating to reach a reward threshold. Uncus volume, obesity, and agentic positive emotionality, a personality measure linked to long-term goal pursuit have all been found to be significantly related to one another (Kennedy et al., 2016). Uncus volume and density reductions may represent problems maintaining the long-term restrictive behaviors necessary to maintain a successful diet.
Structural differences related to BMIz were also found in some regions not usually linked to eating behavior or where the observed relationship isn’t well understood. The frontal pole is involved in understanding emotional states (Gilbert et al., 2006). Alexithymia, or difficulty recognizing internal emotional states, is associated with problems regulating emotion (Pandey et al., 2011) and predicts emotional eating in obese women with a binge-eating disorder (Pinaquy et al., 2012). Eating is one method of emotional regulation (Macht, 2008); difficulty processing emotional states related to atypical frontal pole structure may lead to problems in emotion regulation, in turn resulting in overuse of food as a regulatory tool. The parahippocampus is functionally associated with food anticipation, with hunger level correlating with the strength of activation (Stice et al., 2013). The cerebellum is primarily known for its role in movement (Manto, 2012) but has also been linked to meal anticipation (Mendoza, 2010). Disruptions to meal anticipatory regions may result in overeating through excess snacking. The fusiform responds to satiation (Hinton, 2004) and is more active to food cues in people with the obesity linked FTO gene (Kuhn, 2016). Reduced fusiform density may inhibit satiation processing, leading to overeating. Obesity related disruptions to white matter in the cerebellar peduncle have been linked to impaired motor competence (Augustijin et al., 2017; Verstynen et al., 2012). People with poor motor competence may struggle with physical activity, creating a disincentive towards exercise, promoting a positive caloric imbalance through inactivity rather than overeating. The periaqueductal gray (PAG) works with the raphe magnus in inhibiting pain responses during ingestion (Mason, 2011). Atypical PAG activity may facilitate overeating by masking feelings of discomfort caused by overeating related gut distention. Again, as we lacked extensive cognitive and behavioral measures, these interpretations are speculative but may provide avenues for future research.
4.3. Heritability and Shared Genetics
Most brain structures phenotypically related to BMIz were significantly heritable and shared significant genetics with BMIz. Genetic correlation analyses suggest that BMIz and heritable brain volume/density associated with BMIz share around one third of their genes, and that around 95% of the observed phenotypic correlation between BMIz and brain structure can be attributed to these shared genetic influences. Our findings suggest that there is a significant common genetic underpinning between BMIz and most of the heritable brain structures related to BMIz. While structural reductions in adulthood are at least partially attributed to the deleterious effects of an obesogenic diet (Mueller et al., 2012; Walther et al., 2010), these results suggest that there are brain regions where BMIz related decreases in adolescent volume or density are genetic.
4.4. Significance and Summary of Findings
These analyses were significant in several ways. They extended previous structural BMIz research to density analyses where adolescent analyses were lacking, established structural reductions in areas important to eating behavior, established that brain structure in several of these regions is significantly heritable, and that the phenotypical relationship in these regions is largely due to genes shared by BMIz and brain structure. To our knowledge, this is the first bivariate heritability analysis of VBM derived brain structure and BMIz, and one of a very few bivariate neuroimaging analyses known to the author not examining summary values (Chen et al., 2011; Hulshoff Pol et al., 2006; Rao et al., 2018; Rimol et al., 2010). Examining neuroimaging data at the vertex or voxel level is important as summary measures ignore local variation in structure and may overlook significant associations spread out across multiple contiguous regions (Betjemann et al., 2010; Glahn et al., 2010; Glahn et al., 2013; Greenspan et al., 2016; Jahanshad et al., 2013; Karlsgodt et al., 2010; Kochunov et al., 2016; Patel et al., 2017; Pinel and Dehaene, 2013; Spieker et al., 2015); important as there is functional variance within regions and trait-associated variance does not necessarily conform to the anatomical boundaries typically used for segmentation or parcellation.
Overall, these analyses suggested there are shared genetic factors between BMIz and brain structure underlying multiple neurobehavioral domains tied to eating behavior. Heritable structural differences in areas related to sensation, caloric and reward evaluation, decision-making, and reward processing could contribute to obesogenic behaviors through impaired inhibition, eating absent hunger, or overeating due to reduced sensation related to interoception or taste. Almost all of the phenotypic relationship between BMIz and these regions is due to genetic factors. This suggests that there is a genetic component to many of the observed neurostructural differences associated with adolescent obesity, though lacking longitudinal data, we can’t tell if these genetic factors cause altered brain structure leading to obesity or cause obesity, leading to altered brain structure. As the analyses presented here were limited to the relationship between BMIz and brain structure, omitting cognitive, behavioral, etc. measures, the interpretations presented here are speculative.
Limitations and future directions
These analyses were limited in some ways. Lacking an extensive consumptive behavior/cognition battery, our interpretations of the possible ways the observed structural differences might manifest were highly speculative. We also lacked sufficient power to examine more complex bivariate ACE or ADE models that would have allowed us to explore shared environment or non-additive genetic effects; however, the univariate BMIz analyses suggested that there was not any shared environment or non-additive effects to analyze. Furthermore, we had to use a voxelwise correction for our bivariate heritability significance rather than use the permutation-null comparison significance method used in APACE as processing the 1000 minimum permutations required would have taken approximately a year.
Post-hoc Analyses
Adult BMI has been linked to increased in-scanner movement (Siegal et al., 2017), meaning structural differences attributed here to BMIz may instead reflect movement artifacts. Post-hoc analyses were conducted on the average density or volume values extracted from the clusters that were genetically linked to BMIz, adding as a covariate mean movement parameters from resting state scans obtained in the same session. Movement and BMIz were significantly related (partial r = 0.359, p < 0.001, controlling for age, sex, ethnicity, and the interaction between BMIz and age), however the relationship between BMIz and brain structure remained significant in all clusters after the addition of mean movement as a covariate. This sample included individuals who were underweight (below a Z of −1.64 or <5th percentile) which is associated with structural differences separate from those found in obesity (Phillipou et al., 2018). All regions with a significant genetic correlation had both significant phenotypic and genetic correlations following post-hoc analyses where three twin pairs that included an underweight member were removed.
Future research using longitudinal data should clarify the direction of causality in the relationship between brain structure and BMIz and test the hypothesis that this relationship is mediated by eating behavior. One important prediction suggested by the present findings is that both brain structure (in the regions identified here) and BMIz would show significant genetic correlations with disordered eating behaviors.
5. Conclusions
These analyses suggested that both BMIz and brain structures related to eating behavior in adolescents are influenced by overlapping genetic factors. This evidence for a genetic predisposition suggests that if a child has a family history of obesity, they may benefit from interventions to compensate for the associated neurobehavioral differences, even if obesity has not manifested. This may take the form of mindfulness training to focus on the sensation of their food and interoceptive signals, interventions to focus on the long-term ramifications of their eating behavior, or digitally assisted calorie counting to compensate for poor internal caloric evaluation. Mindfulness and calorie counting have demonstrated some efficacy in altering eating behavior or promoting weight loss in people already obese (Alberts et al., 2012; Katterman et al., 2014; Mason et al., 2016; Mendes et al., 2017). As the health consequences of early life obesity can persist after weight loss (Reilly and Kelly, 2011), it is important to address these behaviors early.
Figure 5:
Literature-based eating and obesity related functions of regions phenotypically and genetically linked to BMIz in these analyses. Significant volume and density clusters in a glass brain color coded by relevant function. Striped clusters associated with multiple relevant functions, colors in striped clusters refer to cluster as a whole, not specific colored region. Black clusters (pulvinar, inferior temporal) have no known functional relevance.
Acknowledgements
Research reported in this publication was supported by Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD) of the National Institutes of Health under award number R01HD083614. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The authors acknowledge organizational and technical support by Lindsey Wold, BS and other project staff. The authors also acknowledge the generous giving of time by the study participants and their families.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declarations of interest: none.
References
- Alberts HJEM, Thewissen R, and Raes L (2012). Dealing with problematic eating behaviour. The effects of a mindfulness-based intervention on eating behaviour, food cravings, dichotomous thinking and body image concern. Appetite, 58, 847–851. [DOI] [PubMed] [Google Scholar]
- Ashburner J (2007). A fast diffeomorphic image registration algorithm. NeuroImage, 1, 95–113. [DOI] [PubMed] [Google Scholar]
- Augustijn MJCM, Deconinck FJA, D’Hondt E, Van Acker L, De Guchtenaere A, Lenoir M, Caeyenberchs K (2017). Reduced motor competence in children with obesity is associated with structural differences in the cerebellar peduncles. Brain Imaging and Behavior, 12(4), 1000–1010. [DOI] [PubMed] [Google Scholar]
- Betjemann RS, Phinney Johnson E, Barnard H, Boada R, Filley CM, Filipek PA, Willcutt EG, DeFries JC, et al. (2010). Genetic Covariation Between Brain Volumes and IQ, Reading Performance, and Processing Speed. Behav Genet, 40, 135–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blokland GAM, de Zubicaray GI, McMahon KL, and Wright MJ (2012). Genetic and Environmental Influences on Neuroimaging Phenotypes: A Meta-Analytical Perspective on Twin Imaging Studies. Twin Res Hum Genet, 15(3), 351–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum K, Thanos PK and Gold MS (2014). Dopamine and glucose, obesity, and reward deficiency syndrome. Frontiers in Psychology, 5, 919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer JA, Garrison KA, and Whitfield-Gabrieli S (2013). What about the “self’ is processed in the posterior cingulate cortex? Frontiers in Human Neuroscience, 7, 647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carnell S, Haworth CMA, Plomin R, and Wardle J (2008). Genetic influence on appetite in children. International Journal of Obesity, 32, 1468–1473. [DOI] [PubMed] [Google Scholar]
- Chen CH, Panizzon MS, Eyler LT, Jernigan TL, Thompson W, Fennema-Notestine C, Jak AJ, Neale MC et al. (2011). Genetic Influences on Cortical Regionalization in the Human Brain. Neuron, 72(4), 537–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Smith S, Viding E, and Nichols TE (2014). APACE: accelerated permutation inference for the ACE model. The 20th Annual Meeting of the Organization for Human Brain Mapping, OHBM, Hamburg, Germany. [Google Scholar]
- Curran JE, McKay DR, Winkler AM, Olvera RL, Carless MA, Dyer TD, Kent JW Jr, Kochunov P et al. (2013). Identification of Pleiotropic Genetic Effects on Obesity and Brain Anatomy. Hum Hered, 75(0), 136–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson TL, Kanoski SE, Shier LA, Clegg DJ, and Benoit SC (2007). A Potential Role for the Hippocampus in Energy Intake and Body Weight Regulation. Curr Opin Pharmacol, 7(6), 613–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Pasquale F, Corbetta M, Betti V, and Della Penna S (2018). Cortical cores in network dynamics. NeuroImage, 180, 370–382. [DOI] [PubMed] [Google Scholar]
- DePasque S and Galvan A (2017). Frontostriatal development and probabilistic reinforcement learning during adolescence. Neurobiology of Learning and Memory, 143, 1–7. [DOI] [PubMed] [Google Scholar]
- Eklund A, Nichols TE, and Knutsson H (2016). Cluster failure: Why fMRI inferences for spatial extent have inflated false-positive rates. PNAS, 113(28), 7900–7905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elks CE, den Hoed M, Zhao JH, Sharp SJ, Wareham NJ, Loos RJF, and Ong KK (2012). Variability in the heritability of body mass index: a systematic review and metaregression. Frontiers in Endocrinology, 3(29), 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friston KJ, Ashburner JT, Kiebel SJ, Nichols TE, and Penny WD (2007). Statistical Parametric Mapping: The Analysis of Functional Brain Images. London: Academic Press/Elsevier. [Google Scholar]
- Gallant AR, Tremblay A, Perusse L, Bouchard C, Despres JP, and Drapeau V (2010). The Three-Factor Eating Questionnaire and BMI in adolescents: results from the Que’bec Family Study. British Journal of Nutrition, 104, 1074–1079. [DOI] [PubMed] [Google Scholar]
- Galvan A, Hare TA, Davidson M, Spicer J, Clover G and Casey BJ (2005). The Role of Ventral Frontostriatal Circuitry in Reward-Based Learning in Humans. Journal of Neuroscience, 25(38), 8650–8656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert SJ, Spengler S, Simons JS, Steele JD, Lawrie SM, Frith CD, and Burgess PW (2006). Functional Specialization within Rostral Prefrontal Cortex (Area 10): A Meta-analysis. Journal of Cognitive Neuroscience, 18(6), 932–948. [DOI] [PubMed] [Google Scholar]
- Glahn DC, Winkler AM, Kochunov P, Almasy L, Duggirala R, Carless MA, Curran JC, Olvera RL (2010). Genetic control over the resting brain. PNAS, 107(3), 1223–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glahn DC, Kent JW Jr, Sprooten E, Diego VP, Winkler AM, Curan JE, McKay DR, Knowles EE et al. (2013). Genetic basis of neurocognitive decline and reduced white-matter integrity in normal human brain aging. PNAS, 110(47), 19006–19011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldsmith HH (1991). A Zygosity questionnaire for young twins: a research note. Behav Genet, 21(3), 257–69. [DOI] [PubMed] [Google Scholar]
- Greenspan KS, Arakelian CR, and van Erp TGM (2016). Heritability of Hippocampal Formation Sub-region Volumes. J Neurol Neurosci, 7(6), 159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayasaka S and Nichols TE (2003). Validating cluster size inference: random field and permutation methods. NeuroImage, 20, 2343–2356. [DOI] [PubMed] [Google Scholar]
- Hinton EC, Parkinson JA, Holland AJ, Arana FS, Roberts AC, and Owen AM (2004). Neural contributions to the motivational control of appetite in humans. European Journal of Neuroscience, 20, 1411–1418. [DOI] [PubMed] [Google Scholar]
- Hulshoff Pol HE, Schnack HG, Posthuma D, Mandl RCW, Baare WF, van Oel C, van Haren NE, Collins DL et al. (2006). Genetic Contributions to Human Brain Morphology and Intelligence. The Journal of Neuroscience, 26(40), 10235–10242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahanshad N, Bhatt P, Hibar DP, Villalon JE, Nir TM, Toga AW, Jack CR Jr, Bernstein MA et al. (2013). Bivariate Genome-Wide Association Study of Genetically Correlated Neuroimaging Phenotypes from DTI and MRI through a Seemingly Unrelated Regression Model In Shen L, Liu T, Yap PT, Huang H, Shen D, and Westin CF Multimodal Brain Image Analysis (pp. 189–201), Dordrecht, London: Springer. [Google Scholar]
- James PT, Leach R, Kalamara E, and Shayechi M (2001). The Worldwide Obesity Epidemic. Obesity Research, 9(4), 228S–233S. [DOI] [PubMed] [Google Scholar]
- Janowitz D, Wittfeld K, Terock J, Freyberger HJ, Hegenscheid K, Volzke H, Habes M, Hosten N et al. (2015). Association between waist circumference and gray matter volume in 2344 individuals from two adult community-based samples. NeuroImage, 122, 149–157. [DOI] [PubMed] [Google Scholar]
- Jansen AG, Mous SE, White T, Posthuma D, and Polderman TJC (2015). What Twin Studies Tell Us About the Heritability of Brain Development, Morphology, and Function: A Review. Neuropsychol Rev, 25, 27–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsgodt KH, Kochunov P, Winkler AM, Laird AR, Almasy L, Duggirala R, Olvera RL, Fox PT et al. (2010). A Multimodal Assessment of the Genetic Control over Working Memory. J Neurosci, 30(24), 8197–8202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katterman SN, Kleinmann BM, Hood MM, Nackers LM, and Corsica JA (2014). Mindfulness meditation as an intervention for binge eating, emotional eating, and weight loss: A systematic review. Eating Behaviors, 15, 197–204. [DOI] [PubMed] [Google Scholar]
- Kennedy J and Dimitropoulos A (2014). Influence of feeding state on neurofunctional differences between individuals who are obese and normal weight: A meta-analysis of neuroimaging studies. Appetite, 75, 103–109. [DOI] [PubMed] [Google Scholar]
- Kennedy JT, Collins PF, and Luciana M (2016). Higher Adolescent Body Mass Index Is Associated with Lower Regional Gray and White Matter Volumes and Lower Levels of Positive Emotionality. Frontiers in Neuroscience, 10, 413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerns JG, Cohen JD, MacDonald III AW, Cho RY, Stenger VA, and Carter CS (2004). Anterior Cingulate Conflict Monitoring and Adjustments in Control. Science, 303(5660), 1023–1026. [DOI] [PubMed] [Google Scholar]
- Keskitalo K, Tuorila H, Spector TD, Cherkas LF, Knaapila A, Kaprio J, Silventoinen K, and Perola M (2008). The Three-Factor Eating Questionnaire, body mass index, and responses to sweet and salty fatty foods: a twin study of genetic and environmental associations. Am J Clin Nutr, 88, 263–71. [DOI] [PubMed] [Google Scholar]
- Kiernan JA (2012). Anatomy of the Temporal Lobe. Epilepsy Research and Treatment, 2012, 176157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S (2013). Neuroscientific model of motivational process. Frontiers in Psychology, 4, 98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishinevsky FI, Cox JE, Murdaugh DL, Stoeckel LE, Cook EW III, Weller RE (2012). fMRI reactivity on a delay discounting task predicts weight gain in obese women. Appetite, 58, 582–592. [DOI] [PubMed] [Google Scholar]
- Kochunov P, Thompson PM, Winkler A, Morrisey M, Fu M, Coyle TR, Du X, Muellerklein F et al. (2016). The common genetic influence over processing speed and white matter microstructure: Evidence from the Old Order Amish and Human Connectome Projects. NeuroImage, 125, 189–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krames ES and Foreman R (2007). Spinal Cord Stimulation Modulates Visceral Nociception and Hyperalgesia via the Spinothalamic Tracts and the Postsynaptic Dorsal Column Pathways: A Literature Review and Hypothesis. Neuromodulation, 10(3), 224–237. [DOI] [PubMed] [Google Scholar]
- Kuhn AB, Feis DL, Schilbach L, Kracht L, Hess ME, Mauer J, Bruning JC, and Tittgemeyer M (2016). FTO gene variant modulates the neural correlates of visual food perception. NeuroImage, 128, 21–31. [DOI] [PubMed] [Google Scholar]
- Kuczmarski RJ, Ogden CL, Guo SS, Grummer-Strawn LM, Flegal KM, Mei Z, Wei R, Curtin LR et al. (2002). “2000 CDC Growth Charts for the United States: methods and development.” National Center for Health Statistics. Vital Health Stat 11(246). [PubMed] [Google Scholar]
- Le DSNT, Pannacciullim N, Chen K, Salbe AD, Hill JO, Wing RR, Reiman EM, and Krakoff J (2007). Less activation in the left dorsolateral prefrontal cortex in the reanalysis of the response to a meal in obese than in lean women and its association with successful weight loss. Am J Clin Nutr, 86(3), 573–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenroot RK, Schmitt JE, Ordaz SJ, Wallace GL, Neale MC, Lerch JP, Kendler KS, Evans AC et al. (2009). Differences in Genetic and Environmental Influences on the Human Cerebral Cortex Associated With Development During Childhood and Adolescence. Human Brain Mapping, 30, 163–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maayan L, Hoogendoorn C, Sweat V, and Convit A (2011). Disinhibited eating in obese adolescents is associated with orbitofrontal volume reductions and executive dysfunction. Obesity (Silver Springs), 19(7), 1382–1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macht M (2008). How emotions affect eating: A five-way model. Appetite, 50, 1–11. [DOI] [PubMed] [Google Scholar]
- Manto M, Bower JM, Conforto AB, Delgado-Garcia JM, da Guarda SNF, Gerwig M, Habas C, Hagura N (2012). Consensus Paper: Roles of the Cerebellum in Motor Control - The Diversity of Ideas on Cerebellar Involvement in Movement. Cerebellum, 11(2), 457–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin LE, Holsen LM, Chambers RJ, Bruce AS, Brooks WM, Zarcone JR, Butler MG, and Savage CR (2010). Neural Mechanisms Associated With Food Motivation in Obese and Healthy Weight Adults. Obesity, 18, 254–260. [DOI] [PubMed] [Google Scholar]
- Mason A, Epel ES, Aschbacher K, Lustig RH, Acree M, Kristeller J, Cohn M, Dallman M et al. (2016). Reduced reward-driven eating accounts for the impact of a mindfulness-based diet and exercise intervention on weight loss: Data from the SHINE randomized controlled trial. Appetite, 100, 86–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason P (2011). FROM DESCENDING PAIN MODULATION TO OBESITY VIA THE MEDULLARY RAPHE. Pain, 152(3), S20–S24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendes MDSD, de Melo ME, Fernandes AE, Fujiwara CTH, Pioltine MB, Teixeira A, Coelho K, Galasso M et al. (2017). Effects of two diet techniques and delivery mode on weight loss, metabolic profile and food intake of obese adolescents: a fixed diet plan and a calorie-counting diet. European Journal of Clinical Nutrition, 71, 549–551. [DOI] [PubMed] [Google Scholar]
- Mendoza J, Pevet P, Felder-Schmittbuhl MP, Bailly Y, and Challet E (2010). The Cerebellum Harbors a Circadian Oscillator Involved in Food Anticipation. Journal of Neuroscience, 30(5), 1894–1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller K, Sacher J, Arelin K, Holiga S, Kratzsch J, Villringer A, and Schroeter ML (2012). Overweight and obesity are associated with neuronal injury in the human cerebellum and hippocampus in young adults: a combined MRI, serum marker and gene expression study. Transl Psychiatry, 2, e200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nangunoori RK, Tomycz ND, Oh MY, and Whiting DM (2016). Deep Brain Stimulation for Obesity: From a Theoretical Framework to Practical Application. Neural Plasticity, 2016, 7971460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NCD Risk Factor Collaboration. (2017). Worldwide trends in body-mass index, underweight, overweight, and obesity from 1975 to 2016: a pooled analysis of 2416 population-based measurement studies in 128·9 million children, adolescents, and adults. Lancet, 390, 2627–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neale MC and Maes HHM (2004). Methodology for Genetic Studies of Twins and Families, Dordrecht, The Netherlands: Kluwer Academic Publishers B.V. [Google Scholar]
- Neale MC, Hunter MD, Pritikin JN, Zahery M, Brick TR, Kirkpatrick RM, Estabrook R, Bates TC et al. (2016). OPENMX 2.0: EXTENDED STRUCTURAL EQUATION AND STATISTICAL MODELING. Psychometrika, 81(2), 535–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey R, Saxena P, and Dubey A (2011). Emotion regulation difficulties in alexithymia and mental health. Europe’s Journal of Psychology, 7(4), 604–623. [Google Scholar]
- Pannacciulli N, Del Parigi A, Chen K, Le DSNT, Reiman EM, and Tataranni PA (2006). Brain abnormalities in human obesity: A voxel-based morphometric study. NeuroImage, 31, 1419–1425. [DOI] [PubMed] [Google Scholar]
- Patel S, Park MTM, Devenyi GA, Patel R, Masellis M, Knight J, and Chakravarty MM (2017). Heritability of Hippocampal Subfield Volumes Using a Twin and Non-Twin Siblings Design. Human Brain Mapping, 38, 4337–4352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peper JS, Schnack HG, Brouwer RM, Van Baal GCM, Pjetri E, Szekely E, van Leeuwen M, van den Bergm SM et al. (2009). Heritability of Regional and Global Brain Structure at the Onset of Puberty: A Magnetic Resonance Imaging Study in 9-Year-Old Twin Pairs. Human Brain Mapping, 30, 2184–2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillipou A, Rossell SL, Gurvich C, Castle DJ, Abel LA, Nibbs RG, and Hughes ME (2018). Differences in regional grey matter volumes in currently ill patients with anorexia nervosa. European Journal of Neuroscience, 47, 177–183. [DOI] [PubMed] [Google Scholar]
- Pinaquy S, Chabrol H, Simon C, Louvet JP, and Barbe P (2012). Emotional Eating, Alexithymia, and Binge-Eating Disorder in Obese Women. Obesity, 11(2), 195–201. [DOI] [PubMed] [Google Scholar]
- Pinel P and Dehaene S (2013). Genetic and environmental contributions to brain activation during calculation. NeuroImage, 81, 306–316. [DOI] [PubMed] [Google Scholar]
- Pothos E, Tapper K, and Calitri R (2009). Cognitive and behavioral correlates of BMI among male and female undergraduate students. Appetite, 52, 797–800. [DOI] [PubMed] [Google Scholar]
- Rao LL, Zhou Y, Zheng D, Yang LQ, and Li S (2018). Genetic Contribution to Variation in Risk Taking: A Functional MRI Twin Study of the Balloon Analogue Risk Task. Psychological Science, 29(10), 1679–1691. [DOI] [PubMed] [Google Scholar]
- Reilly JJ and Kelly J (2011). Long-term impact of overweight and obesity in childhood and adolescence on morbidity and premature mortality in adulthood: systematic review. International Journal of Obesity, 35, 891–898. [DOI] [PubMed] [Google Scholar]
- Rimol LM, Panizzon MS, Fennema-Notestine C, Eyler LT, Fischl B, Franz CE, Hagler DJ, Lyons MJ et al. (2010). Cortical Thickness is Influenced by Regionally-Specific Genetic Factors. Biol Psychiatry, 67(5), 493–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolls ET (2016). Functions of the anterior insula in taste, autonomic, and related functions. Brain and Cognition, 110, 4–19. [DOI] [PubMed] [Google Scholar]
- Rushworth MFS (2008). Intention, Choice, and the Medial Frontal Cortex. Annals of the New York Academy of Sciences, 1124(1), 181–207. [DOI] [PubMed] [Google Scholar]
- Sassi F, Devaux M, Cecchini M, Rusticelli E (2009). “The Obesity Epidemic: Analysis of Past and Projected Future Trends in Selected OECD Countries”, OECD Health Working Papers, No. 45, OECD Publishing. [Google Scholar]
- Siegel JS, Mitra A, Laumann TO, Seitzman BA, Raichle M, Corbetta M, and Snyder AZ (2017). Data Quality Influences Observed Links Between Functional Connectivity and Behavior. Cerebral Cortex, 27, 4492–4502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small DM, Zatorre RJ, Dagher A, Evans AC, and Jones-Gotman M (2001). Changes in brain activity related to eating chocolate: From pleasure to aversion. Brain, 124, 1720–1733. [DOI] [PubMed] [Google Scholar]
- Smith KS and Berridge KC (2005). The Ventral Pallidum and Hedonic Reward: Neurochemical Maps of Sucrose “Liking” and Food Intake. Journal of Neuroscience, 25(38), 8637–8649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowell ER, Thompson PM, Tessner KD, Toga AW (2001). Mapping Continued Brain Growth and Gray Matter Density Reduction in Dorsal Frontal Cortex: Inverse Relationships during Postadolescent Brain Maturation. Journal of Neuroscience, 21(22), 8819–8829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spieker EA, Kochunov P, Rowland LM, Sprooten E, Winkler AM, Olvera RL, Almasy L, Duggirala R et al. (2015). Shared genetic variance between obesity and white matter integrity in Mexican Americans. Frontiers in Genetics, 6(26), 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stice E, Spoor S, Bohon C, Small DM (2008). Relation Between Obesity and Blunted Striatal Response to Food Is Moderated by TaqIA A1 Allele. Science, 322, 449–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stice E, Burger K, Yokum S (2013). Caloric Deprivation Increases Responsivity of Attention and Reward Brain Regions to Intake, Anticipated Intake, and Images of Palatable Foods. NeuroImage, 67, 322–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Y, Jackson T, Wei D, Qiu J, and Chen H (2017). Regional Gray Matter Volume Is Associated with Restrained Eating in Healthy Chinese Young Adults: Evidence from Voxel-Based Morphometry. Frontiers in Psychology, 8, 443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung J, Lee K, Song YM (2009). Relationship of eating behavior to long-term weight change and body mass index: the Healthy Twin study. Eat Weight Disord, 14(2–3), 98–105. [DOI] [PubMed] [Google Scholar]
- Sung J, Lee K, Song YM, Lee MK, and Lee DH (2010). Heritability of Eating Behavior Assessed Using the DEBQ (Dutch Eating Behavior Questionnaire) and Weight-related Traits: The Healthy Twin Study. Obesity, 18, 1000–1005. [DOI] [PubMed] [Google Scholar]
- Swagerman SC, Brouwer RM, de Geus EJC, Holshoff, Pol HE, and Boomsma DI (2014). Development and heritability of subcortical brain volumes at ages 9 and 12. Genes, Brain, and Behavior, 13, 733–742. [DOI] [PubMed] [Google Scholar]
- Tholin S, Rasmussen F, Tynelius P, and Karlsson J (2005). Genetic and environmental influences on eating behavior: the Swedish Young Male Twins Study. Am J Clin Nutr, 81, 564–9. [DOI] [PubMed] [Google Scholar]
- Tindell AJ, Smith KS, Pecina S, Berridge KC, and Aldridge JW (2006). Ventral Pallidum Firing Codes Hedonic Reward: When a Bad Taste Turns Good. J Neurophysiol, 96, 2399–2409. [DOI] [PubMed] [Google Scholar]
- Tisdall MD, Hess AT, Reuter M, Meintjes EM, Fischl B, and van der Kouwe AJW (2012). Volumetric Navigators (vNavs) for Prospective Motion Correction and Selective Reacquisition in Neuroanatomical MRI. Magn Reson Med, 68(2), 389–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuulari JJ, Karlsson HK, Antikainen O, Hirvonen J, Pham T, Salminen P, Helmio M, Parkkola R et al. (2016). Bariatric Surgery Induces White and Grey Matter Density Recovery in the Morbidly Obese: A Voxel-Based Morphometric Study. Human Brain Mapping, 37, 3745–3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Berg SM, Setiawan A, Bartels M, Polderman TJC, van der Vaart AW, and Boomsma DI (2006). Individual Differences in Puberty Onset in Girls: Bayesian Estimation of Heritabilities and Genetic Correlations. Behavior Genetics, 36(2), 261–270. [DOI] [PubMed] [Google Scholar]
- van der Laan LN, Charbonnier L, Griffioen-Roose S, Kroese FM, van Rijn I, Smeets PAM (2016). Supersize my brain: A cross-sectional voxel-based morphometry study on the association between self-reported dietary restraint and regional grey matter volumes. Biological Psychology, 117, 108–116. [DOI] [PubMed] [Google Scholar]
- van der Lee SJ, Roshchupkin GV, Adams HHH, Schmidt H, Hofer E, Saba Y, Schmidt R, Hofman A et al. (2017). Gray Matter Heritability in Family-Based and Population-Based Studies Using Voxel-Based Morphometry. Human Brain Mapping, 38, 2408–2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verstynen TD, Wenstein AM, Schneider WW, Jakicic JM, Rofey DL, and Erickson KI (2012). Increased Body Mass Index Is Associated With a Global and Distributed Decrease in White Matter Microstructural Integrity. Psychosomatic Medicine, 74, 682–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther K, Birdsill AC, Glisky EL, Ryan L (2010). Structural Brain Differences and Cognitive Functioning Related to Body Mass Index in Older Females. Human Brain Mapping, 31, 1052–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weise CW, Piaggi P, Reinhardt M, Chen K, Savage CR, Krakoff J, and Pleger B (2017). The obese brain as a heritable phenotype – A combined morphometry and twin study. Int J Obes, 41(3), 458–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westenhoefer J, Broeckmann P, Munch AK, and Pudel V (1994). Cognitive control of eating behaviour and the disinhibition effect. Appetite, 23(1), 27–41. [DOI] [PubMed] [Google Scholar]
- Willette AA and Kapogiannis D (2015). Does the brain shrink as the waist expands? Ageing Res Rev, 20, 86–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler AM, Ridgway GR, Webster MA, Smith SM, and Nichols TE (2014). Permutation inference for the general linear model. NeuroImage, 92, 381–397. [DOI] [PMC free article] [PubMed] [Google Scholar]