Abstract
Corticotropin-releasing hormone (CRH) is an essential, evolutionarily-conserved stress-neuropeptide. In addition to hypothalamus, CRH is expressed in brain regions including amygdala and hippocampus where it plays crucial roles in modulating the function of circuits underlying emotion and cognition. CRH+ fibers are found in nucleus accumbens (NAc), where CRH modulates reward/motivation behaviors. CRH actions in NAc may vary by the individual’s stress history, suggesting roles for CRH in neuroplasticity and adaptation of the reward circuitry. However, the origin and extent of CRH+ inputs to NAc are incompletely understood. We employed viral genetic approaches to map both global and CRH+ projection sources to NAc in the mouse. We injected into NAc variants of a new designer adeno-associated virus that permits robust retrograde access to NAc-afferent projection neurons. Cre-dependent viruses injected into CRH-Cre mice enabled selective mapping of CRH+ afferents. We employed anterograde AAV1-directed axonal tracing to verify NAc CRH+ fiber projections and established the identity of genetic reporter–labeled cells via validated antisera against native CRH. We quantified the relative contribution of CRH+ neurons to total NAc-directed projections. Combined retrograde and anterograde tracing identified the paraventricular nucleus of the thalamus, bed nucleus of stria terminalis, basolateral amygdala and medial prefrontal cortex as principal sources of CRH+ projections to NAc. CRH+ NAc afferents were selectively enriched in NAc-projecting brain regions involved in diverse aspects of the sensing, processing and memory of emotionally salient events. These findings suggest multiple, complex potential roles for the molecularly-defined, CRH-dependent circuit in modulation of reward and motivation behaviors.
Keywords: nucleus accumbens, circuitry, viral tracing, CRH, transgenic mice, stress, reward, molecular-specific pathways, RRID: IMSR_JAX:012704, RRID: AB_2716806, RRID: AB_2313584, corticotropin-releasing factor
Graphical Abstract
Novel viral genetic studies employing retrograde and anterograde tracing demonstrate corticotropin releasing hormone- expressing afferent projection to the nucleus accumbens (NAc). A novel basolateral amygdala to NAc projection is identified as well as a surprising enrichment of projections from brain regions involved in stress, anxiety and emotional memory (paraventricular thalamus).
1. INTRODUCTION
Corticotropin-releasing hormone (CRH) is an essential, evolutionarily conserved neuropeptide, evolved to convey, process and modulate responses to potential threats (Joëls & Baram, 2009; Vale et al., 1983). Originally, the CRH-expressing neurons in the paraventricular nucleus of the hypothalamus were identified; these neurons execute the peptide’s primary neuro-endocrine function in the regulation of the hormonal response to stress (Swanson, Sawchenko, Lind, & Rho, 1987; Swanson, Sawchenko, Rivier, & Vale, 1983). Subsequently, CRH-expressing neurons and receptors have been identified throughout selective regions of the brain, where they orchestrate the peptide’s influence on numerous brain functions, typically in response to stress or threat (Joëls & Baram, 2009). For examples, amygdalar CRH has been implicated in fear and anxiety (Pomrenze et al., 2019; Regev et al., 2011; Regev, Tsoory, Gil, & Chen, 2012; Sanford et al., 2017; Wang et al., 2013); hippocampal CRH contributes to stress-related memory changes (Y. Chen et al., 2010; Maras & Baram, 2012); CRH terminals in the locus ceruleus contribute to stress-related neurotransmission (Van Bockstaele & Valentino, 2013); and CRH-positive fibers in the VTA influence dopamine release (George, Le Moal, & Koob, 2012).
A role for CRH, released from axons terminals and acting on its cognate receptors, in the function of the intricate pleasure / reward circuitry has been increasingly apparent. For examples, CRH release in medial prefrontal cortex (mPFC) and central nucleus of the amygdala (CeA) is triggered by rewards such as food consumption (Merali, McIntosh, & Anisman, 2004; Merali, McIntosh, Kent, Michaud, & Anisman, 1998). CRH expression is upregulated in the amygdala by cocaine administration (Koob & Le Moal, 2001). Microinjections of CRH into ventrolateral BNST can trigger drug self-administration reinstatement (Garavan et al., 2000), and dopamine / CRH neurons in the ventral tegmental area influence reward behavior (Grieder et al., 2014). In addition, increased expression of CRH in the central nucleus of the amygdala seems to interfere with the pleasure / reward circuit function, because partial silencing of the augmented expression of the peptide after early-life stress reversed anhedonia in adult rodents (Bolton et al., 2018).
Within the nucleus accumbens (NAc), CRH-expressing cells have been described (Kono et al., 2017; Merchenthaler, 1984; Merchenthaler, Vigh, Petrusz, & Schally, 1982), and robust CRH immunoreactive fibers are found in the core and shell (Swanson et al., 1983). In addition, the CRH receptors CRFR1 and CRFR2 (Justice, Yuan, Sawchenko, & Vale, 2008; Lemos et al., 2012; Lim et al., 2007) are detected within the NAc. These seem to reside in tyrosine-hydroxylase-positive axon terminals, a location enabling the peptide to influence dopamine release in the NAc (Lemos et al., 2012). This effect contributes to the modulation of reward and motivation behavior (Lemos et al., 2012). Thus, blocking CRH receptors modulates partner preference in prairie voles (Lim et al., 2007) and alters nicotine-use behaviors (Marcinkiewcz et al., 2009). In contrast, CRH microinjections in the NAc amplify incentive salience of Pavlovian cues for rewards in rodents (Peciña et al., 2006), a potential mechanism for stress-triggered drug relapse and stress-related binge eating (Schroeder et al., 2017). In stress-naïve rodents, microinjections of CRH into the NAc induces a conditioned place preference and increases local dopamine release (Lemos et al., 2012). However, in an individual with a history of traumatic stress, intra-NAc CRH provokes conditioned place avoidance and dopamine release is no longer induced; notably, these effects persist for months (Lemos et al., 2012). Thus, there is cumulative evidence for a complex and nuanced role for CRH in reward processing that is strongly governed by an individual’s history of stress.
Surprisingly, relatively little is known about the origin and circuitry of CRH+ inputs to the NAc, and the potential contribution of several distinct molecularly-specified afferent pathways to context-induced neuroplasticity in the NAc. Such information is crucial to enable targeted and focused interventions for certain emotional problems, providing impetus to uncover the origin and circuitry of NAc CRH+ inputs. In the present work, we used new viral genetic mapping approaches (Oh et al., 2014; Tervo et al., 2016) to uncover the origin and circuitry of NAc-directed CRH+ afferents and compared them with overall sources of input to this nucleus.
2. MATERIALS AND METHODS
2.1. Subjects
All experiments were conducted according to the National Institutes of Health guidelines for animal care and use and were approved by the Institutional Animal Care and Use Committee of the University of California, Irvine (UCI). B6(Cg)-Crhtm1(cre)Zjh/J (Crh-IRES-Cre) mice (either sex, Jax, Stock No: 012704) were used to trace CRH+ inputs to the NAc (n = 5), and an additional 10 mice were employed for anterograde tracing (see below). Wild type C57BL/J6 mice (either sex) acquired from the Jackson Laboratory were used to trace all (global) inputs to the NAc (n = 5). Animals were group housed with littermates in standard conditions at 72º F with 42% humidity and a 12-hour light-dark cycle (lights on at 6:30 am, lights off at 6:30 pm). Mice used in the experiments were 13–41 weeks old and had ad libitum access to food and water in their home cages before and after surgeries. Only mice with correct targeted injection locations were included for analysis (see “2.5.3 Quality control” below).
2.2. Viral injections
To perform stereotaxic viral injections into the brain, mice were anesthetized under 1.5% isoflurane with a 0.8 L/min oxygen flow rate using an isoflurane table top unit (HME109, Highland Medical Equipment). Mice were then placed in a rodent stereotactic apparatus (Leica Angle Two for mouse) with continuous 1–1.5% isoflurane anesthesia with the head secured. Under aseptic conditions a small incision was made in the shaved and sterilized scalp, the skin reflected, and the skull exposed to show the landmarks of bregma and lambda, and the injection sites were located.
A three-axis micromanipulator (Leica Angle Two system) was used to determine coordinates for the injection sites. Small craniotomies were made in the skull over the injection site, exposing the pia surface. A pulled glass pipette (tip diameter ~20 µm) was loaded with viruses and then lowered into the brain with the appropriate coordinates. A Picospritzer (General Valve) was used to pulse the virus into the brain. Viruses were injected into the brain at a rate of 20–30 nL/min, with 10-ms pulse duration. To prevent virus backflow after the delivery, the pipette was not moved for 5 min after completion of the injection. Once the injection pipette was withdrawn, the incision was closed with tissue adhesive (3M Vetbond), an injection of carprofen (5 mg/kg, i.p., Patterson Veterinary) was administered as an analgesic, and the mouse was removed from the stereotactic apparatus. Mice were monitored in an empty cage on a heating pad until the isoflourane anesthesia effects wore off (typically 5–20 minutes). Once fully ambulatory, mice were returned to their group housed home cages for recovery.
2.2.1. Retrograde viral labeling of CRH+ NAc inputs
To selectively target CRH+ inputs to the NAc, we used Crh-IRES-Cre mice (Y. Chen, Molet, Gunn, Ressler, & Baram, 2015; Wamsteeker Cusulin et al., 2013), and infused 0.2 µl of a Cre-driven adeno-associated virus (AAV) (AAV2-retro-CAG-FLEX-tdTomato-WPRE, 5.71 × 1013 genomic copies (GC)/ml, packaged by Vigene Biosciences) into the NAc of the left hemisphere (Target coordinates relative to the Bregma, 2 of 5 mice: AP 1.18 mm, ML −0.83 mm, DV −4.54 mm; 2 of 5 mice: AP 1.18 mm, ML −0.66 mm, DV −4.55 mm; 1 of 5 mice: AP 1.18 mm, ML −0.95 mm, DV −4.29 mm). Post-surgery, the AAV virus was allowed to retrogradely transport from targeted CRH-Cre+ terminals in the NAc for 21 days before the mice were deeply anesthetized and perfused and brain were removed. Whereas rAAV2-retro approach does not indicate the presence of synapses onto neurons in the NAc, it reflects the presence of axons from projecting neurons. Monosynaptic rabies tracing approaches (Sun et al., 2014; Sun, Nitz, Holmes, & Xu, 2018) in conjunction with specific labeling may be used to determine whether NAc neurons receive direct synaptic inputs from CRH neurons.
2.2.2. Retrograde viral labeling of all NAc inputs
To target global inputs to the NAc, we employed wild-type C57BL/6J mice and infused 0.2 µl of adeno-associated virus (AAV) (rAAV2-retro-CAG-tdTomato, 1.8 × 1012 genomic copies (GC)/ml, Janelia Virus Services) into the NAc of the left hemisphere (Target coordinates relative to the Bregma, 2 of 5 mice: AP 1.18 mm, ML −0.83 mm, DV −4.54 mm; 3 of 5 mice: AP 1.18 mm, ML −0.95 mm, DV −4.29 mm). We have estimated the spread of the injections to be AP: 126 ± 22 µm, ML: 150 ± 40 µm, DV: 198 ± 36 µm (all coordinates expressed as mean ± SEM relative to Bregma). See Tervo et al. (2016) for rAAV2-retro design. Post-surgery, the AAV virus was allowed to retrogradely transport from terminals in the NAc for 21 days before the mice were deeply anesthetized and perfused.
2.2.3. Anterograde viral labeling of CRH+ projections from BLA, CeA and PVT to NAc
To better delineate the neuroanatomical distribution of the novel apparent CRH+ connection between NAc and BLA, we injected 0.2–0.3 µl of a Cre-driven AAV anterograde tracer (AAV1-DIO-tdTomato, UPenn Vector Core) into the left BLA (AP: −1.34 mm, ML: −2.92 mm, DV: −4.66 mm; all values given relative to the Bregma) of five CRH-Cre mice. To assess the specificity of BLA-NAc projections, we examined for potential CeA-origin NAc afferents. Specifically, we injected 0.2–0.3 µl of a Cre-driven AAV anterograde tracer (AAV1-DIO-tdTomato, UPenn Vector Core) into the left CeA (AP: −1.70 mm, ML: −2.50 mm, DV: −4.30 mm). Similarly, to better delineate and confirm a second apparent major source of CRH+ NAc inputs, we injected 0.4 µl of the same anterograde tracer into the left PVT (AP: −0.94 mm, ML: −0.16 mm, DV: −3.08 mm; all values given relative to the Bregma) of another five CRH-Cre mice.
2.3. Perfusion and brain sectioning
Mice were deeply anaesthetized using isoflurane and transcardially perfused via the ascending aorta with 5 ml of phosphate buffered saline (0.01 M PBS) followed by 25–50 ml of 4% paraformaldehyde (PFA) in 0.1 M phosphate buffer (PB, pH 7.4, 4°C). Brains were removed and postfixed with the same fixative overnight (4°C), and then immersed in 15%, followed by 30% sucrose in 0.1 M PB for cryoprotection (4°C). Brains were sectioned coronally at 30 µm thickness using a freezing microtome (Leica SM2010R, Nussloch, Germany). The data shown in Figure 6 employed 20 µm sections obtained using a cryostat (Leica). Sections were stored at 4°C until their processing: a 1:3 series (every third slice) was wet-mounted on microscope slides and cover-slipped with mounting medium (Vectashield; H-1000, Vector, Burlingame, CA).
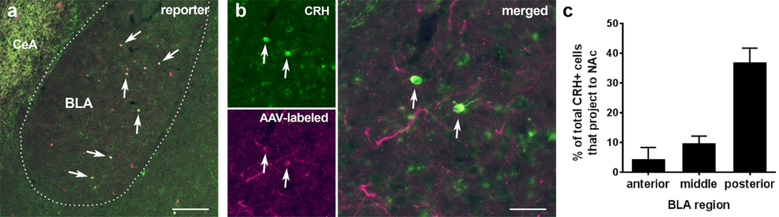
Figure 6. Concordant viral-genetic reporter visualization and endogenous CRH immunohistochemistry identify an anterior-posterior gradient of NAc-projecting CRH neurons in the BLA.
(a) In the basolateral amygdala (BLA) of adult Crh-IRES-Cre;Ai14 mice, there is a population of neurons that co-express the endogenous CRH (green) and the Ai14 reporter (tdTomato; magenta). Arrows indicate the co-localization of endogenous CRH and the Ai14 reporter signal. Over 70% of all tdTomato+ neurons in BLA expressed endogenous CRH. CeA: central amygdala, a region rich in CRH expression. (b) When an rAAV2-retrovirus (rAAV2-retro DIO-CAG-tdTomato) was injected into the nucleus accumbens (NAc) of Crh-IRES-Cre mice, retrogradely labeled cells were detected in the ipsilateral BLA. The representative images show virus-labeled cells immunopositive for CRH. These CRH+ NAc projecting BLA cells tended to largely reside in the posterior BLA. Arrows indicate the colocalization of CRH (green) and reporter (magenta). Bar = 150 µm in A and 37.5 µm in B. (c) Among rAAV2-retro-labeled (tdTomato expressing) cells in posterior BLA (AP: −2.06~−2.78 mm), ~59% cells (58.93 ± 5.97 %) co-expressed endogenous CRH identified via CRH antibody immunostaining as shown in b. (d) In the posterior BLA a large fraction of CRH-expressing cells projected to the NAc. The BLA was divided into anterior (AP: −0.58~−1.30 mm), middle (AP: −1.32~−2.04 mm), and posterior (AP: −2.06~−2.78 mm) subdivisions. Data are derived from 6 sections per area per mouse; n = 3 mice.
2.4. CRH immunostaining
To prepare tissue for the CRH staining in Figure 1, two CRH-Cre mice that had been injected with the Cre-driven AAV retrograde tracer in the NAc also received a single intracerebroventricular injection of colchicine (Tocris Bioscience, Bristol, UK, 30 µg/0.5 µl saline) 24 hours prior to perfusion to enhance CRH peptide localization in neuronal cell bodies (Dabrowska, Hazra, Guo, Dewitt, & Rainnie, 2013; Swanson et al., 1987). Coronal sections (30 µm) were subjected to CRH immunostaining. Briefly, sections were blocked with 5% normal donkey serum in PBS for 2 hours to prevent non-specific binding. After rinsing with PBS three times, 15 minutes per wash, on a shaker at 95 rpm, sections were incubated for 48 hours at 4°C with rabbit anti-CRH primary antibody (courtesy of Paul E. Sawchenko, Salk Institute, PBL#rC68) in the blocking solution (dilution factor 1:10,000). The sections were rinsed with PBS three times, 15 minutes per wash, followed by incubation in an Alexa Fluor (AF) 488-conjugated donkey-anti-rabbit secondary antibodies in the blocking solution (dilution 1:200, Jackson ImmunoResearch, 711–545-152) for 2 hours. All the sections were rinsed with PBS three times, 5 minutes each. Sections were stored at 4°C, counter-stained with 10 μM DAPI (Sigma, D-9542), then wet-mounted on microscope slides and cover-slipped with mounting medium (Vectashield).
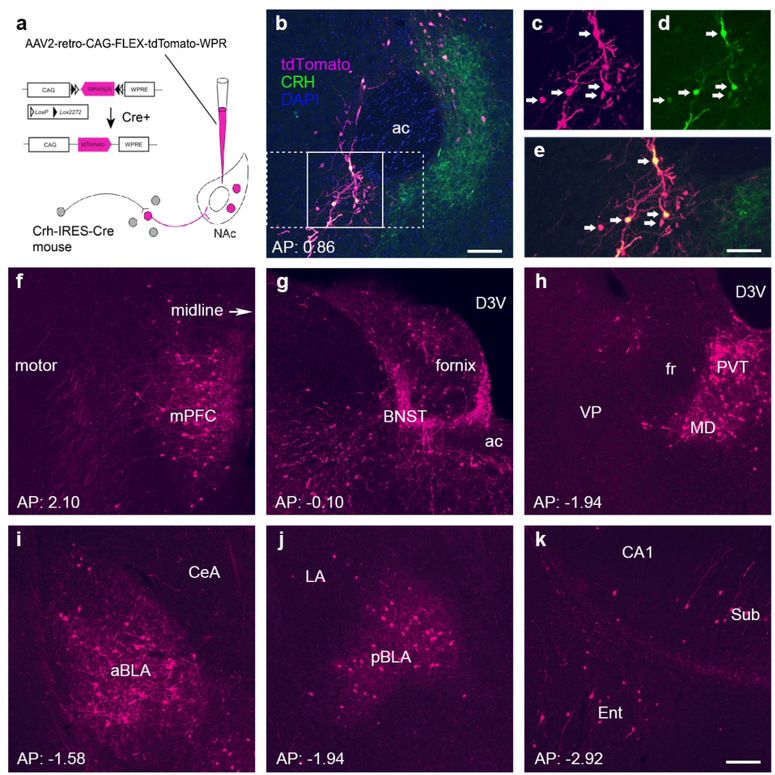
Figure 1. The major CRH-expressing afferents to the nucleus accumbens originate in the paraventricular nucleus of the thalamus, the bed nucleus of the stria terminalis, the medial prefrontal cortex and the basolateral amygdala.
(a) Schematic of the retrograde-labeling virus and the location of the injection site. Specifically, a Cre-driven adeno-associated virus (AAV; AAV2-retro-CAG-FLEX-tdTomato-WPRE) was injected into the left hemisphere NAc in CRH-IRES-Cre mice. (b–e) Endogenous CRH+ cells in the nucleus accumbens (NAc) were infected by the rAAV2-retro. (b) Low magnification image from a representative injection site, the NAc in the left hemisphere (n = 5 mice). The labeling of cell bodies and fiber terminals in the NAc is apparent. The tdTomato reporter for the virus is shown in magenta, whereas immunostaining to confirm local CRH+ neurons is shown in green. The section was counterstained with DAPI (blue). Framed areas in b were magnified in c–e to demonstrate virus-labeled CRH cells (c), antibody-immunolabeled CRH cells (d), and the co-expression of virus label and endogenous CRH (e). (f–k) Anterior-posterior series of panels demonstrating significant sources of retrogradely labeled CRH+ NAc afferents. Labeled cells and fibers were primarily found in the ipsilateral (left) hemisphere. (f) The medial prefrontal cortex (mPFC) including prelimbic and infralimbic regions contributed 5.75% of CRH+ projections to the NAc. (g) 9.27% of CRH+ inputs originated from the bed nucleus of the stria terminalis (BNST). (h) Thalamic nuclei were a major source of CRH projections to the NAc, contributing over a third (34%). Among them, the paraventricular nucleus (PVT) accounted for a third of all thalamic NAc-projecting cells (9.40% of the total). (i–j) 8.57% of CRH+ projections came from amygdala nuclei, and over half from the BLA. (k) CRH+ inputs arise from the subiculum and the entorhinal cortex. ac: anterior commissure; arrows in c-e indicate dual-labeled cell bodies of CRH cells. ac = anterior commissure; aBLA = anterior basolateral amygdala; BNST = bed nucleus of the stria terminalis; CeA = central amygdala; D3V = dorsal third ventricle; Ent = entorhinal cortex; LA = lateral amygdala; mPFC = medial prefrontal cortex; pBLA = posterior basolateral amygdala; PVT = paraventricular nucleus of the thalamus; Sub = subiculum. Bar = 100 µm in b, 63 µm in c–e and 100 µm for f–k.
To assess the fidelity of CRH and the reporter, and to enable visualization of potentially low levels of CRH in reporter-expressing cells (Figure 6), concurrent visualization of CRH and tdTomato was performed using the tyramide signal amplification (TSA) technique (Y. Chen, Brunson, et al., 2004; Y. Chen et al., 2015). Sections (20 µm) were treated in 0.3% H2O2/PBS-T for 20–30 min, rinsed in PBS-T for 30 minutes and then blocked with 5% normal goat serum in PBS-T for 2 hours. After rinsing of 15 minutes in PBS-T, sections were incubated with CRH rabbit antiserum (1:20,000) for 3 days (4°C), and then incubated in HRP conjugated anti-rabbit IgG (1:1,000; Perkin Elmer, Boston, MA) for 1.5 hours. Fluorescein-conjugated tyramide was diluted (1:150) in amplification buffer (Perkin Elmer, Boston, MA), and was applied in the dark for 5–6 min on ice.
2.5. Image acquisition, data quantification, quality control, and statistical analysis
2.5.1. Brain-wide imaging and quantification
Virus labeled sections were scanned under a 10× objective of a fluorescent microscope (Olympus BX 61) equipped with a high-sensitive CCD camera and Metamorph software for brain-wide analysis of NAc inputs (Figures 1–5). We employed confocal microscopy for imaging virus-labeled neurons after anterograde tracing (Figure 7; LSM 700, Carl Zeiss). Most images were obtained using the Metamorph image acquisition software (Molecular Devices, Sunnyvale, CA) and analyses were done using Adobe Photoshop (CS4).
Figure 5. CRH+ basolateral amygdala projections to the NAc, and their selectivity, are confirmed using anterograde tracers.
(a) Schematic of methodology for tracing CRH+ BLA projections to NAc in Crh-IRES-Cre mice. The Cre driven AAV1 was injected into the BLA or the CeA. (b) The injection of AAV1-DIO-tdTomato into central and medial amygdala (left panel) of Crh-IRES-Cre mice did not lead to CRH+ axonal labeling in the NAc (right panel). This demonstrates the specificity of BLA-origin CRH+ projections to the NAc that are shown in c. (c) Photomicrographs of the injection sites, labeling CRH+ cell bodies in the BLA (top panels) and of CRH+ fibers in the NAc (bottom panels) after BLA injections of anterograde-tracing virus in 5 mice. ac: anterior commissure. Bar = 100 µm.
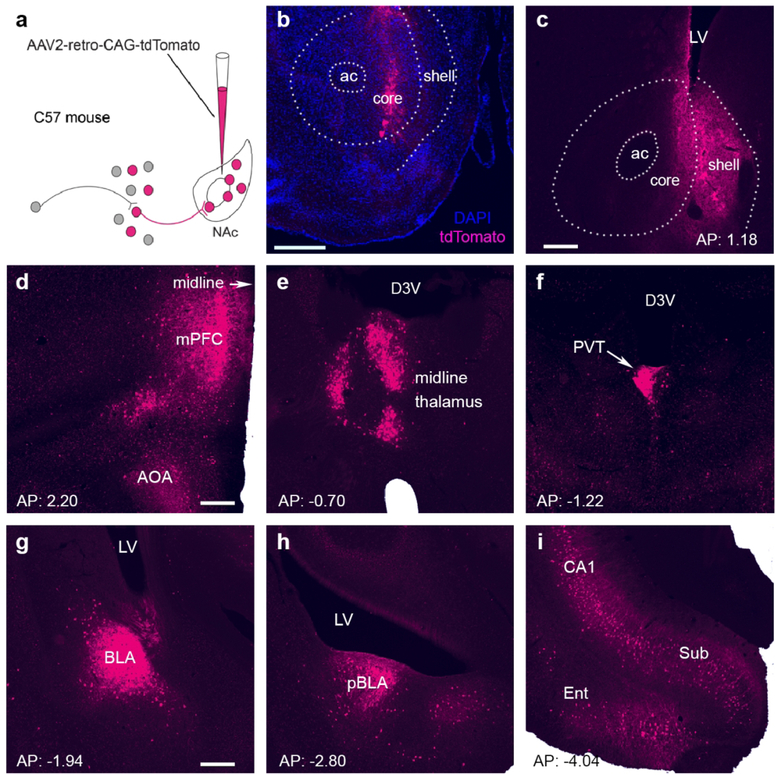
Figure 7. The regional origins of global afferents into the nucleus accumbens.
(a) Schematic of the design of a non-Cre driven rAAV2-retro-tdTomato that permits strong retrograde-labeling of cell bodies and processes. The virus was delivered into the NAc of C57BL adult male mice. (b,c) Two representative injection sites in the NAc (left hemisphere). Numerous cells in the nucleus were infected by the rAAV2-retrovirus, apparent from the red fluorescent reporter (represented by magenta). The section in b was counterstained with DAPI (blue). ac: anterior commissure; LV: lateral ventricle. Bar = 500 µm in b and 200 µm in c. (d–i) Antero-posterior series of panels demonstrating significant sources of retrogradely labeled NAc afferents in the ipsilateral hemisphere. (d) The labeling of cell bodies and processes were apparent in the anterior olfactory area (AOA) and medial prefrontal cortex (mPFC). These contributed 9.68% and 7.68% of all global projections to the NAc, respectively. (e, f) Thalamic nuclei contributed 6% of global inputs to the NAc, of which half (3%) originated from the paraventricular thalamic nucleus (PVT). (g,h) Amygdala nuclei were the sources of 14.14% of NAc inputs; the BLA contributed 6.73%. (i) NAc-directed projections from the entorhinal cortex (Ent) comprised 8.33% of the total, and the subiculum (Sub) accounted for 6%. See Figure 2 for abbreviations; LV = lateral ventricle. Bar = 200 µm for d–i.
Section images were overlaid on corresponding atlas maps. This enabled us to outline and determine the borders of the different brain regions. Immuno-fluorescent neurons were counted manually using the Photoshop counting tool. Because labeling was almost entirely unilateral, counts were only done for the hemisphere ipsilateral to the injection. A total of 90,402 neurons in 541 sections from 10 mice were quantified (1,753.2 ± 221.6 cells per CRH+ mouse and 16,337 ± 5,318.6 cells per CRH-non-selective mice). The percentage of inputs was calculated as the ratio of the number of labeled neurons from a given brain region over the total number of input neurons counted for each mouse.
2.5.2. Quantification of co-localization of the virus-reporter-tdTomato or Crh-IRES-Cre reporter and CRH-labeled cells in the BLA
Crh-IRES-Cre mice were injected with rAAV2-retro-FLEX-tdTomato in the NAc to label NAC-directed projecting CRH+ cells with tdTomato. The numbers of CRH+, tdTomato+, and CRH-tdTomato dual-labeled cells in BLA (Figure 6) were quantified based on unbiased stereological principles (Sterio, 1984; West, 1999). Systematic series of sections (1 in 6) throughout the entire anteroposterior extent of BLA (AP: −0.58~−2.78 mm) was harvested and 2 sets of section series were subjected to CRH-tyramide staining and cell counting. BLA was operationally defined as anterior (AP: −0.58~−1.30 mm), middle (AP: −1.32~−2.04 mm), and posterior (AP: −2.06~−2.78 mm) subdivisions. Cell nuclei were counted using the “optical dissector” technique (West, 1999) relying on the leading edges of nuclei in each section. Confocal images were taken using an LSM-510 confocal microscope (Zeiss, Göttingen, Germany) with an Apochromat 63× oil objective (numeric aperture = 1.40). Virtual z-sections of <1 µm were taken at 0.5 µm intervals. Image frame was digitized at 12 bit using a 1,024 × 1,024 pixel frame size. To prevent bleed-through in dual-labeling experiments, images were scanned sequentially (using the “multi-track” mode) by two separate excitation laser beams: an Argon laser at a wavelength of 488 nm and a He/Ne laser at 543 nm. Z-stack reconstructions and final adjustments of image brightness were performed using ImageJ software (version 1.43, NIH). For the counting of cells in the BLA, with the aid of a square lattice system, we used 20× confocal images, and cells further verified under 63× magnification. For each animal, 6 sections per BLA area were counted, and a total of 3 Crh-IRES-Cre mice injected with rAAV2-retro-FLEX-tdTomato were used to calculate the cell numbers and overlap ratios.
2.5.3. Quality control and technical considerations
All viral injection sites were verified posthoc. Data from mice where injections fell outside of the specified target ranges were excluded from the study. Neuroanatomical locations were verified by two observers. For assessment of the congruence of viral tracing and the native peptide, a CRH antiserum was used, a generous gift from Dr. Sawchenko, Salk Institute. The antiserum’s selectivity to the native peptide was established using CRH-null mice (Y. Chen et al., 2015).
These studies rely on innovative viral methodologies that still require comprehensive substantiation. Specifically, we employed a novel rAAV2-retro virus described recently (Tervo et al., 2016), and use the term “inputs” to describe the retrograde labeling finding with an assumption that rAAV2-retro labeled neurons in other brain regions project to and form synapses in the NAc. The method does introduce a degree of uncertainty: Whereas considerable efforts have been made to engineer AAV variants to allow improved viral tracing, several basic aspects of AAV cellular entry and tropism are still poorly understood (e.g., Pillay et al., 2016). Tervo et al. have provided empirical evidence to support the rAAV2-retro entry through axonal terminals rather than axons of passage, and stated that rAAV2-retro-mediated labeling of unexpected pathways had not been reported. In addition, the AAV tracing might only label a subset of CRH+ neurons, though this is rather unlikely in view of the broad tropism of the virus (Tervo et al., 2016). Clearly, additional studies will be necessary to fully rule out the ability of the virus to infect axons of passage. In support of the novel pathways we describe, we verified the rAAV2-retro CRH+ retrograde results with anterograde axonal tracing that employ an independent and different virus, AAV1. These experiments confirmed NAc inputs from BLA and PVT, supporting the retrogradely identified CRH+ inputs to NAc.
2.5.4. Statistical tests
All analyses were performed without knowledge of group. The relative enrichment of CRH was assessed using Student’s t test and 2-way ANOVA as appropriate. Posthoc analysis was done with Bonferroni’s Multiple Comparison Test.
3. RESULTS
3.1. The major CRH-expressing afferents to the NAc originate in the paraventricular nucleus of the thalamus, bed nucleus of the stria terminalis, medial prefrontal cortex and the basolateral amygdala
We evaluated and quantified input-mapped neurons that were labeled retrogradely from the NAc in five mice with verified NAc injections sites. These adult CRH-Cre mice were injected with a Cre-driven variant of a new designer rAAV2-retro (Tervo et al., 2016) to map CRH+ inputs to the NAc. As shown in Figure 1 a–e, tdTomato-expressing virus label in the NAc was predominantly co-localized with cells and processes expressing the endogenous CRH, as verified by dual immuno-labeling. This indicates that the novel retrograde AAV faithfully expresses in CRH+ neurons.
Inspection of anterior-posterior series of sections throughout the brain revealed a remarkable selectivity of CRH+ inputs to the NAc. As shown in Figure 2, injection of the Cre-driven rAAV2-retro that traces CRH+ NAc inputs, led to clear ‘hot-spots’ of afferent origins in a representative CRH-Cre mouse. Therefore, these regions were examined in detail.
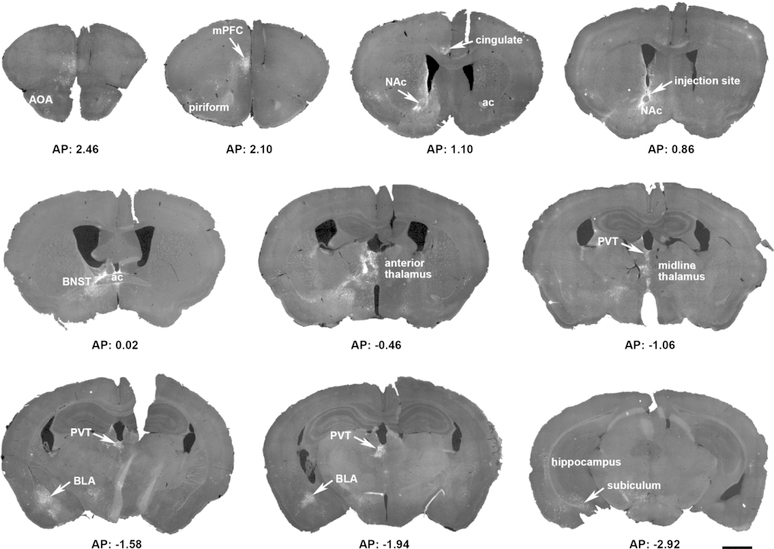
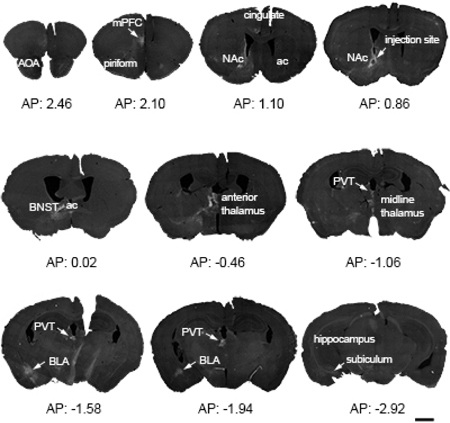
Figure 2. Overview of a representative mouse brain, highlighting the sparse, selective origins of CRH+ NAC-directed inputs.
A Cre-driven adeno-associated virus (AAV; AAV2-retro-CAG-FLEX-tdTomato-WPRE) was injected into the left hemisphere nucleus accumbens (NAc) in a CRH-IRES-Cre mouse. The 15 coronal sections (30 µm) demonstrate the location of reporter-labeled cells and fibers through the rostro-caudal axis of the brain. Sections were obtained from 2.8 mm to −4.72 mm AP relative to the Bregma. ac = anterior commissure; AOA = anterior olfactory are; BLA = basolateral amygdala; BNST = bed nucleus of the stria terminalis; mPFC = medial prefrontal cortex; PVT = paraventricular nucleus of the thalamus. Bar = 1 mm.
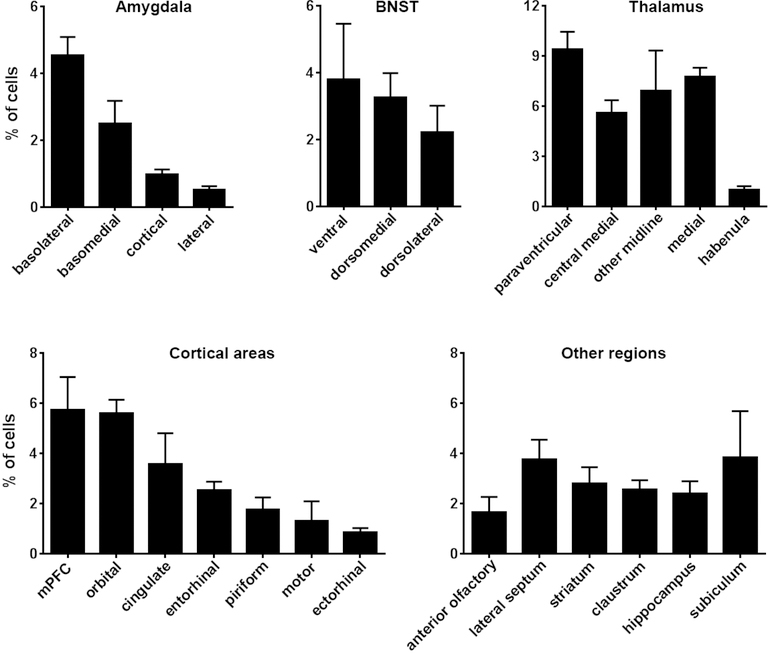
A major origin of CRH+ projections to the NAc was the thalamus. Several thalamic nuclei contributed over a third (34%) of all afferents across the five mice. The paraventricular nucleus (PVT) accounted for a third of all thalamic inputs, or 9.40% of all counted CRH+ brain inputs to the NAc (Figure 1h and Figure 3). A second key source of CRH+ projections to the NAc was the bed nucleus of the stria terminalis (BNST; Figure 1g; 9.27%). A rich source of CRH+ NAc afferents arose in the amygdalar complex (Figure 1i,j). The amygdalar complex accounted for 8.60% of CRH+ inputs to the NAc, and the primary nucleus of origin was the BLA, which contributed ~5% of inputs (Figure 3). Other significant sources of CRH+ NAc inputs included the medial prefrontal cortex (Figure 1f; 5.75%). Here, both prelimbic (PL) and infralimbic (IL) cortex contributed NAc-projecting cells. Whereas the PL was the source of 3.92 ± 0.44% of CRH+ inputs, the IL contributed 3.00 ± 1.34%. The subiculum and entorhinal cortex contributed 3.9% and 2.5%, respectively (Figure 1k and Figure 3).
Figure 3. Quantification of CRH-expressing cells projecting to the nucleus accumbens.
The graphs show the average contribution, depicted as %, of CRH+ cells in several key brain regions that provide inputs to the NAc. The relative contribution is defined as the percentage of the number of labeled neurons in a specified structure versus all quantified CRH+ afferent cells. Data are presented as mean ± SEM. Five mice were quantified with NAc viral injection. A total of 8,766 cells were counted manually from 206 sections (33–65 sections and 1,278–2,389 cells per mouse, an average of 1753.2 cells ± 221.6 SEM per mouse). Consistent with illustrations in Figure 2, the majority of CRH+NAC-targeting origin cells resided in the amygdala, BNST, thalamus (mainly PVT) and cortex (mainly mPFC). Please see Figure 2 for abbreviations.
To better characterize the origins of CRH+ NAc inputs, we quantified the cells of origin throughout the brain as shown in Figure 3. Specifically, we examined in detail CRH+ cells of origin in five mice upon injection of AAV2-retro-FLEX-tdTomato in the NAc. A total of 8,766 cells were counted manually from 206 sections (33–65 sections and 1,278–2,389 cells per mouse; 1,753.2 ± 221.6 each). Figure 3 demonstrates the predominant contribution of four principal sources of CRH+ afferents to the overall CRH+ projections to the NAc. Indeed, the thalamic nuclei, BNST, amygdala and mPFC combined accounted for ~60% of CRH+ inputs to the NAc, with the PVT and BLA contributing a sixth of all CRH+ NAc afferents. Because of the well-established involvement of these two nuclei in the circuitry underlying the stress, fear/anxiety and emotional salience, we proceeded to further establish retrogradely-identified CRH expressing NAc projections from these structures.
3.2. Anterograde tracing affirms a robust CRH+ NAc-directed projection from the PVT
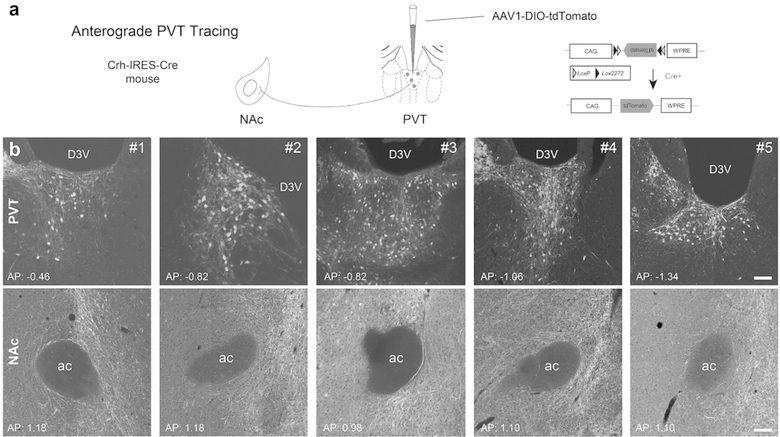
To better delineate and enhance the rigor for establishing the novel CRH+ connection between NAc and the PVT, we injected a Cre-driven AAV1 into the left PVT of five CRH-Cre mice. We then examined and quantified the PVT-origin pathway to the NAc, as shown schematically in Figure 4a. In each of the five mice depicted in the paired panels in Figure 4b, CRH+ fibers clearly emerge from the PVT and reach the NAc. CRH+ cell bodies in the PVT injection site sent axons that were distributed throughout the NAc. NAc-projecting CRH+ cells spanned the anteroposterior length of the PVT (from 0.46 to −1.34 mm, in reference to Bregma), with potentially a higher density of projecting cells posteriorly (Figure 4b). Together, the combined retrograde and anterograde approaches established a novel CRH+ PVT-NAc projection.
Figure 4. Anterograde tracing affirms a robust CRH+ NAc-directed projection from the PVT.
(a) Schematic of the methodology employed for anterograde tracing of CRH+ PVT projections to NAc in Crh-IRES-Cre mice. A Cre-driven AAV1 virus, with the construct shown in the schematic was injected into the PVT. Five mice were employed and representative images for each animal were displayed in b. (b) Injection sites labeling CRH+ cell bodies in the PVT and CRH+ fibers in the NAc from the PVT projections. D3V: dorsal third ventricle; ac: anterior commissure. Bar = 100 µm.
3.3. CRH+ amygdala projections to the NAc are confirmed using anterograde axonal tracing
To better delineate the neuroanatomical distribution of the novel CRH+ connection between NAc and the BLA, we injected the Cre-driven AAV1 into the left BLA of five CRH-Cre mice. The experimental design, analogous to that employed for the PVT, is schematically shown in Figure 5a. The paired panels for each of the mice (Figure 5c) demonstrate the cells of origin in the BLA as well as the distribution of labeled BLA-origin fibers and terminals within the NAc. A slight preponderance of their termination in the ventromedial aspect of the NAc was noted. Finally, anterograde tracing after injecting the virus into the central amygdala nucleus (CeA), a subnucleus rich in CRH somata and fibers, failed to identify axons and terminals in the NAc, demonstrating the specificity of the BLA-NAc projection (Figure 5b). Thus, converging evidence from retrograde and anterograde tracing with the use of CRH+ neuron-targeting mice and viruses identified a novel robust CRH+ projection from BLA to the NAc.
3.4. Molecular and neuroanatomical characteristics of the novel BLA-NAc CRH+ pathway
Fiber projections and monosynaptic connections from BLA to NAc have been described (Stuber et al., 2011). These consist of glutamatergic neurons and are considered to augment the pleasure/reward functions of the NAc (Stuber et al., 2011). The results above established the presence of a novel CRH+ fiber projection from BLA onto the NAc. This was a surprising discovery, because the BLA is classically considered poor in CRH-expressing cells as the large majority of amygdalar CRH+ neurons reside in the central nucleus (Y. Chen et al., 2015; Sanford et al., 2017; Swanson et al., 1983). A CRH+ BLA-NAc connection may be poised to convey stress-related information from BLA to NAc, with potential negative effects on NAc reward/pleasure functions. These facts prompted us to further characterize this pathway, by examining the molecular identity of cells expressing the viral-genetic reporters. We labeled these cells for endogenous CRH, employing an antiserum that was tested and validated in CRH-null mice (Y. Chen et al., 2015).
First, as shown in Figure 6a, the BLA of adult Crh-IRES-Cre;Ai14 mice harbors a robust population of neurons that co-express the endogenous CRH (green) and the reporter tdTomato (magenta). The arrows in the figure indicate the co-localization of endogenous CRH and the Crh-Cre-driven reporter signal. Quantification demonstrated that 70.04 ± 2.66% of all tdTomato expressing cells co-expressed endogenous CRH. Thus the remaining neurons (not marked with arrows) that express the reporter but not endogenous CRH might represent ‘legacy expression’: We use a mouse genetic approach of Crh-Ires-Cre; crossed with a reporter mouse. If there is a developmental transient CRH expression, the Cre will be turned on to mediate cre recombination and induce the tdTomato reporter expression which is long lasting. Figure 6a distinguishes this from endogenous CRH expression (CRH immunostaining in tdTomato expressing cells, arrows) in the BLA of the transgenic mouse.
When rAAV2-retro-FLEX-tdTomato virus was injected into the NAc of Crh-IRES-Cre mice, retrogradely labeled cells were detected in the ipsilateral BLA (Figure 6b). There was a good congruence of the retrograde reporter and endogenous CRH: ~59% of cells co-expressed endogenous CRH, identified via CRH antibody immunostaining, and the viral reporter (Figure 6c). Notably, retrogradely-labeled CRH+ cells tended to reside in the posterior BLA, as 40% of all CRH-expressing cells in this region projected to the NAc. Notably, both CRH (Itoi et al., 2014) and CRH mRNA (Kono et al., 2017) have been observed in mice and specifically in transgenic mice (CRH-Cre and a Venus reporter), using tools that are completely independent from those employed here.
3.5. CRH+ inputs into the NAc are not a mere reflection of global afferents to this nucleus
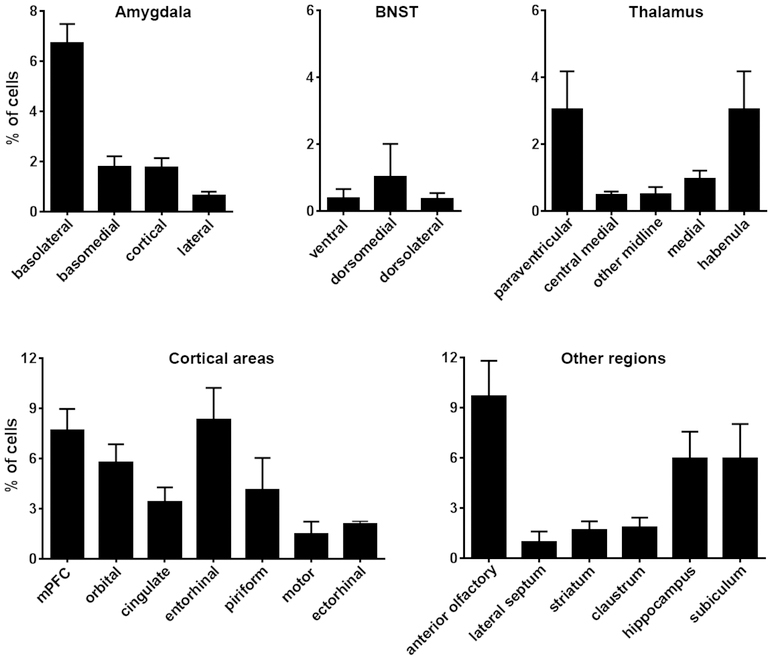
The findings above demonstrated that relatively few brain regions accounted for the majority of the origins of CRH+ projections to the NAc. This distribution might merely reflect the overall repertoire of NAc regions contributing to global NAc afferents. Alternatively, some but not all sources of NAc innervation might be specifically enriched in CRH-expressing fibers. To distinguish between these two possibilities, we examined and quantified all inputs to the NAc. We employed wild-type C57BL/6J mice and injected rAAV2-retro-tdTomato into the NAc of the left hemisphere, of five mice as shown in Figure 7a. Cell bodies were labeled in the sources of origin, and enabled visualization and quantification of NAc inputs (Figure 7b–7i). We found ~10% of all afferents to the NAc arise from the anterior olfactory area, and ~9% of all inputs to the NAc originated from the entorhinal cortex (Figure 8). The medial prefrontal cortex, hippocampus proper and subiculum accounted for 6–7% of NAc inputs each. In the mPFC, both PL and IL projected to the NAc, contributing 1.01 ± 0.51% and 0.46 ± 0.25% of inputs, respectively.
Figure 8. Quantification of global projection sources into the nucleus accumbens.
The graphs show the average contribution, depicted as %, of cells in several key brain regions that project to the NAc. The relative contribution is defined as the percentage of the number of labeled neurons in a specified structure versus all quantified reporter-labeled afferent cells. Please note different Y-axis scales in the bottom vs. top panels. Data are presented as mean ± SEM. Five mice were quantified. A total of 81,636 cells were counted manually from 341 sections (41–95 sections and 2,975–30,542 cells per mouse, an average of 16,337 cells ± 5,318.6 SEM per mouse). A large portion of afferents originated in cortical and limbic regions, including the anterior olfactory area, entorhinal cortex, hippocampus and subiculum, with moderate input from amygdala and low contribution from thalamus and BNST. Please see figure 2 for abbreviations.
Surprisingly, only 6% of NAc inputs originated from thalamic nuclei and the PVT accounted for only 3% of the total. Amygdala nuclei contributed 14% of inputs of which the BLA accounted for ~50% (7% of the total). These values diverged significantly from the distribution of CRH+ afferents to the NAc (compare Figures 8 and 3). Therefore, we compared for each of the major NAc-afferent regions and its contribution to the global and the CRH-specific innervation of the NAc (Figure 9). As apparent from Figure 9a, several brain regions represent the majority of global projections to the NAc, but they did not contribute a large proportion of CRH+ inputs. These regions include the anterior olfactory area, the entorhinal and piriform cortices and the habenula. In contrast, several regions were relatively enriched in CRH-specific NAc-directed fiber projections (Figure 9b). Among them, the PVT stood out: while accounting for only 3% of the global NAc input, the PVT provided 10% of CRH+ afferents to the NAc. Similarly, the BNST contributed ~10% of CRH+ projections and only ~2% of global projections to the NAc. Other regions contributing to circuitry involved with the sensing or processing of stress/fear were also relatively enriched in CRH+ NAc afferents, including the BLA and mPFC.
Figure 9: Comparisons of global and CRH-expressing NAc afferents uncover an enrichment of NAc-directed CRH+ projections by stress-related brain regions.
The panels in a and b depict the relative contribution of a given brain region to global (in black) or CRH-expressing (gray) NAc afferents. Data are expressed as % contribution (numbers of reporter-labelled cells) over the total cells comprising 100% of global or CRH-specific neurons projecting to the NAc. (a) Several brain regions represent the majority of global NAc input, with much less representation in CRH+ inputs. These regions include the anterior olfactory area, the entorhinal and piriform cortices and the habenula. (b) In contrast, several regions are relatively enriched in CRH-specific NAc-directed cell bodies. Among them, the PVT accounts for only 3% of the global NAc input yet provides 10% of CRH+ afferents to the NAc. The BNST contributes ~10% of CRH+ projections to the NAc and only ~2% of global input to the NAc. Other regions relatively enriched in CRH+ NAc afferents include the BLA and the mPFC. Please see abbreviations in Figure 2.
4. DISCUSSION
Our present study reports two principal discoveries. First, we provide novel information about the CRH+ projections to the NAc. We employ innovative retrograde and anterograde viral genetic approaches, and validate these with conventional immunolabeling. We thus delineate the origins and pathways of molecularly-defined inputs to the NAc, identifying a novel CRH+ circuitry. Notably, the specific molecule expressed in these projections is the neuropeptide CRH, which contributes to neuromodulation and neuroplasticity related to stress, anxiety and fear. The second discovery centers on the enrichment of CRH+ afferents to the NAc from distinct regions that play a role in the integration of salience, including of adverse or stressful information. This finding suggests a role of the CRH+ afferents in conveying diverse and context-dependent information from distinct brain regions to influence NAc circuitry and function.
First, we employed novel retrograde viruses to assess the origin of long-range afferents to the NAc. Classically known inputs to the NAc are glutamatergic projections from PVT (Papp et al., 2012; Pinto, Jankowski, & Sesack, 2003), mPFC, hippocampus, and BLA, as well as dopaminergic projections from the VTA (Russo & Nestler, 2013). It is also hypothesized that oxytocin neurons from the PVN project onto anandamide-producing neurons within the NAc to drive local endocannabinoid signaling (Wei, Allsop, Tye, & Piomelli, 2017). However, the sources of CRH+ inputs to the NAc have not been previously elucidated.
Employing a brain-wide mapping approach, we established the origins of CRH+ NAc afferents. As apparent from the overview in Figure 2, there afferent projections to the NAc were limited to relatively few brain regions. Importantly, the regions contributing richly to CRH+ projections to the NAc are mainly the regions involved in the processing and integration of salient and context-dependent signals (Agoglia & Herman, 2018; Do-Monte, Quiñones-Laracuente, & Quirk, 2015; Liberzon & Abelson, 2016; Zhu et al., 2018), where activation of endogenous CRH-expressing cells in these regions is typically induced by adverse emotional or physical events (e.g., Gunn et al., 2017; Marcinkiewcz et al., 2009).
The strong contribution of the PVT to NAc afferents is intriguing. Pioneering studies by Bhatnagar & Dallman demonstrated that the PVT is activated during acute stress selectively in rats that had sustained prior stress (Bhatnagar & Dallman, 1998; Hsu, Kirouac, Zubieta, & Bhatnagar, 2014). This was established by demonstrating selective Fos expression in animals experiencing restraint stress who have had prior chronic intermittent cold stress, compared to those experiencing restraint stress without a prior history of chronic stress. PVT projects primarily to the amygdala, BNST and the NAc, as well as limbic and many hypothalamic nuclei (Berendse & Groenewegen, 1990; Freedman & Cassell, 1994; Li & Kirouac, 2008; Moga, Weis, & Moore, 1995; Pinto et al., 2003). It is interesting to note that the PVT not only harbors CRH+ cells, but is also rich in CRH receptors, both type 1 (Y. Chen, Brunson, Müller, Cariaga, & Baram, 2000; Van Pett et al., 2000) and type 2 (Eghbal-Ahmadi et al., 1998).
The data above are consistent with the notion that PVT is involved in the storage and integration of ‘stress memory’. A projection from PVT to NAc might thus relay this ‘history’ to influence NAc function. In support of the role of PVT in modulating reward decisions as a result of long-term salient memory, Do monte et al. have demonstrated roles for amygdala microcircuitry and the mPFC in the retrieval of fear memory, whereas the PVT was required for retrieval of older events (Do-Monte et al., 2015). Indeed, Fenoglio et al. have demonstrated engagement (Fos activation) of PVT neurons in emotional events occurring during the neonatal period that induce resilience to stress during adulthood (Fenoglio, Chen, & Baram, 2006; Singh-Taylor et al., 2018). Quite recently, Zhu et al., demonstrated that the PVT was required for both appetitive and aversive associative learning and reward extinction (Zhu et al., 2018). In view of the fact that activation of CRFR1 by endogenous CRH typically takes place during emotional or aversive / stressful events, the PVT-NAc CRH+ pathway is poised to influence hedonic / pleasure functions orchestrated by the NAc (Bolton et al., 2018; Peciña et al., 2006).
A second finding is the novel and substantial CRH+ BLA-NAc projection. The BLA has been considered poor in CRH+ cells, and the canonical pathway connecting BLA to NAc is glutamatergic. Whereas this glutamatergic pathway promotes pleasure (Stuber et al., 2011), the potential function of the CRH+ BLA-NAc pathway will require future studies. Our experiments also uncovered a significant contribution of the mPFC to the spectrum of CRH+ inputs to the NAc. The mPFC is a well-established regulator of NAc (Russo & Nestler, 2013), and altered mPFC-NAc signaling has been implicated in pathological conditions involving the reward / pleasure circuitry such as addiction, alcoholism, anhedonia and PTSD (Garavan et al., 2000; Goldstein & Volkow, 2002; Kalivas, Volkow, & Seamans, 2005; Sailer et al., 2008). Putative roles of CRH+ mPFC-origin NAc afferents may thus contribute to stress-related pathologies such as relapse in alcoholism or drug use disorders (Koob & Le Moal, 2001; Valentino & Volkow, 2018).
The fundamental novel concept emerging from our studies is that several brain regions with CRH+ neurons innervate the NAc. These regions (BnST, PVT, mPFC, BLA) contribute to the numerous and nuanced influences of emotional salience, including stress, on reward-related decisions and behaviors (Diehl et al., 2018; Do-Monte et al., 2015; Knudsen, 2018; Linnman et al., 2012; Zhu et al., 2018). These brain structures contribute different functional output to the overall reward circuitry and to the NAc. The identification of CRH+ pathways from several of these circuit nodes to the NAc raises intriguing question about the relative contributions of each pathway to pleasure and reward behaviors and to psychopathology. It is likely that each of the CRH projections might modulate different cell population within the NAc and/or exert a distinct effect (inhibition, excitation, modulation). The combinatorial output of these novel CRH+ afferents to the overall circuit function and the modulation of the output by prior or concurrent stress or other kinds of emotional salience (Zhu et al., 2018), remain to be unraveled.
The structure of the Nac and the distributions of CRH-expressing axon terminals in it pose additional complexity: The NAc is comprised of two regions, the shell and the core that are believed to involve distinct cell populations, circuitries and functions (Reynolds & Berridge, 2002). Indeed, mapping of CRH cells in mouse NAc suggested that they might be more abundant in the shell (Kono et al., 2017), as was the distribution of the CRFR1 receptor (Justice, Yuan, Sawchenko, & Vale, 2008). Here, we aimed our retrograde injection sites to the NAc core (in 4/5 mice). In addition, our analyses of anterograde labelling from the PVT and BLA centered on regions close to the anterior commissure (Figures 4,5), and for technical reasons (fluorescence fading) cannot be extended. Thus, further future fine mapping of distinct CRH+ connectivity of the NAc core versus shell regions will be of interest.
In summary, the discoveries presented here support major CRH+ fiber projections that are afferent to the Nac.. CRH is a peptide released by stress in several circuit nodes throughout the brain (Y.-W. Chen, Rada, Bützler, Leibowitz, & Hoebel, 2012; Y. Chen, Bender, et al., 2004; George et al., 2012; Grieder et al., 2014; Gunn et al., 2017; Van Bockstaele & Valentino, 2013; Wanat, Bonci, & Phillips, 2013; Zhao-Shea et al., 2015) and influences circuit function. We find here enrichment of CRH+ NAc input from anxiety/stress-related brain regions, suggesting novel neuroanatomical circuit mechanisms for major potential influences of stress / fear on NAc function. These influences are important, because there are well-established and serious effects of adversity on NAc function and reward-circuit output that contribute to addiction, depression, and potentially PTSD.
Acknowledgements:
This work was supported by US National Institutes of Health grants MH105427 (XX), MH73136 (TZB), MHP50 096889 (TZB), NS078434 (XX), T32-NS45540 (CI, TZB, PI). Access to the confocal facility was funded by the Optical Biology Shared Resource of the Cancer Center Support Grant (CA-62203) at the University of California-Irvine.
Footnotes
Statement of conflict of interests: All authors declare no conflict of interests.
REFERENCES
- Agoglia AE, & Herman MA (2018). The center of the emotional universe: Alcohol, stress, and CRF1 amygdala circuitry. Alcohol, 72, 61–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berendse HW, & Groenewegen HJ (1990). Organization of the thalamostriatal projections in the rat, with special emphasis on the ventral striatum. The Journal of Comparative Neurology, 299, 187–228. [DOI] [PubMed] [Google Scholar]
- Bhatnagar S, & Dallman M (1998). Neuroanatomical basis for facilitation of hypothalamic-pituitary-adrenal responses to a novel stressor after chronic stress. Neuroscience, 84, 1025–1039. [DOI] [PubMed] [Google Scholar]
- Bolton JL, Molet J, Regev L, Chen Y, Rismanchi N, Haddad E, … Baram TZ. (2018). Anhedonia following early-life adversity involves aberrant interaction of reward and anxiety circuits and is reversed by partial silencing of amygdala corticotropin-releasing hormone gene. Biological Psychiatry, 83, 137–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y-W, Rada PV, Bützler BP, Leibowitz SF, & Hoebel BG (2012). Corticotropin-releasing factor in the nucleus accumbens shell induces swim depression, anxiety, and anhedonia along with changes in local dopamine/acetylcholine balance. Neuroscience, 206, 155–166. [DOI] [PubMed] [Google Scholar]
- Chen Y, Bender RA, Brunson KL, Pomper JK, Grigoriadis DE, Wurst W, & Baram TZ (2004). Modulation of dendritic differentiation by corticotropin-releasing factor in the developing hippocampus. Proceedings of the National Academy of Sciences, 101, 15782–15787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Brunson KL, Adelmann G, Bender RA, Frotscher M, & Baram TZ (2004). Hippocampal corticotropin releasing hormone: pre- and postsynaptic location and release by stress. Neuroscience, 126, 533–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Brunson KL, Müller MB, Cariaga W, & Baram TZ (2000). Immunocytochemical distribution of corticotropin-releasing hormone receptor type-1 (CRF(1))-like immunoreactivity in the mouse brain: light microscopy analysis using an antibody directed against the C-terminus. The Journal of Comparative Neurology, 420, 305–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Molet J, Gunn BG, Ressler K, & Baram TZ (2015). Diversity of reporter expression patterns in transgenic mouse lines targeting corticotropin-releasing hormone-expressing neurons. Endocrinology, 156, 4769–4780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Rex CS, Rice CJ, Dubé CM, Gall CM, Lynch G, & Baram TZ (2010). Correlated memory defects and hippocampal dendritic spine loss after acute stress involve corticotropin-releasing hormone signaling. Proceedings of the National Academy of Sciences of the United States of America, 107, 13123–13128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dabrowska J, Hazra R, Guo J-D, Dewitt S, & Rainnie DG (2013). Central CRF neurons are not created equal: phenotypic differences in CRF-containing neurons of the rat paraventricular hypothalamus and the bed nucleus of the stria terminalis. Frontiers in Neuroscience, 7, 156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diehl MM, Bravo-Rivera C, Rodriguez-Romaguera J, Pagan-Rivera PA, Burgos-Robles A, Roman-Ortiz C, & Quirk GJ (2018). Active avoidance requires inhibitory signaling in the rodent prelimbic prefrontal cortex. ELife, 7, e34657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Do-Monte FH, Quiñones-Laracuente K, & Quirk GJ (2015). A temporal shift in the circuits mediating retrieval of fear memory. Nature, 519, 460–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eghbal-Ahmadi M, Hatalski CG, Lovenberg TW, Avishai-Eliner S, Chalmers DT, & Baram TZ (1998). The developmental profile of the corticotropin releasing factor receptor (CRF2) in rat brain predicts distinct age-specific functions. Developmental Brain Research, 107, 81–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenoglio KA, Chen Y, & Baram TZ (2006). Neuroplasticity of the hypothalamic-pituitary-adrenal axis early in life requires recurrent recruitment of stress-regulating brain regions. The Journal of Neuroscience, 26, 2434–2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freedman LJ, & Cassell MD (1994). Relationship of thalamic basal forebrain projection neurons to the peptidergic innervation of the midline thalamus. The Journal of Comparative Neurology, 348, 321–342. [DOI] [PubMed] [Google Scholar]
- Garavan H, Pankiewicz J, Bloom A, Cho JK, Sperry L, Ross TJ, … Stein EA. (2000). Cue-induced cocaine craving: neuroanatomical specificity for drug users and drug stimuli. The American Journal of Psychiatry, 157, 1789–1798. [DOI] [PubMed] [Google Scholar]
- George O, Le Moal M, & Koob GF (2012). Allostasis and addiction: role of the dopamine and corticotropin-releasing factor systems. Physiology & Behavior, 106, 58–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein RZ, & Volkow ND (2002). Drug addiction and its underlying neurobiological basis: neuroimaging evidence for the involvement of the frontal cortex. The American Journal of Psychiatry, 159, 1642–1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grieder TE, Herman MA, Contet C, Tan LA, Vargas-Perez H, Cohen A, … George O. (2014). VTA CRF neurons mediate the aversive effects of nicotine withdrawal and promote intake escalation. Nature Neuroscience, 17, 1751–1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunn BG, Cox CD, Chen Y, Frotscher M, Gall CM, Baram TZ, & Lynch G (2017). The endogenous stress hormone CRH modulates excitatory transmission and network physiology in hippocampus. Cerebral Cortex, 27, 4182–4198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu DT, Kirouac GJ, Zubieta J, & Bhatnagar S (2014). Contributions of the paraventricular thalamic nucleus in the regulation of stress, motivation, and mood. Frontiers in Behavioral Neuroscience, 8, 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoi K, Talukder AH, Fuse T, Kaneko T, Ozawa R, Sato T, … Sakimura K. (2014). Visualization of corticotropin-releasing factor neurons by fluorescent proteins in the mouse brain and characterization of labeled neurons in the paraventricular nucleus of the hypothalamus. Endocrinology, 155, 4054–4060. [DOI] [PubMed] [Google Scholar]
- Joëls M, & Baram TZ (2009). The neuro-symphony of stress. Nature Reviews Neuroscience, 10, 459–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Justice NJ, Yuan ZF, Sawchenko PE, & Vale W (2008). Type 1 corticotropin-releasing factor receptor expression reported in BAC transgenic mice: Implications for reconciling ligand-receptor mismatch in the central corticotropin-releasing factor system. The Journal of Comparative Neurology, 511, 479–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalivas PW, Volkow N, & Seamans J (2005). Unmanageable motivation in addiction: a pathology in prefrontal-accumbens glutamate transmission. Neuron, 45, 647–650. [DOI] [PubMed] [Google Scholar]
- Knudsen EI (2018). Neural circuits that mediate selective attention: a comparative perspective. Trends in Neurosciences, 41, 789–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kono J, Konno K, Talukder AH, Fuse T, Abe M, Uchida K, … Itoi K. (2017). Distribution of corticotropin-releasing factor neurons in the mouse brain: a study using corticotropin-releasing factor-modified yellow fluorescent protein knock-in mouse. Brain Structure and Function, 222, 1705–1732. [DOI] [PubMed] [Google Scholar]
- Koob GF, & Le Moal M (2001). Drug addiction, dysregulation of reward, and allostasis. Neuropsychopharmacology, 24, 97–129. [DOI] [PubMed] [Google Scholar]
- Lemos JC, Wanat MJ, Smith JS, Reyes BAS, Hollon NG, Van Bockstaele EJ, … Phillips PEM. (2012). Severe stress switches CRF action in the nucleus accumbens from appetitive to aversive. Nature, 490, 402–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, & Kirouac GJ (2008). Projections from the paraventricular nucleus of the thalamus to the forebrain, with special emphasis on the extended amygdala. The Journal of Comparative Neurology, 506, 263–287. [DOI] [PubMed] [Google Scholar]
- Liberzon I, & Abelson JL (2016). Context processing and the neurobiology of post-traumatic stress disorder. Neuron, 92, 14–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim MM, Liu Y, Ryabinin AE, Bai Y, Wang Z, & Young LJ (2007). CRF receptors in the nucleus accumbens modulate partner preference in prairie voles. Hormones and Behavior, 51, 508–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linnman C, Zeidan MA, Furtak SC, Pitman RK, Quirk GJ, & Milad MR (2012). Resting amygdala and medial prefrontal metabolism predicts functional activation of the fear extinction circuit. American Journal of Psychiatry, 169, 415–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maras PM, & Baram TZ (2012). Sculpting the hippocampus from within: stress, spines, and CRH. Trends in Neurosciences, 35, 315–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcinkiewcz CA, Prado MM, Isaac SK, Marshall A, Rylkova D, & Bruijnzeel AW (2009). Corticotropin-releasing factor within the central nucleus of the amygdala and the nucleus accumbens shell mediates the negative affective state of nicotine withdrawal in rats. Neuropsychopharmacology, 34, 1743–1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merali Z, McIntosh J, & Anisman H (2004). Anticipatory cues differentially provoke in vivo peptidergic and monoaminergic release at the medial prefrontal cortex. Neuropsychopharmacology, 29, 1409–1418. [DOI] [PubMed] [Google Scholar]
- Merali Z, McIntosh J, Kent P, Michaud D, & Anisman H (1998). Aversive and appetitive events evoke the release of corticotropin-releasing hormone and bombesin-like peptides at the central nucleus of the amygdala. The Journal of Neuroscience, 18, 4758–4766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchenthaler I (1984). Corticotropin releasing factor (CRF)-like immunoreactivity in the rat central nervous system. Extrahypothalamic distribution. Peptides, 5, 53–69. [DOI] [PubMed] [Google Scholar]
- Merchenthaler I, Vigh S, Petrusz P, & Schally AV (1982). Immunocytochemical localization of corticotropin-releasing factor (CRF) in the rat brain. American Journal of Anatomy, 165, 385–396. [DOI] [PubMed] [Google Scholar]
- Moga MM, Weis RP, & Moore RY (1995). Efferent projections of the paraventricular thalamic nucleus in the rat. The Journal of Comparative Neurology, 359, 221–238. [DOI] [PubMed] [Google Scholar]
- Oh SW, Harris JA, Ng L, Winslow B, Cain N, Mihalas S, … Zeng H (2014). A mesoscale connectome of the mouse brain. Nature, 508, 207–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papp E, Borhegyi Z, Tomioka R, Rockland KS, Mody I, & Freund TF (2012). Glutamatergic input from specific sources influences the nucleus accumbens-ventral pallidum information flow. Brain Structure and Function, 217, 37–48. [DOI] [PubMed] [Google Scholar]
- Peciña S, Schulkin J, Berridge KC, Koob GF, Bloom FE, Dunn AJ, … Raine L (2006). Nucleus accumbens corticotropin-releasing factor increases cue-triggered motivation for sucrose reward: paradoxical positive incentive effects in stress? BMC Biology, 4, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillay S, Meyer NL, Puschnik AS, Davulcu O, Diep J, Ishikawa Y, … Carette JE (2016). An essential receptor for adeno-associated virus infection. Nature, 530, 108–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto A, Jankowski M, & Sesack SR (2003). Projections from the paraventricular nucleus of the thalamus to the rat prefrontal cortex and nucleus accumbens shell: Ultrastructural characteristics and spatial relationships with dopamine afferents. The Journal of Comparative Neurology, 459, 142–155. [DOI] [PubMed] [Google Scholar]
- Pomrenze MB, Tovar-Diaz J, Blasio A, Maiya R, Giovanetti SM, Lei K, … Messing RO (2019). A corticotropin releasing factor network in the extended amygdala for anxiety. The Journal of Neuroscience, 39, 1030–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regev L, Neufeld-Cohen A, Tsoory M, Kuperman Y, Getselter D, Gil S, & Chen A (2011). Prolonged and site-specific over-expression of corticotropin-releasing factor reveals differential roles for extended amygdala nuclei in emotional regulation. Molecular Psychiatry, 16, 714–728. [DOI] [PubMed] [Google Scholar]
- Regev L, Tsoory M, Gil S, & Chen A (2012). Site-specific genetic manipulation of amygdala corticotropin-releasing factor reveals its imperative role in mediating behavioral response to challenge. Biological Psychiatry, 71, 317–326. [DOI] [PubMed] [Google Scholar]
- Reynolds SM, & Berridge KC (2002). Positive and negative motivation in nucleus accumbens shell: bivalent rostrocaudal gradients for GABA-elicited eating, taste “liking”/”disliking” reactions, place preference/avoidance, and fear. Journal of Neuroscience, 22, 7308–7320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo SJ, & Nestler EJ (2013). The brain reward circuitry in mood disorders. Nature Reviews Neuroscience, 14, 609–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sailer U, Robinson S, Fischmeister FPS, König D, Oppenauer C, Lueger-Schuster B, … Bauer H (2008). Altered reward processing in the nucleus accumbens and mesial prefrontal cortex of patients with posttraumatic stress disorder. Neuropsychologia, 46, 2836–2844. [DOI] [PubMed] [Google Scholar]
- Sanford CA, Soden ME, Baird MA, Miller SM, Schulkin J, Palmiter RD, … Zweifel LS (2017). A central amygdala CRF circuit facilitates learning about weak threats. Neuron, 93, 164–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder M, Jakovcevski M, Polacheck T, Lebow M, Drori Y, Engel M, … Chen A (2017). A methyl-balanced diet prevents CRF-induced prenatal stress-triggered predisposition to binge eating-like phenotype. Cell Metabolism, 25, 1269–1281.e6. [DOI] [PubMed] [Google Scholar]
- Singh-Taylor A, Molet J, Jiang S, Korosi A, Bolton JL, Noam Y, … Baram TZ (2018). NRSF-dependent epigenetic mechanisms contribute to programming of stress-sensitive neurons by neonatal experience, promoting resilience. Molecular Psychiatry, 23, 648–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterio DC (1984). The unbiased estimation of number and sizes of arbitrary particles using the disector. Journal of Microscopy, 134, 127–136. [DOI] [PubMed] [Google Scholar]
- Stuber GD, Sparta DR, Stamatakis AM, van Leeuwen WA, Hardjoprajitno JE, Cho S, … Bonci A (2011). Excitatory transmission from the amygdala to nucleus accumbens facilitates reward seeking. Nature, 475, 377–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Nguyen AQ, Nguyen JP, Le L, Saur D, Choi J, … Xu X (2014). Cell-type-specific circuit connectivity of hippocampal CA1 revealed through cre-dependent rabies tracing. Cell Reports, 7, 269–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Nitz DA, Holmes TC, & Xu X (2018). Opposing and complementary topographic connectivity gradients revealed by quantitative analysis of canonical and noncanonical hippocampal CA1 inputs. ENeuro, 5, ENEURO.0322–17.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson LW, Sawchenko PE, Lind RW, & Rho JH (1987). The CRH motoneuron: differential peptide regulation in neurons with possible synaptic, paracrine, and endocrine outputs. Annals of the New York Academy of Sciences, 512, 12–23. [DOI] [PubMed] [Google Scholar]
- Swanson LW, Sawchenko PE, Rivier J, & Vale WW (1983). Organization of ovine corticotropin-releasing factor immunoreactive cells and fibers in the rat brain: an immunohistochemical study. Neuroendocrinology, 36, 165–186. [DOI] [PubMed] [Google Scholar]
- Tervo DGR, Hwang B-Y, Viswanathan S, Gaj T, Lavzin M, Ritola KD, … Karpova AY (2016). A designer AAV variant permits efficient retrograde access to projection neurons. Neuron, 92, 372–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vale W, Rivier C, Brown MR, Spiess J, Koob G, Swanson L, … Rivier J (1983). Chemical and biological characterization of corticotropin releasing factor. Recent Progress in Hormone Research, 39, 245–270. [DOI] [PubMed] [Google Scholar]
- Valentino RJ, & Volkow ND (2018). Untangling the complexity of opioid receptor function. Neuropsychopharmacology : Official Publication of the American College of Neuropsychopharmacology, 43, 2514–2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Bockstaele EJ, & Valentino RJ (2013). Neuropeptide regulation of the locus coeruleus and opiate-induced plasticity of stress responses. In Advances in pharmacology (Vol. 68, pp. 405–420). San Diego, CA: Elsevier. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Pett K, Viau V, Bittencourt JC, Chan RK, Li HY, Arias C, … Sawchenko PE (2000). Distribution of mRNAs encoding CRF receptors in brain and pituitary of rat and mouse. The Journal of Comparative Neurology, 428, 191–212. [DOI] [PubMed] [Google Scholar]
- Wamsteeker Cusulin JI, Füzesi T, Watts AGA, Bains JJS, Joels M, Baram TZ, … Tian L (2013). Characterization of corticotropin-releasing hormone neurons in the paraventricular nucleus of the hypothalamus of Crh-IRES-Cre mutant mice. PLoS ONE, 8, e64943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanat MJ, Bonci A, & Phillips PEM (2013). CRF acts in the midbrain to attenuate accumbens dopamine release to rewards but not their predictors. Nature Neuroscience, 16, 383–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XD, Su YA, Wagner KV, Avrabos C, Scharf SH, Hartmann J, … Schmidt MV (2013). Nectin-3 links CRHR1 signaling to stress-induced memory deficits and spine loss. Nature Neuroscience, 16, 706–713. [DOI] [PubMed] [Google Scholar]
- Wei D, Allsop S, Tye K, & Piomelli D (2017). Endocannabinoid signaling in the control of social behavior. Trends in Neurosciences, 40, 385–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West MJ (1999). Stereological methods for estimating the total number of neurons and synapses: issues of precision and bias. Trends in Neurosciences, 22, 51–61. [DOI] [PubMed] [Google Scholar]
- Zhao-Shea R, DeGroot SR, Liu L, Vallaster M, Pang X, Su Q, … Tapper AR (2015). Increased CRF signalling in a ventral tegmental area-interpeduncular nucleus-medial habenula circuit induces anxiety during nicotine withdrawal. Nature Communications, 6, 6770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Nachtrab G, Keyes PC, Allen WE, Luo L, & Chen X (2018). Dynamic salience processing in paraventricular thalamus gates associative learning. Science, 362, 423–429. [DOI] [PMC free article] [PubMed] [Google Scholar]