Capsule summary:
This study reports a homozygous mutation in REL abrogating c-Rel protein expression in a patient with combined immunodeficiency characterized by susceptibility to Mycobacterium tuberculosis, Salmonella, Cryptosporidium, and cytomegalovirus.
Keywords: c-Rel, combined immunodeficiency, mycobacteria
To the Editor:
c-Rel is a transcription factor in the NF-κB family, critical for T and B cell function. c-Rel binds to the promoters of genes that encode cytokines important for immunity against infectious pathogens, including IL-2, IFN-γ, IL-12, IL-21, and IL-23.1 Rel-deficient mice have impaired T and B cell proliferation, hypogammaglobulinemia, and cytokine secretion.2–5 Human patients with mutations in c-Rel have not been previously published. Here, we describe a homozygous mutation abrogating c-Rel expression in a patient with impaired T and B cell activation, susceptibility to opportunistic infections, and Mycobacterium tuberculosis.
The patient is the son of first-degree cousins of Arabic descent. He received live vaccinations against poliovirus, measles, and Bacille Calmette-Guerin (BCG) without clinical sequelae. At 30 months of age, he developed chronic diarrhea associated with multiple infections, including Salmonella enteritica, cytomegalovirus (CMV), and Cryptosporidium. In the setting of chronic cryptosporidiosis, he developed hepatomegaly and sclerosing cholangitis. Immunologic evaluation at six years of age was notable for leukocytosis, lymphocytosis, and thrombocytosis (Table 1). He had increased numbers of CD4+ and CD8+ T cells with decreased memory CD4+CD45RO+ T cells, and reduced proliferation to phytohemagglutinin (PHA) (Table 1). He had B cell lymphopenia, impaired B cell proliferation upon stimulation with CD40 ligand + IL-21, reduced IgG and undetectable IgA levels, and non-protective titers to the tetanus and diphtheria vaccines despite booster vaccinations (Table 1). Despite vaccination with BCG, he developed femoral Mycobacterium tuberculosis osteomyelitis at 7 years of age that was successfully treated with isoniazid, rifampin, pyrazinamide, and ethambutol. He also developed one lobar pneumonia confirmed by chest X-ray. Additional immunophenotyping done at 9 years of age revealed a predominance of naïve CD19+IgD+CD27− B cells (85%; normal 47.3 – 77%) and reduced percentages of switched memory CD19+CD27+IgD− B cells (0.3%; reference range 10.0 – 30.4). He is currently treated with intravenous immunoglobulin and prophylactic antibiotics while undergoing evaluation for hematopoietic stem cell transplantation.
Table 1.
Immunological profile of the proband at 6 years of age. Bold values are outside the normal range. Age-matched normal ranges are shown in parentheses.
| Hemogram 103 cells/μL | 6 years |
| White blood cells | 12,600 (5,700 – 9,900) |
| Neutrophils | 5,800 (2,800 – 6,300) |
| Lymphocytes | 5,900 (1,200 – 4,700) |
| Platelets | 447,000 (227,000 – 350,000) |
| Lymphocyte subpopuations | |
| CD3+, 103 cells/μL | 4,969 (1,200 – 2,600) |
| CD3+CD45RO+, % CD3+ | 10.3% (15 – 41) |
| CD3+CD4+, 103 cells/μL | 1,843 (650 – 1,500) |
| CD3+CD8+, 103 cells/μL | 2,483 (370 – 1,100) |
| CD19+, 103 cells/μL | 103 (270 – 860) |
| CD3− CD56+, 103 cells/μL | 70 (100 – 480) |
| T cell proliferation, % control | |
| PHA | 47.3% |
| PMA + ionomycin | 108% |
| B cell proliferation, % control | |
| CD40L + IL21 | 6.4% |
| Serum Immunoglobulins mg/dL (normal) | |
| IgG | 150 (650-1,150) |
| IgM | 150 (50-150) |
| IgA | Undetectable (40-160) |
| Anti-diphtheria, IU/mL | 0.015 (>0.1 IU/mL) |
| Tetanus, IU/mL | 0.041 (>0.1 IU/mL) |
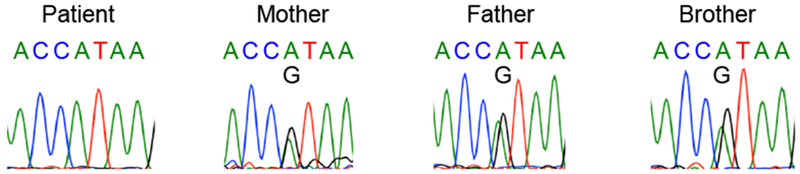
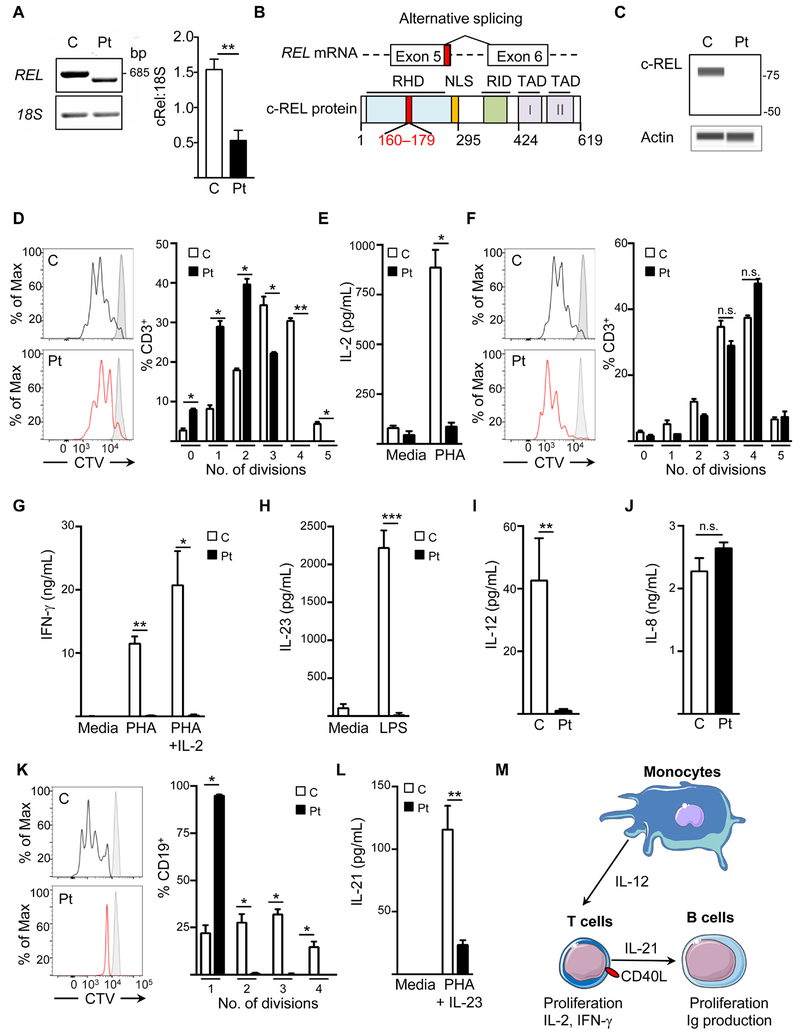
Whole-exome sequencing of genomic DNA from the proband identified 80 rare variants in the proband which were absent from the gnomAD and 1000 genomes databases (Table E1 in this article’s Online Repository). Of these, a homozygous mutation in the canonical donor splice-site after exon 5 of REL (NM_002908.3: c.535+1G>A) was considered the most likely pathogenic variant due to the known contribution of c-Rel in T and B cell activation and cytokine secretion. Sanger sequencing confirmed that the mutation is homozygous in the proband and heterozygous in his parents and healthy brother (Fig E1 in this article’s Online Repository). Amplification of the patient’s REL cDNA using primers specific to sequences in exons 2 and 7 identified an alternatively spliced transcript with lower expression than that of wild-type REL from control cDNA (Fig 1). Sanger sequencing of the patient’s mutant REL transcript indicated that the mutant utilizes a cryptic donor splice site in exon 5 (NM_002908.3: c.483) and a cryptic acceptor splice site near the start of exon 6 (NM_002908.3: c.538) (Fig 1B, top). The mutant transcript lacks 54 nucleotides encoding 18 residues within the Rel homology domain (Fig 1B, bottom). If expressed, the molecular mass of the mutant c-Rel protein is predicted to be 66 kilodaltons. However, immunoblotting using an antibody specific to the protein’s N-terminus demonstrated the absence of c-Rel protein in the patient’s peripheral blood mononuclear cells (PBMCs) with no detectable truncation products, indicating that the mutation abrogates protein expression (Fig 1C).
Table E1.
Variants identified by WES in the proband which are absent from the gnomAD and 1000 genomes databases. The mutation in REL is bolded.
| Gene | Chr:Pos | Ref/Alt | Zygosity | Alteration(s) |
|---|---|---|---|---|
| TTLL10 | 1:1120348 | G/A | Heterozygous | NM_001130045.1:c.1261-1G>A |
| PRDM16 | 1:3347585 | C/T | Heterozygous | NM_022114.3:c.3434C>T |
| MST1L | 1:17085015 | C/T | Heterozygous | NM_001271733.1:c.1460G>A |
| IPO13 | 1:44433161 | A/T | Heterozygous | NM_014652.3:c.2788A>T |
| BRDT | 1:92457845 | C/T | Heterozygous | NM_001242806.2:c.2101C>T |
| OR10J1 | 1:159409676 | T/C | Heterozygous | NM_012351.2:c.128T>C |
| IGSF8 | 1:160061714 | A/G | Heterozygous | NM_052868.5:c.1741T>C |
| PLEKHA6 | 1:204192629 | C/A | Heterozygous | NM_014935.4:c.3116G>T |
| MARK1 | 1:220809217 | C/G | Heterozygous | NM_001286124.1:c.1319C>G |
| OBSCN | 1:228525005 | C/T | Heterozygous | NM_001271223.2:c.19592C>T |
| HEATR1 | 1:236721531 | G/C | Heterozygous | NM_018072.5:c.5204+6C>G |
| REL | 2:61144153 | G/A | Homozygous | NM_002908.3:c.535+1G>A |
| ANKRD36C | 2:96616506 | G/C | Heterozygous | NM_001310154.1:c.1286C>G |
| ANKRD36C | 2:96616507 | C/A | Heterozygous | NM_001310154.1:c.1285G>T |
| SSFA2 | 2:182756857 | C/T | Heterozygous | NM_001130445.2:c.64C>T |
| GIGYF2 | 2:233712226 | CACAGCAGCAGCAGCAGCAGC/- | Heterozygous | NM_001103147.1:c.3693_3713del |
| COL6A5 | 3:130110144 | G/C | Heterozygous | NM_001278298.1:c.2539G>C |
| NMD3 | 3:160939747 | TTTTTTTTTTTTTTTTT/- | Heterozygous | NM_001320227.1:c.-20-22_-20-6del |
| CYTL1 | 4:5018591 | T/C | Heterozygous | NM_018659.2:c.299A>G |
| SORCS2 | 4:7436509 | C/T | Heterozygous | NM_020777.2:c.548+38427C>T |
| CEP135 | 4:56835228 | G/C | Heterozygous | NM_025009.4:c.1045-1G>C |
| THEGL | 4:57467169 | C/A | Heterozygous | NM_001256475.1:c.1177C>A |
| CEP44 | 4:175223340 | A/G | Heterozygous | NM_001145314.1:c.237+3A>G |
| CNPY3 | 6:42902265 | G/T | Heterozygous | NM_001318842.1:c.204G>T |
| IMPG1 | 6:76744476 | A/T | Heterozygous | NM_001563.3:c.330T>A |
| MDN1 | 6:90494802 | G/C | Heterozygous | NM_014611.2:c.1378C>G |
| TIAM2 | 6:155450909 | C/A | Homozygous | NM_012454.3:c.552C>A |
| GET4 | 7:916358 | G/A | Heterozygous | NM_015949.2:c.76G>A |
| DNAH11 | 7:21599405 | G/T | Heterozygous | NM_001277115.1:c.877G>T |
| SEMA3A | 7:83590893 | T/C | Heterozygous | NM_006080.2:c.2110A>G |
| RP1L1 | 8:10480232 | C/A | Heterozygous | NM_178857.5:c.480G>T |
| SH2D4A | 8:19251032 | G/A | Heterozygous | NM_022071.3:c.1252G>A |
| LYN | 8:56866432 | A/G | Heterozygous | NM_002350.3:c.679A>G |
| ZNF517 | 8:146033370 | G/A | Heterozygous | NM_213605.2:c.1069G>A |
| RIC1 | 9:5763776 | T/C | Heterozygous | NM_020829.3:c.2749T>C |
| OR1L1 | 9:125424679 | C/A | Heterozygous | NM_001005236.3:c.685C>A |
| SLC16A12 | 10:91198403 | T/A | Heterozygous | NM_213606.3:c.986A>T |
| CPEB3 | 10:94000082 | T/C | Heterozygous | NM_014912.4:c.26A>G |
| TACC2 | 10:123848028 | C/T | Homozygous | NM_206862.3:c.5495C>T |
| MUC6 | 11:1016613 | G/C | Heterozygous | NM_005961.2:c.6188C>G |
| OR52J3 | 11:5068002 | A/G | Heterozygous | NM_001001916.2:c.247A>G |
| OR56A3 | 11:5969049 | T/A | Heterozygous | NM_001003443.2:c.473T>A |
| ARNTL | 11:13393880 | GGG/- | Heterozygous | NM_001297719.1:c.992_994delGGG |
| LMO2 | 11:33886347 | C/T | Heterozygous | NM_005574.3:c.265G>A |
| LRRC4C | 11:40136226 | C/G | Heterozygous | NM_020929.2:c.1617G>C |
| CCDC85B | 11:65658456 | C/T | Heterozygous | NM_006848.2:c.202C>T |
| SHANK2 | 11:70332235 | G/A | Heterozygous | NM_133266.4:c.2399C>T |
| APLP2 | 11:129940533 | C/G | Heterozygous | NM_001642.2:c.105+556C>G |
| VWF | 12:6167043 | G/C | Heterozygous | NM_000552.4:c.1701C>G |
| ATN1 | 12:7050180 | C/T | Heterozygous | NM_001007026.1:c.3352C>T |
| PIK3C2G | 12:18762523 | A/T | Heterozygous | NM_001288772.1:c.4142A>T |
| CELA1 | 12:51740388 | CTGGACCATATCC/- | Heterozygous | NM_001971.5:c.16+7_16+19delGGATATGGTCCAG |
| CELA1 | 12:51740403 | TT/- | Heterozygous | NM_001971.5:c.16+3_16+4delAA |
| G2E3 | 14:31061589 | C/T | Heterozygous | NM_017769.4:c.298C>T |
| GPATCH2L | 14:76633107 | G/A | Heterozygous | NM_017926.3:c.727+37G>A |
| SLC24A4 | 14:92790151 | G/C | Heterozygous | NM_153647.3:c.-23-1G>C |
| WARS | 14:100820895 | T/G | Heterozygous | NM_004184.3:c.431A>C |
| PKM | 15:72492836 | C/T | Heterozygous | NM_001206796.2:c.1690G>A |
| CD276 | 15:74005281 | G/A | Heterozygous | NM_001024736.1:c.1589G>A |
| BCL2A1 | 15:80253513 | T/C | Heterozygous | NM_004049.3:c.424A>G |
| CEMIP | 15:81224250 | A/C | Heterozygous | NM_018689.2:c.2663A>C |
| NME4 | 16:448386 | G/T | Heterozygous | NM_005009.2:c.92-604G>T |
| PARN | 16:14704541 | -/T | Heterozygous | NM_002582.3:c.514dupA |
| GTF3C1 | 16:27476729 | G/C | Heterozygous | NM_001520.3:c.5207C>G |
| CNGB1 | 16:57945772 | T/C | Heterozygous | NM_001297.4:c.2377A>G |
| MRM3 | 17:685929 | T/C | Heterozygous | NM_018146.3:c.311T>C |
| ZBTB4 | 17:7385592 | C/T | Heterozygous | NM_020899.3:c.-81+1731G>A |
| ATAD5 | 17:29196590 | A/G | Heterozygous | NM_024857.4:c.3533A>G |
| MYO1D | 17:30986172 | A/C | Heterozygous | NM_015194.2:c.2306T>G |
| SOCS7 | 17:36523760 | G/A | Heterozygous | NM_014598.3:c.1192-5G>A |
| KRTAP4-8 | 17:39254133 | G/T | Heterozygous | NM_031960.2:c.204C>A |
| MRPS23 | 17:55926758 | C/T | Heterozygous | NM_016070.3:c.58G>A |
| TNFAIP8L1 | 19:4651887 | C/A | Heterozygous | NM_152362.2:c.6C>A |
| SH2D3A | 19:6753521 | G/A | Heterozygous | NM_005490.2:c.1516C>T |
| ZNF492 | 19:22817275 | CG/- | Heterozygous | NM_020855.2:c.-95_-94delCG |
| FCGBP | 19:40364256 | G/C | Heterozygous | NM_003890.2:c.14386C>G |
| LILRB2 | 19:54783690 | G/T | Heterozygous | NM_005874.4:c.311C>A |
| CHODL | 21:19618290 | G/A | Heterozygous | NM_024944.2:c.79+671G>A |
| IL17RA | 22:17566097 | G/A | Heterozygous | NM_014339.6:c.116G>A |
| CELSR1 | 22:46768834 | C/G | Heterozygous | NM_014246.1:c.7705G>C |
Figure E1.
Chromatograms of Sanger sequencing of the REL mutation in the patient, his parents, and healthy brother.
Figure 1.
A. Representative electrophoresis (left) and densitometry quantification of REL cDNA from EBV-transformed lymphoblastoid cell lines (EBV-LCLs) from controls and the patient. B. Schematic of REL cDNA and protein with the patient’s deleted nucleotides and residues in red. C. Representative immunoblot of c-Rel in lysates of peripheral blood mononuclear cells (PBMCs) from the patient and controls. D. CD3+ T cell proliferation to phytohemagglutinin (PHA), measured by CellTrace Violet (CTV) dilution. E. IL-2 secretion from patient and control PBMCs, with or without PHA stimulation. F. CD3+ T cell proliferation to PHA+IL-2. G, H. Secretion of IFN-γ (G) and IL-23 (H) by PBMCs from the patient and controls cultured with the indicated stimuli. I, J. Secretion of IL-12 (I) and IL-8 (J) by EBV-LCLs from the patient and controls stimulated with PDBu. K. CD19+ B cell proliferation to anti-CD40+IL-21. L. IL-21 secretion by PHA blasts from the patient or controls with or without PHA+IL-23 stimulation. M. Schematic of the contribution of c-Rel to adaptive and innate immunity. *p<0.05, **p<0.01. Data in A, E, G – J, and L are pooled from two independent experiments; data in C, D, F, K are representative of two to three independent experiments.
The patient’s history of infections with CMV and cryptosporidiosis with secondary sclerosing cholangitis indicated defective T cell function. Mice deficient in c-Rel have impaired T cell proliferation and defective IL-2 secretion; the addition of exogenous IL-2 normalizes T cell proliferation.2,3 After PHA stimulation, the patient’s T cells had reduced proliferation, as demonstrated by minimal proliferation beyond the third round of division, and negligible IL-2 secretion (Fig 1D and E). As has been seen in c-Rel-deficient mice, addition of IL-2 normalized the proliferation of the patient’s T cells stimulated with PHA (Fig 1F).
c-Rel binds to the promoters of the genes encoding IFN-γ, IL-23, and IL-12, all of which contribute to host defense against mycobacteria and Salmonella.1,6 IFN-γ was undectable after PHA stimulation of the patient’s T cells and failed to increase with the addition of IL-2 (Fig 1G). Antigen presenting cells are the predominant sources of IL-12 and IL-23.6 Stimulation of the patient’s PBMCs with LPS did not result in IL-23 secretion, as compared to the control (Fig 1H). Similarly, the patient’s EBV-LCLs secreted minimal amounts of IL-12 after phorbol 12, 13-dibutyrate stimulation (PDBu) (Fig 1I). In contrast, secretion of IL-8, a cytokine independent of c-Rel, was comparable between control and patient EBV-LCLs (Fig 1J). Although the patient demonstrated susceptibility to extrapulmonary Mycobacterium tuberculosis, he had no evidence of complications from the less virulent, attenuated Mycobacterium bovis BCG strain. The residual T cell proliferation in the patient may have helped to prevent complications from the BCG vaccine.
In B cells, c-Rel is among the first NF-kB transcription factors activated after CD40 stimulation.4 c-Rel-deficient mice have severely impaired B cell proliferation following CD40 ligation.2,4 Similarly, after anti-CD40 stimulation, nearly all of the patient’s CD19+ B cells were limited to only 1 round of division, compared to up to 4 rounds of division for the control (Fig 1K). In addition to its role in B cell proliferation, c-Rel also binds to the promoter of the gene encoding IL-21, a cytokine important for the development of germinal center B cells, isotype switching, and the antibody response to T dependent antigens.7 In vitro stimulation of PHA blasts with PHA+IL-23 revealed defective secretion of IL-21 in the patient compared to control (Fig 1L). Our patient shares features found in individuals with deficiency of IL-21 or the IL-21 receptor that include hypogammaglobulinemia, recurrent gastrointestinal infections, pneumonia, and a predominance of naïve B cells.7 In addition to its role in B cell function, IL-21 enhances the effector function of CD8+ T cells by increasing the expression of IFN-γ, perforin, and granzyme, all of which facilitate the elimination of Cryptosporidium.7
The patient’s combined immunodeficiency mirrors the defects in T cell activation, B cell proliferation, innate immunity and humoral immunity seen in c-Rel-deficient mice.2–5 His susceptibility to mycobacteria, Salmonella, and opportunistic pathogens illustrate the pleiotropic contributions of c-Rel to host immunity in humans, as illustrated in Fig 1M. Small molecule inhibitors of c-Rel are under investigation as therapies for lymphoid tumors and graft versus host disease, but the potential risks of infections with these drugs have not yet been studied.8 This novel immunodeficiency highlights the potential susceptibility to a broad range of pathogens with therapeutic strategies targeting c-Rel.
Materials and Methods
Study oversight.
Written informed consent was obtained from the patient’s parents. This study was approved by the Institutional Review Board of Boston Children’s Hospital.
Immunophenotyping.
Antibodies and viability dyes used for flow cytometry studies of T and B cells are as follows: Zombie Aqua™ Fixable Viability Kit, anti-human CD4-APC/Cy7 (clone OKT4), anti-human CD8-PE (clone SK1), anti-human CD3-FITC (clone OKT3), Annexin V, Alexa Fluor® 647, anti-human CD27-APC/Cy7 (clone O323), anti-human CD19-FITC (clone HIB19), all from Biolegend, Dedham, MA, USA). The analysis of flow cytometric data was performed with FlowJo software version 10.5.0 (TreeStar).
Cell cultures.
T cell proliferation was quantified using dilution of CellTrace™ Violet Cell Proliferation Kit (Invitrogen, Waltham, MA, USA) after stimulation with PHA (5 μg/mL; Sigma Aldrich, St. Louis, MO, USA) ± IL-2 (40 U/mL; R&D Systems, Minneapolis, MN, USA) for 2.5 days, at which point cell culture supernatants were also harvested for measure of IFN-γ and IL-2. B cell proliferation was quantified using dilution of CellTrace Violet after stimulation with anti-CD40 monoclonal antibody (clone G28-5, at a concentration of 10 μg/mL, Enzo Biochem, Farmingdale, NY, USA) + IL-21 (50 ng/mL, Cell Sciences, Newburyport, MA, USA) for 3 days. For IL-21 secretion, PBMCs were stimulated with PHA for 7 days, followed by PHA + IL-23 (100 ng/mL, R&D Systems, Minneapolis, MN, USA) for 72 hours. For IL-23 secretion, patient and control PBMCs were stimulated with LPS (100 ng/mL, InvivoGen, San Diego, CA, USA) for 18 hours. For IL-12 secretion, live B-LCLs were isolated after eliminating dead cells using the Dead Cell Removal Kit (Miltenyi Biotec GmbH, Bergisch Gladbach, Germany) per the manufacturer’s guidelines. Remaining live B-LCLs were stimulated with pDBU (10−7 M, Sigma Aldrich, St. Louis, MO, USA) for 18 hours as previously described.1
Cytokine quantification.
Concentrations of IL-21, IFN-γ, IL-2, and IL-12 in cell culture supernatants were measured by Cytometric Bead Array (BD Biosciences, Singapore) per the manufactuer’s guidelines. IL-23 was measured in cell culture supernatants by ELISA.
Whole exome sequencing (WES).
Library preparation for WES was performed using Agilent SureSelect. WES was performed on genomic DNA from the proband using the Illumina HiSeq-2000 (Illumina Inc., San Diego, CA). The average coverage of the exome was 53x. The Burrows-Wheeler Aligner (BWA) was used to map reads to the human reference genome assembly GRCh37.2 Variants were called using the Genome Analysis Toolkit (GATK).3
Primers.
Sanger sequencing of the proband’s genomic DNA for the c-REL mutation was performed with the following primers: (F: 5′-GATATTGAAGATTGTGACCTCA-3′, R: 5′- TATTTCTCTAATGTCTGGAAGT-3′). RT-PCR was performed using the following primers: c-REL (F: 5′-GCACAGACAACAACCGAA-3′, R: 5′- GTCAGAAGGTCTCCGCA-3′); 18S (F: 5′-CGGCTACCACATCCAAGGAA-3′, R: 5′- GCTGGAATTACCGCGGCT-3′). ImageJ analyzer software (1.48v) for quantitation.
Capillary immunoassay.
Control or patient PBMCs were lysed in sample buffer (Pierce, Appleton, WI, USA) and 2-mercaptoethanol (Sigma Aldrich, St. Louis, MO, USA Electrophoresis) and transferred to 12 – 230 kDa Wes Separation Modules (Protein Simple) per the manufacter’s guidelines with the following antibodies, as indicated in figure legends: anti-c-REL (clone D4Y6M, Cell Signaling Technology, Danvers, Massachusetts, USA), and anti-β-actin (Sigma Aldrich, St. Louis, MO, USA). Densitometry was performed using the Compass Software for Simple Western.
Statistics.
Student’s t-test was used for comparisons of two groups; one- or two-way ANOVA with the with Holm-Šídák post-comparison test was used for the comparison of more than 2 groups, as indicated. Data are graphed as mean with SEM. All experiments were performed with two to three controls. Statistical analysis was performed using GraphPad Prism.
Acknowledgments
Supported by: 5T32AI007512-33 (S.B.C), 5K08AI116979-04 (J.C.), 1R01AI139633-01 (RSG), Perkin Fund (RSG)
Abbreviations:
- BCG
Bacille Calmette-Guerin
- CMV
cytomegalovirus
- EBV-LCLs
Epstein Barr virus-transformed lymphoblastoid cell lines
- LPS
lipopolysaccharide
- NF-κB
nuclear factor-kappa B
- PDBu
phorbol 12, 13-dibutyrate
- PBMCs
peripheral blood mononuclear cells
- PHA
phytohemagglutinin
- PMA
phorbol 12-myristate 13-acetate
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest: The authors declare no conflicts of interest.
References
- 1.Gilmore TD, Gerondakis S. The c-Rel Transcription Factor in Development and Disease. Genes Cancer. 2011;2:695–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kontgen F, Grumont RJ, Strasser A, Metcalf D, Li R, Tarlinton D, et al. Mice lacking the c-rel proto-oncogene exhibit defects in lymphocyte proliferation, humoral immunity, and interleukin-2 expression. Genes Dev. 1995;9:1965–77. [DOI] [PubMed] [Google Scholar]
- 3.Liou H-C, Jin Z, Tumang J, Andjelic S, Smith KA, Liou M-L. c-Rel is crucial for lymphocyte proliferation but dispensable for T cell effector function. Int Immunol. 1999;11:361–71. [DOI] [PubMed] [Google Scholar]
- 4.Milanovic M, Heise N, De Silva NS, Anderson MM, Silva K, Carette A, et al. Differential requirements for the canonical NF-κB transcription factors c-REL and RELA during the generation and activation of mature B-cells. Immunol Cell Biol. 2017;95:261–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zheng Y, Ouaaz F, Bruzzo P, Singh V, Gerondakis S, Beg AA. NF-kappa B RelA (p65) is essential for TNF-alpha-induced fas expression but dispensable for both TCR-induced expression and activation-induced cell death. J Immunol Baltim Md 1950. 2001;166:4949–57. [DOI] [PubMed] [Google Scholar]
- 6.Casanova J-L. Severe infectious diseases of childhood as monogenic inborn errors of immunity. Proc Natl Acad Sci. 2015;201521651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tangye SG. Advances in IL-21 biology—enhancing our understanding of human disease. Curr Opin Immunol. 2015;34:107–15. [DOI] [PubMed] [Google Scholar]
- 8.Kaltschmidt B, Greiner JFW, Kadhim HM, Kaltschmidt C. Subunit-Specific Role of NF-κB in Cancer. Biomedicines. 2018;6:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
References for online methods
- 1.Picard C, Fieschi C, Altare F, Al-Jumaah S, Al-Hajjar S, Feinberg J, et al. Inherited Interleukin-12 Deficiency: IL12B Genotype and Clinical Phenotype of 13 Patients from Six Kindreds. Am J Hum Genet. 2002;70:336–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li H, Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 2009;25:1754–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nature Genetics. 2011;43:491–8. [DOI] [PMC free article] [PubMed] [Google Scholar]