Abstract
Cholesterol is an important lipid molecule and is needed for all mammalian cells. In various cell types, excess cholesterol is stored as cholesteryl esters; acyl-CoA:cholesterol acyltransferase 1 (ACAT1) plays an essential role in this storage process. ACAT1 is located at the endoplasmic reticulum and has nine transmembrane domains (TMDs). It is a member of the membrane-bound O-acyltransferase (MBOAT) family, in which members contain multiple TMDs and participate in a variety of biological functions. When solubilized in the zwitterionic detergent CHAPS, ACAT1 can be purified to homogeneity with full enzyme activity and behaves as a homotetrameric protein. ACAT1 contains two dimerization motifs. The first motif is located near the N-terminus and is not conserved in MBOATs. Deletion of the N-terminal dimerization domain converts ACAT1 to a dimer with full catalytic activity; therefore, ACAT1 is a two-fold dimer. The second dimerization domain, located near the C-terminus, is conserved in MBOATs; however, it was not known whether the C-terminal dimerization domain is required for enzyme activity. Here we show that treating ACAT1 with non-ionic detergent, Triton X-100 or octyl glucoside, causes the enzyme to become a two-fold monomer without any enzymatic activity. Detergent exchange of Triton X-100 with CHAPS restores ACAT1 to a two-fold dimer but fails to restore its enzymatic activity. These results implicate that ACAT1 requires hydrophobic subunit interactions near the C-terminus in order to remain active as a two-fold dimer. Our results also caution the use of Triton X-100 or octyl glucoside to purify other MBOATs.
Keywords: ACAT, SOAT, MBOAT, Cholesteryl esters, Detergent, Solubilization
Graphical Abstract
1. Introduction
Cholesterol is an important lipid molecule that is required for the growth, maintenance, and viability of all mammalian cells. Excess cholesterol is stored as cholesteryl esters. The enzymes acyl-coenzyme A: cholesterol acyltransferases (ACATs), also called sterol O :acyltransferases (SOATs), catalyze the cholesterol storage process by utilizing long chain fatty coenzyme A as the fatty acid donor and convert cholesterol to cholesteryl esters. There are two isoenzymes, ACAT1 and ACAT2, that are encoded by two different genes [1–4]. ACAT1 is expressed ubiquitously in essentially all tissues examined, including the brain, while ACAT2 is only highly expressed in the intestinal enterocytes and in hepatocytes [5, 6], reviewed in [7]. Both enzymes contain multi-span membrane domains and both are located at the endoplasmic reticulum; neither enzyme is transcriptionally regulated by the master transcription factor SREBP2, which controls the expression of many genes in the cholesterol biosynthesis pathway. Instead, both ACAT1 and ACAT2 are allosterically regulated by their own sterol substrate [8].
ACAT1 and ACAT2 are members of the membrane-bound O-acyltransferase (MBOAT) enzyme family [9]. MBOATs contain multi-span membrane domains, with an invariant first active site histidine within a long stretch of hydrophobic residues, and an almost invariant second active site asparagine. Several MBOAT enzymes including ACAT1, ACAT2, diacylglycerol acyltransferase 1 (DGAT1), Porcupine (PORCN), and ghrelin O:acyltransferase (GOAT) are potential drug targets [10]. As a prerequisite for rational drug design, it is essential that these enzymes are solubilized in detergents and purified to homogeneity with retention of enzymatic activity, so biochemical and structural work can be conducted. However, a major hurdle for taking this approach is that MBOATs have been notoriously resistant to biochemical purification. At present, only a few MBOATs have been purified to homogeneity with retention of enzymatic activity [11–14].
ACAT1 contains nine transmembrane domains (TMDs) and behaves as a homotetramer in intact cells and in vitro [15]. More detailed analysis showed that ACAT1 contains two dimerization domains. The first dimerization domain is a hydrophilic α-helical-rich (coiled-coil) segment near the N-terminus [16]. This domain is not conserved within MBOAT. Deletion of the N-terminal dimerization domain converts ACAT1 to a homodimer, with full retention of enzymatic activity. A second dimerization domain exists near its C-terminus. Results of site-specific mutagenesis analyses suggested that the second dimerization domain involves the long hydrophobic stretch that encompasses the 7th and 8th TMDs [17], with the invariant histidine active site located near the end of the 7th TMD [18].
Currently, it is not known how disruption of the C-terminal dimerization domain would affect enzymatic activity. When the zwitterionic detergent 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate (CHAPS) was used for enzyme solubilization, the tagged recombinant ACAT1 expressed in Chinese hamster ovary (CHO) cells or in human embryonic kidney 293 (HEK293) cells could be purified to homogeneity with retention of enzymatic activity. Previously, it was shown that the non-ionic detergent Triton X-100 could solubilize the enzyme, but also caused inactivation of the ACAT enzyme activity [11, 19]. It was not clear whether Triton X-100 inactivated the ACAT1 enzyme by specifically inhibiting the enzyme activity or by dissociating it into a two-fold monomer with no enzymatic activity. Here we designed experiments to distinguish between these possibilities and report our results.
2. Materials and Methods
2.1. Antibodies and reagents
The rabbit polyclonal antibodies (DM102) against the N-terminal fragment (1–131) of human ACAT1 was described previously [20]. THE™ DYKDDDDK (FLAG) tag mouse monoclonal antibody was from GenScript (Piscataway, NJ, USA). Horseradish peroxidase (HRP)-conjugated goat anti-rabbit and HRP-conjugated goat anti-mouse was from Bio-Rad Laboratories (Hercules, CA, USA). Triton X-100, anti-FLAG M2 affinity gel, FLAG peptide, oleic acid, coenzyme A tri-lithium salt, sodium taurocholate, egg phosphatidycholine (PC), cholesterol, cholesteryl oleate, fatty acid-free bovine serum albumin, aldolase, β-amylase, catalase, thyroglobulin, and protease inhibitor cocktail were from MilliporeSigma (St. Louis, MO, USA). CHAPS and octyl glucoside were from Anatrace (Maumee, OH, USA). Peroxide-free Triton X-100 (proteomics grade) was from VWR (Solon, OH, USA). HisPur Ni-NTA resin was from Thermo Fisher Scientific (Rockford, IL, USA). Fugene 6 was from Promega (Madison, WI, USA). [3H]Oleoyl-CoA was synthesized as described previously [21].
2.2. Cell culture
All cell lines were maintained at 37°C under humidified conditions and 5% CO2. HEK293 cells were maintained in DMEM medium (Corning, Manassas, VA, USA) supplemented with 10% newborn calf serum (Atlanta Biologicals, Flowery Branch, GA, USA). CHO cells were maintained in 50% F-12 medium (Corning, Manassas, VA, USA) and 50% DMEM medium supplemented with 10% calf serum (Atlanta Biologicals, Flowery Branch, GA, USA).
2.3. Purification of human ACAT1 with hexa histidine-tag at the N-terminus and FLAG-tag at the C-terminus (HisACAT1/FLAG)
The construction, expression, and purification of HisACAT1/FLAG plasmid was described previously with slight modification [12]. HisACAT1/FLAG was stably expressed in HEK293 cells. Cells were grown as monolayers in 150 mm plates. Once confluent, cells were rinsed with 2x cold PBS before being solubilized in buffer A (1 M KCl, 40.7 mM (or 2.5%) CHAPS, 50 mM Tris-HCl pH 7.8, 10% glycerol). Lysate was homogenized in a stainless steel Dounce tissue grinder and subjected to a 100,000 × g spin for 1 hour at 4°C. The solubilized enzyme was loaded onto a 10 ml resin bed of HisPur Ni-NTA resin and flowed over the resin for 3 hours at 4°C. The column was washed with 2 column volumes (CVs) of 20 mM imidazole in buffer B (0.5 M KCl, 8.1 mM (or 0.5%) CHAPS, 50 mM Tris pH 7.8, 10% glycerol). The protein was eluted with 2 CVs of 100 mM EDTA in buffer B. The eluate was loaded onto a 1 ml resin bed of anti-FLAG M2 affinity gel and flowed over the resin for 0.5 hours at 4°C. The column was washed with 10 CVs of buffer B. HisACAT1/FLAG was eluted with 3 CVs of 300 μg/ml FLAG peptide in buffer B. The eluates were aliquoted as 100 μl portions into pre-chilled low adhesion microcentrifuge tubes and stored at −80°C.
2.4. Expression and purification of human ACAT1 lacking the first 65 amino acids, with a N-terminal hexa histidine-tag and C-terminal FLAG-tag (Δ1–65 HisACAT1/FLAG)
The plasmid construction of Δ1–65 HisACAT1 was described previously [16]. Δ1–65 HisACAT1/FLAG cDNA was ligated into the mammalian expression vector pAG3Zeo [22] and transfected into ACAT1-deficent CHO (AC29) cells using Fugene 6. The pAG3Zeo construct has an improved promoter, compared to pcDNA3, and provides zeocin resistance. This construct was created to produce a cell line with more stable and greater expression of Δ1–65 HisACAT1/FLAG. Individual stable clones were isolated by selecting cells resistant to zeocin at 300 μg/ml. Purification followed the before mentioned HisPur Ni–NTA protocol and anti-FLAG M2 affinity gel protocol. The eluates were aliquoted as 100 μl portions into pre-chilled low adhesion microcentrifuge tubes and stored at −80°C.
2.5. Liposomal ACAT activity assay
The procedure was adapted from [11]. This assay was routinely used to measure ACAT activity in vitro. Samples were diluted into 60 μl of preformed liposomes (consisting of 11.3 mM PC, 9.3 mM taurocholate, 1.8 mM cholesterol) and 10 nmoles of [3H]oleoyl-CoA. To assess effects on HisACAT1/FLAG enzymatic activity, varying concentrations of KCl, CHAPS, Triton X-100, and octyl glucoside were added to the reaction as indicated. The reactions were assayed in duplicates or triplicates for 10–30 mins at 37°C.
2.6. Dynamic light scattering
The particle size distribution of PC, taurocholate, cholesterol, and detergent (Triton X-100, octyl glucoside, or CHAPS) solutions were determined with a Malvern Panalytical Zetasizer Nano-ZS using 10 mm path length disposable cuvettes. The samples were prepared fresh prior to experimentation and centrifuged at 12,000 × g at room temp for 5 minutes prior to measurement. Each sample was measured in triplicate at 25°C.
2.7. Sucrose density gradient centrifugation
The procedure was adapted from [15]. A 5–30% linear sucrose gradient was formed in a 11 × 60 mm ultra-Clear tube (Beckman #344062) with 6 layers varied by 5% sucrose increments. Each layer was 0.6 ml with a total volume of 3.6 ml. The sucrose gradients were created in buffers containing 1 M KCl, 50 mM Tris-HCl pH 7.8, and 8.1 mM CHAPS, or 1.7 mM Triton X-100, or 20 mM octyl glucoside. A 200 μl sample of purified ACAT1 protein was layered on top of the gradient. The centrifugations were carried out using a Beckman SW 60 Ti rotor in a Beckman model L8–70M ultracentrifuge. Centrifugations were performed at 50,000 rpm for 10 hrs at 4°C. At the end of the centrifugation, fractions were collected at 100 μl per fraction.
The migration profiles of the standard protein size markers (BSA, aldolase, β-amylase, catalase, and thyroglobulin) were determined by running fractions on 10% SDS-PAGE gel and stained by GelCode Blue (Thermo Fisher, 24590). The migration profiles of HisACAT1/FLAG and Δ1–65 HisACAT1/FLAG were determined by running sucrose fractions on 10% SDS-PAGE gel and transferred to polyvinylidene difluoride (PVDF) membranes (GE Healthcare Life Sciences, 10600023). The membranes were incubated with primary anti-ACAT1 antibodies DM102, then with HRP-conjugated anti-rabbit secondary antibodies, and detected by using SuperSignal West Pico PLUS Chemiluminescent Substrate kit (Thermo Fisher, 34578).
2.8. Temperature dependent inactivation of HisACAT1/FLAG
Purified HisACAT1/FLAG was incubated with HisPur Ni-NTA resin for an hour at 4°C with gentle mixing on an orbital shaker. The resin was washed with 2 CVs of buffer A (1 M KCl, 8.1 mM CHAPS, 50 mM Tris-HCl pH 7.8), or with buffer B (1 M KCl, 1.7 mM peroxide-free Triton X–100, 50 mM Tris-HCl pH 7.8), or with preformed liposomes (9.3 mM taurocholate, 11.3 mM PC, 1.8 mM cholesterol). Then the resin was resuspended into a 50/50 slurry, in the respective buffers, and an aliquot was withdrawn to determine the input activity of HisACAT1/FLAG. The remaining slurry was subjected to 4°C or 37°C for an indicated amount of time. Enzymatic activity was determined by using the liposomal ACAT activity assay, using 10 μl of resin suspension that contained HisACAT1/FLAG retained on the resin as the enzyme source.
2.9. Detergent exchange of sucrose density gradient fractions with Microsep filters
The Triton X–100 containing sucrose gradient fractions #10–20 were first concentrated in 10K Microsep Advance Centrifugal Devices with Omega Membrane (Pall Corporation, MCP010C41) by spinning at 3,200 × g at 4°C. Once concentrated to 250 μl, a 1 ml solution of 1 M KCl, 8.1 mM CHAPS, 50 mM Tris-HCl pH 7.8 was added to the filter unit to exchange detergents. This exchange was repeated three times until total volume was 250 μl. Afterwards, 200 μl of the concentrated sample was loaded onto a 5–30% linear sucrose gradient that contained 1 M KCl, 8.1 mM CHAPS, 50 mM Tris-HCl pH 7.8 and subjected to ultracentrifugation analysis.
2.10. Detergent exchange with HisACAT1/FLAG bound to Ni-NTA resin
Purified HisACAT1/FLAG was incubated with HisPur Ni-NTA resin for an hour at 4°C with gentle mixing on an orbital shaker. The resin was washed with 2 CVs of buffer A (1 M KCl, 8.1 mM CHAPS, 50 mM Tris-HCl pH 7.8) and resuspended into a 50/50 slurry in buffer A. An aliquot was collected to determine the input activity of HisACAT1/FLAG. The remaining resin was washed with 2 CVs of buffer B (1 M KCl, 1.7 mM peroxide-free Triton X–100, 50 mM Tris-HCl pH 7.8) and resuspended into a 50/50 slurry in buffer B. The buffer B slurry was placed at 37°C for 2 minutes and an aliquot was collected to determine the enzymatic activity after Triton X–100 treatment. Next, Triton X–100 was exchanged by washing the resin with 10 CVs of buffer A or of preformed liposomes (9.3 mM taurocholate, 11.3 mM PC, 1.8 mM cholesterol). The remaining resin was resuspended into a 50/50 slurry in buffer A or in preformed liposomes. Enzymatic activity was determined by using the liposomal ACAT activity assay, using 10 μl of resin suspension that contained HisACAT1/FLAG retained on the resin as the enzyme source.
2.11. Statistical analyses
Results are reported as means ± standard deviation (SD). GraphPad Prism 5 software was used for plotting data and statistical analysis. Statistical comparisons for data were performed using unpaired t-test and p-value < 0.05 was considered statistically significant.
3. Results
3.1. CHAPS concentrations greater than its critical micelle concentration inhibits HisACAT1/FLAG enzyme activity in the liposomal ACAT activity assay
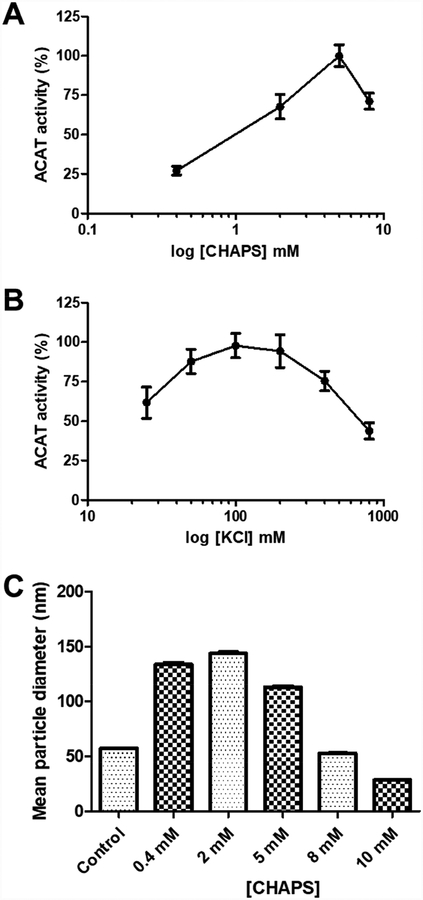
The zwitterionic detergent CHAPS was used to solubilize and purify recombinant ACAT1 enzyme [11]; however, only 7% of the total ACAT activity was recovered [12]. We suspect that during the enzyme purification process, CHAPS may cause partial inhibition of ACAT1 enzymatic activity. To test this possibility, we added varying concentrations of CHAPS to the liposomal ACAT activity assay. Recombinant HisACAT1/FLAG, purified as described in the Materials and Methods section, was used as the enzyme source. At a constant 100 mM of KCl, HisACAT1/FLAG activity was greatest at 5 mM of CHAPS, with a decrease in activity at either lower or higher concentrations (Fig. 1A). A similar effect was seen by varying the concentration of KCl at a constant concentration of CHAPS (5 mM) (Fig. 1B). For the rest of the work described here, we modified the liposomal ACAT activity assay by maintaining 100 mM KCl and 5 mM CHAPS as the final concentrations.
Fig. 1. The effect of CHAPS and KCl concentration on the enzymatic activity of HisACAT1/FLAG.
(A) The effect of varying CHAPS concentration on the enzymatic activity of HisACAT1/FLAG in the liposomal ACAT activity assay (n = 3). KCl concentration was 100 mM. (B) The effect of varying KCl concentration on the enzymatic activity of HisACAT1/FLAG in the liposomal ACAT activity assay (n = 3). CHAPS concentration was 5 mM. ACAT activity (%) was determined by normalization of each experimental trial to background [3H]cholesteryl oleate (0%) and maximal activity (100%) of that trial. (C) The mean diameter (Z-average) of particles found in solution as determined by dynamic light scattering. Control is preformed liposomes (11.2 mM PC, 9.3 mM taurocholate, 1.8 mM cholesterol). The remaining samples have a final concentration of 100 mM KCl, 50 mM Tris-HCl pH 7.8, 6.3 mM PC, 5.2 mM taurocholate, and 1 mM cholesterol with various concentrations of CHAPS.
It was reported that the critical micelle concentration (CMC) of CHAPS is 3–10 mM and the addition of salt, such as KCl, reduces its CMC [23, 24]. Thus, the result presented in Fig. 1A and 1B suggest that CHAPS micelles, present in the final reaction mixture, inhibit ACAT1 activity. To examine the size distribution of lipid particles in the assay mixture, dynamic light scattering (DLS) measurements were performed. The result showed that the 11.2 mM PC, 9.2 mM taurocholate, and 1.8 mM cholesterol solution, previously referred to as “mixed micelles” [11], actually forms a mixture of micelles and liposomes (Fig. 1C, Supplemental table 1, Supplemental fig. 1A); henceforth, this solution is referred to as “preformed liposomes”. When the preformed liposomes are diluted in 100 mM KCl, 50 mM Tris-HCl pH 7.8, and increasing concentrations of CHAPS, the mean particle diameter of the mixture becomes larger until CHAPS reaches 5 mM (Fig. 1C), which is within the CMC of CHAPS (3–10 mM). At 5 mM CHAPS, the DLS result showed that the solution is comprised of micelles, liposomes, and aggregates (Supplemental table 1, Supplemental fig. 1D, Supplemental fig. 2D). With 8 or 10 mM CHAPS present, the solutions have an increased population of micelles (Supplemental table 1, Supplemental fig. 1E, F), and a reduced mean particle diameter (Fig. 1C). When CHAPS was added to the preformed liposomes, the diameter of the particles populating the second intensity peak (i.e. liposomes) stay relatively constant (Supplemental table 1, Supplemental fig. 1B–F). We speculate that the micelles formed upon the addition of CHAPS past its CMC (i.e. 5 mM) may be composed of the detergent alone and inhibit ACAT1 enzymatic activity.
3.2. Triton X–100 or octyl glucoside inhibit HisACAT1/FLAG activity around their CMCs
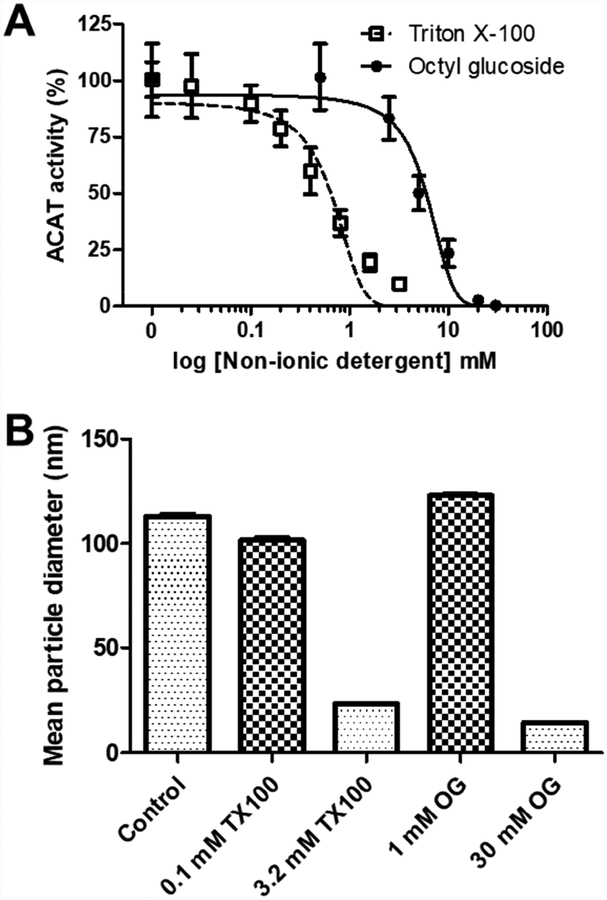
Detergents are needed to solubilize membranes. However, when Triton X–100 was used to purify ACAT1, it severely inhibited ACAT activity [11, 19]. Here we examined the abilities of Triton X–100 and octyl glucoside, another non-ionic detergent commonly used for solubilization, to inactivate purified HisACAT1/FLAG at varying detergent concentrations in the liposomal ACAT activity assay. To perform these experiments, we added 10 μl of stock non-ionic detergent to reach the desired final concentration in the liposomal ACAT activity assay or 10 μl of water as the control. As shown in Fig. 2A, both detergents severely inhibited HisACAT1/FLAG enzyme activity. Statistical analysis, by two-way ANOVA, showed no significant difference between untreated and peroxide-free Triton X–100 (data not shown); therefore, experimental trials were grouped. Triton X–100 has an IC50 of 0.65 mM (Fig. 2A), which is above its CMC of 0.25 mM [24]. Octyl glucoside has an IC50 of 6.0 mM (Fig. 2A), which is below its CMC of 19–25 mM [24, 25]. These results imply that both non-ionic detergents negatively affect ACAT1 activity at concentrations below or above their CMCs.
Fig. 2. Non-ionic detergents inhibit HisACAT1/FLAG activity at and above their CMCs.
The enzymatic inhibition of purified HisACAT1/FLAG (liposomal ACAT activity assay) by Triton X–100 (n = 3) and octyl glucoside (n = 3). Liposomal ACAT activity assay was performed in 100 mM KCl and 5 mM CHAPS. ACAT activity (%) was determined by normalization of each experimental trial to background [3H]cholesteryl oleate (0%) and activity at 100 mM KCl, 5 mM CHAPS (100%) of that trial. (B) The mean diameter (Z-average) of particles, as determined by dynamic light scattering, found in 100 mM KCl, 5 mM CHAPS, 50 mM Tris-HCl pH 7.8, 6.3 mM PC, 5.2 mM taurocholate, and 1 mM cholesterol with added concentrations of Triton X–100 (TX100) or octyl glucoside (OG).
To examine how non-ionic detergents affect lipid particle size within the liposomal ACAT activity assay, we used DLS to measure the particle size distribution upon the addition of Triton X–100 or octyl glucoside to the assay mixture. The results showed that at 100 mM KCl, 5 mM CHAPS, and 50 mM Tris-HCl pH 7.8 buffer (6.3 mM PC, 5.2 mM taurocholate, 1 mM cholesterol), the solution produces a 112.90 nm mean particle diameter (Fig. 2B, Supplemental table 1). Upon the addition of 0.1 mM Triton X–100, there is a slight change in diameter, 101.50 nm; however, at 3.2 mM Triton X–100, there is a reduction in the mean particle diameter, 23.18 nm, (Fig. 2B, Supplemental table 1) which correlates to an increase in the micelle population (Supplemental Fig. 2F). As seen in Supplemental Fig. 2D–H, the height of the micelle intensity peak increased as non-ionic detergents were added to the solution; whereas, the liposome peak was reduced but not to the same extent as micelle formation. We speculate that the micelles formed upon the addition of Triton X–100 or octyl glucoside are composed of the non-ionic detergents alone. We also speculate that under these conditions the micelles formed by Triton X–100 or octyl glucoside cause rapid inactivation of ACAT1 enzymatic activity, as shown in Fig. 2A.
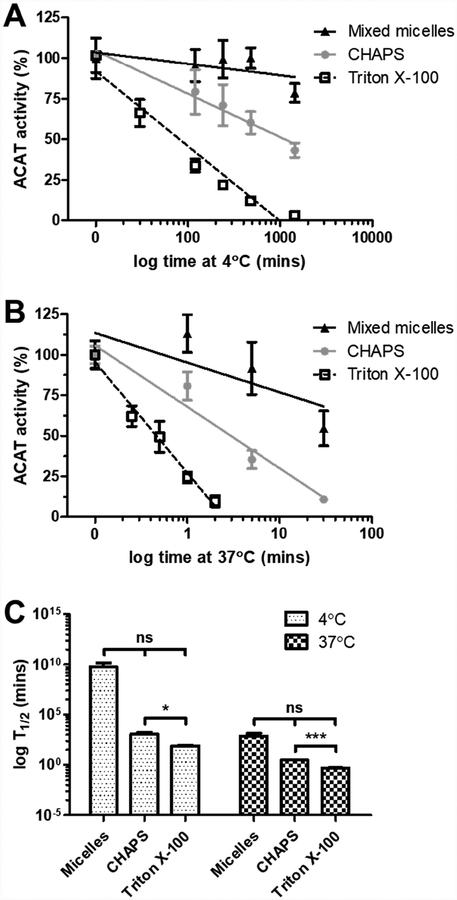
3.3. Triton X–100 inactivates HisACAT1/FLAG enzymatic activity significantly faster than CHAPS at 4°C and 37°C
The significant loss of enzymatic activity during the purification process may be aided by temperature. To test this possibility, we examined the effect of temperature on the inhibition of HisACAT1/FLAG enzymatic activity by 8.1 mM CHAPS and 1.7 mM Triton X–100. The results showed that when compared with preformed liposomes (9.3 mM taurocholate, 11.3 mM PC, 1.8 mM cholesterol), both CHAPS and Triton X–100 inactivate ACAT1 at 4°C (Fig. 3A) or at 37°C (Fig. 3B). The inactivation by Triton X–100 occurs much more rapidly when compared to CHAPS at both 4°C and 37°C (Fig. 3C). These results imply that the purification of ACAT1 in CHAPS should proceed quickly at 4°C as to minimize the gradual inactivation of ACAT1 by CHAPS.
Fig. 3. Triton X–100 inhibits HisACAT1/FLAG activity faster than CHAPS at 4°C and 37°C.
The enzymatic inhibition of purified HisACAT1/FLAG (liposomal ACAT activity assay) by 1 M KCl, 50 mM Tris-HCl pH 7.8, and 8.1 mM CHAPS, or 1.7 mM peroxide-free Triton X–100 at (A) 4°C (n = 3) or (B) 37°C (n = 3). 9.3 mM taurocholate, 11.3 mM phosphatidylcholine, 1.8 mM cholesterol preformed liposomes were used as a control (n = 2). ACAT activity (%) was determined by normalization of each experimental trial to background [3H]cholesteryl oleate (0%) and activity at 0 mins 100 mM KCl, 5 mM CHAPS (100%) of that trial. (C) The amount of time (T1/2) to reach 50% inhibition of HisACAT1/FLAG enzymatic activity. ns = not significant; * p = ≤ 0.05; *** p = ≤ 0.001
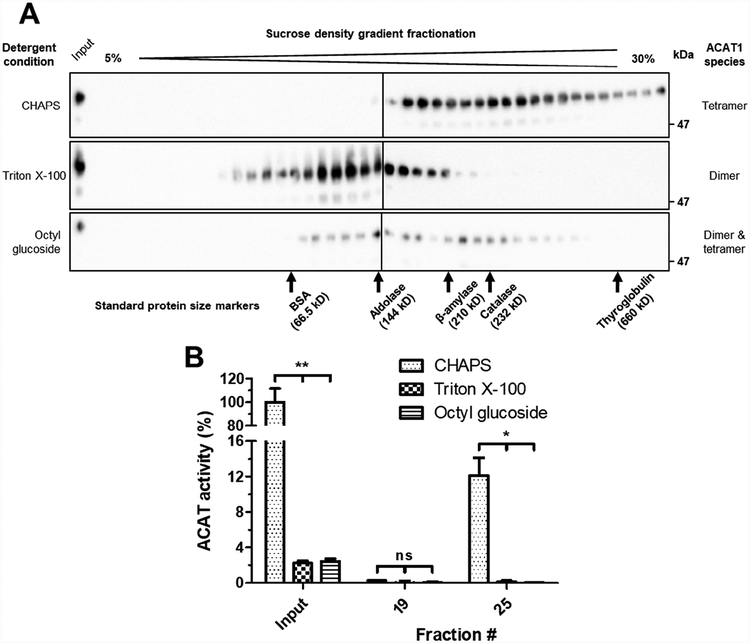
3.4. Triton X–100 or octyl glucoside dissociates HisACAT1/FLAG to form a dimeric species
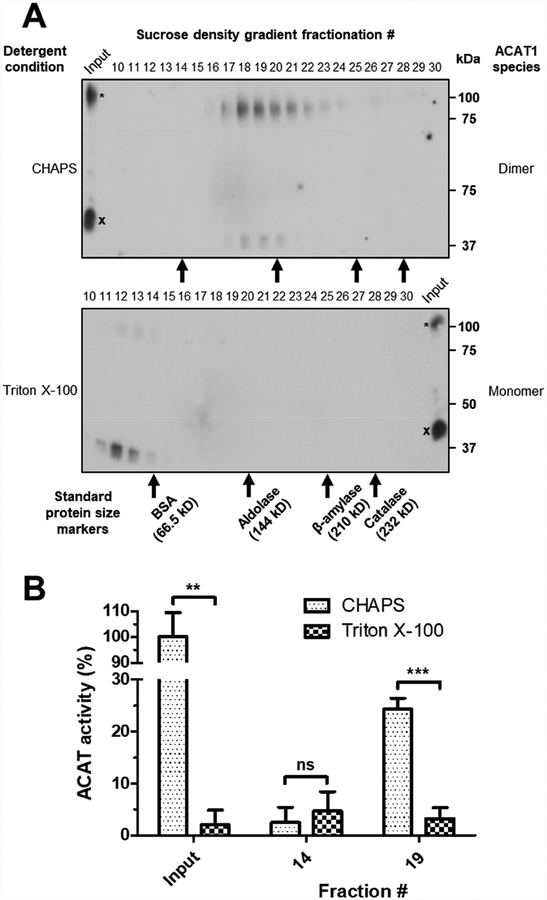
A monomer of HisACAT1/FLAG is 70 kDa, but the apparent molecular mass by SDS-PAGE is 56 kDa. Here we monitored the molecular mass of HisACAT1/FLAG in different detergents by sucrose density gradient centrifugation. Previously, it was shown that when CHAPS was used to solubilize recombinant ACAT1 one can estimate its size by using a set of standard protein size markers [15, 16]. These standard protein size markers include: BSA (66.5 kDa), aldolase (144 kDa), β-amylase (210 kDa), catalase (232 kDa), and thyroglobulin (660 kDa). As shown in Fig. 4A, and consistent with previous work [15], under the condition of the 1 M KCl, 8.1 mM CHAPS, the sedimentation profile of HisACAT1/FLAG shows that it mainly forms a homotetramer. Under both 1 M KCl, 1.7 mM Triton X–100 and 1 M KCl, 20 mM octyl glucoside conditions, most of the HisACAT1/FLAG sedimented to earlier fractions (Fig. 4A), with an apparent molecular weight consistent of a homodimer. These earlier fractions also exhibit a complete loss of ACAT activity (Fig. 4B). Overall, these results suggest that Triton X–100 or octyl glucoside inactivate ACAT1 by promoting its dissociation from a tetrameric to a dimeric conformation.
Fig. 4. Triton X–100 and octyl glucoside causes HisACAT1/FLAG to decompose into fractions of lower density.
(A) Representative Western blot analysis (using anti-ACAT1 antibodies, DM102) of 5–30% sucrose gradients of purified HisACAT1/FLAG in 1 M KCl, 50 mM Tris-HCl pH 7.8, and 8.1 mM CHAPS, or 1.7 mM Triton X–100, or 20 mM octyl glucoside. (B) HisACAT1/FLAG enzymatic activity (liposomal ACAT activity assay) of peak fractions as determined from Western blots. ACAT activity (%) was determined by normalization to background [3H]cholesteryl oleate (0%) and activity of input HisACAT1/FLAG in 1 M KCl, 8.1 mM CHAPS, 50 mM Tris-HCl pH 7.8 (100%). ns = not significant; * p = ≤ 0.05; ** p = ≤ 0.01
3.5. CHAPS can replace Triton X–100 to restore HisACAT1/FLAG to a homotetramer, but cannot restore enzyme activity
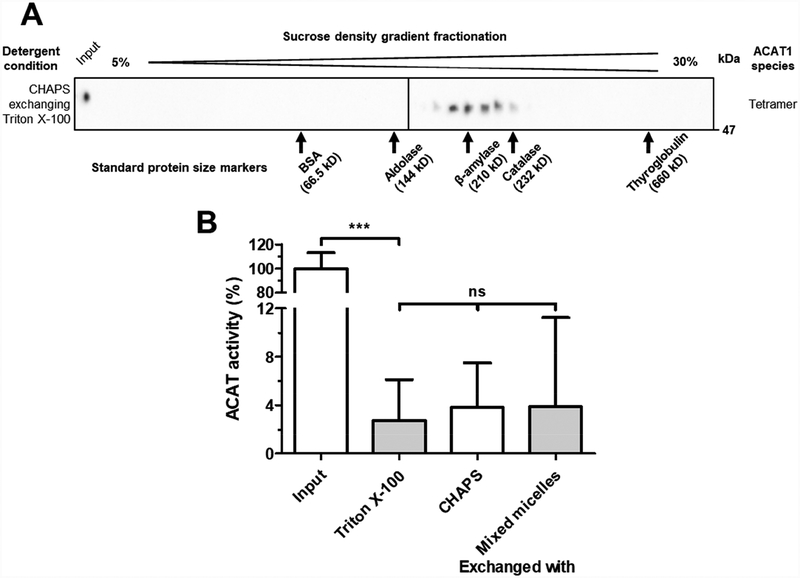
After treating HisACAT1/FLAG with Triton X–100, detergent exchange experiments were performed to determine if HisACAT1/FLAG tetrameric state and/or enzymatic activity could be restored by replacing Triton X–100 with CHAPS or with preformed liposomes. The results showed that the exchange of Triton X–100 with CHAPS did restore HisACAT1/FLAG to a homotetrameric complex (Fig. 5A). However, this procedure, as well as an additional attempt by using preformed liposomes to replace Triton X–100, did not restore HisACAT1/FLAG enzymatic activity (Fig. 5B). It is possible that Triton X–100 causes an irreversible conformational change that inactivates ACAT1, or Triton X–100 replaces a co-factor that is required for enzymatic activity.
Fig. 5. Triton X–100 can be exchanged with CHAPS to recover HisACAT1/FLAG homotetramer, but not enzymatic activity.
(A) Representative Western blot analysis (using anti-ACAT1 antibodies, DM102) of concentrated HisACAT1/FLAG from fractions #10–20 of 1 M KCl, 1.7 mM Triton X–100 sucrose gradients exchanged with 1 M KCl, 8.1 mM CHAPS in Microsep filters. (B) The enzymatic activity of purified HisACAT1/FLAG (liposomal ACAT activity assay) after exposure to 1 M KCl, 1.7 mM peroxide-free Triton X–100 for 2 mins at 37°C and the detergent exchange of Triton X–100 with 1 M KCl, 8.1 mM CHAPS or preformed liposomes (Input and Triton X–100 n = 5; CHAPS n = 3; preformed liposomes n = 2). ACAT activity (%) was determined by normalization of each experimental trial to background [3H]cholesteryl oleate (0%) and activity of input HisACAT1/FLAG in 1 M KCl, 8.1 mM CHAPS, 50 mM Tris-HCl pH 7.8 (100%) of that trial. ns = not significant; *** p = ≤ 0.001
3.6. Triton X–100 converts an N-terminal truncated ACAT1 dimer to a monomer without enzyme activity
The first dimerization domain of ACAT1 is a hydrophilic α-helical-rich (coiled-coil) segment near the N-terminus [16]. Deletion of the N-terminal dimerization domain, designated as Δ1–65 HisACAT1/FLAG, converts ACAT1 to a homodimer with full retention of enzymatic activity [16]. A second dimerization domain exists near the C-terminus and involves a long hydrophobic stretch that encompasses the 7th and 8th TMDs [17]. It is possible that Triton X–100 inactivates ACAT1 through perturbation of subunit interaction(s) within this long, hydrophobic domain. To test this possibility, we used Δ1–65 HisACAT1/FLAG purified from AC29 cells, a CHO cell mutant which is devoid of native ACAT1 [20], and sucrose density gradient centrifugation to monitor C-terminal dimerization.
A monomer of Δ1–65 HisACAT1/FLAG is 62.5 kDa, but the apparent molecular mass by SDS-PAGE is 46 kDa. Consistent with our previous report [16], in 8.1 mM CHAPS and 1 M KCl, Δ1–65 HisACAT1/FLAG sediments as a species with a molecular weight resembling a homodimer (Fig. 6A). When subjected to 1.7 mM Triton X–100 and 1 M KCl, Δ1–65 HisACAT1/FLAG sediments to earlier fractions with a molecular weight resembling a monomer (Fig. 6A). Additional results show that treatment of Δ1–65 HisACAT1/FLAG with Triton X–100 completely abolishes enzymatic activity (Fig. 6B). These results show that Triton X–100 inactivates ACAT1 by affecting its dimerization domain near the C-terminus.
Fig. 6. Triton X–100 causes Δ1–65 HisACAT1/FLAG to decompose into fractions of lower density.
(A) Representative Western blot analysis (using anti-FLAG antibody) of 5–30% sucrose gradients of purified Δ1–65 HisACAT1/FLAG preparations in 1 M KCl, 50 mM Tris-HCl pH 7.8, and 8.1 mM CHAPS, or 1.7 mM Triton X–100. Indicated throughout are dimeric Δ1–65 HisACAT1/FLAG (*) and monomeric Δ1–65 HisACAT1/FLAG (X). (B) Δ1–65 HisACAT1/FLAG activity (liposomal ACAT activity assay) of peak fractions as determined from Western blots. ACAT activity (%) was determined by normalization of each experimental trial to background [3H]cholesteryl oleate (0%) and activity of input Δ1–65 HisACAT1/FLAG in 1 M KCl, 8.1 mM CHAPS, 50 mM Tris-HCl pH 7.8 (100%) of that trial. ns = not significant; ** p = ≤ 0.01; *** p = ≤ 0.001
4. Discussion
Our current study explored how different detergents affect ACAT1 enzymatic activity and its oligomeric state. As assessed by the liposomal ACAT activity assay, HisACAT1/FLAG was most active at 100 mM KCl and 5 mM CHAPS (Fig. 1A, B). Higher concentrations of KCl or CHAPS reduced enzymatic activity (Fig. 1A, B) which was attributable to the formation of CHAPS micelles (Fig. 1C). When HisACAT1/FLAG was exposed to non-ionic detergents, such as Triton X–100 or octyl glucoside, above their CMC the enzyme was inactivated (Fig. 2A). This inactivation also appears to be attributable to the formation of non-ionic detergent micelles in the liposomal ACAT activity assay (Fig. 2B). Unlike the zwitterionic detergent CHAPS, the non-ionic detergent Triton X–100 caused rapid inactivation of HisACAT1/FLAG enzymatic activity at 4°C (Fig. 3A) and at 37°C (Fig. 3B). The Triton X–100-mediated inactivation is not due to impurities because inactivation occurs irrespective of Triton X–100 purity.
We performed protein size determination by using sucrose density gradient centrifugation and found that treating HisACAT1/FLAG with either Triton X–100 or octyl glucoside converted the mainly tetrameric species to a mainly dimeric species (Fig. 4A). The dimeric species formed by Triton X–100 could be exchanged with CHAPS to recover the HisACAT1/FLAG tetramer (Fig. 5A). Several attempts were made to regain enzymatic activity by methods that involved the exchange of detergent bound to the enzyme; however, the attempts were unsuccessful (Fig. 5B).
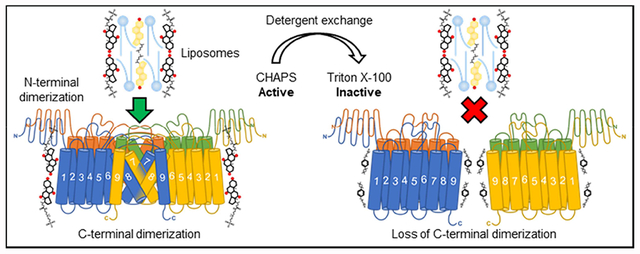
We had previously reported that ACAT1 lacking the N-terminus dimerization domain, designated Δ1–65 HisACAT1/FLAG, is dimeric and retains full enzymatic activity [16]. To validate the finding described in Fig. 4, Δ1–65 HisACAT1/FLAG was purified by a two-step process using Ni-NTA immobilized metal affinity chromatography (IMAC) followed by anti-FLAG M2 affinity gel and the purified protein was subjected to Triton X–100 treatment. The results showed that Triton X–100 converted Δ1–65 HisACAT1/FLAG to a size resembling a monomeric species (Fig. 6A) and completely inactivated its enzymatic activity (Fig. 6B). These results support the interpretation that ACAT1 contains a second dimerization domain near its C-terminus and that adding Triton X–100 or octyl glucoside interrupts this dimerization domain. These observations could be due to the differences in the molecular structures of the detergents. Whereas Triton X–100 penetrates the lipid membrane bilayer, CHAPS does not, rather, it binds to the membrane surface in a flat position [26]. The ability of Triton X–100 to penetrate the lipid bilayer could explain why it can infiltrate the C-terminal dimerization domain of ACAT1.
The question remains as to what causes the dimeric Δ1–65 HisACAT1/FLAG to appear in the Western blots of Fig. 6A. The dimeric species is detectable in variable amount in different preparations. Perhaps the dimeric Δ1–65 HisACAT1/FLAG is caused by the high concentration of SDS present in the loading dye added to the samples before being loaded into the SDS-PAGE gel. Irrespective of the mechanism for dimer formation of Δ1–65 HisACAT1/FLAG in SDS-PAGE, Triton X–100 causes a shift in ACAT1 size as analyzed by sucrose density gradient centrifugation.
Currently, the exact location of the second dimerization domain is unknown. We speculate that the long hydrophobic region flanking the invariant histidine (460 for human ACAT1) may play a critical role in forming the dimerization motif. In ACAT1, these residues constitute the hydrophobic sides of the 7th and 8th TMDs [17]. Further investigation is required to validate this possibility.
5. Conclusions
Based on these results, we conclude that Triton X–100 and octyl glucoside inactivate ACAT1 by dissociating it from a two-fold dimer to a two-fold monomer; in addition, the two-fold dimer complex is required for catalysis. In ACAT1, the disruption of the hydrophobic subunit interactions within the long hydrophobic stretch flanking the active site histidine are most likely affected by Triton X–100. This domain is conserved in many other MBOAT members. Our current study suggests that, for the purification of other MBOATs, the use of Triton X–100 or octyl glucoside should be avoided. Instead, the use of CHAPS, the inclusion of KCl at high concentrations, and a rapid purification procedure may be considered for optimal solubilization and purification of other MBOAT enzymes.
Supplementary Material
Supplemental table 1 The average values of three dynamic light scattering measurements. The Z-average is the intensity-weighted mean diameter of particles found in solution. The polydispersity index (PdI) is a measure of the broadness of the size distribution ranging from 0 to 1, with anything greater than 0.7 being considered too polydispersed. The peak intensity measurement is the average diameter of individual peaks (1- micelles, 2- liposomes, 3- aggregates), as determined by the intensity particle size distribution (PSD). The number measurement is the average diameter of particles as determined from the number PSD. We use intensity PSD to report the size of each peak in the distribution and number PSD to report the size of the most abundant particle found in the distribution.
Supplemental fig. 1 The intensity particle size distributions for (A) preformed liposomes (11.2 mM PC, 9.3 mM taurocholate, 1.8 mM cholesterol) and diluted liposomes (100 mM KCl, 50 mM Tris-HCl pH 7.8, 6.3 mM PC, 5.2 mM taurocholate, 1 mM cholesterol) at various concentrations of CHAPS (B) 0.4 mM, (C) 2 mM, (D) 5 mM, (E) 8 mM, and (F) 10 mM.
Supplemental fig. 2 The intensity particle size distributions for (A) buffer (100 mM KCl, 5 mM CHAPS, 50 mM Tris-HCl pH 7.8), (B) buffer + 3.2 mM Triton X–100, (C) buffer + 30 mM octyl glucoside, (D) diluted liposomes (100 mM KCl, 5 mM CHAPS, 50 mM Tris-HCl pH 7.8, 6.3 mM PC, 5.2 mM taurocholate, 1 mM cholesterol), (E) diluted liposomes + 0.1 mM Triton X–100, (F) diluted liposomes + 3.2 mM Triton X–100, (G) diluted liposomes + 1 mM octyl glucoside, and (H) diluted liposomes + 30 mM octyl glucoside.
Highlights.
ACAT1 is a two-fold dimer and has two dimerization domains.
The N-terminal dimerization domain is not required for enzyme activity.
Triton X-100 disrupts C-terminal dimerization and inactivates the enzyme.
The C-terminal dimerization domain is required for enzyme activity.
Acknowledgements
We thank members of the Chang lab for helpful discussion during the course of this work and Sarah Valles for careful editing of the manuscript. We also thank Dr. Jack Hoopes and Dr. Shan Zhao of Geisel School of Medicine at Dartmouth for providing access and training on the Malvern Panalytical Zetasizer Nano-ZS instrument to perform DLS experiments.
Funding
This work was supported by NIH grants RO1 AG037609 and AG063544 to T.Y. Chang and Catherine Chang.
Abbreviations
- ACAT
acyl-coenzyme A:cholesterol acyltransferase
- CHAPS
3-[(3-Cholamidopropyl)dimethylammonio]-1-propanesulfonate
- CMC
critical micelle concentration
- MBOAT
membrane-bound O-acyltransferase
- PC
phosphatidylcholine
- SOAT
sterol O:acyltransferase
- TMDs
transmembrane domains
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
6. References
- [1].Chang CC, Huh HY, Cadigan KM, Chang TY, Molecular cloning and functional expression of human acyl-coenzyme A:cholesterol acyltransferase cDNA in mutant Chinese hamster ovary cells, The Journal of biological chemistry 268(28) (1993) 20747–55. [PubMed] [Google Scholar]
- [2].Anderson RA, Joyce C, Davis M, Reagan JW, Clark M, Shelness GS, Rudel LL, Identification of a form of acyl-CoA:cholesterol acyltransferase specific to liver and intestine in nonhuman primates, The Journal of biological chemistry 273(41) (1998) 26747–54. [DOI] [PubMed] [Google Scholar]
- [3].Cases S, Novak S, Zheng YW, Myers HM, Lear SR, Sande E, Welch CB, Lusis AJ, Spencer TA, Krause BR, Erickson SK, Farese RV Jr., ACAT-2, a second mammalian acyl-CoA:cholesterol acyltransferase. Its cloning, expression, and characterization, The Journal of biological chemistry 273(41) (1998) 26755–64. [DOI] [PubMed] [Google Scholar]
- [4].Oelkers P, Behari A, Cromley D, Billheimer JT, Sturley SL, Characterization of two human genes encoding acyl coenzyme A:cholesterol acyltransferase-related enzymes, The Journal of biological chemistry 273(41) (1998) 26765–71. [DOI] [PubMed] [Google Scholar]
- [5].Chang CC, Lin S, Sakashita N, Chang TY, Distinct intracellular locations of ACAT-1 and ACAT-2 in differentiated Caco-2 cells (Abstract), AHA Scientific Sessions, New Orleans, 2000. [Google Scholar]
- [6].Lee RG, Willingham MC, Davis MA, Skinner KA, Rudel LL, Differential expression of ACAT1 and ACAT2 among cells within liver, intestine, kidney, and adrenal of nonhuman primates, J. Lipid Res. 41 (2000) 1991–2001. [PubMed] [Google Scholar]
- [7].Chang TY, Li BL, Chang CC, Urano Y, Acyl-coenzyme A:cholesterol acyltransferases, American journal of physiology. Endocrinology and metabolism 297(1) (2009) E1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Rogers MA, Liu J, Kushnir MM, Bryleva E, Rockwood AL, Meikle AW, Shapiro D, Vaisman BL, Remaley AT, Chang CC, Chang TY, Cellular pregnenolone esterification by acyl-CoA:cholesterol acyltransferase, The Journal of biological chemistry 287(21) (2012) 17483–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Hofmann K, A superfamily of membrane-bound O-acyltransferases with implications for wnt signaling, Trends Biochem Sci 25(3) (2000) 111–2. [DOI] [PubMed] [Google Scholar]
- [10].Chang CC, Sun J, Chang TY, Membrane bound O-acyltransferases (MBOAT), Frontiers in Biology 6 (2011) 177–182. [Google Scholar]
- [11].Chang CC, Lee CY, Chang ET, Cruz JC, Levesque MC, Chang TY, Recombinant acyl-CoA:cholesterol acyltransferase-1 (ACAT-1) purified to essential homogeneity utilizes cholesterol in mixed micelles or in vesicles in a highly cooperative manner, The Journal of biological chemistry 273(52) (1998) 35132–41. [DOI] [PubMed] [Google Scholar]
- [12].Chang CC, Miyazaki A, Dong R, Kheirollah A, Yu C, Geng Y, Higgs HN, Chang TY, Purification of Recombinant Acyl-Coenzyme A:Cholesterol Acyltransferase 1 (ACAT1) from H293 Cells and Binding Studies between the Enzyme and Substrates Using Difference Intrinsic Fluorescence Spectroscopy, Biochemistry 49 (2010) 9957–9963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Ma D, Wang Z, Merrikh CN, Lang KS, Lu P, Li X, Merrikh H, Rao Z, Xu W, Crystal structure of a membrane-bound O-acyltransferase, Nature 562(7726) (2018) 286–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Lee CJ, Rana MS, Bae C, Li Y, Banerjee A, In vitro reconstitution of Wnt acylation reveals structural determinants of substrate recognition by the acyltransferase human Porcupine, The Journal of biological chemistry 294(1) (2019) 231–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Yu C, Chen J, Lin S, Liu J, Chang CC, Chang TY, Human acyl-CoA:cholesterol acyltransferase-1 is a homotetrameric enzyme in intact cells and in vitro, The Journal of biological chemistry 274(51) (1999) 36139–45. [DOI] [PubMed] [Google Scholar]
- [16].Yu C, Zhang Y, Lu X, Chen J, Chang CC, Chang TY, Role of the N-terminal hydrophilic domain of acyl-coenzyme A:cholesterol acyltransferase 1 on the enzyme’s quaternary structure and catalytic efficiency, Biochemistry 41(11) (2002) 3762–9. [DOI] [PubMed] [Google Scholar]
- [17].Guo ZY, Chang CC, Chang TY, Functionality of the seventh and eighth transmembrane domains of acyl-coenzyme A:cholesterol acyltransferase 1, Biochemistry 46(35) (2007) 10063–71. [DOI] [PubMed] [Google Scholar]
- [18].Guo ZY, Lin S, Heinen JA, Chang CC, Chang TY, The active site His-460 of human acyl-coenzyme A:cholesterol acyltransferase 1 resides in a hitherto undisclosed transmembrane domain, The Journal of biological chemistry 280(45) (2005) 37814–26. [DOI] [PubMed] [Google Scholar]
- [19].Kaduce TL, Schmidt RW, Spector AA, Acylcoenzyme A:cholesterol acyltransferase activity: solubilization and reconstitution in liposomes, Biochemical and biophysical research communications 81(2) (1978) 462–8. [DOI] [PubMed] [Google Scholar]
- [20].Chang CC, Chen J, Thomas MA, Cheng D, Del Priore VA, Newton RS, Pape ME, Chang TY, Regulation and immunolocalization of acyl-coenzyme A: cholesterol acyltransferase in mammalian cells as studied with specific antibodies, The Journal of biological chemistry 270(49) (1995) 29532–40. [DOI] [PubMed] [Google Scholar]
- [21].Bishop JE, Hajra AK, A method for the chemical synthesis of 14C-labeled fatty acyl coenzyme A’s of high specific activity, Anal. Biochem 106 (1980) 344–350. [DOI] [PubMed] [Google Scholar]
- [22].Ikeuchi T, Dolios G, Kim SH, Wang R, Sisodia SS, Familial Alzheimer disease-linked presenilin 1 variants enhance production of both Abeta 1–40 and Abeta 1–42 peptides that are only partially sensitive to a potent aspartyl protease transition state inhibitor of “gamma-secretase”, The Journal of biological chemistry 278(9) (2003) 7010–8. [DOI] [PubMed] [Google Scholar]
- [23].Chattopadhyay A, Harikumar KG, Dependence of critical micelle concentration of a zwitterionic detergent on ionic strength: implications in receptor solubilization, FEBS Lett 391(1–2) (1996) 199–202. [DOI] [PubMed] [Google Scholar]
- [24].le Maire M, Champeil P, Moller JV, Interaction of membrane proteins and lipids with solubilizing detergents, Biochimica et biophysica acta 1508(1–2) (2000) 86–111. [DOI] [PubMed] [Google Scholar]
- [25].Mukerjee P, Chan CC, Effects of High Salt Concentrations on the Micellization of Octyl Glucoside: Salting-Out of Monomers and Electrolyte Effects on the Micelle−Water Interfacial Tension1, Langmuir 18(14) (2002) 5375–5381. [Google Scholar]
- [26].Rodi PM, Bocco Gianello MD, Corregido MC, Gennaro AM, Comparative study of the interaction of CHAPS and Triton X–100 with the erythrocyte membrane, Biochimica et biophysica acta 1838(3) (2014) 859–66. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental table 1 The average values of three dynamic light scattering measurements. The Z-average is the intensity-weighted mean diameter of particles found in solution. The polydispersity index (PdI) is a measure of the broadness of the size distribution ranging from 0 to 1, with anything greater than 0.7 being considered too polydispersed. The peak intensity measurement is the average diameter of individual peaks (1- micelles, 2- liposomes, 3- aggregates), as determined by the intensity particle size distribution (PSD). The number measurement is the average diameter of particles as determined from the number PSD. We use intensity PSD to report the size of each peak in the distribution and number PSD to report the size of the most abundant particle found in the distribution.
Supplemental fig. 1 The intensity particle size distributions for (A) preformed liposomes (11.2 mM PC, 9.3 mM taurocholate, 1.8 mM cholesterol) and diluted liposomes (100 mM KCl, 50 mM Tris-HCl pH 7.8, 6.3 mM PC, 5.2 mM taurocholate, 1 mM cholesterol) at various concentrations of CHAPS (B) 0.4 mM, (C) 2 mM, (D) 5 mM, (E) 8 mM, and (F) 10 mM.
Supplemental fig. 2 The intensity particle size distributions for (A) buffer (100 mM KCl, 5 mM CHAPS, 50 mM Tris-HCl pH 7.8), (B) buffer + 3.2 mM Triton X–100, (C) buffer + 30 mM octyl glucoside, (D) diluted liposomes (100 mM KCl, 5 mM CHAPS, 50 mM Tris-HCl pH 7.8, 6.3 mM PC, 5.2 mM taurocholate, 1 mM cholesterol), (E) diluted liposomes + 0.1 mM Triton X–100, (F) diluted liposomes + 3.2 mM Triton X–100, (G) diluted liposomes + 1 mM octyl glucoside, and (H) diluted liposomes + 30 mM octyl glucoside.