Capsule summary:
A novel STAT2 variant causing complete STAT2 protein abrogation presents with hemophagocytic lymphohistiocytosis (HLH). This is the first report of HLH in association with STAT2 deficiency.
Keywords: STAT2, HLH, Immunodeficiency, mumps
To the Editor:
Frequent viral infections in infancy may signal a primary defect of innate immunity. Signal transducer and activator of transcription 2 (STAT2) is a transcription factor involved in type I interferon signaling. Type I and III interferons are secreted immediately following pathogen exposure by most cells, while type II interferons are secreted primarily by activated T cells, NK cells, plasmacytoid dendritic cells, and macrophages. Activation of STAT2 leads to the expression of genes important for immunity against viral infections.1 Patients with homozygous mutations in STAT2 are susceptible to severe recurrent viral infections, including vaccine-strain viruses.2 Those who survive childhood may mount sufficient anti-viral responses after maturation of their adaptive immune system.1 Secondary hemophagocytic lymphohistiocytosis (HLH) is a sequela of uncontrolled immune activation typically triggered by infection. HLH is rarely associated with disorders of innate immunity other than chronic granulomatous disease, but has been reported in a single patient with STAT1 deficiency and in two kindreds with IFNGR2 deficiency.3–6 Here, we describe a patient with a novel STAT2 variant that abrogates protein expression who presented with secondary HLH in the setting of vaccine-strain mumps meningitis.
A non-consanguineous Nepali boy, with normal newborn screen for severe combined immune deficiency (T-cell receptor excision circles >252 copies/µL), was hospitalized five times in the first year of life with dehydration due to common viral illnesses, including respiratory syncytial virus, norovirus, and coxsackie virus. At 12 months of age, he was hospitalized with fever, cough, and emesis occurring seven days after vaccination with the measles, mumps, rubella, and varicella vaccines. He rapidly developed respiratory insufficiency and abdominopelvic ascites accompanied by diffuse adenopathy. Laboratory evaluation did not suggest a cellular or humoral immunodeficiency (Table I). He developed pancytopenia, decreased fibrinogen, elevated ferritin, and increased circulating levels of soluble IL-2 receptor, meeting clinical criteria for HLH (Table E1 in this article’s Online Repository).6 Meningitis due to vaccine-strain mumps was confirmed by PCR of cerebral spinal fluid. High-dose intravenous immunoglobulin (IVIG) treatment (2 g/kg) has been reported to be useful in the management of a STAT2-deficient patient with severe viral infections.2 Treatment of our patient with high-dose IVIG led to prompt resolution of HLH, with defervescence and normalization of ferritin level and cell counts. He was subsequently started on IVIG (0.5 g/kg) every 3 weeks and now avoids all live viral vaccines. The patient is now two years old and has not had any further infections, hospitalizations, or episodes of HLH.
Table I.
Immunologic profile of the patient.
| Variable | Age at time of testing* | |
|---|---|---|
| Hemogram 103 cells/µL (normal) | 12 months | 14 months |
| White blood cells (7.73–13.12) | 3.71 | 13.06 |
| Neutrophils (2.47–6.41) | 0.91 | 4.35 |
| Lymphocytes (2.32–5.49) | 2.36 | 6.99 |
| Monocytes (0.25–1.15) | 0.20 | 0.46 |
| Hemoglobin (10.4–12.5) | 7.9 | 10.2 |
| Hematocrit (30.5–36.4) | 23.6 | 29.8 |
| Platelets (223–461) | 89 | 474 |
| Lymphocyte subsets | ||
| CD3+, 103 cells/µL (1,900–6,200) | 804 | 3,651 |
| CD3+CD4+, 103 cells/µL (1,300–3,400) | 261 | 1,070 |
| CD45RA+CCR7+, % CD4+ (66.3–89.4) | 66.8 | 52.4 |
| CD45RA−CCR7+, % CD4+ (9.2–22.4) | 22.6 | 28.2 |
| CD45RA−CCR7−, % CD4+ (1.3–9.4) | 9.5 | 18.1 |
| CD45RA+CCR7−, % CD4+ (0.2–29) | 0.9 | 15 |
| CD3+CD8+, 103 cells/µL (620–2,000) | 505 | 2,328 |
| CD45RA+CCR7+, % CD8+ (57.8–82.9) | 55.9 | 70.4 |
| CD45RA−CCR7+, % CD8+ (1.7–8.5) | 6.5 | 1.5 |
| CD45RA−CCR7−, % CD8+ (5.1–25.1) | 9.5 | 13.2 |
| CD45RA+CCR7−, % CD8+ (6.4–20.8) | 14 | 15 |
| CD19+, 103 cells/µL (610–2,600) | 465 | 2,375 |
| CD27 − IgD+, % CD19+ (76.5–94.7) | ND | 90.9 |
| CD27+IgD+, % CD19+ (3–10.7) | ND | 4.5 |
| CD27+IgD−, % CD19+ (1.4–11.9) | ND | 2.7 |
| CD3 − CD56+, 103 cells/µL (160–1100) | 216 | 1,136 |
| Immunoglobulins (mg/dL) | ||
| IgG, (300–1500) | 884 | 1,386 |
| IgM, (25–115) | 80 | 87 |
| IgA, (16–100) | 52 | 28 |
| Proliferation (counts per minute) | ||
| Concavalin A (65,6999–239,344) | 34,503 | |
| Phytohemagglutinin (96,090–358,179) | 229,096 | |
| Anti-CD3 (62,927–217,761) | 76,757 | |
| T cell mitogen Background (204–2,104) | 264 | |
| Tetanus (8,544–102,895) | 5,530 | |
| Candida (6,231–197,940) | 25,213 | |
| T cell antigen Background (689–9,034) | 3,502 | |
At 12 months of age, testing was done while he was acutely ill and before starting IVIG replacement; at 14 months of age testing was done after recovery from HLH and after IVIG was initiated. Proliferation was not performed at 12 months due to lymphopenia and critically ill state. Bold values are outside the normal range.
Prior to intravenous immunoglobulin replacement
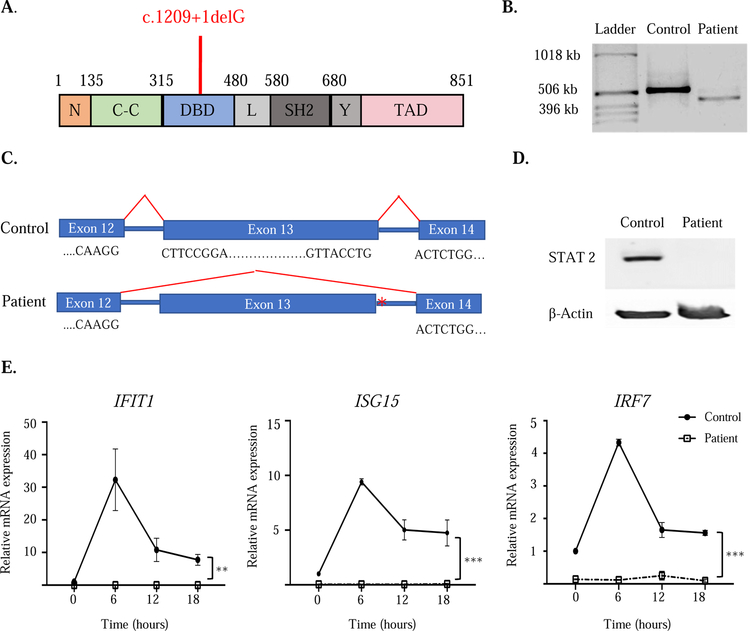
Targeted next-generation sequencing of the patient’s genomic DNA for 207 genes associated with primary immunodeficiency (Primary Immunodeficiency Panel, Invitae Corporation, San Francisco, CA) identified a novel homozygous splice donor variant in STAT2(c.1209+1delG) (Figure IA). His parents are heterozygous for the variant (see Figure E1 in the Online Repository). No mutations were identified in other genes associated with primary HLH or primary immune deficiency. To determine whether the mutation affected splicing of STAT2, reverse transcription polymerase chain reactions (RT-PCR) were performed on mRNA from control and patient EBV-transformed B-lymphoblastoid cell lines (BLCLs) (Figure IB). The 533 base pair fragment surrounding the mutation site was identified in the control. In contrast, a lower molecular weight band of approximately 437 base pairs was identified in cDNA from the patient’s BLCLs. Sanger sequencing demonstrated abnormal splicing in the patient’s cDNA amplicon, resulting in deletion of exon 13 and in-frame splicing of exons 12 and 14 (Figure IC). Immunoblotting of lysates from BLCLs demonstrated abrogation of STAT2 protein expression in our patient without expression of any truncated protein products (Figure ID).
Figure I. The STAT2 homozygous splice donor variant mutation abrogates protein expression.
A. Linear map of STAT2 with the patient’s variant (c.1209+1delG)indicated in red. B. RT-PCR amplification of STAT2 mRNA encompassing the patient mutation site. C. Sanger sequencing of cDNA in the region surrounding the mutation in patient and control. Asterisk indicates mutation location. D. STAT2 protein expression in lysates from BLCLs from patient and control. E. Quantitative PCR analysis of IFIT1, ISG15 and IRF7 mRNA at baseline and after IFN-α stimulation of EBV-transformed BLCLs from the patient and controls (n=3). Values were normalized to GAPDH and expressed relative to the mean of unstimulated controls. Symbols and bars in E represent mean and SEM. ** p<0.01, *** p<0.001. Similar results were obtained in two independent experiments in B, D and E.
STAT2 deficiency in humans and mice is characterized by defective type I interferon signaling.1,2,9 The infection and subsequent transformation of B cells with EBV activates STAT2 and upregulates expression of interferon-stimulated genes, including IFIT1, ISG15 and IRF7, which are further increased after exposure to type I interferons.7.8 Expression of IFIT1, ISG15, and IRF7 were severely depressed in the patient’s BLCLs compared to controls (time 0 in Figure IE). Furthermore, the patient BLCLs cells failed to upregulate IFIT1, ISG15, and IRF7 after IFN-α stimulation (Figure IE), indicating a lack of STAT2-driven signaling in response to type I interferons.
Mouse models of STAT2 deficiency have demonstrated defective responses to viral infections both in vitro and in vivo.1 Human STAT2 deficiency has been associated with development of recurrent and severe viral illnesses. After MMR vaccination, one patient developed disseminated vaccine strain measles virus,1 another patient developed meningitis with vaccine strain mumps virus,9 and four patients developed post-MMR febrile syndromes with negative viral studies.1, 2, 9
This is the first report of HLH in association with STAT2 deficiency. We identified a novel STAT2 variant in our patient, resulting in abrogation of protein expression and type I interferon response. This report, as well as previously published reports of patients with defects in STAT1 and IFNGR2, demonstrate that secondary HLH can occur with defects in either type I or type II interferon signaling..4, 5 The rapid improvement of our patient after high-dose IVIG suggests the utility of this intervention in treating secondary HLH triggered by infection. The beneficial effect of IVIG may be ascribed to passive immunization and possibly to its potential anti-inflammatory effect. Our report and other literature2 suggests patients with STAT2 deficiency may benefit from monthly immunoglobulin replacement during childhood, particularly if they have received live vaccines prior to their molecular diagnosis, until their adaptive immune system matures.
Supplementary Material
Acknowledgments
Supported by: T32AI007512 (MCM),1R01AI139633–01 (RSG), the Perkin Fund (RSG), 5K08AI116979–04 (JC) and K23AI143962–01 (LMB).
Abbreviations:
- STAT2
signal transduction and activator of transcription 2
- HLH
hemophagocytic lymphohistiocytosis
- MMR
measles, mumps, rubella
- RT-PCR
Reverse transcription polymerase chain reaction
- BLCLs
B-lymphoblastoid cell lines
- IVIG
intravenous immunoglobulin
Footnotes
Conflict of interest: The authors declare no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hambleton S, Goodbourn S, Young DF, Dickinson P, Mohamad SM, Valappil M, et al. STAT2 deficiency and susceptibility to viral illness in humans. Proc Natl Acad Sci U S A 2013; 110:3053–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moens L, Van Eyck L, Jochmans D, Mitera T, Frans G, Bossuyt X, et al. A novel kindred with inherited STAT2 deficiency and severe viral illness. J Allergy Clin Immunol 2017; 139:1995–7.e9. [DOI] [PubMed] [Google Scholar]
- 3.Bode SF, Ammann S, Al-Herz W, Bataneant M, Dvorak CC, Gehring S, et al. The syndrome of hemophagocytic lymphohistiocytosis in primary immunodeficiencies: implications for differential diagnosis and pathogenesis. Haematologica 2015:haematol. 2014.121608. [DOI] [PMC free article] [PubMed]
- 4.Faitelson Y, Bates A, Shroff M, Grunebaum E, Roifman CM, Naqvi A. A mutation in the STAT1 DNA-binding domain associated with hemophagocytic lymphohistocytosis. LymphoSign Journal 2014; 1:87–95. [Google Scholar]
- 5.Tesi B, Sieni E, Neves C, Romano F, Cetica V, Cordeiro AI, et al. Hemophagocytic lymphohistiocytosis in 2 patients with underlying IFN-γ receptor deficiency. Journal of Allergy and Clinical Immunology 2015; 135:1638–41. e5. [DOI] [PubMed] [Google Scholar]
- 6.Henter JI, Horne A, Arico M, Egeler RM, Filipovich AH, Imashuku S, et al. HLH-2004: Diagnostic and therapeutic guidelines for hemophagocytic lymphohistiocytosis. Pediatr Blood Cancer 2007; 48:124–31. [DOI] [PubMed] [Google Scholar]
- 7.Zhang L, Hong K, Zhang J, Pagano JS. Multiple signal transducers and activators of transcription are induced by EBV LMP-1. Virology 2004; 20;323(1):141–52. [DOI] [PubMed] [Google Scholar]
- 8.Zhang Jun, Das Subash C., Kotalik Catherine, Pattnaik Asit K., and Zhang Luwen. The Latent Membrane Protein 1 of Epstein-Barr Virus Establishes an Antiviral State via Induction of Interferon-stimulated Genes. J Biol Chem 2006;281(14):9163–9. [DOI] [PubMed] [Google Scholar]
- 9.Shahni R, Cale CM, Anderson G, Osellame LD, Hambleton S, Jacques TS, et al. Signal transducer and activator of transcription 2 deficiency is a novel disorder of mitochondrial fission. Brain 2015; 138:2834–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.