Abstract
Background
Arsenic (As) is an unwanted toxic mineral that threatens the major rice-growing regions in the world, especially in South Asia. Rice production in Bangladesh and India depends on As-contaminated groundwater sources for irrigating paddy fields, resulting in elevated amounts of As in the topsoil. Arsenic accumulating in rice plants has a significant negative effect on human and animal health. Here, we present a quantitative trait locus (QTL) mapping study to identify candidate genes conferring As toxicity tolerance and accumulation in rice (Oryza sativa L.) seedlings. An early backcross breeding population consisting of 194 lines derived from a cross between WTR1 (indica) and Hao-an-nong (japonica) was grown in hydroponics for 25 days, from the seventh day exposed to an environmentally relevant concentration of 10 ppm As.
Results
Arsenic toxicity leads to significantly negative plant responses, including reduced biomass, stunted plant growth, reduced leaf chlorophyll content, and increased shoot As concentration ranging from 9 to 20 mg kg− 1. Marker-trait association was determined for seven As-related traits using 704 single nucleotide polymorphism (SNP) markers generated from a 6 K SNP-array. One QTL was mapped on chromosome 1 for relative chlorophyll content, two QTLs for As content in roots were mapped on chromosome 8, and six QTLs for As content in shoots were mapped on chromosomes 2, 5, 6, and 9. Using the whole-genome sequence of the parents, we narrowed down the number of candidate genes associated with the QTL intervals based on the existence of a non-synonymous mutation in genes between the parental lines. Also, by using publicly available gene expression profiles for As stress, we further narrowed down the number of candidate genes in the QTL intervals by comparing the expression profiles of genes under As stress and control conditions. Twenty-five genes showing transcription regulation were considered as candidate gene nominees for As toxicity-related traits.
Conclusions
Our study provides insight into the genetic basis of As tolerance and uptake in the early seedling stage of rice. Comparing our findings with the previously reported QTLs for As toxicity stress in rice, we identified some novel and co-localized QTLs associated with As stress. Also, the mapped QTLs harbor gene models of known function associated with stress responses, metal homeostasis, and transporter activity in rice. Overall, our findings will assist breeders with initial marker information to develop suitable varieties for As-contaminated ecosystems.
Electronic supplementary material
The online version of this article (10.1186/s12284-019-0321-y) contains supplementary material, which is available to authorized users.
Keywords: Rice, Arsenic toxicity, Single nucleotide polymorphisms, Quantitative trait loci, Candidate genes
Background
Rice (Oryza sativa. L) is one of the world’s most important staple crops, consumed by more than half of the world’s population, and it plays a major role in the entry of mineral nutrients into the food chain (Global Rice Science Partnership 2013; Yorobe et al. 2016). Traditionally, rice is grown in flooded paddy fields, which can harbor unwanted toxic heavy metals such as arsenic, cadmium, mercury, and lead, which represents a threat to human and cattle health (Norton et al. 2010; Pandey et al. 2015). Compared with the other major cereals, rice takes up large amounts of arsenic (As), a highly toxic class I carcinogenic metalloid, which naturally occurs worldwide in the environment and is widely distributed in Earth’s crust (Liu et al. 2012a). Rivers originating from the greater Himalayas carry As from their rock sediments to the densely populated rice-growing plains of South and Southeast Asia, making the major rice-growing belt of Asia vulnerable to As contamination (Sohn 2014; Zhou et al. 2017). In addition, climate change is threatening rice production in these areas. With frequent occurrence of drought and saltwater intrusion in the Ganges-Brahmaputra deltas of India and Bangladesh, farmers depend on groundwater for irrigation of their crops (Guan et al. 2010; Marcaida et al. 2014; Wang et al. 2015). This groundwater is an additional source of As discharged from the naturally As-rich aquifers (Nickson et al. 1998). Rice production in the major regions of Bangladesh and India uses As-contaminated groundwater for irrigation. Typically, 4 to 8 mg kg− 1 of As occurs in flooded paddy soil, but the concentration can reach 83 mg kg− 1 in some parts of Bangladesh and West Bengal regions of India (Zhang et al. 2008; Norton et al. 2010).
Rapidly growing populations need more food despite the increased labor cost, reduced farmland, and water scarcity that threaten rice production. Long-term use of contaminated groundwater for irrigating crops has resulted in a significant increase in As in the topsoil, thereby contaminating the food chain (Pandey et al. 2015). Governments of developing Asian nations are encouraging farmers to adopt modern technologies such as direct-seeded rice (DSR) and alternate wetting-drying (AWD) as a cost-efficient sustainable method to reduce the environmental footprint of rice production (Fischer et al. 2007; Singh Chauhan et al. 2015). Toxic levels of As in the topsoil can potentially affect the performance of DSR by hindering the most sensitive rice germination process and early seed establishment of the rice growth cycle (Abedin and Meharg 2002; Shri et al. 2009). Water management using AWD was proposed as one of the strategies to control As bioavailability in the soil-plant system (Mitra et al. 2017). However, rice varieties suitable for the AWD system under As-toxic soil have not been developed and tested in field conditions for implementation (Suriyagoda et al. 2018). Therefore, it is necessary to reveal the genetic basis of As tolerance and accumulation in the early seedling stage of rice, which will help rice breeders to develop As-tolerant varieties suitable for modern rice production technologies (Tuli et al. 2010).
As toxicity in rice plants triggers various symptoms, including low germination rate, poor seed establishment, reduced photosynthetic rates, stunted plant growth, low biomass production, sterility-related yield loss, and a physiological disorder called straighthead disease. These symptoms are often confounded with other soil-related problems associated with rice (Abedin et al. 2002a; Azizur Rahman et al. 2008). The toxic effects of As in rice vary upon thier chemical form, with the inorganic As species being more toxic than the organic species (Abedin et al. 2002b; Tripathi et al. 2013). The inorganic species of As found in the rice-growing environment are arsenite (As(III)) and arsenate (As(V)). In anaerobic flooded soil conditions such as submerged paddy fields, the reduced form As(III) dominates and in aerobic soil conditions such as upland paddy fields its oxidized form As(V) dominates (Tripathi et al. 2013; Pandey et al. 2015). Both forms are toxic to rice plants (Batista et al. 2011). Rice plants do not posses naturally evolved As transporters (Pandey et al. 2015), instead, As competes with chemically similar essential minerals to enter the plant system (Stoeppler 2003). As(III) is physiochemically similar to the essential mineral silicon (Si) and thus competes with the Si uptake pathway. Alternatively, A(V) is physicochemically similar to essential mineral phosphorous (P) and uses P acquisition pathways to enter the root system, and for efflux toward the xylem and various tissues (Clemens 2006; Zhao et al. 2009; Yang et al. 2018). Most rice genotypes possess a mechanism to retain much of the toxic As burden in the roots. However, a genotype-dependent proportion of As is translocated into the shoots and other tissues, including grains of the rice plant (Carey et al. 2010; Pandey et al. 2015). Since 35–55% of rice is produced in irrigated conditions (Ali et al. 2018b), As(III) contributes the major As species loaded into rice plants (Zhao et al. 2010b). As(III), which exists as the neutral molecule As (OH)3, enters rice root cells through nodulin 26-like intrinsic proteins (NIPs), belonging to the aquaporin family of major intrinsic proteins (MIPs), which are non-permeable to As(V) (Ma et al. 2008; Zhao et al. 2009). Widely spread NIP aquaporins also mediate the transport of a range of neutral molecules, including ammonia, urea, boric acid, and silicic acid (Zhao et al. 2010a). The silicon transporter in rice root OsLSI1 has been suggested as the main As(III) uptake protein, while As(III) efflux from rice root cells to the xylem takes place through OsLSI2 silicon-mediated transporter (Ma et al. 2008; Zhao et al. 2010b). However, none of the above genes associated with the As(III) and As(V) tolerance-low uptake genes were functionally characterized to use in breeding programs for the development of As-safe varieties.
Mapping QTLs to identify causative genes in populations is essential for trait improvement in breeding programs (Würschum 2012). Using backcross breeding populations has proven to be an effective strategy to dissect the genetic factors that underly the phenotypic complexity of nutrient-related traits with a simultaneous focus on varietal development (Xue et al. 2006; Ali et al. 2013; Wu et al. 2014). Although As interactions with rice have been well documented over the past two decades, only a limited number of QTL studies and genes associated with As have been reported for the development of As-safe varieties (Dasgupta et al. 2004; Zhang et al. 2008; Chen et al. 2013; Zhang et al. 2014). For As(V), AsTol on chromosome 6 was the first QTL reported in rice for root tolerance and it is located close to a phosphate uptake QTL (Dasgupta et al. 2004). For As(III), the first QTLs reported in rice are located on chromosomes 2 and 3 for As content in shoot and root, respectively, and chromosome 6 and 8 for As content in brown rice (Zhang et al. 2008). In this current study, as a component of the ongoing Green Super Rice (GSR) backcross breeding program at the International Rice Research Institute (IRRI) for the development of multi-tolerance rice varieties for the resource-poor farmers of Asia and Africa (Li and Ali 2017), we try to understand the genetic and molecular mechanisms underlying As tolerance and accumulation in rice seedlings to accelerate the development of As-tolerant and -safe varieties for contaminated ecosystems. The objective of the study has been the following: 1) to screen for As toxicity tolerance in early backcross breeding populations, 2) to identify QTLs related to As tolerance and accumulation using high-density SNP markers, and 3) to identify candidate genes for As-related traits using the whole-genome sequence approach.
Results
Performance of parental lines and BRILs
Exposure to 10 ppm of As for 18 days induced a significantly negative response in parents and the mapping population. WTR1 showed more tolerance than Hao-an-nong in terms of chlorophyll content and biomass accumulation. For root length and plant height, Hao-an-nong showed higher tolerance. The parents WTR1 and Hao-an-nong showed highly significant differences in accumulating As between shoot and root tissue (Fig. 1). When averaged over all backcross recombinant inbred lines (BRILs), the mean values of shoot dry weight, plant height, and chlorophyll content decreased considerably in the As stress, except for root biomass and root length (Table 1). As content in BRILs ranged from 9.10 mg kg− 1 (AsG-195) to 20.99 mg kg− 1 (AsG-227) and 68.62 mg kg− 1 (AsG-185) to 197.10 mg kg− 1 (AsG-226) in shoot and root, respectively. Compared with the parents, 15 BRILs showed the lowest uptake of As concentration (≤10.00 mg kg− 1 in shoots and ≤ 80.00 mg kg− 1 in roots). In particular, two genotypes, AsG-187 (GSR IR2–1-R14-N2-N5-N21-N42) and AsG-189 (GSR IR2–1-Y4-N4-N2-N3-N2), showed better performances in low uptake of As content in roots and shoots (Additional file 1: Table S1). Most of the traits measured in the control and treatment conditions appeared to be normally distributed (Fig. 2). The overall results indicated that adequate genetic variability is present in the parental material and populations studied for QTL analysis.
Fig. 1.
Phenotypic trait variation in parents of mapping population under As toxicity stress for 18 days. (A) Chlorophyll content; (B) plant height; (C) root length; (D) shoot dry weight; (E) root dry weight; (F) arsenic content in shoot and root tissue. Vertical bars represent standard errors of means (n = 3). Different letters above the data points indicate significant differences between genotypes by LSD test (P < 0.05)
Table 1.
Descriptive statistics and ANOVA results for different phenotypes
| Trait | Control | Treatment 10 ppm As | ANOVA result | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Min | Max | Mean | Min | Max | Mean | G | T | G*T | |
| Chlorophyll content (SPAD) | 29.93 | 40.72 | 35.31 | 16.15 | 31.45 | 23.27 | *** | *** | NS |
| Plant height (cm) | 39.33 | 63.17 | 48.25 | 23.13 | 43.50 | 33.31 | NS | *** | *** |
| Root length (cm) | 10.83 | 19.83 | 15.86 | 9.00 | 18.17 | 13.00 | *** | *** | *** |
| Shoot dry weight (g) | 0.91 | 2.22 | 1.56 | 0.82 | 1.15 | 0.98 | *** | *** | *** |
| Root dry weight (g) | 0.31 | 0.72 | 0.53 | 0.20 | 0.70 | 0.45 | *** | *** | NS |
| Arsenic content of shoots (mg kg−1) | ND | ND | ND | 9.10 | 20.99 | 15.48 | *** | NA | NA |
| Arsenic content of roots (mg kg− 1) | ND | ND | ND | 68.62 | 197.10 | 130.36 | *** | NA | NA |
Significance levels are indicated at P < 0.001/*** Treatment and genotype treatment interaction effects were not analyzed for As content traits since in control conditions arsenic is not measured
ND not determined, NA not significant, NA not applicable, G genotype, T treatment
Fig. 2.
Box plot distribution of measured As-related stress in the population; control with no As and treatment with 10 ppm As a substitution for 18 days. Control: without As substitution, Treatment: with 10 ppm As substitution
Correlation among measured traits
The study of correlation coefficient among various measured As-related traits is of great importance to predict the relationship between complex quantitative As content traits and simple quantitative morphological traits associated with As toxicity. For As content in shoots, the data showed a significantly negative correlation with relative plant height, chlorophyll content, and root length (Fig. 3). For As content in roots, there was a significant negative correlation with relative root length and plant height. A significant and positive correlation coefficient was recorded between As content in shoots and roots, relative root length and relative shoot length, and relative shoot biomass and relative root biomass.
Fig. 3.

Heat map is showing the correlation coefficient of As-related traits. RCC: relative chlorophyll content; RPH: relative plant height; RRL: relative root length; RSBM: relative shoot biomass; RRBM: relative root biomass; AsS: As content in shoots; AsR: As content in roots
SNP markers generated by 6 K SNP-array for QTL mapping
A customized 6 K SNP-Beadchip was used. It consists of 4606 high-quality SNP markers, which detected 1068 polymorphic sites between the parents and BC1F6 BRILs. A total of 704 polymorphic SNPs remained after pairwise comparison of SNPs between the parents for missing and heterozygous SNPs. These 704 SNPs were unevenly distributed across the genome, ranging from 84 SNPs on chromosome 4 to 41 on chromosome 9, with an average spacing of ∼524 kb between the SNPs, ranging from 393.9 kb on chromosome 7 to 797.4 kb on chromosome 2 (Additional file 2: Figure S1). There were 42 large gaps (> 2 Mb) across the genome in the distribution of SNPs generated by the 6 K SNP-array as a result of monomorphic SNP markers shared between parents in these regions. Gaps ranging from 4 Mb on chromosome 2 to 8 Mb on chromosome 11 were observed in the generated physical map of the rice genome using the 704 SNP marker data (Fig. 4). These 704 filtered SNPs were used to analyze the association between markers and As-related traits.
Fig. 4.
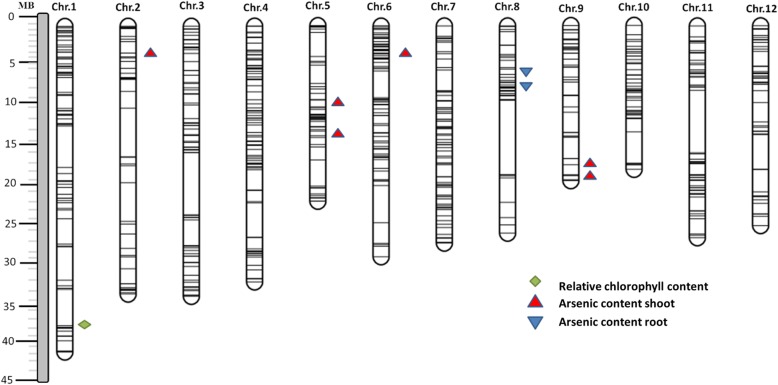
Chromosomal distribution of polymorphic single nucleotide polymorphisms (SNPs) and quantitative trait loci (QTLs) detected in this study. QTLs were located on the chromosome based on the physical position of the SNP marker
Identification of QTLs for as-related traits
As tolerance are a non-target trait that was not considered during the development of the BRILs population used in this study. Therefore, the selected BRILs population could be considered as a random segregation population for mapping As related QTLs, assuming As tolerance was not correlated with those selected traits used in population development (yield under irrigated, rainfed, drought and low input). In total, 40 SNPs showed significant marker-trait association for the As-related traits (Table 2). Nine QTLs were defined by assuming that closely linked significant markers are in the QTL region. Among the nine QTLs, six were identified for As content in shoots, two were identified for As content in roots, and one tolerant QTL was identified for relative chlorophyll content. QTLs for As content in shoots (qAsS2, qAsS5.1, qAsS5.2, qAsS6, qAsS9.1, and qAsS9.2) were mapped on chromosomes 2, 5, 6, and 9, respectively. QTLs for As content in roots (qAsR8.1 and qAsR8.2) were mapped on chromosome 8 and relative chlorophyll content QTL qRChlo1 was mapped on chromosome 1 (Fig. 4). All the QTLs detected (P < 0.01) had a –log p(F) ≥3.3 and phenotypic variance explained by the QTLs ranged from 8.6% to 12.6%, with QTLs qAsS5.2, qAsS6, and qAsR8.1 explaining major phenotypic variance of more than 10% and QTLs qRChlo1, qAsR8.2, qAsS2, qAsS5.1, qAsS9.1, and qAsS9.2 explaining minor phenotypic variance of less than 10% (Table 2).
Table 2.
QTLs associated with As-related traits after 18 days of 10 ppm As stress in WTR1× Hao-an-nong population by single marker regression
| QTLa | Trait | Chr | Positionb | Associated markerc | -log p(F)d | R2 (%)e | Additive effectf | Tolerance alleleg |
|---|---|---|---|---|---|---|---|---|
| qRChlo1 | Relative chlorophyll content | 1 | 39.2829 | 66SNP_1_39282883 | 3.7781 | 8.6216 | 4.0676 | WTR1 |
| 1 | 39.3692 | 67SNP_1_39369209 | 3.7781 | 8.6216 | 4.0676 | WTR1 | ||
| 1 | 39.4208 | 68SNP_1_39420824 | 3.7781 | 8.6216 | 4.0676 | WTR1 | ||
| qAsR8.1 | Arsenic content root | 8 | 6.0577 | 487SNP_8_6057678 | 4.652 | 10.5069 | − 11.7883 | WTR1 |
| qAsR8.2 | Arsenic content root | 8 | 7.854 | 492SNP_8_7854002 | 3.821 | 8.7155 | −10.6686 | WTR1 |
| qAsS2 | Arsenic content shoot | 2 | 4.3429 | 88SNP_2_4342883 | 4.3275 | 9.8119 | −1.1264 | WTR1 |
| 2 | 4.9307 | 89SNP_2_4930742 | 4.1432 | 9.4147 | −1.1015 | WTR1 | ||
| 2 | 5.8303 | 90SNP_2_5830265 | 4.0335 | 9.1768 | −1.0944 | WTR1 | ||
| 2 | 6.4799 | 91SNP_2_6479920 | 4.1741 | 9.4812 | −1.1054 | WTR1 | ||
| 2 | 7.0233 | 92SNP_2_7023295 | 4.1741 | 9.4812 | −1.1054 | WTR1 | ||
| 2 | 7.0767 | 93SNP_2_7076671 | 4.332 | 9.8214 | −1.1248 | WTR1 | ||
| 2 | 7.1037 | 94SNP_2_7103684 | 4.332 | 9.8214 | −1.1248 | WTR1 | ||
| 2 | 7.1708 | 95SNP_2_7170842 | 4.332 | 9.8214 | −1.1248 | WTR1 | ||
| 2 | 7.2388 | 96SNP_2_7238793 | 4.332 | 9.8214 | −1.1248 | WTR1 | ||
| 2 | 7.2775 | 97SNP_2_7277487 | 4.1741 | 9.4812 | −1.1054 | WTR1 | ||
| qAsS5.1 | Arsenic content shoot | 5 | 10.8863 | 287SNP_5_10886331 | 3.7203 | 8.4964 | −1.0489 | WTR1 |
| 5 | 13.3255 | 288SNP_5_13325546 | 3.7203 | 8.4964 | −1.0489 | WTR1 | ||
| 5 | 13.7687 | 289SNP_5_13768744 | 3.7203 | 8.4964 | −1.0489 | WTR1 | ||
| 5 | 14.0797 | 290SNP_5_14079677 | 3.7203 | 8.4964 | −1.0489 | WTR1 | ||
| 5 | 14.644 | 291SNP_5_14643984 | 4.1216 | 9.3682 | −1.1564 | WTR1 | ||
| qAsS5.2 | Arsenic content shoot | 5 | 15.4693 | 292SNP_5_15469279 | 4.0303 | 9.1701 | −1.0871 | WTR1 |
| 5 | 15.556 | 293SNP_5_15556017 | 4.0303 | 9.1700 | −1.0871 | WTR1 | ||
| 5 | 16.4598 | 294SNP_5_16459802 | 4.0303 | 9.1701 | −1.0871 | WTR1 | ||
| 5 | 16.5851 | 295SNP_5_16585060 | 4.0303 | 9.1700 | −1.0871 | WTR1 | ||
| 5 | 16.8086 | 296SNP_5_16808642 | 5.6649 | 12.6433 | −1.3646 | WTR1 | ||
| qAsS6 | Arsenic content shoot | 6 | 0.4008 | 329SNP_6_400753 | 4.1072 | 9.3372 | 1.3763 | Hao-an-nong |
| 6 | 0.6469 | 330SNP_6_646915 | 4.4046 | 9.9782 | 1.4682 | Hao-an-nong | ||
| 6 | 0.8342 | 332SNP_6_834170 | 4.1072 | 9.3372 | 1.3763 | Hao-an-nong | ||
| 6 | 1.5219 | 335SNP_6_1521855 | 3.5808 | 8.1922 | 1.3782 | Hao-an-nong | ||
| 6 | 1.768 | 336SNP_6_1768006 | 5.323 | 11.929 | 1.4867 | Hao-an-nong | ||
| 6 | 1.9284 | 337SNP_6_1928403 | 4.0138 | 9.1355 | 1.2804 | Hao-an-nong | ||
| 6 | 1.9783 | 338SNP_6_1978288 | 4.0138 | 9.1355 | 1.2804 | Hao-an-nong | ||
| 6 | 2.0107 | 339SNP_6_2010737 | 4.5116 | 10.208 | 1.3335 | Hao-an-nong | ||
| 6 | 2.0256 | 340SNP_6_2025629 | 4.5116 | 10.208 | 1.3335 | Hao-an-nong | ||
| qAsS9.1 | Arsenic content shoot | 9 | 18.3666 | 551SNP_9_18366555 | 3.884 | 8.8531 | −1.085 | WTR1 |
| 9 | 18.3891 | 552SNP_9_18389054 | 3.884 | 8.8531 | −1.085 | WTR1 | ||
| 9 | 19.2081 | 553SNP_9_19208050 | 3.7654 | 8.5948 | −1.0579 | WTR1 | ||
| qAsS9.2 | Arsenic content shoot | 9 | 20.587 | 555SNP_9_20587039 | 3.9802 | 9.0617 | −1.0806 | WTR1 |
| 9 | 21.2154 | 557SNP_9_21215424 | 3.6667 | 8.379 | −1.0403 | WTR1 | ||
| 9 | 21.3489 | 558SNP_9_21348882 | 4.368 | 9.8987 | −1.1294 | WTR1 |
aClosely linked markers are assumed as the same QTL. bPhysical position of markers on chromosomes
cMarker associated with QTL. dF-statistical analysis indicates association between markers and trait
eProportion of phenotypic variance explained. fPositive/negative values indicate that WTR1/ Hao-an-nong can increase trait values
gTolerance allele provided by parental line
Selection of candidate genes associated with as-related traits
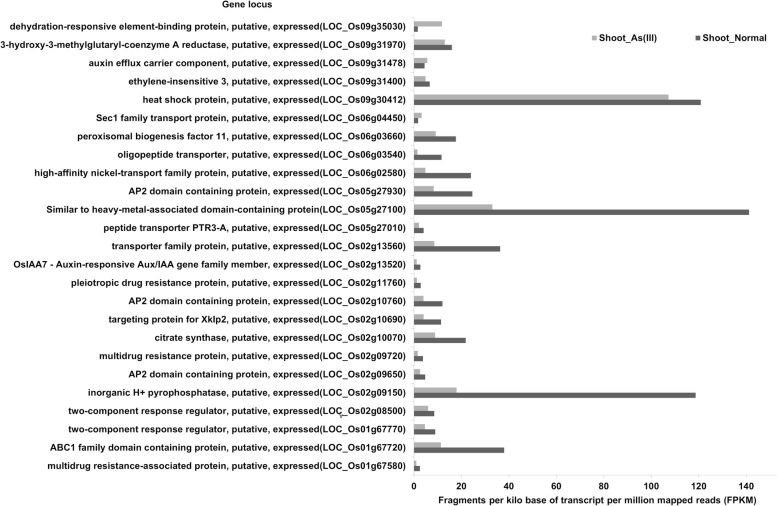
In the confidence interval of the identified QTLs, 35,767 and 440 SNPs were identified between the parents in the Rice SNP-Seek Database and tGBS® dataset, respectively. Among those identified SNPs, 35% of the SNPs showed polymorphism between the parents in 1309 genes, and most of these polymorphisms (93.9%) were synonymous mutations, with non-synonymous mutations (6.1%) between the parents in 676 genes (Table 3 and Additional file 3: Table S2). Among those 676 genes, 35 genes were associated with metal transport, response, and metal homeostasis and thus considered as the most likely candidate genes (Additional file 4: Table S3). Expression data of these genes were retrieved from a study by Yu et al. (2012) using the web application Rice Expression Database (Xia et al. 2017). Twenty-five genes out of the 35 most likely candidate genes showed expression responses to the sodium arsenite (AsIII) stress treatment according to expression data (Yu et al. 2012). Among the 25 genes showing expression responses, 3 genes were upregulated, and 22 showed significant down-regulation (Fig. 5).
Table 3.
SNPs identified in the QTL regions using tGBS and 3 K RGP
| QTLs | Chromosome | No. of SNPs in tGBS | No. of SNPs in 3 K RGP | Polymorphic SNPs | Non-synonymous SNPs | Total no. of genes | Genes with non-synonymous SNPs |
|---|---|---|---|---|---|---|---|
| qRChlo1 | 1 | 4 | 479 | 202 | 63 | 20 | 20 |
| qAsR8.1 | 8 | 34 | 2761 | 757 | 148 | 47 | 27 |
| qAsR8.2 | 8 | 65 | 3070 | 1110 | 179 | 61 | 42 |
| qAsS2 | 2 | 79 | 11,705 | 4542 | 821 | 382 | 225 |
| qAsS5.1 | 5 | 46 | 2346 | 677 | 108 | 94 | 45 |
| qAsS5.2 | 5 | 103 | 2166 | 976 | 192 | 113 | 67 |
| qAsS6 | 6 | 86 | 6789 | 1932 | 357 | 240 | 98 |
| qAsS9.1 | 9 | 11 | 6154 | 2214 | 309 | 326 | 141 |
| qAsS9.2 | 9 | 12 | 297 | 142 | 16 | 26 | 11 |
| Total | 440 | 35,767 | 12,552 | 2193 | 1309 | 676 |
Fig. 5.
Transcriptional regulation of genes contained in identified QTL regions. Only genes containing non-synonymous SNPs showing significant expression response under As(III) stress are plotted. Gene expression data adopted from Yu et al. (2012)
Discussion
In this study, we exposed two parental lines (WTR1/ Hao-an-nong) and 194 BC1F6 lines to a concentration of 10 ppm As(III) at the seedling stage for 18 days in a hydroponic-based experimental setup. Similar concentrations frequently occur in the topsoil of the rice-growing regions of Bangladesh and West Bengal of India (Abedin et al. 2002a; Zhang et al. 2008). When exposed to 10 ppm of As(III), a wide range of morpho-physiological changes occurred in the plants, leading to inhibited plant growth with a rapid decline in biomass and reduced chlorophyll content when compared to the control (Fig. 1) (Wu et al. 2011; Pandey et al. 2015). In our current experiment, the distribution of As showed similar patterns (root>shoot), in agreement with the previously reported genotype-dependent As accumulation and distribution, as the results suggested that the indica recipient parent (WTR1) accumulated less As in shoots than its japonica donor parent (Hao-an-nong) (Rahman et al. 2007; Bhattacharya et al. 2010; Wu et al. 2011). Correlation among the traits revealed that plant performance under As stress depended on As concentration in the tissue, with increased uptake in shoots reducing the plant’s overall performance. Arsenic translocation to shoots was positively correlated with As content in roots, which indicates that avoiding As translocation to shoot tissue will, in turn, increase plant performance by an As avoidance mechanism. In one earlier study, a doubled-haploid (DH) population derived from cross CJ06 × TN1 (japonica × indica) cultivars was used to evaluate the toxic effect of 10 ppm As(III) in a soil-based controlled greenhouse experiment. The correlation in that study revealed results similar to those of the current study, clearly indicating that the performance of plants under As stress is inversely related to the As content in shoots (Fig. 3) (Zhang et al. 2008). Arsenic toxicity traits observed in the rice population in our experimental conditions signify that As tolerance and accumulation in rice seedlings is a quantitatively inherited trait controlled by multiple genes. Overall, selecting lines with the shoot avoidance mechanism would be appropriate for As-contaminated regions since farmers in the major rice-growing regions of India and Bangladesh use rice straw to feed farm cattle (Azizur Rahman et al. 2008). Some of the lines from the population showing high tolerance and low accumulation of As in shoots could be used in breeding programs aiming to identify genes that could increase As tolerance with low accumulation (Additional file 1: Table S1) (Tuli et al. 2010).
When using a 6 K SNP-array for genotyping, the average number of polymorphic markers derived across diverse germplasm was quite high (Thomson et al. 2017). However, only 1068 SNP sites were identified in this study, with 704 polymorphic sites remaining after removing the monomorphic sites. As the population used in the current study was advanced using the backcross breeding approach, only a single fragment or a small number of genomic introgression fragments from a donor parent were retrieved in the population, leading to the lower number of polymorphic sites present in the near-isogenic lines (Li and Ali 2017; Ali et al. 2018a). QTL mapping was performed using 704 SNP markers, and a total of 9 tightly linked QTLs associated with As-related traits were mapped using marker-trait association. The putative QTL regions detected for As-related traits in our study were compared with previous reports based on the physical positions of the associated markers in the Nipponbare genome (International Rice Genome Sequencing Project, http://rgp.dna.affrc.go.jp/IRGSP/). Associated SNPs mapped on chromosomes 2, 6, and 8 were consistent with previous studies for As content QTLs in rice (Dasgupta et al. 2004; Zhang et al. 2008; Chen et al. 2013). On the other hand, the QTLs mapped on chromosomes 1, 5, and 9 were reported for the first time in the As(III)-toxic treatment. Detection of multiple QTLs for traits in stable bi-parental populations illustrates the complexity of As(III)-induced traits (Pascual et al. 2016).
We employed a whole-genome sequencing approach of parents to narrow down the number of genes in the QTL intervals since QTLs mapped on the bi-parental populations are the only loci involved in a gene pool limited to the founder parents of the populations studied (Xu et al. 2017). By analyzing non-synonymous mutations in genes between the parental sequence in the QTL interval, we were able to narrow down the number of candidate genes from 1306 to 676. In order to further narrow down the number of candidate genes, we analyzed transcriptional regulation in response to As(III)-toxic conditions based on the data adopted from a previous expression study (Xia et al. 2017) (Fig. 5). Transcriptional analysis revealed As(III) stress-responsive gene expression in the identified QTL regions, loci LOC_Os05g27100, LOC_Os02g09150, and LOC_Os06g03540 present in the QTL regions qAsS5.2, qAsS2, and qAsS6, respectively, which showed significant down-regulation in gene expression after 24-h As(III) treatment. All three highly down-regulated gene expressions carry one non-synonymous mutation between the parents in the coding region of the gene. Gene loci LOC_Os09g35030 and LOC_Os01g67580 present in the QTL regions qAsS9.1 and qRChlo1, respectively, showed significant up-regulation after 24-h As(III) treatment. Gene locus LOC_Os01g67580 was highly polymorphic with seven non-synonymous mutations present in the coding region of the gene and locus LOC_Os09g35030 carried one non-synonymous mutation between the parents in the gene-coding region (Additional file 3: Table S2). Overall, the whole-genome sequence of parents and gene expression analysis proved to be an effective strategy to narrow down the candidate genes in the QTL intervals.
Plants have evolved versatile detoxification systems to counter the wide variety of phytotoxic compounds present in the natural environment (Coleman et al. 1997; Shamsi et al. 2008). In the region 39.28–39.42 Mb on chromosome 1, QTL qRChlo1 was located. Of the four candidate genes, the most plausible one was LOC_Os01g67580 (OsMRP2), which encodes for a multidrug resistance-associated protein (MRP, ABCC1), a subfamily of ABC transporters. Members of the MRP ATP-binding cassette transporters were originally proposed to be primarily involved in the vacuolar sequestration of potentially toxic metabolites. MRPs exist in all organisms and are known to play key roles in the efflux of xenobiotic compounds (Paumi et al. 2009). In one study involving Arabidopsis thaliana double knockout mutants of ABCC- type transporters (AtABCC1 and AtABCC2), the mutant plants exhibited a very low residual As(III)–PC2 transport activity (Song et al. 2010). In another study, involving the same double knockout mutant lines (AtABCC1 and AtABCC2), tolerance of mercury and cadmium toxicity was conferred by sequestering in the large vacuole (Park et al. 2012). Another study, involving an AtABCC3 transporter mutant in A. thaliana, also confirmed increasing sensitivity to cadmium toxicity in the seedlings (Brunetti et al. 2015). The gene OsMRP2, an ABCC1 transporter identified in the QTL region of qRChlo1, may possibly be involved in As transport and partitioning processes such as vacuolar sequestration of toxic metabolites by the action of ATP-driven efflux pumps (Klein et al. 2006; Jeandroz and Lamotte 2017).
QTLs associated with shoot As content included genes of complex physiological and molecular networks, ranging from metal transporters, sequestration and partitioning of As, and synthesis of detoxification proteins and cheaters that promote shoot-to-root mobility of As (Tripathi et al. 2013; Pandey et al. 2015). In the region of 10.88–14.64 Mb on chromosome 5, QTL qAsS5.1 was located. Of the three candidate genes, the most likely one was LOC_Os05g27100, annotated as heavy metal-associated domain-containing protein (HMA). HMAs are metallochaperones that contain a metal binding domain for safe transport of metallic ions inside and outside of the cell, primarily involved in detoxification mechanism (De Abreu-Neto et al. 2013). In rice, gene OsHMA9 is a metal efflux HMA protein known to play a key role in metal homeostasis of copper, zinc, lead, and cadmium (Lee et al. 2007). In the case of As toxicity in rice, the role of HMA is not yet defined. In the region of 4.34–7.22 Mb on chromosome 2, QTL qAsS2 was located. Of the 13 candidate genes, the most likely one was LOC_Os02g09150, which encodes for inorganic H+ vacuolar pyrophosphates (V-PPase). Plant V-PPase is a specific class of multi-subunit pumps that play an essential role in the productivity of higher plants, generation of proton gradients across tonoplast endomembranes, and their ability to buffer changes in the concentrations of essential and toxic ions (Bak et al. 2013). A study involving A. thaliana double knockout mutant lines of V-PPase (vha-a2 and vha-a3) showed reduced zinc tolerance with no response to salt stress (Krebs et al. 2010). Another study involving the same double knockout mutant lines (vha-a2 and vha-a3) displayed functional regulation of intracellular ion homeostasis by ion compartmentalization of calcium ion (Tang et al. 2012). However, their functional relation and relative contributions to ion storage and detoxification are unclear in rice. In the region of 0.40–2.02 Mb on chromosome 6, QTL qAsS2 was located. Of the six candidate genes, the most likely one was LOC_Os06g03540 (OsOPT2) annotated as oligopeptide transporter family (OPT), which plays an important and diverse role in plant growth and development. It is widely accepted that OPTs are proton-coupled symporters that translocate their substrates in the cytosolic direction with the possible function of long-distance metal distribution, nitrogen mobilization, heavy metal sequestration, and glutathione transport (Lubkowitz 2011). Studies suggest that 16 OPT gene motifs are distributed in the rice genome, and OsOPT2 showed transcriptional accumulation in all tissues, with varying amounts under normal conditions and significantly down-regulated under salinity and cold stress and up-regulated under drought stress, suggesting that OsOPT2 may be involved in diverse plant regulation functions (Liu et al. 2012b). However, OsOPT2 was not functionally characterized in rice.
In most cases, including root-related metal concentration traits, excess mineral attached to the root surface may interfere with the measurement. This measurement is not considered as a good-quality trait for understanding mineral uptake and metabolism in rice (Zhang et al. 2008). Two QTLs identified for root As content, qAsR8.1 (5.84–6.28 Mb) and qAsR8.2 (7.33–8.38 Mb), were located 1 Mb apart from each other on chromosome 8, and co-localized with a QTL previously reported for As content in brown rice (Zhang et al. 2008). Within the region was harbored the well-studied rice gene (OsZIP4) responsible for zinc transport and distribution in rice (Ishimaru et al. 2005, 2007). Moreover, genes present in the root QTL intervals for As content did not show any response in the gene expression analysis.
Reducing the levels of the carcinogenic As in rice is a major health goal. To the best of our knowledge, this study is the first to use the breeding population to map QTLs associated with As toxicity stress in rice. For the QTLs identified in this study to make an impact on adaptive rice breeding, it is necessary to evaluate both parents (WTR1 and Hao-an-nong) and BRILs under high As contaminated soil to understand the As tolerance and accumulation. Further, low As accumulating BRILs (AsG187 and ASG189) identified through this study needs to be tested under high As contaminated soil for As content in mature brown rice and straw before any varietal nomination in the contaminated region. For the development of As-safe rice, the candidate genes nominated in this study need to be verified and functionally characterized.
Conclusions
This study provides novel insights into the genetic basis of AsIII toxicity tolerance and accumulation in rice. Through marker-trait association analysis, a total of nine QTLs were identified on the different chromosomes in rice. Genetically dissecting As toxicity tolerance and accumulation is complex because it is a quantitatively inherited trait controlled by multiple genes. This study also demonstrates that using the whole-genome sequencing of parents and expression analysis provides useful information about the candidate genes present in the QTL intervals, and a total of 25 promising candidate genes for 9 QTL regions were determined. The candidate genes associated with As toxicity tolerance and accumulation identified in this study provide a valuable basis for future functional gene characterization and improvement of rice varieties for As-contaminated ecosystems.
Methods
Plant materials and phenotypic screening
A population of 230 BC1F6 backcross recombinant inbred lines (BRILs) derived from a cross between indica line WTR1 and japonica cultivar Hao-an-nong was produced using single seed descent at the International Rice Research Institute (IRRI), Los Baños, Philippines. For detailed information regarding the population development, see (Jewel et al. 2019). As tolerance and accumulation was a non-target trait that was not considered during the development of the BRILs population which was used in this study. Thus, the selected BRILs population could be considered as a random segregating population for mapping As related QTLs, assuming As tolerance and accumulation was not correlated with those selected traits used in population development (yield under irrigated, rainfed, drought and low input condition). For the hydroponic screening experiment, 194 lines from the developed population were used, and the experiment was carried out in a controlled phytotron glasshouse of IRRI. Optimum rice-growing conditions were maintained throughout the experiment: 29/21 °C (day/night), 70% relative humidity, and natural light. Seeds of the 194 lines were oven-dried for 5 days at 60 °C to break any residual seed dormancy and incubated at 30 °C for 48 h for germination. One seedling per line was transferred per hole with 1 cm diameter on a Styrofoam seedling float with size of 28 × 32 × 1.25 cm having 100 holes (10 × 10) with a nylon net bottom-fixed in a dark plastic tray containing 8 L of full-strength Yoshida nutrient solution (Jodal et al. 1975; Wu et al. 2017) Throughout the experiment, pH was maintained in a range of 5.1 to 5.4 in the nutrient solution. On the seventh day, 10 ppm As was supplied in the form of sodium-(meta)-arsenite (AsIII) (AsNaO2, Sigma-Aldrich, MO, USA). A control treatment without As was maintained throughout the experiment. Plants were grown in As-toxic conditions for 18 days, and nutrient solutions were renewed once every 7 days. The experiment was set up as a complete randomized design with three independent replicates, and five repeats per line in each replicate, leading to a total of 59 hydroponic tanks, each accommodating up to 100 seedlings. To maintain uniform As concentration across the different hydroponic tanks, nutrient solutions were prepared in a single tank having 1000 L capacity and non-segregating parents (WTR1 and Hao-an-nong) were grown as checks in each hydroponic tanks. Any positional effect within the phytotron glasshouse was minimized by altering the position of hydroponic tanks every second day.
Chlorophyll concentration and seedling growth parameters
Leaf yellowing and senescence were observed in plants on the 12th day post-As treatment (PAsT). To compare the differences among the lines, the relative chlorophyll concentrations were measured non-destructively from the base, middle, and tip of the top leaves of each individual plant, and the average values were expressed as SPAD units (SPAD-502 chlorophyll meter, Minolta Camera Co., Ltd., Japan) as the indicator of leaf senescence caused by toxic As treatment. Plants’ responses to the As treatment were evident after 18 days PAsT. Changes in the shoot and root length response to the As treatment were measured for each entry at 18 days PAsT. Shoot length was measured from the base of the plant to the tip of the longest leaf, while root length was measured from the base of the plant to the root tip. Three plants per entry per replicate were rinsed with deionized water to remove culture solution sticking to the surface of the root and then oven-dried for 3 days at 70 °C to remove moisture. Before sample processing for As content analysis, dry biomass was recorded.
Sample preparation and analysis of as concentration
Arsenic content in roots and shoots was determined for parents and BRILs grown in As-toxic conditions at 18 days PAsT. Dried samples were thoroughly homogenized by an ultracentrifugal mill (ZM100, Retsch, Haan, Germany) modified with a tungsten blade to avoid any cross-contamination. Samples ground to 0.5 g were added to a closed vessel digester and 5 mL of high-purity 69% concentrated nitric acid (HNO3) followed by 2 mL hydrogen peroxide (H2O2), and 1 mL of deionized water were added and predigested overnight in the fume hood (Amaral et al. 2013). The following day, the samples were digested using a heating block at 150–155 °C for 3 h under the fume hood. The digested tissue was diluted to a final volume of 25 mL using deionized water and total As was determined using Graphite Furnace Atomic Absorption Spectrophotometry (GFAAS-7000F, SHIMADZU Corporation, Kyoto, Japan) (Hirohashi 2001). A deuterium lamp background correction with high-performance As hollow cathode lamps (193.7 nm) was used to quantify the As content in the standards and samples. An aliquot of 20 μL of the digested sample and 10 μL palladium (100 ppm) as matrix modifier were injected into the pyrol-coated graphite tube with the aid of an autosampler unit. A seven-stage furnace cycle program was adopted with atomization temperature of 2200 °C at the sixth stage used as a standard parameter to determine As content in the sample and expressed in milligrams per kilogram (mg kg− 1).
DNA extraction and genotyping
Genomic DNA was extracted from the seedlings of 194 BRILs and two parents (WTR1 and Hao-an-nong) using DNeasy Plant Mini Kit following the manufacturer’s protocol (QIAGEN, Germantown, MD, USA). High-throughput SNP genotyping was carried out using a custom-design Illumina Infinium rice 6 K SNP-array containing 4606 SNPs covering all 12 rice chromosomes in the Genotyping Services Laboratory facility at IRRI (Thomson 2014; Thomson et al. 2017). A rice 6 K-SNP chip was scanned using an Illumina bead array reader, and automatic allele calling was achieved using Illumina Genome Studio data analysis software (V2010.1. Illumina Inc.) (Thomson et al. 2017). Scoring of alleles in the BRILs at each SNP locus was carried out by comparing parental alleles at the respective SNP locus as co-dominant markers.
Statistical analysis and QTL mapping
Two-way ANOVA was conducted before obtaining the relative phenotypic trait values to observe the effects of lines, treatment (control and As treatment), and line-treatment interaction for different measured traits. Multiple comparison analysis was performed on parents following the least significant difference (LSD) post hoc test. Relative phenotypic value of morphological traits was determined for each line, that is, the phenotypic value obtained from the As treatment divided by the control value (Dasgupta et al. 2004; Shrestha et al. 2018). Pair-wise Pearson‘s correlation analysis was carried out among the As-related traits, in which P- value was two-tailed with two significant levels using P = 0.05 and P = 0.01, and a heat map was generated using corrplot package in R studio. All the statistical analyses were conducted using PBTools package of R (R Core Team 2015). For QTL mapping in the WTR1 × Hao-an-nong backcross breeding population, a physical map consisting of 704 SNP markers covering all 12 chromosomes based on the physical locations of the markers was constructed. QTL mapping was performed by single marker regression analysis using the function single marker analysis (SMA) of iciMapping program v.4.0 (www.isbreeding.net/software/?type=detail&id=18). The threshold (−log p(F) ≥3.3) to declare significant association was set based on a permutation test (1000 permutations, P = 0.01) for each trait (Crump and Llewellyn-Thomas 2011). Tightly linked SNP markers showing significant associations were assumed as the same QTL and named following the standard protocol (McCouch et al. 1997).
Candidate gene identification
For the identified QTLs governing As-related traits, the gene models located in the QTL intervals were searched from the MSU Rice Genome Annotation Database (Ouyang et al. 2007). In the identified QTL regions, loci showing non-synonymous polymorphism between the parents were considered for nomination as candidate genes (Pang et al. 2017). Polymorphisms between parental lines in QTL regions were retrieved from the Rice SNP-Seek Database (http://snp-seek.irri.org) (Alexandrov et al. 2015). Additional genotype information on parents was obtained from the earlier Tunable Genotyping-By-Sequencing (tGBS®) project using 10 ion proton runs, resulting in 794,297 polymorphic SNPs (10.7910/DVN/RRXCR3) (Ali et al. 2018a). Raw sequence reads of parents obtained from the tGBS® project were aligned to the public reference rice genome Osativa_204_v7.0.fa, downloaded from the Phytozome website (https://phytozome.jgi.doe.gov/pz/portal.html#!info?alias=Org_%20Osativa). By using the Genomic Short-read Nucleotide Alignment program (GSNAP) (http://research-pub.gene.com/gmap/), short reads were confidently mapped to the best location in the reference genome by allowing ≤2 mismatches every 36 base pairs (bp), and fewer than 5 bases for every 75 bp as tails were used as criteria. Aligned reads of the parents were compared with the reference genome sequence for polymorphism sights and compared among the parents to obtain SNP information (Wu and Nacu 2010; Ott et al. 2017). Then, we selected gene models in the QTL intervals showing non-synonymous polymorphism associated with metal homeostasis as our most likely candidate genes. Expression profiles of the most likely candidates in the identified QTL regions were extracted from a previously published study in which rice cultivar Nipponbare (Oryza sativa L. subsp. japonica) had been exposed to sodium arsenite (AsIII) stress for 24 h in the seedling stage (Yu et al. 2012). Data from that expression profile experiment were publicly available (Xu et al. 2018) (Xia et al. 2017).
Additional files
Table S1. Mean relative phenotypic values of traits measured under As stress. (XLSX 32 kb)
Figure S1. Distribution of SNPs on the chromosome. (PNG 49 kb)
Table S2. Total number of genes and SNPs identified in the candidate region. (XLSX 110 kb)
Table S3. Candidate genes present in the QTL intervals. (XLSX 12 kb)
Acknowledgments
We thank and acknowledge IRRI Communications for English language editing.
Abbreviations
- As
Arsenic
- AWD
Alternate wetting drying
- BRILs
Backcross recombinant inbred lines
- DSR
Direct seeded rice
- GSR
Green super rice
- PAsT
Post arsenic treatment
- QTL
Quantitative trait locus
- SNP
Single nucleotide polymorphism
- tGBS
Tunable Genotyping by Sequencing
Authors’ contributions
VM was responsible for conducting the As phenotypic experiment of the study and worked on the data analysis and drafting of the manuscript; ZJ, VM, and JA contributed to the development of breeding materials; VM, UA, YP, CM, and AM were involved in the collection of phenotypic data and statistical analysis of the phenotypic and genotypic information. VM, AM, JA, ZL, LBW, and MF helped in reviewing and improving the draft manuscript. JA and ZL conceived the basic idea, implemented research, acquired funding, and corrected the manuscript. All authors read and approved this final version of the research article.
Funding
The authors would like to thank and acknowledge the Bill & Melinda Gates Foundation (BMGF) for providing a research grant to ZL for the Green Super Rice project under ID OPP1130530. We would also like to thank the Department of Agriculture (DA), Philippines, for providing funds to JA under the Next-Gen project.
Availability of data and materials
The comprehensive collected data supporting the conclusions of this review article are provided as figures, tables, and supplementary tables.
Ethics approval and consent to participate
The authors declare that this research review was conducted in the absence of any commercial or economic associations that could be construed as potential conflicts of interest.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Abedin J, Cresser MS, Meharg AA, et al. Arsenic accumulation and metabolism in rice (Oryza sativa L.) Environ Sci Technol. 2002;36:962–968. doi: 10.1021/es0101678. [DOI] [PubMed] [Google Scholar]
- Abedin MJ, Feldmann J, Meharg AA. Uptake kinetics of arsenic species in rice plants. Plant Physiol. 2002;128:1120–1128. doi: 10.1104/pp.010733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abedin MJ, Meharg AA. Relative toxicity of arsenite and arsenate on germination and early seedling growth of rice (Oryza sativa L.) Plant Soil. 2002;243:57–66. doi: 10.1023/A:1019918100451. [DOI] [Google Scholar]
- Alexandrov N, Tai S, Wang W, et al. SNP-seek database of SNPs derived from 3000 rice genomes. Nucleic Acids Res. 2015;43:D1023–D1027. doi: 10.1093/nar/gku1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali J, Aslam UM, Tariq R, et al. Exploiting the genomic diversity of rice (Oryza sativa L.): SNP-typing in 11 early-backcross introgression-breeding populations. Front Plant Sci. 2018;9:1–10. doi: 10.3389/fpls.2018.00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali J, Jewel ZA, Mahender A, et al. Molecular genetics and breeding for nutrient use efficiency in rice. Int J Mol Sci. 2018;19:1–27. doi: 10.3390/ijms19061762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali J, Xu JL, Gao YM, et al. Breeding for yield potential and enhanced productivity across different rice ecologies through green super rice (GSR) breeding strategy. In: Muralidharan K, Siddiq EA, et al., editors. International Dialogue Perception and Prospects of Designer Rice Society for Advancement of Rice research, Directorate of Rice Research, Hyderabad 500 030, India. 2013. pp. 60–68. [Google Scholar]
- Amaral CDB, Nóbrega JA, Nogueira ARA. Sample preparation for arsenic speciation in terrestrial plants - a review. Talanta. 2013;115:291–299. doi: 10.1016/j.talanta.2013.04.072. [DOI] [PubMed] [Google Scholar]
- Azizur Rahman M, Hasegawa H, Mahfuzur Rahman M, et al. Arsenic accumulation in rice (Oryza sativa L.): human exposure through food chain. Ecotoxicol Environ Saf. 2008;69:317–324. doi: 10.1016/j.ecoenv.2007.01.005. [DOI] [PubMed] [Google Scholar]
- Bak G, Lee E-J, Lee Y, et al. Rapid structural changes and acidification of guard cell vacuoles during stomatal closure require phosphatidylinositol 3,5-bisphosphate. Plant Cell. 2013;25:2202–2216. doi: 10.1105/tpc.113.110411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batista BL, Souza JMO, De Souza SS, Barbosa F. Speciation of arsenic in rice and estimation of daily intake of different arsenic species by Brazilians through rice consumption. J Hazard Mater. 2011;191:342–348. doi: 10.1016/j.jhazmat.2011.04.087. [DOI] [PubMed] [Google Scholar]
- Bhattacharya P, Samal AC, Majumdar J, Santra SC. Accumulation of arsenic and its distribution in rice plant (Oryza sativa L.) in Gangetic West Bengal, India. Paddy Water Environ. 2010;8:63–70. doi: 10.1007/s10333-009-0180-z. [DOI] [Google Scholar]
- Brunetti P, Zanella L, De Paolis A, et al. Cadmium-inducible expression of the ABC-type transporter AtABCC3 increases phytochelatin-mediated cadmium tolerance in Arabidopsis. J Exp Bot. 2015;66:3815–3829. doi: 10.1093/jxb/erv185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey A-M, Scheckel KG, Lombi E, et al. Grain unloading of arsenic species in rice. Plant Physiol. 2010;152:309–319. doi: 10.1104/pp.109.146126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Fu S, Zhang P, et al. Proteomic analysis of a disease-resistance-enhanced lesion mimic mutant spotted leaf 5 in rice. Rice. 2013;6:1–10. doi: 10.1186/1939-8433-6-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemens S. Toxic metal accumulation, responses to exposure and mechanisms of tolerance in plants. Biochimie. 2006;88:1707–1719. doi: 10.1016/j.biochi.2006.07.003. [DOI] [PubMed] [Google Scholar]
- Coleman JOD, Blake-Kalff MMA, Davies TGE. Detoxification of xenobiotics by plants: chemical modification and vacuolar compartmentation. Trends Plant Sci. 1997;2:144–151. doi: 10.1016/S1360-1385(97)01019-4. [DOI] [Google Scholar]
- Crump T, Llewellyn-Thomas HA. Assessing medicare beneficiaries’ strength-of-preference scores for health care options: how engaging does the elicitation technique need to be? Health Expect. 2011;14:33–45. doi: 10.1111/j.1369-7625.2010.00632.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasgupta T, Hossain SA, Meharg AA, Price AH. An arsenate tolerance gene on chromosome 6 of rice. New Phytol. 2004;163:45–49. doi: 10.1111/j.1469-8137.2004.01109.x. [DOI] [PubMed] [Google Scholar]
- De Abreu-Neto JB, Turchetto-Zolet AC, De Oliveira LFV, et al. Heavy metal-associated isoprenylated plant protein (HIPP): characterization of a family of proteins exclusive to plants. FEBS J. 2013;280:1604–1616. doi: 10.1111/febs.12159. [DOI] [PubMed] [Google Scholar]
- Fischer G, Tubiello FN, van Velthuizen H, Wiberg DA. Climate change impacts on irrigation water requirements: effects of mitigation, 1990-2080. Technol Forecast Soc Change. 2007;74:1083–1107. doi: 10.1016/j.techfore.2006.05.021. [DOI] [Google Scholar]
- Global Rice Science Partnership . Rice almanac. Los Baños: International Rice Research Institute; 2013. [Google Scholar]
- Guan YS, Serraj R, Liu SH, et al. Simultaneously improving yield under drought stress and non-stress conditions: a case study of rice (Oryza sativa L.) J Exp Bot. 2010;61:4145–4156. doi: 10.1093/jxb/erq212. [DOI] [PubMed] [Google Scholar]
- Hirohashi T. Maize non-photosynthetic ferredoxin precursor is mis-sorted to the intermembrane space of chloroplasts in the presence of light. Plant Physiol. 2001;125:2154–2163. doi: 10.1104/pp.125.4.2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishimaru Y, Masuda H, Suzuki M, et al. Overexpression of the OsZIP4 zinc transporter confers disarrangement of zinc distribution in rice plants. J Exp Bot. 2007;58:2909–2915. doi: 10.1093/jxb/erm147. [DOI] [PubMed] [Google Scholar]
- Ishimaru Y, Suzuki M, Kobayashi T, et al. OsZIP4, a novel zinc-regulated zinc transporter in rice. J Exp Bot. 2005;56:3207–3214. doi: 10.1093/jxb/eri317. [DOI] [PubMed] [Google Scholar]
- Jeandroz S, Lamotte O. Editorial: plant responses to biotic and abiotic stresses: lessons from cell signaling. Front Plant Sci. 2017;8:1. doi: 10.3389/fpls.2017.01772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jewel ZA, Ali J, Pang Y, et al. Developing green super rice varieties with high nutrient use efficiency by phenotypic selection under varied nutrient conditions. Crop J. 2019;7:368–377. doi: 10.1016/j.cj.2019.01.002. [DOI] [Google Scholar]
- Jodal M, Hallbäck DA, Svanvik J, Lundgren O. A method for the continuous study of net water transport in the feline small bowel. Acta Physiol Scand. 1975;95:441–447. doi: 10.1111/j.1748-1716.1975.tb10072.x. [DOI] [PubMed] [Google Scholar]
- Klein M, Burla B, Martinoia E. The multidrug resistance-associated protein (MRP/ABCC) subfamily of ATP-binding cassette transporters in plants. FEBS Lett. 2006;580:1112–1122. doi: 10.1016/j.febslet.2005.11.056. [DOI] [PubMed] [Google Scholar]
- Krebs M, Beyhl D, Gorlich E, et al. Arabidopsis V-ATPase activity at the tonoplast is required for efficient nutrient storage but not for sodium accumulation. Proc Natl Acad Sci. 2010;107:3251–3256. doi: 10.1073/pnas.0913035107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Kim Y-Y, Lee Y, An G. Rice P1B-type heavy-metal ATPase, OsHMA9, is a metal efflux protein. Plant Physiol. 2007;145:831–842. doi: 10.1104/pp.107.102236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Ali J (2017) Breeding green super rice (GSR) varieties for sustainable rice cultivation In: Muralidharan K, Siddiq EA. 2013. International Dialogue on Perception and Prospects of Designer Rice. Society for Advancement of Rice Research, Directorate of Rice Resea. Hyderabad. 109–129
- Liu P, He K, Li Y, et al. Exposure to mercury causes formation of male-specific structural deficits by inducing oxidative damage in nematodes. Ecotoxicol Environ Saf. 2012;79:90–100. doi: 10.1016/j.ecoenv.2011.12.007. [DOI] [PubMed] [Google Scholar]
- Liu T, Zeng J, Xia K, et al. Evolutionary expansion and functional diversification of oligopeptide transporter gene family in rice. Rice. 2012;5:12. doi: 10.1186/1939-8433-5-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubkowitz M. The oligopeptide transporters: a small gene family with a diverse group of substrates and functions? Mol Plant. 2011;4:407–415. doi: 10.1093/mp/ssr004. [DOI] [PubMed] [Google Scholar]
- Ma JF, Yamaji N, Mitani N, et al. Transporters of arsenite in rice and their role in arsenic accumulation in rice grain. Proc Natl Acad Sci. 2008;105:9931–9935. doi: 10.1073/pnas.0802361105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcaida M, Li T, Angeles O, et al. Biomass accumulation and partitioning of newly developed green super Rice (GSR) cultivars under drought stress during the reproductive stage. F Crop Res. 2014;162:30–38. doi: 10.1016/j.fcr.2014.03.013. [DOI] [Google Scholar]
- McCouch SR, Cho YG, Yato M, et al. Report on QTL nomenclature. Rice Genet Newsl. 1997;14:11–13. [Google Scholar]
- Mitra A, Chatterjee S, Moogouei R, Gupta D. Arsenic accumulation in rice and probable mitigation approaches: a review. Agronomy. 2017;7:67. doi: 10.3390/agronomy7040067. [DOI] [Google Scholar]
- Nickson R, McArthur J, Burgess W, et al. Arsenic poisoning of Bangladesh groundwater. Nature. 1998;395:338. doi: 10.1038/26387. [DOI] [PubMed] [Google Scholar]
- Norton GJ, Deacon CM, Xiong L, et al. Genetic mapping of the rice ionome in leaves and grain: identification of QTLs for 17 elements including arsenic, cadmium, iron and selenium. Plant Soil. 2010;329:139–153. doi: 10.1007/s11104-009-0141-8. [DOI] [Google Scholar]
- Ott A, Liu S, Schnable JC, et al. tGBS® genotyping-by-sequencing enables reliable genotyping of heterozygous loci. Nucleic Acids Res. 2017;45:e178. doi: 10.1093/nar/gkx853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang S, Zhu W, Hamilton J, et al. The TIGR rice genome annotation resource: improvements and new features. Nucleic Acids Res. 2007;35:8–11. doi: 10.1093/nar/gkl976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey S, Rai R, Rai LC (2015b) in Microbes and Plants. In: SJS Flora (ed) Handbook of Arsenic Toxicology, 1st edn. Elsevier, Amsterdam, pp 627–674.
- Pang Y, Chen K, Wang X, et al. Simultaneous improvement and genetic dissection of salt tolerance of rice (Oryza sativa L.) by designed QTL pyramiding. Front Plant Sci. 2017;8:1–11. doi: 10.3389/fpls.2017.01275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J, Song WY, Ko D, et al. The phytochelatin transporters AtABCC1 and AtABCC2 mediate tolerance to cadmium and mercury. Plant J. 2012;69:278–288. doi: 10.1111/j.1365-313X.2011.04789.x. [DOI] [PubMed] [Google Scholar]
- Pascual L, Albert E, Sauvage C, et al. Dissecting quantitative trait variation in the resequencing era: complementarity of bi-parental, multi-parental and association panels. Plant Sci. 2016;242:120–130. doi: 10.1016/j.plantsci.2015.06.017. [DOI] [PubMed] [Google Scholar]
- Paumi CM, Chuk M, Snider J, et al. ABC transporters in Saccharomyces cerevisiae and their interactors: new technology advances the biology of the ABCC (MRP) subfamily. Microbiol Mol Biol Rev. 2009;73:577–593. doi: 10.1128/MMBR.00020-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team . A language and environment for statistical computing. Vienna: R Foundation for Statistical Computing; 2015. [Google Scholar]
- Rahman MA, Hasegawa H, Rahman MM, et al. Accumulation of arsenic in tissues of rice plant (Oryza sativa L.) and its distribution in fractions of rice grain. Chemosphere. 2007;69:942–948. doi: 10.1016/j.chemosphere.2007.05.044. [DOI] [PubMed] [Google Scholar]
- Shamsi IH, Jilani G, Zhang GP, Kang W. Cadmium stress tolerance through potassium nutrition in soybean. Asian J Chem. 2008;20:1099–1108. [Google Scholar]
- Shrestha A, Dziwornu AK, Ueda Y, et al. Genome-wide association study to identify candidate loci and genes for Mn toxicity tolerance in rice. PLoS One. 2018;13:1–15. doi: 10.1371/journal.pone.0192116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shri M, Kumar S, Chakrabarty D, et al. Effect of arsenic on growth, oxidative stress, and antioxidant system in rice seedlings. Ecotoxicol Environ Saf. 2009;72:1102–1110. doi: 10.1016/j.ecoenv.2008.09.022. [DOI] [PubMed] [Google Scholar]
- Singh Chauhan B, Opeña J, Ali J. Response of 10 elite “green super rice” genotypes to weed infestation in aerobic rice systems. Plant Prod Sci. 2015;18:228–233. doi: 10.1626/pps.18.228. [DOI] [Google Scholar]
- Sohn E. Contamination: the toxic side of rice. Nature. 2014;514:62–63. doi: 10.1038/514S62a. [DOI] [PubMed] [Google Scholar]
- Song W-Y, Park J, Mendoza-Cozatl DG, et al. Arsenic tolerance in Arabidopsis is mediated by two ABCC-type phytochelatin transporters. Proc Natl Acad Sci. 2010;107:21187–21192. doi: 10.1073/pnas.1013964107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoeppler M. Heavy metals in soils. TrAC Trends Anal Chem. 2003;15:8–9. [Google Scholar]
- Suriyagoda LDB, Dittert K, Lambers H. Arsenic in Rice soils and potential agronomic mitigation strategies to reduce arsenic bioavailability: a review. Pedosphere. 2018;28:363–382. doi: 10.1016/S1002-0160(18)60026-8. [DOI] [Google Scholar]
- Tang RJ, Liu H, Yang Y, et al. Tonoplast calcium sensors CBL2 and CBL3 control plant growth and ion homeostasis through regulating V-ATPase activity in Arabidopsis. Cell Res. 2012;22:1650–1665. doi: 10.1038/cr.2012.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson MJ. High-throughput SNP genotyping to accelerate crop improvement. Plant Breed Biotechnol. 2014;2:195–212. doi: 10.9787/PBB.2014.2.3.195. [DOI] [Google Scholar]
- Thomson MJ, Singh N, Dwiyanti MS, et al. Large-scale deployment of a rice 6 K SNP array for genetics and breeding applications. Rice. 2017;10:40. doi: 10.1186/s12284-017-0181-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi P, Tripathi RD, Singh RP, et al. Arsenite tolerance in rice (Oryza sativa L.) involves coordinated role of metabolic pathways of thiols and amino acids. Environ Sci Pollut Res. 2013;20:884–896. doi: 10.1007/s11356-012-1205-5. [DOI] [PubMed] [Google Scholar]
- Tuli R, Chakrabarty D, Trivedi PK, Tripathi RD. Recent advances in arsenic accumulation and metabolism in rice. Mol Breed. 2010;26:307–323. doi: 10.1007/s11032-010-9412-6. [DOI] [Google Scholar]
- Wang W, Fu B, Ali J, et al. Genome-wide responses to selection and genetic networks underlying submergence tolerance in Rice. Plant Genome. 2015;8:13. doi: 10.3835/plantgenome2014.10.0066. [DOI] [PubMed] [Google Scholar]
- Wu C, Ye Z, Shu W, et al. Arsenic accumulation and speciation in rice are affected by root aeration and variation of genotypes. J Exp Bot. 2011;62:2889–2898. doi: 10.1093/jxb/erq462. [DOI] [PubMed] [Google Scholar]
- Wu L-B, Shhadi MY, Gregorio G, et al. Genetic and physiological analysis of tolerance to iron pulse stresses in rice. Rice. 2014;7:1–12. doi: 10.1186/s12284-014-0008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu LB, Ueda Y, Lai SK, Frei M. Shoot tolerance mechanisms to iron toxicity in rice (Oryza sativa L.) Plant Cell Environ. 2017;40:570–584. doi: 10.1111/pce.12733. [DOI] [PubMed] [Google Scholar]
- Wu TD, Nacu S. Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics. 2010;26:873–881. doi: 10.1093/bioinformatics/btq057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Würschum T. Mapping QTL for agronomic traits in breeding populations. Theor Appl Genet. 2012;125:201–210. doi: 10.1007/s00122-012-1887-6. [DOI] [PubMed] [Google Scholar]
- Xia L, Zou D, Sang J, et al. Rice expression database (RED): An integrated RNA-Seq-derived gene expression database for rice. J Genet Genomics. 2017;44:235–241. doi: 10.1016/j.jgg.2017.05.003. [DOI] [PubMed] [Google Scholar]
- Xu X, Hao L, Zhu J, et al. Database resources of the BIG data center in 2018. Nucleic Acids Res. 2018;46:14–20. doi: 10.1093/nar/gkx897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Li P, Yang Z, Xu C. Genetic mapping of quantitative trait loci in crops. Crop J. 2017;5:175–184. doi: 10.1016/j.cj.2016.06.003. [DOI] [Google Scholar]
- Xue Y, Wan JM, Jiang L, et al. QTL analysis of aluminum resistance in rice (Oryza sativa L.) Plant Soil. 2006;287:375–383. doi: 10.1007/s11104-006-9086-3. [DOI] [Google Scholar]
- Yang Y, Zhang A, Chen Y, et al. Impacts of silicon addition on arsenic fractionation in soils and arsenic speciation in Panax notoginseng planted in soils contaminated with high levels of arsenic. Ecotoxicol Environ Saf. 2018;162:400–407. doi: 10.1016/j.ecoenv.2018.07.015. [DOI] [PubMed] [Google Scholar]
- Yorobe JM, Ali J, Pede VO, et al. Yield and income effects of rice varieties with tolerance of multiple abiotic stresses: the case of green super rice (GSR) and flooding in the Philippines. Agric Econ (United Kingdom) 2016;47:261–271. [Google Scholar]
- Yu LJ, Luo YF, Liao B, et al. Comparative transcriptome analysis of transporters, phytohormone and lipid metabolism pathways in response to arsenic stress in rice (Oryza sativa) New Phytol. 2012;195:97–112. doi: 10.1111/j.1469-8137.2012.04154.x. [DOI] [PubMed] [Google Scholar]
- Zhang J, Zhu YG, Zeng DL, et al. Mapping quantitative trait loci associated with arsenic accumulation in rice (Oryza sativa) New Phytol. 2008;177:350–355. doi: 10.1111/j.1469-8137.2007.02267.x. [DOI] [PubMed] [Google Scholar]
- Zhang M, Pinson SRM, Tarpley L, et al. Mapping and validation of quantitative trait loci associated with concentrations of 16 elements in unmilled rice grain. Theor Appl Genet. 2014;127:137–165. doi: 10.1007/s00122-013-2207-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao FJ, Ago Y, Mitani N, et al. The role of the rice aquaporin Lsi1 in arsenite efflux from roots. New Phytol. 2010;186:392–399. doi: 10.1111/j.1469-8137.2010.03192.x. [DOI] [PubMed] [Google Scholar]
- Zhao FJ, Ma JF, Meharg AA, McGrath SP. Arsenic uptake and metabolism in plants. New Phytol. 2009;181:777–794. doi: 10.1111/j.1469-8137.2008.02716.x. [DOI] [PubMed] [Google Scholar]
- Zhao F-J, McGrath SP, Meharg AA. Arsenic as a food chain contaminant: mechanisms of plant uptake and metabolism and mitigation strategies. Annu Rev Plant Biol. 2010;61:535–559. doi: 10.1146/annurev-arplant-042809-112152. [DOI] [PubMed] [Google Scholar]
- Zhou Q, Teng Y, Liu Y. A study on soil-environmental quality criteria and standards of arsenic. Appl Geochem. 2017;77:158–166. doi: 10.1016/j.apgeochem.2016.05.001. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Mean relative phenotypic values of traits measured under As stress. (XLSX 32 kb)
Figure S1. Distribution of SNPs on the chromosome. (PNG 49 kb)
Table S2. Total number of genes and SNPs identified in the candidate region. (XLSX 110 kb)
Table S3. Candidate genes present in the QTL intervals. (XLSX 12 kb)
Data Availability Statement
The comprehensive collected data supporting the conclusions of this review article are provided as figures, tables, and supplementary tables.