Abstract
Background
CircRNAs involved in the development of many diseases have been recently identified. However, the possible role of circRNAs in human hepatocellular carcinoma (HCC) remains to be investigated.
Objective
This study aimed to identify the expression of hsa_circ_0085616 in different HCC cell lines and its function in HCC tumorigenesis.
Methods
The expression of hsa_circ_0085616 was first measured in 68 pairs of HCC tissues and matched normal adjacent tissues. Further analysis was performed in different HCC cell lines and human normal hepatic cell lines. Moreover, we down- or upregulated its expression by cell transfection in vitro. Cell proliferation, migration, invasion and apoptosis were measured, and proliferation-related signaling pathway proteins were also analyzed.
Results
We found that hsa_circ_0085616 was highly expressed in all HCC cell lines compared to the normal liver cell line. Upregulation or downregulation of hsa_circ_0085616 expression could strengthen or weaken the proliferative ability of HCC cells in vitro. Moreover, the protein levels of β-catenin, p-ERK, and p-AKT, which play important roles in the typical proliferation pathway, were also affected by the expression of hsa_circ_0085616.
Conclusion
Our study indicated that hsa_circ_0085616 might be a potential biomarker and a new therapeutic target for HCC.
Keywords: circular RNA, hepatocellular carcinoma, tumor biomarker, hsa_circ_0085616
Introduction
Hepatocellular carcinoma (HCC) is the fifth most common cancer and the second most frequent cause of cancer-related death globally.1 Despite significant progress in detection and treatment of this disease, HCC still has a poor prognosis due to early recurrence.2 Nearly 40% of patients develop recurrences within the first year after hepatectomy.3 Thus, the development of new effective prognostic markers and therapeutic targets is urgently needed.
Circular RNA (circRNA) is a new type of noncoding RNA that was first discovered by Sanger in a virus using electron microscopy in the 1970s.4 Previous studies have shown that circRNAs exhibit cell-type- and developmental-stage-specific expression.5 CircRNAs might act as miRNA sponges and mature miRNAs, which have important regulatory functions in cell proliferation, differentiation and apoptosis. Therefore, circRNAs are believed to be involved in important cellular processes and tumor pathogenesis.6 In recent years, an increasing number of studies have revealed that circRNAs are involved in many malignant diseases.7–10 Recently, circRNAs were also shown to be involved in the development of HCC.11,12 However, the function of circRNAs in HCC remains poorly characterized.
In this study, we randomly selected four differentially expressed circRNAs (hsa_circ_0002124, hsa_circ_0000467, hsa_circ_0001459, hsa_circ_0085616) from the circRNA database CircBase (http://circbase.org/), which had been characterized by preliminary broad sequencing analysis (Table 1). We then focused our attention on circRNA hsa_circ_0085616, which is encoded by the ArfGAP With SH3 Domain, Ankyrin Repeat And PH Domain 1 (ASAP1) gene that is located at chr8:116700000–145138636. We selected hsa_circ_0085616 for investigation based on the high expression in tumor tissues and different tumor cell lines, while the three other circRNAs were not highly expressed. We found that circRNA hsa_circ_0085616 was differentially expressed between human liver tumor tissues and tumor adjacent tissues. We then observed high expression levels of hsa_circ_0085616 in all HCC cell strains (Figure 1). To further understand the role of hsa_circ_0085616 in the development of HCC, the function of hsa_circ_0085616 in the proliferation, migration and invasion of HCC and its possible molecular mechanisms/regulatory pathways in vitro were explored.
Table 1.
The four differently expressed circRNAs in human HCC samples
| CircRNA | Gene name | Chrom | HCC samples expression (TPM) | Normal samples expression (TPM) | Log2 ratio (T/N) | Up or down regulation | FDR |
|---|---|---|---|---|---|---|---|
| hsa_circ_0002124 | NUSAP1 | 15 | 135.364 | 9.723 | 3.799 | Up | 0.0002 |
| hsa_circ_0000467 | SKA3 | 13 | 485.719 | 14.585 | 5.058 | Up | 0.0000 |
| hsa_circ_0001459 | NEIL3 | 4 | 270.729 | 34.031 | 2.992 | Up | 0.0000 |
| hsa_circ_0085616 | ASAP1 | 8 | 621.083 | 170.156 | 1.868 | Up | 0.0000 |
Abbreviation: TPM, Transcripts per million.
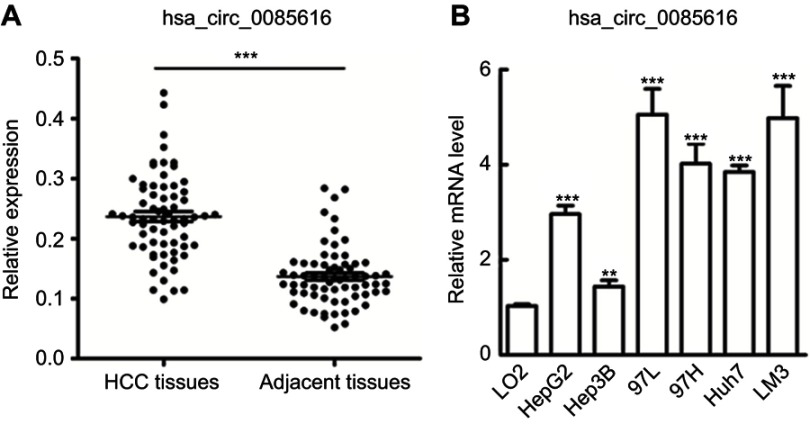
Figure 1.
Expression of hsa_circ_0085616 in clinical tissue samples and HCC cell lines. (A) Hsa_circ_0085616 was detected in 68 pairs of liver cancer tissues by RT-PCR. (B) Hsa_circ_0085616 was detected in different HCC cell lines and normal liver cell lines by RT-PCR. **p<0.01 and ***p<0.001 compared to controls.
Materials and methods
Clinical samples
The clinical specimens were retrospectively collected from the sample bank of Sun Yat-Sen Memorial Hospital of Sun Yat-Sen University (Guangzhou, China). Tumor tissues and tumor adjacent tissues were from 68 HCC patients who underwent surgical resection without any radiotherapy and chemotherapy before the surgery from July 2016 to December 2018. HBV infection was confirmed in all HCC patients.This study was approved by the Ethical Committee of Sun Yat-Sen Memorial Hospital (No. SYSU-IACUC-2019-B403). All patients provided written informed consent in accordance with the Declaration of Helsinki.
Cell lines and culture
The HCC cell lines HepG2, MHCC-97L, MHCC-97H, Huh7, and LM3 and the normal liver cell line L02 were purchased from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). Hep3B cells were obtained from the American Type Culture Collection (Manassas, VA, USA). All cell lines were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM; Gibco, USA) supplemented with 10% fetal bovine serum (FBS, Gibco USA), 100 U/mL penicillin, and 100 U/mL streptomycin and incubated at 37 °C under an atmosphere containing 5% CO2.
RNA extraction and qRT-PCR analyses
Total RNA was extracted by TRIzol reagent (Thermo Fisher Scientific, Inc., Waltham, MA, USA) in accordance with the manufacturer’s instructions. The expression levels of circRNAs and mRNAs were evaluated by qPCR using the SYBR Select Master Mix kit (Thermo Fisher Scientific, Inc., Waltham, MA, USA), and β-actin was used as the internal standard.
The following primers were used:
hsa_circ_0085616-CF 5ʹ-CACCGAGGACTACAACTCGC-3ʹ
hsa_circ_0085616-CR 5ʹ-GCCTCAGTGAAAACCATCTGC-3ʹ
β-actin-CF 5ʹ-AACTACCTTCAACTCCATCA-3ʹ
β-actin-CR 5ʹ-GAGCAATGATCTTGATCTTCA-3ʹ
Cell transfection
Among all the tumor cell lines, MHCC-97L and Hep3B were used for cell transfection. The small interfering RNAs (siRNAs) and plasmid (PcDNA3.1+) used for cell transfection were synthesized by General Biosystems (Hefei, China) and GenePharma Co., Ltd. (Shanghai, China). Target sequences of siRNAs were as follows: SiRNA#1 (5ʹ-3ʹ) GCTTCCTGGTCTCTGATAT, SiRNA#2 (5ʹ-3ʹ) GGTCTCTGATATCAGCTCT. Plasmids and siRNAs were transfected into MHCC-97L or Hep3B cells using Lipofectamine 3000 Transfection Reagent (Life Technologies). Then, the cellular functions, including cell viability, metastasis ability and apoptosis, were evaluated.
MTS cell proliferation and transwell assays
Cell proliferation was measured using the MTS CellTiter 96 AQueous One Solution Cell Proliferation Assay (Promega, Madison, WI, USA). Cells were seeded at 1×104 per well in 96-well plates after transfection and incubated for 24, 48, and 72 hrs, and then, the absorbance at 490 nm was measured according to the manufacturer’s protocol.
Cell migration and invasion were determined using a transwell system (Corning, NY, USA). MHCC-97L and Hep3B cells (1.0 × 104) were transfected with siRNAs or plasmid for 24 h and then seeded in the upper chamber and allowed to migrate or invade toward the chamber. After 12 h (for migration assays without Matrigel coating) or 24 h (for invasion assays with Matrigel coating), the migrated or invaded cells on the underside of the membrane were counted, and the relative migration or invasion was determined.
Cell apoptosis
After transfection, tumor cells were washed with 10% PBS twice and were switched to 1×106/mL binding buffer before resuspension. Cells were incubated under dark conditions for 8 min after adding 7-AAD and Annexin V (BD Biosciences, Heidelberg, Germany) and then examined by flow cytometry on an Accuri C6 Flow Cytometer (BD Biosciences, Heidelberg, Germany).
Western blotting
Cultured cells were collected and lysed with RIPA buffer containing the protease inhibitors on ice for 30 min. Protein was separated by SDS-PAGE, transferred onto a nitrocellulose membrane (Pall Co., USA) and probed with primary antibodies, including GAPDH as the internal standard, and then with horseradish peroxidase-labeled secondary antibodies. The blots were incubated with antibodies against β-catenin (1:5000, rabbit monoclonal, ab32572, Abcam, Inc.), p-ERK (1:500, rabbit polyclonal, ab79483, Abcam, Inc.), ERK (1:1000, rabbit monoclonal, ab32537, Abcam, Inc.), p-AKT (1:5000, rabbit monoclonal, ab81283, Abcam, Inc.), AKT (1:5000, rabbit monoclonal, ab179463, Abcam, Inc.). GAPDH (1:5000, rabbit monoclonal, ab181602, Abcam, Inc.) was used as a loading control. Protein levels were detected by a chemiluminescence approach according to the protocol (ECL, Forevergen, Guangzhou, China). Images were analyzed using ImageJ software (NIH, USA). The proteins were represented by using GAPDH loading as normalization.
Statistical analysis
Statistical analyses were performed using GraphPad Prism version 5.00 (GraphPad Software, San Diego, CA). Student’s t–test was performed when two groups were compared. One-way ANOVA was applied to compare differences among multiple groups. p-values lower than 0.05 were considered significantly different.
Results
Expression of hsa_circ_0085616 in HCC and normal liver tissues and HCC cell lines
A published study showed that increased expression of the ASAP1 gene was associated with the proliferation and invasion of HCC with potential oncogenic activity.13 The hsa_circ_0085616 expression profile was first measured in 68 pairs of HCC tumor tissues and pair-matched normal adjacent tissues. We observed upregulation of hsa_circ_0085616 in tumor tissues compared to the adjacent tissues (Figure 1A). We further observed the expression of hsa_circ_0085616 in the normal cell line L02 and HCC cell lines, including HepG2, MHCC-97L, MHCC-97H, Huh7, LM3, and Hep3B. RT-PCR results showed that all the liver cancer cell lines expressed higher levels of hsa_circ_0085616 than the L02 normal liver cell line. Meanwhile, Hep3B showed the lowest level of hsa_circ_0085616 in all cancer cell lines, while the MHCC-97L and LM3 cell lines showed higher expression levels (Figure 1B).
Correlation between the expression of hsa_circ_0085616 and clinical characteristics in patients
The clinicopathological characteristics of the 68 patients and the relative expression levels of hsa_circ_0085616 are presented in Table 2. The expression of hsa_circ_0085616 was detected in 68 pairs of HCC tissues and adjacent noncancerous tissues by qRT-PCR, and the relative expression was the expression level of cancer tissue compared with that of normal tissue. The data showed that hsa_circ_0085616 obviously increased in patients with higher TNM stage (p=0.0071) and poor differentiation (p=0.0053). However, no association between its expression and other clinical characteristics was observed.
Table 2.
Clinicopathological characteristics of the patients with hepatocellular carcinoma (n=68) and relative expression levels of hsa_circ_0085616
| N=68 | Relative expression | p-value (*p<0.05) | |
|---|---|---|---|
| Age (years) | |||
| <60 | 23 | 1.674 | 0.775 |
| ≥60 | 45 | 1.699 | |
| Gender | |||
| Female | 12 | 1.644 | 0.802 |
| Male | 56 | 1.673 | |
| Tumor size(cm) | |||
| ≥5 | 30 | 1.662 | 0.884 |
| <5 | 38 | 1.651 | |
| Cirrhosis | |||
| Positive | 57 | 1.681 | 0.653 |
| Negtive | 11 | 1.647 | |
| TNM stage | |||
| T1+T2 | 32 | 1.589 | 0.0071* |
| T3+T4 | 36 | 1.752 | |
| Differentiation | |||
| Well | 7 | 1.582 | 0.0053* |
| Moderate | 38 | 1.654 | |
| Poor | 23 | 1.761 |
Downregulated expression of hsa_circ_0085616 suppressed tumor cell aggressive characteristics
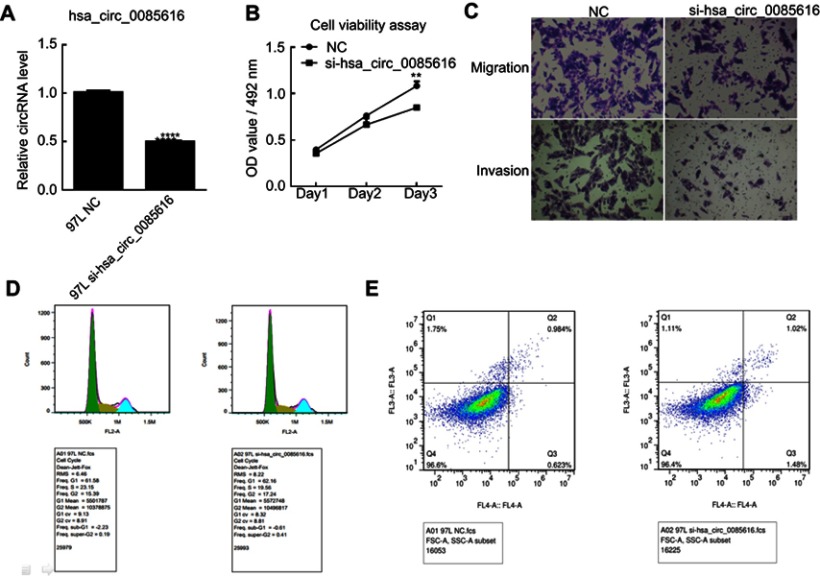
To investigate the role of has_circ_0085616 in HCC cancer cell growth, the MHCC-97L (97L) cell line was chosen according to the RT-PCR results. The cells were transfected with siRNA using Lipofectamine 3000 Transfection Reagent. The RT-PCR results indicated that hsa_circ_0085616 was effectively downregulated in the 97L cells (Figure 2A). Then, the cellular functions, including cell viability, metastasis ability and apoptosis, were evaluated. The MTS assay indicated that si-hsa_circ_0085616 visibly downregulated the proliferation of 97L cells compared to the normal control (Figure 2B). The results of the transwell assay showed that the migration of 97L cells was reduced in transfected cells compared to negative control cells. Similarly, the invasiveness of transfected cells was weakened (Figure 2C). Furthermore, we investigated the effect of hsa_circ_0085616 on the cell cycle and apoptosis and measured the percentage of tumor cells in all cell cycle stages, including G0, G1, S G2, and M. Downregulation of hsa_circ_0085616 decreased the percentage of cells in G2/M and S, while it increased the percentage of cells in G1/G0 stage (Figure 2D). For the apoptosis of transfected cells, apoptosis increased only slightly, with no significant difference compared to the negative control (1.02% vs 0.984, p>0.05, Figure 2E).
Figure 2.
Expression of hsa_circ_0085616 in the MHCC 97L cell line was downregulated by siRNA. (A) MHCC 97L cells transfected with si-hsa_circ_0085616 had significantly decreased expression of hsa_circ_0085616 compared with the negative control. (B) The proliferation of 97L cells decreased when cells were transfected with si-hsa_circ_0085616. (C) The transwell assay showed that the migration and invasion of 97L cells were suppressed when cells were transfected with si-hsa_circ_0085616. (D) Flow cytometry results showed that more 97L transfected cells stayed at G1/G0 stage than S and G2/M stage. (E) Flow cytometry results showed that 97L cell apoptosis increased after transfection with siRNA. **p<0.01 and ****p<0.0001 compared to controls.
Abbreviation: NC, Negative control.
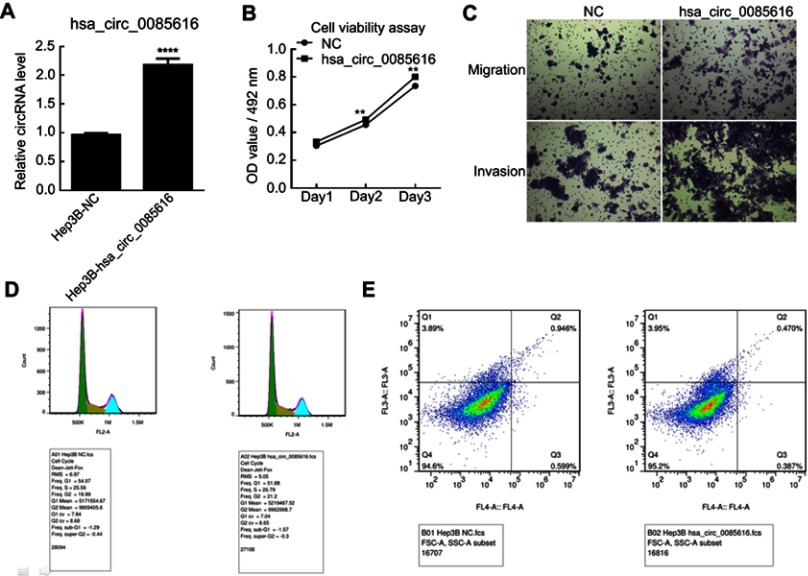
Overexpression of hsa_circ_0085616 promotes HCC cell line proliferation and migration
We observed that hsa_circ_0085616 was effectively upregulated in Hep3B cells after transfection by plasmid according to the RT-PCR results (Figure 3A). The MTS assay also showed that the proliferation of Hep3B cells increased after transfection (Figure 3B). The results of the transwell assay demonstrated that upregulated hsa_circ_0085616 could enhance the migration and invasion potential of HCC cells (Figure 3C). Next, we examined the effects of upregulated hsa_circ_0085616 on apoptosis and the cell cycle. We found that the percentage of tumor cells in G1/G0 was reduced, while the percentage of cells in S and G2/M conversely increased (Figure 3D). Apoptosis of transfected Hep3B cells was significantly reduced compared to the normal control group (0.470% vs 0.946%, p<0.05, Figure 3E).
Figure 3.
The expression of hsa_circ_0085616 in the Hep3B cell line was upregulated by plasmid. (A) Hep3B cells transfected with plasmid had significantly increased expression of hsa_circ_0085616 compared with the negative control. (B) The proliferation of Hep3B cells increased when cells showed overexpression of hsa_circ_0085616. (C) The transwell assay showed that the migration and invasion of transfected Hep3B cells were promoted when compared to the negative control. (D) Flow cytometry results showed that more Hep3B-transfected cells stayed at S and G2/M stage than G1/G0 stage. (E) Flow cytometry results showed that Hep3B cell apoptosis decreased after transfection with plasmid. **p<0.01 and ****p<0.0001 compared to controls.
Abbreviation: NC, Negative control.
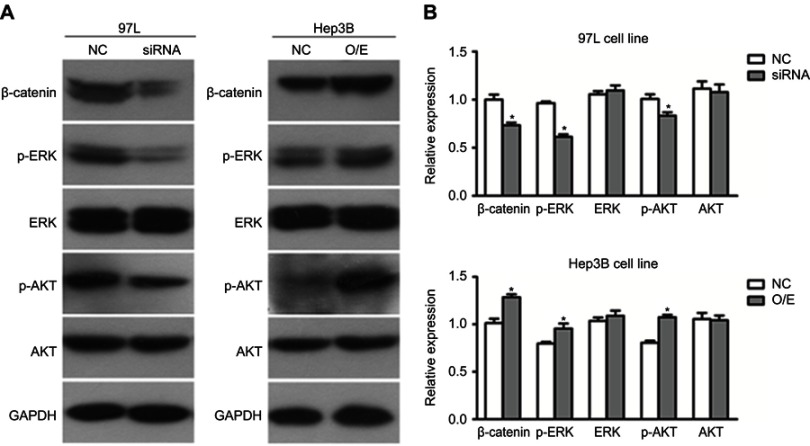
Potential signaling pathways of hsa_circ_0085616 to promote tumor cell proliferation
To further explore the underlying molecular mechanism and illustrate the possible function of hsa_circ_0085616 in HCC, we also tested some important signaling pathways of cell proliferation at the protein level by Western blot analysis (Figure 4A). Suppressed expression of hsa_circ_0085616 decreased the protein levels of β-catenin, p-ERK, and p-AKT, while upregulation of hsa_circ_0085616 increased the levels (Figure 4B). These results indicated that hsa_circ_0085616 may ultimately promote cell proliferation through these typical signaling pathways.
Figure 4.
Hsa_circ_0085616 modified growth and migration-associated proteins. Proteins highly associated with cell growth and migration were detected by Western blot. (A) Western blotting analysis showed that the protein levels of p-ERK, p-AKT and β-catenin were affected in 97L Hep3B cells after transfection. (B) Relative expression of proteins of the 97L and Hep3B cell lines. *p<0.05 compared to controls.
Abbreviations: NC, Negative control; O/E, Overexpression.
Discussion
In our present study, the results indicated that hsa_circ_0085616 expression was remarkably higher in HCC cancer tissues than matched adjacent normal tissues using RT-PCR. High expression of hsa_circ_0085616 was further identified in different HCC cell lines. After downregulation and overexpression of hsa_circ_0085616, we further confirmed its role in cellular functions and the associated underlying mechanism in HCC.
CircRNAs are mostly generated from exons of protein-coding genes, and in many cases, they are more abundant than the linear product from their host gene. The importance of circRNAs in gene regulation and normal cellular homeostasis is now beginning to be recognized.14 CircRNAs are believed to play very important roles in different diseases, especially tumorigenesis.15 There is increasing interest in exploring the potential role of circRNAs as novel biomarkers in human cancers.16 Recently, some studies have examined the role of circRNAs in the survival rate of patients with HCC.17,18 However, the use of circRNAs for the diagnosis and prognosis of patients with HCC remains largely unknown.
Here, by assessing clinical specimens, including tumor tissues and tumor adjacent tissues, of 68 HCC patients, we identified and explored the role of hsa_circ_0085616 in the development of HCC in vitro. To date, emerging studies have focused on the association between HCC and circRNAs,11,19,20 but no previous studies have reported the role of hsa_circ_0085616 in any disease. Our studies showed that overexpression of hsa_circ_0085616 could promote the proliferation, migration, and invasion of HCC cancer cells in vitro. Meanwhile, overexpression of hsa_circ_0085616 could inhibit the apoptosis of HCC and drove the cell cycle into S/G2 phase. When the expression of hsa_circ_0085616 was suppressed by siRNA, all of these tumor cell aggressive characteristics were weakened. These findings suggested that hsa_circ_0085616 is a potential marker that will benefit HCC cancer diagnosis and therapy.
To further understand the mechanism, we investigated whether several typical tumor growth signaling pathways were associated with hsa_circ_0085616 at the protein level in this study. We observed that β-catenin, p-ERK, and p-AKT protein levels were closely associated with hsa_circ_0085616 expression. These results indicated that hsa_circ_0085616 may ultimately promote tumor cell growth through typical MAPK/ERK, P13K/AKT or Wnt/beta-catenin pathways. The PI3K/AKT signaling pathway is the most common pathway in cell proliferation, migration and invasion.21 The MAPK pathway, which regulates cell growth, division and death, represents a cascade of phosphorylation events, including three pivotal kinases, namely, Raf, MEK (MAP kinase kinase), and ERK (MAP kinase).22 Wnt signaling was identified for its role in carcinogenesis; it controls body axis patterning, cell fate specification, cell proliferation and cell migration.23
Our results first demonstrated that hsa_circ_0085616 may play an essential role in the development of HCC progression. Notably, this study presents several limitations. In vitro experiments cannot fully explain circRNA function, and in vivo animal experiments are needed. In particular, a knockdown mouse model may provide more reliable and solid evidence for this conclusion. In addition, the upstream molecular mechanisms need to be further explored.
Conclusion
Overall, we observed high expression of hsa_circ_0085616 in both liver cancer tissues and HCC cell lines, which might affect liver cancer cell proliferation, migration, invasion and apoptosis. Hsa_circ_0085616 could play an essential role in the development of HCC, indicating it may become a new effective therapy target or prognostic marker for HCC.
Acknowledgments
This work was supported by grants from the Natural Science Foundation of Guangdong Province, China (No. 2018A030310118); the Natural Science Foundation of Guangdong Province, China (No. 2016A030310207); and the National Science Foundation for Young Scientists of China (No. 81702406).
Abbreviations
ERK, extracellular-signal-regulated kinase; AKT, protein kinase B.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.European Association for the Study of the Liver. Electronic address eee, European association for the study of the L. EASL clinical practice guidelines: management of hepatocellular carcinoma. J Hepatol. 2018;69(1):182–236. doi: 10.1016/j.jhep.2018.03.019 [DOI] [PubMed] [Google Scholar]
- 2.Balogh J, Victor D 3rd, Asham EH, et al. Hepatocellular carcinoma: a review. J Hepatocell Carcinoma. 2016;3:41–53. doi: 10.2147/JHC.S61146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fan ST, Lo MC, Poon RT, et al. Continuous improvement of survival outcomes of resection of hepatocellular carcinoma: a 20-year experience. Ann Surg. 2011;253(4):745–758. doi: 10.1097/SLA.0b013e3182111195 [DOI] [PubMed] [Google Scholar]
- 4.Sanger HL, Klotz G, Riesner D, Gross HJ, Kleinschmidt AK. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc Natl Acad Sci U S A. 1976;73(11):3852–3856. doi: 10.1073/pnas.73.11.3852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Qu S, Yang X, Li X, et al. Circular RNA: a new star of noncoding RNAs. Cancer Lett. 2015;365(2):141–148. doi: 10.1016/j.canlet.2015.06.003 [DOI] [PubMed] [Google Scholar]
- 6.Yang Z, Xie L, Han L, et al. Circular RNAs: regulators of cancer-related signaling pathways and potential diagnostic biomarkers for human cancers. Theranostics. 2017;7(12):3106–3117. doi: 10.7150/thno.19016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu ZQ, Yang MG, Liu HJ, Su CQ. Circular RNA hsa_circ_0003221 (circPTK2) promotes the proliferation and migration of bladder cancer cells. J Cell Biochem. 2018;119(4):3317–3325. doi: 10.1002/jcb.26492 [DOI] [PubMed] [Google Scholar]
- 8.Xu XW, Zheng BA, Hu ZM, et al. Circular RNA hsa_circ_000984 promotes colon cancer growth and metastasis by sponging miR-106b. Oncotarget. 2017;8(53):91674–91683. doi: 10.18632/oncotarget.21748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luo YH, Zhu XZ, Huang KW, et al. Emerging roles of circular RNA hsa_circ_0000064 in the proliferation and metastasis of lung cancer. Biomed Pharmacother. 2017;96:892–898. doi: 10.1016/j.biopha.2017.12.015 [DOI] [PubMed] [Google Scholar]
- 10.Huang M, He YR, Liang LC, Huang Q, Zhu ZQ. Circular RNA hsa_circ_0000745 may serve as a diagnostic marker for gastric cancer. World J Gastroenterol. 2017;23(34):6330–6338. doi: 10.3748/wjg.v23.i34.6330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kou P, Zhang C, Lin J, Wang H. Circular RNA hsa_circ_0078602 may have potential as a prognostic biomarker for patients with hepatocellular carcinoma. Oncol Lett. 2019;17(2):2091–2098. doi: 10.3892/ol.2018.9863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang C, Zhang C, Lin J, Wang H. Circular RNA Hsa_Circ_0091579 serves as a diagnostic and prognostic marker for hepatocellular carcinoma. Cell Physiol Biochem. 2018;51(1):290–300. doi: 10.1159/000495230 [DOI] [PubMed] [Google Scholar]
- 13.Wang ZG, Zheng H, Gao W, et al. eIF5B increases ASAP1 expression to promote HCC proliferation and invasion. Oncotarget. 2016;7(38):62327–62339. doi: 10.18632/oncotarget.11469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haque S, Harries LW. Circular RNAs (circRNAs) in Health and Disease. Genes (Basel). 2017;8(12):353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Patop IL, Kadener S. circRNAs in cancer. Curr Opin Genet Dev. 2018;48:121–127. doi: 10.1016/j.gde.2017.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang M, Yang Y, Xu J, Bai W, Ren X, Wu H. CircRNAs as biomarkers of cancer: a meta-analysis. BMC Cancer. 2018;18(1):303. doi: 10.1186/s12885-018-4242-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qiu LP, Wu YH, Yu XF, Tang Q, Chen L, Chen KP. The emerging role of circular RNAs in hepatocellular carcinoma. J Cancer. 2018;9(9):1548–1559. doi: 10.7150/jca.24566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fu L, Jiang Z, Li T, Hu Y, Guo J. Circular RNAs in hepatocellular carcinoma: functions and implications. Cancer Med. 2018. doi: 10.1002/cam4.1574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang X, Zhou H, Jing W, et al. The circular RNA hsa_circ_0001445 regulates the proliferation and migration of hepatocellular carcinoma and may serve as a diagnostic biomarker. Dis Markers. 2018;2018:3073467. doi: 10.1155/2018/3073467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang M, Yu F, Li P. Circular RNAs: characteristics, function and clinical significance in hepatocellular carcinoma. Cancers (Basel). 2018;10(8):258. doi: 10.3390/cancers10110400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hemmings BA, Restuccia DF. The PI3K-PKB/Akt pathway. Cold Spring Harb Perspect Biol. 2015;7:4. doi: 10.1101/cshperspect.a026609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hilger RA, Scheulen ME, Strumberg D. The Ras-Raf-MEK-ERK pathway in the treatment of cancer. Onkologie. 2002;25(6):511–518. doi: 10.1159/000068621 [DOI] [PubMed] [Google Scholar]
- 23.Klaus A, Birchmeier W. Wnt signalling and its impact on development and cancer. Nat Rev Cancer. 2008;8(5):387–398. doi: 10.1038/nrc2389 [DOI] [PubMed] [Google Scholar]