Abstract
Purpose:
Prior molecular profiling of hepatocellular carcinoma (HCC) has identified actionable findings that may have a role in guiding therapeutic decision-making and clinical trial enrollment. We implemented prospective next-generation sequencing (NGS) in the clinic to determine whether such analyses provide predictive and/or prognostic information for HCC patients treated with contemporary systemic therapies.
Experimental Design:
Matched tumor/normal DNA from patients with HCC (N = 127) were analyzed using a hybridization capture-based NGS assay designed to target 341 or more cancer-associated genes. Demographic and treatment data were prospectively collected with the goal of correlating treatment outcomes and drug response with molecular profiles.
Results:
WNT/β-catenin pathway (45%) and TP53 (33%) alterations were frequent and represented mutually exclusive molecular subsets. In sorafenib-treated patients (n = 81), oncogenic PI3K-mTOR pathway alterations were associated with lower disease control rates (DCR, 8.3% vs. 40.2%), shorter median progression-free survival (PFS; 1.9 vs. 5.3 months), and shorter median overall survival (OS; 10.4 vs. 17.9 months). For patients treated with immune checkpoint inhibitors (n = 31), activating alteration WNT/β-catenin signaling were associated with lower DCR (0% vs. 53%), shorter median PFS (2.0 vs. 7.4 months), and shorter median OS (9.1 vs. 15.2 months). Twenty-four percent of patients harbored potentially actionable alterations including TSC1/2 (8.5%) inactivating/truncating mutations, FGF19 (6.3%) and MET (1.5%) amplifications, and IDH1 missense mutations (<1%). Six percent of patients treated with systemic therapy were matched to targeted therapeutics.
Conclusions:
Linking NGS to routine clinical care has the potential to identify those patients with HCC likely to benefit from standard systemic therapies and can be used in an investigational context to match patients to genome-directed targeted therapies.
Introduction
Hepatocellular carcinoma (HCC), a leading worldwide cause of cancer-related morbidity and mortality, frequently presents as incurable liver-limited or widespread metastatic disease (1, 2). Systemic treatment options include several multitargeted tyrosine kinase inhibitors (TKI) with meaningful survival advantages over best supportive care (3–5), as well as immune checkpoint inhibitors with documented durable antitumor activity (6). Nevertheless, the majority of patients will not respond to standard systemic agents or ultimately progress on these therapies. Thus, the development and validation of biomarkers to aid in treatment selection for this heterogenous disease remains of critical importance.
The genetic landscape of hepatocellular carcinoma has been studied extensively (7–16). This body of work has identified recurrent somatic oncogenic drivers or tumor suppressors in several cellular pathways (i.e., telomere maintenance, WNT signaling, cell-cycle control, chromatin remodeling, TP53, PI3K-TOR and MAPK pathways), differential genomics subsets, etiologic-dependent genomic heterogeneity, and several potential therapeutic targets. The vast majority of profiled tumor specimens though were collected from patients with early-stage disease, lack response, and disease-specific outcome data following treatment with contemporary therapies (i.e., TKIs and immune checkpoint inhibitors), or were not analyzed in real-time, and thus could not have been used to inform clinical management.
The purpose of this study was to demonstrate the clinical utility of prospective next-generation sequencing in patients with advanced HCC, to explore potential predictive and prognostic genomic biomarkers of treatment response in this population, and to define the value of sequencing in matching patients to molecularly driven therapeutics.
Materials and Methods
Patient selection
Patients were identified from August 2014 through August 2017 and were eligible if they had a confirmed histologic diagnosis of conventional HCC. Those with fibrolamellar variant or biphenotypic morphology such as mixed HCC-cholangiocarcinomas were excluded. Written informed consent for tumor profiling was obtained from each patient on a prospective genotyping protocol (clinicaltrials.gov, NCT01775072) that was approved by the Memorial Sloan Kettering Cancer Center (MSKCC) Institutional Review Board. The study was conducted in accordance with the U.S. Common Rule.
The electronic medical record was reviewed to extract information on patient sex, date of birth, race, HCC etiologic factor [hepatitis B virus (HBV), hepatitis C virus (HCV), and nonviral], date of diagnosis, tumor histology, histologic grade, specimen location (liver, local recurrence, or extrahepatic metastasis), extent of disease [Barcelona Clinic Liver Cancer (BCLC) staging and proportion with intrahepatic, extrahepatic, and/or gross vascular invasion], treatment history (transplantation, surgical resection, and/or regional therapy), Child-Pugh (CP) classification, alphafetoprotein (AFP, ng/mL), type, number and dates of systemic therapy annotated with radiographic response per the treating physician,thelastdateof follow-up ordateof death, andvital status.
Genomic analysis
All tumor samples were reviewed and confirmed as HCC by at least two gastrointestinal pathologists and with diagnostically challenging cases review was completed at gastrointestinal pathology consensus conference. After pathologic review, formalin-fixed paraffin-embedded tissue blocks were sectioned (5–20 μm) and macrodissected when appropriate. Tissue was deparaffinized and DNA extracted using DNeasy Blood & Tissue Kit (Qiagen).
Genomic DNA from tumor tissue and patient-matched normal blood was subjected to targeted NGS using MSK-IMPACT, a custom, deep-coverage targeted sequencing assay authorized by the New York State Department of Health and the FDA as a clinical test (17). The standard input of DNA was 250 ng, and a DNA minimum input of 50 ng was used in cases where DNA quantity was limited. Briefly, barcoded DNA libraries from tumor and normal samples were captured using custom oligonucleotide probes, sequenced on an Illumina HiSeq 2500 platform, and subjected to a custom analysis pipeline to identify single-nucleotide variants, small indels (<30 bp), copy number alterations (CNA), and selected structural rearrangements in all exons and select introns of 341 or more known cancer-associated genes, as described previously (17). As the assay was expanded during the study period, 341, 410, or 468 genes were interrogated in 21, 70, and 36 cases, respectively (Supplementary Table S1). All candidate variants were manually reviewed using the Integrative Genomics Viewer (18). Testing was performed in a CLIA-certified clinical laboratory.
Tumor mutation burden (TMB) per sample was calculated as the total number of nonsynonymous mutations divided by the actual number of bases analyzed. MSIsensor scores were calculated for all cases and represent the percentage of unstable microsatellites of all tested microsatellites (19). MSIsensor interrogates the aligned sequencing data for available microsatellite regions with sufficient coverage in a tumor/normal pair where it identifies deletion length variation. χ2 test is used to identify significantly varied loci, and the percentage of unstable loci, after multiple testing correction on the P values, is reported as a MSIsensor score. Tumors with values ≥10 were defined as microsatellite instability-high (MSI-H) status.
Genomic alterations were filtered for oncogenic variants using OncoKB (20), a precision oncology knowledge-base that tracks the effects of cancer variants and their potential clinical action-ability (http://oncokb.org). Genomic alterations were classified as actionable using a level of evidence scale of 1 to 4, where Level 1 −2A alterations indicated a standard therapeutic intervention and levels 2B-4 included investigational therapeutic alterations, which may direct a patient toward a clinical trial relevant to that biomarker.
All data are available for visualization and analysis via the cBioPortal for Cancer Genomics (http://cbioportal.org/study?id=hcc_mskimpact_2018; refs. 21, 22), and variant calls can be downloaded from the cBioPortal DataHub (https://github.com/cBioPortal/datahub).
Biostatistics
All HCC tumor samples sequenced over the prospective sampling period were included in the analysis. Two-sided Fisher exact test was performed to compare the frequency of oncogenic alterations to clinicopathologic characteristics within the MSK cohort and to those of The Cancer Genome Atlas (TCGA) for HCC (7). Mutual exclusivity was assessed by calculating the OR and FDR- corrected P value for each gene alteration. In a descriptive fashion, TMB for all solid tumors in cBioPortal for Cancer Genomics was determined to illustrate the relative degree of hypermutation for HCC in comparison with other solid tumors.
Radiographic response was adjudicated by the treating physician in the case of patients treated with sorafenib and by RECIST version 1.1 for patients treated with immune checkpoint inhibitors, everolimus, or other investigational agents (23). Patients who received concurrent regional therapy such as embolization or radiotherapy were excluded from clinical correlation due to an inability to attribute objective response to systemic therapy. The objective response rate (ORR) was defined as the proportion of confirmed complete responses (CR) or partial responses (PR) in all evaluable patients for a specific therapy. The disease control rate (DCR) was defined as the proportion of CR, PR, and stable disease (SD) for ≥ 4 months in all evaluable patients for a specific therapy. Fisher exact test was performed to assess the impact of select clinicopathologic features and gene pathway alterations on response. Wilcoxon rank-sum test was used to assess the impact of median TMB on objective response to immune checkpoint inhibitors.
Progression-free survival (PFS) and overall survival (OS) for patients treated with sorafenib and immune checkpoint inhibitors was calculated from the date of start of treatment to the date of radiographic disease progression, death, or last evaluation. PFS and OS were calculated using Kaplan-Meier methodology. Given the sample size of our cohort against the total number of evaluated genes (>341), multiple comparisons for each altered gene would diminish power and interfere with meaningful interpretation. We therefore grouped genes into known oncogenic signaling pathways operant in HCC, specifically the TP53 pathway, WNT signaling, cell-cycle control, chromatin remodeling, PI3K-TOR, and the MAPK pathways (24), and investigated associations between these pathways and PFS and OS by log-rank test. HRs were report with 95% confidence interval (95% CI). P value of ≤0.05 were considered significant. Statistical analysis was completed using Rv3.4.3 (https://www.R-project.org).
Results
Landscape of genomic alterations in HCC
Over the study period, a total of 134 HCC samples were analyzed from 127 unique HCC patients. In comparison with the TCGA dataset of surgically resected, liver-limited tumors, the MSK cohort was enriched with specimens from patients who were not eligible for curative resection or transplantation (61%), and included, a subset of locally recurrent/extrahepatic (16%) tumors (Fig. 1A; Table 1; ref. 7). Similar to theTCGA, the majority of the MSK cohort were male (79%) of non-Asian descent (76%) with equal proportions of virally mediated (51%) and nonvirally associated (49%) HCC.
Figure 1.
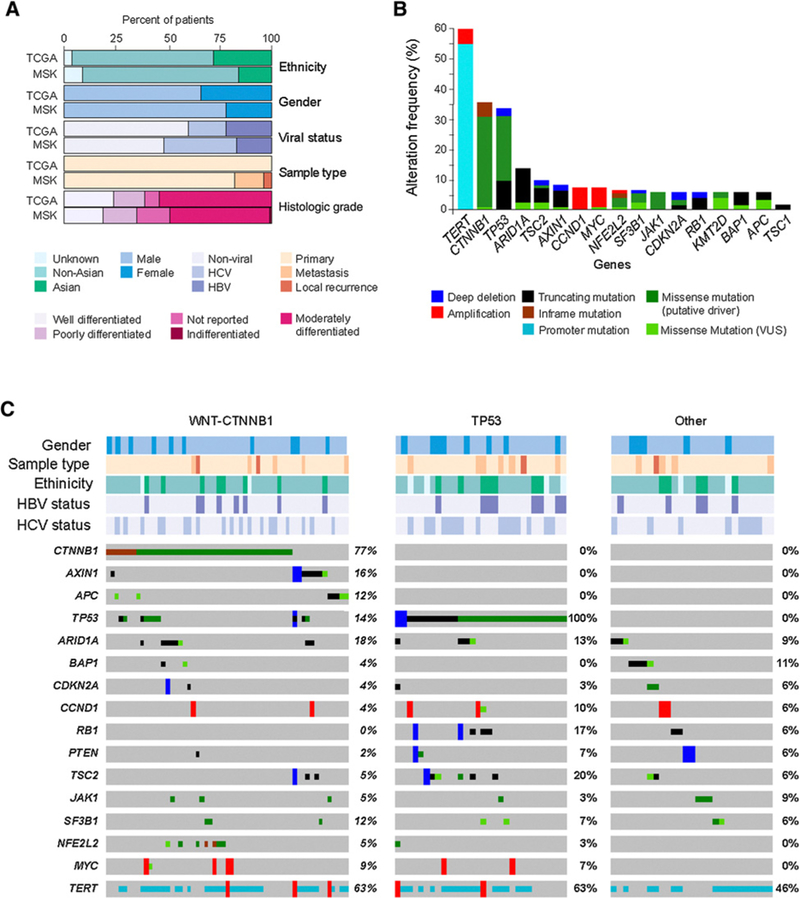
Landscape of genomic alterations in HCC. A, Comparison of the clinical characteristics of the MSK HCC (N = 127) and the TCGA HCC cohorts (N = 196). B, The most common genomic alterations in liver cancer listed in order of frequency. C, Oncoprint of commonly altered genes. Tumors are split into three groups (WNT/β-catenin pathway altered, TP53 altered, and other). Genes were grouped by pathway/function, and select clinicopathologic parameters are shown for all tumors. Clinicopathologic features and mutations were color coded by type. VUS, variant of unknown significance.
Table 1.
Cohort demographics (N = 127)
| Age, median, (range) | 65 (15–85) |
| Sex, male, N (%) | 100 (0.79) |
| Ethnicity, N (%) | |
| Asian | 20 (0.16) |
| Non-Asian | 96 (0.76) |
| Unknown | 12 (0.09) |
| Etiology, N (%) | |
| HBV | 22 (0.17) |
| HCV | 45 (0.35) |
| Nonviral | 62 (0.49) |
| BCLC stage, N (%) | |
| A | 37 (0.29) |
| B | 51 (0.40) |
| C | 38 (0.30) |
| Unknown | 2 (0.02) |
| Histologic grade, N (%) | |
| Well | 24 (0.19) |
| Moderate | 61 (0.48) |
| Poor | 20 (0.16) |
| Undifferentiated | 1 (0.01) |
| Not reported | 22 (0.17) |
| HCC-directed therapy, N (%) | |
| Hepatic allograft | 5 (0.04) |
| Surgical resection | 45 (0.35) |
| Regional therapy | 96 (0.76) |
| Systemic therapy | 86 (0.67) |
| Tumor purity, median (range) | 60 (10–90) |
| Tumor site, N (%) | |
| Liver | 107 (0.84) |
| Local recurrence | 4 (0.03) |
| Extrahepatic metastasis | 16 (0.13) |
Eight-four (63%) of 134 specimens were obtained from core biopsies, while the remaining samples were obtained from resection or hepatic explants. Median estimated tumor purity for sequenced samples was 60% (range, 10%−90%) and a mean sequencing coverage of 690× was achieved (minimum 120×). We identified an average of 4.7 nonsynonymous mutations per tumor sample (range, 1–14).
Genomic alterations were observed in 127 of 134 unique samples (95%). The most frequently mutated genes occurring in ≥10% of all samples included TERT (55.6%), CTNNB1 (35.7%), TP53 (32.5%), ARID1A (12.7%), and TSC2 (10.6%; Fig. 1B). In addition to mutations in CTNNB1, alterations in canonical members of the WNT pathway such as AXIN1 (6.3%), AXIN2 (0.05%), and APC (5.2%) were also identified. CNA included amplifications in CCND1/FGF19 (7%), MYC (6.1%), MET (1.5%) and VEGFA (<1%), and loss in HLA-B (2.7%). Structural alterations, specifically fusion events, were uncommon. Median TMB in HCC was 4.08. When compared with other solid histologies, the variance in TMB was low (SD = 2.71), second only to germ cell tumors (SD = 1.75), and MSI-H and hypermutation were not observed in this tumor type in the MSK cohort (Supplementary Fig. S1). Despite differences in the baseline clinical characteristics between the MSK and TCGA cohorts, a comparison of mutational frequencies did not reveal a significant difference between the relative proportion of altered genes (Supplementary Table S2).
Consistent with prior tumor genomic profiling efforts, TP53 mutations were rarely seen in conjunction with WNT pathway alterations (P = 0.003), effectively delineating at least two major genomic subsets (Fig. 1C). Clinicopathologic features, including sex, ethnicity, specimen location (liver limited, local recurrence, and extrahepatic metastasis), and histologic grade did not associate with genomic alterations. Virally mediated HCC, when compared with nonvirally mediated HCC, was enriched for TP53 mutations (45% vs. 21%, P = 0.001) and was numerically less commonly associated with WNT pathway alterations (41% vs. 60%, P = 0.7).
Genomic determinants of response to sorafenib
Of the 127 unique HCC patients, 87 (68.5%) received systemic therapy while 38 (29.9%) were not candidates for systemic treatment as these patients were free of disease, undergoing serial embolization, or were unfit for systemic treatment. Two patients (1.6%) had missing data regarding systemic therapy. Eighty-one patients were treated with sorafenib, a multitargeted TKI, as a standard of care or in the context of a clinical trial (Table 2). Two patients were excluded from analysis as they were lost to follow- up. The median sorafenib PFS for the cohort overall was 4.8 months, which is similar to contemporary large first-line pivotal studies of sorafenib (3.6 months; ref. 5), and the median OS was a favorable 16.4 months (12.2 months; ref. 5).
Table 2.
Demographics of patients with advanced HCC treated with sorafenib (N = 81)
| Age, median, (range) | 65.2 (16–91) |
| Sex, male, N (%) | 61 (0.75) |
| Race, N (%) | |
| Asian | 15 (0.19) |
| Non-Asian | 66 (0.81) |
| Etiology, N (%) | |
| HCV | 26 (0.32) |
| HBV | 16 (0.20) |
| Nonviral | 40 (0.49) |
| BCLC stage, N (%) | |
| A | 0 (0.00) |
| B | 14 (0.17) |
| C | 67 (0.83) |
| Extent of disease, N (%) | |
| Intrahepatic | 81 (1.00) |
| Extrahepatic | 48 (0.59) |
| Vascular involvement | 35 (0.43) |
| AFP, ng/ML, median (range) | 86.8 (1.8–128876.1) |
| Child-Pugh, N (%) | |
| A | 78 (0.96) |
| B | 2 (0.02) |
| Unknown | 1 (0.01) |
| Prior treatment, N (%) | |
| Surgical resection | 31 (0.38) |
| Regional therapy | 59 (0.73) |
| Hepatic allograft | 3 (0.04) |
| Systemic, N (%) | |
| Sorafenib: first line | 75 (0.93) |
| Sorafenib: second line | 6 (0.07) |
To identify potential genomic biomarkers of response to sor- afenib, we correlated genomic pathway alterations with treatment response and outcomes. We observed that patients whose tumors harbored activating mutations in the PI3K-mTOR pathway exhibited lower rates of clinical benefit (DCR 8.3% vs. 40.2%, P = 0.05; Fig. 2A) from sorafenib treatment and had a shorter median PFS [1.9 vs. 5.3 months, HR 3.8 (95% CI, 2.0–7.5) P < 0.0001; Fig. 2B] and median OS [10.4 vs. 17.9 months, HR 2.5 (95% CI 1.21–5.31), P =0.01; Fig. 2C] when compared with patients without such mutations. Mutations predicted to activate the WNT or MAPK pathway (including alterations in receptor tyrosine kinases that are potential targets of sorafenib) as well as theTP53 pathway, cell-cycle control, and chromatin remodeling had no effect on outcomes (Supplementary Table S3). Finally, VEGFA amplification,aproposedbiomarkerforextremesorafenib responders (25), was rarely observed (3.9%) and at least anecdotally did not appear to associate with clinical benefit (Fig. 2A).
Figure 2.
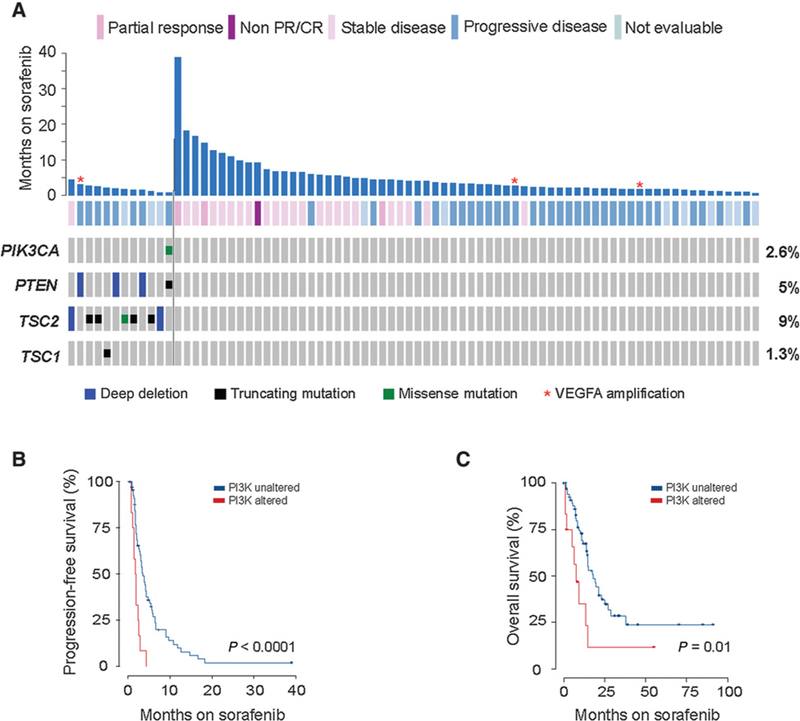
Genomic determinants of response to sorafenib in patients with advanced HCC. A, Months of treatment (y-axis) for each patient annotated with gene alteration and objective response. B, Kaplan-Meier PFS on sorafenib therapy for patients with PI3K-mTOR-activated tumors (N = 12) versus non-PI3K-mTOR tumors (N = 67), demonstrating shorter PFS in PI3K-mTOR activated HCCs. C, Kaplan-Meier OS on first-line sorafenib therapy for patients with PI3K-mTOR- activated tumors (N = 12) versus non-PI3K-mTOR tumors (N = 67), demonstrating a shorter OS in PI3K-mTOR-activated HCCs.
Genomic determinants of response to immune checkpoint inhibitors
As emerging data indicate that a subset of patients with liver cancer have durable responses to immune checkpoint inhibitors, we sought to explore genomic biomarkers of responsiveness or resistance in the 31 patients within the 127 patient MSK cohort treated with immune checkpoint inhibitors (Table 3). Patients received immunotherapy either as standard of care, on a compassionate use program, or in the context of a clinical trial. Of the 31 patients, 1 patient received anti–CTLA-4 monotherapy, 25 patients received monotherapy with a mAb targeting PD-1 or PD-L1, and 5 patients received an anti-PD-1 antibody in combination with antibodies targeting CTLA-4, KIR, or LAG-3. Twenty-seven patients were evaluable for radiographic response; 1 was excluded due to concomitant treatment with radioembolization, and 3 were not evaluable due to incomplete imaging or nonmeat-surable disease. In total for evaluable patients, the ORR was 11.1%. We observed 1 (3.7%) CR, 2 (7.4%) PR, 10 (37%) SD, and 14 (51.9%) progression of disease as best response. The median PFS and OS from the start of immune checkpoint inhibitor therapy was of 5.4 and 12.9 months, respectively.
Table 3.
Demographics of advanced HCC patients treated with immune checkpoint inhibitors (N = 31)
| Age, median, (range) | 66 | (25–85) |
| Sex, male, N (%) | 19 | (0.61) |
| Race, N (%) | ||
| Asian | 7 | (0.23) |
| Non-Asian | 24 | (0.77) |
| Etiology, N (%) | ||
| HCV | 5 | (0.16) |
| HBV | 7 | (0.23) |
| Nonviral | 19 | (0.61) |
| BCLC stage, N (%) | ||
| A | 0 | 0.00 |
| B | 9 | (0.29) |
| C | 22 | (0.71) |
| Extent of disease, N (%) | ||
| Intrahepatic | 31 | (1.00) |
| Extrahepatic | 20 | (0.65) |
| Vascular involvement | 9 | (0.29) |
| AFP, ng/ML, median (range) | 412 | (1.7–25338) |
| Child-Pugh, N (%) | ||
| A | 25 | 0.81 |
| B | 3 | 0.10 |
| Unknown | 3 | 0.10 |
| Prior treatment, N (%) | ||
| Hepatic resection | 16 | 0.52 |
| Regional therapy | 20 | 0.65 |
| Sorafenib | 28 | 0.90 |
| Immunotherapy, N (%) | ||
| Anti-CTLA-4 monotherapy | 1 | 0.03 |
| Anti-PD/PD-L1 monotherapy | 25 | 0.81 |
| Anti-PD-1/PD-L1 + immune checkpoint inhibitor | 5 | 0.16 |
| + Anti-CTLA-4 | 1 | 0.03 |
| + Anti-LAG3 | 2 | 0.065 |
| + Anti-KIR | 2 | 0.065 |
As WNT pathway activation results in T-cell exclusion and innate resistance to immune checkpoint blockade in vivo (26, 27), we correlated radiographic response to immunotherapy with the presence of activating mutations in the WNT pathway. Of 10 patients with WNT pathway alterations, none achieved clinical benefit (i.e., all patients had progression of disease at first interval scan), whereas 9 (53%) of 17 non-WNT pathway-altered patients had durable stable disease (≥ 4 months) or better as best response (P = 0.009; Fig. 3A). To further explore this finding, we correlated patient outcomes with genomic pathway alterations. When comparing WNT-activated tumors to nonaltered tumors, we found a shorter median PFS (2.0 vs. 7.4 months, HR 9.2; 95% CI: 2.928.8, P < 0.0001; Fig. 3B) and a numerically shorter median OS (9.1 vs. 15.2 months, HR 2.6; 95% CI 0.76–8.7, P = 0.11). As noted above, activating WNT pathway alterations had no effect on sorafenib median PFS (4.5 for WNT activating vs. 5.1 for non- WNT patients; Fig. 3C). No other pathways correlated with responsiveness or resistance to checkpoint inhibitor treatment.
Figure 3.
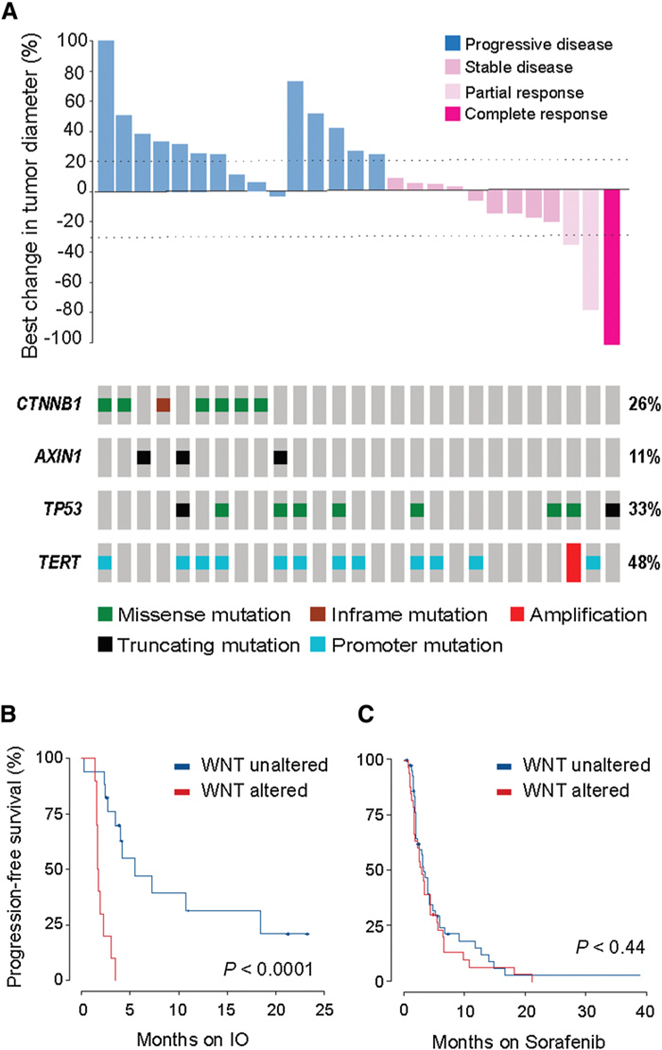
Genomic determinants of response to immune checkpoint blockade in patients with advanced HCC. A, Waterfall plot for 27 evaluable patients treated with immune checkpoint blockade showing best change in target lesions for each patient, annotated with response and most frequent genomic alterations. B, Kaplan-Meier PFS on immune oncology agent (IO) for patients with WNT- activated tumors versus non-WNT- activated tumors, demonstrating shorter PFS in WNT-activated HCCs. C, Kaplan-Meier PFS on sorafenib therapy for patients with WNT-activated tumors versus non- WNT-activated tumors, demonstrating equivalent PFS in WNT-activated HCCs.
Mutational burden has been shown to correlate with clinical benefit from treatment with immune checkpoint inhibitors in patients with melanoma, lung, and other cancer types. For the 27 evaluable patients treated with immune checkpoint inhibitors, the median TMB was 3.8—only 1 patient in the cohort had a TMB of>10.ThemedianTMBwas3.8 and 3.9 (P = 0.98) for responders and nonresponders, respectively. The median PFS stratified by median TMB was similar (TMB <3.8: 3.1 months; TMB ≤3.8: 3.0 months, P = 0.21).
Prospective genotyping and matching to genome-driven therapy
In total, 24% of patients harbored at least one actionable mutation as annotated by OncoKB (Fig. 4A; ref. 20). Consistent with the lack of validated predictive biomarkers of drug response in this disease type, no patient had a level 1 or 2A alteration. Potentially actionable mutations included truncating and inactivating mutations in the mTOR modulators, TSC1 (<1%) and TSC2 (7%), truncating or homozygous deletions in PTEN (3.9%), FGF19 (6.3%), and MET amplifications (1.5%), and oncogenic missense mutations in HRAS, NRAS, and PI3KCA (all <1%).
Figure 4.
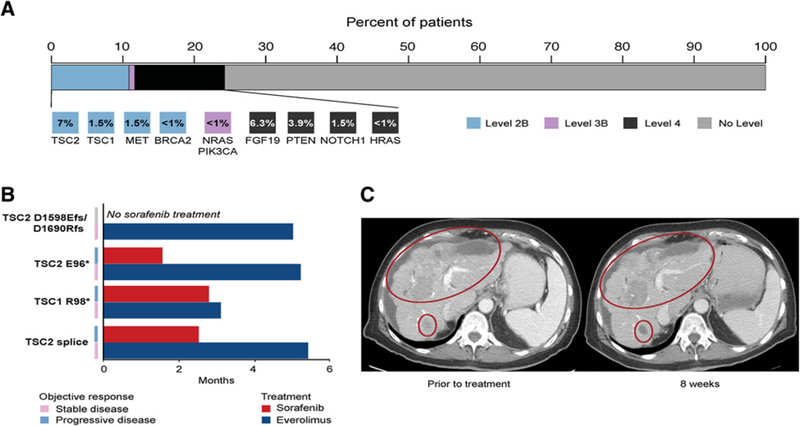
Prospective genotyping of HCC and matching to genome-driven therapy. A, Highest level ofclinical actionability across thecohort, as defined by OncoKB. Standard therapeutic implications include FDA-recognized or National Comprehensive Cancer Network (NCCN) guideline listed biomarkers that are predictive of response to an FDA-approved drug in a specific indication (level 1). Investigational therapeutic implications include FDA-approved biomarkers predictive of response to an FDA-approved drug detected in an off-label indication (level 2B), FDA- or non-FDA-recognized biomarkers that are predictive of response to novel targeted agents that haveshown promising results in clinical trials (level 3B),and non-FDA-recognized biomarkersthatare predictive of responseto novel targeted agentson the basisof compelling preclinical data (level 4). B, Duration of treatment for 4 patients with TSC-mutant HCC treated with sorafenib and mTOR inhibitors. C, A patient with autoimmune hepatitis with Child-Pugh A hepatic function and a TSC2 E95*-mutant HCC who had rapid progression on prior sorafenib with stabilization ofdisease with minor tumoral shrinkage on everolimus.
Five (5.7%) of 87 patients who received systemic therapy were matched to treatments targeting mTOR or MET based on the clinical genomic report. Four (40%) of 10 patients who harbored alterations in TSC1 /2 received everolimus, an mTOR inhibitor, off-label or on an investigational study (Fig. 4B). Of the 2 patients with MET amplifications, one patient with a 3.7-fold VEGFA and 2.1-fold MET-amplified tumor received mAbs to VEGFR2 and MET in the context of a clinical trial. For 4 of these 5 patients, first-line sorafenib treatment was ineffective with radiographic progression of disease within 1.5 to 2.5 months. Second-line targeted therapy resulted in stabilization of disease at first interval scan in all 4 patients—3 of 4 patients had clinical benefit for 5 months including 1 patient who rapidly progressed on prior sorafenib who had clinical improvement and disease stabilization with some areas of tumoral shrinkage (Fig. 4C). The fifth patient received ever- olimus as a first-line treatment for 5.0 months until progression of disease.
Discussion
In this study, we demonstrate the utility of prospective sequencing of advanced liver-limited or metastatic HCC in clinical practice. We confirm genomic findings derived from retrospectively analyzed cohorts of surgically-resected, early-stage, liver-limited HCCs and extend these observations to an advanced patient population more representative of those treated as part of routine clinical practice. As our sampling and analysis strategy occurred in tandem with clinical care, we were able to explore potential predictive and prognostic genetic factors to contemporary therapies, such as sorafenib and immune checkpoint inhibitors. We found that mutations predicted to activate the PI3K-mTOR pathway were associated with poor outcomes in sorafenib-treated patients and that mutations predicted to activate the WNT pathway were associated with innate resistance to immune checkpoint blockade. Furthermore, potentially actionable mutations were identified in a small subset of patients and NGS permitted real-time interpretation and utilization of results in clinical practice.
Our data are consistent with prior reports regarding the frequency and overall distribution of oncogenic alterations in HCC. Specifically, we confirm that mutations predicted to result in WNT/β-catenin activation are present in nearly half of all HCCs, and represent a unique biological subset (28). While other series have identified a MSI-H/mismatch repair-deficient phenotype and/or a high-to-moderate TMB in a small minority of patients, we did not observe any such cases in this prospective series. This may have been due to demographic differences in the populations studied or to the advanced nature of our patient cohort, as MSI-H has been shown to be a favorable prognostic indicator in other cancer types. Finally, except for slight enrichment of TP53 mutations in virally mediated HCCs, genomic alterations did not correlate with specific HCC etiologic factors (7–16).
Multitargeted TKIs have altered the natural history of HCC, and sorafenib is an FDA-approved first-line standard systemic therapy (3, 5, 29). Aside from VEGFA amplifications, which are rare, few genomic biomarkers of drug response have been identified in retrospective studies of multitargeted TKIs. As a signaling pathway downstream from the putative targets of sorafenib, PI3K-mTOR activation, either through primary genomic alterations or negative feedback loops, could abrogate the dependence of HCCs on upstream kinases, and thus may mediate resistance to multitargeted TKIs (30–33). Indeed, preclinical models indicate that sorafenib-resistant HCCs utilize PI3K-mTOR as one potential bypass pathway, and PI3K pathway inhibition via RNA interference or small-molecule inhibitors, restores sorafenib responsiveness (34–36). In clinical samples, mTOR activation has been associated with resistance to sorafenib and our data corroborate these findings (30). mTOR activation also portends worse clinical outcomes postresection in patients with localized HCC (37) and in the metastatic setting (33, 38). Thus, pharmacologic interference with mTOR signaling (37–42) may have clinical import in this HCC molecular subset (38), and several platform/umbrella studies, such as NCI-MATCH (NCT02465060) andASCOTAPUR (NCT02693535), are evaluating the clinical utility of PI3K- mTOR blockade across cancer histologies.
Despite the clear efficacy of immune checkpoint inhibitors in HCC, few patients respond to this treatment. Our data suggest that TMB (43–45) and mismatch repair deficiency/microsatellite instability (46, 47), are unlikely to be major determinants of response to immunotherapy in this disease. On the other hand, the prevalence of WNT/β-catenin alterations in HCC may have important implications for immune-based treatment. Rendering mutant tumors immunologically cold, oncogenic WNT signaling leads to T-lymphocyte exclusion (26, 27) and insensitivity to combination anti-PD-L1 and anti-CTLA-4 mAb therapy in vivo (26). Herein, we present clinical data indicating that patients with HCC with mutations predicted to result in WNT pathway activation are innately resistant to immune checkpoint blockade. In our series, all patients with tumors harboring activating CTNNB1 or inactivating AXIN1 mutations had progressive disease as best response whereas patients with non-WNT pathway-altered tumors were significantly more likely to respond or derive clinical benefit from immunotherapy. As WNT pathway alterations did not correlate with sorafenib PFS and OS, WNT activation may indeed be a predictive biomarker of resistance to immune checkpoint blockade in HCC. Acknowledging these data are preliminary and may be confounded by both known and unknown clinicopathologic factors not controlled for due to the small sample size of the cohort, we believe our clinical findings will prompt further preclinical investigations that will further elucidate the biologic underpinnings of these intriguing initial observations. Moreover, larger retrospective datasets and prospective evaluation of this preliminary observation are imperative, and if validated, this finding would be highly impactful for both patient care and clinical trial design. We also note that other factors, such as HLA-I heterogeneity (48), may also contribute to innate resistance, and such factors also deserve further exploration.
Although the direct clinical impact of prospective NGS for HCC clearly has less utility than in other solid tumors such as lung cancer and melanoma (49), 24% of tumors had potentially actionable alterations that could be targets for currently available FDA-approved drugs, or agents in active clinical development. As an example, our data and other series specifically support continued investigation of mTOR inhibitors in tumors with TSC1/2 alterations (50). Albeit, it is important to recognize that at least in this small case series, mTOR inhibition appeared to have modest cytostatic benefit at best. Emerging data also indicate that an amplicon that includes the FGF19 gene may contribute to HCC oncogenesis, and that inhibition of this pathway with FGF19 mAbs or with highly selective inhibitors of FGFR4, the receptor for FGF19, could be clinically effective in patients with HCC (51). Indeed, several compounds are now in clinical development, with promising preliminary data. Although rare, IDH altered HCCs (<1%) also likely represent a unique biologic subset that exhibit clinical and genetic features similar to cholangiocarcinomas (7), and such tumors may be susceptible to selective small-molecule inhibitors of IDH1 (52). Although initial studies targeting MET (1.5%) failed to demonstrate meaningful clinical activity (53), more potent and specific MET inhibitors are being evaluated, and such efforts have been bolstered by recent positive data with cabozantinib.
Our study has several notable limitations. First, the targeted capture approach used in this study could not by definition detect alterations in genes not included in the assay design, epigenetic mechanisms of gene suppression, or viral HBV DNA sequences. Second, tumor heterogeneity is well documented in HCC and sampling a single site of disease can never fully assess clonal complexity in patients with multisite metastatic disease. To address the issue of tumor heterogeneity, future studies in patients with HCC could seek to include an assessment of genomic alterations in circulating cell-free DNA or in single cells. Third, given the small sample size of the current study, potential confounding variables, such as HCC etiology, AFP levels, extrahepatic spread/vascular invasion, should be evaluated in future studies to assure such factors are not the predominant determinants of therapeutic outcome. To address this limitation, all clinical and genomic data from this study have been made available through the cBioPortal for Cancer Genomics to facilitate future metaanalyses. Finally, the favorable OS observed in the sorafenib cohort compared with the published literature, suggest a selection bias but may also be due in part to access to novel therapies and immunotherapies at our center as well as the high proportion of patients with intact hepatic function in the cohort. Despite these limitations, our data represent the first attempt to link real-time NGS to clinical practice and provide needed insight into feasibility and the potential impact of this investigational modality to direct patient care.
Supplementary Material
Translational Relevance.
Few translational studies have attempted to prospectively genotype patients with advanced hepatocellular carcinoma (HCC) to help inform clinical decision-making and to propose tumoral biomarkers for contemporary systemic therapies. With the implementation of a next-generation sequencing platform in the clinic, we prospectively interrogated 127 patients with HCC treated at our center. HCCs harboring oncogenic PI3K-mTOR alterations had significantly worse outcomes on sorafenib treatment, while the presence of an activating WNT/β-catenin mutation was associated with innate resistant to immune checkpoint inhibitors. Finally, we observed that a subset of HCC patients with mutations in mTOR and MET were matched to appropriate targeted therapeutics and thus provide needed insight into the utility and the potential impact of next-generation sequencing to direct patient care.
Acknowledgments
This work was funded in part by a Cancer Center Support Grant (P30 CA008748), the Marie-Josee and Henry R. Kravis Center for Molecular Oncology, Cycle for Survival, Robertson Foundation (to N. Schultz), and the NCI (U24 CA220457).
J.J. Harding reports receiving commercial research grants from Bristol-Myers Squibb, and is a consultant/advisory board member for Bristol-Myers Squibb, CytomX, Eisai, and Eli Lilly. J.F. Hechtman reports receiving speakers bureau honoraria from Medscape. M.F. Berger is a consultant/advisory board member for Roche. D.M. Hyman reports receiving commercial research grants from AstraZeneca, Loxo Oncology, and Puma Biotechnology, and is a consultant/ advisory board member for AstraZeneca, Atara Biotherapeutics, Bayer, Boeh-ringerlngelheim, ChugaiPharma, CytomXTherapeutics, Genentech, andPfizer. D.S. Klimstra holds ownership interest (including patents) in Paige.AI, is a consultant/advisory board member for Merck and Paige.AI, and received royalties from American Registry of Pathology and UpToDate. Y.Y. Janjigian reports receiving commercial research grants from Bayer, BoehringerIngelheim, Bristol-Myers Squibb, Eli Lilly, Genentech/Roche, and Merck, and is a consul- tant/advisoryboardmemberforBayer, Bristol-Myers Squibb, EliLilly, Imugene, Merck, Merck Serono, and Pfizer. D.B. Solit is a consultant/advisory board member for Illunina, Loxo Oncology, and Pfizer. G.K. Abou-Alfa is a consultant/ advisory board member for 3DMedcare, Agios, Alignmed, Amgen, Antengene, Aptus, Aslan, Astellas, AstraZeneca, Bayer, Beigene, Bioline, Boston Scientific, BridgeBio, Bristol-Myers Squibb, Carsgen, Celgene, Casi, Cipla, CytomX, Daiichi, Debio, Delcath, Eisai, Exelixis, Genoscience, Gilead, Halozyme, Hengrui, Incyte, Inovio, Ipsen, Jansen, Jazz, Kyowa, Kirin, Lilly, Loxo, Merck, Mina, Newlink Genetics, Novella, Onxeo, PCI Biotech, Pfizer, Pharmacyte, Pharmacyclics, Pieris, QED, Redhill, Sanofi, Servier, Silenseed, Sillajen, Sobi, Targovax, Tekmira, Twoxar, Vicus, Yakult, and Yiviva.
Footnotes
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed by the other authors.
Note: Supplementary data for this article are available at Clinical Cancer Research Online (http://clincancerres.aacrjournals.org/).
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin 2015;65:87–108. [DOI] [PubMed] [Google Scholar]
- 2.Altekruse SF, Henley SJ, Cucinelli JE, McGlynn KA. Changing hepatocellular carcinoma incidence and liver cancer mortality rates in the United States. Am J Gastroenterol 2014;109:542–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Llovet JM, Ricci S, Mazzaferro V, Hilgard P, Gane E, Blanc JF, et al. Sorafenib in advanced hepatocellular carcinoma. N Engl J Med 2008; 359:378–90. [DOI] [PubMed] [Google Scholar]
- 4.Bruix J, Qin S, Merle P, Granito A, Huang YH, Bodoky G, et al. Regorafenib for patients with hepatocellular carcinoma who progressed on sorafenib treatment (RESORCE): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet 2016;389: 56–66. [DOI] [PubMed] [Google Scholar]
- 5.Kudo M, Finn RS, Qin S, Han KH, Ikeda K, Piscaglia F, et al. Lenvatinib versus sorafenib in first-line treatment of patients with unresectable hepatocellular carcinoma: a randomised phase 3 non-inferiority trial. Lancet 2018;391:1163–73. [DOI] [PubMed] [Google Scholar]
- 6.El-Khoueiry AB, Sangro B, Yau T, Crocenzi TS, Kudo M, Hsu C, et al. Nivolumab in patients with advanced hepatocellular carcinoma (CheckMate 040): an open-label, non-comparative, phase 1/2 dose escalation and expansion trial. Lancet 2017;389:2492–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cancer Genome Atlas Research Network. Electronic address: wheeler@bcm.edu & Cancer Genome Atlas Research Network. Comprehensive and integrative genomic characterization ofhepatocellular carcinoma. Cell 2017;169:1327–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fujimoto A, Totoki Y, Abe T, Boroevich KA, Hosoda F, Nguyen HH, et al. Wholegenome sequencing ofliver cancers identifies etiological influences on mutation patterns and recurrent mutations in chromatin regulators. Nat Genet 2012;44:760–4. [DOI] [PubMed] [Google Scholar]
- 9.Guichard C, Amaddeo G, Imbeaud S, Ladeiro Y, Pelletier L, Maad IB, et al. Integrated analysis of somatic mutations and focal copy-number changes identifies key genes and pathways in hepatocellular carcinoma. Nat Genet 2012;44:694–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang J, Deng Q, Wang Q, Li KY, Dai JH, Li N, et al. Exome sequencing of hepatitis B virus-associated hepatocellular carcinoma. Nat Genet 2012;44:1117–21. [DOI] [PubMed] [Google Scholar]
- 11.Schulze K, Imbeaud S, Letouzé E, Alexandrov LB, Calderaro J, Rebouissou S, et al. Exome sequencing of hepatocellular carcinomas identifies new mutational signatures and potential therapeutic targets. Nat Genet 2015;47:505–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Totoki Y, Tatsuno K, Covington KR, Ueda H, Creighton CJ, Kato M, et al. Trans-ancestry mutational landscape of hepatocellular carcinoma genomes. Nat Genet 2014;46:1267–73. [DOI] [PubMed] [Google Scholar]
- 13.Ahn SM, Jang SJ, Shim JH, Kim D, Hong SM, Sung CO, et al. Genomic portrait of resectable hepatocellular carcinomas: implications of RB1 and FGF19 aberrations for patient stratification. Hepatology 2014;60: 1972–82. [DOI] [PubMed] [Google Scholar]
- 14.Kan Z, Zheng H, Liu X, Li S, Barber TD, Gong Z, et al. Whole-genome sequencing identifies recurrent mutations in hepatocellular carcinoma. Genome Res 2013;23:1422–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sung WK, Zheng H, Li S, Chen R, Liu X, Li Y, et al. Genome-wide survey of recurrent HBV integration in hepatocellular carcinoma. Nat Genet 2012;44:765–9. [DOI] [PubMed] [Google Scholar]
- 16.Cleary SP, Jeck WR, Zhao X, Chen K, Selitsky SR, Savich GL, et al. Identification of driver genes in hepatocellular carcinoma by exome sequencing. Hepatology 2013;58:1693–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cheng DT, Mitchell TN, Zehir A, Shah RH, Benayed R, Syed A, et al. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): a hybridization capture-based next-generation sequencing clinical assay for solid tumormolecular oncology. J Mol Diagn 2015;17:251–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thorvaldsdóttir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinformatics 2013;14:178–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Niu B, Ye K, Zhang Q, Lu C, Xie M, McLellan MD, et al. MSIsensor: microsatellite instability detection using paired tumor-normal sequence data. Bioinformatics 2014;30:1015–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chakravarty D, Gao J, Phillips SM, Kundra R, Zhang H, Wang J, et al. Oncokb: a precision oncology knowledge base. JCO Precis Oncol 2017;2017. doi: 10.1200/PO.17.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, AksoyBA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012;2:401–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013;6:pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eisenhauer EA, Therasse P, Bogaerts J, Schwartz LH, Sargent D, Ford R, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer 2009;45:228–47. [DOI] [PubMed] [Google Scholar]
- 24.Sanchez-Vega F, Mina M, Armenia J, Chatila WK, Luna A, La KC, et al. Oncogenic signaling pathways in The Cancer Genome Atlas. Cell 2018;173:321–37.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Horwitz E, Stein I, Andreozzi M, Nemeth J, Shoham A, Pappo O, et al. Human and mouse VEGFA-amplified hepatocellular carcinomas are highly sensitive to sorafenib treatment. Cancer Discov 2014;4:730–43. [DOI] [PubMed] [Google Scholar]
- 26.Spranger S, Bao R, Gajewski TF. Melanoma-intrinsic β-catenin signalling prevents anti-tumour immunity. Nature 2015;523:231–5. [DOI] [PubMed] [Google Scholar]
- 27.Xiao Q, Wu J, Wang WJ, Chen S, Zheng Y, Yu X, et al. DKK2 imparts tumor immunity evasion through β-catenin-independent suppression of cytotoxic immune-cell activation. Nat Med 2018;24:262–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lachenmayer A, Alsinet C, Savic R, Cabellos L, Toffanin S, Hoshida Y, et al. Wnt-pathway activation in two molecular classes of hepatocellular carcinoma and experimental modulation by sorafenib. Clin Cancer Res 2012;18:4997–5007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wilhelm SM, Carter C, Tang L, Wilkie D, McNabola A, Rong H, et al. BAY 43–9006 exhibits broad spectrum oral antitumor activity and targets the RAF/MEK/ERK pathway and receptor tyrosine kinases involved in tumor progression and angiogenesis. Cancer Res 2004;64:7099–109. [DOI] [PubMed] [Google Scholar]
- 30.Masuda M, Chen WY, Miyanaga A, Nakamura Y, Kawasaki K, Sakuma T, et al. Alternative mammalian target of rapamycin (mTOR) signal activation in sorafenib-resistant hepatocellular carcinoma cells revealed by array-based pathway profiling. Mol Cell Proteomics 2014;13:1429–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lindblad O, Cordero E, Puissant A, Macaulay L, Ramos A, Kabir NN, et al. Aberrant activation of the PI3K/mTOR pathway promotes resistance to sorafenib in AML. Oncogene 2016;35:5119–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dong J, Zhai B, Sun W, Hu F, Cheng H, Xu J. Activation of phosphatidy-linositol 3-kinase/AKT/snail signaling pathway contributes to epithelialmesenchymal transition-induced multi-drug resistance to sorafenib in hepatocellular carcinoma cells. PLoS One 2017;12:e0185088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guan DX, Shi J, Zhang Y, Zhao JS, Long LY, Chen TW, et al. Sorafenib enriches epithelial cell adhesion molecule-positive tumor initiating cells and exacerbates a subtype of hepatocellular carcinoma through TSC2-AKT cascade. Hepatology 2015;62:1791–803. [DOI] [PubMed] [Google Scholar]
- 34.Chen KF, Chen HL, Tai WT, Feng WC, Hsu CH, Chen PJ, et al. Activation of phosphatidylinositol 3-kinase/Akt signaling pathway mediates acquired resistance to sorafenib in hepatocellular carcinoma cells. J Pharmacol Exp Ther 2011;337:155–61. [DOI] [PubMed] [Google Scholar]
- 35.Zhang H, Wang Q, Liu J, Cao H. Inhibition of the PI3K/Akt signaling pathway reverses sorafenib-derived chemo-resistance in hepatocellular carcinoma. Oncol Lett 2018;15:9377–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Serova M, de Gramont A, Tijeras-Raballand A, Dos Santos C, Riveiro ME, Slimane K, et al. Benchmarking effects of mTOR, PI3K, and dual PI3K/mTOR inhibitors in hepatocellular and renal cell carcinoma models developing resistance to sunitinib and sorafenib. Cancer Chemother Pharmacol 2013;71:1297–307. [DOI] [PubMed] [Google Scholar]
- 37.Villanueva A, Chiang DY, Newell P, Peix J, Thung S, Alsinet C, et al. Pivotal role of mTOR signaling in hepatocellular carcinoma. Gastroenterology 2008;135:1972–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huynh H, Hao HX, Chan SL, Chen D, Ong R, Soo KC, et al. Loss of tuberous sclerosis complex 2 (TSC2) is frequent in hepatocellular carcinoma and predicts response to mTORC1 inhibitor everolimus. Mol Cancer Ther 2015;14:1224–35. [DOI] [PubMed] [Google Scholar]
- 39.Thomas HE, Mercer CA, Carnevalli LS, Park J, Andersen JB, Conner EA, et al. mTOR inhibitors synergize on regression, reversal of gene expression, and autophagy in hepatocellular carcinoma. Sci Transl Med 2012;4:139ra84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Buitrago-Molina LE, Pothiraju D, Lamlé J, Marhenke S, Kossatz U, Breu-hahn K, et al. Rapamycin delays tumor development in murine livers by inhibiting proliferation of hepatocytes with DNA damage. Hepatology 2009;50:500–9. [DOI] [PubMed] [Google Scholar]
- 41.Voss MH, Hakimi AA, Pham CG, Brannon AR, Chen YB, Cunha LF, et al. Tumor genetic analyses of patients with metastatic renal cell carcinoma and extended benefit from mTOR inhibitor therapy. Clin Cancer Res 2014; 20:1955–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kwiatkowski DJ, Choueiri TK, Fay AP, Rini BI, Thorner AR, de Velasco G, et al. Mutations inTSC1, TSC2, and MTOR are associated with response to rapalogs in patients with metastatic renal cell carcinoma. Clin Cancer Res 2016;22:2445–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hellmann MD, Nathanson T, Rizvi H, Creelan BC, Sanchez-Vega F, Ahuja A, et al. Genomic features of response to combination immunotherapy in patients with advanced non-small-cell lung cancer. Cancer Cell 2018; 33:843–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Snyder A, Makarov V, Merghoub T, Yuan J, Zaretsky JM, Desrichard A, et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N Engl J Med 2014;371:2189–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rizvi NA, Hellmann MD, Snyder A, Kvistborg P, Makarov V, Havel JJ, et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 2015;348: 124–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Le DT, Durham JN, Smith KN, Wang H, Bartlett BR, Aulakh LK, et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science 2017;357:409–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Le DT, Uram JN, Wang H, Bartlett BR, Kemberling H, Eyring AD, et al. PD-1 blockade in tumors with mismatch-repair deficiency. N Engl J Med 2015;372:2509–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chowell D, Morris LGT, Grigg CM, Weber JK, Samstein RM, Makarov V, et al. Patient HLA class I genotype influences cancer response to checkpoint blockade immunotherapy. Science 2018;359:582–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zehir A, Benayed R, Shah RH, Syed A, Middha S, Kim HR, et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med 2017;23:703–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Iyer G, Hanrahan AJ, Milowsky MI, Al-Ahmadie H, Scott SN, Janakiraman M, et al. Genome sequencing identifies a basis for everolimus sensitivity. Science 2012;338:221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hagel M, Miduturu C, Sheets M, Rubin N, Weng W, Stransky N, et al. First selective small molecule inhibitor of FGFR4 for the treatment of hepatocellular carcinomas with an activated FGFR4 signaling pathway. Cancer Discov 2015;5:424–37. [DOI] [PubMed] [Google Scholar]
- 52.Popovici-Muller J, Lemieux RM, Artin E, Saunders JO, Salituro FG, Travins J, et al. Discovery of AG-120 (Ivosidenib): a first-in-class mutant IDH1 inhibitor for the treatment of IDH1 mutant cancers. ACS Med Chem Lett 2018;9:300–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rimassa L, Assenat E, Peck-Radosavljevic M, Pracht M, Zagonel V, Mathurin P, et al. Tivantinib for second-line treatment of MET-high, advanced hepatocellular carcinoma (METIV-HCC): a final analysis of a phase 3, randomised, placebo-controlled study. Lancet Oncol 2018; 19:682–93. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


