Abstract
Background
The methylation of microtubule-associated protein tau (MAPT) was first described in patients with Alzheimer’s disease. In this study, we aim to determine if MAPT promoter CpG island is hypermethylated and whether this signature could work as a prognostic marker for patients with stage II colorectal cancer (CRC).
Methods
MAPT methylation level and CpG island methylator phenotype (CIMP) status were examined. The prognostic value of MAPT methylation was analyzed using Cox regression analysis.
Results
Amongst stage II CRC patients (n=107), hypermethylation of MAPT promoter CpG island was seen in 23.4% of them. MAPT methylation was much more frequent in patients with age ≥60 compared to age <60 (P<0.001). MAPT were preferentially methylated among proximal colon tumors or CIMP high tumors (both P<0.001). Five-year overall survival (OS) rates were 57.1% and 79.4% for patients with and without MAPT hypermethylation, respectively, HR=2.33 (95% CI, 1.19–4.57; P=0.014). MAPT hypermethylation remained an important prognostic variable for OS in multivariate analysis with a HR of 2.29 (95% CI, 1.01–5.18; P=0.047).
Conclusion
Our findings suggest that MAPT is frequently methylated and hypermethylation is associated with worse prognosis in patients with stage II CRC.
Keywords: colorectal cancer, methylation, MAPT, prognosis
Introduction
Colorectal cancer (CRC), the second and third most common cancer in women and men, respectively, causes 832,000 deaths in 2015 worldwide.1 CRCs develop as a result of the accumulation of genetic and epigenetic alterations. CpG island methylator phenotype (CIMP) is one of the main mechanisms underlying CRC progression, which is characterized by the simultaneous methylation of cytosine residues in CpG islands from the promoter regions of multiple cancer-specific genes.2 Widespread changes in DNA methylation are common in CRC and may confer to oncogenesis through transcriptional silencing of tumor-suppressor genes.3
Microtubule-associated protein tau (MAPT) is a gene located on chromosomal subband 17q21.4 Tau protein encoded by MAPT is among the best characterized microtubule-associated protein that promotes microtubule assembly and reduces microtubule instability.5 In addition to neurons, it is expressed at low levels in several non-neuronal cells. It has been shown to be involved in a number of neurodegenerative disorders including Alzheimer’s disease, Parkinson’s disease, and progressive supranuclear palsy, etc. The ability of tau to induce microtubule polymerization6 and its interaction with actin7 indicates a possible role for tau in tumor cell migration and invasion. However, the function of tau in cancer remains relatively unexplored.
Recently, methylation of the MAPT promoter has been reported in a number of human diseases, including Alzheimer’s disease,8 Parkinson’s,9 and prostate cancer.10 However, the methylation status of MAPT and its influence on prognosis in CRC remains unclear. The goal of this study was to assess the methylation of MAPT promoter CpG island to determine its prognostic value in a cohort with 107 stage II CRC patients.
Materials and methods
Subjects for methylation analysis
Colorectal tissue samples were acquired by surgical resection from 107 patients (47 male and 60 female) with stage II CRC at the Johns Hopkins Hospital and the Johns Hopkins Bayview Hospital between 1995 and 2009. The median age was 67.8 years (range, 37–90 years). Tissue samples were available from all patients for molecular analysis; in 50 of these samples, a matched nontumor colorectal tissue was also obtained that was at least 2 cm distant from the tumor and in which cancer cell infiltration was ruled out by histologic review. Patients who were willing to participate provided written informed consent. This study was with approval from the Institutional Review Board and in accordance with Health Insurance Portability and Accountability Act regulations.
MAPT methylation analysis by quantitative real-time methylation-specific PCR (Q-MSP)
Q-MSP was used to assess MAPT methylation in primary tumors. Primers were designed using MSPprimer.11 Sequences are listed in Table 1. DNA was extracted and 1 µg was used for bisulfite conversion with the EZ DNA methylation Kit (Zymo Research). The Q-MSP was carried out in duplicates in 96-well plates using a total reaction volume of 20 μL including 10.0 μL of 2× Power SYBR Green PCR Master Mix (Applied Biosystems), 2.5 pmol each of forward and reverse primers, and 2 μL of DNA template. Fragments were amplified at 95°C for 10 mins, and 40 cycles of 95°C for 15 s followed by 60°C for 60 s using the Applied Biosystems 7500 One-Step Plus Real-Time PCR System. Dissociation curve analysis was performed to confirm the specificity of amplicons. Cycle threshold (Ct) values were used to calculate methylation index (MI) using the following formula: MI=100/[1+2(Ctm − Ctu)]. Ctm and Ctu indicate Ct value of the probes specific for the methylated and unmethylated states, respectively.
Table 1.
Primers for quantitative real-time methylation-specific PCR of MAPT
| Unmethylation | Methylation | ||
|---|---|---|---|
| Forward primer sequence | Reverse primer sequence | Forward primer sequence | Reverse primer sequence |
| GTTTTTGTGGAGGTTGTGTTGTTTG | ACCAACAAAAAAAACACATTCCTAAAACCAACA | CGGAGGTCGCGTTGTTC | AAACGCGTTCCTAAAACCGACG |
CIMP methylation status analysis by methylight
A 5-gene signature including CACNA1G, IGF2, NEUROG1, RUNX3, and SOCS1 was used to assess CIMP methylation status in tumor tissues.12 Methylation was quantified using MethyLight.12 Alu was used as a normalization control. A 5′ FAM fluorophore, a 3′ IBFQ quencher, and an internal ZEN quencher (Integrated DNA Technologies) were used for all CIMP probes.13 DNA methylation was calculated as percent of methylated reference (PMR)=100×[(methylated reaction/Alu)sample/(methylated reaction/Alu)M.SssI-reference)]. Genes were considered methylated when PMR≥0. When at least three of the five studied markers were methylated, the samples were considered CIMP high.12
Statistical analysis
For statistical analyses, the SPSS 18.0 (SPSS, Chicago, IL, USA) was used. Determination of cutoff values was made by the receiver-operator characteristics (ROC) curve. The area under the curve and the best sensitivity and specificity were then computed. The comparison of clinicopathologic factors was analyzed using the χ2 tests. Results were considered significant when P<0.05. The Kaplan–Meier method and log-rank test were used to estimate survival. Univariate and multivariate hazard ratios (HRs) were determined by using cox proportional hazard regression models. Overall survival (OS) was defined as the time from cancer diagnosis until death of all causes.
Results
Methylation analysis
DNA extraction and methylation analyses were successful in all 107 patients. Cutoff value of MI to separate all 107 cases into two groups (low-risk group and high-risk group) was calculated by using ROC analysis. Optimal cutoff was determined by maximizing the sensitivity and specificity. Therefore, tumors were dichotomized into both a unhypermethylated group (MI<10) and a hypermethylated group (MI≥10). Samples were hypermethylated in 25 cases (23.4%) of the 107 patients. To confirm that these methylation consequences were specific to tumor tissue, we re-measured methylation levels of MAPT in 50 tumor samples in parallel with their paired normal colonic mucosa tissues. MAPT was hypermethylated in 11 of these tumors and no methylation was found in matched normal sample. The MI values of tumor were 28.4, 18.4, 74.7, 14.6, 10.0, 27.2, 31.0, 13.9, 28.3, 34.7, and 93.7.
Methylation and clinicopathological features
Associations between MAPT methylation status and patient clinicopathological features were examined. No significant difference was found in the distribution of patients with positive or negative hypermethylation of MAPT in terms of gender, lymph nodes examined, tumor differentiation, or pT4. However, MAPT hypermethylation was much more common in patients with age ≥60 (P<0.001, Table 2) and in proximal colon tumors (P<0.001, Table 2). MAPT hypermethylation was also strongly associated with CIMP status (P<0.001, Table 2).
Table 2.
Differential clinicopathologic features of colorectal cancer according to MAPT methylation status
| Variables | Total n=107 (%) |
MAPT-M (%) |
MAPT-U (%) |
P-value | |
|---|---|---|---|---|---|
| Age (years) | <60 | 33 (30.8) | 1 (4.0) | 32 (39.0) | 0.000 |
| ≥60 | 74 (69.2) | 24 (96.0) | 50 (61.0) | ||
| Sex | Male | 47 (43.9) | 8 (32.0) | 39 (47.6) | 0.170 |
| Female | 60 (56.1) | 17 (68.0) | 43 (52.4) | ||
| Location | Proximal | 55 (51.4) | 21 (84.0) | 34 (41.5) | 0.000 |
| Distal | 52 (48.6) | 4 (16.0) | 48 (58.5) | ||
| Lymph nodes examined | ≥12 | 74 (69.2) | 20 (80.0) | 54 (65.9) | 0.180 |
| <12 | 33 (30.8) | 5 (20.0) | 28 (34.1) | ||
| Differentiation | Well to moderate | 89 (83.2) | 20 (80.0) | 69 (84.1) | 0.628 |
| Poor | 18 (16.8) | 5 (20.0) | 13 (15.9) | ||
| pT4 | No | 96 (89.7) | 24 (96.0) | 72 (87.8) | 0.452 |
| Yes | 11 (10.3) | 1 (4.0) | 10 (12.2) | ||
| CIMP | Low | 83 (77.6) | 9 (36.0) | 74 (90.2) | 0.000 |
| High | 24 (22.4) | 16 (64.0) | 8 (9.8) |
Abbreviations: CIMP, CpG island methylator phenotype; MAPT-U, MAPT-unhypermethylated; MAPT-M, MAPT-hypermethylated.
Methylation and survival
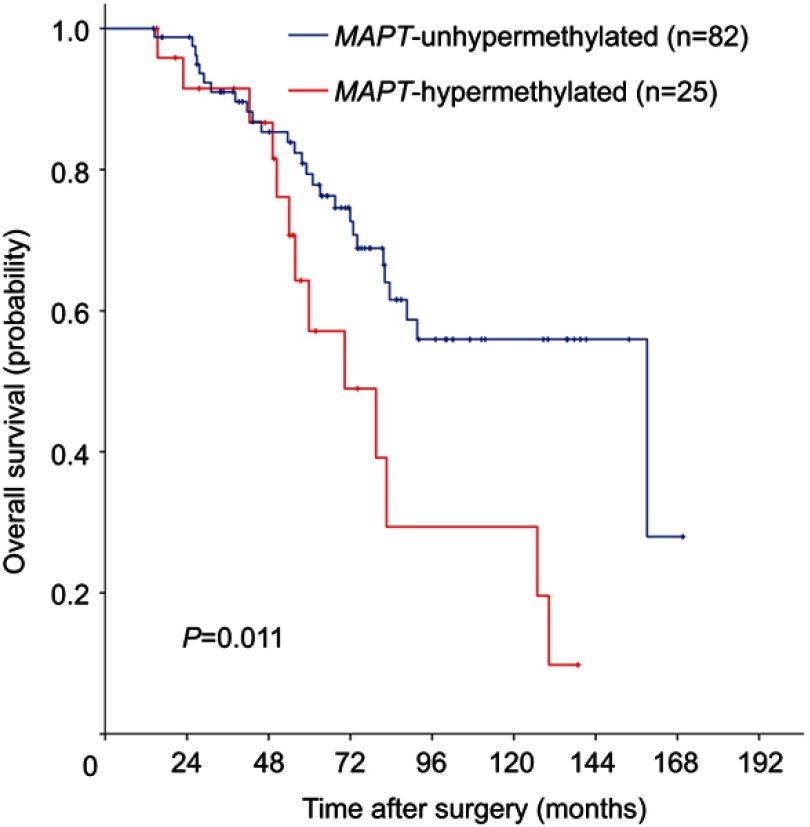
Survival analyses were conducted to assess the prognostic role of MAPT methylation. Among 107 eligible patients with adequate follow-up, there were 40 deaths (37.4%). Five-year OS was significantly lower in MAPT-hypermethylated cases than MAPT-unhypermethylated cases (57.1% vs 79.4%; log-rank P=0.011) in Kaplan–Meier analysis (Figure 1). MAPT hypermethylation was associated with a significant decrease in OS (HR: 2.33; 95% CI: 1.19–4.57; P=0.014) in univariate Cox regression analysis. MAPT hypermethylation remained significantly associated with OS (HR: 2.29; 95% CI: 1.01–5.18; P=0.047) in multivariate analysis (Table 3).
Figure 1.
Kaplan–Meier survival estimates of overall survival between groups classified by MAPT methylation status in patients with stage II colorectal cancer. P-values were based on the log-rank test.
Table 3.
Univariate and multivariate Cox proportional hazard analysis of overall survival
| Variables | Total n | OS | |||
|---|---|---|---|---|---|
| Univariate | Multivariate | ||||
| HR (95% CI) | P-value | HR (95% CI) | P-value | ||
| Age (years) | 0.072 | 0.234 | |||
| <60 | 33 | 1.00 (Referent) | 1.00 (Referent) | ||
| ≥60 | 74 | 2.04 (0.94–4.45) | 1.68 (0.72–3.92) | ||
| Sex | 0.824 | ||||
| Male | 47 | 1.00 (Referent) | |||
| Female | 60 | 0.93 (0.50–1.74) | |||
| Location | 0.439 | 0.656 | |||
| Proximal | 55 | 1.00 (Referent) | 1.00 (Referent) | ||
| Distal | 52 | 0.78 (0.42–1.46) | 1.18 (0.56–2.48) | ||
| Lymph nodes examined | 0.514 | 0.433 | |||
| ≥12 | 74 | 1.00 (Referent) | 1.00 (Referent) | ||
| <12 | 33 | 1.24 (0.65–2.38) | 1.33 (0.65–2.71) | ||
| Differentiation | 0.463 | 0.804 | |||
| Well to moderate | 89 | 1.00 (Referent) | 1.00 (Referent) | ||
| Poor | 18 | 0.70 (0.28–1.80) | 0.88 (0.31–2.49) | ||
| pT4 | 0.326 | 0.689 | |||
| No | 96 | 1.00 (Referent) | 1.00 (Referent) | ||
| Yes | 11 | 0.55 (0.17–1.80) | 0.77 (0.21–2.83) | ||
| MAPT hypermethylation | 0.014 | 0.047 | |||
| No | 82 | 1.00 (Referent) | 1.00 (Referent) | ||
| Yes | 25 | 2.33 (1.19–4.57) | 2.29 (1.01–5.18) | ||
Discussion
In this study, we examined the methylation status of MAPT promoter CpG island and its possible prognostic value in patients with stage II CRC. We demonstrate here for the first time that MAPT CpG island of promoter is hypermethylated in 23.4% of patients with stage II CRC and MAPT methylation was absent from a subgroup of paired normal colorectal mucosa, confirming that this methylation is tumor specific. Importantly, this event was unequally distributed among different ages; MAPT methylation was much more common in patients with age ≥60 than in patients with age <60. Furthermore, MAPT methylation was strongly associated with tumor location and CIMP status. MAPT appears to be preferentially methylated among proximal colon tumors or CIMP high tumors. Most important is that we highlight the negative and independent prognostic effect of MAPT hypermethylation on prognosis in patients with CRC using qMSP.
Pusztai et al previously assessed tau protein expression in primary tumors from 1942 patients with breast cancer by using tissue microarrays. They found 43% of patients were tau positive. Tau positivity was correlated with lower histologic grade and associated with better disease-free survival and OS both in univariate and multivariate analyses.14 Hristodorov et al cloned tau in-frame with EGF using EGF as a binding component to test the efficacy of tau. The tau-mediated and proliferation-dependent antitumor activity was demonstrated in vitro using EGF receptor-overexpressing breast cancer cell lines and in vivo using a mouse xenograft model.15 This holds true in lymphomas by using similar technique, in which study they noted tau could induce apoptosis when delivered to rapidly proliferating cancer cells.16 Collectively, these observations provide strong evidence that MAPT is a candidate tumor-suppressor gene.
In contrast, an analysis of tau expression in 102 primary breast carcinomas and matched metastases reveals that 52% hold tau expression in metastases and 26% show significantly elevated tau expression during disease progression. They also demonstrated that endogenic tau localized to microtentacles and was both essential and adequate to promote microtentacle extension in detached breast tumor cells, and tau-induced microtentacles enhanced the suspended cells to reattach and the circulating tumor cells to retain in lung capillaries.17 These results suggest that tau may augment, rather than inhibit tumor development.
Previous studies showed that tau was a multifunctional protein, whose role depended on its localization. For example, it mediates microtubule polymerization and stabilization in the cytoskeleton,18 while involved in DNA protection and the promotion of chromosomal stability within the nucleus.19,20 In addition, the MAPT primary transcript contains 16 exons, alternative splicing of these exons results in at least six tau isoforms.21,22 Each of these isoforms is likely to have specific physiological roles since they are differentially expressed over development. Moreover, the longest tau isoform have 80 putative Ser or Thr phosphorylation sites. Phosphorylation of these sites differentially influences its biological function.23 Taken together, localizations, differential splicing, and phosphorylation may give rise to a complex pattern of interacting tau isoforms with tumor-suppressor or oncogenic functions. Therefore, for facilitating our comprehension of tau’s function in cancer, further studies should investigate its isoforms and the effect of phosphorylation of tau as the microarray probes or the antibody used were directed against shared domains and were not aimed at phosphorylation status.
Traditional risk factors for CRC include <12 lymph nodes examined, poor histologic differentiation, pT4 lesions, intestinal obstruction or perforation, and lymphovascular invasion. These high-risk features are also recommended for clinical decisions regarding adjuvant therapy in stage II CRC. However, these features have mainly been accepted from indirect evidence in stage III CRC studies and their clinical performance has limitations. As shown in our present study, none of the risk factors including <12 lymph nodes examined, tumor differentiation, or pT4 lesions, was associated with OS in patients with stage II CRC.
To our knowledge, the expression and methylation of MAPT in colon tissue or CRC has not been reported to date. However, a study analyzing tau levels in tissue samples from patients with Alzheimer’s disease revealed that total tau protein has the highest levels in brain, followed by submandibular gland, sigmoid colon, liver, scalp, and abdominal skin.24 We searched the Cancer Genome Atlas (TCGA) database (https://cancergenome.nih.gov/) and found that 434 cases of primary CRC samples had both MAPT gene expression and methylation data. MAPT gene was differentially expressed and methylated (data not shown). In our series, MAPT promoter CpG island is frequently methylated in stage II CRC. Poorer prognosis is found in patients with MAPT hypermethylated tumors regardless of several clinicopathological parameters. It is also important to consider that the inclusion of other methylated genes in addition to MAPT in a multigene or multimolecular prediction score can improve its prognostic values. Further studies to evaluate the function of tau in CRC are currently in progress.
Acknowledgments
We thank Kathy Bender and Joann Murphy at the Johns Hopkins Hospital for administrative support. We also thank Sharon Metzger-Gaud, Theresa Sanlorenzo-Caswell, and the Johns Hopkins Cancer Registry for assistance with the primary cancer databases. This work was supported by the Scientific Research Foundation for the Returned Overseas Chinese Scholars, State Education Ministry of China.
Abbreviations
MAPT, microtubule-associated protein tau; CRC, colorectal cancer; OS, overall survival; Ct, threshold cycles; FFPE, formalin-fixed and paraffin-embedded.
Disclosure
Nita Ahuja has received grant funding from Cepheid and Astex and has served as consultant to Ethicon. She has licensed methylation biomarkers to Cepheid and has a patent Predicting Response to Epigenetic Drug Therapy pending to JHU, a patent Diagnostic Test for the Early Detection of Pancreatic Cancer with royalties paid to Cepheid, and a patent Markers for Improved Detection of Breast Cancer issued. The authors report no other conflicts of interest in this work.
References
- 1.Global Burden of Disease Cancer C, Fitzmaurice C, Allen C, et al. Global, regional, and national cancer incidence, mortality, years of life lost, years lived with disability, and disability-adjusted life-years for 32 Cancer Groups, 1990 to 2015: a systematic analysis for the Global Burden of Disease Study. JAMA Oncol. 2017;3:524–548. doi: 10.1001/jamaoncol.2016.5688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Toyota M, Ahuja N, Ohe-Toyota M, Herman JG, Baylin SB, Issa JP. CpG island methylator phenotype in colorectal cancer. Proc Natl Acad Sci. 1999;96:8681–8686. doi: 10.1073/pnas.96.15.8681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berman BP, Weisenberger DJ, Aman JF, et al. Regions of focal DNA hypermethylation and long-range hypomethylation in colorectal cancer coincide with nuclear lamina-associated domains. Nat Genet. 2011;44:40–46. doi: 10.1038/ng.969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Neve RL, Harris P, Kosik KS, Kurnit DM, Donlon TA. Identification of cDNA clones for the human microtubule-associated protein tau and chromosomal localization of the genes for tau and microtubule-associated protein 2. Brain Res. 1986;387:271–280. doi: 10.1016/0169-328x(86)90033-1 [DOI] [PubMed] [Google Scholar]
- 5.Arendt T, Stieler JT, Holzer M. Tau and tauopathies. Brain Res Bull. 2016;126:238–292. doi: 10.1016/j.brainresbull.2016.08.018 [DOI] [PubMed] [Google Scholar]
- 6.Drubin DG, Kirschner MW. Tau protein function in living cells. J Cell Bio. 1986;103:2739–2746. doi: 10.1083/jcb.103.6.2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yu JZ, Rasenick MM. Tau associates with actin in differentiating PC12 cells. Faseb J. 2006;20:1452–1461. doi: 10.1096/fj.05-5206com [DOI] [PubMed] [Google Scholar]
- 8.Iwata A, Nagata K, Hatsuta H, et al. Altered CpG methylation in sporadic Alzheimer’s disease is associated with APP and MAPT dysregulation. Hum Mol Genet. 2014;23:648–656. doi: 10.1093/hmg/ddt451 [DOI] [PubMed] [Google Scholar]
- 9.Coupland KG, Mellick GD, Silburn PA, et al. DNA methylation of the MAPT gene in Parkinson’s disease cohorts and modulation by vitamin E in vitro. Mov Disord. 2014;29:1606–1614. doi: 10.1002/mds.25784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shui IM, Wong CJ, Zhao S, et al. Prostate tumor DNA methylation is associated with cigarette smoking and adverse prostate cancer outcomes. Cancer. 2016;122:2168–2177. doi: 10.1002/cncr.30045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brandes JC, Carraway H, Herman JG. Optimal primer design using the novel primer design program: mSPprimer provides accurate methylation analysis of the ATM promoter. Oncogene. 2007;26:6229–6237. doi: 10.1038/sj.onc.1210433 [DOI] [PubMed] [Google Scholar]
- 12.Weisenberger DJ, Siegmund KD, Campan M, et al. CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat Genet. 2006;38:787–793. doi: 10.1038/ng1834 [DOI] [PubMed] [Google Scholar]
- 13.Fu T, Pappou EP, Guzzetta AA, et al. CpG island methylator phenotype-positive tumors in the absence of MLH1 methylation constitute a distinct subset of duodenal adenocarcinomas and are associated with poor prognosis. Clin Cancer Res. 2012;18:4743–4752. doi: 10.1158/1078-0432.CCR-12-0707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pusztai L, Jeong JH, Gong Y, et al. Evaluation of microtubule-associated protein-tau expression as a prognostic and predictive marker in the NSABP-B 28 randomized clinical trial. J Clin Oncol. 2009;27:4287–4292. doi: 10.1200/JCO.2008.21.6887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hristodorov D, Mladenov R, Pardo A, et al. Microtubule-associated protein tau facilitates the targeted killing of proliferating cancer cells in vitro and in a xenograft mouse tumour model in vivo. Br J Cancer. 2013;109:1570–1578. doi: 10.1038/bjc.2013.457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hristodorov D, Nordlohne J, Mladenov R, et al. Human microtubule-associated protein tau mediates targeted killing of CD30(+) lymphoma cells in vitro and inhibits tumour growth in vivo. Br J Haematol. 2014;164:251–257. doi: 10.1111/bjh.12626 [DOI] [PubMed] [Google Scholar]
- 17.Matrone MA, Whipple RA, Thompson K, et al. Metastatic breast tumors express increased tau, which promotes microtentacle formation and the reattachment of detached breast tumor cells. Oncogene. 2010;29:3217–3227. doi: 10.1038/onc.2010.68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Drechsel DN, Hyman AA, Cobb MH, Kirschner MW. Modulation of the dynamic instability of tubulin assembly by the microtubule-associated protein tau. Mol Bio Cell. 1992;3:1141–1154. doi: 10.1091/mbc.3.10.1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rossi G, Dalpra L, Crosti F, et al. A new function of microtubule-associated protein tau: involvement in chromosome stability. Cell Cycle. 2008;7:1788–1794. doi: 10.4161/cc.7.12.6012 [DOI] [PubMed] [Google Scholar]
- 20.Violet M, Delattre L, Tardivel M, et al. A major role for Tau in neuronal DNA and RNA protection in vivo under physiological and hyperthermic conditions. Front Cell Neurosci. 2014;8:84. doi: 10.3389/fncel.2014.00052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goedert M, Spillantini MG, Potier MC, Ulrich J, Crowther RA. Cloning and sequencing of the cDNA encoding an isoform of microtubule-associated protein tau containing four tandem repeats: differential expression of tau protein mRNAs in human brain. Embo J. 1989;8:393–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goedert M, Wischik CM, Crowther RA, Walker JE, Klug A. Cloning and sequencing of the cDNA encoding a core protein of the paired helical filament of Alzheimer disease: identification as the microtubule-associated protein tau. Proc Natl Acad Sci. 1988;85:4051–4055. doi: 10.1073/pnas.85.11.4051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sergeant N, Delacourte A, Buee L. Tau protein as a differential biomarker of tauopathies. Biochim Biophys Acta. 2005;1739:179–197. doi: 10.1016/j.bbadis.2004.06.020 [DOI] [PubMed] [Google Scholar]
- 24.Dugger BN, Whiteside CM, Maarouf CL, et al. The presence of select tau species in human peripheral tissues and their relation to Alzheimer’s disease. J Alzheimer’s Dis. 2016;54:1249. doi: 10.3233/JAD-169007 [DOI] [PubMed] [Google Scholar]