Abstract
Objective:
To provide an update of currently recognized clinically relevant candidate and known genes for human reproduction and related infertility plotted on high resolution chromosome ideograms (850 band level) and represented alphabetically in tabular form.
Method:
Descriptive authoritative computer-based website and peer-reviewed medical literature searches used pertinent keywords representing human reproduction and related infertility along with genetics and gene mutations. A master list of genes associated with human reproduction and related infertility was generated with a visual representation of gene locations on high resolution chromosome ideograms. GeneAnalytics pathway analysis was carried out on the resulting list of genes to assess underlying genetic architecture for infertility.
Results:
Advances in genetic technology have led to the discovery of genes responsible for reproduction and related infertility. Genes identified (N = 371) in our search primarily impact ovarian steroidogenesis through sex hormone biology, germ cell production, genito-urinary or gonadal development and function, and related peptide production, receptors and regulatory factors.
Conclusions:
The location of gene symbols plotted on high resolution chromosome ideograms forms a conceptualized image of the distribution of human reproduction genes. The updated master list can be used to promote better awareness of genetics of reproduction and related infertility and advance discoveries on genetic causes and disease mechanisms.
Keywords: Human reproduction and related infertility, genes, Genetic pathways, Meiosis, Steroidogenesis, Chromosome ideograms
1. Introduction
Infertility is the inability of couples who are sexually active and not taking contraceptives to achieve a pregnancy within one year. About 70 million infertile couples are noted worldwide and 10%–15% of the US population experiences infertility (Silber and Barbey, 2012; Ombelet et al., 2008). Male infertility accounts for about 50% of infertile cases and can be multifactorial in origin; however, structural chromosomal and mitochondrial DNA abnormalities and hormonal/endocrine disturbances are also known. Non-genetic causes can include abnormal testicular development or descent, genital tract abnormalities, infection, age-related factors, chronic illnesses, impotence, medication and immunity status (Shah et al., 2003). Female infertility is caused by hormonal, anatomical, genetic or environmental factors such as fibroids, tubal blockage, cervical mucus or chromosome abnormalities, pelvic inflammatory disease and age-related factors (Olooto et al., 2012). Women beginning in their 30s will experience a greater than 25% chance of becoming infertile (Silber and Barbey, 2012; Ombelet et al., 2008).
An increased prevalence of human infertility and reproductive problems have been observed in westernized societies and is often related to a selected delay in establishing pregnancies by older women and the ensuing increase in genetic and hormonal imbalance with advancing age in both men and women. The increasing worldwide obesity epidemic further impacts reproduction by disturbing energy balance and expenditure often influenced by genetic and hormonal factors contributing to ovulation dysfunction, spontaneous abortions and overall infertility (Silber and Barbey, 2012; Marsh and Hecker, 2014; Pandy et al., 2010; Balen et al., 2006; Gesink Law et al., 2007; Jungheim et al., 2012; Ogden et al., 2006). Genes encode proteins affecting reproductive organs and function, germ cell production and hormonal factors as well as those contributing to caloric intake (i.e., food seeking, eating behavior), body composition (fat and fat-free mass) and energy storage and utilization (physical activity and metabolism) (Jungheim et al., 2012; Choquet and Meyre, 2011a, 2011b; Zaadstra et al., 1993; Rich-Edwards et al., 1994; Metwally et al, 2007). Obesity often complicates reproduction by affecting genetic factors that are known to play a role in the coordination of many encoded proteins released at selected intervals for the establishment of pregnancies (Jungheim er al., 2012; Metwally et al., 2007). For example, genetic-related conditions that correlate with decreased reproduction (e.g., fewer reproductive cycles) include women with polycystic ovarian syndrome (PCOS) and female carriers of the fragile X syndrome (i.e., FMR1 gene mutation) (Pasquali and Gambineri, 2006; Peng et al., 2014; Ehrmann, 2005). About 5 to 10% of women from the general population are diagnosed with PCOS and over 40 genes are known to play a role in this disorder (Venkatesh et al., 2014).
A recent computer-based search of websites and peer-reviewed medical literature sources (e.g., www.pubmed.org;www.ncbi.nlm.nih.gov) using keywords related to obesity and genetics identified 370 clinically relevant candidate and known genes for obesity and similarly 153 human infertility and reproduction genes when searching keywords related to infertility and reproduction (Butler et al., 2015). Among them, at least 21 genes were found to be associated with both obesity and human infertility. We now report an updated list of clinically relevant and known genes for human reproduction and related infertility with the display of gene symbols on high resolution (850 band level) chromosome ideograms and also alphabetically arranged in a tabular form. This illustrative effort of showing the location of genes involved with human infertility and reproduction on chromosome ideograms brings a better awareness of the gene(s) that might be involved in a given chromosomal aberration in an infertile couple and thereby render a more precise genetic basis for infertility, enabling a specific gene-based personalized medicine approach to treatment and genetic counseling. Additionally, GeneAnalytics (http://geneanalytics.genecards.org/) was used to assess the underlying genetic architecture of human reproduction and related infertility by mapping the identified genes to tissues and cells, diseases, phenotypes, molecular pathways and biological processes with the greatest overlap and probable relevance.
2. Materials and methods
Our study involved a computer-based internet approach by searching peer-reviewed articles published in the medical literature [e.g., PubMed (www.pubmed.org)] and other relevant computer-based authoritative websites including Online Mendelian Inheritance in Man (www.OMIM.org); GeneCards (www.genecards.org); and the National Center for Biotechnology Information.
(www.ncbi.nlm.nih.gov). We searched keywords including human infertility, reproduction, meiosis, azoospermia, premature ovarian failure, primary ovarian insufficiency, endometriosis, diminished ovarian reserve and estrogen combined with other keywords such as genes, genetics, mutations or gene variants in order to identify sources with evidence for reproductive genetic factors that are supported by clinical, functional or experimental data for the causation of human infertility in both sexes. The title of the research articles found through this web-based search often contained the keywords of human infertility and genes (or genetics). The research articles were then examined for evidence of involvement of genes or genetic factors playing a role in human infertility or reproduction. Specifically, reviews of whole-genome-wide association studies (GWAS), gene linkage or expression patterns with DNA sequencing of infertile individuals identified a list of multiple genes which were compiled to develop an updated master list of such genes. We then plotted the recognized symbols for each gene that is known or is clinically relevant for causing human infertility and/or involved in reproduction onto high resolution (850 band level) chromosome ideograms (Butler et al., 2015; Shaffer et al., 2013). The master list of all identified genes was then alphabetized by gene symbol, full name of the gene and chromosome band location in a tabular form.
Genome-wide pathway analysis was carried out for the derived gene master list and all mapped genomic variants associated with human reproduction and related infertility using GeneAnalytics (http://geneanalytics.genecards.org/), a commercially available software program based on proprietary, comprehensive and organized databases from the LifeMap suite. Mammalian genes were organized into functional categories based upon tissues and cells, diseases, pathways, GO-biological processes, GO-molecular function, phenotypes and compounds (endogenous or exogenous) with functional link to the queried gene set. Gene ontology (GO) terms (e.g., superpathways and GO-biological function) were scored based upon transformation of the binomial p-value which is equivalent to a corrected p-value with significance defined at p < 0.0001. Disease-matching scores were derived based upon the number of overlapping genes and the nature of the gene-disease association. Tissues and cells were scored using a matching algorithm that weighs tissue specificity, abundance and function of the gene. Related pathways were then grouped into superpathways to improve inferences, pathway enrichment, reduce redundancy, and rank genes within a biological mechanism via the multiplicity of constituent pathways (Belinky et al., 2015).
3. Results and discussion
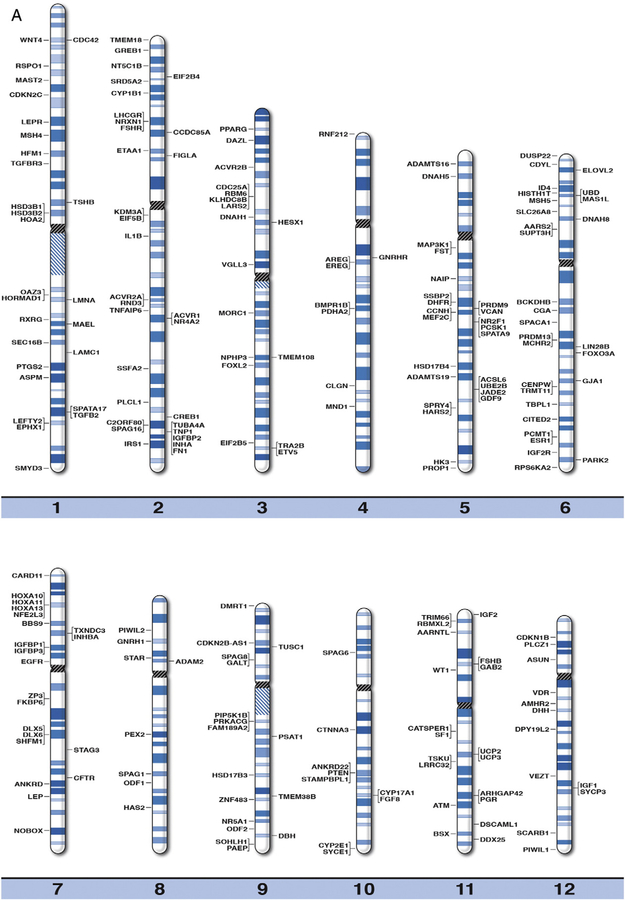
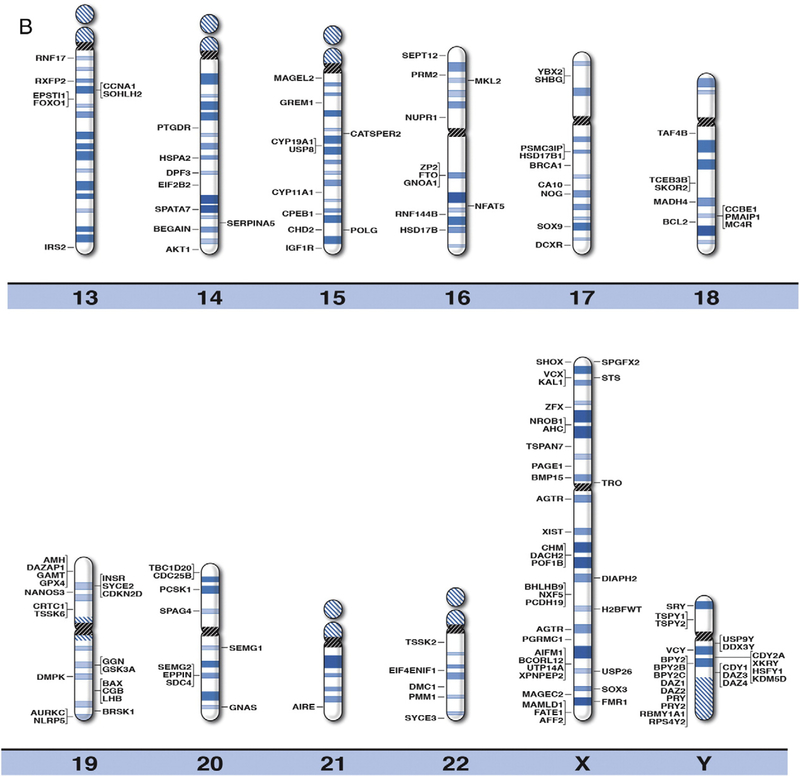
We developed high resolution chromosome ideograms at the 850 band level and plotted the location of the gene symbol representing each gene onto the ideogram at the precise chromosome band or subband level for each of the 371 genes reported to play a role in human infertility and/or reproduction. The genes were identified by searching the medical literature and computer-based websites. Clinically relevant or known human infertility and/or reproductive genes were found to be distributed on all chromosomes (Fig. 1). Not surprisingly, the largest percentage of human reproduction and related infertility genes (i.e., 59 of 371 genes or 16%) were located on the sex chromosome pair (i.e., X and Y) in relationship to the autosomes or non-sex chromosomes. The gene symbol, expanded name and chromosome location are listed in alphabetical order in Table 1 for each of the 371 human infertility and/or reproductive genes. These genes represented a wide range of functions including sex hormone, peptide receptors, organ development, growth, gene transcription or translational factors, germ cell production and metabolic or neuronal influences (Venkatesh et al., 2014; Layman, 2002; Okada et al., 2010, Hu et al., 2014; Kosova et al., 2012; El Inati et al., 2012; Qiu et al., 2013; Ferfouri et al., 2013; Albertsen et al., 2013; Brannian and Hansen, 2002; Fragouli et al., 2014; Bulun, 2014; Greene et al., 2014; Qin et al., 2014; Abid et al., 2013; Bergh et al., 1993).
Fig. 1.
High resolution human chromosome ideograms (850 band level) with gene symbols representing genetic biomarkers for infertility and reproduction plotted on chromosome bands for each of the 371 genes. The centromere area is highlighted in black which separates the upper short ‘p’ arm from the lower long ‘q’ arm for each chromosome. The gene symbols are arranged in alphabetical order with the expanded name and precise chromosome band location listed in Table 1.
Table 1.
Currently recognized genes for human reproduction and infertility with their chromosome locations.
| GENE SYMBOL | GENE NAME | BAND |
|---|---|---|
| AARS2 | Leukoencephalopathy, progressive with ovarian failure; LKENP | 6p21:1 |
| ACSL6 | Acyl-CoA synthetase long-chain family member 6 | 5q31.1 |
| ACVR1 | Activin A receptor, type I | 2q24.1 |
| ACVR2A | Activin A receptor, type IIA | 2q22.3 |
| ACVR2B | Activin A receptor, type IIB | 3p22.2 |
| ADAM2 | A disintegrin and metalloproteinase domain 2 | 8p11.22 |
| ADAMTS16 | ADAM metalopeptidase with thrombospondin type I motif, 16 | 5p15.32 |
| ADAMTS19 | ADAM metalopeptidase with thrombospondin type I motif, 19 | 5q23.3 |
| AFF2 | Fragile X mental retardation 2 | Xq28 |
| AGTR2 | Angiotensin lireceptor, type2 | Xq23 |
| AHC | Adrenal hypoplasia, congenital | Xp21.2 |
| AIFM1 | Apoptosis-inducing factor, mitochondrion-associated, 1 | Xq26.1 |
| AIRE | Autoimmune regulator | 21q22.3 |
| AKT1 | V-Akt murine thymoma viral oncogene homolog 1 | 14q32.33 |
| AMH | Anti-mullerian hormone | 19p13.3 |
| AMHR2 | Anti-mullerian hormone receptor, type II | 12q13.13 |
| ANKRD7 | Ankyrin repeat domain 7 | 7q31.31 |
| ANKRD22 | Ankyrin repeat domain 22 | 10q23.31 |
| AR | Androgen receptor | Xq12 |
| AREG | Amphiregulin | 4q13.3 |
| ARHGAP42 | Rho GTPase-activating protein 42 | 11q22.1 |
| ARNTL | Aryl hydrocarbon receptor nuclear translocator-like | 11p15.2 |
| ASPM | ASP (abnormal spindle) homolog, microcephaly associated (Drosophila) | lq31.3 |
| ASUN | Asunder spermatogenesis regulator | 12p11.23 |
| ATM | Ataxia-telangiectasia mutated | 11q22.3 |
| AURKC | Aurora kinase C | 19q13.43 |
| BAX | BCL2-associated X protein | 19q13.33 |
| BBS9 | Parathyroid hormone- responsive B1 | 7p14.3 |
| BCKDHB | Branched-chain keto acid dehydrogenase E1, beta polypeptide | 6q14.1 |
| BCL2 | B-cell CLL/lymphoma 2 | 18q21.33 |
| BCORL1 | BCL6 corepressor-like 1 | Xq26.1 |
| BECAIN | Brain-enriched granylate kinase-associated | 14q32.2 |
| BHLHB9 | Basic helix-loop-helix domain- containing protein, class B9 | Xq22.1 |
| BMP15 | Bone morphogenetic protein 15 | Xq11.22 |
| BMPR1B | Bone morphogenetic protein receptor, type 1B | 4q22.3 |
| BPY2 | Basic charge, Y-linked, 2 | Yq11.223 |
| BPY2B | Basic charge, Y-linked, 2B | Yq11.223 |
| BPY2C | Basic charge, Y-linked, 2C | Yq11.223 |
| BRCA1 | Breast cancer 1, early onset | 17q21.31 |
| BRDT | Bromodomain, testis-specific | 1p22.1 |
| BRSK1 | BR serine/threonine kinase 1 | 19ql3.42 |
| BSX | Brain specific homeobox | 11q24.1 |
| CA10 | Carbonic anhydrase X | 17q21.33 |
| CATSPFR1 | Cation channel, sperm associated 1 | 11q13.1 |
| CATSPER2 | Cation channel, sperm associated 2 | 15q15.3 |
| CARD11 | Caspase recruitment domain family, member 11 | 7p22.2 |
| CCBE1 | Collagen and calcium-binding EGF domain containing protein 1 | 18q21.32 |
| CCDC85A | Coiled-coil domain containing 85A | 2p16.1 |
| CCNA1 | Cyclin A1 | 13q13.3 |
| CCNH | Cyclin H | 5q14.3 |
| CDC25A | Cell division cycle 25A | 3p21.31 |
| CDC25B | Cell division cycle 25B | 20p13 |
| CDC42 | Cell division cycle 42 | 1p3612. |
| CDC6 | Cell division cycle 6 | 17q21.2 |
| CDKN1B | Cyclin-dependent kinase inhibitor 1B | 12p13.1 |
| CDKN2B-AS1 | CDKN2B antisense RNA 1 | 9p21.3 |
| CDKN2C | Cyclin-dependent kinase inhibitor 2C (p18, inhibits CDK4) | 1p32.3 |
| CDKN2D | Cyclin-dependent kinase inhibitor 2D (p19, inhibits CDK4) | 19p13.2 |
| CDY1 | Chromodomain protein, Y- linked, 1 | Yq11.23 |
| CDY2A | Chromodomain protein, Y- linked, 2A | Yq11.222 |
| CDYL | Chromodomain protein, Y-like | 6p25.1 |
| CENPW | Centromeric protein W | 6q22.32 |
| CFTR | Cystic fibrosis transmembrane conductance regulator | 7q31.2 |
| CGA | Chorionic gonadotropin, alpha chain | 6q14.3 |
| CGB | Chorionic gonadotropin, beta polypeptide | 19q13.33 |
| CHD2 | Chromodomain helicase DNA- binding protein 2 | 15q26.1 |
| CHM | Choroideremia | Xq21.2 |
| CITED2 | CBP/p300-interacting transactivator, with glu/asp- rich C-terminal domain 2 | 6q24.1 |
| CLGN | Calmegin | 4q31.1 |
| CPEB1 | Cytoplasmic polyadenylation element-binding protein 1 | 15q25.2 |
| CREB1 | cAMP response element- binding protein 1 | 2q33.3 |
| CRTC1 | CREB-regulated transcription co-activator 1 | 19p13.11 |
| CTNNA3 | Catenin (Cadherin-associated protein), alpha 3 | 10q21.3 |
| CYP11A1 | Cytochrome p450, family 11, subfamily A, polypeptide 1 | 15q24.1 |
| CYP17A1 | Cytochrome p450, family 17, subfamily A, polypeptide 1 | 10q24.32 |
| CYP19A1 | Cytochrome p450, family 19, subfamily A, polypeptide 1 | 15q21.2 |
| CYP1B1 | Cytochrome p450, family 1, subfamily B, polypeptide 1 | 2p22.2 |
| CYP2E1 | Cytochrome p450, family 2, subfamily E, polypeptide 1 | 10q26.3 |
| C20RF80 | Chromosome 2 open reading frame 80 | 2q34 |
| DACH2 | Dachshund, Drosophila, homolog 2 | Xq21.2 |
| DAZ1 | Deleted in azoospermia 1 | Yq11.223 |
| DAZ2 | Deleted in azoospermia 2 | Yq11.223 |
| DAZ3 | Deleted in azoospermia 3 | Yq11.23 |
| DAM | Deleted in azoospermia 4 | Yq11.23 |
| DAZAP1 | DAZ associated protein 1 | 19p13.3 |
| DAZL | Deleted in azoospermia-Like | 3p24.3 |
| DBH | Dopamine α-hydroxylase | 9q34.2 |
| DCXR | Dicarbonyl/L-xylulose reductase | 17q25.3 |
| DDX25 | DEAD (asp-glu-ala-asp) box helicase 25 | 11q24.2 |
| DDX3Y | DEAD (asp-glu-ala-asp) box helicase 3, Y-linked | Yq11.21 |
| DHFR | Dihydrofolate reductase | 5q14.1 |
| DHH | Desert hedgehog | 12q13.l2 |
| DIAPH2 | Diaphanous, Drosophila, homolog of, 2 | Xq21.33 |
| DLX5 | Distal-less homeobox 5 | 7q21.3 |
| DLX6 | Distal-less homeobox 6 | 7q21.3 |
| DMC1 | Disrupted meiotic cDNA 1 | 22q13.1 |
| DMPK | Dystrophia myotonica-protein kinase | 19q13.32 |
| DMRT1 | Doublesex -and MAB3- related transcription factor 1 | 9p24.3 |
| DNAH1 | Dynein, axonemal, heavy chain 1 | 3p21.1 |
| DNAH5 | Dynein, axonemal, heavy chain 5 | 5p15.2 |
| DNAH8 | Dynein, axonemal, heavy chain 8 | 6p21.2 |
| DPF3 | D4, zinc, and double PHD fingers, family, member 3 | 14q24.2 |
| DPY19L2 | DPY19-like 2 | 12q14.2 |
| DSCAML1 | Down syndrome cell adhesion molecule-like 1 | 11q23.3 |
| DUSP22 | Dual specificity phosphatase 22 | 6p25.3 |
| EGFR | Epidermal growth factor receptor | 7p11.2 |
| EIF2B2 | Eukaryotic translation initiation factor 2b, subunit 2 | 14q24.3 |
| EIF2B4 | Eukaryotic translation initiation factor 2b, subunit 4 | 2p23.3 |
| E1F2B5 | Eukaryotic translation initiation factor 2b, subunit 5 | 3q27.1 |
| EIF5B | Eukaryotic translation initiation factor 5b | 2q11.2 |
| EIF4ENIF1 | Eukaryotic translation initiation factor 4E nuclear import factor 1 | 22q12.2 |
| EL0VL2 | Elongation of very long chain fatty acids-like 2 | 6p24.2 |
| EPHX1 | Epoxide hydrolase 1 | 1q42.l2 |
| EPPIN | Epididymal peptidase inhibitor | 20q13.12 |
| EPSTI1 | Epithelial stromal interaction 1 | 13q14.11 |
| EREG | Epiregulin | 4q13.3 |
| ESR1 | Estrogen receptor 1 | 6q25.1 |
| ETAA1 | Ewing tumor-associated antigen 1 | 2p14 |
| ETV5 | ETS variant gene 5 | 3q27.2 |
| FAM189A2 | Family with sequence similarity 189, member A2 | 9q21.11 |
| FATE1 | Fetal and adult testis expressed 1 | Xq28 |
| FCF8 | Fibroblast growth factor 8 | 10q24.32 |
| F1GLA | Folliculogenesis specific basic helix-loop-helix | 2p13.3 |
| FKBP6 | FK506 binding protein 6 | 7q11.23 |
| FMN2 | Formin 2 | 1q43 |
| FMR1 | Fragile X mental retardation 1 | Xq27.3 |
| FM | Fibronectin 1 | 2q35 |
| FOXL2 | Forkhead box L2 | 3q22.3 |
| FOXO1 | Forkhead box O1 | 13q14.11 |
| FOXO3A | Forkhead box O3A | 6q21 |
| FST | Follistatin | 5q11.2 |
| FSHB | Follicle stimulating hormone, beta polypeptide | 11p14.1 |
| FSHR | Follicle stimulating hormone receptor | 2pl6.3 |
| FTO | Fat mass-and obesity- associated gene | 16q12.2 |
| GAB2 | GRB2-associated binding protein 2 | UqU.1 |
| GALT | Galactose 1 phosphate uridylyl transferase | 9p13.3 |
| GAMT | Guanidinoacetate N-methyl transferase | 19p13.3 |
| GDF9 | Growth/differentiation factor 9 | 5q31.7 |
| GGN | Gametogenetin | 19q13.2 |
| GJA1 | GAP junction protein, alpha −1 | 6q22.31 |
| GNAO1 | Guanine nucleotide-binding protein, alpha activating activity polypeptide O | 16q12.2 |
| GNAS | Guanine nucleotide-binding protein, alpha-stimulating activity polypeptide 1 | 20q13.32 |
| GNRH1 | Gonadotropin-releasing hormone 1 | 8p21.2 |
| GNRHR | Gonadotropin-releasing hormone receptor | 4q13.2 |
| GPR3 | G protein-coupled receptor 3 | 1p36.11 |
| GPX4 | Glutathione peroxidase 4 | 19p13.3 |
| GREB1 | Growth regulation by estrogen in breast cancer 1 | 2p25.1 |
| GREM1 | Gremlin 1 homolog, cystine knot superfamily | 15q13.3 |
| GSK3A | Glycogen synthase kinase 3 alpha | 19q13.2 |
| H2BFWT | H2B histone family, member W, testis-specific | Xq22.2 |
| HAO2 | Hydroxyacid oxidase 2 | 1p12 |
| HARS2 | Histidyl-tRNA synthetase2 | 5q31.3 |
| HAS2 | Hyaluronan synthase 2 | 8q24.13 |
| HESX1 | HESX homeobox 1 | 3p14.3 |
| HFM1 | Premature ovarian failure 9; POF9 | 1p22.2 |
| HIST1H1T | Histone gene cluster 1, HI histone family, member T | 6p22.2 |
| HK3 | Hexokinase 3 | 5q35.2 |
| HORMAD1 | HORMA domain containing 1 | 1q21.3 |
| HOXA10 | Homeobox A10 | 7p15.2 |
| HOXA11 | Homeobox A11 | 7p15.2 |
| HOXA13 | Homeobox A13 | 7p15.2 |
| HSD3B1 | 3-beta-hydroxysteroid dehydrogenase I | 1p12 |
| HSD3B2 | 3-beta-hydroxysteroid dehydrogenase II | 1p12 |
| HSD17B1 | 17-beta-hydroxysteroid dehydrogenase 1 | 17q21.2 |
| HSD17B2 | 17-beta-liydroxysteroid dehydrogenase II | 16q23.3 |
| HSD17B3 | 17-beta-hydroxysteroid dehydrogenase III | 9q22.32 |
| HSD17B4 | 17-beta-hydroxysteroid dehydrogenase IV | 5q23.1 |
| HSFY1 | Heat shock transcription factor, Y-linked 1 | Yq11.222 |
| HSPA2 | Heat shock 70kDa protein 2 | 14q23.3 |
| ID4 | Inhibitor of DNA binding 4 | 6p22.3 |
| IGF1 | Insulin-like growth factor 1 | 12q23.2 |
| IGF1R | Insulin-like growth factor 1 receptor | 15q26.3 |
| IGF2 | Insulin-like growth Factor 2 | 11p15.5 |
| IGF2R | Insulin-like growth factor 2 receptor | 6q25.3 |
| IGFBP1 | Insulin-like growth factor- binding protein 1 | 7p12.3 |
| IGFBP2 | Insulin-like growth factor- binding protein 2 | 2q35 |
| IGFBP3 | Insulin-like growth factor- binding protein 3 | 7p12.3 |
| IL1B | Interleukin 1, beta | 2q13 |
| INCENP | Inner centromere protein antigens 135/155kDa | 11q12.3 |
| INHA | Inhibin alpha | 2q35 |
| INHBA | Inhibin, beta A | 7p14.1 |
| INSR | Insulin receptor | 19p13.2 |
| IRS1 | Insulin receptor substrate 1 | 2q36.3 |
| IRS2 | Insulin receptor substrate 2 | 13q34 |
| JADE2 | PHD finger protein 15 | 5q31.1 |
| KALI | Kallmann syndrome 1 | Xp22.31 |
| KDM3A | Lysine (K)-specific demethylase 3A | 2p11.2 |
| KDM5D | Lysine (K)-specific demethylase 5D | Yq11.222 |
| KLHDC8B | Kelch domain-containing protein 8B | 3p21,31 |
| LAMC1 | Laminin, gamma 1 | 1q25.3 |
| LARS2 | Leucyl-tRNA synthetase 2 | 3p21.31 |
| LEFTY2 | Left-right determination factor 2 | 1q42.12 |
| LEP | Leptin | 7q32.1 |
| LEPR | Leptin receptor | 1p31.3 |
| LHB | Luteinizing hormone beta polypeptide | 19q13.33 |
| LHCGR | Luteinizing hormone/choriogonadotropin receptor | 2p16.3 |
| L1N28B | LIN 28, C. elegans, homolog of, B | 6q16.3 |
| LMNA | Lamin A | 1q22 |
| LRRC32 | Leucin-rich repeat containing protein 32 | 11q13.5 |
| MADH4 | Mothers against decapentaplegic homolog 4 | 18q21.2 |
| MAGEC2 | Melanoma antigen, family C, 2 | Xq27.2 |
| MAEL | Maelstrom spermatogenic transposon silencer | 1q24.1 |
| MAGEL2 | MAGE (melanoma-associated antigen)-like 2 | 15q11.2 |
| MAMLD1 | Mastermind-like domain- containing protein 1 | Xq28 |
| MAP3K1 | Mitogen-activated kinase kinase kinase 1 | 5q11.2 |
| MAS1L | MAS! oncogene-like | 6p22.1 |
| MAST2 | Microtubule associated serine/threonine kinase 2 | 1p34.1 |
| MCHR2 | Melanin-concentrating hormone receptor 2 | 6q16.2 |
| MC4R | Melanocortin 4 receptor | 18q2l.32 |
| MEF2C | MADS box transcription enhancer factor 2, polypeptide C | 5q14.3 |
| MKL2 | MKL/myocardin-like 2 | 16p13.12 |
| MND1 | Meiotic nuclear divisions 1 homolog (S. cerevisia) | 4q31.3 |
| MORC1 | MORC family CW-type zinc finger 1 | 3q13.13 |
| MSH4 | MutS homolog 4 | 1p31.1 |
| MSH5 | MutS homolog 5 | 6p21,33 |
| NA1P | NLR family, apoptosis inhibitory protein | 5q13.2 |
| NANOS3 | NANOS, Drosophila, homolog 3 | 19p13.13 |
| NFAT5 | Nuclear factor of activated T cells 5 | 16q22.1 |
| NFE2L3 | Nuclear factor erythroid 2-like 3 | 7p15.2 |
| NLRP5 | NLR family, Pyrin domain containing 5 | 19q13.43 |
| NOBOX | Newborn ovary homeobox | 7q35 |
| NOG | Noggin | 17q22 |
| NPHP3 | Nephronopthisis 3 | 3q22.1 |
| NR0B1 | Nuclear receptor subfamily 0, group B, member 1 | Xp21.2 |
| NR2F1 | Nuclear receptor subfamily 2, group F, member 1 | 5q15 |
| NR4A2 | Nuclear receptor subfamily 4, group A, member 2 | 2q24.1 |
| NR5A1 | Nuclear receptor subfamily 5, group A, member 1 | 9q33.3 |
| NRXN1 | Neurexin | 2p16.3 |
| NT5C1B | 5’-nucleotidase, cytosolic IB | 2p24.2 |
| NUPR1 | Nuclear protein, transcriptional regulator, 1 | 16p11.2 |
| NXF5 | Nuclear RNA export factor, 5 | Xq22.1 |
| OAZ3 | Ornithine decarboxylase antizyme 3 | 1q21.3, |
| ODF1 | Outer dense fiber of sperm tails 1 | 8q22.3 |
| ODF2 | Outer dense fiber of sperm tails 2 | 9q34.11 |
| PAEP | Progestagen-associated endometrial protein | 9q34.3 |
| PACE1 | P antigen family, member 1 (prostate associated) | Xp11.23 |
| PARK2 | Parkin RBR E3 ubiquitin protein ligase | 6q26 |
| PCDH19 | Protocadherin 19 | Xq22.1 |
| PCMT1 | Protein-L-isoaspartate (D- Aspartate) O- methvltransferase | 6q25.1 |
| PCSK1 | Proprotein convertase, subtilisin/kexin-type 1 | 5qJ5 |
| PCSK2 | Proprotein convertase, subtilisin/kexin-type 2 | 20p12.1 |
| PDHA2 | Pyruvate dehydrogenase, alpha-2 | 4q22.3 |
| PGR | Progesterone receptor | 11q22.1 |
| PGRMC1 | Progesterone receptor membrane component 1 | Xq24 |
| PIP5K1B | Phosphatidylinositol 4- phosphate 5-kinase, type1, beta | 9q21.11 |
| PIWIL1 | PlWI-like RNA-mediated gene silencing 1 | 12q24.33 |
| PIWIL2 | PlWI-like RNA-mediated gene silencing 2 | 8p21.3 |
| PLCL1 | Phospholipase C-like 1 | 2q33.1 |
| PLCZ1 | Phospholipase C, zeta 1 | 12p12.3 |
| PMAIP1 | Phorbol-12-myristate-13- acetate-induced protein 1 | 18q21.32 |
| PMM1 | Phosphomannomutase | 22q13.2 |
| POF1B | Premature ovarian failure, IB | Xq21.2 |
| POLG | Polymerase, DNA, gamma | 15q26.1 |
| PPARG | Peroxisome proliferator- activated receptor-gamma | 3p25.2 |
| PRDM9 | PR domain containing 9 | 5p14.2 |
| PRDM13 | PR domain containing 13 | 6q16.2 |
| PRKACG | Protein kinase, cAMP- dependent, catalytic, gamma | 9q21.11 |
| PRM2 | Protamine 2 | 16p13.13 |
| PROP1 | PROP paired-like homeobox 1 | 5q35.3 |
| PRY | PTPBL-related gene on Y | Yq11.223 |
| PRY2 | PTPBL-related gene on Y, 2 | Yq11.223 |
| PSAT1 | Phosphoserine aminotransferase 1 | 9q21.2 |
| PSMC3IP | Proteasome 26S subunit, ATPase, 3-interacting protein | 17q21.2 |
| PTEN | Phosphatase and tensin homolog | 10q23.31 |
| PTGDR | Prostaglandin D2 receptor | 14q22.1 |
| PTGS2 | Prostaglandin-endoperoxide synthase 2 | 1q31.1 |
| PEX2 | Peroxisomal biogenesis factor 2 | 8q2l.l1 |
| RBM6 | RNA-binding motif protein 6 | 3p21.31 |
| RBMXL1 | RNA binding motif protein, X- linked-like 1 | 1p22.2 |
| RBMXL2 | RNA binding motif protein, X- linked like 2 | 11p15.4 |
| RBMY1A1 | RNA binding motif protein, Y- linked, family 1, member A1 | Yq11.223 |
| RND3 | RHO family GTPase 3 | 2q23.3 |
| RNF144B | Ring finger protein 144B | 6p22.3 |
| RNFI7 | Ring finger protein 17 | 13q12.12 |
| RNF212 | Ring finger protein 212 | 4p16.3 |
| RPS4Y2 | Ribosome protein S4,Y-linked 2 | Yq11.223 |
| RPS6KA2 | Ribosomal protein S6 kinase 90kDa, polypeptide 2 | 6q27 |
| RSPO1 | R-spondin 1 | 1p34.3 |
| RXFP2 | Relaxin/insulin-like family peptide receptor 2 | 13q13.1 |
| RXRG | Retinoid X receptor, gamma | 1q23.3 |
| SCARB1 | Scavenger receptor class B, member 1 | 12q24.31 |
| SDC4 | Syndecan 4 | 20q13.12 |
| SEC16B | SEC16 homolog B | 1q25.2 |
| SEMG1 | Semenogelin I | 20q13.12 |
| SEMG2 | Semenogelin II | 20q13.12 |
| SEPT12 | Septin 12 | 16p13.3 |
| SERP1NA5 | SERP1N peptidase inhibitor, clade A, member 5 | 14q32.13 |
| SF1 | Splicing factor 1 | 11q13.1 |
| SHBG | Sex hormone-binding globulin | 17p13A |
| SHFM1 | Split hand/foot malformation (ectrodactyly) type 1 | 7q21.3 |
| SHOX | Short stature homeobox | Xp22.33 |
| SKOR2 | SKI family transcriptional corepressor 2 | 18q21.1 |
| SLC26A8 | Solute carrier family 26 (anion exchanger), member 8 | 6p21.31 |
| SMYD3 | SET and MYND domain containing protein 3 | 1q44 |
| SOHLH1 | Spermatogenesis and oogenesis-specific basic helix- loop-helix protein 1 | 9q34.3 |
| SOHLH2 | Spermatogenesis and oogenesis specific basic helix- loop-helix 2 | 13q13.3 |
| SOX3 | SRY (sex determining region Y) -box 3 | Xq27.1 |
| SOX9 | SRY-box 9 | 17q24.3 |
| SPACA1 | Sperm acrosome associated 1 | 6q15 |
| SPAG1 | Sperm associated antigen 1 | 8q22.2 |
| SPAG4 | Sperm associated antigen 4 | 20q11.22 |
| SPAG6 | Sperm associated antigen 6 | 10p12.2 |
| SPAG8 | Sperm associated antigen 8 | 9p13.3 |
| SPAG16 | Sperm associated antigen 16 | 2q34 |
| SPATA7 | Spermatogenesis associated 7 | 14q31.3 |
| SPATA9 | Spermatogenesis associated 9 | 5q15 |
| SPATA17 | Spermatogenesis associated 17 | 1q41 |
| SPGFX2 | Spermatogenic failure, X- linked, 2 | Xp22.33 |
| SPRY4 | Sprouty homolog 4 (Drosophila) | 5q31.3 |
| SRD5A2 | Steroid 5-alpha-reductase 2 | 2p23.1 |
| SRY | Sex-determining region Y | Yp11.31 |
| SSBP2 | Single-stranded DNA binding protein 2 | 5q14.1 |
| SSFA2 | Sperm specific antigen 2 | 2q31.3 |
| STAG3 | Stromal antigen 3 | 7q22.1 |
| STAMBPL1 | STAM binding peotein -like 1 | 10q23.31 |
| STAR | Steroidogenic acute regulatory protein | 8p11.23 |
| STS | Steroid sulfatase (microsomal), isozyme S | Xp22.31 |
| SUPT3H | Suppressor of Ty 3, S. cerevisiae homolog | 6p21.1 |
| SYCE1 | Synaptonemal complex central element protein 1 | 10q26.3 |
| SYCE2 | Synaptonemal complex central element protein 2 | 19p13.2 |
| SYCE3 | Synaptonemal complex central element protein 3 | 22q13.33 |
| SYCP3 | Synaptonemal complex protein 3 | 12q23.2 |
| TAF4B | TAF4B RNA polymerase II, TATA box-binding protein- associated factor | 18q11.2 |
| TBC1D20 | TBC1 domain family, member 20 | 20p13 |
| TBPL1 | TBP-like 1 | 6q23.2 |
| TCEB3B | Transcription elongation factor B, polypeptide 3B | 18q21.1 |
| TGFB2 | Transforming growth factor, beta 2 | 1q41 |
| TGFBR3 | Transforming growth factor- beta receptor, type III | 1p22.1 |
| TMEM18 | Trans membrane protein 18 | 2p25.3 |
| TMEM38B | Transmembrane protein 38B | 9q31.2 |
| TMEM108 | Transmembrane protein 108 | 3q22.1 |
| TNFAIP6 | Tumor necrosis factor-alpha- induced protein 6 | 2q23.3 |
| TNP1 | Transition protein 1 | 2q35 |
| TRA2B | Transformer 2, Drosophila, homolog of beta | 3q27.2 |
| TR1M66 | Tripartite motif-containing protein 66 | 11p15.4 |
| TRMT11 | tRNA methyl transferase 11 | 6q22.32 |
| TRO | Trophinin | Xp11.21 |
| TSHB | Thyroid-stimulating hormone, beta chain | 1p13.2 |
| TSKU | Tsukushin, small leucine rich proteoglycan | 11q13.5 |
| TSPAN7 | Tetraspanin 7 | Xp11.4 |
| TSPY1 | Testis-specific protein, Y- l inked 1 | Yp11.2 |
| TSPY2 | Testis-specific protein,Y-linked 2 | Yp11.2 |
| TSSK2 | Testis-specific serine kinase 2 | 22q11.21 |
| TSSK6 | Testis-specific serine kinase 6 | 19p13.11 |
| TUBA4A | Tubulin, alpha-4A | 2q35 |
| TUSC1 | Tumor suppressor candidate 1 | 9p21.2 |
| TXNDC3 | Thioredoxin domain- containing protein 3 | 7pl4.1 |
| UBD | Ubiquitin D | 6p22.1 |
| UBE2B | Ubiquitin-conjugating enzyme E2B | 5q31.1 |
| UCP2 | Uncoupling protein 2 | 11q13.4 |
| UCP3 | Uncoupling protein 3 | 11q13.4 |
| USP8 | Ubiquitin-specific protease 8 | 15q21.2 |
| USP26 | Ubiquitin-specific protease 26 | Xq26.2 |
| USP9Y | Ubiquitin-specific protease9, Y-linked | Yq11.21 |
| UTP14A | U3 small nucleolar ribonucleoprotein, homolog A | Xq26.1 |
| VCAN | Versican (chondroitin sulfate proteoglycan 2) | 5q14.2 |
| VCX | Variable charge, X-linked | Xp22.31 |
| VCY | Variable charge, Y-linked | Yq11.221 |
| VDR | Vitamin D receptor | 12q13.11 |
| VEZT | Vezatin, adherens junctions transmembrane protein | 12q22 |
| VCLL3 | Vestigial-like 3 | 3p12.1 |
| WNT4 | Wingless-type MMTV integration site family, member 4 | 1p36.12 |
| WT1 | Wilms tumor 1 | 11p13 |
| XIST | X inactivation-specific transcript | Xq13.2 |
| XKRY | XK, Kell blood group complex subunit-related, Y-linked | Yq11.222 |
| XPNPEP2 | X-propyl aminopeptidase 2 | Xq26.1 |
| YBX2 | Y box binding protein 2 | 17p13.1 |
| ZFX | Zink finger protein, X-linked | Xp22.11 |
| ZNF483 | Zinc finger protein 483 | 9q31.3 |
| ZP2 | Zona pellucida glycoprotein 2 (sperm receptor) | 16p12.2 |
| ZP3 | Zona pellucida glycoprotein 3 (sperm receptor) | 7q11.23 |
The GeneAnalytics program identified 5 superpathways with a significant number of overlapping genes in the compiled list of human reproduction and related infertility genes. The Ovarian Steroidogenesis superpathway received the most significant score (score = 80.2) with 23 out of a total gene set of 51 included on our master list of reproduction genes. Genes associated with 16 disease processes showed significant overlap. The top five diseases were Prostate Cancer (32 out of 313, score = 28.6), Infertility (21 out of 22, score = 25.5), Breast Cancer (32 out of 781, score = 24.8), Obesity (29 out of 576, score = 21.7) and Lung Cancer (30 out of 622, score = 21.0). Significant overlap related to tissue and cell type was found for the testis (124 out of 3818, score = 16.6). Multiple overlapping pathways for biological (27 pathways) and molecular (5 pathways) processes were identified that significantly impacted spermatogenesis, male gonad development, cell differentiation and organismal development involving steroidogenesis, hormone receptor activity, sequence-specific DNA and protein binding. These pathways point to functional roles in cellular growth and development targeting gonadal development, maturation and function as key underlying genetic architecture for reproduction and related infertility.
We summarized evidence from the peer-reviewed medical literature using computer-based search engine websites for genes playing a definitive role in human infertility and reproduction. We thus provided an update on the list of 153 relevant genes in human reproduction and/or infertility that was reported previously (Butler et al., 2015). This updated list includes a total of 371 clinically relevant and known genes for human infertility and/or reproduction. We also provided a pictorial image of the location and distribution of genes on high resolution chromosome ideograms. Not surprisingly, a preponderance of human reproduction and related infertility genes were found on the sex chromosome pair in relationship to the 22 pairs of autosomes which include 68 genes primarily impacting testis formation or sperm production with 24 located on the Y chromosome and 5 on the X chromosome. Genes that directly influence reproduction or infertility are also involved with testes function and spermatogonial cell production (e.g., ETV5, SPACA1, TSSK6), premature ovarian failure (e.g., FMR1) or primary ovarian insufficiency (e.g., SHOX, CCNH, HSD3B2, GNAS), development of obesity or susceptibility (e.g., FTO, PCSIK,STAR), gene expression or transcription activators (e.g., NFE2L3) or other transcription and translation factors (e.g., CRTC1, TCEB3B, NUPR1, EIF2B2) required for the proper development of somatotrophs, thyrotrophs and gonadotrophs (e.g., PROP1), organization or function of the endoplasmic reticulum and protein export (e.g., SEC16B) or recycling (e.g., MAGEL2), testes-specific RNA splicing factors (e.g., TRA2B) and transcription factors involved with genito-urinary or gonad development (e.g., WT1, CFTR) [genes reviewed in Online Mendelian Inheritance in Man (www.OMIM.org) and in Gene Cards (www.genecards.org)]. Collectively, our list of reproduction and infertility genes impact common hormone pathways encompassing ovarian steroidogenesis along with related peptide production and their receptors, specifically those required for hormone storage and transport, gonadal structure and function and germ cell development as noted in Table 1. These roles also directly influence cellular growth and development relevant to disease states involving cancer risk for multiple cell types (prostate, breast, lung and colon).
Data from genome-wide association studies (GWAS) have shown that the onset of menarche in females is influenced by at least 35 individual genes (Qiu et al., 2013; Montgomery et al., 2014). These include FTO, TRA2B, ETV5, TMEM18 and SEC16B which are also known as obesity-related genes (Choquet and Meyre, 2011a, 2011b; Speliotes et al., 2010; Scherag et al., 2010). Pathways analysis of our tabulated list of human reproduction and infertility genes also identified significant overlap with the Obesity disease state. An example of an obesity-related disorder that is characterized by menstrual irregularities, hyperandrogenism and subfertility is polycystic ovary syndrome (PCOS). About 50% of women with PCOS are also obese (Pasquali and Gambineri, 2006; Erhmann, 2005) with several genes implicated (Venkatesh et al., 2014). Factors contributing to human reproduction and/or infertility identified in our review of involved genes include obesity, hormonal imbalance and protein-based disturbances such as leptin which is a key regulator of appetite produced by adipose tissue and known to inhibit ovarian steroidogenesis (Moschos et al., 2002). This process influences sex hormone binding and androgen receptor sensitivity (Peng et al., 2014) with increased levels of androgen resulting in apoptosis of granulosa cells with conversion of androgens peripherally to estrogens in fat cells. This inhibits gonadotrophin secretion and its level thereby impacting the hormone balance and fertility status of women with obesity (Balen et al., 2006; Metwally et al., 2007; Fragouli et al., 2014; Bergh et al., 1993; Zaadstra et al., 1993). Obesity impacts fertility in women by also decreasing the conception rate with a relative risk for anovulatory infertility estimated at 2.7 (Gesink Law et al., 2007; Wise et al., 2010; Rich-Edwards et al., 1994). Spontaneous conceptions are also known to decrease with subsequent increases in body mass index (BMI) in women. Obesity, therefore, impacts fertility and reproduction by overlapping shared genetic factors implicated or involved in perturbed metabolic and hormonal function (Venkatesh et al., 2014).
Infertility-related genes are noted to be involved with spermatogenesis (e.g.,ACVR2A,AR,ARNTL), testes (e.g., ANKRD7, BAX, BCL2) or ovarian follicle development (e.g., AMH, BMP15, DMC1), premature ovarian failure (e.g., FMR1) or primary ovarian insufficiency (e.g., SHOX, CCNH, HSD3B2, GNAS), development of obesity or susceptibility (e.g., FTO, PCSK1, STAR), gene expression or transcription activators (e.g., NFE2L3) or other transcription and translation factors (e.g., CRTC1, TCEB3B, NUPR1, EIF2B2) that are required for the proper development of somatotrophs, thyrotrophs and gonadotrophs (PROP1), organization or function of the endoplasmic reticulum and protein export (e.g., SEC16B) or recycling (e.g., MAGEL2), neuronal influences on body weight regulation (e.g., TMEM18), testes-specific RNA splicing factors (e.g., TRA2B) and transcription factors involved with genito-urinary or gonad development (e.g., WT1, CFTR) [genes reviewed in OMIM-(www.omim.org) and Gene Cards (www.genecards.org)]. Several obesity-related genes are clearly known to influence infertility by impacting hormonal status and related peptide production. Examples of obesity-related hormonal genes are LEP, LHB, AMH, INHA, GNRH1, IGF1, FSHB, FST, EPPIN, SHBG and IGF2 and examples of obesity-related peptide producing genes and receptors are AR, ESR1, INSR, FSHR, AMHR2, LHCGR, ACVR1, PPARG, STS, LEPR, VDR and IGF1R (Butler et al., 2015). Thus, infertility susceptibility genes are known to affect common hormonal pathways along with related peptide production and their receptors, specifically those required for hormone storage and transport, gonadal structure and function and germ cell development, as noted in Table 1.
Advances in genetic technology using next generation sequencing of DNA exome or RNA expression data should allow for the discovery of hither to unknown disease-causing genes and their functional regulatory sequences, and thus enable a holistic understanding of commonly disturbed mechanisms in the development of human reproduction, ovulation, sperm production and infertility. This complete systems biological understanding has the potential to lead to targeted avenues of novel treatments and management in a significant number of infertile individuals. Therefore, molecular signatures of human reproduction and infertility-based gene profiles and coding expression patterns with overlap in interconnected disturbed gene pathways of infertility will be important to decipher and study. Further deciphering of infertility genomic perturbation and the resultant changes in the functional hormonal pathways will help characterize the disease mechanisms and processes to provide new targets for drug design and therapy. Characterization of these perturbations on an individual basis should not only lead to targeted more effective treatment modalities, but pave the way for prevention of infertility in the human population.
4. Conclusions
We compiled an updated master list of clinically relevant genes for human reproduction and/or infertility by an extensive search of keywords related to human infertility, reproduction and genetics from peer-reviewed medical research articles and related nationally sponsored computer-based websites. The symbols for 371 genes were then plotted on high resolution human chromosome ideograms at precise chromosome band locations thereby producing a convenient visual image of the distribution of genetic factors contributing to human infertility and reproduction with alphabetical listing of genes in a tabular form allowing comparison to guide diagnosis, research, counseling and treatment particularly in individuals with chromosomal and/or genomic aberrations. The current number of genes identified in this report will vary in the future when stimulated by the latest advances in genomic technology and augmented by an increased number of subjects analyzed. The authors encourage the use of this current updated collection of clinically relevant candidate and known genes in the evaluation of patients and families in the clinical setting. This genetic information will in-turn encourage additional basic and translational research in human infertility and reproduction.
Acknowledgments
We thank Carla Meister and Tiffany Embry for their expert preparation of the manuscript and Lorie Gavulic for excellent artistic design and preparation of chromosome ideograms. Partial funding support was provided by the Prader–Willi Syndrome Association (USA) (QB860480), the Headley Family Scholarship 40130x and the National Institute of Child Health and Human Development (NICHD) grant HD02528.
Abbreviations:
- DNA
deoxyribonucleic acid
- RNA
ribonucleic acid
- PCOS
polycystic ovarian syndrome
- GWAS
genome-wide association studies
- OMIM
Online Mendelian Inheritance in Man
Footnotes
Conflict of interest
None.
References
- Abid S, Sagare-Patil V, Gokral J, Modi D, 2013. Cellular ontogeny of RBMY during human spermatogenesis and its role in sperm motility. J. Biosci 38, 85–92. [DOI] [PubMed] [Google Scholar]
- Albertsen HM, Chettier R, Farrington P, Ward K, 2013. Genome-wide association study link novel loci to endometriosis. PLoS One 8, e58257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balen AH, Platteau P, Andersen AN, Devroey P, Sorensen P, Helmgaard L, 2006. The influence of body weight on response to ovulation induction with gonadotrophins in 335 women with World Health Organization group II anovulatory infertility. BJOG 113,1195–1202. [DOI] [PubMed] [Google Scholar]
- Belinky F, Nativ N, Stelzer G, Zimmerman S, Iny Stein T, Safran M, Lancet D, 2015. PathCards: multi-source consolidation of human biological pathways. Database 10.1093/database/bav006 [epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergh C, Carlsson B, Olsson JH, Selleskog U, Hillensjo T, 1993. Regulation of androgen production in cultured human thecal cells by insulin-like growth factor I and insulin. Fertil. Steril 59, 323–331. [DOI] [PubMed] [Google Scholar]
- Brannian JD, Hansen KA, 2002. Leptin and ovarian folliculogenesis: implications for ovulation induction and ART outcomes. Semin. Reprod. Med 20,103–112. [DOI] [PubMed] [Google Scholar]
- Bulun SE, 2014. Aromatase and estrogen receptor alpha deficiency. Fertil. Steril 101, 323–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler MG, McGuire A, Manzardo AM, 2015. Clinically relevant known and candidate genes for obesity and their overlap with human infertility and reproduction. J. Assist. Reprod. Genet 32, 495–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choquet H, Meyre D, 2011a. Molecular basis of obesity: current status and future prospects. Curr. Genet 12,154–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choquet H, Meyre D, 2011b. Genetics of obesity: what have we learned? Curr. Genet 12,169–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrmann DA, 2005. Polycystic ovary syndrome. N. Engl. J. Med 352,1223–1236. [DOI] [PubMed] [Google Scholar]
- El Inati E, Muller J, Viville S, 2012. Autosomal mutations and human spermatogenic failure. Biochim. Biophys. Acta 1822,1873–1879. [DOI] [PubMed] [Google Scholar]
- Ferfouri F, Boitrelle F, Ghout I, Albert M, Molina Gomes D, Wainer R, 2013. A genome-wide DNA methylation study in azoospermia. Andrologie 1, 815–821. [DOI] [PubMed] [Google Scholar]
- Fragouli E, Lalioti MD, Wells D, 2014. The transcriptome of follicular cells: biological insights and clinical implications for the treatment of infertility. Hum. Reprod. Update 20,1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gesink Law DC, Maclehose RF, Longnecker MP, 2007. Obesity and time to pregnancy. Hum. Reprod 22, 414–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene AD, Patounakis G, Segars JH, 2014. Genetic associations with diminished ovarian reserve: a systematic review of the literature. J. Assist. Reprod. Genet 31, 935–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z, Li Z, Yu J, Tong C, Lin Y, Guo X, 2014. Association analysis identifies new risk loci for non-obstructive azoospermia in Chinese men. Nat. Commun 5,3857. [DOI] [PubMed] [Google Scholar]
- Jungheim ES, Travieso JL, Carson KR, Moley KH, 2012. Obesity and reproductive function. Obstet. Gynecol. Clin. N. Am 39, 479–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosova G, Scott NM, Niederberger C, Prins GS, Ober C, 2012. Genome-wide association study identifies candidate genes for male fertility traits in humans. Am. J. Hum. Genet 90, 950–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Layman LC, 2002. Human gene mutations causing infertility. J. Med. Genet 39, 153–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh CA, Hecker E, 2014. Maternal obesity and adverse reproductive outcomes: reducing the risk. Obstet. Gynecol. Surv 69, 622–628. [DOI] [PubMed] [Google Scholar]
- Metwally M, Li TC, Ledger WL, 2007. The impact of obesity on female reproductive function. Obes. Rev 8, 515–523. [DOI] [PubMed] [Google Scholar]
- Montgomery GW, Zondervan KT, Nyholt DR, 2014. The future for genetic studies in reproduction. Mol. Hum. Reprod 20,1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moschos S, Chan JL, Mantzoros CS, 2002. Leptin and reproduction: a review. Fertil. Steril 77, 433–444. [DOI] [PubMed] [Google Scholar]
- Ogden CL, Carroll MD, Curtin LR, McDowell MA, Tabak CJ, Flegal KM, 2006. Prevalence of overweight and obesity in the United States. 1999–2004.J. Am. Med. Assoc 295, 1549–1555. [DOI] [PubMed] [Google Scholar]
- Okada Y, Tateishi K, Zhang Y, 2010. Histone demethylase JHDM2A is involved in male infertility and obesity. J. Androl 31, 75–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olooto WE, Amballi AA, Banjo TA, 2012. A review of female infertility: important etiological factors and management. J. Microbiol. Biotech. Res 2, 379–385. [Google Scholar]
- Ombelet W, Cooke I, Dyer S, Serour G, Devroey P, 2008. Infertility and the provision of infertility medical services in developing countries. Hum. Reprod. Update 14, 605–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey S, Maheshwari A, Bhattacharya S, 2010. The impact of female obesity on the outcome of fertility treatment. J. Hum. Reprod. Sci 3, 62–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquali R, Gambineri A, 2006. Polycystic ovary syndrome: a multifaceted disease from adolescence to adult age. Ann. N. Y. Acad. Sci 1092,158–174. [DOI] [PubMed] [Google Scholar]
- Peng CY, Xie HJ, Guo ZF, Nie YL, Chen J, Zhou JM, 2014. The association between androgen receptor gene CAG polymorphism and polycystic ovary syndrome: a case-control study and meta-analysis. J. Assist. Reprod. Genet 31,1211–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Y, Ji J, Du G, Wu W, Dai J, Hu Z, 2014. Comprehensive pathway-based analysis identifies associations of BCL2, GNAOl and CHD2 with non-obstructive azoospermia risk. Hum. Reprod 29, 860–866. [DOI] [PubMed] [Google Scholar]
- Qiu R, Chen C, Jiang H, Shen L, Wu M, Liu C, 2013. Large genomic region free of GWAS-based common variants contains fertility-related genes. PLoS ONE 8, e61917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rich-Edwards JW, Goldman MB, Willett WC, Hunter DJ, Stampfer MJ, Colditz GA, 1994. Adolescent body mass index and infertility caused by ovulatory disorder. Am. J. Obstet. Gynecol 171–77. [DOI] [PubMed] [Google Scholar]
- Scherag A, Dina C, Hinney A, Vatin V, Scherag S, Vogel CI, Muller TD, 2010. Two new loci for body-weight regulation identified in a joint analysis of genome-wide association studies for early-onset extreme obesity in French and German study groups. PLoS Genet 6, el000916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaffer LG, McGowan-Jordan J, Schmid M, 2013. Unionville (CT). In: Karger S. (Ed.), International System for Human Cytogenetic Nomenclature (ISCN). [Google Scholar]
- Shah K, Sivapalan G, Gibbons N, Tempest H, Griffin DK, 2003. The genetic basis of infertility. Reproduction 126,13–25. [DOI] [PubMed] [Google Scholar]
- Silber SJ, Barbey N, 2012. Scientific molecular basis for treatment of reproductive failure in the human: an insight into the future. Biochim. Biophys. Acta 1822,1981–1996. [DOI] [PubMed] [Google Scholar]
- Speliotes EK, Wilier CJ, Berndt SI, Monda KL, 2010. Thorleifsson G, Jackson AU, et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat Genet. 42, 937–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatesh T, Suresh PS, Tsutsumi R, 2014. New insights into the genetic basis of infertility. Appl. Clin. Genet 7, 235–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise LA, Rothman KJ, Mikkelsen EM, Sorensen HT, Riis A, Hatch EE, 2010. An internet-based prospective study of body size and time-to-pregnancy. Hum. Reprod 25, 253–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaadstra BM, Seidell JC, Van Noord PA, te Velde ER, Habbema JD, Vrieswijk B, 1993. Fat and female fecundity: prospective study of effect of body fat distribution on conception rates. BMJ 306,484–487. [DOI] [PMC free article] [PubMed] [Google Scholar]