Abstract
Background:
Scavenger receptors play a significant role in clearing aged proteins from the plasma, including the large glycoprotein coagulation factors von Willebrand factor (VWF) and factor VIII (FVIII). A large GWAS meta analysis has identified genetic variants in the gene SCARA5, which encodes the class A scavenger receptor SCARA5, as being associated with plasma levels of VWF and FVIII.
Objectives:
The ability of SCARA5 to regulate the clearance of VWF-FVIII was characterized.
Methods:
VWF-FVIII interactions with SCARA5 were evaluated by solid phase binding assays and in vitro cell based assays. The influence of SCARA5 deficiency on VWF:Ag and half-life was assessed in a murine model. The expression pattern of SCARA5 and its co-localization with VWF was evaluated in human tissues.
Results:
VWF and the VWF-FVIII complex bound to human recombinant SCARA5 in a dose- and calcium-dependent manner. SCARA5 expressing HEK 293T cells bound and internalized VWF and the VWF-FVIII complex into early endosomes. In vivo, SCARA5 deficiency had a modest influence on the half-life of human VWF. mRNA analysis and immunohistochemistry determined that human SCARA5 is expressed in kidney podocytes and the red pulp, white pulp and marginal zone of the spleen. VWF was found to co-localize with SCARA5 expressed by littoral cells lining the red pulp of the human spleen.
Conclusions:
SCARA5 is an adhesive and endocytic receptor for VWF. In human tissues, SCARA5 is expressed by kidney podocytes and splenic littoral endothelial cells. SCARA5 may have a modest influence on VWF clearance in humans.
Keywords: von Willebrand factor, factor VIII, receptors scavenger, GWAS, endocytosis, endothelial cells
Introduction:
Scavenger receptors are a superfamily of membrane receptors characterized by the ability to bind and internalize an array of endogenous and exogenous ligands and pathogens. Scavenger receptors are classified by domain structure and ligand binding properties into 12 separate families (classes A-L) and have been shown to regulate physiological processes such as clearance of apoptotic and necrotic cellular materials, aged plasma proteins, and cholesterol [1]. A number of pathophysiological conditions are linked to disordered or dysfunctional scavenger receptor activity including infection, cancer, and atherosclerosis [2,3]. Importantly, while scavenger receptors interface directly with and regulate constituents of the blood or plasma, the influence of this receptor family on the hemostatic system is largely uncharacterized.
Under physiological conditions, blood is maintained in a fluid state by a balance between pro- and anticoagulant pathways that rapidly activate to prevent blood loss in response to vascular injury but prevent pathological thrombus development. Quantitative abnormalities in plasma levels of coagulation factors, including the multimeric glycoprotein von Willebrand factor (VWF) can associate with either bleeding disorders (von Willebrand disease, VWD) or increase the risk for thrombosis [2,4]. Plasma levels of VWF can be modified by pathways that regulate its synthesis or secretion from endothelial cells or megakaryocytes, or clearance, which involves a series of semi-selective ligand-receptor mediated interactions involving hepatocytes, macrophages, and endothelial cells in the liver and/or spleen [5,6].
To date, receptors belonging to four separate scavenger receptor families, SR-A1 (class A), asialoglycoprotein receptor (ASGPR, class E), stabilin-2 (class H), and low density lipoprotein receptor-related protein 1 (LRP-1, class L) have been shown to bind and regulate the clearance of VWF [7–10]. While the associations between VWF and the receptors SR-A1, ASGPR, and LRP-1 were characterized in candidate gene rationalized studies, the influence of stabilin-2 on VWF clearance was first suggested by the CHARGE genome wide association study (GWAS) meta-analysis, which associated common variants with plasma levels of VWF or its binding partner the coagulation cofactor factor VIII (FVIII) [11,12]. In follow up studies, rare and common STAB2 gene variants have been associated with VWF plasma levels in normal individuals and patients with type 1 VWD or low VWF, or an increased risk for venous thromboembolism [8,13,14]. In addition to the STAB2 locus, the CHARGE GWAS identified variants in the genes encoding two other cell surface receptors, the endocytic lectin receptor CLEC4M and the scavenger receptor SCARA5 (Scavenger Receptor Class A Member 5, or SR-A5), as being associated with plasma VWF and/or FVIII [12,15].
Similar to SR-A1, SCARA5 is a type II membrane protein and displays homology with other class A scavenger receptors. SCARA5 is comprised of a C-terminus intracellular region, a transmembrane region, and extracellular spacer domain, a collagenous domain and a scavenger receptor cysteine rich (SRCR) domain at the N-terminus [16]. SCARA5 assembles as a trimer at the cell surface, and has been reported to bind lipopolysaccharide, ferritin, polyanions, and microbes, but not modified low-density lipoproteins [16,17]. In mice, SCARA5 is expressed by epithelial cells and fibroblasts in the testis, heart, brain, and spleen [16,18]. In contrast, the expression pattern of SCARA5 in human tissues is largely uncharacterized, although SCARA5 mRNA is expressed in CD31+CD34−CD8α+ littoral cells (LCs), a sub-type of splenic sinusoidal endothelial cells [19]. In this report, we describe the ability of SCARA5 to bind and/or internalize VWF using cell expression and solid phase binding assays. We determine the influence of SCARA5 deficiency on VWF plasma levels and half-life in a mouse model. Finally, we investigate the anatomic sites of SCARA5 mRNA and protein expression, and its co-localization with VWF in human tissues.
Materials and Methods
Recombinant SCARA5 production and purification:
HEK293 EBNA cells secreting the HIS- and AVI-tagged soluble human SCARA5 protein (structure and sequence provided in Supplemental Figures 2 and 3) into cell culture media were cultured in DMEM with Glutamax-1 supplemented with 10% fetal calf serum, penicillin-streptomycin, sodium pyruvate, and selective antibiotics (250 μg/ml G418, 125 μg/ml zeosin) and a daily addition of 100 μg/ml ascorbic acid. Upon confluency, the media was replaced with serum-free DMEM/F-12 (1:1) supplemented with glutamax-1, penicillin-streptomycin, sodium pyruvate and ascorbic acid. The serum-free cell culture media was harvested after 3–10 days of culture. SCARA5 purification was performed on an ÄKTA pure 25 system by immobilized metal affinity chromatography using a HisTrap Excel column followed by anion-exchange using a HiTrap Q HT column. The fractions containing the desired protein were identified by SDS-PAGE and Western blotting, pooled and concentrated with Amicon Ultra Centrifugal filter, 30 kDa cut-off and then run through a Superdex 200 size-exclusion chromatography column. Finally, the fractions containing the purified protein were identified by SDS-PAGE and Western blotting, pooled, and concentrated for later use. Western blot analysis of the recombinant soluble SCARA5 under non-denaturing conditions indicated that the majority of the protein migrated in a trimeric form (data not shown), presumably mediated through natural cross-linking via modified lysine residues in the collagenous region [20].
Solid phase VWF-SCARA5 binding assays:
The interaction between recombinant SCARA5 and VWF/FVIII was measured as described [15] with several modifications. Recombinant human SCARA5 was coated in carbonate buffer at 5 μg/mL on Maxisorp plates (Nunc, Rochester, USA). VWF binding to SCARA5 was detected by an HRP-conjugated rabbit anti-VWF antibody (Dako, Agilent, Santa Clara, USA), FVIII binding to SCARA5 was detected by an HRP-conjugated sheep anti-FVIII antibody (Affinity Biologicals, Ancaster, Canada). For all experiments, binding was compared with background binding to a BSA-coated negative control. For some experiments, immobilized SCARA5 was preincubated with polyinosinic acid potassium salt (PolyI), poly guanylic acid (PolyG), dextran sulfate, chondroitin sulfate (Sigma Aldrich, St. Louis, USA), polyphosphate (PolyP) (courtesy of Dr. JH Morrissey, University of Michigan), or the following anti-SCARA5 antibodies: PA5–23551 (Invitrogen), LS-B5013 (LS Bio), AF4900 (R&D), or mouse anti-human SCARA5 αh-SR5.2. αh-SR5.2 was produced and purified as previously described [18] using a human SCARA5 peptide (amino acids 356–495) in an pET28a(+) vector. Antibody binding sites can be found in Supplemental Figure 2.
Imaging Studies:
Antibodies used include: sheep anti-FVIII (Affinity Biologicals), rabbit anti-VWF (Dako), sheep anti-VWF (Abcam, Cambridge, UK), rabbit anti-EEA1 (Cell Signalling Technology, Beverly, MA, USA), rabbit anti-SCARA5 (HPA024661, Sigma Aldrich), mouse anti-SCARA5 (αh-SR5.2), anti-human CD31 (Dako), anti-human CD68 (Santa Cruz Biotechnology, Dallas, USA), anti-human synaptopodin (R&D, Minneapolis, USA), mouse anti-human CD34 (QBEnd-10, ThermoFisher Scientific, Waltham, USA) and mouse anti-human CD8α (4B11, ThermoFisher Scientific).
Immunocytochemistry:
HEK 293T cells were transiently transfected with the full-length human SCARA5 cDNA by lipofectamine (ThermoFisher Scientific). A 10–20% transfection efficiency was routinely observed. 24 hours post-transfection, cells were washed and exposed to VWF and/or FVIII in binding buffer (10 mM HEPES pH 7.4, 135 mM NaCl, 10 mM KCl, 5 mM CaCl2, 2 mM MgSO4) for 15 – 60 minutes. Cells were prepared for immunofluorescent imaging as previously described [15].
Immunohistochemistry:
DAB peroxidase IHC analysis of SCARA5 expression by normal human tissues was performed using a rabbit anti-human SCARA5 antibody (Sigma Aldrich) on a Ventana Discovery Immunostainer (Ventana Medical System, Tucson, AZ) and imaged using an Aperio ScanScope SC slide scanner. Immunofluorescent staining was performed as previously described and imaging was performed with a Leica SP8 laser scanning confocal microscope using 63X oil immersion objective [8]. All IHC conditions were compared with isotype controls (data not shown). All images were analyzed using ImageJ or FIJI software (NIH, Bethesda, USA).
Animal model and clearance studies:
All animal experiments were approved by the Queen’s University Animal Care Committee (Kingston, ON, Canada).
Plasma collection from SCARA5 KO mice:
Blood was collected from age and sex-matched 8–10 week old normal C57Bl/6 mice from our colony and SCARA5 KO mice [18] on a C57Bl/6 background via inferior vena cava into 10% buffered citrate. Platelet poor plasma was prepared by centrifugation at 10,000xg for 10 minutes and stored at −80˚C until analysis
VWF half-life studies:
Murine plasma-derived VWF was generated by hydrodynamic tail vein injection of the murine VWF cDNA into VWF/FVIII DKO mice. Blood was collected 3 days post-injection, plasma VWF was quantified and diluted to 20 U/mL in VWF/FVIII DKO plasma. Human plasma-derived VWF (FVIII-free) was kindly provided by Biotest (Munich, Germany). C57Bl/6 SCARA5 KO were crossed with C57BL/6 VWF KO mice to generate VWF/SCARA5 double knockout (DKO) mice. VWF was administered via tail vein injection at 200 U/kg. Blood was collected via retro-orbital plexus into 10% buffered citrate. VWF clearance statistics were determined by one phase exponential decay or AUC analysis in Graph Pad Prism (San Diego, USA).
Measurement of VWF:Ag and VWFpp.
VWF:Ag was determined by ELISA using Dako antibodies (A0082 and P0226). Murine VWF propeptide (VWFpp) levels were measured by ELISA as described [21] using 349.2 and 349.3 antibodies kindly provided by Dr. S. Haberichter (Blood Research Institute, Blood-Center of Wisconsin, Milwaukee, USA).
Human SCARA5 mRNA quantification:
RNA extracted from human liver, spleen, kidney and testes was purchased from Cell Applications Inc. (San Diego, CA, USA), and transfected HEK 293T and whole blood RNA was isolated by RNAeasy Mini Kit (Qiagen, Hilden, Germany). RNA was reverse transcribed by random hexamer primers using SuperScriptIII reverse transcriptase in accordance with manufacturer’s protocols. SCARA5 and GAPDH gene expression was quantified using TaqMan® Gene Expression Assays with TaqMan Fast Advanced Mastermix (ThermoFisher Scientific).
Statistical analysis:
T-tests or one-way ANOVA was performed on experiments with n≥3 using GraphPad Prism software. Values are expressed as mean ± standard error. Figures denote P<0.05 with * and P <.001 with **.
Results:
SCARA5 directly binds to and mediates the intracellular internalization of VWF:
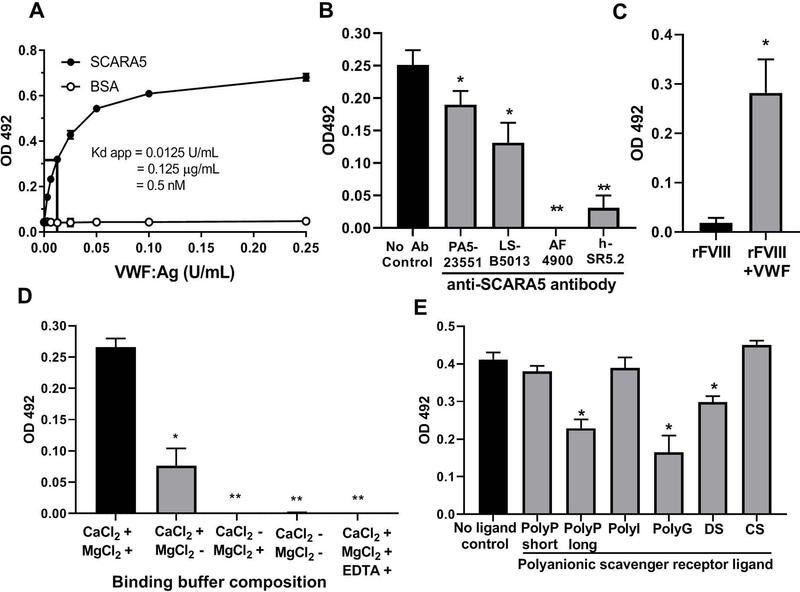
We first assessed the ability of recombinant human soluble SCARA5 to bind directly to VWF using a solid phase binding assay. Recombinant human SCARA5 was adsorbed directly to a microtitre dish and exposed to combinations of human VWF or FVIII-containing products in a calcium-supplemented buffer. VWF or FVIII binding was measured using an HRP-conjugated anti-VWF or anti-FVIII polyclonal antibody. We observed that human plasma-derived VWF bound directly to immobilized SCARA5 in a saturable, dose-dependent manner, as compared with a BSA control (Figure 1A). Here, we estimated the apparent Kd as 0.125 ± 0.01 μg/mL (0.5 nM) confirming that this binding interaction is of high affinity.
Figure 1. Recombinant soluble SCARA5 binds to VWF.
(A) Dose-dependent binding of soluble human plasma-derived VWF to immobilized recombinant, purified soluble human SCARA5 by solid phase assay. (B) Binding of soluble human plasma-derived VWF (1 U/mL) to immobilized recombinant human SCARA5 (5 μg/mL) that was pre-incubated for 30 minutes at room temperature with anti-human SCARA5 antibodies PA5–23551 (1:5 dilution), LS-B5013 (100 ug/mL), AF4900 (100 ug/mL), and αh-SR5.2 (40 ug/mL). (C) Binding of soluble human recombinant FVIII (10 U/mL) to immobilized recombinant human SCARA5 (5 μg/mL) in the presence or absence of human plasma-derived VWF (1 U/mL). (D) Binding of soluble human plasma-derived VWF (1 U/mL) to immobilized recombinant, purified human SCARA5 (5 μg/mL) in the presence or absence of calcium (2 mM), magnesium (2 mM), and EDTA (10 mM). (E) Binding of soluble human plasma-derived VWF (0.5 U/mL) to immobilized recombinant human SCARA5 (5 μg/mL) preincubated with scavenger receptor ligands short or long-chain PolyP (100 μg/mL), PolyI (100 μg/mL), PolyG (1 mg/mL), dextran sulfate (DS) (2 mg/mL) or chondroitin sulfate (CS) (2 mg/mL) for 30 minutes at 37°C.
Binding between SCARA5 and VWF may be mediated by either the collagenous or SRCR domains of the SCARA5 protein. The SRCR domain contains 6 cysteine residues that are likely to participate in intra-chain disulfide bonds that conserve protein conformation and ligand binding affinity. We confirmed the specificity of SCARA5-VWF binding in this assay by blocking immobilized SCARA5 with anti-SCARA5 antibodies prior to the addition of soluble VWF (Figure 1B). We observe that antibodies that bind to full length SCARA5, the collagenous domain and/or the SRCR domain significantly attenuate VWF binding to SCARA5 in this system. We next compared the binding of human rFVIII to immobilized SCARA5 in the presence or absence of human plasma-derived VWF to a BSA control. Here we observed that the presence of VWF significantly enhanced FVIII binding, suggesting that FVIII may interact with SCARA5 through VWF (Figure 1C) but that SCARA5 does not bind FVIII directly.
Previous studies of the SRCR domain of glycoprotein 340 suggest that the presence of calcium promotes the conformational stability of the SRCR domain and enhances ligand binding [22], and similarly, the binding between VWF and SR-A1 is also calcium-dependent [7]. Using EDTA supplemented or calcium/magnesium-deficient buffers, we demonstrate that SCARA5-VWF binding is inhibited, confirming that this interaction is also calcium-dependent and partially magnesium-dependent (Figure 1D). Class A scavenger receptors have been previously reported to bind polyanionic motifs in their respective ligands through cationic collagenous or cysteine-rich domains [23,24]. We pre-incubated immobilized recombinant SCARA5 with the polyanionic ligands short or long-chain PolyP, PolyI, PolyG, dextran sulfate (DS) or chondroitin sulfate (CS). Long-chain PolyP, PolyG and dextran sulfate partially attenuated the interaction between VWF and SCARA5 when compared with a BSA control (Figure 1E). This observation is consistent with previous studies documenting that PolyG can inhibit the interaction between SCARA5-expressing cells and labeled S. aureus and E. coli bioparticles [16].
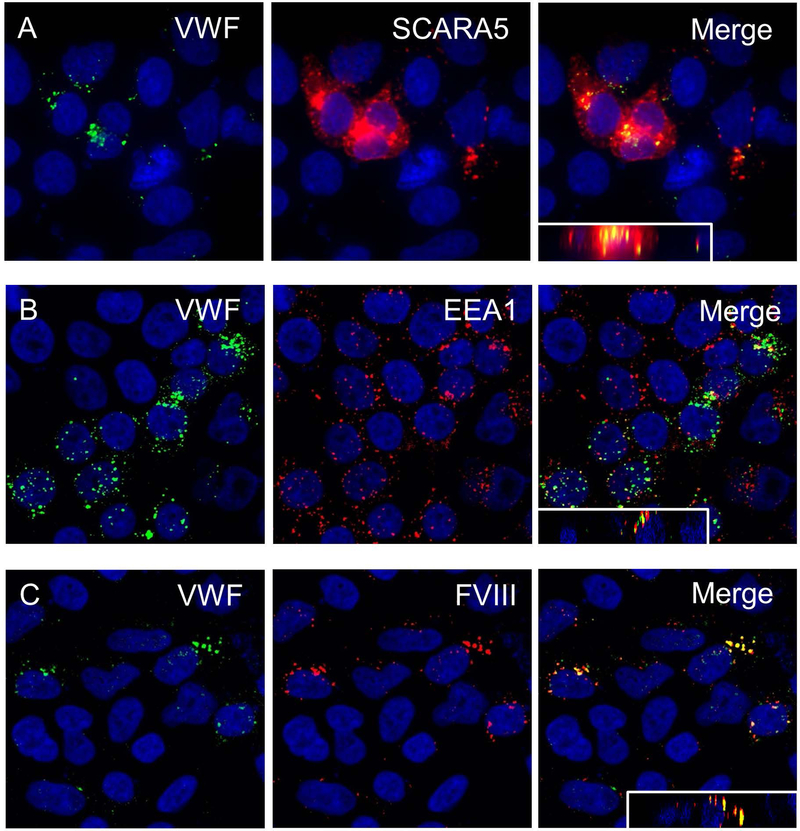
We next determined the ability of SCARA5-expressing cells to bind and internalize VWF. HEK 293T cells were transiently transfected with the human SCARA5 cDNA. 24 hours post-transfection, cells were washed and exposed to VWF in a calcium-supplemented binding buffer for 15–60 minutes. Cells were washed and processed for immunofluorescent imaging as previously described. We observed that human plasma-derived VWF was bound to and internalized by HEK 293T cells (Figure 2A) expressing the human SCARA5 cDNA but not to control cells that did not express SCARA5 including HEK 293T cells transiently transfected with vector backbone (data not shown). VWF partially co-localized with areas of SCARA5 expression, which is likely related to the disengagement of receptor-ligand interaction upon transportation of the complex to the acidic endosomal compartments, and the potential re-exportation of internalized SCARA5 to the cell membrane by recycling endosomes. We confirmed that SCARA5 expressing HEK 293T cells were able to internalize VWF into early endosomes, by demonstrating that VWF partially co-localized with early endosomal antigen-1 (EEA1) within 15 minutes of exposure to VWF (Figure 2B). Finally, we demonstrate that when SCARA5-expressing HEK 293T cells were exposed to the VWF-FVIII complex, internalized VWF and FVIII partially co-localized, confirming that SCARA5 can mediate internalization of FVIII in a VWF-dependent manner (Figure 2C). For all merged images an orthogonal view is inset. No FVIII binding to SCARA5-expressing HEK 293T cells was observed in the absence of VWF (data not shown).
Figure 2. SCARA5 expressing cells bind and internalize VWF-FVIII.
HEK 293T cells were transiently transfected with the full-length human SCARA5 cDNA. 24 hours post transfection cells were exposed to human pdVWF for 15–60 minutes, washed, and processed for immunocytochemistry. (A) Human pdVWF bound to and internalized by HEK 293T cells transiently expressing human SCARA5. (B) Human pdVWF internalized by HEK 293T cells transiently expressing human SCARA5 partially co-localized with EEA1 after 15 minutes. (C) Human pdVWF internalized by HEK 293T cells transiently expressing human SCARA5 partially co-localized with human FVIII. For merged images an orthogonal view is inset. For all images, DAPI = blue.
SCARA5 deficiency does not strongly modify VWF levels or half-life in vivo:
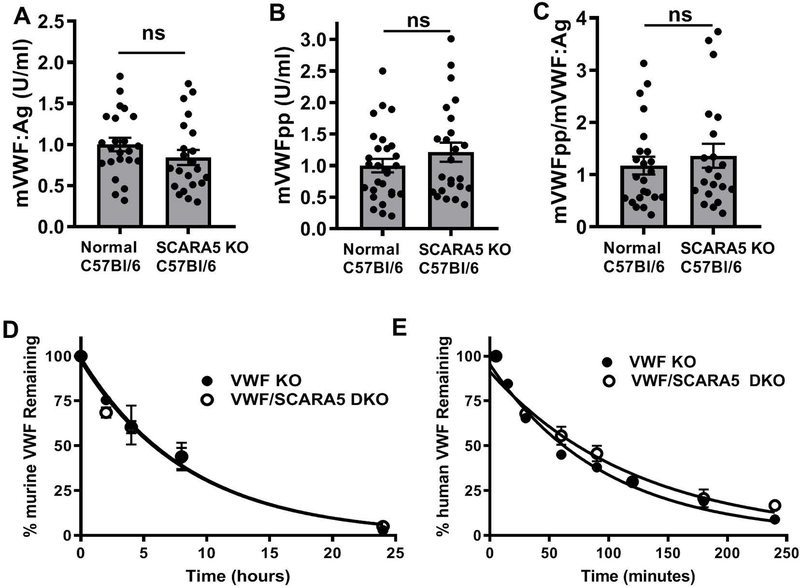
We next assessed the influence of SCARA5 deficiency on plasma VWF levels and half-life in a mouse model. We observed no significant differences in VWF:Ag, VWFpp, or VWFpp/VWF:Ag, between control C57Bl/6 and SCARA5 KO mice (Figure 3A–C). We then generated VWF/SCARA5 DKO mice and measured the half-life of human and murine VWF (administered by tail vein injection at 200 U/kg) in these animals as compared with VWF KO mice. We observed no significant difference in the half-life of plasma-derived murine VWF in the absence of SCARA5 expression (Figure 3D). In contrast, we observed an increase in the half-life of human plasma-derived VWF in VWF/SCARA5 DKO mice as compared with VWF KO controls (84.9 minutes versus 66.8 minutes, p=0.026) (Figure 3E). Area Under Curve (AUC) analysis did not find a significant difference between VWF/SCARA5 DKO mice as compared with VWF KO controls for human plasma-derived VWF (p=0.127) or murine plasma-derived VWF (p=0.982).
Figure 3. Influence of SCARA5 deficiency on VWF and FVIII in vivo.
Plasma VWF:Ag levels (A), VWFpp (B), and VWFpp/VWF:Ag (C)) were measured in C57Bl/6 SCARA5 KO mice as compared with wild-type C57Bl/6 mice. VWF/SCARA5 DKO mice were generated and VWF clearance was measured in comparison with VWF KO mice. (D) Clearance of murine plasma-derived VWF in the absence of murine SCARA5 expression. (E) Clearance of human plasma-derived VWF in the absence of murine SCARA5 expression.
Previous studies performed by van Schooten et al. [25] using radiolabelled VWF have shown that VWF can be taken up by cells in the liver, spleen and kidneys. Given its relative size and proportionate blood flow it is presumed that the liver is the major site of VWF clearance in vivo. We infused VWF KO and VWF/SCARA5 DKO mice with 200 U/kg of human plasma-derived VWF, perfused and collected liver, spleen and kidney tissues for IHC after 30 minutes, and stained the tissues using an anti-VWF antibody. VWF stained predominantly sinusoidal endothelial cells and macrophages in the liver, cells within the marginal zone and red pulp of the spleen, and within the mesangial cells of the kidney glomerulus. However, no significant differences were observed in the pattern of VWF staining between the organs from VWF KO and VWF/SCARA5 DKO mice (Supplemental Figure 1). Collectively, this data suggests that the predominantly epithelial and fibroblast expression of SCARA5 in mouse tissues does not significantly contribute to VWF clearance in this animal model.
SCARA5 is expressed in the human kidney and spleen:
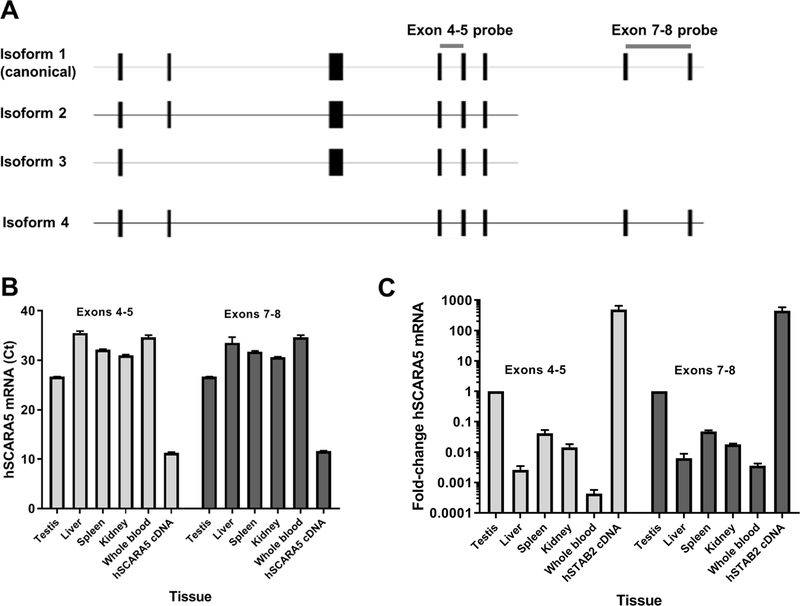
We finally assessed the site of SCARA5 expression in human tissues using qRT-PCR, and IHC. There are four alternative splice isoforms of the human SCARA5 gene that have been identified. Isoform 1 is the canonical splice form and encodes the full-length SCARA5 protein, while the other three isoforms lack structural elements including the transmembrane region and SRCR domains that might impair SCARA5 function. We utilized two sets of probes for our quantitative analysis - one designed to detect all four SCARA5 isoforms (spanning exons 4 and 5), and one designed to detect isoforms 1 and 4 which are capable of encoding the SRCR domain (spanning exons 7 and 8) (Figure 4A). We quantified SCARA5 mRNA levels from human testes, liver, spleen, kidney and whole blood as compared with a GAPDH control. We report the Ct values for each sample (using equimolar amounts of starting cDNA for each reaction) (Figure 4B) and ΔΔCt values for SCARA5 levels as compared to the GAPDH control and to the results obtained from the testes samples (Figure 4C). We observed a high level of SCARA5 expression in our positive control (HEK 293T cells transfected with the full length human SCARA5 cDNA). Compared with SCARA5 levels in the human testes, we observed moderate expression in the spleen and kidney, and lower expression in the liver and whole blood RNA extracts.
Figure 4. SCARA5 mRNA expression in human tissues.
(A) Diagram of human SCARA5 splicing isoforms and probe coverage. SCARA5 expression in human tissues analyzed by (B) Ct (cycle threshold) and (C) 2–∆∆Ct analysis relative to GAPDH controls.
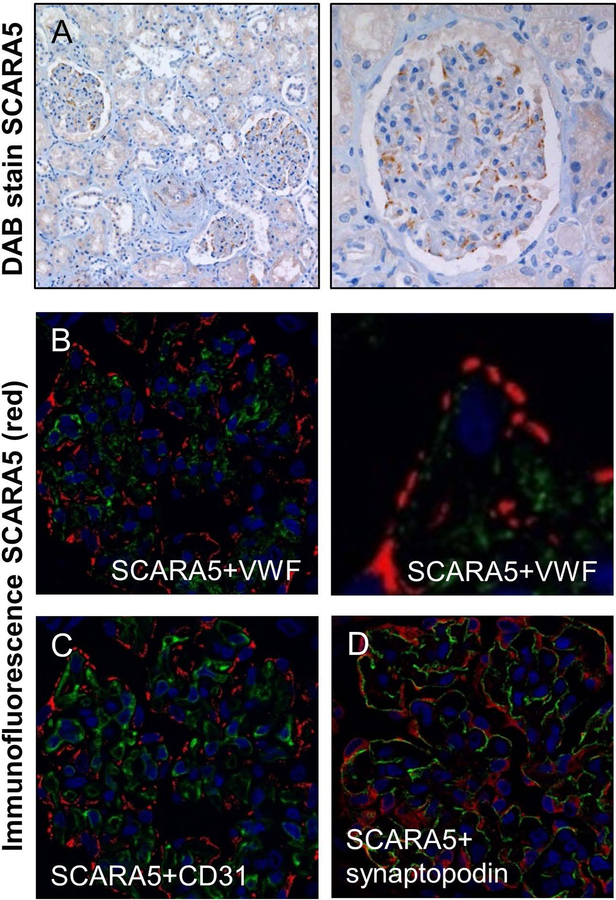
We next characterized the expression pattern of SCARA5 and its localization with VWF and other cellular markers in human tissues using an antibody that binds within the spacer region of human SCARA5 (Supplemental Figure 2). This antibody therefore likely recognizes SCARA5 protein isoform 1 (full length), as well as isoforms 2 and 3 but not isoform 4 (Supplemental Figure 2). We confirmed the ability of this antibody to recognize full-length human SCARA5 by immunocytochemistry on HEK 293T cells transfected with the full-length human SCARA5 cDNA (data not shown) and then performed IHC in human liver, kidney and spleen tissues. Using IHC DAB staining in the human liver, we were unable to detect any SCARA5 signal when compared with an isotype control (data not shown), which is consistent with the published data concerning the expression pattern of murine SCARA5 in the liver [16,18]. In the human kidney, we observed the majority of SCARA5 staining was localized within the podocytes of the glomerulus (Figure 5A) and confirmed this by co-staining with the podocyte marker synaptopodin (Figure 5D). Both VWF and CD31 were expressed by endothelial cells in the glomerulus which are separated from SCARA5-expressing podocytes by a basement membrane (Figure 5B and 5C). No VWF was found to associate with SCARA5-expressing podocytes (Figure 5B).
Figure 5. SCARA5 protein expression in the human kidney.
(A) DAB stain of SCARA5 expression in the human kidney (brown). (B) Immunofluorescent stain of SCARA5 (red) and VWF (green) in the glomerulus. (C) SCARA5 (red) and CD31 (green) immunofluorescent imaging in the glomerulus. (F) SCARA5 (red) and synaptopodin (green) immunofluorescent imaging in the glomerulus. For all images DAPI = blue.
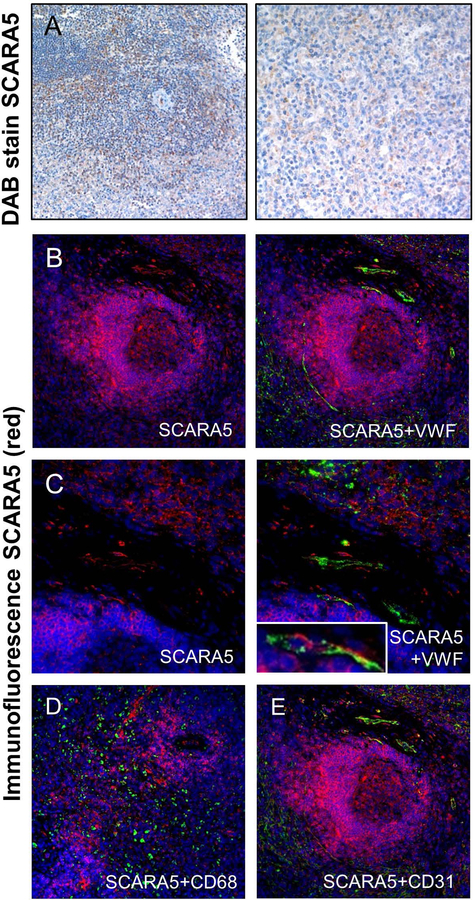
In the human spleen, we have previously detected VWF associating with CD31 and stabilin-2 expressing endothelial cells and CD68-expressing macrophages [8]. Additionally, human VWF infused into VWF KO mice associates predominantly with CD31-expressing endothelial cells, although some F4/80-expressing macrophages also associated with infused VWF. In these studies, IHC analysis of the human spleen demonstrated that cells within the red pulp, marginal zone, and white pulp of the spleen express SCARA5 (Figure 6A). Immunofluorescent analysis of this tissue demonstrated some colocalization between SCARA5 and VWF that occurs in CD31-expressing endothelial cells (Figure 6B, 6C, 6E) but not CD68-expressing macrophages (Figure 6D).
Figure 6. SCARA5 protein expression in the human spleen.
(A and B) DAB stain of SCARA5 expression in the human spleen (brown). (C) Immunofluorescent stain of SCARA5 in the spleen (red) (20×). (D) VWF (green) and SCARA5 (red) immunofluorescent imaging in the spleen (20×). (C) Immunofluorescent staining of SCARA5 (red) in the spleen (40×). (D) VWF (green) and SCARA5 (red) immunofluorescent imaging in the spleen (40×). (E) SCARA5 (red) and CD68 (green) immunofluorescent imaging in the spleen (20×). (D) SCARA5 (red) and CD31 (green) immunofluorescent imaging in the spleen (20×). For all images, DAPI = blue.
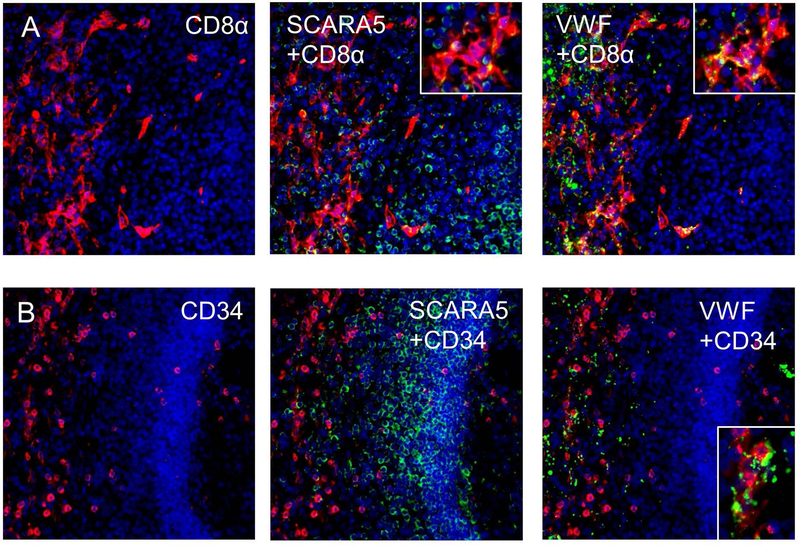
Previously the mRNA expression of SCARA5 in splenic littoral (CD31+CD34−CD8α+) but not splenic vascular endothelial cells (CD31+CD34+CD8α−) has been reported [19]. Utilizing the same CD8α and CD34 antibodies that have been previously validated for the isolation of these two cell populations by FACs, we performed IHC on human spleen samples and co-stained for SCARA5 and VWF. We observed some co-localization between SCARA5 and CD8α expressing cells in the splenic red pulp within endothelial cell-like structures (Figure 7A). This same population of cells was also found to associate with VWF staining. Conversely, we observed little co-localization between CD34 and SCARA5 expressing cells in the splenic marginal zone (Figure 7B), although VWF also localized to CD34-expressing cells. Additional overlays of these images can be found in Supplemental Figures 3 and 4.
Figure 7. SCARA5 expression in splenic microvascular endothelial cells and littoral cells.
(A) Immunofluorescent stain of littoral cell marker CD8α (red) and co-localization with SCARA5 and VWF (green). (B) Immunofluorescent stain of littoral cell marker CD32 (red) and co-localization with SCARA5 and VWF (green). For all images, DAPI = blue.
Discussion:
The association between common SCARA5 gene variants and plasma levels of VWF:Ag or FVIII:C was first identified by the CHARGE GWAS meta analysis in a population of 23,000 individuals of European ancestry and a subsequent replication cohort of 7600 additional subjects [12]. In this study, the SCARA5 intronic variants rs2726953 and rs9644133 were shown to associate with a 4.5% (p=1.3×10−16) increase and a 4.1% (p=4.4×10−15) decrease in plasma VWF and FVIII levels respectively. Regional plot analysis of the SCARA5 gene revealed several other SCARA5 gene variants that associate with plasma VWF:Ag below a genome-wide significance threshold of p=5.0×10−8. The observation was confirmed in a recently published follow-up GWAS involving a multi-ethnice population of over 46,000 normal individuals [11]. Analysis using rAggr software under default settings did not identify any variants in linkage disequilibrium with the CHARGE SNVs outside the SCARA5 gene (Supplemental Tables 1 and 2), indicating that these signals are likely associated with the SCARA5 gene.
However, while the association between CHARGE variants in VWF, CLEC4M, STXBP5, STX2, and STAB2 and plasma VWF levels have been verified in separate smaller populations of normal individuals or those with quantitative VWF abnormalities, the association between SCARA5 gene variants and VWF-FVIII is less clear [8,15,26–29]. One analysis of the Guttenberg Health Study found an association between the SCARA5 SNV rs9644133 and plasma FVIII:C in female but not male subjects [30], and the mechanistic basis for a sex-specific effect is not currently understood. A separate GWAS meta analysis found an association between a SCARA5 SNV and plasma VWF:Ag in one out of three patient cohorts, although this association failed to reach a study-wide or GWAS significance level [31]. Other studies involving normal individuals and patient-based cohorts have failed to identify an association between SCARA5 gene variants and plasma VWF:Ag levels in patients with type 1 VWD [27]. In a type 1 VWD study that includes both Canadian and American subjects, we have previously found that variants in both CLEC4M, and STAB2 associate with plasma VWF levels [8,32]. However, in the same subjects, no association between SCARA5 gene variants and plasma VWF levels was observed (Supplemental Table 3). Thus, we undertook these experimental studies to to rule out the possibility of a false positive GWAS result, identify the mechanistic basis by which the GWAS-identified SCARA5 variants convey their effects on VWF, and to better understand pathobiological pathways that contribute to VWF clearance.
We first began by determining the ability of SCARA5 to bind VWF using solid phase binding assays (Figure 1). We determined that SCARA5 can bind to VWF with a high affinity in a calcium- and magnesium-dependent manner. While the predominant ligand binding-region of the class A scavenger receptors has been previously thought to be the collagenous domain [33], the SRCR domain, which mediates ligand binding in a number of protein families, is thought to be the primary ligand-binding domain in the class A scavenger receptor MARCO [23,34]. As the ligand-binding properties of the SRCR domain of glycoprotein 340 are enhanced in the presence of calcium [22], this suggests involvement of the SRCR domain in mediating the interaction between SCARA5 and VWF. Previous studies have determined that the VWF A1 domain is essential for mediating binding to SR-A1 and LRP-1, however the role of the A1 domain involved in regulating binding to SCARA5 is currently unknown [7,35] Further studies are required to better understand the mechanistic basis of this interaction. Next, using a heterologous system in which we transfected the human SCARA5 cDNA into HEK 293T cells, we observed that SCARA5 transfected cells were able to bind to and internalize VWF alone or the VWF-FVIII complex and transport its VWF cargo to early endosomes in a manner similar to other VWF clearance receptors (Figure 2). Collectively, this data suggests that SCARA5 is capable of binding to and mediating the intracellular internalization of VWF.
We next wanted to determine if SCARA5 is capable of regulating VWF clearance using an in vivo mouse model of SCARA5 deficiency. We observed that there was no evidence of murine VWF clearance in SCARA5 KO mice, and a modest influence on human VWF clearance (Figure 3). In some cases, animal models may be only a partially informative tool for understanding the pathways that regulate VWF-FVIII clearance. For example, the VWF clearance receptor CLEC4M does not have a murine ortholog, and stabilin-2 clears only the human but not the murine form of the VWF protein [8,36]. Additionally, some forms of the VWF molecule exhibit different clearance profiles in humans when compared to animal models. ABO glycans can regulate VWF clearance in human but not in mouse models, while the common VWF variants p.T789A/p.Y795= are thought to have a small influence on VWF clearance in humans but exhibit a large influence in a mouse model [28,37]. Although murine SCARA5 has a ~88% amino acid identity to human SCARA5, the relative affinity of murine SCARA5 for human and murine VWF is unknown. However, murine SCARA5 has only been reported to be expressed on epithelial cells or fibroblasts in the murine testis, heart, brain, and spleen that do not come into direct contact with the blood [16,18], and not by macrophages, hepatocytes, or endocytic endothelial cells which are thought to clear the majority of human VWF [8,25].
As the GWAS data indicated an association between human VWF and human SCARA5, we next sought to determine the anatomic site and cell-specific expression pattern of human SCARA5. We began by analyzing SCARA5 expression in commercially available mRNA extracts from human tissues (Figure 4). We observed that the highest SCARA5 expression was found in the testes, followed by the spleen, and kidney, while SCARA5 mRNA levels in the liver and peripheral blood mononuclear cells was low, consistent with previously described mouse data [16,18]. We then performed IHC analysis on healthy human liver, spleen, and kidney tissues using an anti-human SCARA5 antibody that recognizes the extracellular spacer region that was validated using immunocytochemistry on HEK 293T cells transiently transfected with the SCARA5 cDNA. SCARA5 expression was highest in kidney glomerular podocytes and cells of the red and white pulp of the spleen but absent in the liver (Figures 5–7).
In the glomerulus of the human kidney, VWF is expressed by CD31-positive microvascular endothelial cells, which are separated by a basement membrane from visceral epithelial cells or podocytes. Here, soluble VWF and other plasma macromolecules circulating in the lumen of the glomerulus are prevented from being lost in the urine by a glomerular filtration barrier comprised of the basement membrane, fenestrated endothelium, and the podocyte slit diaphragm. While endocytic receptors expressed on podocytes have been shown to regulate the clearance of plasma macromolecules from the basement membrane including FcRn-mediated clearance of IgG and the scavenger receptor CXCL16-mediated clearance of oxLDL [38,39], it is unclear how podocyte-expressed SCARA5 may interact with plasma VWF in the absence of renal glomerular disease. Importantly, we observed no VWF within the SCARA5-expressing cells in the human kidney, and infusion of human plasma-derived VWF into VWF KO mice demonstrated that VWF was taken up by kidney mesangial cells but not podocytes (Figure 5 and Supplemental Figure 1). Additionally, SCARA5 expression was not previously observed in the glomeruli of the murine kidney [16,18].
Splenic endothelium is comprised of both vascular and fenestrated sinusoidal endothelium which has endocytic and phagocytic capabilities. Importantly, we have previously demonstrated that infused human VWF is taken up by murine splenic sinusoidal endothelium, which expresses stabilin-2, and that in human splenic tissues VWF co-localizes with CD31 and stabilin-2 expressing endothelium [8]. Here, we observed that SCARA5 was expressed predominantly in the white pulp of the human spleen, but also expressed by CD31-positive endothelial cells in the splenic red pulp, but not by CD68-expressing macrophages (Figure 6). In a systematic characterization of CD45-depleted and FACs purified human splenic microvascular endothelial cells, CD31+CD34−CD8α+ littoral cells, have been previously demonstrated to express a series of scavenger receptors including stabilin-1, stabilin-2, the macrophage mannose receptor CD206, and SCARA5 by mRNA analysis [19]. Consistent with this observation, both VWF and SCARA5 co-localized with CD8α expressing cells within the red pulp of the spleen in sinusoidal-like structures (Figure 7A). While CD8α is also expressed on subsets of T-cells which can be localized within the splenic white pulp, T zone, and red pulp, immunohistochemistry analysis with other T-cell markers (CD3) does not produce the same pattern characteristic of the total CD8α antigen stain, indicating that both T-cells and endothelial cells are identified by the CD8α marker [40].
Together, this evidence supports a role for littoral cells as specialized endothelium with scavenger capabilities within the human spleen, although VWF binding assays performed ex vivo using littoral endothelial cells derived from the human spleen would be required for confirmation. Importantly, murine spleens lack sinusoidal structures and given the distinct patterns of expression between splenic SCARA5 expression in human (sinusoidal endothelium) and murine tissues (interstitial fibroblasts), this may account for the weak clearance phenotype of human VWF in SCAR5 KO mice [23]. Although the human IHC analysis cannot definitively demonstrate if VWF/SCARA5 co-expressing cells are producing and/or clearing this VWF, our VWF infusion data suggests that at least a portion of the VWF observed in these cells has been endocytosed. Moreover, a previously published microarray analysis of total RNA from FACS-isolated littoral cells did not identify VWF expression as being either significantly upregulated or downregulated when compared with a reference pool [19]. It is therefore plausible that along with stabilin-2 and possibly other receptors, expression of SCARA5 in the splenic sinusoids contributes to the clearance of VWF in humans.
Trait mapping studies such as GWAS have begun to elucidate the genetic architecture that regulates complex quantitative traits such as plasma VWF levels, however validation of these observations using further genetic, cell, and animal models is essential to confirm the mechanistic basis for these associations. While often the data supporting these associations, such as that for VWF levels and ABO, STXBP5, and STAB2 are robust and rational, other associations, such as for SCARA5, remain somewhat equivocal. Indeed, it seems plausible that there is a true biological interaction between SCARA5 and VWF in humans, but that the magnitude of this effect on VWF clearance and thus VWF plasma levels is not currently understood. Further human genetic data, or humanized animal models may be required to improve our understanding of the regulation of VWF clearance by SCARA5, and how genetic variants at the SCARA5 locus modify plasma VWF levels.
Supplementary Material
Essentials:
SCARA5 was previously identified as a potential clearance receptor for VWF-FVIII by GWAS
SCARA5 expressing cells bind to and internalize VWF and the VWF-FVIII complex
SCARA5 deficiency weakly modifies the half-life of human VWF in a mouse model
VWF co-localizes with SCARA5 expressed by human splenic littoral endothelial cells
Acknowledgements:
This research was supported by grants from the Canadian Institute of Health Research (FDN 154285), and the National Institutes of Health Zimmerman Program for the Molecular and Clinical Biology of VWD (HL081588). We thank L. Boudreau, C. Dwyer, P. Lima, S. Tinlin, and A. S. Paine for technical assistance. We thank P. James, N. Renwick, B. Montgomery, J. Morrissey, S. Haberichter for provisions of reagents, patient DNA, and tissue samples. We thank K. Song, D. Hurlbut, and I. Young for helpful discussions of immunohistochemical analyses. We thank W. Hopman for performing statistical analyses.
DL receives grant support from Bayer, Bioverativ, CSL, and Octapharma. OR* receives grant support and honoraria from CSL.
Footnotes
Conflict-of-interest disclosure: The remaining authors declare no competing financial interests.
References:
- 1.PrabhuDas MR, Baldwin CL, Bollyky PL, Bowdish DME, Drickamer K, Febbraio M, Herz J, Kobzik L, Krieger M, Loike J, McVicker B, Means TK, Moestrup SK, Post SR, Sawamura T, Silverstein S, Speth RC, Telfer JC, Thiele GM, Wang X-Y, et al. A Consensus Definitive Classification of Scavenger Receptors and Their Roles in Health and Disease. J Immunol 2017; 198: 3775–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kzhyshkowska J, Neyen C, Gordon S. Role of macrophage scavenger receptors in atherosclerosis. Immunobiology 2012; 217: 492–502. [DOI] [PubMed] [Google Scholar]
- 3.Yu X, Guo C, Fisher PB, Subjeck JR, Wang X-Y. Scavenger Receptors: Emerging Roles in Cancer Biology and Immunology. Adv Cancer Res 2015; 128: 309–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Swystun LL, Lillicrap D. How much do we really know about von Willebrand disease? Curr Opin Hematol 2016; 23: 471–8. [DOI] [PubMed] [Google Scholar]
- 5.Chen J, Chung DW. Inflammation, von Willebrand factor, and ADAMTS13. Blood 2018; 132: 141–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.O’Sullivan JM, Ward S, Lavin M, O’Donnell JS. von Willebrand factor clearance – biological mechanisms and clinical significance. Br J Haematol 2018; 183: 185–95. [DOI] [PubMed] [Google Scholar]
- 7.Wohner N, Muczynski V, Mohamadi A, Legendre P, Proulle V, Aymé G, Christophe OD, Lenting PJ, Denis CV., Casari C Macrophage scavenger receptor SR-A1 contributes to the clearance of von willebrand factor. Haematologica 2018; 103: 728–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Swystun LL, Lai JD, Notley C, Georgescu I, Paine AS, Mewburn J, Nesbitt K, Schledzewski K, Géraud C, Kzhyshkowska J, Goerdt S, Hopman W, Montgomery R, James PD, Lillicrap D. The endothelial cell receptor stabilin-2 regulates VWF-FVIII complex half-life and immunogenicity. J Clin Investig 2018; 128: 4057–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grewal PK, Uchiyama S, Ditto D, Varki N, Le DT, Nizet V, Marth JD. The Ashwell receptor mitigates the lethal coagulopathy of sepsis. Nat Med 2008; 14: 648–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rastegarlari G, Pegon JN, Casari C, Odouard S, Navarrete A-M, Saint-Lu N, van Vlijmen BJ, Legendre P, Christophe OD, Denis CV, Lenting PJ. Macrophage LRP1 contributes to the clearance of von Willebrand factor. Blood 2012; 119: 2126–34. [DOI] [PubMed] [Google Scholar]
- 11.Sabater-Lleal, Huffman JE, de Vries P, Marten J Genome-Wide Association Transethnic Meta-Analyses Identifies Novel Associations Regulating Coagulation Factor VIII and von Willebrand Factor Plasma Levels. Circulation 2019; 139: 620–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smith NL, Chen M-H, Dehghan A, Strachan DP, Basu S, Soranzo N, Hayward C, Rudan I, Sabater-Lleal M, Bis JC, de Maat MPM, Rumley A, Kong X, Yang Q, Williams FMK, Vitart V, Campbell H, Mälarstig A, Wiggins KL, Van Duijn CM, et al. Novel associations of multiple genetic loci with plasma levels of factor VII, factor VIII, and von Willebrand factor: The CHARGE (Cohorts for Heart and Aging Research in Genome Epidemiology) Consortium. Circulation 2010; 121: 1382–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Desch KC, Ozel A, Halvorsen M, Michels A, Swystun L, Mokry L, Richards B, Germain M, Tregouet DA, Reitsma PH, Kearon K, Li JZ, Goldstein D., Lillicrap D, Ginsburg D Exome sequencing studies identify mutations in STAB2 as a genetic risk for venous thromboembolic disease. Blood (ASH Annu Meet Abstr) 2017; 126. [Google Scholar]
- 14.Desch KC, Ozel AB, Halvorsen M, Jacobi PM, Germain M, Tregouet DA, Reitsma PH, Goldstein D, Ginsburg D. Exome Sequencing in venous thromboembolic disease identifies excess mutation burden in PROS1, PROC, SERPINC1, STAB2. Blood (ASH Annu Meet Abstr) 2016; 128: 3794. [Google Scholar]
- 15.Rydz N, Swystun LL, Notley C, Paterson AD, Riches JJ, Sponagle K, Boonyawat B, Montgomery RR, James PD, Lillicrap D. The C-type lectin receptor CLEC4M binds, internalizes, and clears von Willebrand factor and contributes to the variation in plasma von Willebrand factor levels. Blood 2013; 121: 5228–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jiang Y, Oliver P, Davies KE, Platt N. Identification and characterization of murine SCARA5, a novel class A scavenger receptor that is expressed by populations of epithelial cells. J Biol Chem 2006; 281: 11834–45. [DOI] [PubMed] [Google Scholar]
- 17.Li JY, Paragas N, Ned RM, Qiu A, Viltard M, Leete T, Drexler IR, Chen X, Sanna-Cherchi S, Mohammed F, Williams D, Lin CS, Schmidt-Ott KM, Andrews NC, Barasch J. Scara5 is a ferritin receptor mediating non-transferrin iron delivery. Dev Cell 2009; 16: 35–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ojala JRM, Pikkarainen T, Elmberger G, Tryggvason K. Progressive reactive lymphoid connective tissue disease and development of autoantibodies in scavenger receptor A5-deficient mice. Am J Pathol 2013; 182: 1681–95. [DOI] [PubMed] [Google Scholar]
- 19.Qiu J, Salama ME, Hu CS, Li Y, Wang X, Hoffman R. The characteristics of vessel lining cells in normal spleens and their role in the pathobiology of myelofibrosis. Blood Adv 2018; 2: 1130–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yamauchi M, Sricholpech M. Lysine post-translational modifications of collagen. Essays Biochem 2012; 52: 113–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pruss CM, Golder M, Bryant A, Hegadorn CA, Burnett E, Laverty K, Sponagle K, Dhala A, Notley C, Haberichter S, Lillicrap D. Pathologic mechanisms of type 1 VWD mutations R1205H and Y1584C through in vitro and in vivo mouse models. Blood 2011; 117: 4358–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Purushotham S, Deivanayagam C. The calcium-induced conformation and glycosylation of scavenger-rich cysteine repeat (SRCR) domains of glycoprotein 340 influence the high affinity interaction with antigen I/II homologs. J Biol Chem 2014; 289: 21877–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ojala JRM, Pikkarainen T, Tuuttila A, Sandalova T, Tryggvason K. Crystal structure of the cysteine-rich domain of scavenger receptor MARCO reveals the presence of a basic and an acidic cluster that both contribute to ligand recognition. J Biol Chem 2007; 282: 16654–66. [DOI] [PubMed] [Google Scholar]
- 24.Doi T, Higashino KI, Kurihara Y, Wada Y, Miyazaki T, Nakamura H, Uesugi S, Imanishi T, Kawabe Y, Itakura H, Yazaki Y, Matsumoto A, Kodama T. Charged collagen structure mediates the recognition of negatively charged macromolecules by macrophage scavenger receptors. J Biol Chem 1993; 268: 2126–33. [PubMed] [Google Scholar]
- 25.van Schooten CJ, Shahbazi S, Groot E, Oortwijn BD, van den Berg HM, Denis CV, Lenting PJ. Macrophages contribute to the cellular uptake of von Willebrand factor and factor VIII in vivo. Blood 2008; 112: 1704–12. [DOI] [PubMed] [Google Scholar]
- 26.Swystun LL, Lillicrap D. Genetic regulation of plasma von Willebrand factor levels in health and disease. J Thromb Haemost 2018; 16: 2375–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sanders YV, van der Bom JG, Isaacs A, Cnossen MH, de Maat MPM, Laros-van Gorkom BAP, Fijnvandraat K, Meijer K, van Duijn CM, Mauser-Bunschoten EP, Eikenboom J, Leebeek FWG. CLEC4M and STXBP5 gene variations contribute to von Willebrand factor level variation in von Willebrand disease. J Thromb Haemost 2015; 13: 956–66. [DOI] [PubMed] [Google Scholar]
- 28.Mufti A, Ogiwara K, Swystun LL, Eikenboom JCJ, Budde U, Hopman WM, Halldén C, Goudemand J, Peake I, Goodeve AC, Lillicrap D, Hampshire DJ. The common VWF single nucleotide variants c.2365A>G and c.2385T>C modify VWF biosynthesis and clearance. Blood Adv 2018; 2: 1585–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Van Loon JE, Leebeek FWG, Deckers JW, Dippel DWJ, Poldermans D, Strachan DP, Tang W, O’Donnell CJ, Smith NL, De Maat MPM. Effect of genetic variations in syntaxin-binding protein-5 and syntaxin-2 on von willebrand factor concentration and cardiovascular risk. Circ Cardiovasc Genet 2010; 3: 507–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hermanns MI, Grossmann V, Spronk HMH, Schulz A, Jünger C, Laubert-Reh D, Mazur J, Gori T, Zeller T, Pfeiffer N, Beutel M, Blankenberg S, Münzel T, Lackner KJ, Cate-Hoek AJT, Cate H Ten, Wild PS Distribution, genetic and cardiovascular determinants of FVIII:c - Data from the population-based Gutenberg Health Study. Int J Cardiol 2015; 187: 166–74. [DOI] [PubMed] [Google Scholar]
- 31.Antoni G, Oudot-Mellakh T, Dimitromanolakis A, Germain M, Cohen W, Wells P, Lathrop M, Gagnon F, Morange P-E, Tregouet D-A. Combined analysis of three genome-wide association studies on vWF and FVIII plasma levels. BMC Med Genet 2011; 12: 102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rydz N, Swystun LL, Notley C, Paterson AD, Riches JJ, Sponagle K, Boonyawat B, Montgomery RR, James PD, Lillicrap D. The C-type lectin receptor CLEC4M binds, internalizes, and clears von Willebrand factor and contributes to the variation in plasma von Willebrand factor levels. Blood 2013; 121: 5228–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rohrer L, Freeman M, Kodama T, Penman M, Krieger M. Coiled-coil fibrous domains mediate ligand binding by macrophage scavenger receptor type II. Nature 1990; 343: 570–2. [DOI] [PubMed] [Google Scholar]
- 34.Novakowski KE, Huynh A, Han S, Dorrington M, Yin C, Tu Z, Pelka P, Whyte P, Guarné A, Sakamoto K. A Naturally-Occurring Transcript Variant of MARCO Reveals the SRCR Domain is Critical for Function. Immunol Cell Biol 2016; 94: 646–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wohner N, Legendre P, Casari C, Christophe O, Lenting P, Denis C. Shear stress-independent binding of von Willebrand factor-type 2B mutants p. R1306Q & p. V1316M to LRP1 explains their increased clearance. J Thromb Haem 2015; 13: 815–20. [DOI] [PubMed] [Google Scholar]
- 36.Tanne A, Ma B, Boudou F, Tailleux L, Botella H, Badell E, Levillain F, Taylor ME, Drickamer K, Nigou J, Dobos KM, Puzo G, Vestweber D, Wild MK, Marcinko M, Sobieszczuk P, Stewart L, Lebus D, Gicquel B, Neyrolles O. A murine DC-SIGN homologue contributes to early host defense against Mycobacterium tuberculosis. J Exp Med 2009; 206: 2205–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Groeneveld DJ, van Bekkum T, Cheung KL, Dirven RJ, Castaman G, Reitsma PH, van Vlijmen B, Eikenboom J. No evidence for a direct effect of von Willebrand factor’s ABH blood group antigens on von Willebrand factor clearance. J Thromb Haemost 2015; 13: 592–600. [DOI] [PubMed] [Google Scholar]
- 38.Gutwein P, Abdel-Bakky MS, Schramme A, Doberstein K, Kämpfer-Kolb N, Amann K, Hauser IA, Obermüller N, Bartel C, Abdel-Aziz AAH, El Sayed ESM, Pfeilschifter J. CXCL16 is expressed in podocytes and acts as a scavenger receptor for oxidized low-density lipoprotein. Am J Pathol 2009; 174: 2061–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Akilesh S, Huber TB, Wu H, Wang G, Hartleben B, Kopp JB, Miner JH, Roopenian DC, Unanue ER, Shaw AS. Podocytes use FcRn to clear IgG from the glomerular basement membrane. Proc Natl Acad Sci 2008; 105: 967–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rivino L, Gruarin P, Häringer B, Steinfelder S, Lozza L, Steckel B, Weick A, Sugliano E, Jarrossay D, Kühl AA, Loddenkemper C, Abrignani S, Sallusto F, Lanzavecchia A, Geginat J. CCR6 is expressed on an IL-10 – producing, autoreactive memory T cell population with context-dependent regulatory function 2010; 207: 565–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.