Abstract
Background
Let-7 microRNAs (miRNAs) have the effects of inhibiting tumor growth and metastasis, however, the research in nasopharyngeal carcinoma (NPC) is limited. This study focused on the effects of Let-7 on NPC migration and invasion and the mechanism of action.
Material/Methods
Plasmid transfection was used to upregulate the expression levels of Let-7g-5p and insulin-like growth factor-1 receptor (IGF-1R). Cell counting kit-8 (CCK-8) assay was applied to test the cell viability. Scratch assay and Transwell assay were performed to detect the migration and invasion abilities. Bioinformatics prediction and luciferase reporter assay were used to determine and verify the downstream target genes for Let-7g-5p. Protein and mRNA were detected by western blot and real-time quantitative polymerase chain reaction (RT-qPCR), respectively.
Results
Let-7g-5p was under-expressed in human NPC cells. Overexpression of Let-7g-5p could inhibit cell viability and inhibit the migration and invasion of SUNE1 cells. The dual-luciferase reporter assay showed that IGF-1R was a direct target gene of Let-7g-5p, which was directly regulated IGF-1R expression by 3′UTR. Let-7g-5p overexpression could inhibit the expression of IGF-1R gene, and upregulation of IGF-1R gene expression reversed the inhibitory effect of Let-7g-5p on cell viability and epithelial-mesenchymal transition processes.
Conclusions
Let-7g-5p is lowly expressed in NPC and it was the first to discover that IGF-1R was a target gene of let-7g-5p in NPC. Upregulation of IGF-1R reversed the inhibitory effect of Let-7g-5p on epithelial-mesenchymal transition.
MeSH Keywords: Epithelial-Mesenchymal Transition; MicroRNAs; Nasopharyngeal Neoplasms; Receptor, IGF Type 1
Background
Nasopharyngeal carcinoma (NPC) is a common head and neck malignancy and the most common malignant tumor in otolaryngology [1]. Studies have shown that in recent years, the incidence of NPC has shown an upward trend due to changes in environment and habits [2]. Radiotherapy, which plays an important role in NPC, is the basis of initial treatment due to the radiosensitive behavior of NPC [3]. Studies have shown that radiotherapy is effective in the treatment of NPC and can improve the survival rate of patients [4,5]. In addition, NPC also has chemosensitive attributes. Chemotherapy is an important method for treating NPC, however, due to the special site of NPC, the incidence of acute toxicity of NPC chemotherapy remains high and it has high recurrence rate and metastasis rate [3,6–9]. Therefore, it is important to find new treatments.
MicroRNAs (miRNAs) are a class of endogenous, non-coding RNA molecules of approximately 18–25 nucleotides in length. MiRNAs can regulate protein expression by inhibiting or inducing the degradation of messenger RNAs (mRNAs) by specifically binding to the 3′ untranslated region (UTR) of the mRNAs [10,11]. The study shows that about 30% of genes receive miRNA regulation and regulate the occurrence and development of many cancers [12,13]. Studies have shown that miRNA expression abnormalities are related to the occurrence and development of cancer [14–17].
About 1100 miRNAs have been identified in Homo sapiens, and the Let-7 family is one of the most widely studied miRNAs [18]. The occurrence, development, and metastasis of NPC are widely regulated by miRNAs [19,20], and it has been found that Let-7 is downregulated in NPC [21]. There are 13 members in the human Let-7 family [22]. It has been found that the expression level of Let-7 is decreased in lung cancer and breast cancer, and increasing the level of Let-7 can inhibit the growth of tumor cells and promote their apoptosis [23,24]. Studies have shown that non-small cell lung cancer patients with low expression of Let-7 have significantly shorter survival time than patients with moderate Let-7 expression [25]. Yan et al. has found that the expression level of Let-7 family member Let-7c had an inhibitory effect on tumor cell migration and invasion [26]. Li et al. found that 7 members of Let-7 family (Let-7a, Let-7b, Let-7c, Let-7d, Let-7e, Let-7f, and Let-7g) were significantly under-expressed in NPC [27]. Studies have shown that miRNAs of Let-7 family suppresses NPC cells proliferation by downregulating c-Myc expression [28]. These studies only examined the expression of Let-7 family in NPC and its effect on proliferation of NPC. However, no research has been conducted on Let-7 affecting NPC migration and invasion.
This study focused on the expression characteristics of the Let-7 family in NPC cells and examined the effect of Let-7g on cell migration and invasion. Through follow-up studies, we found and verified that insulin-like growth factor-1 receptor (IGF-lR) was the target gene of Let-7g in NPC cells. We also explored its mechanism of cell migration and invasion. Our study provides a theoretical basis for future development of NPC treatment with Let-7g as a pathway.
Material and Methods
Cells culture and transfection
The nasopharyngeal epithelial cell line NP69, NPC cell lines SUNE1, C666-1, 6-10B, and HNE-3 were purchased from ATCC (USA). Cells were cultured in DMEM medium containing 10% FBS and 1% penicillin-streptomycin at 37°C in an incubator with 5% CO2. Culture related reagents were purchased from GIBCO Invitrogen (USA).
The targets of Let-7g-5p predicted by computer-aided algorithms were obtained from TargetScan, microRNA and miRBase. The Let-7g-5p mimics were transfected into the cells by Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) following the manufacturer’s protocol. Empty vector was used as negative control.
The IGF-lR cDNA was subcloned into pcDNA3.1 (Sangon Biotech, China) to construct a pcDNA-IGF-1R expression vector. IGF-1R transfection was performed using Lipofectamine 2000 following the manufacturer’s protocol.
Cell counting kit-8 (CCK-8) assay
The CCK-8 assay was applied to test the cell viability and the kit was purchased from Tongren (Japan). We explored the time point by pre-experiment, and 0, 24, and 48 hours were selected as the optimal time point for detection, according to the growth status of cells and the significance and stability of optical density (OD) value. The cells were pre-incubated at 37°C in an incubator with 5% CO2. CCK-8 reagent was added and cultured at 37°C with 5% CO2 for 4 hours. The OD of each well at 450 nm was measured using a microplate reader (ELX 800, Bio-Teck, USA).
Scratch assay
A black Marker pen was used to draw a horizontal line across the hole behind the 6-well plate. 5×105 cells were added to each well and incubated overnight. The tip of the pipette was used for vertical scratching and incubated in serum-free medium (Oringeng, USA) for 24 hours. After taking a photo, the image was processed using ImageJ software (Rawak Software, Inc., Germany) and the percentage of wound width was calculated.
Transwell assay
Transwell chamber and related reagents were purchased from Corning (USA). 500 pL of whole culture was added in the lower chamber, and then 5 × 104 cells were added and cultured for 4 hours. The upper chamber was placed in the lower chamber. Finally, a cell suspension containing 5×104 cells was contained and dropped vertically into the upper chamber. The cells were cultured at 37°C with 5% CO2 for 6–8 hours. After staining with Giemsa staining (Shanghai Gefan Biotechnology Co., Ltd., China), 5 high power fields were randomly select under a microscope to count the migration rate.
Dual-luciferase reporter assay
TargetScan, miRDB and TargetMiner were applied to predict the target of Let-7g-5p, and the target genes predicted by the 3 online tools were intersected to obtain the common target. For the dual-luciferase assay, the wild-type of the 3′UTR segment of the IGF-1R gene containing the Let-7g-5p binding sequencing was amplified by PCR. A mutant type of IGF-1R 3′-UTR lacking complementarity with Let-7g-5p sequence was constructed using the Site-Directed Mutagenesis kit (Stratagene, La Jolla, CA, USA). Then, the wild-type or mutant type of IGF-1R 3′-UTR was cloned into the pisCHECK-2 vector (Promega, Madison, WI, USA). Glioma cells were co-transfected with the luciferase reporters together with Let-7g-5p mimics by using Lipofectamine 2000. Then 48 hours after transfection, luciferase experiment was performed using a dual-luciferase reporter assay kit (Promega), according to the instructions.
Real-time quantitative polymerase chain reaction (RT-qPCR) analysis
Real-time quantitative polymerase chain reaction (RT-qPCR) was used to detect the IGF-1R, E-cadherin, N-cadherin, vimentin, and matrix metalloproteinase 2 (MMP2) mRNA expression levels. The cells were triturated and lysed, and then the RNA was extracted by RNA extraction kit (Promega, Beijing, China). Reverse transcription kit (TaKaRa, Japan) was used to synthesize cDNA. Reverse transcription reaction conditions were set at 37°C for 15 minutes and reverse transcriptase inactivation condition was at 85°C for 15 seconds. RT-qPCR was performed with the RT-qPCR kit (TaKaRa, Japan). PCR was performed by activating the DNA polymerase at 95°C for 5 minutes, followed by 40 cycles of 2-step PCR (95°C for 10 seconds and 60°C for 30 seconds) and a final extension at 75°C for 10 minutes and held at 4°C. RNase-free water was used as the templates of negative control experiences. All primers were obtained from Genewiz (Suzhou, Jiangsu, China) and listed in Table 1. The formula 2−ΔΔCT was used to analyze the mRNA expression levels.
Table 1.
The sequences of primers.
| Primer name | Sequence (5′-3′) | Product size (bp) |
|---|---|---|
| Let-7b-5p-forward | UGAGGUAGUAG GUUGUGUGGUU | 108 |
| Let-7b-5p-reverse | CGGTGAGGTCT TTGGTTCATAACCAC | |
| Let-7c-5p-forward | GAGGTAGTAGGTTGTATGGTTG | 124 |
| Let-7c-5p-reverse | GCAGGGTCCGAGGTATTC | |
| Let-7f-5p-forward | GCCGTGAGGTAGTAGATTGTAT | 114 |
| Let-7f-5p-reverse | GTGCAGGGTCCGAGGT | |
| Let-7g-5p-forward | GAGTTCCTCCAGCGCTCCGT | 94 |
| Let-7g-5p-reverse | GATGAGCAGGGTGACGCCAT | |
| IGF1R-forward | GAGGCTGTGCTGGACGCTCTAG | 212 |
| IGF1R-reverse | TACAATGTTTGTTTGATTTCATTG | |
| E-cadherin-forward | TCCCATCAGCTGCCCAGAAA | 321 |
| E-cadherin-reverse | ATTGTCCTTGTGTCCTCAGT | |
| Vimentin-forward | CAGCCATGACCACCAGG | 140 |
| Vimentin-reverse | AAGGTCAAGACGTGCCAG | |
| MMP-2-Forward | TGATGGTGTCTGCTGGAAAG | 90 |
| MMP-2-reverse | GACACCTGAAAAGTGCCTG | |
| GAPDH-forward | CCATCTTCCAGGAGCGAGAT | 222 |
| GAPDH-reverse | TGCTGATGATCTTGAGGCTG |
Western blot
Western blot was applied to test IGF-1R, E-cadherin, N-cadherin, vimentin, and MMP2 protein expression levels. The cells were lysed, and the supernatant was collected by centrifuging the cells at 12 000 rpm at 4°C for 15 minutes. Bicinchoninic acid (BCA) assay was used to determine the protein concentration. Sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) was prepared and applied to electrophoresis. The Polyvinylidene fluoride or polyvinylidene difluoride (PVDF) membrane (Bio-Rad, USA) was transferred by a Trans-Blot Transfer Slot (Bio-Rad, USA) and blocked with 5% fat-free milk for 2 hours at room temperature. The primary antibodies (anti-IGF-1R, Abcam, ab100546, dilution: 1: 800; anti-E-cadherin, Abcam, ab6528, dilution: 1: 800; anti-N-cadherin, Abcam, ab18203, dilution: 1: 800; anti-vimentin, Abcam, ab92547, dilution: 1: 600; anti-MMP-2, Abcam, ab168864, dilution: 1: 800) were added according to the kit instruction, and the samples were shaken at room temperature for 2 hours and incubated at 4°C for 12 hours. The secondary antibodies (mouse anti-human IgG, Abcam, ab1927, dilution: 1: 10 000; rabbit anti-human IgG, Abcam, ab6759, dilution: 1: 8000; rabbit anti-goat IgG, Abcam, ab6741, dilution: 1: 10 000; goat anti-rabbit IgG, Abcam, ab6721, dilution: 1: 8000) were added and incubated at room temperature for 1.5 hours. Chemiluminescence detection was carried out using enhanced chemiluminescence (ECL) reagent (Huiying, Shanghai, China).
Results
Expression characteristics of Let-7 in NPC cell lines
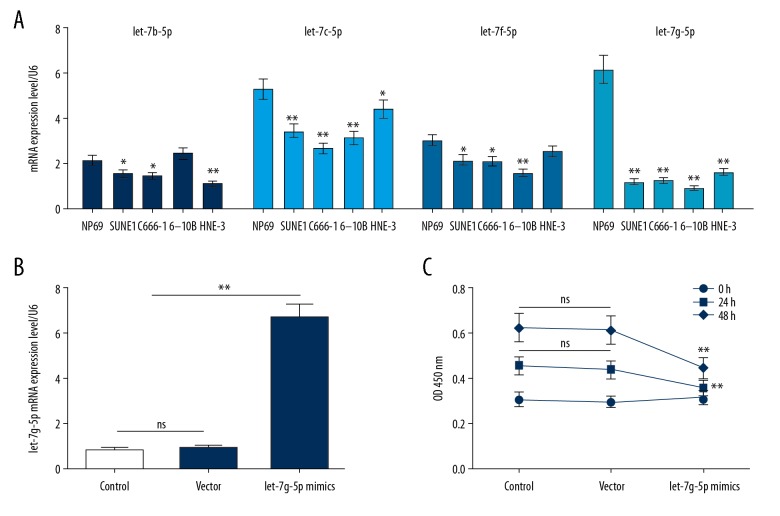
The expression levels of Let-7b-5p, Let-7c-5p, Let-7f-5p, and Let-7g-5p in NPC cell lines SUNE1, C666-1, 6-10B, and HNE-3 were significantly lower than those in NP69 cell line. Among them, Let-7g-5p was most significantly downregulated in the SUNE1 and 6-10B cell lines (Figure 1A). This indicated that Let-7g-5p was under-expressed in human NPC. SUNE1 cells are commonly used in NPC related studies, and the SUNE1 cell line was selected for subsequent experiments.
Figure 1.
Expression of Let-7 in NPC cell lines and the effects of Let-7g-5p on NPC cells. (A) RT-qPCR was used to test the Let-7b-5p, Let-7c-5p, Let-7f-5p and Let-7g-5p expression levels in NP69, SUNE1, C666-1, 6-10B, and HNE-3 cell lines. (B) RT-qPCR was applied to detect Let-7g-5p mimics plasmid transfection efficiency. (C) The effect of Let-7g-5p overexpression on the viability of SUNE1 cells was detected using CCK-8 assay. * P<0.05, ** P<0.01, versus control group. NPC – nasopharyngeal carcinoma; RT-qPCR – real-time quantitative polymerase chain reaction; CCK-8 – cell counting kit-8.
To investigate the effect of Let-7g-5p on cells, transfection technology was used to upregulate the expression level of Let-7g-5p, and the viability of SUNE1 cells was tested by CCK-8 assay. The results of RT-qPCR showed that the expression level of Let-7g-5p mRNA was significantly upregulated after transfection, suggesting a successful transfection (Figure 1B). The results showed that the OD value decreased significantly after 48 hours of transfection, suggesting that overexpression of Let-7g-5p could inhibit cells viability (Figure 1C).
Effect of Let-7g-5p on the migration and invasion of SUNE1 cells
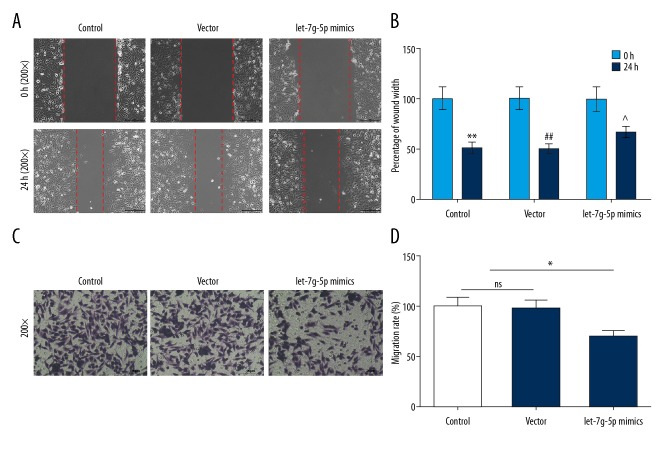
The effects of Let-7g-5p on the migration and invasion of NPC cells were detected using scratch assay and Transwell assay, respectively. The results of the study showed that the percentage of wound width was significantly higher than those in control group and vector group, and that migration rate in the Let-7g-5p mimics group was significantly lower than that in the control group and vector group (Figure 2A–2D), indicating that upregulation of Let-7g-5p expression level might inhibit the migration and invasion in SUNE1 cells.
Figure 2.
Effects of Let-7g-5p overexpression on cell migration and invasion. (A–D) The percentage of wound width and migration rate of SUNE1 cells were detected using scratch assay and Transwell assay, respectively. * P<0.05, ** P<0.01, versus control group.
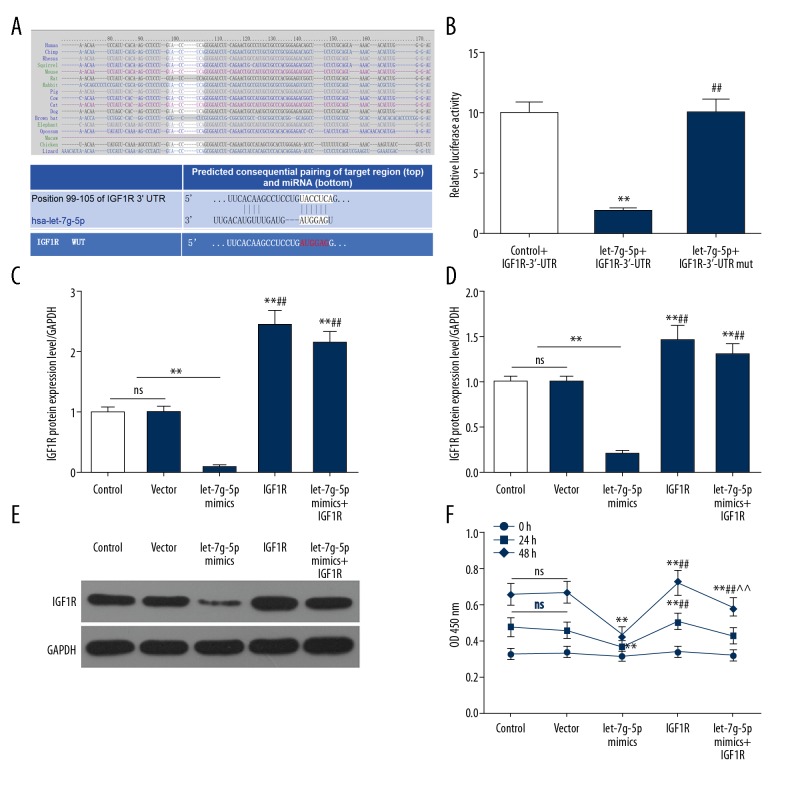
Searching and verification of IGF-1R as one of the downstream target genes for Let-7g-5p in NPC cells
After searching for the target gene of Let-7g-5p in the database TargetScan, miRDB, TargetMiner, we found a potential binding site between the 3′UTR of IGF-1R and Let-7g-5p (Figure 3A). The dual-luciferase reporter assay showed that the relative luciferase activity was significantly decreased after co-transfection with the wild-type IGF-1R expression vector and Let-7g-5p, while the relative luciferase activity was not significantly changed after co-transfection with the mutant IGF-1R expression vector and Let-7g-5p (Figure 3B). This demonstrated that IGF-1R was a direct target gene for Let-7g-5p, which directly regulated IGF-1R expression by 3′UTR.
Figure 3.
Prediction and validation of Let-7g-5p potential target genes and the effects on SUNE1 cells. (A) Searching for the target gene of Let-7g-5p in the database TargetScan, miRDB, TargetMiner. (B) Dual-luciferase reporter assay was used to verify that Let-7g-5p directly regulated IGF-1R expression by 3′UTR. (C–E) RT-qPCR and western blot were used to detect the expression levels of IGF-1R mRNA and protein in the 5 groups. (F) CCK-8 assay was used to detect the cell viability in the 5 groups. * P<0.05, ** P<0.01, versus control group; # P<0.05, ## P<0.01, versus Let-7g-5p mimics group; ^ P<0.05, ^^ P<0.01, versus IGF-1R group. RT-qPCR – real-time quantitative polymerase chain reaction; CCK-8 – cell counting kit-8.
Effects of Let-7g-5p on IGF-1R gene expression and cell viability
To explore the effect of Let-7g-5p on IGF-1R gene expression, the regulation of IGF-1R expression by Let-7g-5p was verified using SUNE1 cells transfected with Let-7g mimics. Control group, vector group, Let-7g-5p mimics group, IGF-1R group, and Let-7g-5p mimics+IGF-1R group were constructed, respectively, and the expression levels of IGF-1R protein and mRNA and cell viability in each group were detected. The results showed that IGF-1R protein and mRNA expression levels and cell viability in Let-7g-5p mimics group were significantly lower than those in control group and vector group, while IGF-1R gene expression level and cell viability in IGF-1R group were increased. IGF-1R gene expression level and cell viability in the Let-7g-5p mimics+IGF-1R group were significantly higher than those in the Let-7g-5p mimics group, and lower than those in IGF-1R group (Figure 3C–3F). This indicated that Let-7g-5p overexpression could inhibited the expression of IGF-1R gene, however, upregulation of IGF-1R gene expression could reverse the tumor suppressor effect of Let-7g-5p overexpression, to some extent, and Let-7g-5p overexpression could partially reverse the IGF-1R upregulation, indicating that IGF-1R was a functional target gene of Let-7g-5p.
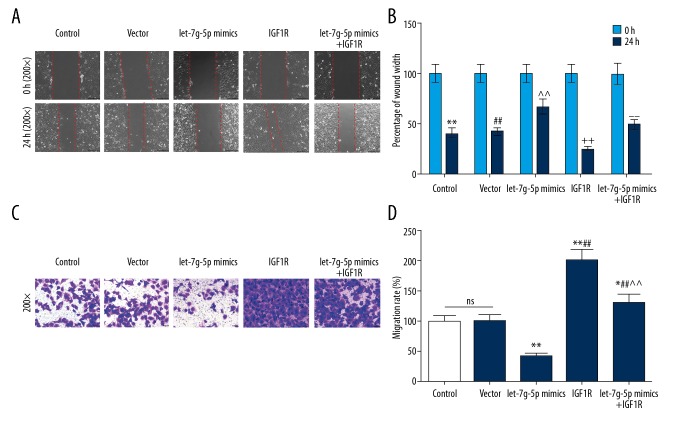
IGF-1R involved in the regulation of cell migration and invasion
To study the effects of IGF-1R acting as a functional target gene of Let-7g-5p on NPC cells, the migration and invasion abilities of the aforementioned 5 groups were examined. The results showed that the percentage of wound width was increased but migration rate was decreased in Let-7g-5p mimics group. However, the percentage of wound width was significantly decreased, and migration rate were significantly increased in the IGF-1R group. The percentage of wound width in Let-7g-5p mimics+IGF-1R group was lower than that in Let-7g-5p mimics group, however, migration rate was higher in Let-7g-5p mimics+IGF-1R group than that in Let-7g-5p mimics group (Figure 4A–4D). This suggested that upregulation of IGF-1R expression reversed the inhibitory effect of Let-7g-5p overexpression on cell migration and invasion.
Figure 4.
Effects of IGF-1R overexpression on the inhibition of migration and invasion by Let-7g-5p. (A–D) The percentage of wound width and migration rate of SUNE1 cells in the five groups were detected using scratch assay and Transwell assay, respectively. * P<0.05, ** P<0.01, versus control group; # P<0.05, ## P<0.01, versus Let-7g-5p mimics group; ^ P<0.05, ^^ P<0.01, versus IGF-1R group.
IGF-1R involved in the regulation of epithelial-mesenchymal transition (EMT)
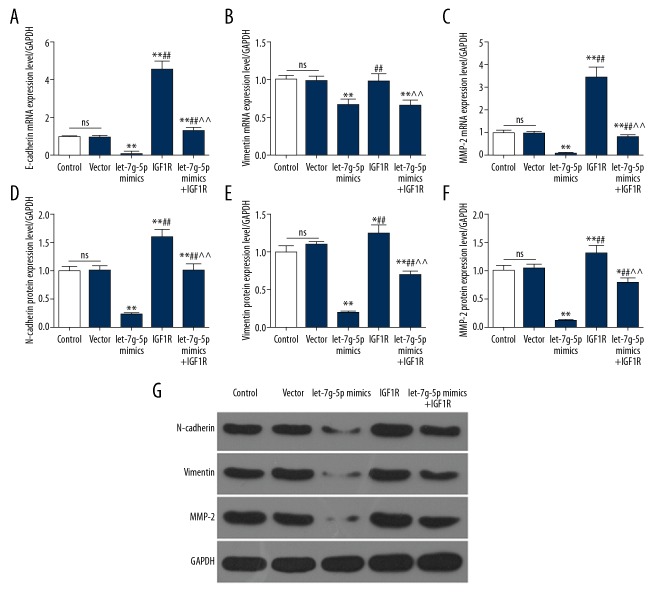
To explore the effects of IGF-1R overexpression on migration and invasion, the expression levels of epithelial-mesenchymal transition (EMT)-related genes were detected in 5 groups. The results showed that the N-cadherin, vimentin, and MMP2 protein and mRNA levels in the Let-7g-5p mimics group were downregulated. The protein and mRNA levels of the 4 genes in the Let-7g-5p mimics+IGF-1R group were significantly higher than those in the Let-7g-5p mimics group (Figure 5A–5G). This suggested that upregulation of IGF-1R could reverse the inhibition of Let-7g-5p overexpression on EMT, thereby promoting migration and invasion of NPC cells.
Figure 5.
Effects of IGF-1R and Let-7g-5p on EMT. (A–G) expression levels of E-cadherin, vimentin, and MMP2 mRNA and protein were detected by RT-qPCR and western blot, respectively. * P<0.05, ** P<0.01, versus control group; # P<0.05, ## P<0.01, versus Let-7g-5p mimics group; ^ P<0.05, ^^ P<0.01, versus IGF-1R group. RT-qPCR – real-time quantitative polymerase chain reaction.
Discussion
Let-7 was first alienated by Reinhartet in Caenorhabditis elegans, and 13 members of the Let-7 family have been discovered so far [22,29]. Let-7 belongs to the tumor suppressor miRNA family, and high levels of let-7 expression have anti-proliferative effects on cancer cells, however, overexpression of Let-7 levels lead to poor prognosis in hepatocellular carcinoma [30]. Let-7 can be used as a predictor of clinical prognosis, and its mechanism in tumorigenesis also can provide a new means for the treatment of tumors [31,32].
Due to the special location of the NPC, the difficulty of treatment after NPC metastasis is greatly increased [33]. Study have shown that Let-7a and Let-7b are under-expressed in NPC cell lines HK1 and HONE1, and it is found that Let-7 has an effect of reducing the proliferation activity of NPC cells [28]. Previous studies have suggested that re-expression of Let-7g miRNAs inhibited the proliferation and migration of hepatocellular carcinoma [34]. The study by Lin et al. also suggested that the ectopic expression of Let-7 family member let-7c could significantly inhibit migration and invasion abilities [35]. Thian et al. found when let-7 was inhibited, the proliferative capacity of NPC cells is reduced and Let-7 inhibited the proliferation of NPC cells by downregulating c-Myc expression [28]. To investigate the effect of Let-7 on NPC migration and invasion, we examined the expression levels of Let-7b-5p, Let-7c-5p, Let-7f-5p, and Let-7g-5p in the NP69, SUNE1, C666-1, 6-10B, and HNE-3 cell lines. The results showed that the 4 types of Let-7 expression level in the NPC cell line were significantly decreased, and the downregulation of Let-7g-5p was most obvious. The experiment also showed that upregulating the level of Let-7g-5p reduced the proliferation ability of the cells. Further experimental results also showed that the upregulation of Let-7g-5p levels by plasmid transfection inhibited the migration and invasion of SUNE1 cells. This indicated that Let-7g-5p could inhibit the migration and invasion of NPC cells.
Gao et al. found that let-7b/IGF-1R-mediated crosstalk between IRS-2/Akt and MAPK was involved in oral squamous cell carcinoma [36]. To further explore the mechanism by which Let-7g-5p affects NPC cell migration and invasion, we found and confirmed that Let-7g-5p directly regulated IGF-1R expression through 3′UTR. Let-7g-5p inhibited the expression level of the IGF-1R gene. Further studies have also shown that a high expression of IGF-1R gene increased the ability of NPC cells to migrate and invade, while upregulation of IGF-1R mRNA and protein expression levels inhibited the ability of Let-7g-5p to resist migration and invasion.
To explore its mechanism, we examined EMT-related proteins, and found that Let-7g-5p reduced the levels of N-cadherin, vimentin, and MMP2 protein and mRNA levels. Upregulation of IGF-1R gene expression would attenuate the inhibitory effect of Let-7g-5p on EMT.
IGF-lR is one of the major members of the tyrosine protein kinase receptor family. Its ligand is insulin like growth factor-1 (IGF-1) [37] and IGF-1 binds to IGF-1R and regulates cell proliferation, apoptosis, and metastasis of NPC via regulating pathways such as MAPK and PI3K/Akt [38,39]. The invasion and migration of tumor cells is related to EMT, and N-cadherin and vimentin are common cell-matrix proteins that are found mainly in cell matrices and act as fixed cells [40,41]. MMP2 is an important component of the matrix metalloproteinase family, which can hydrolyze intercellular matrix protein levels and enable cells to metastasize [42]. The aforementioned proteins are mainly the proteins involved in EMT, and previous studies have shown that EMT is regulated by the IGF-IR gene [43,44]. In recent years, study has found that IGF1R is a direct target of let-7i in T cells of patients with ankylosing spondylitis, and let-7i could inhibit the expression of IGF1R [45]. Fawzy et al. also find that let-7i can control the occurrence of hepatocellular carcinoma by inhibiting IGF1R [46]. Let-7g-5p is under-expressed in glioblastoma, and let-7g-5p inhibits the EMT process (let-7g-5p inhibited) by targeting the VSIG4 gene [47]. This study suggests that in NPC, IGF-1R is the target of let-7g-5p and upregulation of IGF-1R reverses the inhibitory effect of Let-7g-5p on EMT.
In the study, the effect and mechanism of Let-7g-5p on the migration and invasion of NPC were mainly discussed based on in vitro cell experiments. However, the study has some limitations, and it is necessary to further explore the effect and mechanism of Let-7g-5p on NPC in vivo experiments. In addition, it is necessary to further investigate the effect of knockdown let-7g-5p on the migration and invasion of NPC. In the study, the product size of IGF-1R, E-cadherin and GAPDH are more than 200 bp, it might be a good idea to verify the expression of IGF-1R, E-cadherin and GAPDH again with appropriate primers, it is also a limitation of the study.
Conclusions
In summary, this study found that Let-7g-5p was lowly expressed in NPC. It was the first to discover that IGF-1R was a target gene of Let-7g-5p in NPC. Upregulation of IGF-1R reversed the inhibitory effect of Let-7g-5p on EMT. Our study provides a new approach to the treatment of NPC.
Footnotes
Source of support: Departmental sources
Conflict of interest
The authors declare no conflicts of interest.
References
- 1.King AD, Ahuja AT, Leung SF, et al. Neck node metastases from nasopharyngeal carcinoma: MR imaging of patterns of disease. Head Neck. 2000;22(3):275–81. doi: 10.1002/(sici)1097-0347(200005)22:3<275::aid-hed10>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 2.Tang LL, Chen WQ, Xue WQ, et al. Global trends in incidence and mortality of nasopharyngeal carcinoma. Cancer Lett. 2016;374(1):22–30. doi: 10.1016/j.canlet.2016.01.040. [DOI] [PubMed] [Google Scholar]
- 3.Blanchard P, Lee A, Marguet S, et al. Chemotherapy and radiotherapy in nasopharyngeal carcinoma: an update of the MAC-NPC meta-analysis. Lancet Oncol. 2015;16(6):645–55. doi: 10.1016/S1470-2045(15)70126-9. [DOI] [PubMed] [Google Scholar]
- 4.Chan AT, Leung SF, Ngan RK, et al. Overall survival after concurrent cisplatin-radiotherapy compared with radiotherapy alone in locoregionally advanced nasopharyngeal carcinoma. J Natl Cancer Inst. 2005;97(7):536–39. doi: 10.1093/jnci/dji084. [DOI] [PubMed] [Google Scholar]
- 5.Wee J, Tan EH, Tai BC, et al. Randomized trial of radiotherapy versus concurrent chemoradiotherapy followed by adjuvant chemotherapy in patients with American Joint Committee on Cancer/International Union against cancer stage III and IV nasopharyngeal cancer of the endemic variety. J Clin Oncol. 2005;23(27):6730–38. doi: 10.1200/JCO.2005.16.790. [DOI] [PubMed] [Google Scholar]
- 6.Komatsu M, Tsukuda M, Matsuda H, et al. Comparison of concurrent chemoradiotherapy versus induction chemotherapy followed by radiation in patients with nasopharyngeal carcinoma. Anticancer Res. 2012;32(2):681–86. [PubMed] [Google Scholar]
- 7.Chen L, Hu CS, Chen XZ, et al. Adjuvant chemotherapy in patients with locoregionally advanced nasopharyngeal carcinoma: Long-term results of a phase 3 multicentre randomised controlled trial. Eur J Cancer. 2017;75:150–58. doi: 10.1016/j.ejca.2017.01.002. [DOI] [PubMed] [Google Scholar]
- 8.OuYang PY, Xie C, Mao YP, et al. Significant efficacies of neoadjuvant and adjuvant chemotherapy for nasopharyngeal carcinoma by meta-analysis of published literature-based randomized, controlled trials. Ann Oncol. 2013;24(8):2136–46. doi: 10.1093/annonc/mdt146. [DOI] [PubMed] [Google Scholar]
- 9.Liang ZG, Chen ZT, Li L, et al. Progresses and challenges in chemotherapy for loco-regionally advanced nasopharyngeal carcinoma. Asian Pac J Cancer Prev. 2015;16(12):4825–32. doi: 10.7314/apjcp.2015.16.12.4825. [DOI] [PubMed] [Google Scholar]
- 10.Nilsen TW. Mechanisms of microRNA-mediated gene regulation in animal cells. Trends Genet. 2007;23(5):243–49. doi: 10.1016/j.tig.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 11.Cullen BR. Transcription and processing of human microRNA precursors. Mol Cell. 2004;16(6):861–65. doi: 10.1016/j.molcel.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 12.Kumar R, Xi Y. MicroRNA, epigenetic machinery and lung cancer. Thorac Cancer. 2011;2(2):35–44. doi: 10.1111/j.1759-7714.2011.00043.x. [DOI] [PubMed] [Google Scholar]
- 13.Deng S, Calin GA, Croce CM, et al. Mechanisms of microRNA deregulation in human cancer. Cell Cycle. 2008;7(17):2643–46. doi: 10.4161/cc.7.17.6597. [DOI] [PubMed] [Google Scholar]
- 14.Karimi Kurdistani Z, Saberi S, Tsai KW, Mohammadi M. MicroRNA-21: Mechanisms of oncogenesis and its application in diagnosis and prognosis of gastric cancer. Arch Iran Med. 2015;18(8):524–36. [PubMed] [Google Scholar]
- 15.Yang Y, Meng H, Peng Q, et al. Downregulation of microRNA-21 expression restrains non-small cell lung cancer cell proliferation and migration through upregulation of programmed cell death 4. Cancer Gene Ther. 2015;22(1):23–29. doi: 10.1038/cgt.2014.66. [DOI] [PubMed] [Google Scholar]
- 16.Wojcicka A, Kolanowska M, Jazdzewski K. Mechanisms in endocrinology: MicroRNA in diagnostics and therapy of thyroid cancer. Eur J Endocrinol. 2016;174(3):R89–98. doi: 10.1530/EJE-15-0647. [DOI] [PubMed] [Google Scholar]
- 17.Zhang L, Nadeem L, Connor K, Xu G. Mechanisms and therapeutic targets of microRNA-associated chemoresistance in epithelial ovarian cancer. Curr Cancer Drug Targets. 2016;16(5):429–41. doi: 10.2174/1568009616666160404121105. [DOI] [PubMed] [Google Scholar]
- 18.Pillai RS, Bhattacharyya SN, Artus CG, et al. Inhibition of translational initiation by Let-7 microRNA in human cells. Science. 2005;309(5740):1573–76. doi: 10.1126/science.1115079. [DOI] [PubMed] [Google Scholar]
- 19.Qi X, Li J, Zhou C, et al. MicroRNA-320a inhibits cell proliferation, migration and invasion by targeting BMI-1 in nasopharyngeal carcinoma. FEBS Lett. 2014;588(20):3732–38. doi: 10.1016/j.febslet.2014.08.021. [DOI] [PubMed] [Google Scholar]
- 20.Zhang P, Hong H, Sun X, et al. MicroRNA-10b regulates epithelial-mesenchymal transition by modulating KLF4/Notch1/E-cadherin in cisplatin-resistant nasopharyngeal carcinoma cells. Am J Cancer Res. 2016;6(2):141–56. [PMC free article] [PubMed] [Google Scholar]
- 21.Pan XM, Jia J, Guo XM, et al. Lack of association between let-7 binding site polymorphism rs712 and risk of nasopharyngeal carcinoma. Fam Cancer. 2014;13(1):93–97. doi: 10.1007/s10689-013-9681-4. [DOI] [PubMed] [Google Scholar]
- 22.Roush S, Slack FJ. The let-7 family of microRNAs. Trends Cell Biol. 2008;18(10):505–16. doi: 10.1016/j.tcb.2008.07.007. [DOI] [PubMed] [Google Scholar]
- 23.Stahlhut C, Slack FJ. Combinatorial action of microRNAs let-7 and miR-34 effectively synergizes with erlotinib to suppress non-small cell lung cancer cell proliferation. Cell Cycle. 2015;14(13):2171–80. doi: 10.1080/15384101.2014.1003008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peng F, Li TT, Wang KL, et al. H19/let-7/LIN28 reciprocal negative regulatory circuit promotes breast cancer stem cell maintenance. Cell Death Dis. 2017;8(1):e2569. doi: 10.1038/cddis.2016.438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xia XM, Jin WY, Shi RZ, et al. Clinical significance and the correlation of expression between Let-7 and K-ras in non-small cell lung cancer. Oncol Lett. 2010;1(6):1045–47. doi: 10.3892/ol.2010.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yan L, Zhou J, Gao Y, et al. Regulation of tumor cell migration and invasion by the H19/let-7 axis is antagonized by metformin-induced DNA methylation. Oncogene. 2015;34(23):3076–84. doi: 10.1038/onc.2014.236. [DOI] [PubMed] [Google Scholar]
- 27.Li T, Chen JX, Fu XP, et al. microRNA expression profiling of nasopharyngeal carcinoma. Oncol Rep. 2011;25(5):1353–63. doi: 10.3892/or.2011.1204. [DOI] [PubMed] [Google Scholar]
- 28.Wong TS, Man OY, Tsang CM, et al. MicroRNA let-7 suppresses nasopharyngeal carcinoma cells proliferation through downregulating c-Myc expression. J Cancer Res Clin Oncol. 2011;137(3):415–22. doi: 10.1007/s00432-010-0898-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Frost RJ, Olson EN. Control of glucose homeostasis and insulin sensitivity by the Let-7 family of microRNAs. Proc Natl Acad Sci USA. 2011;108(52):21075–80. doi: 10.1073/pnas.1118922109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shi W, Zhang Z, Yang B, et al. Overexpression of microRNA let-7 correlates with disease progression and poor prognosis in hepatocellular carcinoma. Medicine (Baltimore) 2017;96(32):e7764. doi: 10.1097/MD.0000000000007764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang BG, Jiang LY, Xu Q. A comprehensive evaluation for polymorphisms in let-7 family in cancer risk and prognosis: A system review and meta-analysis. Biosci Rep. 2018;38(4) doi: 10.1042/BSR20180273. pii: BSR20180273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dou R, Nishihara R, Cao Y, et al. MicroRNA let-7, T cells, and patient survival in colorectal cancer. Cancer Immunol Res. 2016;4(11):927–35. doi: 10.1158/2326-6066.CIR-16-0112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ke L, Xiang Y, Xia W, et al. A prognostic model predicts the risk of distant metastasis and death for patients with nasopharyngeal carcinoma based on pre-treatment interleukin 6 and clinical stage. Clin Immunol. 2016;164:45–51. doi: 10.1016/j.clim.2016.01.004. [DOI] [PubMed] [Google Scholar]
- 34.Chen KJ, Hou Y, Wang K, et al. Reexpression of Let-7g microRNA inhibits the proliferation and migration via K-Ras/HMGA2/snail axis in hepatocellular carcinoma. Biomed Res Int. 2014;2014 doi: 10.1155/2014/742417. 742417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lin Y, Jiang G, Liu L, et al. Management of thoracic esophageal perforation. World J Surg. 2014;38(5):1093–99. doi: 10.1007/s00268-013-2371-4. [DOI] [PubMed] [Google Scholar]
- 36.Gao L, Wang X, Wang X, et al. IGF-1R, a target of let-7b, mediates crosstalk between IRS-2/Akt and MAPK pathways to promote proliferation of oral squamous cell carcinoma. Oncotarget. 2014;5(9):2562–74. doi: 10.18632/oncotarget.1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Buck E, Gokhale PC, Koujak S, et al. Compensatory insulin receptor (IR) activation on inhibition of insulin-like growth factor-1 receptor (IGF-1R): Rationale for cotargeting IGF-1R and IR in cancer. Mol Cancer Ther. 2010;9(10):2652–64. doi: 10.1158/1535-7163.MCT-10-0318. [DOI] [PubMed] [Google Scholar]
- 38.Zuo Q, Luo RC. [Relationship between the insulin-like growth factor 1 receptor signaling pathway and the resistance of nasopharyngeal carcinoma to cetuximab]. Zhonghua Zhong Liu Za Zhi. 2010;32(8):575–79. [PubMed] [Google Scholar]
- 39.Yuan YL, Zhou XH, Song J, et al. Dual silencing of type 1 insulin-like growth factor and epidermal growth factor receptors to induce apoptosis of nasopharyngeal cancer cells. J Laryngol Otol. 2008;122(9):952–60. doi: 10.1017/S0022215107000606. [DOI] [PubMed] [Google Scholar]
- 40.Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119(6):1420–28. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang N, Hui L, Wang Y, et al. Overexpression of SOX2 promotes migration, invasion, and epithelial-mesenchymal transition through the Wnt/beta-catenin pathway in laryngeal cancer Hep-2 cells. Tumour Biol. 2014;35(8):7965–73. doi: 10.1007/s13277-014-2045-3. [DOI] [PubMed] [Google Scholar]
- 42.Li X, Zhou Q, Tao L, Yu C. MicroRNA-106a promotes cell migration and invasion by targeting tissue inhibitor of matrix metalloproteinase 2 in cervical cancer. Oncol Rep. 2017;38(3):1774–82. doi: 10.3892/or.2017.5832. [DOI] [PubMed] [Google Scholar]
- 43.Li H, Batth IS, Qu X, et al. IGF-IR signaling in epithelial to mesenchymal transition and targeting IGF-IR therapy: Overview and new insights. Mol Cancer. 2017;16(1):6. doi: 10.1186/s12943-016-0576-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang R, Li H, Guo X, et al. IGF-I induces epithelial-to-mesenchymal transition via the IGF-IR-Src-microRNA-Ana-E-cadherin pathway in nasopharyngeal carcinoma cells. Oncol Res. 2016;24(4):225–31. doi: 10.3727/096504016X14648701447931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hou C, Zhu M, Sun M, Lin Y. MicroRNA let-mi induced autophagy to protect T cell from apoptosis by targeting INFAR. Biochem Biophys Res Commun. 2014;453(4):728–34. doi: 10.1016/j.bbrc.2014.10.002. [DOI] [PubMed] [Google Scholar]
- 46.Fawzy IO, Hamza MT, Hosny KA, et al. Abrogating the interplay between IGF2BP1, 2 and 3 and IGF1R by let-7i arrests hepatocellular carcinoma growth. Growth Factors. 2016;34(1–2):42–50. doi: 10.3109/08977194.2016.1169532. [DOI] [PubMed] [Google Scholar]
- 47.Zhang XH, Qian Y, Li Z, et al. Let-7g-5p inhibits epithelial-mesenchymal transition consistent with reduction of glioma stem cell phenotypes by targeting VSIG4 in glioblastoma. Oncol Rep. 2016;36(5):2967–75. doi: 10.3892/or.2016.5098. [DOI] [PubMed] [Google Scholar]