Abstract
Ovarian cancer is the most deadly malignant tumor. MicroRNA-27a-3p (miR-27a-3p) was a tumor oncogene in various cancers. However, the role and mechanism of miR-27a-3p in ovarian cancer are still unknown. In this study, we found that miR-27a-3p over-expression could significantly promote the viability of SK-OV-3 cells, enhance cell migration and invasion, and reduce cell apoptosis. Besides, results from western blot assay showed that miR-27a-3p over-expression could increase Bcl-2 protein expression and decrease Bax protein expression. Furthermore, TargetScan and the dual luciferase reporter gene assay revealed that BTG anti-proliferation factor 1 (BTG1) was a direct target of miR-27a-3p. In addition, we found that miR-27a-3p down-regulation suppressed SK-OV-3 cell viability, migration and invasion, and promoted cell apoptosis. All the effects of miR-27a-3p down-regulation on SK-OV-3 cells were reversed by BTG1-siRNA. Therefore, miR-27a-3p/BTG1 axis may be a new potential target for the treatment of ovarian cancer.
Keywords: miR-27a-3p, BTG1, Ovarian cancer, Biological behaviors
1. Introduction
Ovarian cancer (OC) is the fifth most common malignancy among women all over the world [1]. Furthermore, OC is the deadliest gynaecological cancer with the highest incidence of cancer mortality among all women [2]. In the early stage of OC, its clinical symptoms are not obvious and it is easy to cause missed diagnosis [3, 4]. Thus, most OC patients are already in advanced stage at the time of diagnosis, which leads to poor prognosis [5, 6, 7]. Therefore, it is necessary to explore the molecular mechanisms of ovarian cancer and to further search for new therapeutic strategies.
MicroRNAs (miRNAs) are highly conservative and short non-coding RNAs with a length of 2024 nucleotides, which can post-transcriptionally regulate gene expression [8, 9, 10, 11]. MiRNAs can mediate degradation or inhibit translation of their targets by directly binding to the 3’untranslated region (3’UTR) of their target mRNA [12, 13]. A growing number of studies have shown that abnormal expression and function of miRNAs play an important role in the pathogenesis of malignant diseases [14, 15]. Currently, miRNAs have also been found to participate in the development and progression of OC [16, 17]. MiRNA-27a (miR-27a) is a crucial regulator of cancer development, including laryngeal squamous cell carcinoma [18]. MiR-27a-3p is a member of the miR-27a family, and it could inhibit inflammatory response to spinal cord injury by decreasing the expression of toll-like receptor 4 (TLR4) [19]. MiR-27a-3p has also been studied in several cancers including nasopharyngeal carcinoma [20], hepatocellular carcinoma [21], and oral squamous carcinoma [22]. Qiao et al reported that miR-27a-3p could promote epitheli-al-mesenchymal transition in oral squamous carcinoma stem cells by targeting secreted frizzled-related protein 1 (SFRP1) [23]. However, the role and function of miR-27a-3p in ovarian cancer remains unclear. Therefore, our main purpose was to reveal the role of miR-27a-3p in ovarian cancer and to explore the molecular mechanism.
2. Materials and methods
2.1. Cell culture and cell transfection
SK-OV-3 cell line was obtained from American Type Culture Collection (ATCC, Manassas, VA) and cultured in RPMI1640 medium (Invitrogen, CA, USA) supplemented with 10 % fetal bovine serum (FBS) (Gibco, Carlsbad, CA, USA) at 37°C with 5% CO2. SK-OV-3 cells were transfected with miR-27a-3p mimic (5’-UUCACAGUGGCUAA-GUUCCGC-3’, Guangzhou RiboBio Co., Ltd.), mimic control (5’-UAUCAGACGGCUAAGUUCCGC-3’, Guangzhou RiboBio Co., Ltd.), inhibitor control (Catalog No.: CmiR-AN0001-AM02, Catalog No.: GeneCopoeia Inc.), miR-27a-3p inhibitor (Catalog No.: HmiR-AN0359-AM02, Gene-Copoeia Inc.), control-siRNA, BTG1-siRNA or miR-27a-3p inhibitor+BTG1-siRNA for 48 h using lipofectamine 2000 reagent (Invitrogen; Thermo Fisher Scientific) in accordance with the manufacturer’s protocol.
2.2. qRTPCR analysis
Total RNA was extracted from ovarian cancer cells using Trizol reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer’s introduction. Complementary DNA (cDNA) was synthesized by the miScript Reverse Transcription kit (Qiagen GmbH, Hilden, Germany). And the reverse transcription was performed using the following parameters: 42°C, 60 min; 75°C, 5 min. Quantitative PCR was carried out using the Quanti-Fast SYBR Green PCR kit (Qiagen GmbH) and the reaction system is also 20 μl. Primer sequences for PCR were as follows:
U6, forward 5’-GCTTCGGCAGCACATATACTAAAAT-3’;
reverse 5’-CGCTTCACGAATTTGCGTGTCAT-3’;
GAPDH, forward 5’-CTTTGGTATCGTGGAAGGACTC-3’;
reverse 5’-GTAGAGGCAGGGATGATGTTCT-3’;
miR-27a-3p, forward 5’-TGCGGTTCACAGTGGCTAAG-3’;
reverse 5’-CTCAACTGGTGTCGTGGA-3’;
BTG1, forward 5′-CATCTCCAAGTTTCTCCGCACC-3′;
reverse 5′-GCGAATACAACGGTAACCCGATC-3′. The qPCR transcription was performed using the following parameters: 95°C for 3 min, 40 cycles of 95°C for 30 s, 56°C for 30 s, and 72°C for 30 s. GAPDH was used as an internal control for BTG1 mRNA expression normalization, and U6 was used as an internal control to normalize miR-27a-3p levels. Relative gene expression was calculated using the 2-ΔΔCt method [24].
2.3. Western blot assay
The cells were lysated by RIPA lysis buffer (Beyotime Institute of Biotechnology) with protease inhibitor PMSF. We used BCA Protein Assay Kit (ThermoFisher, Waltham, MA) to measure protein concentration. 20 μg of protein was subjected to SDS-PAGE electrophoresis and transferred to PVDF membranes. Then, the membranes were blocked with 5% skim milk/PBST. After 1 h, the membranes were probed with primary antibodies: Bcl-2 (1:1,000; cat. no. 4223; Cell Signaling Technology Inc., Danvers, MA, USA), Bax (1:1,000; cat. no. 5023; Cell Signaling Technology Inc.), BTG1 (1:1,000; cat. no. ab151740; Abcam, USA),w and β-actin (1:1,000; cat. no. 4970; Cell Signaling Technology Inc.), at 4°C overnight. The next day, the membranes were washed three times with PBST buffer and then incubated with horseradish peroxidase-conjugated anti-rabbit IgG secondary antibodies (1:2,000; cat. no. 7074; Cell Signaling Technology Inc.). The protein band was visualized by enhanced chemiluminescence method (ECL, Millipore, Billerica, MA). β-actin served as loading control for normalization.
2.4. Dual-luciferase reporter assay
TargetScan program (www.targetscan.org/vert_71) was used to predict the targets of miR-27a-3p. The wild type (WTBTG1) and mutant (MUTBTG1) 3′UTRs of BTG1 were cloned into a pmiRRBReportTM dual luciferase reporter gene plasmid vector (Guangzhou RiboBio Co., Ltd., Guangzhou, China). Briefly, miR-27a-3p mimic, mimic control, and luciferase reporter plasmids containing the wild-type or mutant 3’-UTR of BTG1 were co-transfected into SK-OV-3 cells in 24-well plate for 48 h using lipofectamine 2000 (Invitrogen). Relative luciferase activity was detected using the dualluciferase assay system (Promega Corporation, Madison, WI, USA) by micro-plate reader (Molecular Devices, Sunnyvale, CA). Renilla luciferase was used for normalization. Each sample was performed three times.
2.5. Cell viability assay
MTT assay was used to detect cell viability. SK-OV-3 cells (1×104) were seeded into a 96-well plate and incubated overnight in the previously described conditions. After 48 h, 20 μl MTT reagent (Beyotime Institute of Biotechnology) was added to each well and the plates were incubated at 37°C for 4 h. Subsequently, DMSO was added to each well to dissolve the formazan, and the absorbance was read at 490 nm using an automatic multi-hole spectrophotometer.
2.6. Flow cytometry assay
We used specific Annexin V-fluorescein isothiocyanate (FITC) Apoptosis Detection Kit I (BD Bioscience, San Diego, CA, USA) to detect cell apoptosis according to the manufacturer’s introduction. In brief, cells were washed twice with 1×PBS. Then, cells were collected, centrifuged with low temperature at high speed, and re-suspended in 100 μl of FITC-binding buffer. Subsequently, the buffer was added with approximately 5 μl ready-to-use Annexin V-FITC (BD Bioscience) and 5 μl PI. In the dark, cells were incubated for 30 min at room temperature. Annexin V-FITC and PI fluorescence were assessed by BD FACSCalibur flow cytometer (BD Technologies).
2.7. Transwell assay
For the migration assay, un-coated transwell chambers (pore size, 8 μm; Costar; Corning Inc., Corning, NY, USA) were utilized in the present study. And 1x105 SK-OV-3 cells were re-suspended in 200 μl serum-free medium and seeded in the upper chamber. 500 μl RPMI-1640 medium containing 20 % FBS was added to the lower chamber. The 24-well plates were plated at 37°C in a 5% CO2 incubator for 48 h. Then, the cells were fixed with 4% polyoxymethylene and stained with 0.1% crystals for 20 minutes at room temperature. The randomly selected area was photoed with an optical microscope to calculate the number of migrated cells. For the invasion assay, the upper chamber was coated with Matrigel.
2.8. Statistical analysis
All data were displayed from three independent experiments in triplicate. We used SPSS 19.0 by Student’s t-test or one-way ANOVA for biostatistical analysis. The data were shown as the mean ± standard deviation (SD), and p<0.05 was considered as significant.
3. Results
3.1. Effect of miR-27a-3p mimic on biological behavior of SK-OV-3 cells
In order to explore the role of miR-27a-3p in ovarian cancer, we first studied the effect of miR-27a-3p mimic on cell viability, apoptosis, migration, and invasion of ovarian cancer SK-OV-3 cells. MiR-27a-3p mimic or mimic control were transfected into SK-OV-3 cells using Lipofectamine 2000 for 48 h. qPCR assay indicated that miR-27a-3p mimic significantly increased the expression of miR-27a-3p in SK-OV-3 cells (Figure 1A) Cell viability was measured by using MTT assay. The result showed that miR-27a-3p mimic could promote the viability of SK-OV-3 cells (Figure 1B) At the same time, FCM assay showed that miR-27a-3p mimic inhibited SK-OV-3 cell apoptosis (Figure 1C) In addition, western blot assay showed that miR-27a-3p mimic increased Bcl-2 protein expression and decreased Bax protein expression (Figure 1D) To determine the effect of miR-27a-3p on OC cell metastasis, we performed transwell assay to detect SK-OV-3 cell migration and invasion. These results demonstrated that miR-27a-3p mimic promoted SK-OV-3 cell migration and invasion (Figure 1E and 1F).
Figure 1.
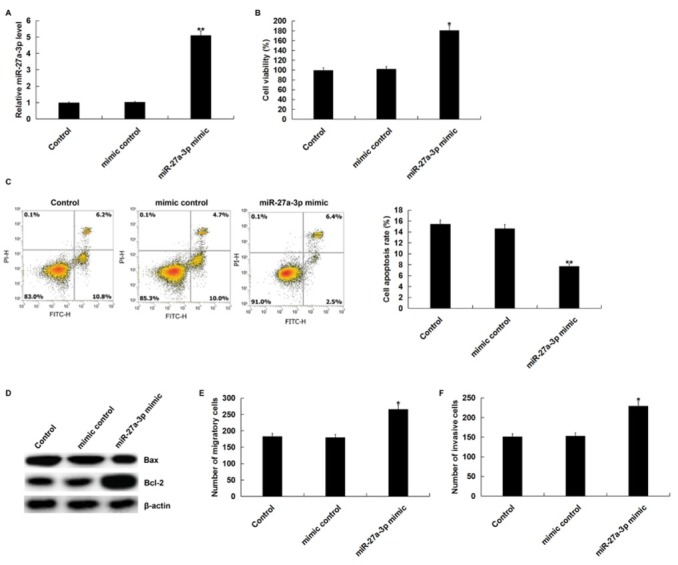
Effect of miR-27a-3p mimic on proliferation, apoptosis, migration, and invasion of SK-OV-3 cells.
(A) The expression level of miR-27a-3p was detected by using qRT-PCR after SK-OV-3 cells were transfected with miR-27a-3p mimic or mimic control using Lipofectamine 2000 for 48 h. (B) MTT assay was performed to detect cell viability. (C) Cell apoptosis was measured by flow cytometry. (D) The protein expression of Bcl-2 and Bax was detected bu using western blot assay. Cell migration (E) and invasion (F) were determined using the Transwell assay. Control: SK-OV-3 cells without any treatment; mimic control: SK-OV-3 cells transfected with mimic control; miR-27a-3p mimic: SK-OV-3 cells transfected with miR-27a-3p mimic. The data were shown as the mean ± SD. *, **p<0.05, 0.01 vs. Control.
3.2. BTG1 was a direct target of miR-27a-3p
To determine the specific mechanism of miR-27a-3p in ovarian cancer, we used the miRNA target prediction tool TargetScan to carry out bioinformation analysis to identify the potential targets of miR-27a-3p. It was found that MiR-27a-3p has hundreds of potential target genes, including BTG1 (Figure 2A) BTG1 belongs to the BTG/Tob anti-proliferation gene family [25, 26]. BTG1 plays an important role in the regulation of various biological and cellular processes including cell cycle, cell proliferation, and cell apoptosis [26, 27]. Besides, study has revealed that BTG1 was down-regulated in OC [28]. However till now, the relationship between miR-27a-3p and BTG1 in OC remains unknown. Therefore, we choose BTG1 for further study, and we speculated that BTG1 was the direct target gene of miR-27a-3p. To further verify the binding sites between BTG1 and miR-27a-3p, a luciferase reporter assay was performed. The results indicated that the relative luciferase activity was significantly reduced in cells co-transfected with wild-type BTG1 3’UTR and miR-27a-3p mimic. However, when miR-27a-3p mimic was co-transfected with mutant BTG1 3’UTR in SK-OV-3 cells, luciferase activity was essentially unchanged (Figure 2B) Taken together, these results suggested that BTG1 was a direct target gene of miR-27a-3p.
Figure 2.
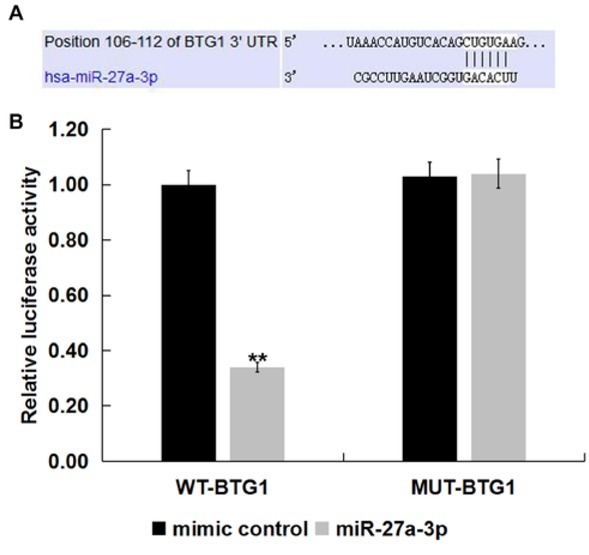
BTG1 was target gene of miR-27a-3p.
TargetScan was used to predict the putative target gene of miR-27a-3p. Predicted binding sites of miR-27a-3p and 3’-UTR of BTG1. (B) Dual-luciferase reporter assays were performed when SK-OV-3 cells were co-transfected with mimic control or miR-27a-3p mimic and a pGL3 construct containing the WT or MUT 3’-UTR of BTG1 for 48 h. The data were shown as the mean ± SD. **p<0.01 vs. mimic control.
We then investigated the effect of miR-27a-3p on BTG1 expression in SK-OV-3 cells. SK-OV-3 cells were transfected with inhibitor control, miR-27a-3p inhibitor, control-siRNA, BTG1-siRNA, or miR-27a-3p inhibitor+BTG1-siRNA. After 48 h, we detected transfection efficiency by qRT-PCR and/or western blot assay. qRT-PCR assay indicated that miR-27a-3p inhibitor significantly reduced the expression of miR-27a-3p in SK-OV-3 cells (Figure 3A) qRT-PCR and western blot assay showed that BTG1-siRNA significantly reduced the mRNA and protein expression of BTG1 in SK-OV-3 cells (Figure 3B and C) We also found that miR-27a-3p inhibitor significantly increased mRNA and protein BTG1 expression, which was reversed by BTG1-siRNA (Figure 3D and E)
Figure 3.
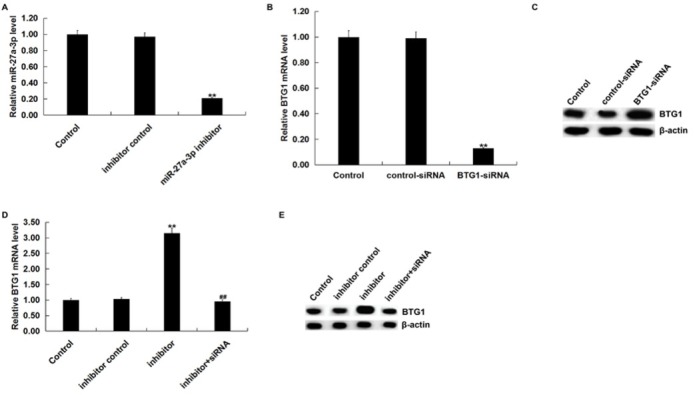
BTG1 was negatively regulated by miR-27a-3p
(A) The expression of miR-27a-3p was detected using qRT-PCR after SK-OV-3 cells were transfected with inhibitor control or miR-27a-3p inhibitor for 48 h. qPCR assay (B) and western blot assay (C) detected the expression of BTG1 after SK-OV-3 cells were transfected with control-siRNA or BTG1-siRNA for 48 h. qPCR assay (D) and western blot assay (F) that the expression of BTG1 after SK-OV-3 cells were transfected with inhibitor control, miR-27a-3p inhibitor or miR-27a-3p inhibitor+BTG1-siRNA for 48 h. Control: SK-OV-3 cells without any treatment; inhibitor control: SK-OV-3 cells transfected with inhibitor control; inhibitor/miR-27a-3p inhibitor: SK-OV-3 cells transfected with miR-27a-3p inhibitor; control-siRNA: SK-OV-3 cells transfected with control-siRNA; BTG1-siRNA: SK-OV-3 cells transfected with BTG1-siRNA; inhibitor+siRNA: SK-OV-3 cells transfected with miR-27a-3p inhibitor+BTG1-siRNA. The data were shown as the mean ± SD. **p<0.01 vs. Control. ##p<0.01 vs. inhibitor.
3.3. Effect of miR-27a-3p inhibitor on biological behavior of SK-OV-3 cells
To further determine the role of miR-27a-3p in SK-OV-3 cells, SK-OV-3 cells were transfected with inhibitor control, miR-27a-3p inhibitor, or miR-27a-3p inhibitor+BTG1-siRNA for 48 h. MTT assay showed that miR-27a-3p inhibitor significantly inhibited the viability of SK-OV-3 cells (Figure 4A) In addition, we performed flow cytometry to measure cell apoptosis. MiR-27a-3p inhibitor induced SK-OV-3 cell apoptosis (Figure 4B) Besides, miR-27a-3p inhibitor decreased Bcl-2 protein expression, and increased Bax protein expression (Figure 4C) Furthermore, transwell assay indicated that miR-27a-3p inhibitor inhibited cell migration and invasion (Figure 4D and E) Importantly, all of these changes were reversed by BTG1-siRNA. Taken together, miR-27a-3p inhibitor inhibited OC cell malignant biological behaviors by targeting BTG1.
Figure 4.
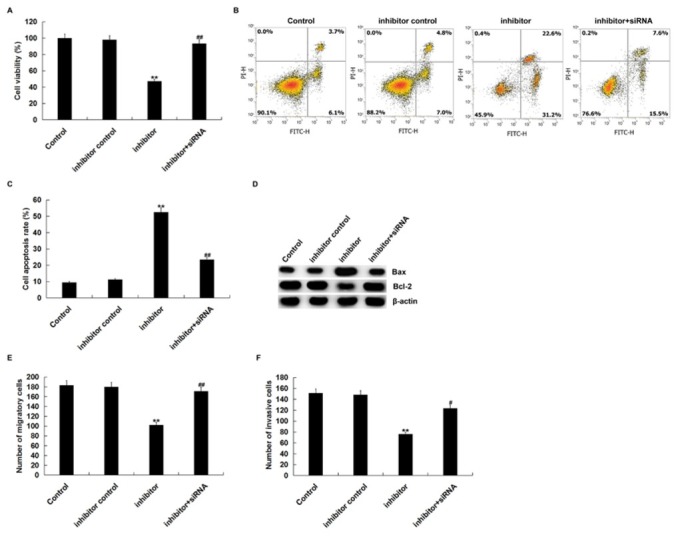
miR-27a-3p inhibitor suppressed cell proliferation, migration and invasion and induced apoptosis.
(A) MTT assay was used to detect cell viability when SK-OV-3 cells were transfected with inhibitor control, miR-27a-3p inhibitor or miR-27a-3p inhibitor+BTG1-siRNA for 48 h. (B and C) Cell apoptosis was measured by flow cytometry. (D) Western blot assay that the protein expression level of Bcl-2 and Bax. The migration (E) and invasion ability (F) was detected by using transwell assay. Control: SK-OV-3 cells without any treatment; inhibitor control: SK-OV-3 cells transfected with inhibitor control; inhibitor: SK-OV-3 cells transfected with miR-27a-3p inhibitor; inhibitor+siRNA: SK-OV-3 cells transfected with miR-27a-3p inhibitor+BTG1-siRNA.The data were shown as the mean ± SD. **p<0.01 vs. Control. #, ##p<0.05, 0.01 vs. inhibitor.
4. Discussion
It has been reported that miRNAs can serve as either oncogenes or tumour suppressors by directly or indirectly modulating cancer genes expression. In our study, we found that miR-27a-3p mimic promoted SK-OV-3 cell viability and decreased SK-OV-3 cell apoptosis. In addition, western blot assay indicated that miR-27a-3p mimic increased Bcl-2 protein expression, and decreased Bax protein expression. Transwell assay showed that miR-27a-3p mimic promoted cell migration and invasion. Furthermore, it was confirmed that BTG1 was target of miR-27a-3p. To further determine the role of miR-27a-3p, SK-OV-3 cells were transfected with miR-27a-3p inhibitor. Results indicated that miR-27a-3p inhibitor suppressed SK-OV-3 cell viability and induced cell apoptosis. In addition, miR-27a-3p inhibitor inhibited cell migration and invasion. These changes caused by miR-27a-3p inhibitor were reversed by BTG1-siRNA.
Ovarian cancer is one of the most deadly gynecologic malignancies and is usually diagnosed at advanced stages. At present, the combination of surgery and chemotherapy has improved the treatment of ovarian cancer, but the successful rate of complete cure is only 30% [29]. In addition, the mechanism of occurrence and development of ovarian cancer is still largely unclear, so we urgently need to find new ways to treat ovarian cancer. Currently, many scholars have demonstrated that miRNA is an important regulator in various types of cancers [30]. Furthermore, it has been reported that miRNA was involved in many biological processes such as cell proliferation, differentiation, metabolism, and apoptosis [31]. MiRNA has also been found to be abnormally expressed in many cancers such as gastric cancer, liver cancer, pancreatic cancer, and colorectal cancer [32, 33, 34, 35]. For instance, miR4903p over-expression inhibited cell proliferation, migration, and invasion of cancer cells by directly targeting CDK1 [36]. MiR-125b suppressed cell proliferation of ovarian cancer cells [37]. MiR-200a promoted ovarian cancer cell invasion and migration by targeting PTEN [30]. However, the role of miR-27a-3p in ovarian cancer remains unknown.
Previous research have shown that miR-27a-3p is highly expressed in peripheral blood mononuclear cells of pancreatic cancer patients [38]. MiR-27a-3p promoted glioma cell proliferation by targeting MXI1 [39]. In addition, Wu et al suggested that miR-27a-3p could promote ESCC cell proliferation [40]. The research of Tang et al has shown that miR-27a up-regulation leads to poor prognosis in breast cancer patients, which indicated that miR-27a can be used as a prognostic indicator for breast cancer progression and patient survival [41]. In our study, we found that miR-27a-3p mimic promoted cell proliferation, migration and invasion, and inhibited cell apoptosis. However, miR-27a-3p inhibitor suppressed cell proliferation, migration and invasion, and induced cell apoptosis.
B cell translocation gene 1 (BTG1), a tumor suppressor gene, is a member of BTG/Tob anti-proliferative protein family [25, 42, 43]. In addition, BTG1 is an important factor that can affect cell proliferation, differentiation, apoptosis, angiogenesis, and survival [44]. BTG1 was also involved in the development of several diseases, including breast cancer, multiple sclerosis, ovarian cancer, and prostate cancer [45, 46, 47, 48, 49]. Furthermore, it has been reported that BTG1 was a target gene of miR-511 [44]. In our study, we found that BTG1 was a target of miR-27a-3p. MiR-27a-3p inhibitor induced changes of biological behaviors of ovarian cancer cells were reversed by BTG1-siRNA. Taken together, these findings provided new insights into the roles of miR-27a-3p in ovarian cancer.
In summary, miR-27a-3p, an oncogene, was involved in the development and progression of ovarian cancer by targeting BTG1. Furthermore, miR-27a-3p regulated the multiple malignant biological behavior of ovarian cancer cells. However, this is only a preliminary study of the role of miR-27a-3p in OC. In order to make the role of miR-27a-3p in OC more convincing, a lot of experimental research is needed. For example, the role of miR-27a-3p in other OC cell lines should be explored. The expression of miR-27a-3p in OC patients and cell lines should be revealed. Besides, the relationship between the expression of miR-27a-3p and the clinical features of OC patients requires further research. Moreover, the role of miR-27a-3p in OC should be investigated in vivo. In the future, we will study these topics.
Footnotes
Disclosures All authors declare no financial competing interests.
References
- [1].Jemal A., Siegel R., Xu J., Ward E.. Cancer statistics. CA. Cancer. J. Clin. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- [2].Siegel R., Ma J., Zou Z., Jemal A.. Cancer statistics, 2014. CA. Cancer. J. Clin. 2014;64:9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- [3].Banerjee S., Kaye S.B.. New strategies in the treatment of ovarian cancer: Current clinical perspectives and future potential. Clin. Cancer. Res. 2013;19:961–968. doi: 10.1158/1078-0432.CCR-12-2243. [DOI] [PubMed] [Google Scholar]
- [4].Mezzanzanica D.. Ovarian cancer: a molecularly insidious disease. Chin. J. Cancer. 2015;34:1–3. doi: 10.5732/cjc.014.10301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Engel J., Eckel R., Schubert-Fritschle G., Kerr J., Kuhn W., Diebold J., Kimmig R.. Moderate progress for ovarian cancer in the last 20 years: prolongation of survival, but no improvement in the cure rate, European Journal of Cancer 38:2440-2445,2000. Eur. J. Cancer. 2002;38:2435–2445. doi: 10.1016/s0959-8049(02)00495-1. [DOI] [PubMed] [Google Scholar]
- [6].Averette H.E., Janicek M.F., Menck H.R.. The national Cancer data base report on ovarian cancer. American College of Surgeons Commission on Cancer and the American Cancer Society, Cancer. 1995;76:1096–1103. doi: 10.1002/1097-0142(19950915)76:6<1096::aid-cncr2820760626>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- [7].Hennessy B.T., Coleman R.L., Markman M.. Ovarian cancer. Lancet. 2009;374:1371–1382. doi: 10.1016/S0140-6736(09)61338-6. [DOI] [PubMed] [Google Scholar]
- [8].Poy M.N., Eliasson L., Krutzfeldt J., Kuwajima S., Ma X., Macdonald P.E.. A pancreatic islet-specifc microRNA regulates insulin secretion. Nature. 2004;432:226–230. doi: 10.1038/nature03076. [DOI] [PubMed] [Google Scholar]
- [9].Wang Q., Liu N., Yang X., Tu L., Zhang X.. Small RNA-mediated responses to low- and high-temperature stresses in cotton. Sci. Rep. 2016;6:35558–35571. doi: 10.1038/srep35558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Lim L.P., Lau N.C., Garrett-Engele P., Grimson A., Schelter J.M., Castle J.. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- [11].Bartel D.P.. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- [12].Wang Y., Chen F., Zhao M., Yang Z., Zhang S., Ye L., Gao H.. MiR-107 suppresses proliferation of hepatoma cells through targeting HMGA2 mRNA 3′UTR. Biochem Biophys Res Commun. 2016;480:455–460. doi: 10.1016/j.bbrc.2016.10.070. [DOI] [PubMed] [Google Scholar]
- [13].Zhu Q., Gong L., Wang J., Tu Q., Yao L., Zhang J.R.. miR-10b exerts oncogenic activity in human hepatocellular carcinoma cells by targeting expression of CUB and sushi multiple domains 1 (CSMD1) Bmc Cancer. 2016;16 doi: 10.1186/s12885-016-2801-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Calin G.A., Croce C.M.. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- [15].Lu J., Getz G., Miska EA., Alvarez-Saavedra E., Lamb J., Peck D.. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- [16].Gun Oh C., Hyo-Sung J., Hyung Soo H., Woong S.J., Yoon Hee L., Dae Gy H.. Differential MicroRNA Expression Profiles in Primary and Recurrent Epithelial Ovarian Cancer. Anticancer Res. 2015;35:2611–2617. [PubMed] [Google Scholar]
- [17].Llaurado M., Majem B., Altadill T., Lanau L., Castellvi J., Sanchez-Iglesias J.L.. MicroRNAs as prognostic markers in ovarian cancer. Mol Cell Endocrinol. 2014;390:73–84. doi: 10.1016/j.mce.2014.03.006. [DOI] [PubMed] [Google Scholar]
- [18].Tian Y., Fu S., Qiu G.B., Xu Z.M., Liu N., Zhang X.W.. MicroRNA-27a promotes proliferation and suppresses apoptosis by targeting PLK2 in laryngeal carcinoma. Bmc Cancer. 2014;14 doi: 10.1186/1471-2407-14-678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Zhang P., Li L.Q., Zhang D., Shen Y.. Over-expressed miR-27a-3p inhibits inflammatory response to spinal cord injury by decreasing TLR4. Eur Rev Med Pharmaco. 2018;22:5416–5423. doi: 10.26355/eurrev_201809_15800. [DOI] [PubMed] [Google Scholar]
- [20].Li L.H., Luo Z.H.. Dysregulated miR-27a-3p promotes nasopharyngeal carcinoma cell proliferation and migration by targeting Mapk10. Oncol Rep. 2017;37:2679–2687. doi: 10.3892/or.2017.5544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Guo D., Li Y., Chen Y., Zhang D., Wang X., Lu G. DANCR promotes HCC progression and regulates EMT by sponging miR-27a-3p via ROCK1/LIMK1/COFILIN1 pathway. Cell Prolif. 2019. p. e12628. [DOI] [PMC free article] [PubMed]
- [22].Zeng G., Xun W., Wei K., Yang Y., Shen H.J.O.R.. MicroRNA-27a-3p regulates epithelial to mesenchymal transition via targeting YAP1 in oral squamous cell carcinoma cells. Oncol Rep. 2016;36 doi: 10.3892/or.2016.4916. [DOI] [PubMed] [Google Scholar]
- [23].Qiao B., He B.X., Cai J.H., Tao Q., Lam A.K.J.S.R.. MicroRNA-27a-3p Modulates the Wnt/β-Catenin Signaling Pathway to Promote Epithelial-Mesenchymal Transition in Oral Squamous Carcinoma Stem Cells by Targeting SFRP1. Sci Rep. 2017;7:44688. doi: 10.1038/srep44688. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [24].Livak K.J., Schmittgen T.D.J.M. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2 (-Delta Delta C(T)) Method. Methods. 2001. [DOI] [PubMed]
- [25].Winkler G.S.. The mammalian anti-proliferative BTG/ Tob protein family. J Cell Physiol. 2010;222:66–72. doi: 10.1002/jcp.21919. [DOI] [PubMed] [Google Scholar]
- [26].Rouault J.P., Rimokh R., Tessa C., Paranhos G., Ffrench M., Duret L.. BTG1, a member of a new family of antiproliferative genes. EMBO J. 1992;11:1663–70. doi: 10.1002/j.1460-2075.1992.tb05213.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Prevot D., Voeltzel T., Birot A.M., Morel A.P., Rostan M. C., Magaud J. P.. The leukemia-associated protein Btg1 and the p53-regulated protein Btg2 interact with the homeoprotein Hoxb9 and enhance its transcriptional activation. J Biol Chem. 2000;275:147–53. doi: 10.1074/jbc.275.1.147. [DOI] [PubMed] [Google Scholar]
- [28].Kim J.Y., Do S.I., Bae G.E., Kim H.S.. B-cell translocation gene 1 is downregulated by promoter methylation in ovarian carcinoma. J Cancer. 2017;8:2669–2675. doi: 10.7150/jca.21037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Zhu C.L., Gao G.S.. miR-200a Overexpression in Advanced Ovarian Carcinomas as a Prognostic Indicator. Asian Pac J Cancer P. 2014;15:8595–8601. doi: 10.7314/APJCP.2014.15.20.8595. [DOI] [PubMed] [Google Scholar]
- [30].Suo H.B., Zhang K.C., Zhao J.. MiR-200a promotes cell invasion and migration of ovarian carcinoma by targeting PTEN. Eur Rev Med Pharmaco. 2018;22:4080–4089. doi: 10.26355/eurrev_201807_15398. [DOI] [PubMed] [Google Scholar]
- [31].Zisoulis D.G., Kai Z.S., Chang R.K., Pasquinelli A.E.. Autoregulation of microRNA biogenesis by let-7 and Argonaute. Nature. 2012;486:541–U140. doi: 10.1038/nature11134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Liu Y.Q., Liu R., Yang F., Cheng R.J., Chen X.R., Cui S.F.. miR-19a promotes colorectal cancer proliferation and migration by targeting TIA1. Mol Cancer. 2017;16 doi: 10.1186/s12943-017-0625-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Han T.S., Hur K., Xu G.R., Choi B., Okugawa Y., Toiyama Y.. MicroRNA-29c mediates initiation of gastric carcinogenesis by directly targeting ITGB1. Gut. 2015;64:203–U30. doi: 10.1136/gutjnl-2013-306640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Chang R.M., Yang H., Fang F., Xu J.F., Yang L.Y.. MicroRNA-331-3p Promotes Proliferation and Metastasis of Hepatocellular Carcinoma by Targeting PH Domain and Leucine-Rich Repeat Protein Phosphatase. Hepatology. 2014;60:1251–1263. doi: 10.1002/hep.27221. [DOI] [PubMed] [Google Scholar]
- [35].Xiao J.J., Lv D.C., Zhou J.Z., Bei Y.H., Chen T., Hu M.R.;. Therapeutic Inhibition of miR-4260 Suppresses Colorectal Cancer via Targeting MCC and SMAD4. Theranostics. 2017;7:1901–1913. doi: 10.7150/thno.19168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Chen S., Chen X., Xiu Y.L., Sun K.X., Zhao Y.. MicroRNA-490-3P targets CDK1 and inhibits ovarian epithelial carcinoma tumorigenesis and progression. Cancer Lett. 2015;362:122–130. doi: 10.1016/j.canlet.2015.03.029. [DOI] [PubMed] [Google Scholar]
- [37].Guan Y., Yao H.L., Zheng Z.H., Qiu G.R., Sun K.L.. MiR-125b targets BCL3 and suppresses ovarian cancer proliferation. Int J Cancer. 2011;128:2274–2283. doi: 10.1002/ijc.25575. [DOI] [PubMed] [Google Scholar]
- [38].Wang W.S., Liu L.X., Li G.P., Chen Y., Li C.Y., Jin D.Y.. Combined Serum CA19-9 and miR-27a-3p in Peripheral Blood Mononuclear Cells to Diagnose Pancreatic Cancer. Cancer Prev Res. 2013;6:331–338. doi: 10.1158/1940-6207.CAPR-12-0307. [DOI] [PubMed] [Google Scholar]
- [39].Xu W.Y., Liu M.F., Peng X.L., Zhou P., Zhou J.W., Xu K.. miR-24-3p and miR-27a-3p promote cell proliferation in glioma cells via cooperative regulation of MXI1. Int J Oncol. 2013;42:757–766. doi: 10.3892/ijo.2012.1742. [DOI] [PubMed] [Google Scholar]
- [40].Wu X.Z., Wang K.P., Song H.J., Xia J.H., Jiang Y., Wang Y.L.. MiR-27a-3p promotes esophageal cancer cell proliferation via F-box and WD repeat domain-containing 7 (FBXW7) suppression. Int J Clin Exp Med. 2015;8:15556–15562. [PMC free article] [PubMed] [Google Scholar]
- [41].Tang W., Zhu J.J., Su S.C., Wu W., Liu Q., Su F.X.;. MiR-27 as a Prognostic Marker for Breast Cancer Progression and Patient Survival. Plos One. 2012;7 doi: 10.1371/journal.pone.0051702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Kanda M., Sugimoto H., Nomoto S., Oya H., Hibino S., Shimizu D.. B-cell translocation gene 1 serves as a novel prognostic indicator of hepatocellular carcinoma. Int J Oncol. 2015;46:641–648. doi: 10.3892/ijo.2014.2762. [DOI] [PubMed] [Google Scholar]
- [43].Xiao F., Deng J.L., Yu J.J., Guo Y.J., Chen S.H., Guo F.F.. A novel function of B-cell translocation gene 1 (BTG1) in the regulation of hepatic insulin sensitivity in mice via c-Jun. Faseb J. 2016;30:348–359. doi: 10.1096/fj.15-278689. [DOI] [PubMed] [Google Scholar]
- [44].Zhang S.Q., Yang Z., Cai X.L., Zhao M., Sun M.M. Li J.. miR-511 promotes the proliferation of human hepatoma cells by targeting the 3’UTR of B cell translocation gene 1 (BTG1) mRNA. Acta Pharmacol Sin. 2017;38:1161–1170. doi: 10.1038/aps.2017.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Camina-Tato M., Morcillo-Suarez C., Navarro A., Fernandez M., Horga A., Montalban X.. Genetic association between polymorphisms in the BTG1 gene and multiple sclerosis. J Neuroimmunol. 2009;213:142–147. doi: 10.1016/j.jneuroim.2009.05.010. [DOI] [PubMed] [Google Scholar]
- [46].Li W., Zou S.T., Zhu R., Wan J.M., Xu Y., Wu H.R.. Bcell translocation 1 gene inhibits cellular metastasisassociated behavior in breast cancer. Mol Med Rep. 2014;9:2374–80. doi: 10.3892/mmr.2014.2118. [DOI] [PubMed] [Google Scholar]
- [47].Wang Y., Shao C., Shi C.H., Zhang L., Yue H.H., Wang P.F.;. Change of the cell cycle after flutamide treatment in prostate cancer cells and its molecular mechanism. Asian J Androl. 2005;7:375–80. doi: 10.1111/j.1745-7262.2005.00031.x. [DOI] [PubMed] [Google Scholar]
- [48].Zhao Y., Gou W.F., Chen S., Takano Y., Xiu Y.L., Zheng H.C.. BTG1 expression correlates with the pathogenesis and progression of ovarian carcinomas. Int J Mol Sci. 2013;14:19670–80. doi: 10.3390/ijms141019670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Zhu R., Li W., Xu Y., Wan J.M., Zhang Z.L.. Upregulation of BTG1 enhances the radiation sensitivity of human breast cancer in vitro and in vivo. Oncol Rep. 2015;34:3017–3024. doi: 10.3892/or.2015.4311. [DOI] [PubMed] [Google Scholar]


