Abstract
The eukaryotic genomes are pervasively transcribed. In addition to protein-coding RNAs, thousands of long noncoding RNAs (lncRNAs) modulate key molecular and biological processes. Most lncRNAs are found in the nucleus and associate with chromatin, but lncRNAs can function in both nuclear and cytoplasmic compartments. Emerging work has found that many lncRNAs regulate gene expression and can affect genome stability and nuclear domain organization both in plant and in the animal kingdom. Here, we describe the major plant lncRNAs and how they act, with a focus on research in Arabidopsis thaliana and our emerging understanding of lncRNA functions in serving as molecular sponges and decoys, functioning in regulation of transcription and silencing, particularly in RNA-directed DNA methylation, and in epigenetic regulation of flowering time.
Keywords: Plant lncRNAs, Noncoding RNAs, Epigenetics, Exosome, FLC, Transcriptional regulation
5.1. Introduction
In eukaryotes, transcriptome studies showed that >90% of the genome is transcribed and a myriad of transcripts corresponds to noncoding RNAs (ncRNAs) [1, 2], including long ncRNAs (lncRNAs), which are classically >200 nt long and have no discernable coding potential [3–5]. Plant genomes produce tens of thousands of lncRNAs from intergenic, intronic, or coding regions. RNA Pol II transcribes most lncRNAs (from the sense or antisense strands); plants also have Pol IV and Pol V, the two plant-specific RNA polymerases that can produce lncRNAs [6, 7]. Majority of described up-to-date plant lncRNAs are polyadenylated, while in yeast and mammals, there are many non-polyadenylated lncRNAs as well [8]. However, there are several well-studied important functional non-polyadenylated lncRNAs [9–11]; and the recent work in Arabidopsis found that abiotic stress induced the production of hundreds of non-polyadenylated lncRNAs [12–14].
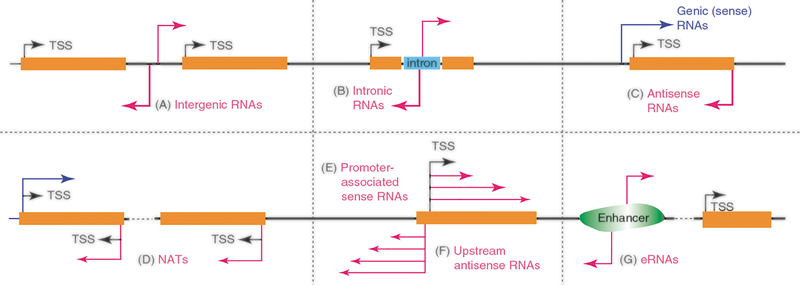
Most lncRNAs can be broadly classified based on their relationships to protein-coding genes: (1) long intergenic ncRNAs (lincRNAs) (Fig. 5.1A); (2) lncRNAs produced from introns (incRNAs), which can be transcribed in any orientation relative to coding genes (Fig. 5.1B); and (3) antisense RNAs and natural antisense transcripts (NATs), which are transcribed from the antisense strand of genes (Fig. 5.1C and D) [15]. Various types of lncRNAs are also transcribed near transcription start sites (TSSs) and transcription termination sites (TTSs) or from enhancer regions (eRNAs) (Fig. 5.1G) and splice sites. For example, yeast produces cryptic unstable transcripts (CUTs) and stable unannotated transcripts (SUTs) from around TSSs [16], Xrn1-sensitive XUTs [17], and Nrd1-dependent NUTs [18, 19], and mammalian cells produce PROMPTs and upstream antisense RNAs (uaRNAs) [20] and others (Fig. 5.1E and F).
Fig. 5.1.
Classification of lncRNAs based on their relationship to protein-coding genes. Orange boxes correspond to the protein-coding genes and pink lines correspond to lncRNAs. Arrows indicate the direction of transcription. Each panel depicts a subtype of lncRNAs: intergenic or long intergenic noncoding RNAs (lincRNAs) (A), intronic RNAs (B), antisense RNAs (C), natural anti-sense transcripts (NATs) (D), promoter-proximal sense (E) and upstream antisense RNAs (F), eRNAs (G)
Information about TSS-proximal lncRNAs in plants remains scant. However, recent analyses of nascent RNA from Arabidopsis seedlings obtained using a combination of global nuclear run-on sequencing (GRO-seq), 5′ GRO-seq, and RNA-seq did not detect upstream antisense TSS-proximal ncRNAs [21]. These data suggest a possibility that divergent transcription is lacking in Arabidopsis (and likely maize), in contrast to the situation in many other eukaryotes, indicating that eukaryotic promoters might be not inherently bidirectional. In Arabidopsis TSS-proximal lncRNAs that were observed in the RNA exosome-deficient lines include the upstream noncoding transcripts (UNTs), which are transcribed as sense RNAs and are colinear with the 5′ ends of the associated protein-coding gene, extending into the first intron. The UNTs resemble yeast CUTs and mammalian PROMPTs [1].
The exosome-sensitive enhancer RNAs (eRNAs) produced from enhancer regions make up a large proportion of non-polyadenylated lncRNAs in mammalian cells (Fig. 5.1G) [8]. However, information about plant enhancers has only recently started to emerge. An analysis of chromatin signatures predicted over 10,000 plant intergenic enhancers [22]. However, their potential roles as transcriptional enhancers in vivo will require follow-up experiments, and eRNAs have not yet been reported in plants.
5.2. Recent Advances in Studying Plant lncRNAs
Mammalian lncRNAs are by far the best-studied. However, in recent years, identification of plant lncRNAs has largely caught up with mammalian field. The plant databases where the information on lncRNAs can be found are summarized in Table 5.1.
Table 5.1.
List of plant lncRNA databases
| Database | Descriptions/features | Website | Ref |
|---|---|---|---|
| The Arabidopsis Information Resource (TAIR) | Comprehensive database of Arabidopsis thaliana genome, including annotated genome sequences (TAIR10), gene structures, and transcriptome data for coding and nonprotein-coding loci. TAIR has multiple analytical tools: interactive genome browser, BLAST, motif analysis, bulk data retrieval, and a chromosome map tool | https://www.arabidopsis.org/ | [23] |
| Araportll | A comprehensive database based on Arabidopsis Col-0 version 11 (Araport11) includes additional coding and noncoding annotations compared to TAIR10, such as lincRNAs, NATs, and other ncRNAs | https://www.araport.org/ | [24] |
| Plant long noncoding RNA database (PLncDB) | This database includes a curated list of >13,000 lincRNAs identified using RNA-seq and tiling array and their organ-specific expression and the differential expression in RdDM mutants. PLncDB has a genome browser for viewing the association of various epigenetic markers | http://chualab.rockefeller.edu/gbrowse2/homepage.html | [3] |
| Green Non-coding Database (GREENC) |
GREENC has >120,000 annotated lncRNAs from 37 plant species and algae. The user can access the coding potential and folding energy for each lncRNA | http://greenc.sciencedesigners.com/wiki/Main_Page | [25] |
| NONCODE v4.0 | NONCODE includes >500,000 lncRNAs from 16 species. Arabidopsis is the only plant species, as NONCODE focuses on non-plant species, including human and mouse | http://www.noncode.org/index.phphttp://www.noncode.org/index.php | [26] |
| CANTATAdb | CANTATAdb contains >45,000 plant lncRNAs from ten model plant species. In addition to tissue-specific expressions and coding potential, each lncRNA is also evaluated based on potential roles in splicing regulation and miRNA modulations | http://cantata.amu.edu.pl/ | [27] |
| Plant ncRNA database (PNRD) | PNRD has >25,000 ncRNAs of 11 different types and from 150 plant species. It also includes analytical tools, such as an miRNA predictor, coding potential calculator, and customized genome browser | http://structuralbiology.cau.edu.cn/PNRD/ | [28] |
| Plant Natural Antisense Transcripts DataBase (PlantNATsDB) |
A database for natural antisense transcripts (NATs) from 70 plant species, associated gene information, small RNA expression, and GO annotation | http://bis.zju.edu.cn/pnatdb/ | [29] |
An examination of >200 transcriptome data sets in Arabidopsis identified ~40,000 candidate lncRNAs; these included NATs (>30,000) and lincRNAs (>6000) [3, 4, 30]. Most of the lincRNAs did not produce smRNAs, and, like mammalian lncRNAs, the lincRNA transcript levels were 30–60-fold lower than that of transcript levels of the associated mRNA. Work in Arabidopsis found that NAT pairs, lncRNAs transcribed from opposite strands, occur widely: ~70% of protein-coding loci in Arabidopsis produce candidate NAT pairs 200–12,370 nt long (average length of 731 nt) [4]. Some NAT pairs show complete overlap (~60%), but others have complementary segments at their 5′ or the 3′ ends.
The expression levels of many lincRNAs differ significantly depending on the tissue and also change during stress; this indicates that lncRNAs undergo dynamic regulation and act in regulation of development and stress responses [30]. The expression levels of many NATs also are tissue-specific and change in response to biotic or abiotic stresses. For example, a recent study identified ~1400 NATs that respond to light; of the NAT pairs, about half respond in the same direction, and half respond in opposite directions. For the light-responsive NATs, the associated genes also showed peaks of histone acetylation; the acetylation levels changed with the changes in NAT expression in response to light [4].
Among the lncRNAs, Arabidopsis and rice have intermediate-sized ncRNAs (im-ncRNAs), which are 50–300 nt long [31, 32] and originate from 5′ UTRs, coding regions, and introns. The genes associated with 5′ UTR im-ncRNAs tended to have higher expression and H3K4me3 and H3K9ac histone marks, which are associated with transcriptional activation. Plants that have reduced levels of some im-ncRNAs showed developmental phenotypes or detectable molecular changes [31].
While we continue to gain better understandings of the mechanisms of lncRNA action, the mechanisms that regulate lncRNAs in plants remain limited. Like all transcripts, lncRNAs undergo transcriptional level regulation and regulation that affects lncRNA biogenesis, processing, and turnover. One of the players in this regulation is the exosome complex, which plays a major role in regulating the quantity, quality, and processing of various transcripts, including lncRNAs. The exosome complex is a conserved machinery with 3′–5′ exoribonuclease activity that consists of nine-subunit core associated with its enzymatic subunits, Rrp44 and Rrp6. The depletion of the Arabidopsis exosome allowed identification of a number of Arabidopsis ncRNAs as well as the genomic regions where the exosome is involved in their metabolism [1].
5.3. Molecular Functions of Plant lncRNAs
lncRNAs are present at low levels and show little sequence conservation compared with mRNAs; therefore, early studies questioned their importance and necessity and also suggested that lncRNAs might result from transcriptional noise. Indeed, considerable debate remains about the functionality of lncRNAs. However, evidence has emerged in recent years to indicate that many lncRNAs function in a large number of diverse molecular processes in eukaryotic cells; these include the regulation of yeast mating type [33, 34] and modulation of embryonic stem cell pluripotency and various diseases [35]. In plants, lncRNAs function in gene silencing, flowering time control, organogenesis in roots, photomorphogenesis in seedlings, abiotic stress responses, and reproduction [5, 11–14, 36–40].
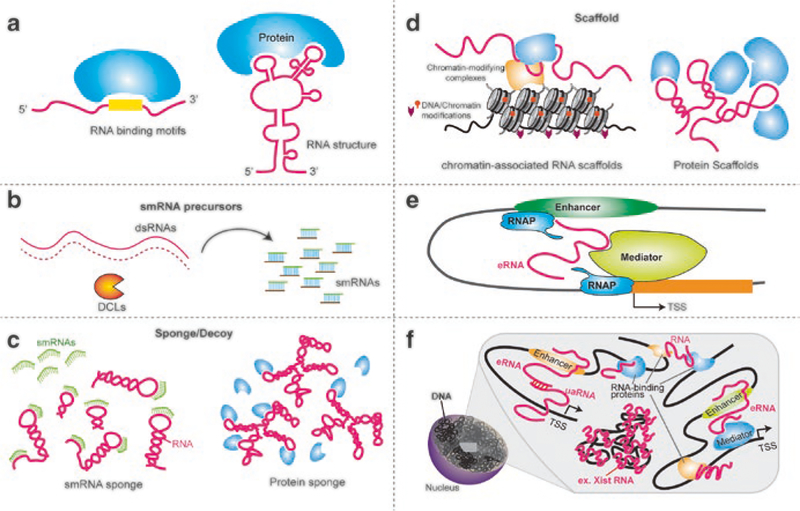
For their effects on gene regulation, lncRNAs act at multiple levels and with simple or complex mechanisms. lncRNAs can act in cis or trans, function by sequence complementarity to RNA or DNA, and be recognized via specific sequence motifs or secondary/tertiary structures (Fig. 5.2a). At the most simple level, lncRNAs can serve as precursors to smRNAs (Fig. 5.2b), as in the case of RNA Pol IV transcripts [6, 42–46]. Some lncRNAs keep regulatory proteins or microRNAs from interacting with their DNA or RNA targets by acting as decoys that mimic the targets (Fig. 5.2c). Some of the plant examples include the Arabidopsis microRNA target mimics IPS1 lncRNA and the decoy ASCO-lncRNA [38, 47].
Fig. 5.2.
Example lncRNAs and the mechanisms of their action. (a) Specific sequence motifs or secondary structures could be required for lncRNA function. (b) lncRNAs, specifically, the double-stranded transcripts, can serve as precursors to smRNAs in the RNA interference (RNAi) pathway. (c) lncRNAs can function as scaffolds for the recruitment of chromatin-modifying factors or as a platform for assembly of protein complexes. (d) lncRNAs can function as molecular sponges or decoys for smRNAs and also act as decoys to titrate away RNA-binding proteins. (e) The eRNAs, which are expressed from enhancers, are regulated by the exosome and can interact with other regions of DNA, such as enhancers or promoters, affecting the topology of the local DNA and thus altering gene expression. Adapted from [41]. (f) lncRNAs that interact with several chromatin-remodeling proteins and chromatin regions could affect higher-order nuclear structure
In animal systems, some lncRNAs directly affect Pol II and its associated transcriptional machinery by promoting phosphorylation of transcription factors (TFs) regulating their DNA-binding activity [48]. Many lncRNAs affect different processes related to transcription, including the initiation and elongation of transcripts, by affecting the pausing of RNA Pol II. Other lncRNAs act as scaffolds to recruit enzymes that remodel chromatin and thus alter chromatin structure and nuclear organization (Fig. 5.2d) (reviewed in [49]). Examples of plant lncRNAs that regulate transcription have started to emerge; for example, HID1 binds to the promoter of PIF3 gene to downregulate its expression [39]. However, no plant lncRNAs have yet been implicated in regulation of transcription elongation or Pol II pausing.
Different types of lncRNAs associate with chromatin and act as scaffolds that allow the assembly of complexes of chromatin-modifying enzymes. Recruitment of these proteins can require small RNAs or not. For example, the siRNA-directed DNA methylation (RdDM) pathway, which occurs specifically in plants, requires small RNAs [37]. Other lncRNAs can recruit complexes of enzymes that remodel chromatin but do not require smRNAs. The mechanism that provides targeting specificity for these lncRNAs remains to be discovered. Work in mammalian systems showed that lncRNAs can interact with proteins of the Trithorax group and activate transcription via trimethylation of histone H3K4 [50]. Other lncRNAs interact with proteins that modify histones with repressive marks, such as Polycomb Repressive Complex 2 (PRC2), to repress transcription via methylation of histone H3K27 [51]. The best-studied RNAi-independent pathway that relies on lncRNAs interacting with Polycomb is epigenetic regulation via histone modifications and expression of Arabidopsis FLOWERING LOCUS C (FLC).
Additional examples include enhancer RNAs (eRNAs), shown to be involved in regulation of transcription initiation. Enhancers are regulatory genomic regions that are shown to be involved in transcriptional regulation through targeting promoters of protein-coding genes in a tissue-specific and developmental manner as well as modulating spatial organization of the genome [52]. Work in mammalian systems has shown that exosome-sensitive eRNAs function in activation of transcription, consistent with the enhancer function. Some eRNAs act in cis to recruit complexes of coactivator proteins that form chromosome loops that connect the enhancer with its promoter, thus activating gene expression (Fig. 5.2e) [41, 53]. However, no eRNAs have not been identified in plants yet. The exosome function of resolving R-loops, which are RNA-DNA triplexes, might also reduce genomic instability in the regions expressing eRNAs [41]. R-loops form during transcription and can persist in regions that are divergently transcribed [54]. These results suggest that the exosome modulates the interactions among the key elements that regulate gene expression and the organization of the nucleus.
The examples of the well-studied plant lncRNAs with established functions and mechanisms of action are listed in Table 5.2.
Table 5.2.
List of plant lncRNAs
| lncRNAs | Description and function | References |
|---|---|---|
| ASCO-lncRNA | Functions in lateral root development in Arabidopsis. Regulator of alternative splicing. Works as a decoy lncRNA | [38] |
| IPS1 | Functions in regulating phosphate balance and phosphate starvation response in Arabidopsis. Competes with PHO2 mRNA for interaction with miR399 and acts as non- cleavable miRNA target | [47] |
| HID1 | Functions in regulation of photomorphogenesis in Arabidopsis seedlings. Trans-acting lncRNA (236 nt) acts by associating with the PIF3 promoter and represses its transcription. Evolutionary conserved in land plants | [39] |
| COOLAIR | Functions in regulation of flowering in Arabidopsis in both vernalization and autonomous pathways. Modulates FLC expression by multiple mechanisms | [55] |
| COLDAIR | Functions in regulation of flowering in Arabidopsis in the vernalization pathway. Associates with Polycomb to mediate silencing of FLC and affects chromatin looping at FLC in response to vernalization | [9] |
| COLDWRAP | Functions in regulation of flowering in Arabidopsis in the vernalization pathway. Participates in and coordinates vernalization-mediated Polycomb silencing of the FLC. Also affects formation of an intragenic chromatin loop that represses FLC | [11] |
| ASL | Functions in regulation of flowering in the autonomous pathway in Arabidopsis. AtRRP6L regulates ASL to modulate H3K27me3 levels. | [10] |
| APOLO | Functions in regulation of auxin signaling outputs in Arabidopsis. Participates in chromatin loop dynamics. Affects formation of a chromatin loop in the PID promoter region | [56] |
| Pol IV transcripts | Technically shorter in length than the standard lncRNAs. Function in silencing of transposons (TEs) and repeats in RdDM pathway. Serve as precursors for siRNAs in RdDM pathway | [57–59] |
| Pol V transcripts | Function in silencing TEs and repeats in RdDM pathway. Serve as a scaffold lncRNAs for assembly of siRNAs and proteins in RdDM pathway | [60] |
| ENOD40 | Functions in regulation of symbiotic interactions between leguminous plants and soil bacteria in Medicago truncatula. Suggested to function in re-localization of proteins in plants | [38, 61] |
| LDMAR | Regulates photoperiod-sensitive male sterility in rice by affecting DNA methylation in the LDMAR promoter region. The precise mechanism of LDMAR function and the interaction between LDMAR and siRNAs remain to be clarified | [62] |
5.4. Plant lncRNAs Functioning as Molecular Sponges and Decoys
Work in Arabidopsis identified lncRNAs that compete with microRNAs (miRNAs) or mimic the targets of miRNAs; similar function was also identified in animal systems. For example, the IPS1 lncRNA plays a role in regulating phosphate balance and uptake by competing for binding the PHO2 mRNA. PHO2 negatively regulates phosphate transporters and is itself downregulated by miR399 cleavage of its mRNA; IPS1 serves as mimic that cannot be cleaved by miR399 due to the mismatch but can titrate off miR399 [47]. Bioinformatics approaches also have predicted many additional miRNA target mimics in Arabidopsis, but the functions of many of these remain to be deciphered [63].
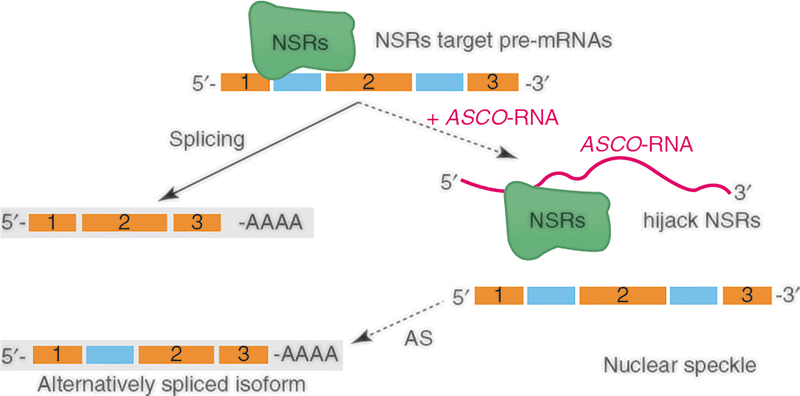
The Arabidopsis ASCO-lncRNA functions as decoy and regulates plant root development. ASCO-RNA competes with the binding of nuclear speckle RNA-binding proteins (NSRs), regulators of alternative splicing, to their targets; hijacking the NSRs changes the splicing patterns of NSR-regulated mRNA targets resulting in the production of alternative splice isoforms and leading to switch of developmental fates in plant roots (Fig. 5.3) [38].
Fig. 5.3.
Plant lncRNAs can affect the expression of proteins that regulate alternative splicing. The ASCO-lncRNA functions as a decoy that competes with mRNAs for binding to NSR splicing regulators
5.5. Plant lncRNAs Functioning in Regulation of Transcription and Silencing
5.5.1. Regulation of PIF3 Transcription by HID1 im-ncRNA
One of the interesting Arabidopsis lncRNAs, HIDDEN TREASURE 1 (HID1), also classified in original study as im-ncRNA with a length of 236 nt, is involved in the regulation of transcription of the transcription factor PIF3, a member of “phytochrome-interacting factors” (PIFs), a family of basic helix-loop-helix (bHLH) transcription factors [39]. HID1 is evolutionarily conserved in land plants and functions in trans as a component of an RNA-protein complex. It interacts with the promoter region of PIF3 and suppresses PIF3 transcription. The HID1 im-ncRNA is among rare examples of lncRNAs for which it was shown that its function requires its secondary structure. The secondary structure of HID1 in Arabidopsis and rice shows substantial conservation and expression of OsHID1 could complement the Arabidopsis hid1 mutant phenotype, indicating its importance in regulation of photomorphogenesis in seedlings.
5.5.2. Role of lncRNAs in RdDM
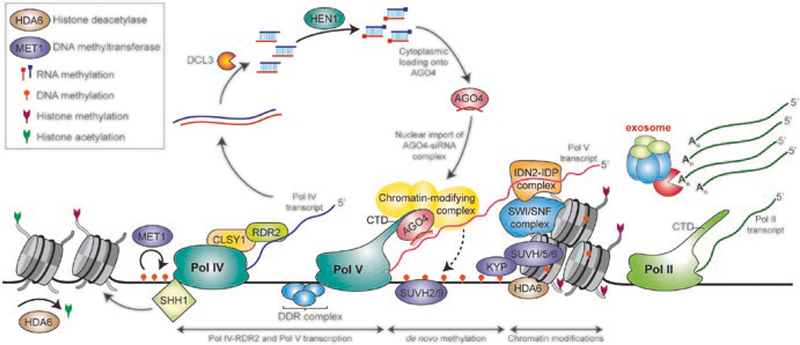
In plants, lncRNAs also function in epigenetic silencing, acting via siRNA-dependent DNA methylation (RdDM) (Fig. 5.4). RdDM in plants has similar mechanisms to gene silencing mediated by siRNAs in S. pombe [64–67]. RdDM primarily functions to repress transcription of transposons and repetitive sequences and requires RNA Pol IV and Pol V, two plant-specific RNA polymerases [6], and perhaps some involvement of RNA Pol II [68]. RNA Pol IV produces ncRNAs that serve as templates for 24 nt siRNAs, and RNA Pol V transcribes lncRNAs, which act as scaffolds that the AGO-siRNA complex recognizes through sequence complementarity (reviewed in [37]). In Arabidopsis, most siRNAs are generated by Pol IV; however, Pol V and Pol II can also make siRNA templates, suggesting additional complexity involved in siRNA biogenesis [69–72].
Fig. 5.4.
LncRNAs participating in the RdDM pathway. Transcripts produced by Pol IV are precursors for 24 nt siRNA; transcripts produced by Pol V are scaffolds and siRNA targets. SHH1 reads the H3K9me status of chromatin and recruits Pol IV; then the chromatin-remodeling protein CLSY1 assists in the passage of Pol IV [73]. Pol IV transcripts are transcribed by RDR2 into double-stranded RNAs (dsRNAs) before they are processed by DCL3 into 24 nt siRNAs and stabilized by methylation at the 3′ end by HEN1. These siRNAs associate with AGO and return to the nucleus as a part of the AGO-siRNA complex, which targets nascent Pol V scaffold transcripts. Pol V is recruited by SUVH2 or SUVH9 to its target genomic loci marked by DNA methylation [74], and Pol V transcription is facilitated by the DDR complex [75].The IDN2-IDP complex binds to Pol V scaffold RNAs and interacts with the SWI/SNF complex, which adjusts the position of nucleosomes [76]. The AGO4-siRNA complex interacts with Pol V; in this interaction, the siRNA in the complex base pairs with the transcript produced by Pol V to target a chromatin-modifying complex that catalyzes de novo methylation at the genomic loci. Then, the silencing mediated by DNA methylation is further amplified by methylation of histone H3K9 by KYP, SUVH5, and SUVH6 (reviewed in [37]). The silencing of solo LTRs requires the exosome, which does not act via siRNAs and DNA methylation. Rather, the exosome interacts with transcripts from a nearby scaffold-producing region and acts in silencing the solo LTR by altering chromatin structure via H3K9 histone methylation, suggesting this may function in parallel with RdDM
Identification of the Pol IV- and particularly Pol V-produced lncRNAs has remained challenging until recently [57–60]. One of the recent genome-wide studies identified Pol IV/RDR2-dependent transcripts (P4RNAs) from thousands of Arabidopsis loci. Interestingly, these P4RNAs are transcribed mainly from intergenic regions; 65% of the P4RNAs overlapped with transposable elements or repeats, and 9% of the RNAs overlapped with genes [57]. The Pol IV/RDR2-dependent transcripts are non-polyadenylated and produced from the sense and antisense DNA strands. Surprisingly, instead of a 5′ triphosphate, the P4RNAs have a monophosphate [57].
Until very recently Pol V transcripts eluded detection on the genome-wide scale due to the very low levels of their accumulation, which made them difficult to detect using RNA-seq. Based on the analysis of the individual transcripts, Pol V lncRNAs are non-polyadenylated and either tri-phosphorylated or capped at the 5′ ends [6]. Recent genome-wide study using RIP-seq identified 4502 individual Pol V-associated transcripts [60]. It was previously annotated that the Pol V-transcribed regions have an average length of 689 nt. Surprisingly, it was found that experimentally identified Pol V lncRNAs are shorter than previously annotated, with their median size ranging from 196 to 205 nt yet spanning the entire region. This data suggested that Pol V might not transcribe the entire regions continuously but is possibly controlled by internal promoters situated within the annotated regions that lead to active Pol V transcription.
Unlike RNA polymerases I, II, and III, which use specific sequence elements that identify their promoters, no specific DNA sequence elements were found in Pol V-transcribed regions. Instead internal repressive chromatin modifications appeared to control Pol V transcription and contribute to initiation by internal promoters. Interestingly, Pol V produces lncRNAs bidirectionally on annotated Pol V transcripts with no correlations in strand preference. However, despite Pol V that transcribes both strands of DNA, a subset of Pol V transcripts on transposons was found to be enriched on one strand in a way that indicated that limited strand preference of Pol V in these loci may be involved in determining boundaries of heterochromatin on transposons.
Previous genome-wide studies using ChIP-seq identified Pol V-associated genomic regions and found Pol V may also function in pathways other than the RdDM pathway [6, 75–80]. About 75% of Pol V-occupied genomic sites are transposons and repetitive sequences that also have 24 nt siRNAs and high levels of DNA methylation, indicating that Pol V induces RdDM at these sites. The other 25% of Pol V-associated sites include many protein-coding genes that have lower methylation levels and do not associate with siRNAs. This indicates that Pol V may also function in other silencing pathways [77]. Pol II also can produce scaffold transcripts that recruit siRNAs bound by AGO [68]. However, it remains unclear how Pol II targets specific intergenic loci and how Pol II interacts with Pol IV and Pol V.
Interestingly, the exosome also appears to play some role in silencing of these regions. A genome-wide study that identified exosome targets found many polyadenylated substrates of the exosome complex that corresponded to ncRNAs from centromeric regions, repetitive sequences, and other siRNA-producing loci and undergo RdDM-mediated silencing [1]. However, when we explored the connection between the two silencing pathways, RdDM and the exosome in Arabidopsis, we found that mutants of the core exosome subunits only produce a small effect on smRNAs [81]. This differs from results found in studies of the exosome in fission yeast, as in this system, the exosome prevents RNAs from spuriously entering into smRNA pathways [65]. Instead, less H3K9me2 was observed at several loci controlled by RdDM in exosome-deficient lines. The exosome interacts genetically with RNA Pol V and physically associates with polyadenylated Pol II transcripts from the regions generating Pol V scaffold RNAs [81]. These observations indicate that the exosome functions in lncRNA metabolism or processing in scaffold-generating regions. The exosome may also mediate the interactions among Pol II, Pol V, and Pol IV, modulating transcriptional repression. One outstanding question is whether and how the exosome (possibly acting through lncRNAs) contributes to silencing of loci via fine-tuning histone modifications and if the same mechanism of action can be observed genome wide.
However, Arabidopsis exosome subunits have diverse functions [1]. The additional enzymatic subunit, AtRRP6L1, is independent of the exosome core functions [10]. Mutations in AtRRP6L1 effect siRNA metabolism and DNA methylation [82]. Therefore, the exosome and the additional enzymatic subunits played an important role in regulation of ncRNAs, including siRNAs, in the RdDM pathway.
5.6. lncRNAs in the Regulation of Flowering
Because of the importance of flowering time regulation for plant adaptation to different latitudes, the lncRNAs that regulate flowering are among the best-studied functional plant lncRNAs. Work in Arabidopsis has shown that these lncRNAs regulate the initiation of flowering by modulating the expression of FLOWERING LOCUS C (FLC), which encodes a MADS-box transcription factor. FLC represses downstream genes required for flowering and thus negatively regulates flowering, acting in a dose-dependent manner. FLC functions in the vernalization pathway, which modulates flowering time in response to prolonged low temperature, and in the autonomous pathway, which modulates flowering time independently of environmental factors [83].
The regulation of flowering time involves epigenetic silencing of FLC, mainly via modification of histones. Repression of FLC requires PRC2, which is recruited to FLC and methylates histone H3K27. Alteration of chromatin, particularly changes in histone modifications that remove H3K4me3, H3K36me3, and H2Bub1 and replace those modifications with H3K27me3, epigenetically represses FLC expression (reviewed in [36]).
The lncRNAs COLDAIR, COLDWRAP, and COOLAIR are transcribed from FLC and function in FLC epigenetic silencing (Fig. 5.5) [9, 11, 84]. Vernalization induces transient transcription of COLDAIR, a 5′ capped, non-polyadenylated lncRNA, transcribed from FLC intron 1, in the same direction as FLC (Fig. 5.5). CURLY LEAF (CLF), a homolog of mammalian EZH2 (an enzymatic component of PRC2), binds to COLDAIR, and knockdown of COLDAIR decreases CLF and H3K27me3 enrichment at FLC in response to cold. This thus hampers the repression of FLC during vernalization and indicates that COLDAIR’s repression of FLC is essential for the vernalization response [9]. Previous work suggested that PRC2 recruitment to FLC requires COLDAIR for the initiation of epigenetic silencing, analogous to the functions of the mammalian lncRNAs HOTAIR and Xist [51]. However, mammalian PRC2 shows high-affinity binding to unrelated RNAs; therefore, other factors, in addition to lncRNAs, may provide the specificity that targets PRC2 to FLC [85].
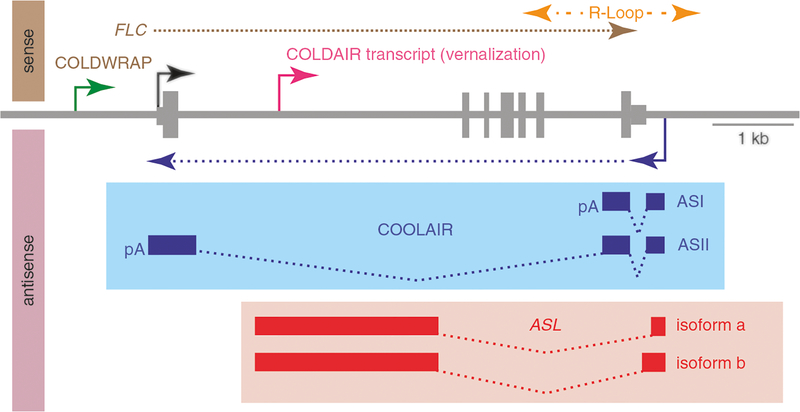
Fig. 5.5.
Regulatory lncRNAs produced from the FLC locus. Diagram of the FLC locus [84]. The FLC transcriptional start site is indicated by black arrow, and the vertical bars indicate exons in the FLC sense transcript. During vernalization, the COLDAIR lncRNA (pink) is transcribed in the sense direction, starting in the first intron of FLC. Another sense lncRNA, COLDWARP, is transcribed from the repressed promoter of FLC (green). The COOLAIR (blue) and ASL (red) lncRNA transcripts are transcribed from the indicated start sites (purple arrow) in the antisense direction; both result from alternative polyadenylation at poly(A) site either in the sense promoter region or intron 6. The ASL lncRNA also undergoes alternative splicing. Blue boxes indicate the exon of COOLAIR; red boxes indicate the exons of AS I and II; dotted lines indicate the spliced regions. ASL covers FLC intron I. Yellow dotted lines indicate the R-loops, in the COOLAIR promoter region, and repress COOLAIR transcription
An additional Polycomb-interacting lncRNA, cold of winter-induced noncoding RNA from the promoter (COLDWRAP), was identified to be expressed from the upstream promoter region of FLC locus and shown to function in repression of FLC (Fig. 5.5) [11]. COLDWRAP is a 316 nt lncRNA that is transcribed in the sense direction with its transcription start located 225 nt upstream from the FLC mRNA. COLDAIR and COLDWRAP both have 5′ caps, but most transcripts of COLDWRAP appear to be non-polyadenylated. Interestingly, association of the Polycomb complex with COLDWRAP appears to be specific, as native CLF binds significantly to the sense strand of COLDWRAP but only weakly to the antisense strand. In addition, the 5′ half of COLDWRAP and several stable secondary structures identified in this region are needed for RNA-protein interactions. Importantly, COLDWRAP working in a cooperative manner with COLDAIR is necessary for vernalization-mediated FLC silencing. COLDWRAP functions to retain Polycomb at the FLC promoter through the formation of a repressive intragenic chromatin loop forming a stable repressive chromatin structure.
The COOLAIR is a set of lncRNAs transcribed from the 3′ end of FLC in the antisense direction, which are alternatively spliced and polyadenylated, proximal AS I and distal AS II [55]. In response to cold, the locus first produces COOLAIR, then COLDAIR, before H3K27me3 accumulates; therefore, initial studies indicated that COOLAIR may act early in vernalization [55]. However, knockdown of COOLAIR did not affect the vernalization response [86]. Rather, COOLAIR increases the rate of FLC transcriptional repression during vernalization, and its function does not require PRC2 or H3K27me3 [36, 87]. The COOLAIR knockdown desynchronized the change from H3K36me to H3K27me3 in FLC; therefore, this switch at FLC may require COOLAIR or transcription in the antisense direction [87].
COOLAIR represses FLC in the vernalization and autonomous pathways. In the autonomous pathway, COOLAIR 3′ end processing affects the FLC chromatin [84]. The autonomous pathway factors FCA, FY, and FPA, along with the polyadenylation cleavage factors CstF64 and CstF77, and the spliceosome component PRP8, favor the production of AS I by increasing usage of the proximal COOLAIR polyadenylation site [84, 88, 89]. This increases levels of the FLOWERING LOCUS D (FLD) histone demethylase at FLC leading to H3K4me2 demethylation of FLC [90].
Unraveling the functional importance of transcription of COOLAIR and the functions of COOLAIR transcripts remains challenging. Since it is difficult to determine whether it is the COOLAIR transcription, COOLAIR transcripts, or both that are functionally important, the secondary RNA structure of COOLAIR was recently determined experimentally [91]. It was found that even despite the relatively low sequence identity between Arabidopsis and evolutionarily divergent Brassicaceae species, the structures showed remarkable evolutionary conservation. This conservation applied to multi-helix junctions and through covariation of a non-contiguous DNA sequence. The observed conservation of COOLAIR lncRNA structure in the Brassicaceae indicates that the COOLAIR lncRNA itself is very likely to function in regulation of FLC, although the process of antisense transcription from FLC may also affect FLC regulation.
Recent work also discovered the Antisense Long (ASL) transcript in early-flowering Arabidopsis ecotypes that do not require vernalization for flowering [10]. In contrast to the other lncRNAs transcribed from FLC, ASL does not get polyadenylated, although it is alternatively spliced. The ASL transcript is >2000 nucleotides long and is transcribed from the antisense strand, starting at the same promoter as COOLAIR. The 5′ regions of COOLAIR and ASL overlap, but ASL spans intron 1 (important for maintenance of FLC silencing) and includes the COLDAIR region, which is transcribed in the sense direction. The ASL transcript physically associates with the FLC locus and H3K27me3 [10], suggesting that ASL and COOLAIR play different roles in FLC silencing and perhaps in the maintenance of H3K27me3.
It is interesting that the exosome again is involved in the regulation of the antisense transcript and does so in a surprising way. Two of the exosome components, RRP6-Like (RRP6L) proteins, are involved in lncRNA-mediated regulation of flowering. RRP6, one of the catalytic subunits, has both core-complex-dependent and core-complex-independent functions [92, 93]. In Arabidopsis, RRP6L1 and RRP6L2 regulate COOLAIR and ASL expression or processing in the exosome core-complex-independent way [10]. Mutations of RRP6L also derepress FLC; this delays flowering. The AS I and II downregulation observed in RRP6Ls multiple mutants resembled the patterns that occur in CstF64 and CstF77 mutants, which are 3′ end processing factors [10, 84], indicating that COOLAIR 3′ end processing may require RRP6Ls.
Very surprisingly, emerging work indicates that RRP6Ls have a major role in regulation of the synthesis or biogenesis of ASL, as RRP6Ls mutants lack (or have minuscule amounts of) ASL transcript. This result finding is unexpected because RRP6 functions as a 3′–5′ exoribonuclease and RRP6 mutants generally fail to degrade or process certain RNAs; thus, these mutants usually overaccumulate certain RNAs. However, recent work found that the abundance of many yeast mRNAs also decreased in the rrp6Δ mutants [19]. Similarly, in humans, inactivation of the RRP6 homolog also causes a dramatic decrease in Xist levels [94].
Another function of RRP6Ls involves affecting the epigenetic marks at FLC; mutants of RRP6L have decreased H3K27me3 levels and decreased density of nucleosomes at FLC. These mutants therefore show increased expression of FLC and delayed flowering. RRP6L1 physically interacts with the ASL RNA and with chromatin at FLC; this indicates that RRP6Ls may regulate ASL to maintain H3K27me3 levels at FLC. Therefore, RRP6Ls regulate FLC lncRNAs, and their regulation of various antisense RNAs may affect FLC silencing [10].
R-loops that form over the COOLAIR promoter region affect COOLAIR transcription, although effects of R-loop formation on FLC expression are not fully unclear [95]. Failure of the termination of transcription can often produce R-loops [96], which can recruit the exosome co-transcriptionally through the noncanonical pathway for 3′ end processing [19]. Work in mammals showed that RRP6 can resolve deleterious R-loops [41]; thus, plant RRP6Ls may affect both the processing and expression of antisense transcripts from FLC in a similar manner.
In mammalian systems, lncRNAs have key roles in molding the three-dimensional organization of the nucleus (Fig. 5.2f) [97–99]. In plants, emerging research is beginning to reveal the role of lncRNAs in architecture of the nucleus, and some RNA studies also indicate that lncRNAs may have similar roles in 3-D nuclear architecture in plants and animals. Several studies have also addressed genome organization using Hi-C approach in Arabidopsis [100–104]. The RdDM pathway likely also affects the higher-order structure of chromatin by acting with MORC proteins. In Arabidopsis, MORC6 may have ATPase activity and interact with the DDR complex component DMS3; the action of this complex may be analogous to that of mammalian cohesin-like proteins that function in inactivation of the X-chromosome in mice. Consistent with this, MORC1 and MORC6 mutant plants have de-condensed pericentromeric heterochromatin [105]. The promoter and 3′ terminator of FLC form gene loops [106, 107], and COLDAIR and COLDWRAP lncRNAs participate in this process [11]. FLC alleles also undergo long-distance interactions, clustering during vernalization-mediated epigenetic silencing. This interaction requires VRN5 and VERNALIZATION 2, two PRC2 trans-acting factors [108]. However, we lack information on how lncRNAs function in long-distance interactions of the chromatin at FLC. As illustrated by FLC, plant lncRNAs carry out diverse, varied, and important functions. Our understanding of lncRNA functions continues to emerge as new studies uncover the mechanisms controlling lncRNA transcription and processing.
5.7. Concluding Remarks
The recent discovery that genomes undergo pervasive transcription opened many questions on the functions of these RNAs. Since then, studies in the various kingdoms of eukaryotes have broadened our understanding of the biogenesis and functions of various lncRNAs. However, although various studies have identified and classified many categories of lncRNAs, the functions of lncRNAs, and how they carry out these functions, remain to be discovered. Work in plants identifying lncRNAs systematically has caught up with work in other systems. Plant studies have also discovered lncRNA functions in controlling flowering time and RdDM-mediated silencing of genes. However, many other lncRNAs remain to be examined. The regulation of plant lncRNA synthesis and biogenesis also will require further work to elucidate. Understanding the mechanisms that control plant lncRNA expression and biogenesis will require integration of bioinformatics, genetic, and biochemical data to provide a complete understanding of lncRNA function and biology. A complete understanding of the various facets of plant lncRNAs will reciprocally advance our understanding of lncRNAs in other species.
References
- 1.Chekanova JA, Gregory BD, Reverdatto SV, Chen H, Kumar R, Hooker T, Yazaki J, Li P, Skiba N, Peng Q, Alonso J, Brukhin V, Grossniklaus U, Ecker JR, Belostotsky DA (2007) Genome-wide high-resolution mapping of exosome substrates reveals hidden features in the Arabidopsis transcriptome. Cell 131(7):1340–1353. doi: 10.1016/j.cell.2007.10.056 [DOI] [PubMed] [Google Scholar]
- 2.Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, Stadler PF, Hertel J, Hackermüller J, Hofacker IL, Bell I, Cheung E, Drenkow J, Dumais E, Patel S, Helt G, Ganesh M, Ghosh S, Piccolboni A, Sementchenko V, Tammana H, Gingeras TR (2007) RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science 316(5830):1484–1488. doi: 10.1126/science.1138341 [DOI] [PubMed] [Google Scholar]
- 3.Jin J, Liu J, Wang H, Wong L, Chua N-H (2013) PLncDB: plant long non-coding RNA database. Bioinformatics 29(8):1068–1071. doi: 10.1093/bioinformatics/btt107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang H, Chung PJ, Liu J, Jang I-C, Kean MJ, Xu J, Chua N-H (2014) Genome-wide identification of long noncoding natural antisense transcripts and their responses to light in Arabidopsis. Genome Res 24(3):444–453. doi: 10.1101/gr.165555.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang Y-C, Liao J-Y, Li Z-Y, Yu Y, Zhang J-P, Li Q-F, Qu L-H, Shu W-S, Chen Y-Q (2014) Genome-wide screening and functional analysis identify a large number of long noncoding RNAs involved in the sexual reproduction of rice. Genome Biol 15(12):512. doi: 10.1186/s13059-014-0512-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wierzbicki AT, Haag JR, Pikaard CS (2008) Noncoding transcription by RNA polymerase Pol IVb/Pol V mediates transcriptional silencing of overlapping and adjacent genes. Cell 135(4):635–648. doi: 10.1016/j.cell.2008.09.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li L, Eichten SR, Shimizu R, Petsch K, Yeh C-T, Wu W, Chettoor AM, Givan SA, Cole RA, Fowler JE, Evans MM, Scanlon MJ, Yu J, Schnable PS, Timmermans MC, Springer NM, Muehlbauer GJ (2014) Genome-wide discovery and characterization of maize long noncoding RNAs. Genome Biol 15(2):R40. doi: 10.1186/gb-2014-15-2-r40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Andersson R, Gebhard C, Miguel-Escalada I, Hoof I, Bornholdt J, Boyd M, Chen Y, Zhao X, Schmidl C, Suzuki T, Ntini E, Arner E, Valen E, Li K, Schwarzfischer L, Glatz D, Raithel J, Lilje B, Rapin N, Bagger FO, Jørgensen M, Andersen PR, Bertin N, Rackham O, Burroughs AM, Baillie JK, Ishizu Y, Shimizu Y, Furuhata E, Maeda S, Negishi Y, Mungall CJ, Meehan TF, Lassmann T, Itoh M, Kawaji H, Kondo N, Kawai J, Lennartsson A, Daub CO, Heutink P, Hume DA, Jensen TH, Suzuki H, Hayashizaki Y, Müller F, FANTOM C, Forrest ARR, Carninci P, Rehli M, Sandelin A (2014) An atlas of active enhancers across human cell types and tissues. Nature 507(7493):455–461. doi: 10.1038/nature12787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heo JB, Sung S (2011) Vernalization-mediated epigenetic silencing by a long intronic noncoding RNA. Science 331(6013):76–79. doi: 10.1126/science.1197349 [DOI] [PubMed] [Google Scholar]
- 10.Shin J-H, Chekanova JA (2014) Arabidopsis RRP6L1 and RRP6L2 function in FLOWERING LOCUS C silencing via regulation of antisense RNA synthesis. PLoS Genet 10(9):e1004612. doi: 10.1371/journal.pgen.1004612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim D-H, Sung S (2017) Vernalization-triggered intragenic chromatin loop formation by long noncoding RNAs. Dev Cell 40(3):302–312.e4. doi: 10.1016/j.devcel.2016.12.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Di C, Yuan J, Wu Y, Li J, Lin H, Hu L, Zhang T, Qi Y, Gerstein MB, Guo Y, Lu ZJ (2014) Characterization of stress-responsive lncRNAs in Arabidopsis thaliana by integrating expression, epigenetic and structural features. Plant J 80(5):848–861. doi: 10.1111/tpj.12679 [DOI] [PubMed] [Google Scholar]
- 13.Yuan J, Zhang Y, Dong J, Sun Y, Lim BL, Liu D, Lu ZJ (2016) Systematic characterization of novel lncRNAs responding to phosphate starvation in Arabidopsis thaliana. BMC Genomics 17(1):655. doi: 10.1186/s12864-016-2929-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li S, Yamada M, Han X, Ohler U, Benfey PN (2016) High-resolution expression map of the Arabidopsis root reveals alternative splicing and lincRNA regulation. Dev Cell 39(4):508–522. doi: 10.1016/j.devcel.2016.10.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mattick JS, Rinn JL (2015) Discovery and annotation of long noncoding RNAs. Nat Struct Mol Biol 22(1):5–7. doi: 10.1038/nsmb.2942 [DOI] [PubMed] [Google Scholar]
- 16.Xu Z, Wei W, Gagneur J, Perocchi F, Keller C, Camblong J, Guffanti E, Stutz F, Huber W, Steinmetz LM (2009) Bidirectional promoters generate pervasive transcription in yeast. Nature 457(7232):1033–1037. doi: 10.1038/nature07728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van Dijk EL, Chen CL, d’Aubenton-Carafa Y, Gourvennec S, Kwapisz M, Roche V, Bertrand C, Silvain M, Legoix-Né P, Loeillet S, Nicolas A, Thermes C, Morillon A (2011) XUTs are a class of Xrn1-sensitive antisense regulatory non-coding RNA in yeast. Nature 475(7354):114–117. doi: 10.1038/nature10118 [DOI] [PubMed] [Google Scholar]
- 18.Schulz D, Schwalb B, Kiesel A, Baejen C, Torkler P, Gagneur J, Soeding J, Cramer P (2013) Transcriptome surveillance by selective termination of noncoding RNA synthesis. Cell 155(5):1075–1087. doi: 10.1016/j.cell.2013.10.024 [DOI] [PubMed] [Google Scholar]
- 19.Fox MJ, Gao H, Smith-Kinnaman WR, Liu Y, Mosley AL (2015) The exosome component Rrp6 is required for RNA polymerase II termination at specific targets of the Nrd1-Nab3 pathway. PLoS Genet 11(2):e1004999. doi: 10.1371/journal.pgen.1004999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Flynn RA, Almada AE, Zamudio JR, Sharp PA (2011) Antisense RNA polymerase II divergent transcripts are P-TEFb dependent and substrates for the RNA exosome. Proc Natl Acad Sci 108(26):10460–10465. doi: 10.1073/pnas.1106630108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hetzel J, Duttke SH, Benner C, Chory J (2016) Nascent RNA sequencing reveals distinct features in plant transcription. Proc Natl Acad Sci 113(43):12316–12321. doi: 10.1073/pnas.1603217113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu B, Zhang W, Zhang T, Liu B, Jiang J (2015) Genome-wide prediction and validation of intergenic enhancers in Arabidopsis using open chromatin signatures. Plant Cell 27(9):2415–2426. doi: 10.1105/tpc.15.00537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lamesch P, Berardini TZ, Li D, Swarbreck D, Wilks C, Sasidharan R, Muller R, Dreher K, Alexander DL, Garcia-Hernandez M, Karthikeyan AS, Lee CH, Nelson WD, Ploetz L, Singh S, Wensel A, Huala E (2012) The Arabidopsis Information Resource (TAIR): improved gene annotation and new tools. Nucleic Acids Res 40(Database issue):D1202–D1210. doi: 10.1093/nar/gkr1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cheng C-Y, Krishnakumar V, Chan AP, Thibaud-Nissen F, Schobel S, Town CD (2017) Araport11: a complete reannotation of the Arabidopsis thaliana reference genome. Plant J 89(4):789–804. doi: 10.1111/tpj.13415 [DOI] [PubMed] [Google Scholar]
- 25.Paytuví Gallart A, Hermoso Pulido A, Anzar Martínez de Lagrán I, Sanseverino W, Aiese Cigliano R (2015) GREENC: a Wiki-based database of plant lncRNAs. Nucleic Acids Res. doi: 10.1093/nar/gkv1215.gkv1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao Y, Li H, Fang S, Kang Y, Wu W, Hao Y, Li Z, Bu D, Sun N, Zhang MQ, Chen R (2016) NONCODE 2016: an informative and valuable data source of long non-coding RNAs. Nucleic Acids Res 44(D1):D203–D208. doi: 10.1093/nar/gkv1252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Szcześniak MW, Rosikiewicz W, Makałowska I (2016) CANTATAdb: a collection of plant long non-coding RNAs. Plant Cell Physiol 57(1):e8–e8. doi: 10.1093/pcp/pcv201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yi X, Zhang Z, Ling Y, Xu W, Su Z (2015) PNRD: a plant non-coding RNA database. Nucleic Acids Res 43(Database issue):D982–D989. doi: 10.1093/nar/gku1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen D, Yuan C, Zhang J, Zhang Z, Bai L, Meng Y, Chen L-L, Chen M (2012) PlantNATsDB: a comprehensive database of plant natural antisense transcripts. Nucleic Acids Res 40(Database issue):D1187–D1193. doi: 10.1093/nar/gkr823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu J, Jung C, Xu J, Wang H, Deng S, Bernad L, Arenas-Huertero C, Chua N-H (2012) Genome-wide analysis uncovers regulation of long intergenic noncoding RNAs in Arabidopsis. Plant Cell 24(11):4333–4345. doi: 10.1105/tpc.112.102855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang Y, Wang X, Deng W, Fan X, Liu T-T, He G, Chen R, Terzaghi W, Zhu D, Deng XW (2014) Genomic features and regulatory roles of intermediate-sized non-coding RNAs in Arabidopsis . Mol Plant 7(3):514–527. doi: 10.1093/mp/sst177 [DOI] [PubMed] [Google Scholar]
- 32.Liu T-T, Zhu D, Chen W, Deng W, He H, He G, Bai B, Qi Y, Chen R, Deng XW (2013) A global identification and analysis of small nucleolar RNAs and possible intermediate-sized non-coding RNAs in Oryza sativa. Mol Plant 6(3):830–846. doi: 10.1093/mp/sss087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van Werven FJ, Neuert G, Hendrick N, Lardenois A, Buratowski S, van Oudenaarden A, Primig M, Amon A (2012) Transcription of two long noncoding RNAs mediates mating-type control of gametogenesis in budding yeast. Cell 150:1170–1181. doi: 10.1016/j.cell.2012.06.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zofall M, Yamanaka S, Reyes-Turcu FE, Zhang K, Rubin C, Grewal SIS (2012) RNA elimination machinery targeting meiotic mRNAs promotes facultative heterochromatin formation. Science 335(6064):96–100. doi: 10.1126/science.1211651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Flynn RA, Chang HY (2014) Long noncoding RNAs in cell-fate programming and reprogramming. Cell Stem Cell 14(6):752–761. doi: 10.1016/j.stem.2014.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Berry S, Dean C (2015) Environmental perception and epigenetic memory: mechanistic insight through FLC. Plant J 83(1):133–148. doi: 10.1111/tpj.12869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Matzke MA, Mosher RA (2014) RNA-directed DNA methylation: an epigenetic pathway of increasing complexity. Nat Rev Genet 15(6):394–408. doi: 10.1038/nrg3683 [DOI] [PubMed] [Google Scholar]
- 38.Bardou F, Ariel F, Simpson CG, Romero-Barrios N, Laporte P, Balzergue S, Brown JWS, Crespi M (2014) Long noncoding RNA modulates alternative splicing regulators in Arabidopsis. Dev Cell 30(2):166–176. doi: 10.1016/j.devcel.2014.06.017 [DOI] [PubMed] [Google Scholar]
- 39.Wang Y, Fan X, Lin F, He G, Terzaghi W, Zhu D, Deng XW (2014) Arabidopsis non-coding RNA mediates control of photomorphogenesis by red light. Proc Natl Acad Sci 111(28):10359–10364. doi: 10.1073/pnas.1409457111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang D, Qu Z, Yang L, Zhang Q, Liu Z-H, Do T, Adelson DL, Wang Z-Y, Searle I, Zhu J-K (2017) Transposable elements (TEs) contribute to stress-related long intergenic noncoding RNAs in plants. Plant J. doi: 10.1111/tpj.13481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pefanis E, Wang J, Rothschild G, Lim J, Kazadi D, Sun J, Federation A, Chao J, Elliott O, Liu Z-P, Economides AN, Bradner JE, Rabadan R, Basu U (2015) RNA exosome-regulated long non-coding RNA transcription controls super-enhancer activity. Cell 161(4):774–789. doi: 10.1016/j.cell.2015.04.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zilberman D, Cao X, Jacobsen SE (2003) ARGONAUTE4 control of locus-specific siRNA accumulation and DNA and histone methylation. Science 299(5607):716–719. doi: 10.1126/science.1079695 [DOI] [PubMed] [Google Scholar]
- 43.Xie Z, Johansen LK, Gustafson AM, Kasschau KD, Lellis AD, Zilberman D, Jacobsen SE, Carrington JC (2004) Genetic and functional diversification of small RNA pathways in plants. PLoS Biol 2(5):E104. doi: 10.1371/journal.pbio.0020104.sg002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zheng X, Zhu J, Kapoor A, Zhu J-K (2007) Role of Arabidopsis AGO6 in siRNA accumulation, DNA methylation and transcriptional gene silencing. EMBO J 26(6):1691–1701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gao Z, Liu H-L, Daxinger L, Pontes O, He X, Qian W, Lin H, Xie M, Lorkovic ZJ, Zhang S, Miki D, Zhan X, Pontier D, Lagrange T, Jin H, Matzke AJM, Matzke M, Pikaard CS, Zhu J-K (2010) An RNA polymerase II- and AGO4-associated protein acts in RNA-directed DNA methylation. Nature 465(7294):106–109. doi: 10.1038/nature09025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Matzke M, Kanno T, Daxinger L, Huettel B, Matzke AJ (2009) RNA-mediated chromatin-based silencing in plants. Curr Opin Cell Biol 21(3):367–376. doi: 10.1016/j.ceb.2009.01.025 [DOI] [PubMed] [Google Scholar]
- 47.Franco-Zorrilla JM, Valli A, Todesco M, Mateos I, Puga MI, Rubio-Somoza I, Leyva A, Weigel D, García JA, Paz-Ares J (2007) Target mimicry provides a new mechanism for regulation of microRNA activity. Nat Genet 39(8):1033–1037. doi: 10.1038/ng2079 [DOI] [PubMed] [Google Scholar]
- 48.Wang P, Xue Y, Han Y, Lin L, Wu C, Xu S, Jiang Z, Xu J, Liu Q, Cao X (2014) The STAT3-binding long noncoding RNA lnc-DC controls human dendritic cell differentiation. Science 344(6181):310–313. doi: 10.1126/science.1251456 [DOI] [PubMed] [Google Scholar]
- 49.Bonasio R, Shiekhattar R (2014) Regulation of transcription by long noncoding RNAs. Annu Rev Genet 48:433–455. doi: 10.1146/annurev-genet-120213-092323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang KC, Yang YW, Liu B, Sanyal A, Corces-Zimmerman R, Chen Y, Lajoie BR, Protacio A, Flynn RA, Gupta RA, Wysocka J, Lei M, Dekker J, Helms JA, Chang HY (2011) A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature 472(7341):120–124. doi: 10.1038/nature09819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E, Chang HY (2010) Long noncoding RNA as modular scaffold of histone modification complexes. Science 329(5992):689–693. doi: 10.1126/science.1192002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li W, Notani D, Rosenfeld MG (2016) Enhancers as non-coding RNA transcription units: recent insights and future perspectives. Nat Rev Genet 17(4):207–223. doi: 10.1038/nrg.2016.4 [DOI] [PubMed] [Google Scholar]
- 53.Lai F, Orom UA, Cesaroni M, Beringer M, Taatjes DJ, Blobel GA, Shiekhattar R (2013) Activating RNAs associate with mediator to enhance chromatin architecture and transcription. Nature 494(7438):497–501. doi: 10.1038/nature11884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Skourti-Stathaki K, Proudfoot NJ (2014) A double-edged sword: R loops as threats to genome integrity and powerful regulators of gene expression. Genes Dev 28(13):1384–1396. doi: 10.1101/gad.242990.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Swiezewski S, Liu F, Magusin A, Dean C (2009) Cold-induced silencing by long antisense transcripts of an Arabidopsis polycomb target. Nature 462(7274):799–802. doi: 10.1038/nature08618 [DOI] [PubMed] [Google Scholar]
- 56.Ariel F, Jegu T, Latrasse D, Romero-Barrios N, Christ A, Benhamed M, Crespi M (2014) Noncoding transcription by alternative RNA polymerases dynamically regulates an auxin-driven chromatin loop. Mol Cell 55(3):383–396. doi: 10.1016/j.molcel.2014.06.011 [DOI] [PubMed] [Google Scholar]
- 57.Li S, Vandivier LE, Tu B, Gao L, Won SY, Li S, Zheng B, Gregory BD, Chen X (2015) Detection of Pol IV/RDR2-dependent transcripts at the genomic scale in Arabidopsis reveals features and regulation of siRNA biogenesis. Genome Res 25(2):235–245. doi: 10.1101/gr.182238.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Blevins T, Podicheti R, Mishra V, Marasco M, Wang J, Rusch D, Tang H, Pikaard CS (2015) Identification of Pol IV and RDR2-dependent precursors of 24 nt siRNAs guiding de novo DNA methylation in Arabidopsis. eLife 4:e09591. doi: 10.7554/eLife.09591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhai J, Bischof S, Wang H, Feng S, Lee T-F, Teng C, Chen X, Park SY, Liu L, Gallego-Bartolome J, Liu W, Henderson IR, Meyers BC, Ausin I, Jacobsen SE (2015) A one precursor one siRNA model for Pol IV-dependent siRNA biogenesis. Cell 163(2):445–455. doi: 10.1016/j.cell.2015.09.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Böhmdorfer G, Sethuraman S, Rowley MJ, Krzyszton M, Rothi MH, Bouzit L, Wierzbicki AT (2016) Long non-coding RNA produced by RNA polymerase V determines boundaries of heterochromatin. eLife 5:1325. doi: 10.7554/eLife.19092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Campalans A, Kondorosi A, Crespi M (2004) Enod40, a short open reading frame-containing mRNA, induces cytoplasmic localization of a nuclear RNA binding protein in Medicago truncatula. Plant Cell 16(4):1047–1059. doi: 10.1105/tpc.019406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ding J, Lu Q, Ouyang Y, Mao H, Zhang P, Yao J, Xu C, Li X, Xiao J, Zhang Q (2012) A long noncoding RNA regulates photoperiod-sensitive male sterility, an essential component of hybrid rice. Proc Natl Acad Sci 109(7):2654–2659. doi: 10.1073/pnas.1121374109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wu H-J, Wang Z-M, Wang M, Wang X-J (2013) Widespread long noncoding RNAs as endogenous target mimics for microRNAs in plants. Plant Physiol 161(4):1875–1884. doi: 10.1104/pp.113.215962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bühler M, Haas W, Gygi SP, Moazed D (2007) RNAi-dependent and -independent RNA turnover mechanisms contribute to heterochromatic gene silencing. Cell 129(4):707–721. doi: 10.1016/j.cell.2007.03.038 [DOI] [PubMed] [Google Scholar]
- 65.Bühler M, Spies N, Bartel DP, Moazed D (2008) TRAMP-mediated RNA surveillance prevents spurious entry of RNAs into the Schizosaccharomyces pombe siRNA pathway. Nat Struct Mol Biol 15(10):1015–1023. doi: 10.1038/nsmb.1481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Reyes-Turcu FE, Zhang K, Zofall M, Chen E, Grewal SIS (2011) Defects in RNA quality control factors reveal RNAi-independent nucleation of heterochromatin. Nat Struct Mol Biol 18(10):1132–1138. doi: 10.1038/nsmb.2122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Holoch D, Moazed D (2015) RNA-mediated epigenetic regulation of gene expression. Nat Rev Genet 16(2):71–84. doi: 10.1038/nrg3863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zheng B, Wang Z, Li S, Yu B, Liu JY, Chen X (2009) Intergenic transcription by RNA polymerase II coordinates Pol IV and Pol V in siRNA-directed transcriptional gene silencing in Arabidopsis. Genes Dev 23(24):2850–2860. doi: 10.1101/gad.1868009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang X, Henderson IR, Lu C, Green PJ, Jacobsen SE (2007) Role of RNA polymerase IV in plant small RNA metabolism. Proc Natl Acad Sci 104(11):4536–4541. doi: 10.1073/pnas.0611456104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lee T-F, Gurazada SGR, Zhai J, Li S, Simon SA, Matzke MA, Chen X, Meyers BC (2012) RNA polymerase V-dependent small RNAs in Arabidopsis originate from small, intergenic loci including most SINE repeats. Epigenetics 7(7):781–795. doi: 10.4161/epi.20290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.You W, Lorkovic ZJ, Matzke AJM, Matzke M (2013) Interplay among RNA polymerases II, IV and V in RNA-directed DNA methylation at a low copy transgene locus in Arabidopsis thaliana. Plant Mol Biol 82(1–2):85–96. doi: 10.1007/s11103-013-0041-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sasaki T, Lee T-F, Liao W-W, Naumann U, Liao J-L, Eun C, Huang Y-Y, Fu JL, Chen P-Y, Meyers BC, Matzke AJM, Matzke M (2014) Distinct and concurrent pathways of Pol II- and Pol IV-dependent siRNA biogenesis at a repetitive trans-silencer locus in Arabidopsis thaliana. Plant J 79(1):127–138. doi: 10.1111/tpj.12545 [DOI] [PubMed] [Google Scholar]
- 73.Law JA, Du J, Hale CJ, Feng S, Krajewski K, Palanca AMS, Strahl BD, Patel DJ, Jacobsen SE (2013) Polymerase IV occupancy at RNA-directed DNA methylation sites requires SHH1. Nature 498(7454):385–389. doi: 10.1038/nature12178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Liu Z-W, Shao C-R, Zhang C-J, Zhou J-X, Zhang S-W, Li L, Chen S, Huang H-W, Cai T, He X-J (2014) The SET domain proteins SUVH2 and SUVH9 are required for Pol V occupancy at RNA-directed DNA methylation loci. PLoS Genet 10(1):e1003948. doi: 10.1371/journal.pgen.1003948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhong X, Hale CJ, Law JA, Johnson LM, Feng S, Tu A, Jacobsen SE (2012) DDR complex facilitates global association of RNA polymerase V to promoters and evolutionarily young transposons. Nat Struct Mol Biol 19(9):870–875. doi: 10.1038/nsmb.2354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhu Y, Rowley MJ, Böhmdorfer G, Wierzbicki AT (2013) A SWI/SNF chromatin-remodeling complex acts in noncoding RNA-mediated transcriptional silencing. Mol Cell 49(2):298–309. doi: 10.1016/j.molcel.2012.11.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wierzbicki AT, Cocklin R, Mayampurath A, Lister R, Rowley MJ, Gregory BD, Ecker JR, Tang H, Pikaard CS (2012) Spatial and functional relationships among Pol V-associated loci, Pol IV-dependent siRNAs, and cytosine methylation in the Arabidopsis epigenome. Genes Dev 26(16):1825–1836. doi: 10.1101/gad.197772.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wierzbicki AT, Ream TS, Haag JR, Pikaard CS (2009) RNA polymerase V transcription guides ARGONAUTE4 to chromatin. Nat Genet 41(5):630–634. doi: 10.1038/ng.365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zheng Q, Rowley MJ, Böhmdorfer G, Sandhu D, Gregory BD, Wierzbicki AT (2012) RNA polymerase V targets transcriptional silencing components to promoters of protein-coding genes. Plant J. doi: 10.1111/tpj.12034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Böhmdorfer G, Rowley MJ, Kuciński J, Zhu Y, Amies I, Wierzbicki AT (2014) RNA-directed DNA methylation requires stepwise binding of silencing factors to long non-coding RNA. Plant J 79(2):181–191. doi: 10.1111/tpj.12563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Shin J-H, Wang H-LV, Lee J, Dinwiddie BL, Belostotsky DA, Chekanova JA (2013) The role of the Arabidopsis exosome in siRNA-independent silencing of heterochromatic loci. PLoS Genet 9(3):e1003411. doi: 10.1371/journal.pgen.1003411.s007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhang H, Tang K, Qian W, Duan C-G, Wang B, Zhang H, Wang P, Zhu X, Lang Z, Yang Y, Zhu J-K (2014) An Rrp6-like protein positively regulates noncoding RNA levels and DNA methylation in Arabidopsis. Mol Cell 54(3):418–430. doi: 10.1016/j.molcel.2014.03.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Amasino RM, Michaels SD (2010) The timing of flowering. Plant Physiol 154(2):516–520. doi: 10.1104/pp.110.161653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Liu F, Marquardt S, Lister C, Swiezewski S, Dean C (2009) Targeted 3′ processing of antisense transcripts triggers Arabidopsis FLC chromatin silencing. Science 327(5961):94–97. doi: 10.1126/science.1180278 [DOI] [PubMed] [Google Scholar]
- 85.Davidovich C, Wang X, Cifuentes-Rojas C, Goodrich KJ, Gooding AR, Lee JT, Cech TR (2015) Toward a consensus on the binding specificity and promiscuity of PRC2 for RNA. Mol Cell 57(3):552–558. doi: 10.1016/j.molcel.2014.12.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Helliwell CA, Robertson M, Finnegan EJ, Buzas DM, Dennis ES (2011) Vernalization-repression of Arabidopsis FLC requires promoter sequences but not antisense transcripts. PLoS One 6(6):e21513. doi: 10.1371/journal.pone.0021513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Csorba T, Questa JI, Sun Q, Dean C (2014) Antisense COOLAIR mediates the coordinated switching of chromatin states at FLC during vernalization. Proc Natl Acad Sci 111(45):16160–16165. doi: 10.1073/pnas.1419030111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hornyik C, Terzi LC, Simpson GG (2010) The Spen family protein FPA controls alternative cleavage and polyadenylation of RNA. Dev Cell 18(2):203–213. doi: 10.1016/j.devcel.2009.12.009 [DOI] [PubMed] [Google Scholar]
- 89.Marquardt S, Raitskin O, Wu Z, Liu F, Sun Q, Dean C (2014) Functional consequences of splicing of the antisense transcript COOLAIR on FLC transcription. Mol Cell 54(1):156–165. doi: 10.1016/j.molcel.2014.03.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Liu F, Quesada V, Crevillen P, Bäurle I, Swiezewski S, Dean C (2007) The Arabidopsis RNA-binding protein FCA requires a lysine-specific demethylase 1 homolog to downregulate FLC. Mol Cell 28(3):398–407. doi: 10.1016/j.molcel.2007.10.018 [DOI] [PubMed] [Google Scholar]
- 91.Hawkes EJ, Hennelly SP, Novikova IV, Irwin JA, Dean C, Sanbonmatsu KY (2016) COOLAIR antisense RNAs form evolutionarily conserved elaborate secondary structures. Cell Rep 16(12):3087–3096. doi: 10.1016/j.celrep.2016.08.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Callahan KP, Butler JS (2008) Evidence for core exosome independent function of the nuclear exoribonuclease Rrp6p. Nucleic Acids Res 36(21):6645–6655. doi: 10.1093/nar/gkn743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kiss DL, Andrulis ED (2010) Genome-wide analysis reveals distinct substrate specificities of Rrp6, Dis3, and core exosome subunits. RNA 16(4):781–791. doi: 10.1261/rna.1906710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ciaudo C, Bourdet A, Cohen-Tannoudji M, Dietz HC, Rougeulle C, Avner P (2006) Nuclear mRNA degradation pathway(s) are implicated in Xist regulation and X chromosome inactivation. PLoS Genet 2(6):e94. doi: 10.1371/journal.pgen.0020094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sun Q, Csorba T, Skourti-Stathaki K, Proudfoot NJ, Dean C (2013) R-loop stabilization represses antisense transcription at the Arabidopsis FLC locus. Science 340(6132):619–621. doi: 10.1126/science.1234848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Skourti-Stathaki K, Kamieniarz-Gdula K, Proudfoot NJ (2014) R-loops induce repressive chromatin marks over mammalian gene terminators. Nature 516(7531):436–439. doi: 10.1038/nature13787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hacisuleyman E, Goff LA, Trapnell C, Williams A, Henao-Mejia J, Sun L, McClanahan P, Hendrickson DG, Sauvageau M, Kelley DR, Morse M, Engreitz J, Lander ES, Guttman M, Lodish HF, Flavell R, Raj A, Rinn JL (2014) Topological organization of multichromosomal regions by the long intergenic noncoding RNA Firre. Nat Struct Mol Biol 21(2):198–206. doi: 10.1038/nsmb.2764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Engreitz JM, Pandya-Jones A, McDonel P, Shishkin A, Sirokman K, Surka C, Kadri S, Xing J, Goren A, Lander ES, Plath K, Guttman M (2013) The Xist lncRNA exploits three-dimensional genome architecture to spread across the X chromosome. Science 341(6147):1237973. doi: 10.1126/science.1237973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Quinodoz S, Guttman M (2014) Long noncoding RNAs: an emerging link between gene regulation and nuclear organization. Trends Cell Biol 24(11):651–663. doi: 10.1016/j.tcb.2014.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Feng S, Cokus SJ, Schubert V, Zhai J, Pellegrini M, Jacobsen SE (2014) Genome-wide Hi-C analyses in wild-type and mutants reveal high-resolution chromatin interactions in Arabidopsis. Mol Cell 55(5):694–707. doi: 10.1016/j.molcel.2014.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Grob S, Schmid MW, Grossniklaus U (2014) Hi-C analysis in Arabidopsis identifies the knot, a structure with similarities to the flamenco locus of Drosophila. Mol Cell 55(5):678–693. doi: 10.1016/j.molcel.2014.07.009 [DOI] [PubMed] [Google Scholar]
- 102.Wang C, Liu C, Roqueiro D, Grimm D, Schwab R, Becker C, Lanz C, Weigel D (2015) Genome-wide analysis of local chromatin packing in Arabidopsis thaliana. Genome Res 25(2):246–256. doi: 10.1101/gr.170332.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Liu C, Weigel D (2015) Chromatin in 3D: progress and prospects for plants. Genome Biol 16(1):170. doi: 10.1186/s13059-015-0738-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Liu C, Wang C, Wang G, Becker C, Zaidem M, Weigel D (2016) Genome-wide analysis of chromatin packing in Arabidopsis thaliana at single-gene resolution. Genome Res. doi: 10.1101/gr204032.116.gr204032116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Moissiard G, Cokus SJ, Cary J, Feng S, Billi AC, Stroud H, Husmann D, Zhan Y, Lajoie BR, McCord RP, Hale CJ, Feng W, Michaels SD, Frand AR, Matteo P, Dekker J, Kim JK, Jacobsen SE (2012) MORC family ATPases required for heterochromatin condensation and gene silencing. Science 336(6087):1448–1451. doi: 10.1126/science.1221472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Crevillen P, Sonmez C, Wu Z, Dean C (2013) A gene loop containing the floral repressor FLC is disrupted in the early phase of vernalization. EMBO J 32(1):140–148. doi: 10.1038/emboj.2012.324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Jegu T, Latrasse D, Delarue M, Hirt H, Domenichini S, Ariel F, Crespi M, Bergounioux C, Raynaud C, Benhamed M (2014) The BAF60 subunit of the SWI/SNF chromatin-remodeling complex directly controls the formation of a gene loop at FLOWERING LOCUS C in Arabidopsis. Plant Cell 26(2):538–551. doi: 10.1105/tpc.113.114454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Rosa S, De Lucia F, Mylne JS, Zhu D, Ohmido N, Pendle A, Kato N, Shaw P, Dean C (2013) Physical clustering of FLC alleles during polycomb-mediated epigenetic silencing in vernalization. Genes Dev 27(17):1845–1850. doi: 10.1101/gad.221713.113 [DOI] [PMC free article] [PubMed] [Google Scholar]