Abstract
A significant challenge for solid tumor treatment is ensuring that a sufficient concentration of therapeutic agent is delivered to the tumor site at doses that can be tolerated by the patient. Biomolecular targeting can bias accumulation in tumors by taking advantage of specific interactions with receptors overexpressed on cancerous cells. However, while antibody-based immunoconjugates show high binding to specific cells, their low dissociation constants (KD) and large Stokes radii hinder their ability to penetrate deep into tumor tissue, leading to incomplete cell killing and tumor recurrence. To address this, we demonstrate the design and production of a photo-cross-linkable affibody that can form a covalent bond to epidermal growth factor receptor (EGFR) under near UV irradiation. Twelve cysteine mutations were created of an EGFR affibody and conjugated with maleimide-benzophenone. Of these only one exhibited photoconjugation to EGFR, as demonstrated by SDS-PAGE and Western blot. Next this modified affibody was shown to not only bind EGFR expressing cells but also show enhanced retention in a 3D tumor spheroid model, with minimal loss up to 24 h as compared to either unmodified EGFR-binding affibodies or nonbinding, photo-cross-linkable affibodies. Finally, in order to show utility of photo-cross-linking at clinically relevant wavelengths, upconverting nanoparticles (UCNPs) were synthesized that could convert 980 nm light to UV and blue light. In the presence of UCNPs, both direct photoconjugation to EGFR and enhanced retention in tumor spheroids could be obtained using near-infrared illumination. Thus, the photoactive affibodies developed here may be utilized as a platform technology for engineering new therapy conjugates that can penetrate deep into tumor tissue and be retained long enough for effective tumor therapy.
Graphical Abstract
INTRODUCTION
One of the major difficulties of cancer treatment is ensuring that a sufficient concentration of the therapeutic agent accumulates in the tumor when systemic dosages are limited by patient tolerance. Targeted therapy by biomacromolecules such as antibodies have shown great promise for improving the efficacy of cancer drugs by biasing uptake at cell receptors overexpressed in tumor tissue. Though antibody therapies have shown excellent affinity for cancer-specific receptors, their efficacy against solid tumors has been limited by poor diffusion of antibodies into the tumor due to several issues.1 First, the large size of the antibody can often hinder diffusion deep within tumors compared to smaller molecules.2 Second, high affinity antibodies have been shown to experience a so-called “barrier effect”, in which the targeting agents become immobilized at the periphery of tumors.3 This effect has been attributed to the low dissociation rate (KD) of targeting ligands bound at the tumor periphery, preventing them from further diffusion into the tumor, as well as their overall size.3 Attempts to overcome this effect by increasing antibody dosage has led to systemic off-target effects, which can be further exacerbated by slow blood clearance of antibodies.4 While antibodies with lower affinities do penetrate further into solid tumors, they can also quickly leave the tumor, and thus they are not likely to be retained long enough for therapeutic effect.
To address the limitations of macromolecular diffusion of monoclonal antibodies, affibodies have been developed as alternate targeting ligands. Affibodies are small, 56-residue proteins that form stable triple α helix bundles derived from a modified B domain (Z domain) of S. aureus protein A. They can be engineered to bind molecular targets such as cell receptors with high specificity and tunable affinity by randomizing 13 amino acids located along two of the three helices, followed by screening for tight binders.5–7 Success in particular has been demonstrated for affibodies that bind to epidermal growth factor receptor (EGFR). The first affibodies to bind the extracellular domain of EGFR were selected through phage display,8 followed by subsequent versions obtained by directed evolution (KD ≈ 2.8 nM).9 Due to its favorable affinity, this affibody structure has been utilized extensively for molecular imaging in vivo.10–14 Other versions have also been used to study the dynamics of EGFR in vitro15–18 or for active targeting of drug carrying nanoparticles.6,19 Unlike antibodies, affibodies are relatively simple to engineer, site-specifically modify, and produce.5,6,20 They do not require the formation of disulfide bridges or glycosylation to maintain their structure or function as antibodies do, but they can be similarly functionalized with effector molecules such as cytokines or radioisotopes and can even be bound to the Fc domain of antibodies to induce a similar immune response.20 Because of their small Stokes radius (1–2 nm as opposed to 5–6 nm), affibodies exhibit better tumor penetration than antibodies.21,22 However, these affibodies also suffer from fast dissociation and thus poorer retention within tumors.23
In the current work, photo-cross-linking was investigated as a method to retain affibodies throughout a tumor by first allowing for sufficient perfusion and then creating a covalent bond to a cell receptor with a focused, externally targeted light source. While photoactivatable functional groups have been genetically or post-translationally incorporated into other proteins for triggered covalent bond formation between those proteins and their native noncovalent binding partners,24,25 photo-cross-linking for targeted therapy applications has, to our knowledge, not been explored. Here, we show that the site-specific addition of photo-cross-linkable groups to EGFR binding affibodies enables the formation of covalent bridges between the affibody and cell receptor upon irradiation. Next, we show that triggering photo-cross-linking allows the affibody to not only diffuse into a 3D tumor spheroid but also promote longer retention times, which can in future work be utilized for prolonged therapeutic activity for improved tumor eradication.
RESULTS AND DISCUSSION
First, affibodies that can form covalent linkages to EGFR upon irradiation were produced by incorporating a photo-cross-linkable functional group at a specific amino acid site. A previously developed EGFR-binding affibody known as ZEGFR1907 (referred to here as wildtype or WT) was genetically modified at its putative binding site.9 Because this affibody was developed from a nonbinding Z domain by mutating 13 amino acids clustered along two of the three alpha helices, amino acid positions within and surrounding this interface were chosen as sites for incorporating the photo-cross-linkable unit, benzophenone (BP). As the WT affibodies do not possess cysteine residues, a cysteine was first inserted at specific amino acid sites, followed by reaction with maleimide-benzophenone to introduce a single BP per affibody. Benzophenone was chosen because it has been previously utilized as a photo-cross-linking agent for proteins upon activation with 365 nm light at intensities that do not significantly damage biological samples.25–27
To produce the constructs, the amino acid sequence of the WT affibody was codon optimized for production in E. coli and inserted into the Pet21b+ vector. Twelve different mutants, each of which contained a single amino acid changed to cysteine, were created by PCR-based site-directed mutagenesis. These constructs were then transformed into E. coli strain BL21 (DE3) to produce affibodies that also possessed a C-terminal 6x histidine tag for purification and an N-terminal T7 epitope tag for later detection by anti-T7 antibodies. After growing transformants, the modified affibodies were collected from the bacterial lysate by association with Ni-NTA beads, followed by immediate addition of maleimide-benzophenone and reaction for 18 h at RT. Excess maleimide-BP was removed by centrifugal filtration. The conjugation of each BP containing affibody mutant was confirmed by ESI mass spectrometry (Figure S1).
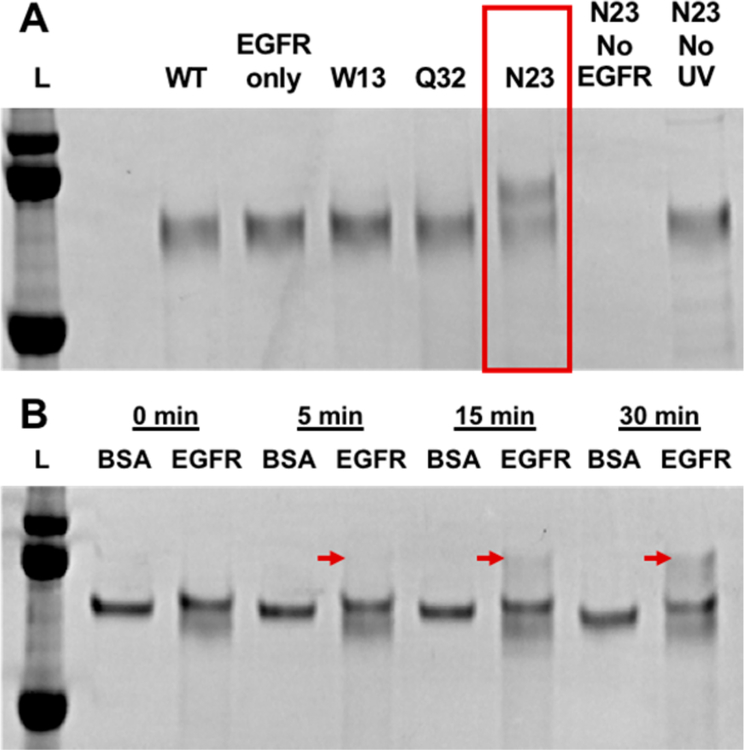
Each BP-modified affibody was next screened for its ability to photo-cross-link to purified EGFR by denaturing sodium dodecyl sulfate (SDS) polyacrylamide gel electrophoresis (PAGE). For this, purified EGFR extracellular domains were mixed with excess WT and BP-modified affibodies and irradiated under low intensity (890 μW/cm2) 365 nm light for 5–30 min, which is well within recommended limits.28 Successful photo-cross-linking of affibody to EGFR was determined by detecting the presence of a higher molecular weight band corresponding to the combined weight of both EGFR (70 kDa) extracellular domain and affibody (10 kDa) in the SDS-PAGE gel. Out of all the different BP-modified mutants tested, only a single mutant, with a cysteine substituted at position N23 (N23BP), was capable of photo-cross-linking to EGFR (Figure 1A). Little to no photo-conjugation was observed for unmodified EGFR binding affibodies (WT) or affibodies that possessed benzophenone (BP) at alternate amino acid sites (Figure S2). Neither increased irradiation time nor affibody concentration improved photo-cross-linking for N23BP or any other mutant. In additional control tests, the N23BP affibody did not show cross-linking to other proteins such as bovine serum albumin (Figure 1B), demonstrating both specificity for the intended cell receptor and its possible utility in vivo. In testing varying irradiation times, cross-linking between EGFR and N23BP affibody was observed to occur within 15 min. Affibodies containing BP at different amino acid sites may not have shown photo-cross-linking to EGFR because the BP group inhibited affibody-EGFR binding or the BP group was not properly oriented relative to EGFR once bound. BP is also capable of cross-linking to the affibody itself once it absorbs a photon, so it is possible that some mutants may have formed intramolecular cross-links rather than to EGFR.
Figure 1.
(A) Denaturing polyacrylamide gel electrophoresis (SDS-PAGE) showing photo-cross-linking products of the affibody as indicated. All samples were treated with PNGase to degloycosylate and visualize EGFR extracellular domain (∼70 kDa) as a distinct band. Each sample contained both an affibody and EGFR fragment unless otherwise indicated. Note that only the N23BP mutant gave a photoproduct, which corresponded to a mass increase of 10 kDa. Ladder proteins are (top to bottom) 100, 75, and 50 kDa. (B) N23BP and EGFR mixed as in (A) or with the equimolar concentration (260 nM) bovine serum albumin (BSA) as indicated and irradiated for the time listed. A band corresponding to the EGFR-affibody conjugate appears at 5, 15, and 30 min (red arrows). Ladder proteins are (top to bottom) 100, 75, and 50 kDa.
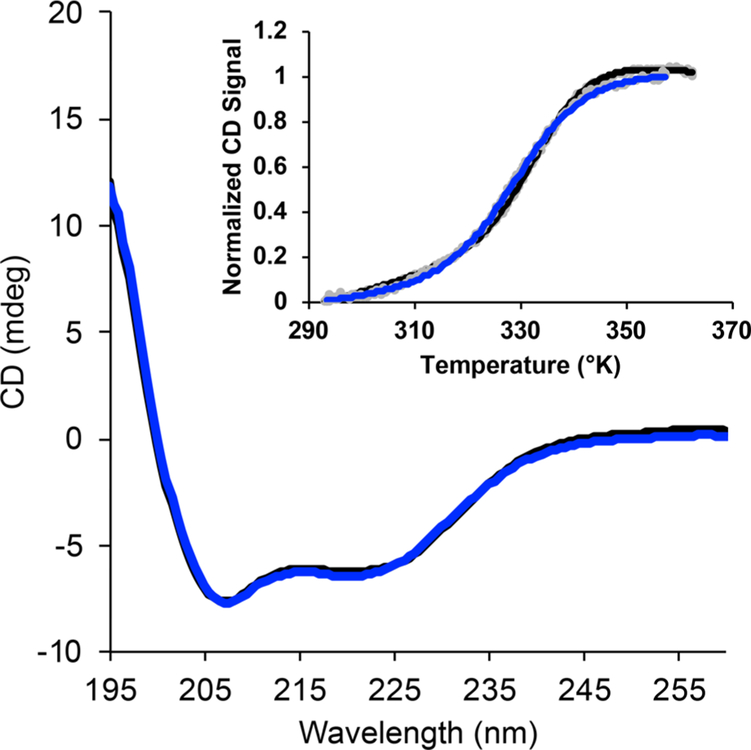
The potential effect of amino acid mutation and BP conjugation on affibody affinity and overall stability was determined next. First, microscale thermophoresis (NanoTemper) was used to measure the affinity of the N23BP affibody to purified EGFR and compared to WT affibody (Figure S3). This technique detects a binding event between two biomolecules through the change in their mobility within a thermal gradient. Pure EGFR extracellular domain was labeled with Alexa Fluor 647 (Thermo Fisher) and mixed with affibody at concentrations ranging from 10 μM to 300 pM. The relative mobility of the EGFR molecule was then measured for each solution and plotted to create a binding curve. From this method, the dissociation constant (KD) between EGFR and WT affibody was measured to be 7.12 ± 3.50 nM, in good agreement with the reported value of 2.8 nM.9 The KD of the N23BP mutant for EGFR was found to be approximately 1–2 orders of magnitude higher than for WT; an exact affinity measurement was hindered by the apparent formation of higher order oligomers between both WT and N23BP affibodies and purified EGFR extracellular domain at μM concentrations of affibody. To determine if mutating the affibody caused any changes to secondary structure, temperature-dependent circular dichroism (CD) studies were also performed with N23BP and compared to WT (Figures 2 and S4). While N23BP showed a possible slight decrease in CD signal at room temperature, potentially due to the achiral nature of the BP group, the N23BP mutant demonstrated only a slightly decreased apparent melting temperature (330.2 ± 0.5 K vs 334.0 ± 1.7 K). These studies show that, while mutating at site N23 and attaching BP did reduce the affibody’s overall affinity, it did not cause significant structural changes.
Figure 2.
Circular dichroism spectra of both the WT (black) and the N23 mutant conjugated with maleimide-benzophenone (blue). Inset: Melting curves showing the normalized circular dichroism signal at 222 nm versus temperature for both WT (black) and N23BP (blue) affibodies.
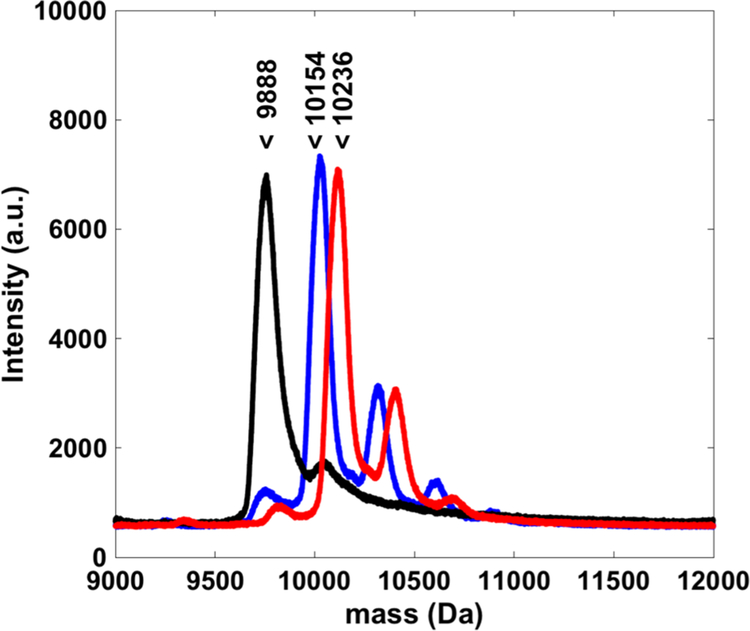
Because one of the hypotheses of this work is that photo-cross-linking would allow increased retention in tumors, in vitro studies were pursued next. In order to establish comparisons for these studies, a benzophenone-containing parent Z domain affibody was prepared as a control affibody that can undergo photo-cross-linking but cannot bind to EGFR. To produce the BP containing Z domain, a cysteine was inserted at position Q32 and reacted with maleimide-benzophenone (BP). In earlier work, these BP containing Z domains have been shown to cause photo-cross-linking between the Z domain and its natural binding partner, antibody Fc domains.27 As with previous affibody mutants, the BP modified Z domains were validated via mass spectroscopy (Figure 3). Finally, to facilitate further studies, all three affibodies (WT, N23BP, and Z) were conjugated with an NHS-activated Rhodamine dye. After removing unbound dye via desalt column, the degree of labeling was determined to be approximately 1 to 1 by UV–vis spectroscopy (Figure S5), and addition of rhodamine did not affect KD of the affibody for EGFR (Figure S6).
Figure 3.
Overlaid MALDI-MS spectra of affibodies (black, WT; blue, N23BP; red, Z) after conjugation with maleimide-benzophenone. Cysteine-containing affibodies show near complete conjugation with one maleimide-benzophenone, and smaller peaks show a second conjugation, likely with a lysine residue. Labels indicate the expected mass of each affibody.
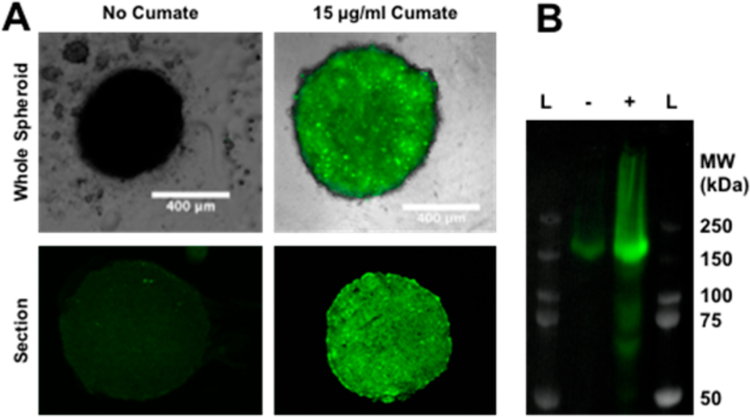
To validate the retention of the photo-cross-linkable affibodies in EGFR-expressing tumors, we next sought to produce 3D tumor spheroid models that could express tunable amounts of EGFR. 3D tumor spheroids were used instead of standard 2D plated cells because spheroids would be expected more closely mimic the morphology and cellular state of cancer cells in vivo, which would affect the availability of EGFR for dictating accumulation in tumor cells. For this, we started with 4T1 mouse mammary carcinoma cells, which have been shown to readily form tumor spheroids in polyhydroxyethyl methacrylate (polyHEMA) coated round-bottom wells.29 However, these 4T1 cells did not produce detectable amounts of EGFR as determined by Western blot with an anti-EGFR antibody, so to induce EGFR expression, these 4T1 cells were next transfected with an inducible human EGFR expression cassette (PBQM812A-1, System Biosciences) in which EGFR is expressed as a fusion to green fluorescent protein (GFP) through a self-cleaving linkage. Coexpressing EGFR with GFP allowed correlation of the amount of EGFR present per spheroid in subsequent studies. Transfected cells were selected with puromycin, induced with cumate and fractionated via fluorescence assisted cell sorting (FACS). As shown in Figure 4, the transfected 4T1 cells were successfully able to form spheroids within a round-bottomed 96-well plate coated with a polyHEMA (see Materials and Methods). As shown in Figure 4 spheroids grown under inducing conditions also showed significantly higher levels of GFP expression throughout and were further analyzed by running a Western blot to detect approximately 5-fold higher levels of EGFR.
Figure 4.
(A) Top: Composite brightfield and fluorescence microscopy images of spheroids formed from transfected 4T1 cells grown either with (right) or without (left) 15 μg/mL cumate to induce EGFR and GFP expression. Bottom: Fluorescence images of cryotome-sectioned similarly prepared spheroids showing the distribution of fluorescence throughout the interior. All images were captured and viewed under identical settings for comparison. (B) Western blot for EGFR expression of the combined lysates of five spheroids grown either with (+) or without (−) 15 μg/mL cumate. Image shows composite overlay of both bright field imaging for display of prestained protein ladders (L) and chemiluminescence imaging for the anti-EGFR antibody. The molecular weight of each ladder band is indicated to the right.
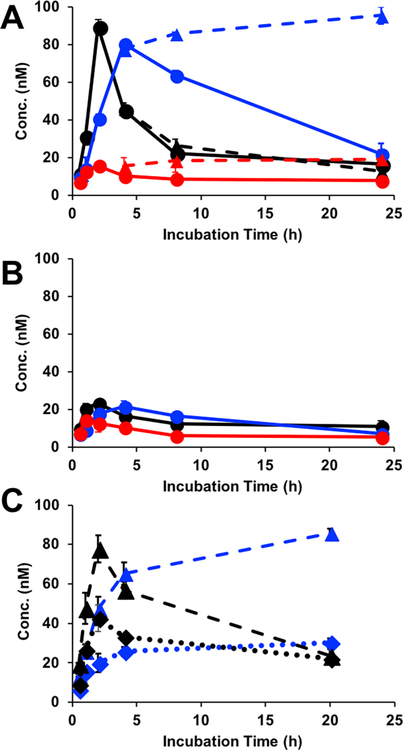
Next, 3D tumor spheroid models were grown with and without cumate in the media and then challenged with one of the three Rhodamine-labeled affibodies described above (WT, N23BP, or Z). After various times of incubation ranging from 0.5 to 24 h, the media was removed, and the spheroids were washed briefly with PBS and lysed (see Materials and Methods). To determine the amount of affibody diffused and retained per spheroid, fluorescence measurements of the lysate were taken at each time point. As shown in Figure 5B, when spheroids were not induced with cumate to express EGFR, little affibody was detected within the spheroids. In contrast, cumate-induced spheroids showed the uptake of both WT and N23BP affibodies over a 2–4 h incubation (Figure 5A). Interestingly, despite its lower affinity, the N23BP mutant reached a similar peak concentration as WT affibody, albeit after a slightly delayed time. After this peak concentration was reached, the concentration of both affibodies began to decrease, which we hypothesize could be due to affibody internalization, subsequent degradation, and release of the Rhodamine dye from the spheroid. As expected, since the Z domain shows no binding to EGFR, little retention in the spheroid was observed.
Figure 5.
Retention of fluorescently labeled affibodies (black, WT; blue, N23BP; red, Z) in 3D tumor spheroids grown from transfected 4T1 cells either (A) induced with 15 μg/mL cumate or uninduced (B). Additional induced spheroids were irradiated with 365 nm light for 30 min after 3.5 h of incubation and again measured for retention (dashed lines, triangles) and compared to nonirradiated spheroids (solid lines, circles). Concentrations of retained affibodies were calculated by comparing spheroid lysate fluorescence to standard curves prepared for each labeled affibody. (C) Retention of fluorescently labeled affibodies (black, WT; blue, N23BP) with (dotted line, diamonds) or without (dashed line, triangles) addition of EGF.
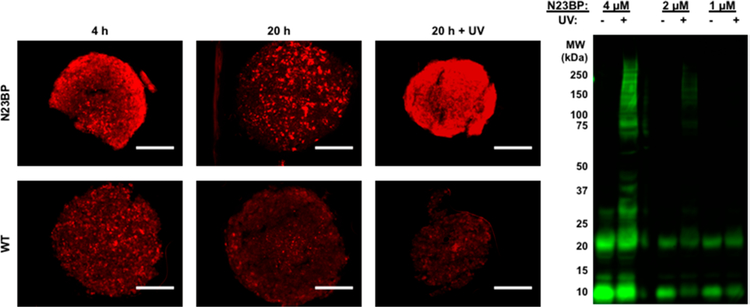
Next, having established the time at which affibodies had diffused into the spheroids, these studies were repeated with 30 min of irradiation with 365 nm light. As shown in Figure 5A, these studies demonstrated that, while irradiation did not significantly change the behavior of WT affibody, the N23BP mutant not only showed retention within the spheroid but also continued to accumulate slowly within the spheroid after irradiation. Spheroids treated with the N23BP affibody and irradiated were further lysed and analyzed via Western blot using an anti-T7 primary antibody to visualize the N23BP photoconjugates. As shown in Figure 6, several bands near the expected molecular weight of the affibody EGFR conjugate were observed with increasing concentrations of N23BP. To further validate the specificity of this study, the experiment was rerun using free epidermal growth factor mixed in a 1:1 ratio with affibody. As a result of EGF addition, the retention of N23BP after 20 h was similar to the WT (Figure 5C). In fact, the retention profile was similar to nonbinding Z domain (Figure 5A), thereby confirming the role of affibody-EGFR interactions in retention in spheroids. Together, these experiments demonstrated the specificity of the photo-cross-linking reaction, even within the complex cellular environment. To determine the distribution of each affibody throughout the spheroid, and how this may be affected by photo-cross-linking, individual spheroids were next sectioned and imaged for both GFP (which correlates to EGFR expression) and Rhodamine (affibodies). As shown in Figure 6, both WT and N23BP affibodies were observed to diffuse throughout the spheroid within 4 h but were largely gone after 20 h. However, irradiation at 3.5 h caused the N23BP mutant to be highly retained throughout the spheroid, even after 20 h total. These results not only support the bulk fluorescence measurements but also demonstrate the ability of the BP-modified affibody to diffuse quickly throughout this tumor model similarly to WT but allow it be to retained for much longer after irradiation.
Figure 6.
Left: Spheroids were treated with 1 μM N23BP (top) or WT (bottom) affibody for a total of 4 h (left column) and 20 h (middle and right columns) and additionally irradiated with 365 nm light for 30 min after 3.5 h incubation (right column). These spheroids were sectioned at approximately the same depth and imaged for Rhodamine and GFP distribution. Micrographs show Rhodamine signal within each section normalized by the average GFP intensity. Scale bars are 200 μm. Right: Five spheroids were grown and irradiated for 1 h with the indicated N23BP affibody concentration in growth media. Affibody containing media was removed, and spheroids were lysed and loaded onto a PAGE gel and probed for affibody conjugates using an anti-T7 antibody. High molecular weight bands around the expected molecular weight of EGFR were observed only when the spheroid-affibody mixtures were irradiated, indicating photo-cross-linking.
A possible explanation for the enhanced retention results could be a combination of binding to membrane-bound EGFR and internalization, along with affibody diffusion between the cells comprising the spheroid. It is likely that internalization is the dominant process because, although binding and diffusion should both increase the amount of retained affibody, neither process should reduce affibody retention. In contrast, affibodies have been shown to be internalized and then secreted with a similar time scale as observed here.15 To quantify the amount of affibody retained within individual cells, the above experiment was repeated with induced cells grown as an adherent monolayer. These cells were similarly incubated with media containing the labeled affibodies for various amounts of time, with and without irradiation. After washing and lysis, the retention in 2D cell culture followed a similar trend to that in spheroids (Figure 7A). Taken together, these experiments suggest that photo-cross-linking may either prevent internalization of the affibody after covalently conjugating to the cell receptor or, if internalized, may prevent affibody secretion.
Figure 7.
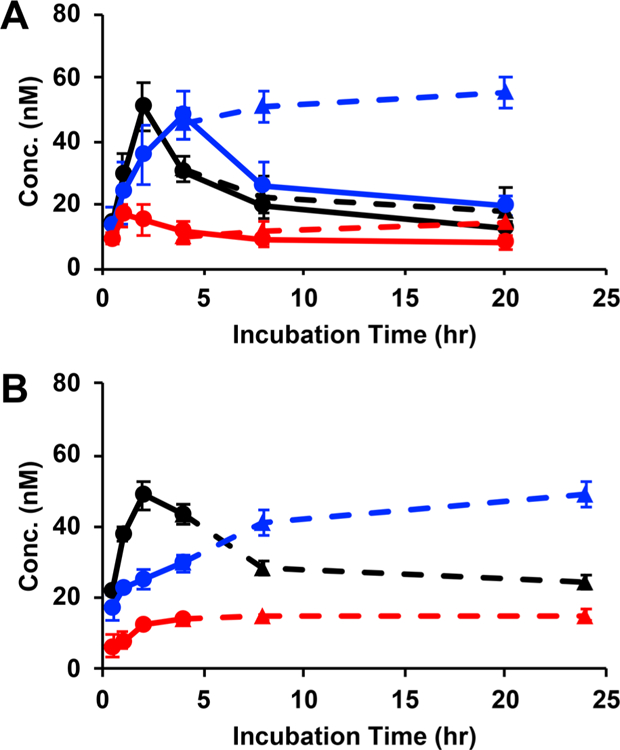
Retention of fluorescently labeled affibodies (black, WT; blue, N23BP; red, Z) in either (A) transfected 4T1 cells induced with 15 μg/mL cumate or (B) MDA-MB-468 cells, each grown as an adherent monolayer of cells. Wells were either irradiated after 3.5 h of incubation for 30 min with 365 nm light (dashed lines, triangles) or kept in the dark (solid lines, circles). Concentrations of retained affibodies were calculated by comparing spheroid lysate fluorescence to standard curves prepared for each labeled affibody.
In addition, to show that enhanced affibody retention was not specific to the transfected 4T1 cell line, the retention experiment was repeated in the human mammary carcinoma cell line MDA-MB-468 in 2D culture. As before, irradiation allowed for increased retention of the N23BP mutant compared to the two control affibodies (Figure 7B), demonstrating the wide applicability of this approach to cells that overexpress EGFR.
Finally, the affibodies were used in conjunction with upconverting nanoparticles (UCNPs) to improve their relevance for clinical applications. While the UV conditions used in these experiments did not appear to cause significant cell toxicity (Figure S7), UV light is readily absorbed and scattered by living tissue. In contrast, near-infrared (NIR) wavelengths can penetrate up to several cm past the skin.30 However, photoreactions that operate at NIR wavelengths are uncommon. Therefore, in this work UCNPs were used to convert incident NIR light to either UV or visible light via a multiphoton absorption mechanism, thereby creating a local source of photons that can induce a photo-cross-linking reaction between BP and EGFR. First, UCNPs were composed of NaYF4 doped with Yb and Tm to promote upconversion from NIR to UV.31–33 Following synthesis, the oleylamine ligands were exchanged to poly(ethylene glycol) to facilitate dispersibility in aqueous media. The resultant nanoparticles had an average diameter of ∼35 nm as measured by TEM (Figure S8), and excitation at 980 nm produced emission peaks at 362 and 475 nm (Figure S9). In particular, the emission wavelength matched well with the absorption range of BP (Figure S5). The ability of UCNPs to facilitate N23BP photo-cross-linking with EGFR was tested by incubating each component in PBS, then irradiating with 980 nm laser light for 30 min, followed by analysis by SDS-PAGE. As shown in Figure 8, the combination of N23BP and 980 nm laser light resulted in the formation of a dark band at the expected molecular weight of EGFR, whereas without irradiation or with WT (no BP), each lane in the gel was similar to background.
Figure 8.
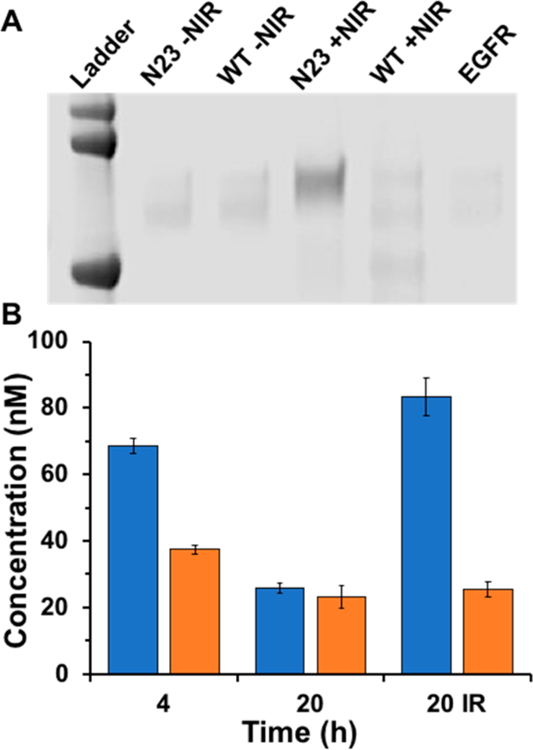
(A) Denaturing polyacrylamide gel electrophoresis (SDS-PAGE) showing photo-cross-linking products of either N23BP or WT to EGFR, with or without irradiation at 980 nm. Note that only 980 nm irradiation of N23BP produced a photoproduct significantly different than free EGFR (right lane). Ladder proteins are (top to bottom) 100, 75, and 50 kDa. (B) Retention of fluorescently labeled affibodies (left, N23BP; right, WT) in 3D tumor spheroids grown from transfected 4T1 cells either induced with 15 μg/mL cumate. Concentrations of retained affibodies were calculated by comparing spheroid lysate fluorescence to standard curves prepared for each labeled affibody. “20 IR” designates that this sample was irradiated at 980 nm after 3 h incubation, then left to grow to a total of 20 h. Lanes without an IR designation were not irradiated.
To validate the ability of UCNPs to facilitate photo-cross-linking and retention in tumor spheroids, the UCNPs were combined with either rhodamine-labeled N23BP or rhodamine-labeled WT affibody. As before (Figure 5), without irradiation, the N23BP affibodies showed high uptake at 4 h but little retention after 20 h. If instead the N23BP affibodies were irradiated with 980 nm laser light for 30 min, the retention after 20 h was >3-fold higher than without irradiation (Figure 8). For WT affibodies without BP, no difference in retention was observed with or without irradiation at 980 nm. Thus, UCNPs facilitated the photo-cross-linking of the N23BP affibodies to EGFR-expressing spheroids using a clinically relevant 980 nm wavelength.
CONCLUSION
In conclusion, we have demonstrated a method to engineer affibodies that can bind to EGFR and form covalent cross-links upon irradiation with 365 nm light. The photo-cross-linkable EGFR binding affibodies (N23BP) showed high specificity for the correct substrate and were able to covalently conjugate to the cell receptor in complex media. Affibodies that did not possess either a benzophenone group or affinity to EGFR showed no photo-cross-linking. In testing the affibodies in 3D tumor spheroids, both the native and BP containing afffibody perfused into the spheroids, but only the photo-cross-linkable affibodies were retained for periods up to 24 h. These results were also confirmed by sectioning. Western blot analysis showed that the photo-cross-linkable affibodies did indeed conjugate to the correct cell receptor demonstrating specificity in a tumor matrix. Lastly, we showed that these photo-cross-linkable affibodies can generally be targeted to cancer cells or tumors that overexpress EGFR.
MATERIALS AND METHODS
Production of Photo-Cross-Linkable EGFR-Targeted Affibodies.
The coding sequence for the EGFR binding affibody designated as ZEGFR:1907 or the nonbinding parent Z domain was synthesized by Integrated DNA Technologies and ligated into the pET21b+ vector, attaching an N-terminal T7 epitope tag and C-terminal 6xHis tag.9 The ZEGFR:1907 construct was further mutated via the Q5 site directed mutagenesis kit (New England Biolabs) to substitute a single codon for cysteine at one of several locations within the affibody sequence. These locations included K4, F5, W10, W13, E15, R17, N23, W25, A29, I31, Q32, and D36.
Recombinant Expression in E. coli.
Expression of affibodies was done by transforming these constructs into a BL21 (DE3) strain of E. coli. Transformants were selected on Luria broth (LB) agar plates containing 100 μg/mL ampicillin and grown overnight in 5 mL of LB containing the same. This culture was diluted 1:100 the following day into 50 mL of ampicillin-containing (Sigma-Aldrich) LB and grown in a 250 mL Erlenmeyer flask until the OD600 was approximately 0.8, at which point affibody expression was induced by adding 1 mM IPTG. After inducing for 3 h, cells were pelleted by centrifuging at 10,000g for 5 min, the media was removed, and the pellet was stored at −20 °C until the following day. Cells were lysed by resuspending in 25 mL of equilibration buffer (PBS, pH 7.4 with 5 mM imidazole) followed by probe sonication on ice. Lysates were centrifuged at 12,000g for 20 min, after which the affibody-containing soluble fraction was removed and reacted with 1 mM TCEP and 25 μL of sedimented Ni-NTA conjugated agarose beads (Thermo Fisher) for 30 min at RT in an end over end mixer. Beads were sedimented by centrifuging at 700g for 2 min and were washed by resuspending in wash buffer (PBS pH 7.4, with 25 mM imidazole and 1 mM TCEP) and vortexing 4 times. Finally, elution buffer (PBS pH 7.4, with 25 mM imidazole and 1 mM TCEP) was incubated with beads for 10–15 min in end over end mixer. Typical affibody yields were approximately 1 mg of protein per 50 mL of culture (200 μL at ∼5 mg/mL).
Synthesis of Affibodies Conjugated to Maleimide-Benzophenone.
Newly purified affibodies were immediately quantified using a NanoDrop Spectrophotometer (NanoDrop Lite, Thermo Fisher), diluted to 50 μM in PBS pH 7.4 and reacted with approximately a 20:1 molar excess of 4N-maleimido-benzophenone (Sigma-Aldrich). This was done by first dissolving the maleimide-benzophenone in DMSO at 10 mM and then mixing this solution at a 1:9 ratio with the 50 mM affibody solution. This mixture was then reacted in the dark overnight at RT, after which excess maleimide-benzophenone was removed through a centrifugal desalt column (Zeba, Themo Fisher).
Testing for Photo-Cross-Linking in Solution.
Affibodies were diluted to a concentration of 33 μM with 260 nM EGFR extracellular domain (Thermo Fisher) in a small volume of 7.5 μL of PBS and irradiated under a near UV (365 nm) lamp at 850 μW/cm2 for 1 h. The photo-cross-linked products were then denatured with heating and SDS and enzymatically deglycosylated with PNGase F (New England Biolabs) at 37 °C for 2 h. After this, samples were mixed with LDS sample buffer, loaded onto 4–12% Bis-Tris polyacrylamide gels (Thermo Fisher), and electrophoresed through the gel for 35 min with 200 V and MES based running buffer (50 mM MES, 50 mM Tris Base, 0.1% SDS, 1 mM EDTA, pH 7.3). The gel was then washed and stained with Coomassie dye for total protein content (Simple Blue Safe Stain, Thermo Fisher) and imaged with a Typhoon FLA 9500 scanner (GE Healthcare).
Microscale Thermophoresis Measurements.
Purified human EGFR extracellular domain (Sino Biologics) was conjugated at a 20:1 ratio with NHS active ester of Alexa Fluor 647 dye (Thermo Fisher) leading to an approximate 1:1 labeling ratio as determined by UV–vis. Sixteen low binding micro tubes were used to mix labeled EGFR at 10 nM with various concentrations of either N23BP or WT unlabeled affibodies in PBS buffer containing 0.025% Tween20 (NanoTemper). These mixtures were then loaded into capillaries and the thermophoresis signal from each was measured using a microscale thermophoresis instrument (NanoTemper). The resulting MST signals were analyzed with respect to affibody concentration and fit to a one to one binding model.
Circular Dichroism (CD) Measurements.
Affibodies were diluted to 10 μM in PBS pH 7.4. For each affibody, the CD signal was monitored at 222 nm using an Applied Photophysics Chirascan Plus CD instrument. Measurements were obtained as temperature was raised from 20 to 90 °C at 2 °C min−1 and monitored within the sample with a small thermocouple probe. Each experiment was repeated several times.
Transfection of 4T1 Mouse Carcinoma Cells for Inducible EGFR Expression.
4T1 cells were procured from the American Type Culture Collection (ATCC) and grown in 75 cm2 culture flasks to 80% confluence in Dulbecco’s Modified Eagle Media with high glucose and glutamine (Thermo Fisher). Cells were then dispersed by removing media, washing briefly with Dulbecco’s PBS and treating with trypsin for 15 min. A modified version of the PBQM812A-1 vector (System Biosciences) was used to insert a cumate inducible expression cassette, along with a puromycin resistance selection marker, into the 4T1 genome using the piggyBAC transposon system. This vector was modified to induce expression of a His-tagged human EGFR fused via to a T2A self-cleaving linker to GFP at its C-terminus. This vector was first complexed with lipofectamine (Thermo Fisher) before being added to the 4T1 cells in suspension. These transfected cells were then plated on 75 cm2 flasks and selected for with puromycin (Puromycin Dihydrochloride, Thermo Fisher) starting at 1 μg/mL for several passages and ending with 2 μg/mL. Transfected cells were additionally induced via 15 μg/mL cumate (Sigma-Aldrich) added to the culture media and then sorted for GFP expression via FACS. A subpopulation consisting of the top 5% most fluorescent cells were used in all future experiments.
Preparation of 4T1 Spheroids.
A stock solution of 30 mg/mL poly hydroxyethyl methacrylate (poly-HEMA, Sigma-Aldrich) was prepared in 95% ethanol. Then 200 μL of this solution was added to each of the wells of a round-bottom, 96 well plate, followed by removal of the ethanol by air drying. The transfected 4T1 cell was cultured at 37 °C with 5% CO2 and DMEM media (High Glucose, Phenol Red, without Sodium Pyruvate, Thermo Fisher) supplemented with 10% fetal bovine serum (FBS, Gibco) and 1% penicillin-streptomycin (Sigma-Aldrich). After the cells reached 80% confluence, the cells were passaged and 5000 of these cells in 200 μL of the same growth media were added to the precoated 96 well plate. The plate was then incubated for an additional 2–3 days to allow the cells to grow into spheroids. Where indicated in the text, 15 μg/mL (final concentration) cumate (Sigma-Aldrich) was added to the spheroid growth media to induce EGFR expression.
Measurement of Affibody Retention in 4T1 Spheroids by Photoluminescence.
Prior to experimentation, affibodies were conjugated with a 20:1 molar excess of NHS-Rhodamine (Thermo Fisher) for 1 h at RT in PBS containing 0.1% (v/v) Tween. Excess Rhodamine was then removed via desalt columns (Thermo Fisher), and the conjugation was assessed by UV–vis spectroscopy.
After spheroids from 4T1 cells were grown for 2 days, the growth media was carefully removed from the wells and replaced with 1 μM of each of the rhodamine conjugated affibodies diluted in fresh growth media. Spheroids were incubated with the labeled affibodies in the CO2 jacketed incubator at 37 °C for 0.5, 1, 2, 4, 8, and 24 h. At each time point, the growth media was removed, spheroids were washed twice with PBS for 5 min each, then 200 μL of lysis buffer (0.1% (v/v) Triton X100 in Tris Buffered Saline) was added, and the solution was vortexed. Absorbance measurements were acquired of the homogeneous lysis solution. For photo-cross-linking studies, the spheroids were irradiated in the affibody containing growth media after 3.5 h incubation for 30 min using a 365 nm lamp with a power output of 850 μW/cm2. The rest of the protocol was the same as above. The fluorescent intensity of the resultant solution was measured from each of the wells by a Tecan Safire II Multimode Plate Reader.
EGF Inhibition Assay.
4T1 spheroids from 5000 cells/well were prepared as described above in 96-well polyHEMA-coated round bottomed plates. Then 1 μM of Rhodamine-conjugated affibody (both WT and N23C) and EGF were incubated with the spheroids in 100 μL of media for different time points (0.5, 1, 2, 4, and 20 h) at 37 °C. After 3.5 h, half of the 4 and 20 h samples were irradiated with UV for 30 min and half were kept in the dark as controls. At each time point, spheroids were washed for 5 min with 100 μL of PBS and then lysed using 200 μL of lysis buffer (150 mM NaCl, 50 mM Tris Base, and 0.1% (v/v) Triton X). The fluorescence from the lysate was then measured at 572 nm on a Spectra Max (Molecular Devices) plate reader. All experiments were performed in triplicate.
Cell Toxicity to UV Irradiation.
4T1 cells were plated in a 96-well plate at approximately 2000 cells/well. The wells were incubated with 1 μM affibody (N23BP, WT, or Z) or no affibody at 37 °C. After 3.5 h, half of the wells were irradiated with UV for 30 min, and the plate was left overnight at 37 °C. The media was then aspirated from each well, cells were washed once with PBS for 5 min, and 80 μL of fresh media was added along with 20 μL freshly prepared 5 mg/mL of 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT, Biosynth). After 4 h, media and MTT were removed carefully without disturbing the crystals, 100 μL of isopropyl alcohol was added to the wells, and the plate was left overnight on a shaker. Absorbance was measured the next day at 570 nm, and absorbance readings were scaled against control wells without either affibody or UV as a measure of 100% viability. Six wells were used for each sample (n = 6).
Synthesis of Upconverting Nanoparticles (UCNPs).
For this purpose, 0.1562 g of YCl3, 0.0503 g of YbCl3, and 0.0055 g of TmCl3 were mixed in 3 mL of oleic acid and 17 mL of octadecene in a three-neck round-bottom flask. The mixture was then heated to 160 °C under an argon atmosphere with continuous stirring to form a homogeneous solution, after which it was allowed to cool to RT. Then 10 mL of methanol containing 0.1 g of NaOH and 0.148 g of NH4F was added dropwise, and the reaction was allowed to proceed for another 40 min. The methanol was then vaporized by maintaining the solution at 110 °C for 20 min. The flask was then refilled with argon and degassed three times further. After this, the temperature of the mixture was raised quickly to 300 °C and allowed to stir at reflux under a water-jacketed condenser for 2 h. The solution was then cooled to RT, and the UCNP’s were precipitated with ethanol by centrifugation at 9000 rpm and then washed three times with 1:1 ethanol and water. The UCNP’s were then suspended in toluene to final concentration of 30 mg/mL.
UCNP Ligand Exchange.
A 30 mg/mL suspension of UCNPs in toluene was dried at 70 °C overnight. Next 15 mg of the dried UCNPs was then dissolved in 500 μL of chloroform. Then 15 mg of C18-PEG-NH2 was dissolved in 1 mL of water, and this solution added dropwise to the UCNP-chloroform suspension. The mixture was stirred at RT overnight while exposed to the surroundings to evaporate the chloroform, and the amine-UCNPs were then collected by centrifugation.
UCNP-Assisted Photo-Cross-Linking of Affibodies to EGFR.
A mixture was prepared containing final concentrations of 60 μM affibody (N23C-BP or WT), 260 nM EGFR extracellular fragment, and 100 μg/mL UCNP. Each mixture was irradiated with a 980 nm laser diode (OEM, CW) of 10 kW/cm2 with a spot size of 1 mm for 30 min. A total of 10 μL of each sample was withdrawn and kept at 70 °C for 10 min to denature the proteins, followed by running these samples by sodium dodecyl sulfate (SDS) polyacrylamide gel electrophoresis (PAGE) and followed by visualization with Coomassie stain (Thermo Fisher).
UCNP-Assisted Affibody Retention in Tumor Spheroids.
GFP-EGFR expressive 4T1 spheroids in a 96-well plate were mixed with either 1 μM Rh-conjugated N23C-BP or Rh-WT affibody and 100 μg/mL of UCNP. After 3 h, each sample was irradiated with 980 nm laser diode (OEM, CW) of 10 kW/cm2 with a spot size of 1 mm for 30 min, after which the samples were left overnight at 37 °C. At the respective time points, the spheroids were washed with PBS for 5 min and lysed in 200 μL of lysis buffer. The fluorescence in the lysis buffer was measured using the plate reader at 572 nm, and affibody concentration was determined by comparison to a calibration curve.
Supplementary Material
ACKNOWLEDGMENTS
Research was primarily supported by NIH DP2EB020401 and NSF DMR 1420736. The Nanotemper Microscale Thermophoresis instrumentation was supported by the instrumentation grant NIH S10OD21603. We would like to thank Scott A. Stewart and the Ahn lab at University of Boulder, Department of Chemistry and Biochemistry for donating the vector used for transfecting 4T1 cells with inducible EGFR expression. Dr. Douglas Chapnick and Prof. Xuedong Liu provided helpful discussions. We would also like to thank Profs. Amy Palmer and Jerome Fox for use of their plate readers. Finally, we would also like to thank the Biochemistry instrument core for the use of their Applied Photophysics Chirascan Plus CD instrument.
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/jacs.8b07601.
Mass spectrometry, gel electrophoresis showing photo-cross-linking to pure EGFR, melting temperature curves, and UV–vis spectra of rhodamine conjugated N23BP (PDF)
The authors declare no competing financial interest.
REFERENCES
- (1).Rudnick SI; Adams GP Cancer Biother.Radiopharm 2009, 24 (2), 155–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Yan L; Ehrlich PJ; Gibson R; Pickett C; Beckman RA mAbs 2009, 1 (1), 67–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Adams GP; Schier R; McCall AM; Simmons HH; Horak EM; Alpaugh RK; Marks JD; Weiner LM Cancer Res 2001, 61 (12), 4750–4755. [PubMed] [Google Scholar]
- (4).Tabrizi M; Bornstein GG; Suria H AAPS J 2010, 12 (1), 33–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Nygren P-A FEBS J 2008, 275 (11), 2668–2676. [DOI] [PubMed] [Google Scholar]
- (6).Löfblom J; Feldwisch J; Tolmachev V; Carlsson J; Ståhl S; Frejd FY FEBS Lett 2010, 584 (12), 2670–2680. [DOI] [PubMed] [Google Scholar]
- (7).Tolmachev V; Tran TA; Rosik D; Sjöberg A; Abrahmsén L; Orlova AJ Nucl. Med 2012, 53 (6), 953–960. [DOI] [PubMed] [Google Scholar]
- (8).Friedman M; Nordberg E; Höidén-Guthenberg I; Brismar H; Adams GP; Nilsson FY; Carlsson J; Ståhl S Protein Eng., Des. Sel 2007, 20 (4), 189–199. [DOI] [PubMed] [Google Scholar]
- (9).Friedman M; Orlova A; Johansson E; Eriksson TLJ; Höidén-Guthenberg I; Tolmachev V; Nilsson FY; Ståhl SJ Mol. Biol 2008, 376 (5), 1388–1402. [DOI] [PubMed] [Google Scholar]
- (10).Tolmachev V; Friedman M; Sandström M; Eriksson TLJ; Rosik D; Hodik M; Ståhl S; Frejd FY; Orlova AJ Nucl. Med 2009, 50 (2), 274–283. [DOI] [PubMed] [Google Scholar]
- (11).Miao Z; Ren G; Liu H; Jiang L; Cheng Z Bioconjugate Chem 2010, 21 (5), 947–954. [DOI] [PubMed] [Google Scholar]
- (12).Miao Z; Ren G; Liu H; Jiang L; Cheng ZJ Biomed. Opt 2010, 15 (3), 036007–036007–7. [DOI] [PubMed] [Google Scholar]
- (13).Jokerst JV; Miao Z; Zavaleta C; Cheng Z; Gambhir SS Small 2011, 7 (5), 625–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Antaris AL; Chen H; Cheng K; Sun Y; Hong G; Qu C; Diao S; Deng Z; Hu X; Zhang B; et al. et al Nat. Mater 2016, 15 (2), 235–242. [DOI] [PubMed] [Google Scholar]
- (15).Nordberg E; Friedman M; Göstring L; Adams GP; Brismar H; Nilsson FY; Ståhl S; Glimelius B; Carlsson J Nucl. Med. Biol 2007, 34 (6), 609–618. [DOI] [PubMed] [Google Scholar]
- (16).Göstring L; Chew MT; Orlova A; Höidén-Guthenberg I; Wennborg A; Carlsson J; Frejd FY Int. J. Oncol 2010, 36 (4), 757–763. [DOI] [PubMed] [Google Scholar]
- (17).Nordberg E; Ekerljung L; Sahlberg SH; Carlsson J; Lennartsson J; Glimelius B Int. J. Oncol 2010, 36 (4), 967–972. [DOI] [PubMed] [Google Scholar]
- (18).Hayashi T; Yasueda Y; Tamura T; Takaoka Y; Hamachi IJ Am. Chem. Soc 2015, 137 (16), 5372–5380. [DOI] [PubMed] [Google Scholar]
- (19).Beuttler J; Rothdiener M; Müller D; Frejd FY; Kontermann RE Bioconjugate Chem 2009, 20 (6), 1201–1208. [DOI] [PubMed] [Google Scholar]
- (20).Rönnmark J; Hansson M; Nguyen T; Uhlén M; Robert A; Ståhl S; Nygren P-ÅJ Immunol. Methods 2002, 261 (1–2), 199–211. [DOI] [PubMed] [Google Scholar]
- (21).Tolmachev V; Rosik D; Wållberg H; Sjöberg A; Sandström M; Hansson M; Wennborg A; Orlova A Eur. J. Nucl. Med. Mol. Imaging 2010, 37 (3), 613–622. [DOI] [PubMed] [Google Scholar]
- (22).Hawe A; Hulse WL; Jiskoot W; Forbes RT Pharm. Res 2011, 28 (9), 2302–2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Schmidt MM; Wittrup KD Mol. Cancer Ther 2009, 8 (10), 2861–2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Hui JZ; Tsourkas A Bioconjugate Chem 2014, 25 (9), 1709–1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Grunbeck A; Huber T; Sachdev P; Sakmar TP Biochemistry 2011, 50 (17), 3411–3413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Chin JW; Martin AB; King DS; Wang L; Schultz PG Proc. Natl. Acad. Sci. U. S. A 2002, 99 (17), 11020–11024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Brasino M; Cha JN Analyst 2017, 142, 91. [DOI] [PubMed] [Google Scholar]
- (28).Council NR Prudent Practices in the Laboratory: Handling and Management of Chemical Hazards, Updated Version; 2011. [PubMed]
- (29).Kuch V; Schreiber C; Thiele W; Umansky V; Sleeman JP Int. J. Cancer 2013, 132 (3), E94–E105. [DOI] [PubMed] [Google Scholar]
- (30).Weissleder R Nat. Biotechnol 2001, 19 (4), 316–317. [DOI] [PubMed] [Google Scholar]
- (31).He L; Dragavon J; Cho S; Mao C; Yildirim A; Ma K; Chattaraj R; Goodwin AP; Park W; Cha JN J. Mater. Chem. B 2016, 4 (25), 4455–4461. [DOI] [PubMed] [Google Scholar]
- (32).He L; Brasino MD; Mao C; Cho S; Park W; Goodwin AP; Cha JN Small 2017, 13 (24), 1700504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Mahalingam V; Vetrone F; Naccache R; Speghini A; Capobianco JA Adv. Mater 2009, 21 (40), 4025–4028. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.