Abstract
Deep learning has shown remarkable improvements in the analysis of medical images without the need for engineered features. In this work, we hypothesize that deep learning is complementary to traditional feature estimation. We propose a network design to include traditional structural imaging features alongside deep convolutional ones and illustrate this approach on the task of imaging-based age prediction in two separate contexts: Tl-weighted brain magnetic resonance imaging (MRI) (N = 5,121, ages 4-96, healthy controls) and computed tomography (CT) of the head (N = 1,313, ages 1 – 97, healthy controls). In brain MRI, we can predict age with a mean absolute error of 4.08 years by combining raw images along with engineered structural features, compared to 5.00 years using image-derived features alone and 8.23 years using structural features alone. In head CT, we can predict age with a median absolute error of 9.99 years combining features, compared to 11.02 years with image-derived features alone and 13.28 years with structural features alone. These results show that we can complement traditional feature estimation using deep learning to improve prediction tasks. As the field of medical image processing continues to integrate deep learning, it will be important to use the new techniques to complement traditional imaging features instead of fully displacing them.
Keywords: Deep Learning, Convolutional Neural Networks, Brain Age, Medical Image Processing
1. Introduction
In a recent special issue, Greenspan et al. reviewed the role of deep learning in medical image analysis, concluding that “In the majority of works presented, use of a deep network is shown to improve over the state-of-the-art. As these improvements seem to be consistent across a large variety of domains, and as is usually the case, development of a deep learning solution is found to be relatively straight-forward, we can view this as a major step forward in the medical computing field.” (1) The authors state that networks excel at tasks of lesion detection, segmentation, registration, and predictive models. A key challenge of deep learning applied to medical imaging in a supervised learning framework, is the need for large training sets with high-quality, expertly labelled features. Generating high-quality labels for medical images, in tasks such as detection, diagnosis, or segmentation, requires expert knowledge which does not scale well to the large number of training examples needed for a robust deep learning algorithm. The authors identify a second key issue with the shifting paradigm towards deep learning: “Can we rely on learned features alone or may we combine them with handcrafted features for the task?”(1).
Advances in deep learning have provided an approach for learning a highly non-linear function of a dataset when an appropriate kernel or feature manifold is not known. Historically, feature extraction in the field of computer vision has relied on automatically detecting intensity patterns or textures in an image. However, recent work on deep convolutional neural networks has shown that an adaptive learning of image features through convolutional filters can results in more accurate results. Unlike many datasets used in computer vision, medical imaging is unique in that there is already extensive a priori expert human knowledge associated with the image, such parcellation of tissues into anatomical or functional units. This knowledge can be formulated as imaging features grounded in medicine and physiology, which can direct computer vision tasks to find better relationships in the data. Decades of work on medical image processing have focused on engineering and refining meaningful features to capture the dimensionality of a small imaging dataset. While deep learning has shown remarkable improvements, it has not been shown whether engineered features are redundant. For example, age prediction from medical imaging is a task that has relied heavily on pre-processed regions of interest (ROIs) from T1w structural brain MRI using the volumes of ROIs such as the white matter, ventricles, and the cortex (2-4). Recently, these measures have become efficiently extractable from a standard T1-weighted (Tlw) brain MRI using a multi-atlas segmentation approach (2).
Recently, Cole and Franke evaluated the clinical utility of predicting brain age from handcrafted imaging features and showed that predicted brain age can be used to better understand differences between individuals during the aging process, understand disease processes, and design treatment strategies (3, 5). The authors pose that the absolute difference between predicted age and chronological age, herein called Brain Age Gap (BAG) biomarker, is a valuable imaging metric, since it has been shown to correlate with aging as well as neurodegenerative diseases (5). A recent study showed a correlation between BAG and mortality in subjects over 73 years old, attributing an increase of 6% to mortality risk for every year predicted older (6). BAG also correlated with common metrics of age such as decreased grip strength, decreased expiratory volume, and slower walking time (6). Moreover, an increased BAG has been shown to correlate with several neurodegenerative diseases like Alzheimer’s disease, bipolar disorder, diabetes, Down syndrome, epilepsy, major depression, mild cognitive impairment, traumatic brain injury, and schizophrenia (3, 5). These clinical correlations suggest common secondary effects on the brain, such as inflammation or oxidative stress (5). Some of the challenges with this technology moving forward include incorporating multimodal data, such as orbital computed tomography (CT) to produce new biomarkers like orbital brain age gap (OrbitBAG), and the use of deep learning, which the authors identify as beneficial due to the “removal [of] the reliance on data pre-processing to extract meaningful features.” (5) Predicting age from neuroimaging may provide a new, noninvasive biomarker of aging as well as a discovery tool for positive and negative effectors on aging.
Some of the best results that predict age report a BAG between 4-5 years (7), but these models have been difficult to generalize due to small sample size (7-9), limited age-range (10, 11), or extensive multimodal data requirements (3, 10, 12). Cole et. al proposed a convolutional deep neural network technique on raw T1w MRI images which showed an mean absolute error of 4.65 years in 2,001 healthy adults ages 18 to 90 years (13). This method showed competitive results in age prediction without any a priori feature extraction or image preprocessing. The work by Cole (13) exemplifies the improved performance of deep networks over handcrafted features. However, it does not explore how using both types of features affect prediction accuracy. Understanding the functional difference between engineered features and machine learning tasks can provide insight into structural and functional changes seen in medical imaging.
Deep learning has shown a remarkable improvement over hand-crafted feature-based learning, but it is still unclear whether deep neural networks capture all the information available from expert features. Deep neural networks learn convolutional filters that minimize an objective loss function, such as mean squared error, but do not enforce specific anatomic of physiological principles present in the image. Conversely, using engineered features based on anatomy or function requires a priori expertise and will necessarily limit the information available in the image to the chosen features. The rationale for merging expert features with deep learning is to leverage the existing knowledge present in expert features to direct learning of the convolutional network towards intensity patterns predictive of age that are not captured in the engineered features. The principal contribution of this method is to show that deep learning can be used to enhance prediction along with engineered features. A strength of deep convolutional networks is the ability to find patterns in the data that are not immediately obvious. Therefore, it is best to leverage this powerful tool to find new patterns instead of enforcing the learning of features that we can already obtain through classical methods.
Herein, we hypothesize that deep learning is complementary to traditional feature estimation. We propose to build upon previously validated network designs to include traditional structural imaging features alongside deep convolutional ones and illustrate this approach on the task of imaging-based age prediction on two separate datasets: T1-weighted brain MRI and CT of the head. We show that deep learning can enhance tasks in medical image processing when learned features are combined with handcrafted features. As the field of medical image processing continues to adopt techniques in deep learning, it will be important to preserve hand-crafted features that enhance the task at hand, instead of replacing the features altogether.
2. Methods
2.1. Imaging Datasets & Preprocessing
The complete MRI cohort aggregates 9 datasets with a total 5,048 T1w 3D images from normal healthy subjects, as curated by (14). This cohort includes subjects marked as controls from nine studies (Table 1). The data include subjects with ages ranging between 4 and 94 years old, with a mean age and standard deviation of 29.1 ± 22.6 years. Of 5,048 subjects, 52.4% were male and 47.6% were female. Data were also acquired from different sites so there is a difference in field strength, of which 77% of scans were acquired at 3 Tesla and 23% were acquired at 1.5 Tesla. ROI volumes, sex, and field strength were all used as input features for age prediction.
Table 1. Demographics for brain MRI cohort per site.
Our study uses brain MRI of subjects marked as healthy controls from nine different sites. Parenthesis indicate number of female subjects.
| Site | Mean Age | Young Age (0 - 30 y/o) |
Middle Aged (30 −50 y/o) |
Older Adult (50 - 96 y/o) |
Site total |
|---|---|---|---|---|---|
| ABIDE | 17.2 ± 7.8 | 523 (95) | 39 (3) | 1 (0) | 563 (98) |
| ADHD-200 | 11.6 ± 3.3 | 950 (367) | - | - | 950 (367) |
| BLSA | 68.1 ± 12.7 | 1 (0) | 61 (31) | 552 (311) | 614 (342) |
| Cutting | 12.5 ± 5.0 | 583 (293) | 3 (1) | - | 586 (294) |
| FCON-1000 | 28.3 ± 13.8 | 823 (469) | 130 (56) | 116 (68) | 1,069 (593) |
| IXI | 48.8 ± 16.4 | 98 (54) | 166 (80) | 259 (162) | 523 (296) |
| NDAR | 11.0 ± 3.8 | 328 (168) | - | - | 328 (168) |
| NKI-Rockland | 33.9 ± 21.5 | 58 (26) | 21 (8) | 24 (14) | 103 (48) |
| OASIS | 45.2 ± 23.8 | 139 (78) | 43 (24) | 130 (93) | 312 (195) |
| Total | 29.1 ± 22.7 | 3503 (1550) | 463 (203) | 1082 (648) | 5,048 (2,401) |
For feature extraction, 45 atlases are non-rigidly registered (15) to a target image and non-local spatial staple (NLSS) label fusion (16) is used to fuse the labels from each atlas to the target image using the BrainCOLOR protocol (17). A total of 132 regional volumes were calculated by multiplying the volume of a single voxel by the number of labeled voxels in original image space. Total intracranial volume (TICV) was calculated using SIENAX (18) and used for volume normalization for a total of 132 raw volumes and 132 normalized volumes.
The cohort of head CT images consists of 1,313 clinically acquired scans as part of a larger study on eye disease. All images were acquired at Vanderbilt University Medical Center (VUMC) with variable imaging protocols and scanners (Table 2). Images were retrieved and deidentified for retrospective study under Institutional Review Board (IRB) approval. Since these subjects were undergoing CT imaging for regular clinical care, it includes patients with eye diseases such as glaucoma, intrinsic optic nerve disease (IOND), optic nerve edema, orbital inflammation, or thyroid eye disease (TED), as well as healthy controls who received imaging but were not clinically diagnosed with these diseases. CT images were normalized to a range between −100 and 200 Hounsfield Units (HU) for optimal visualization of orbital structures. The CT images were processed for structural features of the orbit using the protocol described in (19). The data includes subjects of ages ranging between 1 and 97 years old, with a mean age and standard deviation of 52.1 +/− 20.7 years. This dataset includes healthy control subjects (70.0%) as well as subjects with glaucoma (7.7%), IOND (15.2%), optic nerve edema (14.7%), orbital inflammation (2.5%), or TED (5.0%). Seventy-five structural metrics were extracted using multi-atlas segmentation and used as input variables along with disease classes for each subject, using the method proposed by Harrigan et. al. (19).
Table 2. Demographics for head CT cohort per disease status.
Our study uses clinically acquired head CT from healthy subjects as well as five different eye disease status. Note: Some of these subjects have multiple diagnoses. IOND: Intrinsic optic nerve disease; TED: Thyroid eye disease.
| Cohort | Mean Age | Young Age (0 - 30 y/o) |
Middle Aged (30 −50 y/o) |
Older Adult (50 - 96 y/o) |
Total |
|---|---|---|---|---|---|
| Healthy Control | 56.5 ± 19.5 | 108 | 107 | 651 | 866 |
| Glaucoma | 61.0 ± 18.7 | 8 | 15 | 78 | 101 |
| IOND | 44.6 ± 20.0 | 54 | 64 | 82 | 200 |
| Optic Nerve Edema | 32.0 ± 14.7 | 97 | 75 | 21 | 193 |
| Orbital inflammation | 46.1 ±21.6 | 9 | 10 | 14 | 33 |
| TED | 53.3 ± 14.0 | 2 | 25 | 38 | 65 |
| Total | 52.1 ±20.7 | 242 | 252 | 819 | 1,313 |
2.2. Model Architecture
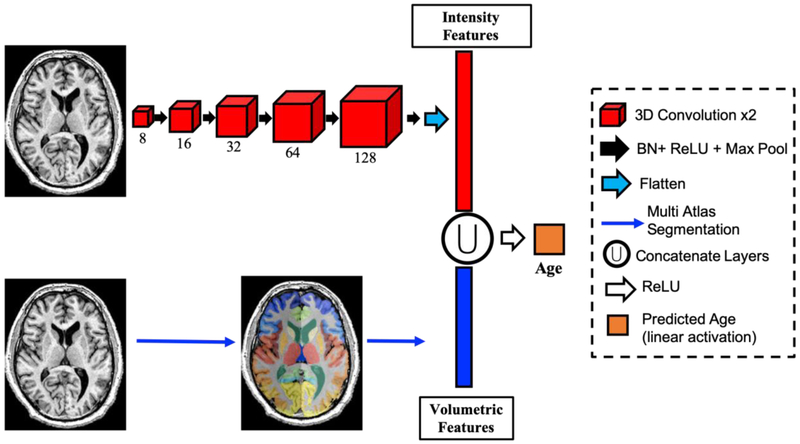
In this work, we adopt the network developed and validated by Cole et. al and extend it to include anatomical features derived from multi-atlas segmentation (13). In Cole’s work, a series of convolutional operations result in a high-dimensional convolutional representation of imaging data, which is then used to directly predict age. In this work, we concatenate the anatomic representation, which consists of volumetric estimates of key ROI’s identified with multi-atlas segmentation. Figure 1 shows the convolutional representation obtained using a 3D convolutional neural network. This consists of 5 layers each with two, 3D convolution operations, ReLU activation, and max pooling; resulting in a representation of the brain imaging with 15,360 features or 3,584 features in the case of orbital CT.
Fig 1.
Pipeline for age prediction. Two sets of features are used: intensity-derived features (red) derived from a convolutional neural network of increasing filter size (red boxes), and structural features (blue) using multiatlas segmentation (bottom). These features are concatenated and used as inputs to directly predict age. BN: Batch Normalization; ReLU: Rectified Linear Unit Activation; Max Pool: Max Pooling Layer.
After concatenation of the convolutional and volumetric features these features undergo ReLU activation and densely connected layer to a single node with linear activation to directly predict age (Figure 1). In addition, we trained two more baseline models for a comparison: 1) the baseline Cole et. al model using our data, and 2) a volumetric features only model, which consisted of two densely-connected layers of 128 nodes. As with previous work, the learning rate was initiated at 0.01 with a 3% decay each epoch. All models were trained using stochastic gradient descent optimization with momentum of 0.9. The loss function was mean absolute error. Training was allowed to continue until the loss function on the validation set did not change by more than 0.1 in 20 epochs. All models were developed using Keras version 2.2.4 with Tensorflow 1.5 and trained on an NVIDIA Titan Graphics Processing Unit (GPU).
2.3. Statistical Analysis
We performed a five-fold cross-validation scheme by withholding 20% of the data for testing while using the remaining 80% for training and validation (70% and 10% respectively). This process was repeated five times until the entire dataset was used for testing only once. Therefore, a total of five networks were trained for each method (ie. volumetric features only, raw image only, or combined) and evaluated on the testing set for each fold. This way, every subject in the data is used as a testing subject only once. All results shown below represent the evaluation of the testing set in each fold grouped together. The BAG biomarker was calculated for all models and compared using a Wilcox signed rank test. We corrected for multiple comparisons using Bonferroni corrections.
Additionally, we test the hypothesis that the BAG biomarker is increased in subjects with existing eye disease compared to healthy controls in the testing set. We tested for significance of the BAG biomarker between each disease group and healthy controls using a linear regression model with disease state as a dummy variable and true age as a covariate. Significance between groups was set to p < 0.05 in the dummy variable.
2.4. Network Visualization with Grad CAM
We use the Gradient Class Activation Maps (Grad CAM) method described by (20) and implemented by the publicly available library keras-vis (21) to visualize the areas of the raw MRI with higher attention in the combined model. We created nine categories based on three age cutoffs for true age and predicted age: young (< 30 years old), middle aged (30 – 50 years old), and older adult (> 50 years old). The nine categories consist of young predicted young, young predicted middle aged, young predicted older adult, middle aged predicted young, middle aged predicted middle aged, middle aged predicted older adult, older adult predicted young, older adult predicted middle aged, and older adult predicted older adult. We randomly selected 10 subjects from each category, calculated the Grad CAM for each and showed the resulting average for each category overlaid on a subject from that sample.
3. Results
In this work, we show that intensity-derived features from a deep convolutional network can enhance learning from structural features by improving the accuracy of age prediction in both brain MRI and head CT datasets.
3.1. Context-Aware Deep Neural Network Best Predicts Age in T1-weighted MRI
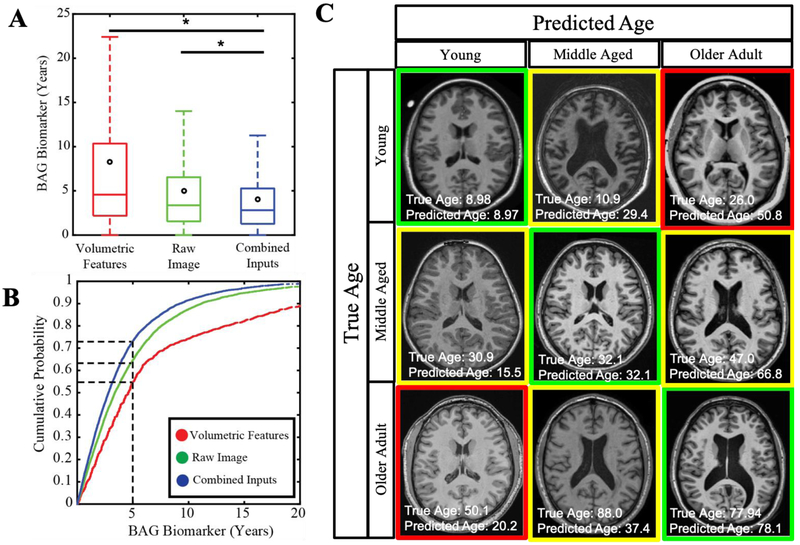
This study investigates the ability to predict age from brain MRI in healthy individuals. We use two sets of inputs to train and validate a fully-connected network: 1) intensity features derived from a convolutional neural network representation, and 2) context features including volumetric estimates for known regions of interest in the brain obtained via atlas-based segmentation. We also introduce sex and scanner field strength with structural features as additional contextual features. Figure 2A shows that intensity-derived features outperform structural features with a mean absolute error of 5.00 vs 8.26 years (p<0.001). However, age prediction improves when both feature sets are used as inputs, resulting in a median absolute of 4.08 years (p<0.001).
Fig2.
Deep Learning improves Age prediction in Brain MRI. Age can be predicted more accurately when using convolutional and structural features on a fully connected network model (A). Most subjects can be predicted within 2.81 years using the combined model (B). Subjects who are predicted old have features of young patients and vice versa (C).
Figure 2B shows the cumulative probability of accurate prediction within an acceptable error range. This shows that 73.2% of subjects fall within an absolute error of 5 years in the combined model. This is an improvement from the intensity features model, which can only predict 64.6% of the subjects within 5 years while the features only can predict 54.9% within 5 years. Figure 2C presents a representative 3-by-3 matrix of subjects where the vertical axis represents true age of a young individual, a middle-aged adult, and an older adult. Some of the canonical features of old age, such as enlarged ventricles are seen in young patients predicted as old. Conversely, anatomical features of young age such as small cortical sulci are apparent in the older subjects.
3.2. Context-Aware Age Prediction Generalizes to Orbital CT
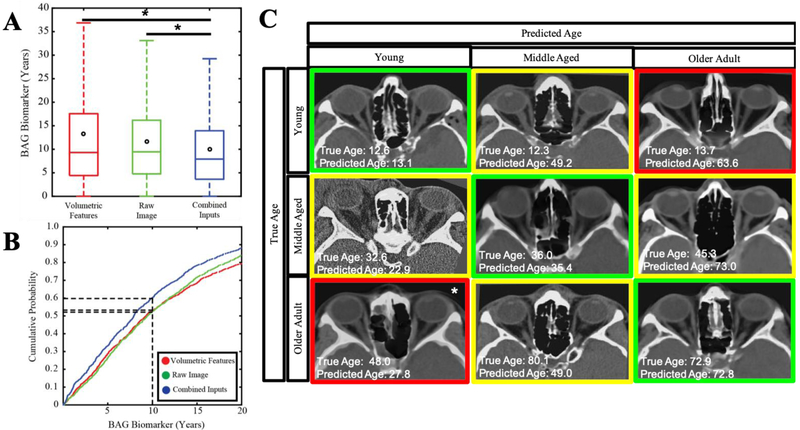
We applied the same network architecture to predict age on a dataset of head CT used in (22), which includes healthy and eye disease populations. Again, we use two inputs: 1) intensity features derived from a convolutional neural network representation, and 2) volumetric estimates of important orbital structures proposed by (19) as well as structural features. Figure 3A shows that intensity-derived features alone outperform hand-crafted features, resulting in a mean absolute error of 11.02 years and 13.28 years respectively (p<0.001). However, using both feature datasets as inputs allows for a significant improvement in prediction, resulting in a median absolute error of 9.99 (p<0.001).
Fig3.
Deep Learning Improves Age Prediction in head CT. Age can be predicted more accurately when using convolutional and structural features on a fully connected network model (A). Most subjects can be predicted within 7.90 years using the combined model (B). Accuracy of prediction does not show characteristic features of aging with our model (C). Note that while the older adult predicted young is 48 years old and technically in the middle aged bin, this was the oldest subject predicted within the young category, so we include it as a proxy.
In the CT dataset, 60.3% of subjects predicted within 10 years (Figure 3B) using the combined model, whereas the 10-year limit only covers 53.2% and 52.5% of the intensity-only or feature-only models, respectively. Representative images are shown in the 3-by-3 matrix in Figure 3C, where the vertical axis represents true age of a teenager, a middle-age adult, and an older adult.
3.3. OrbitBAG as a marker of disease
We further extend the development of a new imaging biomarker and validate it against 5 different diagnoses of eye disease. We estimate OrbitBAG for healthy controls in the testing set as well as unseen subjects with glaucoma, intrinsic optic nerve disease, orbit nerve edema, orbital inflammation, or thyroid eye disease. Figure 4 shows that the OrbitBAG biomarker is significantly elevated in patients with intrinsic optic nerve disease (p < 0.001) and orbital edema (p < 0.001) when controlling for true age. A trend of increased OrbitBAG is observed with glaucoma, orbital inflammation, and thyroid eye disease, albeit not significant (p = 0.48, p = 0.51, p=10, respectively). In the case of orbital edema and thyroid eye disease, OrbitBAG biomarker was not markedly elevated. OrbitBAG values for each condition are shown in Table 2.
Fig 4.
OrbitBAG biomarker for respective cohorts within the Orbital CT dataset. Intrinsic optic nerve disease (IOND) shows a significant increase in OrbitBAG. A similar trend is observed in glaucoma and orbital inflammation, but not statistically significant. Edema and thyroid eye disease (TED) do not show an increase in OrbitBAG.
4. Discussion
In the task of age prediction, deep learning performance can be enhanced by providing context features, such as volumetric estimates of important anatomical structures. We provide a more accurate method of age prediction compared to the literature. Additionally, we show that the proposed method of integrating deep learning with anatomical, hand-crafted features is not unique to brain MRI, but generalizes to orbital CT. We provide a new biomarker OrbitBAG based on orbital CT to estimate age and show it is elevated in patients with intrinsic optic nerve disease. Together, these results provide an accurate biomarker of aging in two different imaging modalities and demonstrate that there is still valuable information in classical image processing that is not being captured by deep learning.
4.1. Image Processing Features Enhance Deep Learning
Several studies have shown that deep neural networks are highly susceptible to changes in output to small adversarial perturbations or irregularities in the data (23-28). For example, scene recognition, a task mastered by deep learning, has been shown to be disrupted by single pixel changes (23, 25). Similarly, Nguyen et. al were able to produce images that are unrecognizable to humans but deemed recognizable by a convolutional neural network with 99.99% confidence (26).In this regard, anatomical features provide a robust representation of the image, compared to the learned convolutional filters. By providing contextual information, the model proposed here can help stabilize the network and capture information not available in the known features. Moreover, the large spatial extent of deep convolutional networks can learn to ignore unnecessary information such as background noise or artifacts. Together, features derived from a convolutional representation as well as features derived from classical image processing can enhance prediction tasks. Here, we see a significant improvement in age prediction accuracy in both brain MRI and head CT, suggesting that the improvement achieved by incorporating anatomical features is not specific to one modality.
Besides a general improvement in accuracy in computer vision tasks like object recognition and segmentation, deep learning has also shown improvements in computational time required for evaluating new images. Although training a deep neural network may take hours to days, evaluating a new image for segmentation or age prediction takes a matter of seconds. This shows a dramatic improvement over common image processing techniques such as multi-atlas segmentation, which can take hours to days to evaluate a single subject. In the work presented here, we propose the incorporation of deep learning with expert features. One limitation of our method is that producing an estimate of age will be dependent on the time needed to craft expert features for a test subject. If these are generated using multi-atlas segmentation, it may take hours. However, new techniques in deep learning are emerging that can replicate common deep learning tasks in a fraction of the time. For instance, Huo et. al. were able to generate a wholebrain volumetric segmentation using deep learning with similar accuracy to multi-atlas segmentation (29). We believe that incorporating anatomical context via hand-crafted features in deep learning pipelines can boost performance and generalizability while still being time-efficient.
Decades of work in image processing before deep learning have developed contextual features such as functional and anatomic regions of interest, surface parcellation, regional connectivity, and deformation models. Unlike the feature maps used in deep learning, these hand-crafted features are often built on underlying principles of anatomy and physiology. A large sample size of these features can possibly capture the entire manifold of possible human values. However, such restrictions do not exist in deep learning and extensive datasets are needed to arrive at plausible solutions. Contextual features could be used to limit the search-space of deep neural networks, diminish the number of parameters needed and avoid overfitting. This work also raises the question of robustness against adversarial attacks when a neural network is grounded by contextual features. Future work may benefit of exploring the role of such features in preventing adversarial attacks in medical imaging.
An alternative method to enhance image processing techniques is to iterate between deep learning and traditional feature engineering. New attention networks like the one used by Huo et. al (30) could help identify key regions of valuable anatomical information for the specified task. These maps can be used along with anatomic and physiological context to craft better manual features. These hand-crafted features can then inform a future network to improve algorithm accuracy and refine feature maps. It is possible that with more data and more complex deep learning models, similar accuracy can be achieved with imaging alone. In this work, we propose a method to guide training with imaging features that are already available. Instead of focusing on improving prediction accuracy, a stable model that integrates clinical and imaging context can be a fruitful ground for inference on key anatomical features preferentially affected by ageing
4.2. Brain Age Gap Used as an Imaging Biomarker of Aging
Previous studies have shown that BAG correlates with aging in a healthy population as well as neurodegenerative diseases in brain MRI (5). In this study, we show that an age prediction biomarker can be developed on a new imaging modality, Orbital CT, and validated against diseased populations. We show that the OrbitBAG biomarker is generally increased in populations that show structural changes such as glaucoma, edema, and orbital inflammation. We also observe a wide distribution of error in age prediction (Figure 2 and 4) in a healthy population. It is possible that these changes are a result of normal anatomical variability or susceptibility of disease. A longitudinal study of these subjects, along with clinical data, would show whether having an outlier BAG is predictive of future disease.
Here, we have demonstrated that while much attention has been dedicated to predicting chronological age from brain MRI, it is also possible to predict age from different imaging modalities obtained from different parts of the human body. With the increase in usage of medical imaging and processing capabilities, it may be possible to predict age from multiple body parts and multiple imaging modalities at once. A whole-body age prediction algorithm may better reflect the state of the entire body and find interesting associations in the aging process of different human organs.
4.3. Network Visualization on Raw MRI Input using Grad CAM
We proposed the integration of contextual anatomical figures along with raw imaging to inform the task of age prediction. A key open question in the field of medical image processing with deep learning is whether deep neural networks capture all available information in an image, or if these algorithms can be better trained by enforcing high-quality a priori information, such as volumetric estimates in regions of interest. It is often difficult to interpret the meaning of each convolutional layer in a deep convolutional network. However, a common tool for data visualization in deep learning are Gradient Class Activation Maps (Grad CAMs), which highlight areas of attention in the input image. Here, we have generated Grad CAMs for the subjects in the testing set for both the MRI and CT task (Figure 5). In the case of MRI, we see wide activation throughout the brain, but particularly centered around head size, the cerebral cortex, and the size of the ventricles. Although volume measurements of the ventricles are included as features in the combined model, the attention maps simultaneously encompass other areas of the brain, suggesting a complex interaction between areas of the brain. Importantly, we do not observe a clear and consistent segmentation of the brain structures which were included as volumes. Interestingly, the subjects off the diagonal, which show the worst prediction of age, have the least activation compared to the other subgroups. In the case of Orbital CT, there is a focus on the skull bones and the nose, which were not included as features, but may be indicative of aging. Interestingly, the activation in younger subjects was more wide-spread than in older subjects, suggesting higher variation across younger subjects, while bone structures may be a stronger indicator of age in older subjects.
Fig5.
Gradient Class Activation Maps (Grad CAM) visualization for raw MRI (left) and raw CT (right) used in the combined networks. Visualizations were binned according to true and predicted age. Ten random subjects were chosen from each category to compute the Grad CAM and the maps were averaged. Activation maps are overlaid over a representative subject from the sample.
Overall, Grad CAMs offer an interesting heuristic for visualization of attention from convolutional neural networks. In both CT and MRI, the attention maps seem to reiterate common changes associated with age such as head size or bone structures. It is reassuring that none of the activation maps uniquely highlight a segmentation corresponding to the volumetric features. However, it is still an open research question on how to interpret these maps and a deeper study is needed.
5. Conclusion
Applications in deep learning have focused on raw images due to the power in automatic feature recognition. However, we show that there remains valuable information in the features derived from image processing. These features with anatomical context can be used to complement deep learning tasks, especially, when there is finite training data. This work has significant implications in the field of medical image processing, as decades of work on feature optimization can be used to improve on already groundbreaking deep learning breakthroughs.
Table 3.
Accuracy of all three models on brain MRI data. MAE: mean absolute error. RMSE: Root mean squared error. R: Pearson correlation coefficient.
| Imaging Modality | Input Data | MAE (yrs) | RMSE | R | R2 |
|---|---|---|---|---|---|
| Brain MRI | Volumetric Features | 8.23 | 12.91 | 0.84 | 0.70 |
| Raw Image (13) | 5.00 | 7.25 | 0.95 | 0.90 | |
| Combined Features | 4.08 | 5.93 | 0.97 | 0.93 | |
| Orbital CT | Volumetric Features | 13.28 | 18.02 | 0.45 | 0.20 |
| Raw Image (13) | 11.02 | 14.11 | 0.75 | 0.56 | |
| Combined Features | 9.99 | 13.19 | 0.76 | 0.58 |
Table 4.
OrbitBAG accuracy results for each disease cohort. MAE: mean absolute error. RMSE: Root mean squared error. R: Pearson correlation coefficient.
| Cohort | MAE (yrs) | RMSE | R | R2 |
|---|---|---|---|---|
| Controls | 9.62 | 12.25 | 0.82 | 0.68 |
| Glaucoma | 9.97 | 12.97 | 0.73 | 0.53 |
| Intrinsic Optic Nerve Disease | 13.92 | 17.12 | 0.66 | 0.44 |
| Optic Nerve Edema | 15.66 | 18.32 | 0.62 | 0.38 |
| Orbital Inflammation | 11.36 | 14.11 | 0.77 | 0.60 |
| Thyroid Eye Disease | 11.69 | 15.03 | 0.43 | 0.19 |
Acknowledgments
This research was supported by NSF CAREER 1452485, NIH grants 1R03EB012461 (Landman), 5-R01-EB017230 (Landman), R01NS095291 (Dawant), U54 HD083211 (Dykens; NIH/NICHD) 5R01 HD044073 (Cutting; NIH/NICHD), 5R01-HD067254 (Cutting; NIH/NICHD), T32-EB021937 (NIH/NIBIB), and T32-GM007347 (NIGMS/NIH). This research was conducted with the support from the Intramural Research Program, National Institute on Aging, NIH. This study was in part using the resources of the Advanced Computing Center for Research and Education (ACCRE) at Vanderbilt University, Nashville, TN. This project was supported in part by ViSE/VICTR VR3029 and the National Center for Research Resources, Grant UL1 RR024975-01, and is now at the National Center for Advancing Translational Sciences, Grant 2 UL1 TR000445-06. This work does not reflect the opinions of the NIH or the NSF. We gratefully acknowledge the support of NVIDIA Corporation with the donation of the Titan X Pascal GPU used for this research.
The NDAR data used in the preparation of this manuscript were obtained from the NIH-supported National Database for Autism Research (NDAR). NDAR is a collaborative informatics system created by the National Institutes of Health to provide a national resource to support and accelerate research in autism. The NDAR dataset includes data from the NIH Pediatric MRI Data Repository created by the NIH MRI Study of Normal Brain Development. This is a multisite, longitudinal study of typically developing children from ages newborn through young adulthood conducted by the Brain Development Cooperative Group and supported by the National Institute of Child Health and Human Development, the National Institute on Drug Abuse, the National Institute of Mental Health, and the National Institute of Neurological Disorders and Stroke (Contract #s N01-HD02-3343, N01-MH9-0002, and N01-NS-9-2314,-2315, −2316, −2317, −2319 and −2320). A listing of the participating sites and a complete listing of the study investigators can be found at http://pediatricmri.nih.gov/nihpd/info/participating_centers.html.
The OASIS data used in the preparation of this manuscript were obtained from the OASIS project funded by grants P50 AG05681, P01 AG03991, R01 AG021910, P50 MH071616, U24 RR021382, R01 MH56584. See http://www.oasis-brains.org/ for more details.
The IXI data used in the preparation of this manuscript were supported by the U.K. Engineering and Physical Sciences Research Council (EPSRC) GR/S21533/02 - http://www.brain-development.org/. The ABIDE data used in the preparation of this manuscript were supported by ABIDE funding resources listed at http://fcon_1000.projects.nitrc.org/indi/abide/. ABIDE primary support for the work by Adriana Di Martino was provided by the NIMH (K23MH087770) and the Leon Levy Foundation
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Greenspan H, van Ginneken B, Summers RM, Guest editorial deep learning in medical imaging: Overview and future promise of an exciting new technique. IEEE Transactions on Medical Imaging 35, 1153–1159 (2016). [Google Scholar]
- 2.Lemaitre H et al. , Normal age-related brain morphometric changes: nonuniformity across cortical thickness, surface area and gray matter volume? Neurobiology of aging 33, 617. e611–617. e619 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kaufmann T et al. , Genetics of brain age suggest an overlap with common brain disorders. bioRxiv, 303164 (2018). [Google Scholar]
- 4.Lewis JD, Evans AC, Tohka J, T1 white/gray contrast as a predictor of chronological age, and an index of cognitive performance. bioRxiv, 171892 (2017). [DOI] [PubMed] [Google Scholar]
- 5.Cole JH, Franke K, Predicting Age Using Neuroimaging: Innovative Brain Ageing Biomarkers. Trends in Neurosciences (2017). [DOI] [PubMed] [Google Scholar]
- 6.Cole JH et al. , Brain age predicts mortality. Molecular Psychiatry (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Franke K, Ziegler G, Klöppel S, Gaser C, A. s. D. N. Initiative, Estimating the age of healthy subjects from T1-weighted MRI scans using kernel methods: exploring the influence of various parameters. Neuroimage 50, 883–892 (2010). [DOI] [PubMed] [Google Scholar]
- 8.Su L, Wang L, Hu D (2012) Predicting the age of healthy adults from structural MRI by sparse representation. in International Conference on Intelligent Science and Intelligent Data Engineering (Springer; ), pp 271–279. [Google Scholar]
- 9.Sabuncu MR, Van Leemput K, A. s. D. N. Initiative, The relevance voxel machine (RVoxM): a self-tuning Bayesian model for informative image-based prediction. IEEE Trans Med Imaging 31, 2290–2306 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brown TT et al. , Neuroanatomical assessment of biological maturity. Curr Biol 22, 1693–1698 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang B, Pham TD, MRI-based age prediction using hidden Markov models. J Neurosci Methods 199, 140–145 (2011). [DOI] [PubMed] [Google Scholar]
- 12.Cherubini A et al. , Importance of Multimodal MRI in Characterizing Brain Tissue and its Potential Application for Individual Age Prediction. IEEE JBiomedHealth Inform (2016). [DOI] [PubMed] [Google Scholar]
- 13.Cole JH et al. , Predicting brain age with deep learning from raw imaging data results in a reliable and heritable biomarker. NeuroImage 163, 115–124 (2017). [DOI] [PubMed] [Google Scholar]
- 14.Huo Y, Aboud K, Kang H, Cutting LE, Landman BA (2016) Mapping Lifetime Brain Volumetry with Covariate-Adjusted Restricted Cubic Spline Regression from Cross-Sectional Multi-site MRI. in International Conference on Medical Image Computing and Computer-Assisted Intervention (Springer; ), pp 81–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Avants BB, Epstein CL, Grossman M, Gee JC, Symmetric diffeomorphic image registration with cross-correlation: evaluating automated labeling of elderly and neurodegenerative brain. Medical image analysis 12, 26–41 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Asman AJ, Dagley AS, Landman BA, Statistical label fusion with hierarchical performance models. Proceedings - Society of Photo-Optical Instrumentation Engineers 9034, 90341E (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klein A et al. (2010) Open labels: online feedback for a public resource of manually labeled brain images. in 16th Annual Meeting for the Organization of Human Brain Mapping. [Google Scholar]
- 18.Smith SM et al. , Accurate, robust, and automated longitudinal and cross-sectional brain change analysis. Neuroimage 17, 479–489 (2002). [DOI] [PubMed] [Google Scholar]
- 19.Harrigan RL et al. , Robust optic nerve segmentation on clinically acquired computed tomography. Journal of Medical Imaging 1, 034006–034006 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Selvaraju RR et al. (2017) Grad-cam: Visual explanations from deep networks via gradient-based localization. in Proceedings of the IEEE International Conference on Computer Vision, pp 618626. [Google Scholar]
- 21.Rac K (2017) keras-vis. GitHub. [Google Scholar]
- 22.Chaganti S et al. , “EMR-Radiological Phenotypes in Diseases of the Optic Nerve and Their Association with Visual Function” in Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support. (Springer, 2017), pp. 373–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moosavi-Dezfooli S-M, Fawzi A, Frossard P (2016) Deepfool: a simple and accurate method to fool deep neural networks. in Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp 2574–2582. [Google Scholar]
- 24.Madry A, Makelov A, Schmidt L, Tsipras D, Vladu A, Towards deep learning models resistant to adversarial attacks. arXivpreprint arXiv:1706.06083 (2017). [Google Scholar]
- 25.Szegedy C et al. , Intriguing properties of neural networks. arXiv preprint arXiv:1312.6199 (2013). [Google Scholar]
- 26.Nguyen A, Yosinski J, Clune J (2015) Deep neural networks are easily fooled: High confidence predictions for unrecognizable images. in Proceedings of the IEEE conference on computer vision and pattern recognition, pp 427–436. [Google Scholar]
- 27.Goodfellow IJ, Shlens J, Szegedy C, Explaining and harnessing adversarial examples. arXiv preprint arXiv: 1412.6572 (2014). [Google Scholar]
- 28.Sharif M, Bhagavatula S, Bauer L, Reiter MK (2016) Accessorize to a crime: Real and stealthy attacks on state-of-the-art face recognition. in Proceedings of the 2016 ACM SIGSAC Conference on Computer and Communications Security (ACM; ), pp 1528–1540. [Google Scholar]
- 29.Huo Y et al. , 3D whole brain segmentation using spatially localized atlas network tiles. NeuroImage (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huo Y et al. , Coronary Calcium Detection using 3D Attention Identical Dual Deep Network Based on Weakly Supervised Learning. arXiv preprint arXiv:1811.04289 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]